Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A new chemo-enzymatic approach to synthesize rare sugars using an engineered glycoside-3-oxidase†
André
Taborda
,
Márcia
Rénio
,
M. Rita
Ventura
 * and
Lígia O.
Martins
* and
Lígia O.
Martins
 *
*
Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Av da República, 2780-157 Oeiras, Portugal. E-mail: lmartins@itqb.unl.pt
First published on 17th December 2024
Abstract
Rare sugars are monosaccharides and disaccharides highly valued for their unique properties and beneficial health effects. Their scarcity has led to inefficient extraction from natural sources, prompting the development of several chemical and enzymatic methods to improve their synthesis. In this study, we aim to optimize a regio- and stereoselective chemo-enzymatic process for synthesizing the rare sugar D-allose. We use a bacterial glycoside-3-oxidase that oxidizes D-Glc at the C2 or C3 position, depending on the presence of a C1 substitution, being converted into the respective keto-derivatives. Through protein engineering, we improve the enzyme's catalytic activity for D-Glc by 20-fold after seven rounds of directed evolution and increase its operational stability by 10-fold. The engineered enzyme uses 1-O-benzyl-D-glucoside as substrate, ensuring regioselective oxidation at the C3 position, followed by a stereoselective chemical reduction and deprotection step, affording D-allose with an overall yield of 81%. This innovative strategy represents a novel, straightforward approach for synthesizing D-allose, avoiding laborious, time-consuming purifications and complicated and lengthy protection–deprotection strategies. Importantly, it shows potential for synthesizing other rare C3 epimers of biomass sugars through eco-friendly and cost-effective processes, with applications in pharmaceuticals and food technology.
1. Introduction
Rare sugars, encompassing mono- and disaccharides and their derivatives, are defined by the International Society of Rare Sugars (ISRS) as carbohydrates found in low abundance in nature. These include twenty hexoses (D-allose, D-allulose and D-tagatose), nine pentoses (D-lyxose and L-xylulose) and disaccharides like turanose and isomaltulose. Rare sugars’ ecological roles and physiological functions remain largely unexplored. Nevertheless, their unique functional properties as food additives, nutraceuticals, and active pharmaceutical ingredients have attracted considerable attention.1–5Various chemical and enzymatic methods have been developed to address their production. These include (i) chemical isomerization, which is typically unselective, resulting in complex product mixtures,6 (ii) selective oxidation of minimally protected sugars followed by reduction,7–10 and (iii) site-selective radical epimerization reactions that produce rare sugars from biomass-derived carbohydrates.11 Enzymatic methods provide regio- and stereo-specificity under milder conditions, making them attractive, sustainable alternatives to traditional chemical synthesis.12 Over the past two decades, most studies on the enzymatic synthesis of rare sugars have utilized a strategy known as Izumoring. This approach enables the cyclical conversion of monosaccharides through enzymatic epimerization, isomerization and oxidation-reduction.13,14 Nevertheless, other non-Izumoring enzymatic techniques have also emerged.12,15 For example, pyranose-2-oxidases (P2Oxs) from the glucose-methanol-choline (GMC) family, can synthesize D-tagatose and D-glucopyranosyl-β-1,6-D-mannopyranose from D-galactose and gentiobiosone by promoting the oxidation of hexoses at the C2 position, respectively, using P2Ox oxidation followed by a chemical reduction.16,17
This work focuses on the chemo-enzymatic synthesis of the rare sugar D-allose, a C3 epimer of D-glucose (D-Glc), as studies exploring the enzymatic synthesis of C3 compared to C2 epimeric rare sugars are more scarce. D-Allose has gained significant attention for its beneficial properties as a non-caloric sweetener,18 anticancer,19–21 anti-oxidant,22 anti-inflammatory,23 anti-hypertensive24 and immunosuppressor agent.25 Despite its various benefits, D-allose remains a minor player in the industrial market due to its limited availability and high production costs.26 Earlier approaches included synthesizing β-D-allose from D-ribose mediated by cyanohydrin27 and later by reducing 1,2:5,6-di-O-isopropylidene-α-D-ribo-hexofuranose-3-ulose hydrate in 68–70% overall yield.28 More recently, the oxidation steps were mediated by other chemicals;7,8,10,29 however, these processes often lead to inconsistent and sometimes very low yields, as well as the formation of undesirable by-products, thus, the need for tedious purification steps, and occasionally the use of highly specialized equipment.
Multi-enzymatic reactions involving isomerases and epimerases starting from D-Glc via a three-pathway enzyme reaction having as intermediates D-fructose and D-psicose have become a common D-allose synthesizing strategy;30 nevertheless, due to the reversibility of these enzymes’ reactions, the resulting final reaction mixtures are complex complicating the target product purification and resulting in low final yields. Recently, an one-pot reaction reportedly produced D-allose from D-fructose using D-allulose 3-epimerase and D-ribose-5-phosphate isomerase with a conversion yield of 13%.31
We have recently characterized a bacterial glycoside-3-oxidase (PsG3Ox)32 that shares high sequence similarities with fungal P2Oxs.33,34 G3Ox oxidizes C-glycosides at the C3 position of the sugar moiety at high catalytic efficiency while exhibiting residual activity for monosaccharides, such as D-Glc.32,35–37 Herein, we engineered G3Ox to improve its catalytic properties towards D-Glc using directed evolution, an approach used successfully in recent years in our group to enhance the properties of various enzymes.38–42 We show that the optimized enzyme can selectively oxidize the C3–OH group of partially protected D-Glc at improved rates and with complete regioselectivity.
Given the enzyme's broad substrate specificity and potential for further engineering, this system is a promising pathway for synthesizing rare sugars, creating economic opportunities for their industrial production and commercialization.
2. Material and methods
2.1 Bacterial strains, plasmids and cultivation medium
Escherichia coli strains, plasmids, and primers used in this work are summarized in Table S1.†E. coli strain DH5α (Novagen) and E. cloni 10G elite (Lucigen corporation-Biosearch technologies) propagated and amplified plasmid constructs. E. coli Rosetta pLysS (DE3, Novagen) was used to express the wild-type PsG3Ox and its variants cloned in the pET-15b plasmid (Novagen). The library screenings used E. coli KRX (Promega) as an expression strain. Luria Bertani medium (LB) was used for cell cultivation, supplemented with 100 μg ml−1 of ampicillin (NZYTech) and, in the case of Rosetta pLysS, also with 20 μg ml−1 of chloramphenicol (NZYTech, Lisbon, Portugal).2.2 Construction of mutant libraries by error-prone PCR
The error-prone PCR (epPCR) of the psg3ox gene was performed using the primers PsG3Ox_GA_FW and PsG3Ox_GA_RV (Table S1†) in a total volume of 50 μL reaction containing 100 ng of DNA template, 1 μM of each primer, 200 μM of dNTPs, 1.5 mM MgCl2, Taq polymerase buffer, 50 μM (or 100 μM) MnCl2 and 2.5 U of Taq polymerase (Thermo Scientific). The PCR program was carried out following program 1 (Table S2†) and purified using Illustra GFX PCR DNA kit (GE Healthcare). The PCR of the vector (pET-15b) was performed with the primer pET15b_GA_FW and pET15b_GA_RV (Table S1†). The PCR was performed in a final volume of 50 μl reaction containing 50 ng of DNA template (pET-15b), 0.5 μM of each primer, 200 μM of dNTPs, 1 U of Q5 high-fidelity DNA polymerase (New England Biolabs) and following the procedure mentioned in program 2 (Table S2†). The PCR product was purified. A digestion of 6 h at 37 °C with DpnI (New England Biolabs) was performed to avoid the presence of the DNA template in the PCR products. The ligation was performed with a ratio gene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) vector 1
vector 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 using the NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs). The ligation was performed at 50 °C for 1 h, and afterward, it was used to transform E. cloni 10 G elite cells, allowing the propagation of the library. The plasmid library was extracted to transform electrocompetent KRX cells in a Gene Pulser Xcell™ (Bio-Rad Laboratories) under defined conditions (C = 10 μF, PC = 600 Ω, V = 1.8 kV). The cell suspension was spread on solid media containing 100 μg ml−1 of ampicillin and 0.1% rhamnose.
10 using the NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs). The ligation was performed at 50 °C for 1 h, and afterward, it was used to transform E. cloni 10 G elite cells, allowing the propagation of the library. The plasmid library was extracted to transform electrocompetent KRX cells in a Gene Pulser Xcell™ (Bio-Rad Laboratories) under defined conditions (C = 10 μF, PC = 600 Ω, V = 1.8 kV). The cell suspension was spread on solid media containing 100 μg ml−1 of ampicillin and 0.1% rhamnose.
2.3 Construction of DNA shuffling libraries
The amplification of the psg3ox genes selected to be shuffled was conducted using the primers pET21_FW and pET21_RV (Table S1†). PCR reactions were performed using the program 3 (Table S2†). The DNA from each amplified gene variant was pooled in the same ratio, totalizing 1400 ng and digested with DNase I (0.15 U ml−1) in 20 μl of 200 mM Tris-HCl, pH 7.0, with 80 mM MnCl2 at 15 °C for 10 min and was stopped by adding 6 μl of 0.5 M of EDTA. The obtained fragments were analyzed in 1% agarose gel, dialyzed using a nitrocellulose membrane and subsequently reassembled in a primerless PCR containing 5 μl of DNA fragments, 10 μM of dNTPs, and 0.5 U of NZYProof DNA Polymerase. The PCR followed program 4 (Table S2†). The final PCR amplification was performed using the primers PsG3Ox_GA_FW and PsG3Ox_GA_RV (Table S1†). The amplification was carried out in a total volume of 20 μl, including 1 μl of the reassembled product, 25 μM of each primer, 10 μM of dNTPs, 200 μM of buffer, and 0.5 U of NZYProof DNA Polymerase. The PCR program followed program 1 (Table S2†). The PCR products were purified using the previously mentioned kit from the agarose gel. The resulting product was ligated in the vector backbone described above for constructing mutant libraries by epPCR, transformed in E. cloni 10 G elite cells to propagate the library and transformed in KRX strain for further screening after plasmid extraction.2.4 Library screenings
For the ‘activity-on-plate’ screening, the colonies resulting from the transformation in KRX strain were transferred onto Whatman chromatography paper, and the plates were re-incubated at 37 °C until the colonies reappeared. Subsequently, the filter papers were soaked in a solution of 20 mM Tris-HCl, pH 7.6, and subjected to 3 cycles of freezing (40 min at −20 °C) followed by thawing (20 min at 37 °C). Following the lysis cycles, the filter papers were soaked in a mixture containing 1 mM 4-aminoantipyrine (AAP, Acros Organics), 10 mM 3,5-dichloro-2-hydroxybenzenesulfonic acid sodium salt (DCHBS, Alfa Aesar), 8 U ml−1 horseradish peroxidase (HRP, Sigma-Aldrich), and 0.5 M D-Glc (PanReac Applichem) in 100 mM phosphate buffer, pH 7.5. The active variants were identified by developing the pink chromogenic compound N-(4-antipyryl)-3-chloro-5-sulfonate-p-benzoquinone-monoimine. The active variants were picked to 96-deep well-plates (VWR) containing 400 μl of LB supplemented with the appropriate antibiotic and left to grow overnight at 37 °C with agitation at 750 rpm (pre-culture). New 96-deep well-plates, containing 900 μl of LB supplemented with ampicillin, were inoculated with 100 μl of the pre-culture and incubated for 3 hours at 37 °C, 750 rpm. Next, the expression of the target gene was induced with 10 mM of rhamnose, and cultures proceeded overnight at room temperature under 750 rpm. Afterwards, the cells were harvested by centrifugation at 4000 rpm (Eppendorf 5810 R centrifuge). The 96-well plates containing the cell pellets underwent three lysis cycles by submersion in liquid nitrogen and thawing at room temperature for 10 minutes. Then, the pellets were resuspended in 200 μl of 20 mM Tris-HCl buffer, pH 7.6, supplemented with 2 mg ml−1 of lysozyme, 1 U ml−1 DNase, 5 mM MgCl2, and protease inhibitors cocktail (3.5 μM antipain and 5 μM leupeptin). This suspension was incubated for 20 minutes at room temperature under agitation at 750 rpm, and afterward, the lysate was clarified by centrifugation at 4000 rpm. The supernatants (cell crude extracts) were collected for enzymatic activity assays in a reaction mixture containing 0.1 mM of AAP, 1 mM of DCHBS, 8 U ml−1 of HRP and 50–500 mM of D-Glc (depending on the round of evolution) in phosphate buffer pH 7.5 at 25 °C. The formation of a pink chromogen at 515 nm (ε515 = 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1) was measured using a Synergy2 microplate reader (BioTek).
000 M−1 cm−1) was measured using a Synergy2 microplate reader (BioTek).
2.5 Site-directed mutagenesis
The primers used to construct SDM variants are listed in Table S1.† PCRs were performed in a thermal cycler in 50 μL reaction volumes containing 100 ng of DNA template, 1 μM of primers, 200 μM of dNTPs (NZYTech), and 1.5 U of NZYProof polymerase (NZYTech). The PCR was performed following program 5 (Table S2†). The amplified product was digested, purified, and then transformed into a cloning strain. DNA sequencing confirmed the presence of the desired mutation(s).2.6 PsG3Ox production, purification and kinetic characterization
The production and purification of PsG3Ox variants were performed as described previously.32 The optimal pH was determined using the Britton–Robinson buffer system (100 mM phosphoric acid, 100 mM boric acid and 100 mM acetic acid mixed with 1 M NaOH to the desired pH) in the pH range 5 to 9. The reaction mixture contains 0.1 mM of AAP, 1 mM of DCHBS, 8 U ml−1 of HRP and 0.3 M of D-Glc. The activity was followed spectrophotometrically at 515 nm, as mentioned above. For steady-state kinetics, the reaction mixture contained 0.1 mM AAP, 1 mM DCHBS, 8 U ml−1 HRP and 0–1.5 M of D-Glc (or 0–50 mM of 1-O-benzyl-D-glucose) in 100 mM sodium phosphate buffer at the optimal pH and 37 °C. The activity was followed spectrophotometrically at 515 nm. Apparent steady-state kinetic parameters were determined by fitting data directly into the Michaelis–Menten equation using Origin-Lab software. All catalytic parameters were calculated using three independent assays in triplicate. The total protein concentration was determined by Bradford assay using bovine serum albumin as standard. Purified preparations’ absorption at 450 nm (εFAD450 nm = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 M−1 cm−1) was measured to assess the functional fraction of enzyme preparations (e.g., for kinetic measurements).
300 M−1 cm−1) was measured to assess the functional fraction of enzyme preparations (e.g., for kinetic measurements).
2.7 Substrate screening
The substrate screening was performed with 100 mM of D(+)Glc (PanReac ApplieChem), D(+)galactose (Sigma-Aldrich), L(+)arabinose (PanReac ApplieChem), 2-deoxy-D-glucose (Sigma-Aldrich), D(−)ribose (VWR), L(+)rhamnose (PanReac ApplieChem), D(+)mannose (Alfa Aesar), D(−)frutose (Sigma-Aldrich), L(−)fucose (Carl Roth), D-lactose (Sigma-Aldrich), D(+)maltose (Sigma-Aldrich), D(+)trehalose (Sigma-Aldrich), D(+)raffinose (Alfa aesar) and D(+)glucosamine (Sigma-Aldrich) in a reaction mixture containing 0.1 mM AAP, 1 mM DCHBS, 8 U ml−1 HRP in 100 mM sodium phosphate buffer at the optimal pH and 25 °C. The activity was followed spectrophotometrically at 515 nm, as mentioned above.2.8 Stability of enzyme variants
The enzymes’ thermostability was determined by steady-state fluorescence in a Cary Eclipse spectrofluorometer (Agilent Technologies). Enzyme preparations of 0.2 mg ml−1 in 20 mM Tris-HCl pH 7.6 with 0.2 M NaCl were placed in a thermostatically controlled block, and a temperature gradient with an increment of 1 °C min−1 was applied up to 70 °C. Structural denaturation was monitored using an excitation wavelength of 296 nm, and the emission was recorded at 340 nm. Protein aggregation was monitored by measuring static light scattering at 500 nm as excitation and emission wavelengths. A pure enzyme preparation was incubated at 40 °C for the kinetic stability assays. Aliquots were withdrawn over time, cooled on ice, and assayed for activity in a reaction containing 0.5 M of D-Glc, 0.1 mM of AAP, 1 mM of DCHBS, 8 U ml−1 of HRP in 100 mM sodium phosphate buffer, pH 7.5. The half-life time, corresponding to the time required for the enzyme to lose half of its initial activity, was estimated using the formula t1/2 = ln(2)/k (where k represents the linear regression slope between ln(activity) and time).2.9 Enzymatic oxidation of benzyl-glucoside by 16F10 variant
The compounds 1-O-benzyl-α/β-D-glucopyranoside and 1-O-benzyl-β-glucopyranoside were synthesized from D-Glc using a reaction procedure previously described.43The enzymatic oxidation of benzyl-glucoside was performed using 20 mg of 1-O-Benzyl-α/β-D-glucopyranoside, 5 U ml−1 of the 16F10 variant and 0.1 mg ml−1 of catalase (Sigma-Aldrich) in a total volume of 1.5 ml in water (adjusted to pH 7.5–8.0). The reaction proceeded for 24 hours at 25 °C under magnetic stirring to facilitate oxygenation of the solution. The reaction progress was monitored by TLC in Merck 60 F254 silica gel plates (90/10 ethyl acetate/methanol). At the end of the reaction time, the enzymes were removed by ultrafiltration using a 30 kDa Amicon (Merck) at 4000 rpm in an Eppendorf centrifuge. The reaction product was lyophilized and purified by flash column (Kieselgel 60, 0.032–0.063 mm) with 100% ethyl acetate to afford 1-O-benzyl-3-keto-α/β-D-glucopyranoside (2α and 2β; 1.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 α/β) in 36% yield as a viscous colourless syrup. 1H NMR (400 MHz, MeOH-d4): δ 7.46–7.27 (m, 10H, Ar–H), 5.28 (d, J = 4.40 Hz, 1H, H1-α), 5.00 (d, J = 11.97 Hz, 1H, CH2–Bn), 4.76 (dd, J = 12.09 Hz, J = 3.00 Hz, 2H, CH2–Bn), 4.61 (d, J = 11.92 Hz, 1H, CH2–Bn), 4.48 (d, J = 7.94 Hz, 1H, H1-β), 4.45 (dd, J = 4.38 Hz, J = 1.24 Hz, 1H, H2-α), 4.29–4.25 (m, 2H, H4-α and H4-β), 4.22 (dd, J = 7.90 Hz, J = 1.65 Hz, 1H, H2-β), 3.99 (dd, J = 12.20 Hz, J = 2.03 Hz, 1H, H6-β), 3.88–3.79 (m, 3H, H6-β and H6-α), 3.75 (ddd, J = 9.70 Hz, J = 4.50 Hz, J = 2.30 Hz, 1H, H5-α), 3.38–3.34 (m, 1H, H5-β). 13C NMR (100 MHz, MeOH-d4): δ 205.7 (C3-α), 205.6 (C3-β), 137.2 (Ar–C-β), 137.0 (Ar–C-α), 128.0, 127.9, 127.8, 127.7, 127.5, 127.4 (Ar–C), 103.3 (C1-β), 100.6 (C1-α), 77.0 (C5-β), 76.9 (C2-β), 75.7 (C5-α), 74.7 (C2-α), 72.3 (C4-α), 72.0 (C4-β), 70.6 (CH2–Bn-β), 69.2 (CH2–Bn-α), 61.2 (C6-β), 60.0 (C6-α) ppm. HRMS: m/z: [M + H2O]+ calcd for C13H20O7 286.1053; found 286.1287.
1 α/β) in 36% yield as a viscous colourless syrup. 1H NMR (400 MHz, MeOH-d4): δ 7.46–7.27 (m, 10H, Ar–H), 5.28 (d, J = 4.40 Hz, 1H, H1-α), 5.00 (d, J = 11.97 Hz, 1H, CH2–Bn), 4.76 (dd, J = 12.09 Hz, J = 3.00 Hz, 2H, CH2–Bn), 4.61 (d, J = 11.92 Hz, 1H, CH2–Bn), 4.48 (d, J = 7.94 Hz, 1H, H1-β), 4.45 (dd, J = 4.38 Hz, J = 1.24 Hz, 1H, H2-α), 4.29–4.25 (m, 2H, H4-α and H4-β), 4.22 (dd, J = 7.90 Hz, J = 1.65 Hz, 1H, H2-β), 3.99 (dd, J = 12.20 Hz, J = 2.03 Hz, 1H, H6-β), 3.88–3.79 (m, 3H, H6-β and H6-α), 3.75 (ddd, J = 9.70 Hz, J = 4.50 Hz, J = 2.30 Hz, 1H, H5-α), 3.38–3.34 (m, 1H, H5-β). 13C NMR (100 MHz, MeOH-d4): δ 205.7 (C3-α), 205.6 (C3-β), 137.2 (Ar–C-β), 137.0 (Ar–C-α), 128.0, 127.9, 127.8, 127.7, 127.5, 127.4 (Ar–C), 103.3 (C1-β), 100.6 (C1-α), 77.0 (C5-β), 76.9 (C2-β), 75.7 (C5-α), 74.7 (C2-α), 72.3 (C4-α), 72.0 (C4-β), 70.6 (CH2–Bn-β), 69.2 (CH2–Bn-α), 61.2 (C6-β), 60.0 (C6-α) ppm. HRMS: m/z: [M + H2O]+ calcd for C13H20O7 286.1053; found 286.1287.
For the conversions with the β anomer, the oxidation reaction was performed using 20 mg of 1-O-benzyl-β-glucopyranoside, 1 U ml−1 of the 16F10 variant, 0.1 mg ml−1 of catalase in a total volume of 1.5 mL in water (adjusted to pH 7.5–8.0). The reaction proceeded for 6 h 30 m at 25 °C with magnetic stirring. The reaction was monitored as above, and the enzyme was removed by ultrafiltration. The 1-O-benzyl-3-keto-β-D-glucopyranoside 2β was lyophilized overnight and obtained in quantitative yield (100%) as a viscous colourless syrup. [α]20D = −75.61 (c 1.2, MeOH). 1H NMR (400 MHz, MeOH-d4): δ 7.45–7.43 (m, 2H, Ar–H), 7.37–7.28 (m, 3H, Ar–H), 5.0 (d, J = 11.93 Hz, 1H, CH2–Bn), 4.75 (d, J = 11.77 Hz, 1H, CH2–Bn), 4.48 (d, J = 7.89 Hz, 1H, H1), 4.27 (dd, J = 10.19 Hz, J = 1.66 Hz, 1H, H4), 4.22 (dd, J = 7.88 Hz, J = 1.73 Hz, 1H, H2), 3.99 (dd, J = 12.01 Hz, J = 2.05 Hz, 1H, H6), 3.85 (dd, J = 12.05 Hz, J = 4.89 Hz, 1H, H6), 3.36 (dd, J = 4.96 Hz, J = 2.08 Hz, 1H, H5) ppm. 13C NMR (100 MHz, MeOH-d4): δ 205.8 (C-3), 137.3 (Ar–C), 128.0, 127.8, 127.5 (Ar–C), 103.3 (C1), 76.9 (C2), 76.8 (C5), 72.3 (C4), 70.7 (CH2–Bn), 61.2 (C6) ppm. HRMS: m/z: [M + H2O]+ calcd for C13H20O7 286.1053; found 286.1050.
2.10 Selective reduction of benzyl-keto-glucoside and removal of the protecting group
LS-selectride 1.0 M in THF (0.47 mmol, 0.5 mL) was added to compound 2β (0.157 mmol, 42 mg) in DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) THF (2
THF (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2 mL) and the reaction was stirred for 2 h at 0 °C. The excess LS-selectride was removed by adding acidic ion exchange resin (Dowex DVB WX8 H+-form), and the mixture was filtered and concentrated under vacuum. The crude product was purified by flash column (Kieselgel 60, 0.032–0.063 mm) with 90
1, 2 mL) and the reaction was stirred for 2 h at 0 °C. The excess LS-selectride was removed by adding acidic ion exchange resin (Dowex DVB WX8 H+-form), and the mixture was filtered and concentrated under vacuum. The crude product was purified by flash column (Kieselgel 60, 0.032–0.063 mm) with 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ethyl acetate/methanol to afford the 1-O-benzyl-β-D-allopyranoside 3 (36 mg) in 86% of yield as a viscous colourless syrup. [α]20D = −66.36 (c 1.2, MeOH). 1H NMR (400 MHz, MeOH-d4): δ 7.45–7.43 (m, 2H, Ar–H), 7.36–7.26 (m, 3H, Ar–H), 4.95 (d, J = 11.74 Hz, 1H, CH2–Bn), 4.76 (d, J = 7.78 Hz, 1H, H1), 4.67 (d, J = 11.74 Hz, 1H, CH2–Bn), 4.07 (t, J = 2.92 Hz, 1H, H3), 3.92–3.87 (m, 1H, H6) 3.74–3.68 (m, 2H, H5 and H6), 3.53 (dd, J = 9.37 Hz, J = 3.01 Hz, 1H, H4), 3.40 (dd, J = 7.99 Hz, J = 3.01 Hz, 1H, H2) ppm. 13C NMR (100 MHz, MeOH-d4): δ 137.9 (Ar–C), 127.9, 127.8, 127.2 (Ar–C), 99.6 (C1), 74.1 (C5), 71.6 (C3), 71.0 (C2), 70.3 (CH2–Bn), 67.6 (C4), 61.8 (C6) ppm. HRMS: m/z: [M + Na]+ calcd for C13H18NaO6 293.0996; found 293.0996.
10 ethyl acetate/methanol to afford the 1-O-benzyl-β-D-allopyranoside 3 (36 mg) in 86% of yield as a viscous colourless syrup. [α]20D = −66.36 (c 1.2, MeOH). 1H NMR (400 MHz, MeOH-d4): δ 7.45–7.43 (m, 2H, Ar–H), 7.36–7.26 (m, 3H, Ar–H), 4.95 (d, J = 11.74 Hz, 1H, CH2–Bn), 4.76 (d, J = 7.78 Hz, 1H, H1), 4.67 (d, J = 11.74 Hz, 1H, CH2–Bn), 4.07 (t, J = 2.92 Hz, 1H, H3), 3.92–3.87 (m, 1H, H6) 3.74–3.68 (m, 2H, H5 and H6), 3.53 (dd, J = 9.37 Hz, J = 3.01 Hz, 1H, H4), 3.40 (dd, J = 7.99 Hz, J = 3.01 Hz, 1H, H2) ppm. 13C NMR (100 MHz, MeOH-d4): δ 137.9 (Ar–C), 127.9, 127.8, 127.2 (Ar–C), 99.6 (C1), 74.1 (C5), 71.6 (C3), 71.0 (C2), 70.3 (CH2–Bn), 67.6 (C4), 61.8 (C6) ppm. HRMS: m/z: [M + Na]+ calcd for C13H18NaO6 293.0996; found 293.0996.
The compound 3 (24 mg, 0.089 mmol) in MeOH (2 mL) was hydrogenated at 50 psi in the presence of Pd/C 10% (0.08 g) overnight. Then, the reaction mixture was filtered through Celite and washed with MeOH; the solvent evaporated to afford D-allose 4 (15 mg) in 94% yield as a viscous colourless syrup, major anomer β. 1H NMR (400 MHz, MeOH-d4): δ 5.06 (d, J = 3.45 Hz, 1H, H1-α), 4.85 (d, J = 7.89 Hz, 1H, H1-β), 4.06 (t, J = 3.00 Hz, 1H, H3-β), 3.85 (dd, J = 11.62 Hz, J = 2.26 Hz, 1H, H6-β), 3.78–3.71 (m, 1H, H5-β), 3.68–3.63 (m, 1H, H6-β), 3.54–3.48 (m, H4-β), 3.26 (dd, J = 7.99 Hz, J = 2.99 Hz, 1H, H2) ppm. 13C NMR (100 MHz, MeOH-d4): δ 94.0 (C1-β), 93.8 (C1-α), 74.1 (C5-β), 72.2 (C2-β), 71.6 (C3-β), 67.6 (C4-β), 61.9 (C6-β) ppm. HRMS: m/z: [M + Na]+ calcd for C6H12NaO6 203.0526; found 203.0528.
2.11 Other methods
All 1H NMR spectra were obtained at 400 MHz in MeOH-d4 using the residual solvent peak as standard, and 13C NMR spectra were obtained at 100.61 MHz in MeOH-d4. Assignments are supported by 2D correlation NMR studies. Specific rotations were measured using a Perkin–Elmer D241 automatic polarimeter and are reported as follows: [α]20D (c g per 100 mL; solvent). ESI-MS determined the molecular mass, and the mass spectra of the samples were acquired in the positive mode in a Bruker microOTOF.3. Results and discussion
3.1 Laboratory evolution of PsG3Ox and kinetic characterization
Directed evolution allows for fine-tuning enzyme properties, yielding tailored enzymes with superior performance44 while circumventing the potentially ineffective rational design. The promiscuous activity of the PsG3Ox for various monosaccharides and C-glycosides32,45 indicates that this enzyme could serve as the starting point for an enzyme engineering strategy to enhance its catalytic efficiency towards D-Glc. Directed evolution using error-prone PCR (epPCR) and DNA shuffling techniques followed by activity screening were applied (Table S3†). After seven iterative rounds of directed evolution, approximately 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 variants were screened, and one hit variant (16F10) was identified with improved activity for D-Glc (Fig. 1 and Table 1).
000 variants were screened, and one hit variant (16F10) was identified with improved activity for D-Glc (Fig. 1 and Table 1).
 | ||
| Fig. 1 Lineage of variants of PsG3Ox generated during the enzyme engineering. The variants’ names are highlighted in a colored box, and the mutations in the variants’ genotypes are listed below. In the initial round, the variant screened with the highest activity, variant 1A1, emerged with a single mutation (G366S). In the second round, the best variant, 5D5, showcases three additional mutations (A75T, A206T, Q295H). In the third round, the selected best variant, 41G6, harbored an additional single mutation (P336S). The fourth round yielded two variants, 2G4 and 7E9, with comparable activities, and their genes underwent epPCR followed by DNA shuffling to retain mutually beneficial mutations. At this point, stability screening was introduced simultaneously with activity screening, leading to the selection of variant 2B9, which exhibited four-fold increased stability at 40 °C compared to variant 7E9. DNA shuffling between 2B9 and the wild-type was performed, eliminating putative deleterious mutations while preserving enzyme properties. The resulting variant, 6E3, bearing only six mutations, displayed similar activity and stability to 2B9 with ten mutations. In a final round of epPCR using 6E3 as a template, variant 16F10, due to the replacement of I357 V and D383N, exhibited a 1.5-fold activity improvement. All screening details are summarized in Table S3.† | ||
| Optimal pH | k cat (s−1) | K m (M) | k cat/Km (M−1 s−1) | |
|---|---|---|---|---|
| Activity was measured using the HRP-AAP/DCHBS coupled assay. All reactions were performed in 100 mM sodium phosphate buffer at the optimal pH and 37 °C. The kinetic parameters were determined by fitting the data directly on the Michaelis–Menten equation using OriginLab. | ||||
| Wild-type | 7.5 | 0.2 ± 0.05 | 0.37 ± 0.13 | 0.5 ± 0.1 |
| 1A1 | 7.5 | 0.4 ± 0.1 | 0.54 ± 0.11 | 0.7 ± 0.1 |
| 5D5 | 8.5 | 1.0 ± 0.2 | 0.33 ± 0.05 | 3.2 ± 0.4 |
| 41G6 | 8.0 | 1.2 ± 0.2 | 0.24 ± 0.05 | 5.1 ± 0.2 |
| 2G4 | 8.0 | 1.9 ± 0.2 | 0.26 ± 0.05 | 6.7 ± 0.8 |
| 7E9 | 8.0 | 2.3 ± 0.3 | 0.27 ± 0.05 | 8.6 ± 0.4 |
| 2B9 | 8.0 | 2.6 ± 0.3 | 0.23 ± 0.06 | 10.6 ± 0.9 |
| 6E3 | 8.0 | 3.0 ± 0.2 | 0.26 ± 0.01 | 11.5 ± 0.6 |
| 16F10 | 8.0 | 4.8 ± 1.0 | 0.37 ± 0.05 | 11.4 ± 0.8 |
All enzyme variants were purified (Table S4†), and the percentage of the functional enzyme (i.e., with a full complement of FAD) ranged from 20 to 60%. The optimal pH (7.5) of wild-type for D-Glc was up-shifted by 0.5 to 1 unit in the second round (variant 5D5). The catalytic performance of the evolutionary intermediates and the hit variant was assessed. We show that throughout the engineering process, the turnover number (kcat) significantly improved, representing the most notable difference in catalytic parameters, positively impacting the catalytic efficiency (kcat/Km) observed for D-Glc among the variants in the later generations (Table 1). In contrast, the Km, a parameter associated with substrate affinity, remained unchanged during the evolutionary pathway. Previous studies have emphasized that these enzymes’ active site topology and oligomeric state are key factors affecting substrate specificity within P2Oxs (tetramer)/G3Oxs (monomer) sub-families despite their high sequence similarity.32,37 Indeed, despite the observed improvements in variants of PsG3Ox, the kcat for D-Glc of the hit variant 16F10 remains 10- to 20-fold lower than those observed for D-Glc in fungal P2Oxs such as TmP2Ox and PcP2Ox, whereas the Km value is three orders of magnitude higher (Table S5†).
The activity of variant 16F10 was also tested against a diverse array of substrates, including different mono-, di- and trisaccharides, as well as one aminosugar, glucosamine (Table 2) and remarkably, it exhibited increased activity for most of the tested substrates. The highest enhancements were measured for D-maltose and D-galactose, with activity increasing ∼35-fold compared to the wild-type. D-Glc and lactose followed, demonstrating around a 20-fold increase than the wild-type.
| Specific activity (nmol min−1 mg−1) | ||
|---|---|---|
| Wild-type | 16F10 | |
| Activity was measured using the HRP-AAP/DCHBS coupled assay. All reactions were performed with 100 mM substrate in 100 mM sodium phosphate buffer at pH 7.5 and 25 °C. Nd – not detected. | ||
| Monosaccharides | ||
| D(+)-Glucose | 36.9 ± 5.7 | 645.4 ± 164.4 |
| D(+)-Galactose | 2.9 ± 0.7 | 97.1 ± 21.6 |
| L(+)-Arabinose | 5.3 ± 0.9 | 68.2 ± 4.8 |
| 2-Deoxy-glucose | 3.2 ± 0.5 | 44.2 ± 7.7 |
| D(−)-Ribose | 2.6 ± 1.8 | 24.3 ± 4.0 |
| L(+)-Rhamnose | 1.4 ± 0.1 | 8.9 ± 1.7 |
| D(+)-Mannose | 0.3 ± 0.2 | 3.2 ± 0.1 |
| D(−)-Fructose | 0.3 ± 0.1 | 1.6 ± 0.5 |
| L(−)-Fucose | 0.5 ± 0.1 | 3.6 ± 1.0 |
| Disaccharides | ||
| D-Lactose | 0.6 ± 0.4 | 10.8 ± 2.8 |
| D(+)-Maltose | 0.2 ± 0.1 | 7.3 ± 1.5 |
| D(+)-Trehalose | 0.2 ± 0.03 | 1.8 ± 1.0 |
| Trisaccharides | ||
| D(+)-Raffinose | 0.2 ± 0.1 | Nd |
| Aminosugars | ||
| D(+)-Glucosamine | 1.3 ± 0.3 | 0.5 ± 0.3 |
3.2 Thermostability of variants
Thermal unfolding profiles were performed to calculate the mid-point thermal temperatures, i.e., melting temperatures (Tm) (Fig. S1†). The unfolding is described as a two-state process where the folded and unfolded states seem to be the only ones that accumulate significantly (Fig S1†). All variants exhibited slightly lower melting temperatures than the wild-type (49 °C); for example, the hit variant showed a Tm around 44 °C (Table 3). However, protein aggregation, assessed by static light scattering (data not shown), reveals the onset of aggregation (Tagg) at similar temperatures. Since the aggregation process irreversibly removes unfolded proteins from the equilibrium between the folded and unfolded states, it may significantly affect the melting temperature measurements, leading to its designation as an apparent value, TAppm.46,47| Variant | T Appm (°C) | T agg (°C) | t 40 °C1/2 (min) |
|---|---|---|---|
| Apparent melting temperatures (TAppm) were measured by steady-state fluorescence following the intrinsic fluorescence of the enzyme, aggregation temperature (Tagg) was monitored by static light scattering and half-life time at 40 °C (t40 °C1/2) was measured by following the loss activity over time. The activity at each time point was measured using the HRP-AAP/DCHBS system. | |||
| Wild-type | 49 ± 2 | 50 | 27 ± 2 |
| 1A1 | 47 ± 3 | 46 | 366 ± 46 |
| 5D5 | 47 ± 3 | 49 | 374 ± 82 |
| 41G6 | 44 ± 1 | 43 | 5 ± 1 |
| 2G4 | 43 ± 1 | 43 | 82 ± 8 |
| 7E9 | 44 ± 2 | 45 | 102 ± 18 |
| 2B9 | 47 ± 2 | 46 | 291 ± 30 |
| 6E3 | 45 ± 2 | 48 | 212 ± 41 |
| 16F10 | 44 ± 2 | 48 | 187 ± 30 |
Regarding the kinetic, so-called, long-term or operational stability, which quantifies the amount of enzyme that loses activity irreversibly during incubation at a specific temperature, the first two variants (1A1 and 5D5) exhibited a remarkable 10-fold improvement in the stability as compared to wild-type (27 min at 40 °C). In contrast, variant 41G6, in the 3rd round, showed a substantial trade-off between activity and stability, with a slight increase in kcat (Table 1) but at a high cost of kinetic stability: ∼100-fold decrease in half-life time (5 min at 40 °C) (Table 3 and Fig. S2†). The kinetic stability quantifies the amount of enzyme that denaturates irreversibly owing to protein aggregation, misfolding, and covalent changes.48 The wild-type and different variants deactivate according to a first-order process, which the classical Lumry can describe–Eyring model applied to the majority of enzymes (N ↔ U → D, where N, U and D are the native, the reversible unfolded and the irreversible denatured enzyme), pointing to a simple pathway of unfolding and denaturation. Since this effect was due to a single mutation, P336S, we have introduced and removed this mutation in different genetic backgrounds. The replacement of P336S in the wild-type has not affected activity but led to a pronounced decrease in the half-life time at 40 °C, making impossible its measurement (Tables S6 and S7†); in contrast, its removal from subsequent variants 2G4, 7E9, and 2B9 (4th and 5th round) result in a significant increase in stability but at expenses of a 2-fold decreased activity (Tables S6 and S7†). These observations suggest that P336S has influenced the evolutionary trajectory of the enzyme towards improved activity for D-Glc and indicate that it exhibits genotype-dependent effects hinting at epistatic interactions.49 The mutations introduced in subsequent rounds of evolution compensated the negative effect of P336S, and all subsequent variants show increased stability, retaining 50% of activity after 80 to 290 min at 40 °C (Table 3). It is important to mention that during the first four generations, the screening of variant libraries during the directed evolution was based solely on activity measurements, which led to the selection of variant 41G6 with unexpectedly low kinetic stability after purification. From the fourth generation onward, we simultaneously screened for activity and stability, selecting only those variants that performed well in both properties. Notably, as mentioned, the hit variant 16F10 exhibited a kcat approximately 20-fold higher, and a half-life time at 40 °C, almost 10-fold higher than the wild-type enzyme (Tables 1 and 3; Fig S1†).
3.3 Molecular details of variants
All mutations present in the hit variant 16F10 and introduced during the evolution are distal to the FADN5 (>10 Å) (Fig. 2A and Fig. S3†). It is now well known that mutations located distantly from the active site can also enhance enzyme performance through allosteric effects, thereby inducing changes at the active site via a network of interactions.50,51 By the combination of site-directed mutagenesis and the analysis of enzyme structures, we hypothesize that the improvement of enzyme activity resulted from the following mutations: G366S (1st round), Q295H (2nd round), P336S (3rd round), A327V (4th round), A335T (5th round), and I357V (7th round); excluding mutations D383N and A206T (Fig. 2A). The visual inspection of the enzyme structure with D-Glc docked in a catalytic competent orientation (PDB 7QFD)32 reveals that the G366S mutation is nearby (∼3.4 Å) to the C4–OH group of D-Glc, arguably forming an additional hydrogen bond through Ser–OH group (Fig. 2B). This mutation is also close to Q297 (∼2.3 Å) and Q340 (∼5.1 Å), which are critical residues for the substrate orientation in the active.32 Mutation Q295H is at ∼4.0 Å to the catalytic residue H440 and also close to Q297 (Fig. 2B). Single mutants A75T, A206T, and Q295H (introduced in the 2nd round) were constructed and whereas A75T and A206T showed similar catalytic properties to wild-type, revealing its minor or null role, the mutation Q295H exhibited a 2-fold higher kcat (Table S8†). The type of amino acid transition Q/H can conceivably change the microenvironment surrounding the residue: the optimal pH (7.5) of wild-type for D-Glc was up-shifted by 0.5 to 1 unit in the second round (variant 5D5), most likely due to the introduction of the Q295H mutation. Interestingly, the combination of Q295H with G366S exhibits an increased activity than the activity sum of individual variants and attains the activity of variant 5D5 (Table S8†). These results demonstrate that the effect of these mutations (Q295H with G366S) is also dependent on the genetic background, i.e., of epistatic interactions,49 previously mentioned in relation to P336S (Table S6†). This residue is positioned in a loop interconnecting β-strands with residues of the active site and its introduction has caused a drastic decrease in kinetic stability (Table 3), likely due to increased loop flexibility (due to the removal of the rigid proline), resulting in a more rapid and irreversible denaturation. The introduction of adjacent A335T mutation in variant 2B9 may have counteracted the destabilizing effect of the P336S, as observed by the increased half-life at 40 °C (from 100 minutes to 290 minutes; Table 3). A327V mutation is close to the β-strand where the substrate-interacting Q340 (at ∼6 Å) belongs and can supposedly induce changes in the positioning or dynamics of this non-catalytic residue. Lastly, I357V is located in a critical structural element, the substrate loop. This loop can adopt different conformations throughout the catalytic cycle,32 and the replacement to a less hydrophobic residue may presumably modulate loop dynamics, impacting catalysis. | ||
| Fig. 2 Visual representation and molecular details of the mutations introduced in the hit variant 16F10. (A) Cartoon representation of wild-type PsG3Ox structure (PDB 7QF8) with all mutations present in 16F10 represented by the Cα as spheres and colored according to the round in which they were introduced (Fig. 1). The insertion-1 region and substrate loop are colored in dark blue and orange, respectively. All mutations are located distal to FADN5 (B) detailed view, close to FAD, with D-Glc docked and the two mutations (G366S, Q295H) closer to the active site. The FAD is colored yellow, the D-Glc is colored orange, the catalytic residues (H440 and N484) are colored light blue, and the non-catalytic interacting residues with D-Glc (R94, T129, Q297, Q340) are colored grey. | ||
3.4 Chemo-enzymatic synthesis of D-allose using the engineered enzyme
D-Allose synthesis involves the regioselective oxidation of D-Glc at the C3 position. Therefore, we have synthesized protected 1-O-benzyl-α/β-D-glucopyranoside (mixture α/β 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and tested the enzymatic activity of wild-type and engineered variant 16F10, which shows higher selectivity for C3 over C2 (Table 4; Fig. S4†). Comparing the catalytic parameters obtained, 16F10 showed a 10-fold increase in the turnover number (kcat). At the same time, the Km remained unchanged as compared to the wild-type enzyme. Interestingly, regarding the Km values, a 25-fold decrease was observed for the substrate 1-O-benzyl-α/β-D-glucopyranoside compared to D-Glc (Table 4). These results are consistent with our previous findings, which highlight the crucial role of an aromatic aglycone in facilitating substrate access and binding to the active site.32
1) and tested the enzymatic activity of wild-type and engineered variant 16F10, which shows higher selectivity for C3 over C2 (Table 4; Fig. S4†). Comparing the catalytic parameters obtained, 16F10 showed a 10-fold increase in the turnover number (kcat). At the same time, the Km remained unchanged as compared to the wild-type enzyme. Interestingly, regarding the Km values, a 25-fold decrease was observed for the substrate 1-O-benzyl-α/β-D-glucopyranoside compared to D-Glc (Table 4). These results are consistent with our previous findings, which highlight the crucial role of an aromatic aglycone in facilitating substrate access and binding to the active site.32
| Variant | k cat (s−1) | K m (mM) | k cat/Km (M−1 s−1) | |
|---|---|---|---|---|
| Activity for D-Glc, 1-O-benzyl-α/β-D-glucopyranoside and 1-O-Benzyl-β-D-glucopyranoside was measured using the HRP-AAP/DCHBS coupled assay. All reactions were performed in 100 mM sodium phosphate buffer at the optimal pH and 37 °C. The kinetic parameters were determined by fitting the data directly on the Michaelis–Menten equation using OriginLab. | ||||
| D-Glc | Wild-type | 0.2 ± 0.05 | 370 ± 130 | 0.5 ± 0.1 |
| 16F10 | 4.8 ± 1.0 | 370 ± 50 | 11.4 ± 0.8 | |
| 1-O-Benzyl-α/β-D-glucopyranoside | Wild-type | 0.06 ± 0.01 | 13.4 ± 3.8 | 5.0 ± 1.0 |
| 16F10 | 0.57 ± 0.05 | 17.4 ± 1.3 | 33.0 ± 1.0 | |
| 1-O-Benzyl-β-D-glucopyranoside | Wild-type | 0.31 ± 0.01 | 16.4 ± 4.0 | 19.3 ± 5.2 |
| 16F10 | 1.26 ± 0.07 | 11.3 ± 1.0 | 111.5 ± 4.0 | |
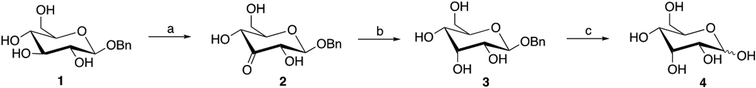
The oxidation product of 1-O-benzyl-α/β-D-glucopyranoside was characterized by NMR, confirming the presence of the ketone at C3 (Fig. S5 and S6†). However, a partial conversion (36% yield) was obtained, even after 24 h reaction. Interestingly, the β-anomer was the predominant isomer present in the NMR spectra, which indicated some stereoselectivity for this isomer. Therefore, the β-isomer (1-O-benzyl-β-D-glucopyranoside) was synthesized,43 and the enzyme's activity was tested towards this substrate. The obtained catalytic parameters confirmed that the β-anomer is more efficiently oxidized than the α-anomer, as observed by the increase of the kcat parameter and also reflected in catalytic efficiency (kcat/Km) (Table 4). A bioconversion reaction using the 1-O-benzyl-β-D-glucopyranoside 1 as substrate (20 mg of the substrate in a volume of 1.5 ml) was performed, and a total conversion (100% yield) was obtained, affording 1-O-benzyl-3-keto-β-D-glucopyranoside 2 (Scheme 1) with complete regioselectivity and without the need of further purification (Fig. S7 and S8†).
The current chemical strategies for the regioselective oxidation of unprotected or semi-protected monosaccharides at the C3 position primarily rely on a Pd/neocuproine catalyst and benzoquinone, yielding good results (Table S9†).7,10 Despite its effectiveness, these methods require a palladium catalyst and a chemical terminal oxidant, which have a significant environmental impact. Also, in the case of some substrates, a mixture of C3/C4 oxidation products was obtained, requiring additional purification steps. A metal-free strategy was also developed using electrochemical oxidation of the C3 position, mediated by quinuclidine.29 However, the obtained yields are lower, specific equipment is required, and, in some cases, it is necessary to protect the C6 position of the pyranosides (Table S9†).
In contrast, the enzymatic oxidation of 1-O-benzyl-β-glucopyranoside at the C3 position is complete and efficient, avoiding hazardous chemicals and preventing the formation of by-products, thereby eliminating the need for extensive purification.
The following step involved the stereoselective reduction of the resulting compound 2 to afford 1-O-benzyl-β-D-allopyranoside 3 (Scheme 1; Fig. S9 and S10†). The allose-derived product has an inverted configuration at the C3 position compared to D-Glc, with the 3-OH group positioned cis to the vicinal 2-OH and 4-OH groups. Different reducing agents were studied to obtain the best yield and cis selectivity (Table 5).
| Reagent | Solvent | Yield (%) | Diastereomeric ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3 |
|---|---|---|---|
| All reactions were performed at 0 °C. The ratio of diastereomers 1 and 3 obtained after reduction with LS-Seletride was determined by 1H NMR. | |||
| NaBH4 | MeOH | 63 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 1.5 |
| K-Selectride® | DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) THF (2 THF (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
64 | 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| LS-Selectride® | DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) THF (2 THF (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
86 | 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Reduction with NaBH4 afforded a mixture of the two diastereomeric alcohols 1 and 3 with very low selectivity towards the cis (alloside 3) product. K- and LS-Selectride provided exclusively the corresponding C3 cis-alcohol 3 (Scheme 1); however, LS-selectride afforded the highest yield (86%, Table 5). The high 1,2-cis stereoselectivity of selectride is well documented. It is attributed to the steric bulk of the reducing agent-inducing attack at the side opposite to a vicinal group in cyclohexanones, which usually forms the axial alcohol predominantly.10,52,53 To finally obtain D-allose, the removal of the anomeric benzyl protecting group was easily accomplished by hydrogenation, affording exclusively D-allose 4 (Scheme 1; Fig. S11 and S12†) in 94% yield, without the need for purification. In conclusion, a new efficient regio- and stereoselective chemo-enzymatic method was developed to synthesize the rare sugar D-allose in 81% overall yield.
Despite some progress, synthesizing D-allose using chemical and enzymatic approaches still presents several challenges (Table S9†). While chemical methods can achieve viable yields, they are often associated with high production costs due to expensive catalysts and environmental hazards from chemical waste.26 The biosynthetic production of D-allose, mainly through the Izumoring strategy, is a more environmentally friendly and scalable alternative that has emerged as the leading industrial method despite the low production allose yields.26 The new chemo-enzymatic approach described here offers several advantages over existing methods: the first enzymatic step is selectively efficient, and no by-products were formed. The two successive chemical reactions are also very selective and effective using common reagents, requiring only a single, straightforward chromatographic purification step after the carbonyl reduction with LS-selectride. Since no by-products were formed, complex purification is unnecessary, contrasting with reported methods. Considering the affordability of the starting material, the completeness and irreversibility of the enzymatic oxidation, and the high yields obtained in the stereoselective reduction, our process has strong potential for scaling up to industrial production of D-allose.
4. Conclusions
Carbohydrates are central biomolecules in numerous biological fields, with their natural availability highly dependent on their chemical structure. Low-abundance sugars, often called ‘rare sugars’, hold significant potential as nutraceuticals and food additives, yet their synthesis poses some challenges. Carbohydrate oxidases, with their high regio- and stereospecificity, enable the selective oxidation of specific hydroxyl groups in carbohydrates into carbonyl groups.54 They use molecular oxygen as a clean oxidant and produce hydrogen peroxide as a by-product. Traditional methods often involve lengthy synthetic routes and complex purification processes, and they can yield low amounts or mixtures of products. The novel chemo-enzymatic approach developed in this study efficiently synthesizes D-allose from minimally protected D-Glc, achieving an overall yield of 81% using an engineered glycoside-3-oxidase variant that catalyzes the initial regioselective oxidation at the C3 of the substrate. Diastereoselective chemical reduction, followed by subsequent hydrogenolysis, yielded the D-allose sugar. This chemo-enzymatic strategy is efficient and rapid and facilitates the isolation of pure products in excellent yields with minimal and straightforward product isolation procedures. Our method can also be adapted to synthesize other C3 epimers, such as the rare sugars D-gulose and D-altrose, starting from D-galactose and D-mannose. Additionally, the 3-keto group formed during the process offers opportunities for further functionalization into various derivatives, an area of ongoing research.Author contributions
A. T.: investigation and writing the original draft; M. R.: investigation and writing – review & editing; M. R. V and L. O. M.: conceptualization, supervision, and writing – review & editing.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
A patent application has been submitted based on data presented in the paper: Martins, L. O., Taborda, A., Ventura, R. M., Rénio, M. Chemo-enzymatic approach for the synthesis of rare C3-sugars using an glycoside 3-oxidase (engineered) enzymes. PT119793, October 25, 2024.Acknowledgements
We thank Diana Santos for the preliminary data and Teresa Silva and Cristina Timóteo (Research Facilities, ITQB-NOVA) for their technical assistance. This work was supported by the Fundação para a Ciência e Tecnologia (FCT), Portugal grant 2022.02027.PTDC, Project MOSTMICRO-ITQB (UIDB/04612/2020 and UIDP/04612/2020) and LS4FUTURE Associated laboratory (LA/P/0087/2020). The NMR data were acquired at CERMAX, ITQB–NOVA, Oeiras, Portugal, with equipment funded by FCT, project AAC 01/SAICT/2016. AT acknowledges FCT for his PhD fellowship 2020.07928.References
- A. Smith, A. Avery, R. Ford, Q. Yang, A. Goux, I. Mukherjee, D. C. A. Neville and P. Jethwa, Br. J. Nutr., 2022, 128, 389–406 CrossRef CAS PubMed.
- M. Bilal, H. M. N. Iqbal, H. Hu, W. Wang and X. Zhang, Crit. Rev. Food Sci. Nutr., 2018, 58, 2768–2778 CrossRef CAS PubMed.
- W. Mu, W. Zhang, Y. Feng, B. Jiang and L. Zhou, Appl. Microbiol. Biotechnol., 2012, 94, 1461–1467 CrossRef CAS PubMed.
- A. Ahmed, T. A. Khan, D. Dan Ramdath, C. W. C. Kendall and J. L. Sievenpiper, Nutr. Rev., 2021, 80, 255–270 CrossRef PubMed.
- A. d. C. Ortiz, S. O. M. Fideles, C. H. B. Reis, B. T. Pagani, L. M. M. Bueno, M. B. M. Moscatel, R. L. Buchaim and D. V. Buchaim, Nutrients, 2024, 16, 1943 CrossRef CAS PubMed.
- S. J. Angyal, in Glycoscience: Epimerisation, Isomerisation and Rearrangement Reactions of Carbohydrates, ed. A. E. Stütz, Springer Berlin Heidelberg, Berlin, Heidelberg, 2001, pp. 1–14, DOI:10.1007/3-540-44422-X_1.
- M. Jäger, M. Hartmann, J. G. de Vries and A. J. Minnaard, Angew. Chem., Int. Ed., 2013, 52, 7809–7812 CrossRef PubMed.
- W. Muramatsu, Org. Lett., 2014, 16, 4846–4849 CrossRef CAS PubMed.
- K. Chung and R. M. Waymouth, ACS Catal., 2016, 6, 4653–4659 CrossRef CAS.
- V. R. Jumde, N. N. H. M. Eisink, M. D. Witte and A. J. Minnaard, J. Org. Chem., 2016, 81, 11439–11443 CrossRef CAS PubMed.
- Y. Wang, H. M. Carder and A. E. Wendlandt, Nature, 2020, 578, 403–408 CrossRef CAS PubMed.
- W. Zhang, T. Zhang, B. Jiang and W. Mu, Biotechnol. Adv., 2017, 35, 267–274 CrossRef CAS PubMed.
- T. B. Granström, G. Takata, M. Tokuda and K. Izumori, J. Biosci. Bioeng., 2004, 97, 89–94 CrossRef PubMed.
- K. Izumori, J. Biotechnol., 2006, 124, 717–722 CrossRef CAS PubMed.
- P. Intasian, K. Prakinee, A. Phintha, D. Trisrivirat, N. Weeranoppanant, T. Wongnate and P. Chaiyen, Chem. Rev., 2021, 121, 10367–10451 CrossRef CAS PubMed.
- S. Freimund, A. Huwig, F. Giffhorn and S. Köpper, J. Carbohydr. Chem., 1996, 15, 115–120 CrossRef CAS.
- F. Giffhorn, S. Ko, A. Huwig and S. Freimund, Enzyme Microb. Technol., 2000, 27, 734–742 CrossRef CAS PubMed.
- A. D. Mooradian, M. Smith and M. Tokuda, Clin. Nutr. ESPEN, 2017, 18, 1–8 CrossRef PubMed.
- L. Sui, Y. Dong, Y. Watanabe, F. Yamaguchi, N. Hatano, I. Tsukamoto, K. Izumori and M. Tokuda, Int. J. Oncol., 2005, 27, 907–912 CAS.
- T. Mitani, H. Hoshikawa, T. Mori, T. Hosokawa, I. Tsukamoto, F. Yamaguchi, K. Kamitori, M. Tokuda and N. Mori, Head Neck, 2009, 31, 1049–1055 CrossRef PubMed.
- N. Kanaji, K. Kamitori, A. Hossain, C. Noguchi, A. Katagi, N. Kadowaki and M. Tokuda, Oncol. Rep., 2018, 39, 1292–1298 CAS.
- A. Murata, K. Sekiya, Y. Watanabe, F. Yamaguchi, N. Hatano, K. Izumori and M. Tokuda, J. Biosci. Bioeng., 2003, 96, 89–91 CrossRef CAS PubMed.
- D. Gao, N. Kawai and T. Tamiya, Med. Hypotheses, 2011, 76, 911–913 CrossRef PubMed.
- S. Kimura, G.-X. Zhang, A. Nishiyama, Y. Nagai, T. Nakagawa, H. Miyanaka, Y. Fujisawa, A. Miyatake, T. Nagai, M. Tokuda and Y. Abe, J. Hypertens., 2005, 23, 1887–1894 CrossRef PubMed.
- M. A. Hossain, H. Wakabayashi, K. Izuishi, K. Okano, S. Yachida, M. Tokuda, K. Izumori and H. Maeta, J. Biosci. Bioeng., 2006, 101, 369–371 CrossRef PubMed.
- X. Tang, Y. Ravikumar, G. Zhang, J. Yun, M. Zhao and X. Qi, Crit. Rev. Food Sci. Nutr., 2024, 1–28 Search PubMed.
- F. L. Humoller, Methods Carbohydr. Chem., 1962, 1, 102–104 Search PubMed.
- D. C. Baker, D. Horton and C. G. Tindall, Carbohydr. Res., 1972, 24, 192–197 CrossRef PubMed.
- M. Kidonakis, A. Villotet, M. D. Witte, S. B. Beil and A. J. Minnaard, ACS Catal., 2023, 13, 2335–2340 CrossRef PubMed.
- Y. R. Lim and D. K. Oh, Appl. Microbiol. Biotechnol., 2011, 91, 229–235 CrossRef PubMed.
- T. E. Lee, K. C. Shin and D. K. Oh, J. Microbiol. Biotechnol., 2018, 28(3), 418–424 CrossRef PubMed.
- A. Taborda, T. Frazão, M. V. Rodrigues, X. Fernández-Luengo, F. Sancho, M. F. Lucas, C. Frazão, E. P. Melo, M. R. Ventura, L. Masgrau, P. T. Borges and L. O. Martins, Nat. Commun., 2023, 14, 7289 CrossRef PubMed.
- L. Sutzl, G. Foley, E. M. J. Gillam, M. Boden and D. Haltrich, Biotechnol. Biofuels, 2019, 12, 118 CrossRef PubMed.
- T. Kumano, S. Hori, S. Watanabe, Y. Terashita, H. Y. Yu, Y. Hashimoto, T. Senda, M. Senda and M. Kobayashi, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2106580118 CrossRef PubMed.
- A. Kostelac, L. Sützl, J. Puc, V. Furlanetto, C. Divne and D. Haltrich, Int. J. Mol. Sci., 2022, 23, 13595 CrossRef PubMed.
- V. Furlanetto, D. C. Kalyani, A. Kostelac, J. Puc, D. Haltrich, B. M. Hällberg and C. Divne, J. Mol. Biol., 2024, 436, 168547 CrossRef PubMed.
- A. Kostelac, A. Taborda, L. O. Martins and D. Haltrich, Appl. Environ. Microbiol., 2024, 90, e0167623 CrossRef PubMed.
- V. Brissos, M. Ferreira, G. Grass and L. O. Martins, ACS Catal., 2015, 5, 4932–4941 CrossRef.
- V. Brissos, N. Goncalves, E. P. Melo and L. O. Martins, PLoS One, 2014, 9, e87209 CrossRef PubMed.
- V. Brissos, P. T. Borges, F. Sancho, M. F. Lucas, C. Frazão, F. Conzuelo and L. O. Martins, JBIC, J. Biol. Inorg. Chem., 2024, 29, 339–351 CrossRef PubMed.
- V. Brissos, D. Tavares, A. C. Sousa, M. P. Robalo and L. O. Martins, ACS Catal., 2017, 7, 3454–3465 CrossRef.
- V. Brissos, M. Renio, M. Lejmel, R. Estevinho, M. P. Robalo, M. R. Ventura and L. O. Martins, ChemSusChem, 2024, e202401386 CrossRef PubMed.
- W. Ye, Q. Tang, T. Zhou, C. Zhou, C. Fan, X. Wang, C. Wang, K. Zhang, G. Liao and W. Zhou, Eur. J. Med. Chem., 2024, 264, 115988 CrossRef PubMed.
- F. H. Arnold, Angew. Chem., Int. Ed., 2018, 57, 4143–4148 CrossRef PubMed.
- S. Mendes, C. Banha, J. Madeira, D. Santos, V. Miranda, M. Manzanera, M. R. Ventura, W. J. H. van Berkel and L. O. Martins, J. Mol. Catal. B:Enzym., 2016, 133, S34–S43 CrossRef.
- M. Rosa, C. J. Roberts and M. A. Rodrigues, PLoS One, 2017, 12, e0176748 CrossRef PubMed.
- Y. K. Ng, N. N. Tajoddin, P. M. Scrosati and L. Konermann, J. Phys. Chem. B, 2021, 125, 13099–13110 CrossRef PubMed.
- J. M. Sanchez-Ruiz, Biophys. Chem., 2010, 148, 1–15 CrossRef PubMed.
- C. M. Miton, K. Buda and N. Tokuriki, Curr. Opin. Struct. Biol., 2021, 69, 160–168 CrossRef PubMed.
- M. Wilding, N. Hong, M. Spence, A. M. Buckle and C. J. Jackson, Biochem. Soc. Trans., 2019, 47, 701–711 CrossRef PubMed.
- V. Brissos, P. T. Borges, R. Núñez-Franco, M. F. Lucas, C. Frazão, E. Monza, L. Masgrau, T. N. Cordeiro and L. O. Martins, ACS Catal., 2022, 12, 5022–5035 CrossRef PubMed.
- H. C. Brown and S. Krishnamurthy, J. Am. Chem. Soc., 1972, 94, 7159–7161 CrossRef.
- S. Krishnamurthy and H. C. Brown, J. Am. Chem. Soc., 1976, 98, 3383–3384 CrossRef.
- S. Savino and M. W. Fraaije, Biotechnol. Adv., 2021, 51, 107634 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc04449e |
| This journal is © The Royal Society of Chemistry 2025 |