Open Access Article
Open Access ArticleComplementary foods in infants: an in vitro study of the faecal microbial composition and organic acid production†
Vitor
Geniselli da Silva
 ab,
Jane Adair
Mullaney
abc,
Nicole Clémence
Roy
abd,
Nick William
Smith
a,
Clare
Wall
be,
Callum James
Tatton
a and
Warren Charles
McNabb
*ab
ab,
Jane Adair
Mullaney
abc,
Nicole Clémence
Roy
abd,
Nick William
Smith
a,
Clare
Wall
be,
Callum James
Tatton
a and
Warren Charles
McNabb
*ab
aRiddet Institute, Massey University, Palmerston North, Manawatu, New Zealand. E-mail: W.McNabb@massey.ac.nz
bHigh-Value Nutrition National Science Challenge, Auckland, New Zealand
cAgResearch, Palmerston North, New Zealand
dDepartment of Human Nutrition, University of Otago, Dunedin, New Zealand
eDepartment of Nutrition and Dietetics, The University of Auckland, Auckland, New Zealand
First published on 11th April 2025
Abstract
The transition from breastmilk to complementary foods is critical for maturing the colonic microbiota of infants. Dietary choices at weaning can lead to long-lasting microbial changes, potentially influencing health later in life. However, the weaning phase remains underexplored in colonic microbiome research, and the current understanding of how complementary foods impact the infant's colonic microbiota is limited. To address this knowledge gap, this study assessed the influence of 13 food ingredients on the in vitro microbial composition and production of organic acids by the faecal microbiota in New Zealand infants aged 5 to 11 months. To better represent real feeding practices, ingredients were combined with infant formula, other complementary foods, or both infant formula and other foods. Among the individual food ingredients, fermentation with peeled kūmara (sweet potato) increased the production of lactate and the relative abundance of the genus Enterococcus. Fermentation with blackcurrants, strawberries, or raspberries enhanced acetate and propionate production. Additionally, fermentation with blackcurrants increased the relative abundance of the genus Parabacteroides, while raspberry fermentation increased the relative abundance of the genera Parabacteroides and Eubacterium. When combined with infant formula or with blackcurrants, fermenting black beans increased butyrate production and stimulated the relative abundance of Clostridium sensu stricto 1. These foods are promising candidates for future clinical trials.
1. Introduction
The large intestine harbours a diverse microbial community that relies on dietary compounds unabsorbed by the host. Numerous studies have highlighted the crucial role of the colonic microbiota in digestion and have demonstrated the impact of microbial metabolites produced in the colon on host health and well-being.1–3 The relationship between colonic commensals and the host is dynamic and mutual, influenced by multiple factors, with diet playing a major role. Notably, disruptions in faecal microbial composition and concentration of organic acids are often observed in individuals experiencing negative health outcomes, ranging from gastrointestinal diseases to neurological disorders, compared to healthy controls.4–6Dysbiosis in the colonic microbiota (an imbalance in the microbiota) is frequently marked by reduced faecal concentration of short-chain fatty acids (SCFAs).4,7 SCFAs are organic acids produced by the microbial metabolism of complex carbohydrates and benefit the host by supporting intestinal barrier integrity, supplying energy, and regulating metabolic functions, among other benefits.8–11 Given the relationship between the colonic microbiota and host physiology, understanding how diet shapes colonic microbes to promote health has attracted great interest recently. However, research in this area often neglects a critical period for the development of the colonic microbiota: infancy.
In early life, breastmilk is the gold standard for nourishing beneficial colonic commensals.12,13 However, little is known about how complementary foods influence the colonic microbiota when infants start consuming solids (weaning). Longitudinal observations demonstrated that the faecal microbiota, as a proxy of the colonic microbiota, is particularly adaptable during weaning, with dietary-induced changes potentially lasting into later life and affecting long-term health.14,15 At this stage, the gastrointestinal tract is still developing, allowing macronutrients from complementary foods to reach the colon and promote the growth of new commensal microbes.16,17 Therefore, a deeper understanding of how foods impact the microbiota of weaning infants is essential for fostering the adequate development of the colonic microbiota from an early age.
Clinical trials allow for assessing dietary interventions on the faecal microbiota and tracking related health outcomes. However, trials involving vulnerable populations, such as infants, can be particularly time-consuming, costly, and ethically complex. In vitro experimental models, while unable to capture the full complexity of host-microbiota interactions, offer a cheaper and less invasive alternative that addresses some of the ethical and logistical challenges associated with clinical trials.18 Among these methods, static in vitro protocols for food digestion and subsequent faecal fermentation of food remnants provide a useful screening approach to evaluate how dietary compounds influence faecal microbes.19,20
This study investigated the effects of complementary foods on the microbial composition and organic acid production of the faecal microbiota in weaning infants after 24 hours of fermentation. Uniquely, food ingredients were combined with infant formula, other foods, or both to better replicate real-life infant feeding patterns. This research aimed to identify in vitro foods that support adequate development of the faecal microbiota in New Zealand weaning infants.
2. Materials and methods
2.1. Food ingredients
A total of 13 food ingredients were used in this study (Table 1). These included vegetables (pumpkin), legumes (black beans, chickpeas, soybeans, and yellow peas), starchy foods (kūmara and couscous), meat (pork), seafood (prawn), berries (blackcurrants, raspberries, strawberries), and infant formula. These foods were identified through an in silico analysis as candidates for promoting changes in the production of SCFAs by the faecal microbiota of New Zealand weaning infants.21 Foods were purchased from local stores and prepared under various conditions. Fruits and infant formula were obtained as dried powders and used as purchased, while the other ingredients were brought fresh, sous-vide cooked, freeze-dried, and ground using a standardised method (see ESI Table 1† for conditions). Potato starch (Sigma-Aldrich, St Louis, MO, USA) was used as a positive control due to its resistant starch content, which serves as a fermentable substrate for faecal microbes. The moisture content of food ingredient powders was determined by the weight difference before and after 48 hours of incubation at 105 °C. Additionally, compositional analyses of freeze-dried and ground food ingredients were performed at the Massey University Nutrition Laboratory. Analyses were conducted in duplicates for carbohydrates, sugar, total dietary fibre, protein, fat, saturated fat, and energy content (ESI Table 2†).| Ingredient | Description | Source |
|---|---|---|
| Black beans | Dried grains of turtle black beans | Davis Food Ingredients, Palmerston North, New Zealand |
| Blackcurrants | Freeze-dried New Zealand-grown blackcurrants | Fresh As, Auckland, New Zealand |
| Chickpeas | Dried grains of chickpeas (garbanzo beans) | Davis Food Ingredients, Palmerston North, New Zealand |
| Couscous | Medium size grains of dried couscous (Durum wheat) | DARI, Salé, Morocco |
| Infant formula | Nestlé NAN SUPREMEpro 2 | Nestlé New Zealand Limited, Auckland, New Zealand |
| Kūmara | Fresh red kūmara | Countdown, Palmerston North, New Zealand |
| Pork | Fresh lean pork fillet (tenderloin) | Online meats, Ōtāhuhu, New Zealand |
| Prawn | Fresh Australian prawn | Solander Seafood & Fishing, Nelson, New Zealand |
| Pumpkin | Fresh crown pumpkin | Countdown, Palmerston North, New Zealand |
| Raspberries | Freeze-dried New Zealand-grown raspberries | Fresh As, Auckland, New Zealand |
| Soybeans | Dehulled grains of soybeans | Jia Hua Asian Mart, Palmerston North, New Zealand |
| Strawberries | Freeze-dried New Zealand-grown strawberries | Fresh As, Auckland, New Zealand |
| Yellow peas | Dried grains of yellow peas | Davis Food Ingredients, Palmerston North, New Zealand |
2.2 Simulated infant digestion of the food ingredients
Food ingredients were digested in vitro either alone, combined with other foods (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 food-food ratio), combined with infant formula (1
1 food-food ratio), combined with infant formula (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 food-formula ratio), or combined with other foods and infant formula (1
4 food-formula ratio), or combined with other foods and infant formula (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 food-food-formula ratio). These ratios were selected to reflect the high intake of infant formula by formula-fed infants at 6 months of age, which accounts for approximately 80% of their caloric intake.22 A total of 53 samples, each with three replicates, were randomised into batches and independently digested using a protocol adapted to mimic the digestion of a 6-month-old infant. Simulated digestive fluids were prepared as described in the adult INFOGEST protocol,20,23 with enzyme concentrations modified according to a dynamic model for infant digestion24 and a static model for newborn digestion.25
8 food-food-formula ratio). These ratios were selected to reflect the high intake of infant formula by formula-fed infants at 6 months of age, which accounts for approximately 80% of their caloric intake.22 A total of 53 samples, each with three replicates, were randomised into batches and independently digested using a protocol adapted to mimic the digestion of a 6-month-old infant. Simulated digestive fluids were prepared as described in the adult INFOGEST protocol,20,23 with enzyme concentrations modified according to a dynamic model for infant digestion24 and a static model for newborn digestion.25
To simulate oral digestion, 1.5 g of food ingredients were homogenised with 5 mL of deionised water and 5 mL of simulated salivary fluids. No mastication was assumed due to the liquid nature of the resulting mixture. The mixture was incubated for 2 minutes at pH 7.0 and 37 °C with 75 U mL−1 of α-amylase under agitation at 150 rpm. The reaction was stopped with concentrated hydrochloric acid, and simulated gastric fluid was added to bring the volume to 20 mL. The mixture was then incubated for 2 hours at pH 3.0 and 37 °C with 500 U mL−1 of porcine pepsin under agitation at 150 rpm. The reaction was stopped with concentrated sodium hydroxide, and simulated intestinal fluid was added to bring the final volume to 40 mL. Intestinal digestion was simulated by incubating the mixture for 2 hours at pH 7.0 and 37 °C with 100 U mL−1 of protease activity of pancreatin, 200 U mL−1 of pancreatic lipase, 100 U mL−1 of amyloglucosidase, and 10 mmol L−1 of bile salts under agitation at 150 rpm. All chemicals and enzymes were purchased from Sigma-Aldrich (St Louis, MO, USA).
Intestinal digestion was stopped by heat treatment (3 minutes at 95 °C). After digestion, nutrient absorption in the large intestine was simulated by placing digested samples into Spectra/Por® cellulose membrane dialysis tubing (Thermo Fisher Scientific, Waltham, MA, USA) for 24 hours, with at least 2 changes of room-temperature deionised water. Post-dialysis samples were stored at −20 °C until fermentation.
2.3. Faecal fermentation of the digested food ingredients
This research was approved by the Massey University Human Ethics Committee Southern A (Application 22/48). A total of six healthy New Zealand infants at weaning age (5–11 months) were recruited for this study after written consent for their participation was obtained from their primary caregivers. The recruited infants were born at over 32 gestational weeks and weighed more than 2.5 kg (ESI Table 3†). None had received antibiotics in the last three weeks and were not consuming prebiotics or probiotics. All infants had already been exposed to complementary foods and had no known medical conditions.Participants donated multiple faecal samples and were provided with scooping-lid plastic containers and written instructions on collecting and storing the stool samples. Samples were preferentially collected fresh after defaecation and transported refrigerated to the laboratory. Alternatively, samples could be stored in the participant's freezer until transportation. Upon arrival at the laboratory, samples were diluted with 50 mM potassium phosphate buffer pH 6.8 to a concentration of 32% (w/v). The resulting faecal slurry was filtered using a filter bag and stored at −80 °C.
Faecal fermentation followed a standard batch protocol with slight modifications.19 Before fermentation, aliquots from different donors were defrosted and pooled in equal proportions to create an inoculum representative of the faecal microbiota of New Zealand weaning infants. Digested food samples were randomised into independent fermentation batches, and 6 mL of each sample was mixed with 2 mL of 0.15M potassium phosphate buffer pH 7.4 in two 16 × 125 mm Hungate tubes. The potassium phosphate buffer was also used as a negative control. The mixture was degassed with nitrogen, and the headspace of the tubes was filled with carbon dioxide. To ensure the absence of oxygen, 100 μL of 3% (w/v) L-cysteine was added to the tubes. Finally, 2 mL of faecal inoculum was added to the tubes, resulting in a total volume of 10.1 mL. Half of the tubes were immediately incubated on ice (time zero), while the remaining tubes were incubated for 24 hours at 37 °C.
2.4. Gas pressure and pH
After 24 hours of fermentation, the gas pressure of the Hungate tubes was measured (in kPa) using the Go Direct® Gas Pressure Sensor and the software Vernier Graphical Analysis (Vernier Science Education, Beaverton, OR, USA). The pH of samples at the start and end of fermentation was measured using the PL-700AL bench meter (Pacific Sensor Technologies, Rowville, VIC, Australia). Results were expressed as a decrease in pH after 24 hours. Additionally, 1 mL aliquots were collected and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 1 min using the Minispin Plus mini centrifuge (Eppendorf, Hamburg, Germany). The supernatants and pellets were recovered and stored at −80 °C for subsequent analysis of organic acids and microbial composition, respectively.
000g for 1 min using the Minispin Plus mini centrifuge (Eppendorf, Hamburg, Germany). The supernatants and pellets were recovered and stored at −80 °C for subsequent analysis of organic acids and microbial composition, respectively.
2.5. Organic acids analysis
Organic acids were extracted and derivatised following a published protocol,26 with slight modifications. Extractions were performed by mixing 450 μL of fermentation supernatant with 50 μL of the internal standard 50 mM 2-ethyl butyric acid (Sigma-Aldrich, St Louis, MO, USA). Then, 1250 μL of diethyl ether and 250 μL of hydrochloric acid (37%) were added to the mixture. Samples were vortexed, and 100 μL of the diethyl ether phase was transferred to a glass vial containing 20 μL of the derivatising agent N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (Sigma-Aldrich, St Louis, MO, USA). Derivatisation occurred by incubating the mixture for 20 minutes at 80 °C, followed by 48 hours at room temperature.Standard solutions of the organic acids formate, acetate, propionate, isobutyrate, butyrate, isovalerate, valerate, hexanoate, heptanoate, lactate, and succinate, containing 5 mM 2-ethyl butyric acid were prepared alongside the samples. The standard solutions at varying concentrations (0.15, 0.25, 0.50, 1, 2.50, 5, 10, and 20 mM) were used to generate a calibration curve for determining the concentration of the organic acids in the samples. The supernatants from samples at the end of fermentation were diluted with 0.15 M potassium phosphate buffer pH 7.4 to ensure that the concentrations fell within the range of the calibration curve. Organic acid production was calculated as the difference between the concentrations at time zero and 24 hours, expressed in mmol g−1 (dry weight) to account for the theoretical dry mass of the fermented sample.
Organic acids were detected using the GC-2010 gas chromatograph system coupled with a flame ionisation detector (Shimadzu, Kyoto, Japan) and fitted with an HP-1 column (30 m × 0.25 mm ID × 0.25 μm) (Agilent Technologies, Santa Clara, CA, USA). Helium was used as carrier gas with a flow rate of 21.2 mL min−1, a pressure of 131.2 kPa, and a split ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The temperature programme began at 70 °C, increasing to 115 °C at a rate of 6 °C min−1, followed by a final increase to 300 °C at 60 °C min−1, holding for 3 minutes. The detector temperature was 310 °C. Data were acquired and processed using the LabSolutions software (version 5.98) (Shimadzu, Kyoto, Japan).
1. The temperature programme began at 70 °C, increasing to 115 °C at a rate of 6 °C min−1, followed by a final increase to 300 °C at 60 °C min−1, holding for 3 minutes. The detector temperature was 310 °C. Data were acquired and processed using the LabSolutions software (version 5.98) (Shimadzu, Kyoto, Japan).
2.6. Microbial compositional analysis
The DNA from fermentation pellets was extracted using the NucleoSpin DNA Soil kit (Macherey-Nagel, Dueren, Germany) according to the manufacturer's instructions. The quantity of extracted DNA was measured using the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and its quality was assessed through gel electrophoresis using a 1% agarose gel and the lambda HindIII DNA marker (Thermo Fisher Scientific, Waltham, MA, USA). Extracted DNA was stored at −80 °C before sequencing. The V3–V4 regions of the 16S rRNA were amplified using the 341 forward (5′-CCTACGGGAGGCAGCAG-3′) and the 806 reverse (5′-GGACTACHVGGGTWTCTAAT-3′) primers with custom barcodes. All samples from the 24 hours of fermentation were sequenced, while only five randomly selected samples from time zero were sequenced due to resource limitations.PCR amplification, amplicon quantification, purification, and sequencing using a MiSeq platform (Illumina, San Diego, CA, USA) with 2 × 250 bp paired-end reads were performed at Magigene Biotechnology Co. Ltd (Guangzhou, China). Raw data was processed using the New Zealand eScience Infrastructure (NeSI) high-performance computing facilities. In short, primers were removed from raw demultiplexed reads using Cutadapt27 (version 2.3), followed by Trimmomatic.28 The DADA2 pipeline (version 1.32)29 was employed for denoising, truncating reads (to 214 bp for forward reads and 195 bp for reverse reads), chimera removal, and inferring amplicon sequence variants (ASVs) in R (version 4.4).30 Taxonomy assignment was performed using the SILVA database (version 138.1).31
Inconsistencies and missing classifications in the ASV data were addressed using the microbiome package (version 1.26)32 by collapsing taxa into higher taxonomic ranks. Microbial alpha diversity analyses were conducted on unfiltered and unrarefied ASVs using the package phyloseq (version 1.48)33 to measure the Chao1 richness estimator, Shannon index, and Simpson index. For microbial beta diversity analysis, samples were rarified to 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 433 reads, and dissimilarities in microbial abundances were assessed using the Bray–Curtis index. Data were ordinated using principal coordinate analysis (PCoA) based on the Bray–Curtis index employing phyloseq. Microbial relative abundance was visualised using the microViz package (version 0.12.4)34 after filtering taxa that were present in at least 10% of samples and had a relative abundance greater than 0.01%.
433 reads, and dissimilarities in microbial abundances were assessed using the Bray–Curtis index. Data were ordinated using principal coordinate analysis (PCoA) based on the Bray–Curtis index employing phyloseq. Microbial relative abundance was visualised using the microViz package (version 0.12.4)34 after filtering taxa that were present in at least 10% of samples and had a relative abundance greater than 0.01%.
2.7. Statistical analysis
All statistical analyses were performed individually for each subset of samples, which were grouped according to their composition: food ingredients alone, foods combined with infant formula, foods combined with other foods, and foods combined with both infant formula and other foods. A one-way analysis of variance (ANOVA) was used to assess the influence of the substrate (food ingredient or food combination) on pH, gas pressure, and organic acid production after 24 hours of fermentation. Differences in absolute pH changes, gas pressure, and organic acid production between samples were determined using the Tukey Honestly Significant Difference (HSD) test with a 95% confidence level to account for multiple comparisons. Results were plotted using the ggplot2 package (version 3.5.1).35The effect of the substrate on the microbial alpha diversity of samples was assessed using the Kruskal–Wallis test. For diversity indices with significant differences, subsequent pairwise comparisons were performed using Dunn's test via the FSA package (version 0.9.5).36 The Benjamini–Hochberg adjustment was employed to control for false discovery rates. Differences in beta diversity between samples were evaluated through a pairwise permutational multivariate analysis of variance (PERMANOVA), with p-values adjusted using the Benjamini–Hochberg method. Analyses were conducted using the adonis2 function from the vegan package with 9999 permutations (version 2.6–6).37
Differential abundance testing was performed for taxa present in more than 5% of the samples using the ANCOM-BC2 package (version 2.6).38 The ANCOM-BC2 global test served as a preliminary approach to identify taxa varying between at least two samples, while sensitivity analyses assessed the reliability of the results. For taxa identified through the global test, abundance log-fold changes (LFC) between samples were evaluated through multiple pairwise comparisons using a Dunnett's type of test, with p-values adjusted using the Holm-Bonferroni method.
Two-sided Spearman's rank correlation tests were performed to assess the strength of the associations between the following pairs: the nutritional composition of food samples and organic acids produced after 24 hours of fermentation; the nutritional composition of food samples and the relative abundance of microbial genera after 24 hours of fermentation; and produced organic acids and the relative abundance of microbial genera at the end of the fermentation. Only genera with more than 0.05% relative abundance were included in the analyses. The Benjamini–Hochberg method was used to control for false discovery rates. Significant correlations (at a false discovery rate-adjusted p < 0.05) were displayed as heatmaps using the corrplot package (version 0.95).39
3. Results
3.1. Changes in pH and gas pressure
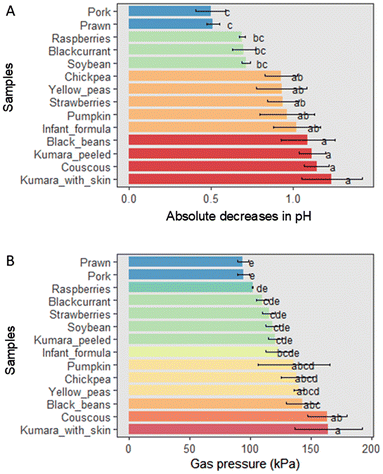
Changes in pH and gas pressure between fermented substrates were only observed for fermentations with food ingredients alone (ANOVA, p < 0.001). Fermentations with kūmara with skin and couscous resulted in the greatest decreases in pH and gas pressures. Fermentation with peeled kūmara also exhibited one of the highest decreases in pH but moderate gas pressure. In contrast, fermenting pork, prawn, raspberries, and blackcurrants promoted the least decreases in pH and the lowest gas pressures (Fig. 1). Fermentation with strawberries showed one of the lowest gas pressures but a moderate decrease in pH. Additionally, combining foods with infant formula resulted in greater pH decreases and increased gas pressures compared to food ingredients alone (ESI Fig. 1 and 2†).3.2. Produced organic acids
After 24 hours of fermentation, fermented food samples produced formate, acetate, propionate, butyrate, isovalerate, lactate, and succinate. Isobutyrate, valerate, hexanoate, and heptanoate were undetected (see ESI Table 4† for detection limits). Individually, fermenting blackcurrants and strawberries resulted in the highest production of organic acids. The same was observed for the fermentation of these berries when combined with infant formula, with each other, or in combination with each other and formula (ESI Fig. 3†).The type of food ingredient significantly influenced the production of formate, acetate, propionate, butyrate, isovalerate, lactate, and succinate, as well as total SCFAs (sum of acetate, propionate, and butyrate) (ANOVA one-way, p < 0.05). Fermentations with blackcurrants, strawberries, and, to a lesser extent, raspberries increased the production of acetate, propionate, and total SCFAs compared to other foods (Tukey HSD, adjusted p < 0.05). The fermentation of kūmara, either peeled or with skin, primarily produced lactate (Fig. 2) (ESI Table 5†).
Fermenting food ingredients combined with infant formula or other foods resulted in fewer differences in organic acid production across samples (ESI Tables 6 and 7†). The type of fermented food-formula combination only influenced butyrate production after adjusting for multiple comparisons, which was highest in fermentation with black beans combined with infant formula (Tukey HSD, adjusted p < 0.005). Similarly, differences between fermented food-food combination samples were only noted for butyrate, with fermentation with black beans combined with blackcurrants yielding the highest production (Tukey HSD, adjusted p < 0.005). No differences in organic acid production were observed between fermentations of food-food-formula combinations after adjusting for multiple comparisons (ESI Table 8†).
3.3. Microbial diversity
Samples at fermentation time zero exhibited higher alpha diversity indices (Chao1, Shannon, and Simpson) compared to those at the end (Kruskal test, p < 0.001) (ESI Fig. 4†). There were no differences in microbial alpha diversity scores between samples at the end of the fermentation after adjusting for multiple comparisons (Dunn's test, adjusted p > 0.05). Samples at fermentation time zero had distinct beta diversity from samples at 24 hours of fermentation, as measured by the Bray–Curtis dissimilarity index (PERMANOVA, p < 0.001) (ESI Fig. 5†). No differences in the Bray–Curtis dissimilarity index were observed between samples at the end of the fermentation after adjusting for multiple comparisons.3.4. Microbial relative abundance
The major phyla present in samples at fermentation time zero were Actinobacteriota, Firmicutes (or Bacillota), Proteobacteria (or Pseudomonadota), and Bacteroidota, with respective relative abundances of 35%, 32%, 20%, and 10%. The most abundant families included Bifidobacteriaceae, Enterobacteriaceae, Lachnospiraceae, and Bacteroidaceae, while the predominant genera were Bifidobacterium, Escherichia-Shigella, Bacteroides, and Veillonella. After 24 hours of fermentation, a shift in the dominant microbial taxa was observed (ESI Tables 9, 10, and 11†). Bacteroidota, followed by Proteobacteria, became the most abundant phyla. The predominant families were Bacteroidaceae, Enterobacteriaceae, Bifidobacteriaceae, and Enterococcaceae, while Bacteroides, Escherichia-Shigella, Bifidobacterium, and Enterococcus became the dominant genera (ESI Fig. 6†).Differential abundance testing for fermentations with food ingredients alone revealed significant changes between samples at the phylum, family, and genus levels (ANCOM-BC2 global test, adjusted p < 0.05) (ESI Tables 9, 10, and 11†). Fermentation with pork, followed by fermentation with raspberries, had the highest relative abundances of the phylum Bacteroidota, the family Bacteroidaceae, and the genus Bacteroides (41% and 40%, respectively). In contrast, fermentation with kūmara with skin exhibited the lowest abundances of these taxa, with Bacteroides accounting for 32%. The family Tannerellaceae and the genus Parabacteroides reached their highest abundances in fermentations with pork and blackcurrants (1% and 0.8%, respectively) and their lowest in fermentations with kūmara both peeled and with skin (0.08%).
The relative abundances of the phylum Actinobacteriota, the family Bifidobacteriaceae, and the genus Bifidobacterium were highest in fermentations with peeled kūmara and prawn (20%) and lowest in fermentations with blackcurrants and strawberries (15%). Fermentations with peeled kūmara, blackcurrants, and raspberries promoted the highest abundances of the phylum Firmicutes, the family Enterococcaceae, and the genus Enterococcus (18%, 17%, and 17%, respectively). In contrast, fermentations with soybeans and chickpeas had the lowest abundances of these taxa, with Enterococcus accounting for 5% of each food. The family Streptococcaceae and the genus Streptococcus were least abundant in fermentations with blackcurrants and strawberries (0.9% each) but showed their highest abundances in fermentation with prawns (2.1%). Additionally, fermentations with kūmara peeled and with skin had the highest abundances of the family Lactobacillaceae and the genus Lacticaseibacillus. Fermentations with raspberries and blackcurrants exhibited the highest abundances of the family Eubacteriaceae and the genus Eubacterium.
Multiple pairwise comparisons against a reference group (ANCOM-BC2 Dunnett-type test) demonstrated that fermentations with blackcurrants and raspberries significantly increased the log-fold change (LFC) in the abundance of the family Tannerellaceae and the genus Parabacteroides and, to a lesser extent Enterococcus, compared to fermentations with other food ingredients (adjusted p < 0.05) (Fig. 3) (ESI Fig. 7†). Importantly, LFC values represent differences in bias-correct abundances between groups and do not directly reflect the relative abundance of taxa. Fermentation with raspberries also exhibited higher LCF values for the phylum Firmicutes, the families Eubacteriaceae and Enterococcaceae, and the genera Sellimonas and Eubacterium compared to fermentations with other foods (ESI Fig. 8†). In contrast, fermentations with kūmara peeled or with skin decreased the LFC in the abundance of the genus Parabacteroides compared to fermentations with other foods (Fig. 3).
Significant differences in taxa relative abundance between fermentations with food-food combinations were at the phylum, family, and genus levels (ANCOM-BC2 global test, p adjusted < 0.05). Fermentation with the couscous-pork combination promoted the highest abundances of the phylum Bacteroidota, the family Bacteroidaceae, and the genus Bacteroides (43%), while fermentation with couscous-pumpkin exhibited the lowest abundance of these taxa (35%). The family Tannerellaceae and the genus Parabacteroides showed the highest relative abundances in the fermentation with pork-raspberries (1.2%) and the lowest in the fermentation with couscous-pork (0.2%).
The phylum Proteobacteria and the family Enterobacteriaceae had the highest abundances in the fermentation with the blackcurrants-strawberries combination (30%) and the lowest in the fermentation with blackcurrants-kūmara with skin (20%). In contrast, the phylum Actinobacteria, the family Bifidobacteriaceae, and the genus Bifidobacterium exhibited the highest abundances in the fermentation with black beans-blackcurrants (19%) and the lowest in the fermentation with blackcurrants-strawberries (14%). Fermentation with blackcurrants-pork had the highest relative abundances of the phylum Verrucomicrobiota, the family Akkermansiaceae, and the genus Akkermansia (1.4%).
The relative abundances of the phylum Firmicutes, the family Enterococcaceae, and the genus Enterococcus were highest in fermentations with blackcurrants combined with kūmara peeled or kūmara with skin (19%). In contrast, the fermentation of the combination blackcurrants-soybean exhibited the lowest abundance of these taxa, with Enterococcus accounting for 6%. The families Eubacteriaceae and Clostridiaceae and the genera Eubacterium and Clostridium sensu stricto 1 had their highest relative abundances in fermentation with black beans-blackcurrants (0.2% each).
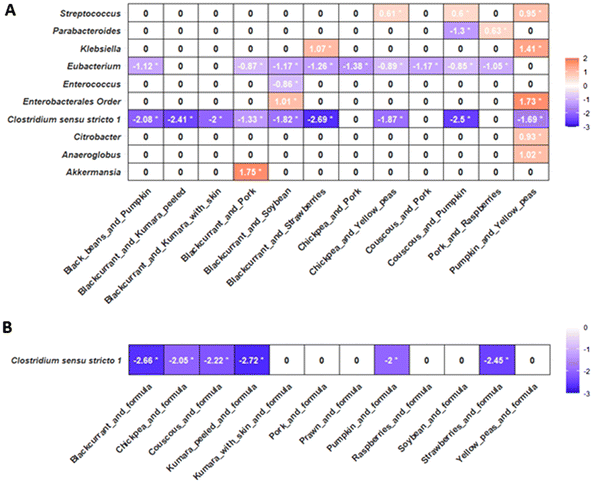
Multiple pairwise comparisons of taxa LFC demonstrated that fermentations with the combination of black beans-blackcurrants had increased LFC values in the abundance of the genera Eubacterium and Clostridium sensu stricto 1 compared to fermentations with other food-food combinations (Fig. 4). Fermentation with black beans-blackcurrant also had higher LFC for the families Eubacteriaceae and Clostridiaceae (ANCOM-BC 2 Dunnett's test, adjusted p < 0.05), while no significant differences were observed at the phylum level (ESI Fig. 9†).
Combining infant formula with food ingredients or food-food combinations reduced the observed differences in microbial relative abundance between fermented samples. No differences in the abundance of bacterial phyla, families, or genera detected between fermentations with food-formula combinations by the ANCOM-BC2 global test passed the sensitivity analyses. This suggests that the variations between fermented samples were likely due to model parameters or assumptions rather than biological differences. Multiple pairwise comparisons, using fermentation with black beans-formula as a reference group due to its increased butyrate produced after 24 hours of fermentation, showed increased LFC in the abundance of the family Clostridiaceae and the genus Clostridium sensu stricto 1 (adjusted p < 0.05), compared to fermentations with other food-formula combinations (Fig. 4).
Significant differences in taxa abundance between fermentations with food-food-formula combinations were observed at the family and genus levels (ANCOM-BC global test, adjusted p < 0.05). The family Bacteroidaceae and the genus Bacteroides had the highest abundances in fermentation with chickpea-yellow peas-formula (43%) and the lowest in fermentation with blackcurrants-kūmara with skin-formula (32%). Additionally, fermentation with chickpea-yellow peas-formula combination exhibited the lowest abundances of the families Streptococcaceae and Eubacteriaceae and the genera Streptococcus and Eubacterium (1.2% and 0.06%, respectively). In contrast, fermentation with couscous-pork-formula promoted the highest abundances of these taxa (1.7% for Streptococcus and 0.1% for Eubacterium). No significant changes in bacterial taxa LFC values between fermentations with food-food-formula combinations were observed after multiple pairwise comparisons using the fermentation with blackcurrants-strawberries-formula combination as a reference group.
3.5. Correlations between food composition, organic acids and microbiota composition
The produced major and total SCFAs exhibited weak positive correlations with the total dietary fibre content across all fermented food samples (Spearman's rank correlation, adjusted p < 0.05). In contrast, fat, energy, protein, and sugar content correlated negatively with the production of these organic acids (Fig. 5). Notably, acetate production had a strong negative correlation with fat and energy content (Spearman's rank correlation coefficient rs values of −0.66 and −0.72, respectively). Lactate production also showed negative correlations with energy, fat, and protein content but had a weak positive correlation with carbohydrate content. Additionally, when analysing individual food ingredients and food-food combinations, lactate production had a strong positive correlation with carbohydrate content and a strong negative correlation with fat content (ESI Fig. 10†).The relative abundances of the genera Veillonella and Enterococcus were positively correlated with the total fibre content in all samples. Trends indicating a weak positive correlation between fibre content and the relative abundance of the genera Parabacteroides and Lacticaseibacillus were also observed (Spearman's rank correlation, adjusted p < 0.1). Furthermore, the relative abundance of Lacticaseibacillus demonstrated a moderate positive correlation with carbohydrate content when analysing only food ingredients (rs = 0.52). In contrast, energy, fat, and sugar content negatively correlated with the relative abundance of Enterococcus and Lacticaseibacillus (rs ranging from −0.23 to −0.52), while they positively correlated with the relative abundance of the genera Streptococcus and Blautia (rs ranging from 0.22 to 0.60). Protein content exhibited weak positive correlations with the relative abundance of the genera Akkermansia, Anaeroglobus, Clostridium sensu stricto 1, and Streptococcus, also showing a trend toward a positive correlation with the abundance of Bacteroides (Fig. 5). A moderate positive correlation was also observed between Clostridium sensu stricto 1 abundance and protein content in food-food combinations (rs = 0.49) (ESI Fig. 10†).
When considering the entire set of samples, the production of acetate, propionate, and total SCFAs positively correlated with the relative abundance of Parabacteroides, Lacticaseibacillus, and Enterococcus, among other genera (Fig. 5). Notably, there were strong correlations between the abundance of Parabacteroides and propionate (rs = 0.50) and between Enterococcus and acetate (rs = 0.43). Enterococcus abundance also showed positive correlations with lactate production, alongside Lacticaseibacillus, as well as with butyrate production in conjunction with Clostridium sensu stricto 1. Additionally, the relative abundance of Lacticaseibacillus demonstrated a moderate positive correlation with lactate production from food ingredients, while Clostridium sensu stricto 1 exhibited a similar correlation with butyrate production from food-formula combinations (ESI Fig. 10†). In contrast, the abundances of Streptococcus and Blautia exhibited negative correlations with the production of major and total SCFAs (rs ranging from −0.57 to −0.24).
4. Discussion
Our study evaluated the effects of complementary foods from various sources, including meat, seafood, starchy foods, and fruits, on the composition and function of the faecal microbiota of New Zealand weaning infants. The transition from breastmilk to solid foods is a critical period for the development of the colonic microbiota of infants, yet it has been traditionally neglected in microbiome research.40 To address this knowledge gap in infant nutrition, this study assessed the fermentation of food ingredients combined with infant formula, other foods, or both other foods and infant formula. This unique aspect of our study aims to better replicate how complementary foods are introduced to infants in real life while also considering the impact of interactions between different dietary compounds on the relative abundance and organic acid production of colonic microbes. Another strength of our study was using standardised protocols and the latest methods for in vitro digestion, faecal fermentation, DNA sequencing, and bioinformatics.Among the food ingredients, fermentation with kūmara (sweet potato) or couscous produced the most gas and promoted the greatest pH changes. In turn, fermentation with pork, followed by fermentation with prawn, blackcurrants, or raspberries, had the lowest values for both measurements. Another in vitro study using faecal inoculum from weaning infants assessed the fermentation of plant-based foods, reporting that fermenting oats, sweetcorn, and carrot produced more gas than apple, blackcurrants, and kiwifruit.41 These findings indicate that the infant microbiota has adapted to fermenting complex carbohydrates rather than sugar or animal protein.
In particular, the soluble fibre content may be a major factor influencing pH and gas production during fermentation. Evidence from swine faecal fermentation of different ratios of soluble to insoluble fibre indicates that a higher proportion of soluble fibre increases total gas production while decreasing pH.42 Additionally, soluble fibre content was associated with higher production of lactate and acetate, whereas insoluble fibres were associated with propionate and butyrate yield.42 However, the study used simple substrates derived from mixes of inulin with non-starch polysaccharides, which did not reflect the complexity of foods. In addition to carbohydrates, foods contain various other components, such as fats and protein, which influence the digestion of other nutrients and impact colonic microbes (see reviews43,44). Furthermore, phytochemicals found in plant-based foods can be metabolised by colonic microbes, generating more absorbable and bioactive molecules,45 or exhibit antimicrobial or prebiotic properties, selectively promoting the growth of certain microbes in the colon.46,47
In our study, fermentation with kūmara produced more lactate than fermentations with other food ingredients. This result is likely an artifact of the in vitro static fermentation, as lactate accumulation is often observed in faecal fermentations due to an excess of fermentable substrates.48 Kūmara is rich in complex carbohydrates, primarily in the form of starch, and contains pectin as a soluble fibre and cellulose, hemicellulose, and lignin as an insoluble fibre.49 Consistent with the increased lactate production, fermentation with peeled kūmara promoted the highest relative abundances of lactic acid bacteria from the genera Bifidobacterium, Enterococcus, and Lacticaseibacillus. Additionally, fermentation with peeled kūmara had higher LFC values for the abundance of the genus Enterococcus compared to other food ingredients. Correlation analyses demonstrated that the carbohydrate content in food ingredients was positively associated with lactate production and the relative abundance of Lacticaseibacillus. Furthermore, Lacticaseibacillus and Enterococcus abundances were positively linked to lactate production and total dietary fibre content.
These bacteria belong to a group of potentially beneficial microbes that produce lactate as their major fermentation product, ultimately contributing to SCFAs produced in the colon through cross-feeding with other microbes.50,51 For instance, the Bifidobacterium genus breaks down carbohydrates, particularly human milk oligosaccharides, through the fructose 6-phosphate pathway to produce acetate and lactate. Most members of the Lacticaseibacillus genus (previously classified under Lactobacillus) are homofermentative, mainly converting carbohydrates into lactate, although some strains are heterofermentative and produce acetate.52
The highest lactate yield in the fermentation with kūmara also explains the greatest pH drop, as lactic acid is a stronger acid than the other major SCFAs. In the colon, lactate can be oxidised to pyruvate and subsequently converted into acetyl-CoA, contributing to the pool of acetate and butyrate,53,54 while propionate can be generated from lactate via methylmalonyl-CoA or acrylyl-CoA pathways.55 Notably, the microbial conversion of lactate into other major SCFAs is sensitive to pH. Evidence in vitro demonstrated that lactate is efficiently transformed into propionate and butyrate at around pH 6.5, whereas at pH 5.5 or lower, its conversion is inhibited, resulting in lactate accumulation and overabundance of Bifidobacteria.56,57 However, it is important to acknowledge that such low pH conditions do not accurately reflect the physiology of the human colon.
In line with those findings, we observed that the fermentation with kūmara peeled or with skin produced some of the lowest amounts of acetate and propionate, also having reduced abundance of the genus Parabacteroides. This suggests that lactate was not efficiently converted into SCFAs and accumulated during fermentation. Our study used a static fermentation protocol, which does not reflect the dynamic inflow and outflow of substances in the human colon. Under more realistic conditions, lactate produced from kūmara fermentation may contribute to greater production of SCFAs, ultimately conferring host benefits.
Research in rats showed that dietary fibre from sweet potatoes stimulated the growth of Bifidobacterium and Lactobacillus during in vitro faecal fermentation.58 Additionally, it increased faecal propionate and butyrate levels in rats that received sweet potato fibre supplementation for four weeks.58 Similarly, faecal fermentation studies using adult inoculum reported that whole sweet potato and its extracted fibre promoted the production of major SCFAs and the abundance of bifidobacteria.59–61 Currently, there is no published research on the effect of kūmara on the colonic microbiota of weaning infants. However, two ongoing clinical trials are evaluating this topic, and their results could provide valuable insights into the field of infant nutrition.62,63
Fermentations of blackcurrants, strawberries and raspberries led to the highest production of acetate and propionate. Consistently, fermentation with raspberries, followed by fermentation with blackcurrants, exhibited the highest abundances of the genus Eubacterium. Additionally, LFC values in the abundance of the genus Parabacteroides were greater in fermentations with raspberries or with blackcurrants compared to other food ingredients. These genera encode carbohydrate-active enzymes (CAZy), allowing them to degrade complex carbohydrates to produce SCFAs.64,65 In contrast, genera colonising the colon in early life, such as Streptococcus and Bifidobacterium, had the lowest abundances in fermentations with blackcurrants or strawberries, suggesting a transition from infant to adult microbiota.
Blackcurrants, strawberries, and raspberries are sources of dietary fibre, particularly insoluble fibre (mostly cellulose), and contain high amounts of polyphenols, mainly anthocyanins, flavonols, ellagitannins, and ellagic acid.66–68 Dietary fibre is fermented by colonic bacteria, primarily producing gases and SCFAs.8,69 In addition, evidence suggests that polyphenols and dietary fibre synergistically affect the colonic microbiota by changing the carbohydrate metabolism of colonic commensals. For instance, cranberry proanthocyanidins enhanced the fermentation of xyloglucans, a type of soluble fibre, by lactic acid bacteria in vitro, leading to increased acetate production.70 Whole-fruit cranberry powder, rather than its fibrous fraction alone, was more efficient in restoring colonic dysbiosis and reducing body weight in obese mice.71
Consistently, the production of major and total SCFAs positively correlated with the dietary fibre content of fermented foods here. The production of acetate, propionate and total SCFAs were also positively associated with a higher relative abundance of Parabacteroides and lower abundances of Streptococcus and Bifidobacterium genera. In agreement with our findings, the faecal fermentation of raspberry using adult inoculum mainly produced acetate and propionate.72 Additionally, the same study demonstrated that polyphenols contributed more to the production of these SCFAs than dietary fibres.72 A four-week raspberry intervention in prediabetic adults reported no changes in faecal microbial alpha and beta diversity compared to baseline values, but an increase in the relative abundance of Eubacterium eligens and Clostridium orbiscindens, as well as reduced plasma total and low-density lipoprotein cholesterol.73
In contrast to our results, another faecal fermentation study using inoculum from weaning infants found no changes in the production of acetate, propionate, and butyrate between blackcurrant and control fermentations.74 However, clinical studies assessing the effect of blackcurrant intervention observed an increase in the faecal abundance of the Ruminococcus genus in postmenopausal women after six months;75 as well as an increase in the faecal abundance of the genera Lactobacillus and Bifidobacterium, alongside a decrease in the abundance of Clostridium and Bacteroides genera after two weeks in healthy adults.76
Little is known about the impact of strawberries on the human colonic microbiota. A study using mice with colitis reported that strawberry supplementation increased the faecal abundance of the genera Bifidobacterium and Lactobacillus, as well as the caecal content of SCFAs.77 Additionally, strawberry supplementation increased the colonic abundance of Bifidobacterium in diabetic mice.78 A four-week trial involving healthy adults who consumed strawberries observed increased faecal abundance of the genera Akkermansia, Bacteroides, and Bifidobacterium but no changes in faecal SCFA levels.79 The evidence above suggests that blackcurrants, strawberries, and raspberries are promising complementary foods for increasing the abundance of SCFA-producing bacteria in the colonic microbiota of infants.
Unlike other major SCFAs, butyrate production did not vary between food ingredient fermentations. However, when black beans were fermented with infant formula or blackcurrants, there was an increase in butyrate production compared to other food-formula or food-food combinations. Similarly, combining black beans with infant formula or blackcurrants in fermentation led to the highest relative abundance of Clostridium sensu stricto 1, a group of bacteria that metabolise carbohydrates and amino acids, producing butyrate via butyryl-CoA and butyrate kinase pathways.80,81 Correlation analyses supported these findings, showing a relationship between higher protein content and increased relative abundance of Clostridium sensu stricto 1, which abundance was also positively associated with butyrate production.
Black beans are a source of protein, dietary fibre, and polyphenols, notably containing high amounts of resistant starch.82 Additionally, soaking and cooking beans before consumption further increases their resistant starch content.83 Traditionally, the colonic fermentation of resistant starch produces butyrate through a cross-feeding mechanism involving key resistant starch degraders, such as Ruminococcus bromii and Bifidobacterium adolescentis, along with butyrate producers from the genera Faecalibacterium, Roseburia, Eubacterium, and Anaerostipes.84,85 However, recent evidence demonstrated that members of Clostridium sensu stricto 1 can also produce butyrate from resistant starch.86
Previous faecal fermentation studies evaluating the effect of black beans on colonic microbes have shown contrasting results. One reported that black beans exhibited a prebiotic effect by increasing the abundance of Bifidobacterium and Lactobacillus genera during fermentation. This increase was associated with a rise in the production of acetate and propionate but a decrease in butyrate levels compared to the fermentation control.87 On the other hand, another study observed that the fermentation of the insoluble indigestible fraction of black beans produced butyrate, as well as acetate and propionate.88 It is important to note that neither study specified the age of the faecal donors nor evaluated changes in the overall composition of the microbiota, limiting comparison with our findings.
Evidence in murine models suggests that consuming black beans benefits the microbiota by increasing the abundance of key taxa producing SCFAs and subsequently leading to greater production of SCFAs. For instance, healthy mice had increased faecal abundance of Prevotella and caecal contents of acetate, propionate, and butyrate after black bean intervention.89 Similarly, rats fed a high fat and sugar diet supplemented with cooked beans exhibited increased faecal abundance of the Clostridia class and the genera Ruminococcus, Coprococcus, and Prevotella, as well as elevated faecal butyrate levels.90 In contrast, a navy bean intervention did not alter faecal SCFA content in overweight adults, while a common bean intervention in weaning infants showed no changes in faecal microbiota diversity of taxa abundance.91,92
Unexpectedly, combining foods with infant formula drastically reduced the variability in organic acid production, taxa abundance, and microbial diversity scores between samples. Since the food-formula combinations consisted of 80% infant formula by mass, this high proportion of formula probably masked the effects of the individual food ingredients on colonic microbes. Similarly, we observed fewer changes in microbiota composition and SCFA production between fermentations with food-food and food-food-formula combinations, suggesting that the impact of specific foods on colonic microbes is less evident when considering the overall dietary pattern. Ultimately, long-term dietary patterns rather than spontaneous consumption of individual foods are more likely to promote notable and lasting changes in colonic commensals.93,94
Nevertheless, our study has limitations. During the transition to solid foods, infants often continue to consume breastmilk.95 However, our study did not evaluate the effect of combining complementary foods with human milk. Instead, breastmilk was replaced with infant formula, which may have influenced the observed effects of complementary foods on the faecal microbiota of weaning infants. This limitation is particularly relevant during the first year of life, as breastfed infants have distinct faecal microbiota and metabolite profiles compared to formula-fed infants.96
Faeces were used due to the ease of collection and non-invasive procedure, which are essential when involving vulnerable participants. However, faecal samples mainly represent microbial communities from the distal colon and do not accurately reflect the microbes that adhere to the mucosa or those found in the proximal colon.97 Due to the screening approach of this study, static protocols were used to simulate infant digestion and subsequent colonic fermentation of foods. These static conditions do not capture the dynamic nature of the gastrointestinal tract of infants. Notably, in static faecal fermentations, microbial metabolites can accumulate, and substrates may become depleted, potentially distorting the microbial community compared to what would be found in a dynamic environment.98
The microbial composition was characterised by 16S rRNA sequencing. While this method is accurate, it has limitations, particularly in resolution and cannot resolve taxonomy at the species level.99 The composition of the identified bacterial taxa was expressed as relative abundances, indicating the proportion of individual microbes within the entire community. Consequently, this may lead to an inaccurate characterisation of the actual microbial community.
Our study evaluated a higher proportion of plant-based foods compared to animal-based foods. This choice was justified by our previous research, which identified complementary foods with the greatest potential for producing SCFAs in silico.21 Although the food ingredients were prepared as similarly as possible to real-life conditions, the preparation likely altered their original structure, ultimately influencing their impact on colonic microbes.100 For instance, cooking and cooling plant-based foods can increase their resistant starch content.83 Finally, while correlation analyses could link changes in the production of SCFAs or microbial relative abundance to protein, fat, and fibre content in the evaluated foods, phytochemicals were not analysed, which is warranted in further research.
Finally, as our study focused on the faecal microbiota of New Zealand weaning infants, our findings may not be directly generalisable to infants from other geographic locations. Geographic location is known to influence the composition of the infant faecal microbiota.101 Furthermore, dietary patterns and eating habits may differ across countries and cultures.102 Consequently, the complementary foods evaluated in our in vitro study may not fully represent those consumed by weaning infants in other parts of the world.
5. Conclusions
This study investigated how various food ingredients and food combinations affect the composition and function of the colonic microbiota in New Zealand weaning infants in vitro. Foods promoting the most favourable changes in the infant microbiota were identified. Notably, fermentation with kūmara, a variety of sweet potatoes rich in complex carbohydrates, effectively promoted lactate production by stimulating the growth of the lactic acid bacteria from the genera Enterococcus and Lacticaseibacillus. Fermentation with blackcurrants, strawberries, and raspberries, notable sources of dietary fibre and polyphenols, increased acetate and propionate production. This increase was linked to a higher relative abundance of Parabacteroides and Eubacterium genera. Additionally, when black beans were fermented with infant formula or blackcurrants, they produced the highest yields of butyrate and increased the abundance of the group Clostridium sensu stricto 1. This is likely due to the high protein and resistant starch content in black beans. Overall, these findings contribute to an under-investigated topic of colonic microbiome research in infants. Kūmara, berries, and black beans are promising candidates for further clinical trials involving infants.Author contributions
Vitor Geniselli: conceptualisation, data curation, formal analysis, investigation, visualisation, and writing – original draft. Jane Mullaney: conceptualisation, data curation, formal analysis, supervision, and writing – review & editing. Nicole Roy: conceptualisation, funding acquisition, supervision, and writing – review & editing. Nick Smith: supervision and writing – review & editing. Clare Wall: funding acquisition, supervision, and writing – review & editing. Callum Tatton: investigation. Warren McNabb: conceptualisation, funding acquisition, project administration, supervision, and writing – review & editing.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
The authors acknowledge the use of New Zealand eScience Infrastructure (NeSI) high-performance computing facilities as part of this research. A thanks to all the participants involved and to Paul MacLean for providing insights into the statistical analyses. Vitor Geniselli is supported by a Riddet Institute PhD Scholarship. The PhD stipend and the project were funded by the High-Value Nutrition National Science Challenge. This study was funded by the New Zealand Ministry for Business, Innovation and Employment (MBIE, Grant No. 3710040) through the High-Value Nutrition National Science Challenge.References
- A. S. Raman, J. L. Gehrig, S. Venkatesh, H.-W. Chang, M. C. Hibberd, S. Subramanian, G. Kang, P. O. Bessong, A. A. M. Lima, M. N. Kosek, W. A. Petri, D. A. Rodionov, A. A. Arzamasov, S. A. Leyn, A. L. Osterman, S. Huq, I. Mostafa, M. Islam, M. Mahfuz, R. Haque, T. Ahmed, M. J. Barratt and J. I. Gordon, A sparse covarying unit that describes healthy and impaired human gut microbiota development, Science, 2019, 365, 6449 CrossRef PubMed.
- A. Velikonja, L. Lipoglavšek, M. Zorec, R. Orel and G. Avguštin, Alterations in gut microbiota composition and metabolic parameters after dietary intervention with barley beta glucans in patients with high risk for metabolic syndrome development, Anaerobe, 2019, 55, 67–77 CrossRef CAS PubMed.
- W. Ma, L. H. Nguyen, M. Song, D. D. Wang, E. A. Franzosa, Y. Cao, A. Joshi, D. A. Drew, R. Mehta, K. L. Ivey, L. L. Strate, E. L. Giovannucci, J. Izard, W. Garrett, E. B. Rimm, C. Huttenhower and A. T. Chan, Dietary fiber intake, the gut microbiome, and chronic systemic inflammation in a cohort of adult men, Genome Med., 2021, 13, 102 CrossRef CAS PubMed.
- S.-J. Chen, C.-C. Chen, H.-Y. Liao, Y.-T. Lin, Y.-W. Wu, J.-M. Liou, M.-S. Wu, C.-H. Kuo and C.-H. Lin, Association of Fecal and Plasma Levels of Short-Chain Fatty Acids With Gut Microbiota and Clinical Severity in Patients With Parkinson Disease, Neurology, 2022, 98, 848–858 Search PubMed.
- L. Zhu, S. S. Baker, C. Gill, W. Liu, R. Alkhouri, R. D. Baker and S. R. Gill, Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH, Hepatology, 2013, 57, 601–609 CrossRef CAS PubMed.
- R. Pittayanon, J. T. Lau, G. I. Leontiadis, F. Tse, Y. Yuan, M. Surette and P. Moayyedi, Differences in Gut Microbiota in Patients With vs Without Inflammatory Bowel Diseases: A Systematic Review, Gastroenterology, 2020, 158, 930–946 CrossRef PubMed.
- S. Sanna, N. R. van Zuydam, A. Mahajan, A. Kurilshikov, A. Vich Vila, U. Võsa, Z. Mujagic, A. A. M. Masclee, D. M. A. E. Jonkers, M. Oosting, L. A. B. Joosten, M. G. Netea, L. Franke, A. Zhernakova, J. Fu, C. Wijmenga and M. I. McCarthy, Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases, Nat. Genet., 2019, 51, 600–605 CrossRef CAS PubMed.
- M. Calatayud, P. Van den Abbeele, J. Ghyselinck, M. Marzorati, E. Rohs and A. Birkett, Comparative Effect of 22 Dietary Sources of Fiber on Gut Microbiota of Healthy Humans in vitro, Front. Nutr., 2021, 8, 700571 CrossRef PubMed.
- H.-B. Wang, P.-Y. Wang, X. Wang, Y.-L. Wan and Y.-C. Liu, Butyrate Enhances Intestinal Epithelial Barrier Function via Up-Regulation of Tight Junction Protein Claudin-1 Transcription, Dig. Dis Sci., 2012, 57, 3126–3135 CrossRef CAS PubMed.
- R. Soret, J. Chevalier, P. D. Coppet, G. Poupeau, P. Derkinderen, J. P. Segain and M. Neunlist, Short-Chain Fatty Acids Regulate the Enteric Neurons and Control Gastrointestinal Motility in Rats, Gastroenterology, 2010, 138, 1772–1782 CrossRef CAS PubMed.
- T. Grüter, N. Mohamad, N. Rilke, A. Blusch, M. Sgodzai, S. Demir, X. Pedreiturria, K. Lemhoefer, B. Gisevius, A. Haghikia, A. L. Fisse, J. Motte, R. Gold and K. Pitarokoili, Propionate exerts neuroprotective and neuroregenerative effects in the peripheral nervous system, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2216941120 CrossRef PubMed.
- B. M. Henrick, L. Rodriguez, T. Lakshmikanth, C. Pou, E. Henckel, A. Arzoomand, A. Olin, J. Wang, J. Mikes, Z. Tan, Y. Chen, A. M. Ehrlich, A. K. Bernhardsson, C. H. Mugabo, Y. Ambrosiani, A. Gustafsson, S. Chew, H. K. Brown, J. Prambs, K. Bohlin, R. D. Mitchell, M. A. Underwood, J. T. Smilowitz, J. B. German, S. A. Frese and P. Brodin, Bifidobacteria-mediated immune system imprinting early in life, Cell, 2021, 184, 3884–3898 CrossRef CAS PubMed.
- A. Marcobal, M. Barboza, J. W. Froehlich, D. E. Block, J. B. German, C. B. Lebrilla and D. A. Mills, Consumption of Human Milk Oligosaccharides by Gut-Related Microbes, J. Agric. Food Chem., 2010, 58, 5334–5340 CrossRef CAS PubMed.
- M. Kalliomäki, M. Carmen Collado, S. Salminen and E. Isolauri, Early differences in fecal microbiota composition in children may predict overweight, Am. J. Clin. Nutr., 2008, 87, 534–538 CrossRef PubMed.
- M.-C. Arrieta, L. T. Stiemsma, P. A. Dimitriu, L. Thorson, S. Russell, S. Yurist-Doutsch, B. Kuzeljevic, M. J. Gold, H. M. Britton, D. L. Lefebvre, P. Subbarao, P. Mandhane, A. Becker, K. M. McNagny, M. R. Sears, T. Kollmann, Child Study Investigators, W. W. Mohn, S. E. Turvey and B. B. Finlay, Early infancy microbial and metabolic alterations affect risk of childhood asthma, Sci. Transl. Med., 2015, 7, 307 Search PubMed.
- E. Fournier, C. Roussel, A. Dominicis, D. Ley, M.-A. Peyron, V. Collado, M. Mercier-Bonin, C. Lacroix, M. Alric, T. Van de Wiele, C. Chassard, L. Etienne-Mesmin and S. Blanquet-Diot, In vitro models of gut digestion across childhood: current developments, challenges and future trends, Biotechnol. Adv., 2022, 54, 107796 CrossRef PubMed.
- C. J. Stewart, N. J. Ajami, J. L. O'Brien, D. S. Hutchinson, D. P. Smith, M. C. Wong, M. C. Ross, R. E. Lloyd, H. Doddapaneni, G. A. Metcalf, D. Muzny, R. A. Gibbs, T. Vatanen, C. Huttenhower, R. J. Xavier, M. Rewers, W. Hagopian, J. Toppari, A.-G. Ziegler, J.-X. She, B. Akolkar, A. Lernmark, H. Hyoty, K. Vehik, J. P. Krischer and J. F. Petrosino, Temporal development of the gut microbiome in early childhood from the TEDDY study, Nature, 2018, 562, 583–588 CrossRef CAS PubMed.
- C. F. Williams, G. E. Walton, L. Jiang, S. Plummer, I. Garaiova and G. R. Gibson, Comparative Analysis of Intestinal Tract Models, Annu. Rev. Food Sci. Technol., 2015, 6, 329–350 CrossRef CAS PubMed.
- S. Pérez-Burillo, S. Molino, B. Navajas-Porras, Á. J. Valverde-Moya, D. Hinojosa-Nogueira, A. López-Maldonado, S. Pastoriza and J. Á. Rufián-Henares, An in vitro batch fermentation protocol for studying the contribution of food to gut microbiota composition and functionality, Nat. Protoc., 2021, 16, 3186–3209 CrossRef PubMed.
- A. Brodkorb, L. Egger, M. Alminger, P. Alvito, R. Assunção, S. Ballance, T. Bohn, C. Bourlieu-Lacanal, R. Boutrou, F. Carrière, A. Clemente, M. Corredig, D. Dupont, C. Dufour, C. Edwards, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. R. Mackie, C. Martins, S. Marze, D. J. McClements, O. Ménard, M. Minekus, R. Portmann, C. N. Santos, I. Souchon, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and I. Recio, INFOGEST static in vitro simulation of gastrointestinal food digestion, Nat. Protoc., 2019, 14, 991–1014 CrossRef CAS PubMed.
- V. G. da Silva, N. W. Smith, J. A. Mullaney, C. Wall, N. C. Roy and W. C. McNabb, Food-breastmilk combinations alter the colonic microbiome of weaning infants: an in silico study, mSystems, 2024, 9, 9 Search PubMed.
- M. Heinig, L. Nommsen, J. Peerson, B. Lonnerdal and K. Dewey, Energy and protein intakes of breast-fed and formula-fed infants during the first year of life and their association with growth velocity: the DARLING Study, Am. J. Clin. Nutr., 1993, 58, 152–161 CrossRef CAS PubMed.
- M. Minekus, M. Alminger, P. Alvito, S. Ballance, T. Bohn, C. Bourlieu, F. Carrière, R. Boutrou, M. Corredig, D. Dupont, C. Dufour, L. Egger, M. Golding, S. Karakaya, B. Kirkhus, S. L. Feunteun, U. Lesmes, A. Macierzanka, A. Mackie, S. Marze, D. J. McClements, O. Ménard, I. Recio, C. N. Santos, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and A. Brodkorb, A standardised static in vitro digestion method suitable for food – an international consensus, Food Funct., 2014, 5, 1113–1124 RSC.
- F. Passannanti, F. Nigro, M. Gallo, F. Tornatore, A. Frasso, G. Saccone, A. Budelli, M. V. Barone and R. Nigro, In vitro dynamic model simulating the digestive tract of 6-month-old infants, PLoS One, 2017, 12, e0189807 CrossRef PubMed.
- O. Ménard, C. Bourlieu, S. C. De Oliveira, N. Dellarosa, L. Laghi, F. Carrière, F. Capozzi, D. Dupont and A. Deglaire, A first step towards a consensus static in vitro model for simulating full-term infant digestion, Food Chem., 2018, 240, 338–345 CrossRef PubMed.
- S. G. Parkar, C. M. H. Jobsis, T. D. Herath, H. M. Stoklosinski, J. W. van Klink, C. E. Sansom, I. M. Sims and D. I. Hedderley, Metabolic and microbial responses to the complexation of manuka honey with α-cyclodextrin after simulated gastrointestinal digestion and fermentation, J. Funct. Foods, 2017, 31, 266–273 CrossRef CAS.
- M. Martin, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet J., 2011, 17, 10–12 CrossRef.
- A. M. Bolger, M. Lohse and B. Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, 30, 2114–2120 CrossRef CAS PubMed.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581–583 CrossRef CAS PubMed.
- R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2021 Search PubMed.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies and F. O. Glöckner, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2013, 41, 590–596 CrossRef PubMed.
- L. Lahti, S. Shetty, et al., Tools for microbiome analysis in R, Microbiome package version 1.26, Bioconductor, 2017 Search PubMed.
- P. J. McMurdie and S. Holmes, phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data, PLoS One, 2013, 8, e61217 CrossRef CAS PubMed.
- D. J. Barnett, I. C. Arts and J. Penders, microViz: an R package for microbiome data visualization and statistics, J Open Source Software, 2021, 6, 3201 CrossRef.
- H. Wickham, ggplot2: Elegant Graphics for Data Analysis, Springer-Verlag, New York, 2016 Search PubMed.
- D. H. Ogle, J. C. Doll, A. P. Wheeler and A. Dinno, FSA: Simple Fisheries Stock Assessment Methods, R package version 0.9.6, 2023 Search PubMed.
- J. Oksanen, G. Simpson, F. Blanchet, R. Kindt, P. Legendre, P. Minchin, R. O'Hara, P. Solymos, M. Stevens, E. Szoecs, H. Wagner, M. Barbour, M. Bedward, B. Bolker, D. Borcard, G. Carvalho, M. Chirico, M. De Caceres, S. Durand, H. Evangelista, R. FitzJohn, M. Friendly, B. Furneaux, G. Hannigan, M. Hill, L. Lahti, D. McGlinn, M. Ouellette, E. Ribeiro Cunha, T. Smith, A. Stier, C. Ter Braak and J. Weedon, vegan: Community Ecology Package, 2024 Search PubMed , Version 2.6-6.
- H. Lin and S. D. Peddada, Analysis of compositions of microbiomes with bias correction, Nat. Commun., 2020, 11, 3514 CrossRef CAS PubMed.
- T. Wei and V. Simko, R package ‘corrplot’: Visualization of a Correlation Matrix, Version 0.96, 2024 Search PubMed.
- V. Biagioli, G. Volpedo, A. Riva, P. Mainardi and P. Striano, From Birth to Weaning: A Window of Opportunity for Microbiota, Nutrients, 2024, 16, 272 CrossRef CAS PubMed.
- S. G. Parkar, J. K. T. Frost, D. Rosendale, H. M. Stoklosinski, C. M. H. Jobsis, D. I. Hedderley and P. Gopal, The sugar composition of the fibre in selected plant foods modulates weaning infants’ gut microbiome composition and fermentation metabolites in vitro, Sci. Rep., 2021, 11, 9292 CrossRef CAS PubMed.
- S. Tao, Y. Bai, X. Zhou, J. Zhao, H. Yang, S. Zhang and J. Wang, In Vitro Fermentation Characteristics for Different Ratios of Soluble to Insoluble Dietary Fiber by Fresh Fecal Microbiota from Growing Pigs, ACS Omega, 2019, 4, 15158–15167 CrossRef CAS PubMed.
- Y. Zheng, C. Qin, M. Wen, L. Zhang and W. Wang, The Effects of Food Nutrients and Bioactive Compounds on the Gut Microbiota: A Comprehensive Review, Foods, 2024, 13, 1345 CrossRef CAS PubMed.
- E. Capuano and A. E. M. Janssen, Food Matrix and Macronutrient Digestion, Annu. Rev. Food Sci. Technol., 2021, 12, 193–212 CrossRef CAS PubMed.
- B. Cerdá, J. C. Espín, S. Parra, P. Martínez and F. A. Tomás-Barberán, The potent in vitro antioxidant ellagitannins from pomegranate juice are metabolised into bioavailable but poor antioxidant hydroxy–6H–dibenzopyran–6−one derivatives by the colonic microflora of healthy humans, Eur. J. Nutr., 2004, 43, 205–220 CrossRef PubMed.
- R. Puupponen-Pimiä, L. Nohynek, C. Meier, M. Kähkönen, M. Heinonen, A. Hopia and K.-M. Oksman-Caldentey, Antimicrobial properties of phenolic compounds from berries, J. Appl. Microbiol., 2001, 90, 494–507 CrossRef PubMed.
- F. Li, M. A. J. Hullar, Y. Schwarz and J. W. Lampe, Human Gut Bacterial Communities Are Altered by Addition of Cruciferous Vegetables to a Controlled Fruit- and Vegetable-Free Diet, J. Nutr., 2009, 139, 1685–1691 CrossRef CAS PubMed.
- C. H. Lifschitz, M. J. Wolin and P. J. Reeds, Characterization of Carbohydrate Fermentation in Feces of Formula-Fed and Breast-Fed Infants, Pediatr. Res., 1990, 27, 165–169 CrossRef CAS PubMed.
- X. Mei, T.-H. Mu and J.-J. Han, Composition and Physicochemical Properties of Dietary Fiber Extracted from Residues of 10 Varieties of Sweet Potato by a Sieving Method, J. Agric. Food Chem., 2010, 58, 7305–7310 CrossRef CAS PubMed.
- R. J. Palframan, G. R. Gibson and R. A. Rastall, Carbohydrate preferences of Bifidobacterium species isolated from the human gut, Curr. Issues Intest. Microbiol., 2003, 4, 71–75 CAS.
- I. E. El-Semman, F. H. Karlsson, S. Shoaie, I. Nookaew, T. H. Soliman and J. Nielsen, Genome-scale metabolic reconstructions of Bifidobacterium adolescentis L2-32 and Faecalibacterium prausnitzii A2-165 and their interaction, BMC Syst. Biol., 2014, 8, 41 CrossRef PubMed.
- J. Zheng, S. Wittouck, E. Salvetti, C. M. A. P. Franz, H. M. B. Harris, P. Mattarelli, P. W. O'Toole, B. Pot, P. Vandamme, J. Walter, K. Watanabe, S. Wuyts, G. E. Felis, M. G. Gänzle and S. Lebeer, A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae, Int. J. Syst. Evol. Microbiol., 2020, 70, 2782–2858 CrossRef CAS PubMed.
- C. Bourriaud, R. J. Robins, L. Martin, F. Kozlowski, E. Tenailleau, C. Cherbut and C. Michel, Lactate is mainly fermented to butyrate by human intestinal microfloras but inter–individual variation is evident, J. Appl. Microbiol., 2005, 99, 201–212 CrossRef CAS PubMed.
- S. H. Duncan, P. Louis and H. J. Flint, Lactate-Utilizing Bacteria, Isolated from Human Feces, That Produce Butyrate as a Major Fermentation Product, Appl. Environ. Microbiol., 2004, 70, 5810–5817 CrossRef CAS PubMed.
- S. Seeliger, P. H. Janssen and B. Schink, Energetics and kinetics of lactate fermentation to acetate and propionate via methylmalonyl-CoA or acrylyl-CoA, FEMS Microbiol. Lett., 2002, 211, 65–70 CrossRef CAS PubMed.
- A. Belenguer, S. H. Duncan, G. Holtrop, S. E. Anderson, G. E. Lobley and H. J. Flint, Impact of pH on Lactate Formation and Utilization by Human Fecal Microbial Communities, Appl. Environ. Microbiol., 2007, 73, 6526–6533 CrossRef CAS PubMed.
- S. P. Wang, L. A. Rubio, S. H. Duncan, G. E. Donachie, G. Holtrop, G. Lo, F. M. Farquharson, J. Wagner, J. Parkhill, P. Louis, A. W. Walker and H. J. Flint, Pivotal Roles for pH, Lactate, and Lactate-Utilizing Bacteria in the Stability of a Human Colonic Microbial Ecosystem, mSystems, 2020, 5, 5 Search PubMed.
- M. Liu, X. Li, S. Zhou, T. T. Y. Wang, S. Zhou, K. Yang, Y. Li, J. Tian and J. Wang, Dietary fiber isolated from sweet potato residues promotes a healthy gut microbiome profile, Food Funct., 2020, 11, 689–699 RSC.
- C. Liu, Y. Miao, J. Zhao, S. Yang, S. Cheng, W. Zhou, W. Guo and A. Li, In vitro simulated digestion of different heat treatments sweet potato polysaccharides and effects on human intestinal flora, Food Chem., 2025, 463, 141190 CrossRef CAS PubMed.
- M. Muchiri and A. L. McCartney, In vitro investigation of orange fleshed sweet potato prebiotic potential and its implication on human gut health, Funct. Foods Health Dis., 2017, 7, 833–848 CAS.
- Y. Cao, B. Tian, Z. Zhang, K. Yang, M. Cai, W. Hu, Y. Guo, Q. Xia and W. Wu, Positive effects of dietary fiber from sweet potato [Ipomoea batatas (L.) Lam.] peels by different extraction methods on human fecal microbiota in vitro fermentation, Front. Nutr., 2022, 7, 986667 CrossRef PubMed.
- C. R. Wall, N. C. Roy, J. A. Mullaney, W. C. McNabb, O. Gasser, K. Fraser, E. Altermann, W. Young, J. Cooney, R. Lawrence, Y. Jiang, B. C. Galland, X. Fu, J. N. Tonkie, N. Mahawar and A. L. Lovell, Nourishing the Infant Gut Microbiome to Support Immune Health: Protocol of SUN (Seeding Through Feeding) Randomized Controlled Trial, JMIR Res Protoc., 2024, 13, e56772 CrossRef PubMed.
- A. L. Lovell, H. Eriksen, S. McKeen, J. Mullaney, W. Young, K. Fraser, E. Altermann, O. Gasser, M. Kussmann, N. C. Roy, W. C. McNabb and C. R. Wall, “Nourish to Flourish”: complementary feeding for a healthy infant gut microbiome—a non-randomised pilot feasibility study, Pilot Feasibility Stud., 2022, 8, 103 CrossRef PubMed.
- J. Xu, M. A. Mahowald, R. E. Ley, C. A. Lozupone, M. Hamady, E. C. Martens, B. Henrissat, P. M. Coutinho, P. Minx, P. Latreille, H. Cordum, A. V. Brunt, K. Kim, R. S. Fulton, L. A. Fulton, S. W. Clifton, R. K. Wilson, R. D. Knight and J. I. Gordon, Evolution of Symbiotic Bacteria in the Distal Human Intestine, PLos Biol., 2007, 5, e156 CrossRef PubMed.
- T. Bhattacharya, T. S. Ghosh and S. S. Mande, Global Profiling of Carbohydrate Active Enzymes in Human Gut Microbiome, PLoS One, 2015, 10, e0142038 CrossRef PubMed.
- N. Baenas, V. Nuñez-Gómez, I. Navarro-González, L. Sánchez-Martínez, J. García-Alonso, M. J. Periago and R. González-Barrio, Raspberry dietary fibre: Chemical properties, functional evaluation and prebiotic in vitro effect, LWT, 2020, 134, 110140 CrossRef CAS.
- R. A. Moyer, K. E. Hummer, C. E. Finn, B. Frei and R. E. Wrolstad, Anthocyanins, Phenolics, and Antioxidant Capacity in Diverse Small Fruits: Vaccinium, Rubus, and Ribes, J. Agric. Food Chem., 2002, 50, 519–525 CrossRef CAS PubMed.
- M. Sójka, E. Klimczak, J. Macierzyński and K. Kołodziejczyk, Nutrient and polyphenolic composition of industrial strawberry press cake, Eur. Food Res. Technol., 2013, 237, 995–1007 CrossRef.
- D. So, K. Whelan, M. Rossi, M. Morrison, G. Holtmann, J. T. Kelly, E. R. Shanahan, H. M. Staudacher and K. L. Campbell, Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis, Am. J. Clin. Nutr., 2018, 107, 965–983 CrossRef PubMed.
- E. Özcan, M. R. Rozycki and D. A. Sela, Cranberry Proanthocyanidins and Dietary Oligosaccharides Synergistically Modulate Lactobacillus plantarum Physiology, Microorganisms, 2021, 9, 656 CrossRef PubMed.
- M.-C. Rodríguez-Daza, M. Roquim, S. Dudonné, G. Pilon, E. Levy, A. Marette, D. Roy and Y. Desjardins, Berry Polyphenols and Fibers Modulate Distinct Microbial Metabolic Functions and Gut Microbiota Enterotype-Like Clustering in Obese Mice, Front. Microbiol., 2020, 11, 2032 CrossRef PubMed.
- V. Núñez-Gómez, M. J. Periago, I. Navarro-González, M. P. Campos-Cava, N. Baenas and R. González-Barrio, Influence of Raspberry and Its Dietary Fractions on the In vitro Activity of the Colonic Microbiota from Normal and Overweight Subjects, Plant Foods Hum. Nutr., 2021, 76, 494–500 CrossRef PubMed.
- X. Zhang, A. Zhao, A. K. Sandhu, I. Edirisinghe and B. M. Burton-Freeman, Red Raspberry and Fructo-Oligosaccharide Supplementation, Metabolic Biomarkers, and the Gut Microbiota in Adults with Prediabetes: A Randomized Crossover Clinical Trial, J. Nutr., 2022, 152, 1438–1449 CrossRef CAS PubMed.
- S. G. Parkar, D. I. Rosendale, H. M. Stoklosinski, C. M. H. Jobsis, D. I. Hedderley and P. Gopal, Complementary Food Ingredients Alter Infant Gut Microbiome Composition and Metabolism In Vitro, Microorganisms, 2021, 9, 2089 CrossRef CAS PubMed.
- B. M. Nosal, S. N. Thornton, M. Darooghegi Mofrad, J. R. Sakaki, K. J. Mahoney, Z. Macdonald, L. Daddi, T. D. B. Tran, G. Weinstock, Y. Zhou, E. C.-H. Lee and O. K. Chun, Blackcurrants shape gut microbiota profile and reduce risk of postmenopausal osteoporosis via the gut-bone axis: Evidence from a pilot randomized controlled trial, J. Nutr. Biochem., 2024, 133, 109701 CrossRef CAS PubMed.
- A.-L. Molan, Z. Liu and G. Plimmer, Evaluation of the Effect of Blackcurrant Products on Gut Microbiota and on Markers of Risk for Colon Cancer in Humans, Phytother. Res., 2014, 28, 416–422 CrossRef CAS PubMed.
- Y. Han, M. Song, M. Gu, D. Ren, X. Zhu, X. Cao, F. Li, W. Wang, X. Cai, B. Yuan, T. Goulette, G. Zhang and H. Xiao, Dietary Intake of Whole Strawberry Inhibited Colonic Inflammation in Dextran-Sulfate-Sodium-Treated Mice via Restoring Immune Homeostasis and Alleviating Gut Microbiota Dysbiosis, J. Agric. Food Chem., 2019, 67, 9168–9177 CrossRef CAS PubMed.
- C. Petersen, U. D. Wankhade, D. Bharat, K. Wong, J. E. Mueller, S. V. Chintapalli, B. D. Piccolo, T. Jalili, Z. Jia, J. D. Symons, K. Shankar and P. V. Anandh Babu, Dietary supplementation with strawberry induces marked changes in the composition and functional potential of the gut microbiome in diabetic mice, J. Nutr. Biochem., 2019, 66, 63–69 CrossRef CAS PubMed.
- Z. Ezzat-Zadeh, S. M. Henning, J. Yang, S. L. Woo, R.-P. Lee, J. Huang, G. Thames, I. Gilbuena, C.-H. Tseng, D. Heber and Z. Li, California strawberry consumption increased the abundance of gut microorganisms related to lean body weight, health and longevity in healthy subjects, Nutr. Res., 2021, 85, 60–70 CrossRef CAS PubMed.
- L. Jia, D. Li, N. Feng, M. Shamoon, Z. Sun, L. Ding, H. Zhang, W. Chen, J. Sun and Y. Q. Chen, Anti-diabetic Effects of Clostridium butyricum CGMCC0313.1 through Promoting the Growth of Gut Butyrate-producing Bacteria in Type 2 Diabetic Mice, Sci. Rep., 2017, 7, 7046 CrossRef PubMed.
- P. A. Lawson and F. A. Rainey, Proposal to restrict the genus Clostridium Prazmowski to Clostridium butyricum and related species, Int. J. Syst. Evol. Microbiol., 2016, 66, 1009–1016 CrossRef CAS PubMed.
- L. Silva-Cristobal, P. Osorio-Díaz, J. Tovar and L. A. Bello-Pérez, Chemical composition, carbohydrate digestibility, and antioxidant capacity of cooked black bean, chickpea, and lentil Mexican varieties Composición química, digestibilidad de carbohidratos, y capacidad antioxidante de variedades mexicanas cocidas de frijol negro, garbanzo, y lenteja, Cienc. Tecnol. Aliment. – J. Food, 2010, 8, 7–14 CAS.
- T. Kutoš, T. Golob, M. Kač and A. Plestenjak, Dietary fibre content of dry and processed beans, Food Chem., 2003, 80, 231–235 CrossRef.
- N. T. Baxter, A. W. Schmidt, A. Venkataraman, K. S. Kim, C. Waldron and T. M. Schmidt, Dynamics of Human Gut Microbiota and Short-Chain Fatty Acids in Response to Dietary Interventions with Three Fermentable Fibers, mBio, 2019, 10, 1 CrossRef PubMed.
- J. Teichmann and D. W. Cockburn, In vitro Fermentation Reveals Changes in Butyrate Production Dependent on Resistant Starch Source and Microbiome Composition, Front. Microbiol., 2021, 12, 640253 CrossRef PubMed.
- T. L. Pickens and D. W. Cockburn, Clostridium butyricum Prazmowski can degrade and utilize resistant starch via a set of synergistically acting enzymes, mSphere, 2023, 9, 1 Search PubMed.
- C. Teixeira-Guedes, T. Sánchez-Moya, C. Pereira-Wilson, G. Ros-Berruezo and R. López-Nicolás, In Vitro Modulation of Gut Microbiota and Metabolism by Cooked Cowpea and Black Bean, Foods, 2020, 9, 861 CrossRef CAS PubMed.
- M. Hernández-Salazar, P. Osorio-Diaz, G. Loarca-Piña, R. Reynoso-Camacho, J. Tovar and L. A. Bello-Pérez, In vitro fermentability and antioxidant capacity of the indigestible fraction of cooked black beans (Phaseolus vulgaris L.), lentils (Lens culinaris L.) and chickpeas (Cicer arietinum L.), J. Sci. Food Agric., 2010, 90, 1417–1422 CrossRef PubMed.
- J. M. Monk, D. Lepp, W. Wu, K. P. Pauls, L. E. Robinson and K. A. Power, Navy and black bean supplementation primes the colonic mucosal microenvironment to improve gut health, J. Nutr. Biochem., 2017, 49, 89–100 CrossRef CAS PubMed.
- M. Sánchez-Tapia, I. Hernández-Velázquez, E. Pichardo-Ontiveros, O. Granados-Portillo, A. Gálvez, A. R. Tovar and N. Torres, Consumption of Cooked Black Beans Stimulates a Cluster of Some Clostridia Class Bacteria Decreasing Inflammatory Response and Improving Insulin Sensitivity, Nutrients, 2020, 12, 1182 CrossRef PubMed.
- M. I. Ordiz, S. Janssen, G. Humphrey, G. Ackermann, K. Stephenson, S. Agapova, O. Divala, Y. Kaimila, K. Maleta, C. Zhong, R. Knight, I. Trehan, P. I. Tarr, B. Rusconi and M. J. Manary, The effect of legume supplementation on the gut microbiota in rural Malawian infants aged 6 to 12 months, Am. J. Clin. Nutr., 2020, 111, 884–892 CrossRef PubMed.
- A. M. Sheflin, E. C. Borresen, J. S. Kirkwood, C. M. Boot, A. K. Whitney, S. Lu, R. J. Brown, C. D. Broeckling, E. P. Ryan and T. L. Weir, Dietary supplementation with rice bran or navy bean alters gut bacterial metabolism in colorectal cancer survivors, Mol. Nutr. Food Res., 2017, 61, 1500905 CrossRef PubMed.
- Z. Miao, W. Du, C. Xiao, C. Su, W. Gou, L. Shen, J. Zhang, Y. Fu, Z. Jiang, Z. Wang, X. Jia, J.-S. Zheng and H. Wang, Gut microbiota signatures of long-term and short-term plant-based dietary pattern and cardiometabolic health: a prospective cohort study, BMC Med., 2022, 20, 204 CrossRef PubMed.
- G. D. Wu, J. Chen, C. Hoffmann, K. Bittinger, Y.-Y. Chen, S. A. Keilbaugh, M. Bewtra, D. Knights, W. A. Walters, R. Knight, R. Sinha, E. Gilroy, K. Gupta, R. Baldassano, L. Nessel, H. Li, F. D. Bushman and J. D. Lewis, Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes, Science, 2011, 334, 105–108 CrossRef CAS PubMed.
- A. A. Roess, E. F. Jacquier, D. J. Catellier, R. Carvalho, A. C. Lutes, A. S. Anater and W. H. Dietz, Food Consumption Patterns of Infants and Toddlers: Findings from the Feeding Infants and Toddlers Study (FITS) 2016, J. Nutr., 2018, 148, 1525S–1535S CrossRef PubMed.
- N. Sillner, A. Walker, M. Lucio, T. V. Maier, M. Bazanella, M. Rychlik, D. Haller and P. Schmitt-Kopplin, Longitudinal Profiles of Dietary and Microbial Metabolites in Formula- and Breastfed Infants, Front. Mol. Biosci., 2021, 8, 660456 CrossRef CAS PubMed.
- K. J. Flynn, M. T. Ruffin IV, D. K. Turgeon and P. D. Schloss, Spatial Variation of the Native Colon Microbiota in Healthy Adults, Cancer Prev. Res., 2018, 11, 393–402 CrossRef PubMed.
- J. Ni, Y. Wang, H. Sun, Z. Chang, R. Wang, Y. Jiang, J. Qin, M. Gao and Z. Li, Comparative study on static and dynamic digest characteristics of oat β-Glucan and β-Gluco-Oligosaccharides, Food Res. Int., 2024, 197, 115153 CrossRef CAS PubMed.
- J. S. Johnson, D. J. Spakowicz, B.-Y. Hong, L. M. Petersen, P. Demkowicz, L. Chen, S. R. Leopold, B. M. Hanson, H. O. Agresta, M. Gerstein, E. Sodergren and G. M. Weinstock, Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis, Nat. Commun., 2019, 10, 5029 CrossRef PubMed.
- A. M. Lerma-Aguilera, S. Pérez-Burillo, B. Navajas-Porras, E. D. León, S. Ruíz-Pérez, S. Pastoriza, N. Jiménez-Hernández, B.-M. Cämmerer, J. Á. Rufián-Henares, M. J. Gosalbes and M. P. Francino, Effects of different foods and cooking methods on the gut microbiota: an in vitro approach, Front. Microbiol., 2023, 14, 1334623 CrossRef PubMed.
- P. P. Echarri, C. M. Graciá, G. R. Berruezo, I. Vives, M. Ballesta, G. Solís, I. V. Morillas, C. G. de los Reyes-Gavilán, A. Margolles and M. Gueimonde, Assessment of intestinal microbiota of full-term breast-fed infants from two different geographical locations, Early Hum. Dev., 2011, 87, 511–513 CrossRef PubMed.
- S. Schiess, V. Grote, S. Scaglioni, V. Luque, F. Martin, A. Stolarczyk, F. Vecchi, B. Koletzko and E. C. O. Project, Introduction of Complementary Feeding in 5 European Countries, J. Pediatr. Gastroenterol. Nutr., 2010, 50, 92–98 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5fo00414d |
| This journal is © The Royal Society of Chemistry 2025 |