 Open Access Article
Open Access ArticleAnalysis of human colostrum reveals differential co-occurrence networks of metabolites, microbiota and cytokines in maternal obesity†
July S.
Gámez-Valdez
 ab,
Karina
Corona-Cervantes
ab,
Karina
Corona-Cervantes
 b,
Erick S.
Sánchez-Salguero
b,
Erick S.
Sánchez-Salguero
 c,
Mario R.
Alcorta-García
c,
Mario R.
Alcorta-García
 de,
Claudia N.
López-Villaseñor
de,
Claudia N.
López-Villaseñor
 df,
Rommel A.
Carballo-Castañeda
df,
Rommel A.
Carballo-Castañeda
 g,
Aldo
Moreno-Ulloa
g,
Aldo
Moreno-Ulloa
 g,
Víctor J.
Lara-Díaz
g,
Víctor J.
Lara-Díaz
 fh,
Marion E. G.
Brunck
ac and
Cuauhtémoc
Licona-Cassani
fh,
Marion E. G.
Brunck
ac and
Cuauhtémoc
Licona-Cassani
 *ab
*ab
aCentro de Biotecnología FEMSA, Escuela de Ingeniería y Ciencias, Tecnologico de Monterrey, N.L., Mexico. E-mail: clicona@tec.mx
bUnidad de Biología Integrativa, Institute for Obesity Research, Tecnologico de Monterrey, N.L., Mexico
cUnidad de Biología Experimental, Institute for Obesity Research, Tecnologico de Monterrey, N.L., Mexico
dHospital Regional Materno Infantil, Servicios de Salud de Nuevo León, OPD, Ciudad Guadalupe, N.L., Mexico
eHospital San José TECSALUD, Monterrey, N.L., Mexico
fEscuela de Medicina y Ciencias de la Salud, Tecnologico de Monterrey, N.L., Mexico
gDepartamento de Innovación Biomédica, Centro de Investigación Científica y de Educación Superior de Ensenada, Baja California (CICESE), Ensenada, Mexico
hFaculty of Medicine, The University of New South Wales, Sydney, Australia
First published on 2nd July 2025
Abstract
Breastmilk is essential for neonatal development, particularly in seeding the gut microbiota and modulating the immune system. This proof-of-concept study explores the systemic nature of colostrum and the influence of maternal obesity on co-occurrences of colostrum bioactives. Using 16S-rRNA sequencing, untargeted metabolomics, and cytokine quantification, we analyzed co-occurring elements in the colostrum of mothers with normal weight (18.5 < BMI < 25) or obesity (BMI > 30). We identified 5 different co-occurrence networks, characterized by positive correlations of taxonomically related bacteria. Our integrative analysis reveals that Aeromonadaceae, Xanthomonadaceae and Staphylococcaceae negatively correlate with pro-inflammatory cytokines TNF-α, IL-6, and IL-12p70 in the colostrum of mothers with obesity (WO). Additionally, lipid mediators, including 15-HEDE and LysoPC (16:00), were associated with cytokines IL-10 and IL-8 and microbiota taxa Burkholderiaceae, Beijerinckiaceae and Planococcaceae – first reported in the colostrum of mothers WO. Our findings suggest a pervasive regulation of bioactives in the colostrum of mothers WO. This may have implications for distinctive neonatal intestine development.
Introduction
Breastmilk is a complex and dynamic fluid that supplies hydration, nutrients, and bioactive molecules essential for the optimal growth and development of neonates.1 The bioactive components of breastmilk regulate vital functions, including passive immunity to pathogens, tolerance to the colonizing microbiota, and other processes necessary to mature the intestinal system.2,3 Multiple maternal factors have been linked to alterations in the breastmilk composition including BMI, age, ethnicity and diet.4–6Most of the current studies draw direct correlations between maternal physiological states and alterations in individual milk components.7 While informative, this approach fails to capture a more comprehensive breadth of how maternal condition correlates to breastmilk bioactives. This becomes evident when considering breastmilk as a complex and dynamic ecological system. As an example, during pregnancy, a clear biological network emerges within the maternal gut, wherein bacterial fermentation produces short-chain fatty acids (SCFAs), triggering the proliferation of regulatory T (Treg) cells and modulating the balance between pro-inflammatory and anti-inflammatory cytokines, thereby promoting an immunotolerant environment that allows the establishment of commensal bacteria.8 Understanding breastmilk as a biological system and how it is influenced by maternal health will provide valuable insights into its potential impacts on neonatal health.9,10
One in three pregnant women worldwide is affected by obesity. This metabolic disorder has been correlated with systemic alterations, including chronic inflammation. The breastmilk bioactive components of mothers with obesity are also altered.5,11,12 For instance, the breastmilk microbiota of mothers with obesity has increased proportions of Staphylococcus and Corynebacterium.13,14 Yet, little is known about the possible co-dependences of these alterations on other breastmilk bioactive components. The breastmilk microbiota is a living component that produces metabolites and other molecules while utilizing maternal factors. Therefore, it is crucial to examine the interactions among soluble bioactives together with the microbiota composition.15–17 Approaching breastmilk as an ecosystem, and more importantly, colostrum – the initial biological message that a lactating mother has formulated for the neonate – will open avenues to explore the complexity and dynamism of its interacting components.
Using co-occurrence networks to analyze the patterns of interactions among bioactives represents a promising approach for exploring the structure of complex biological systems and their interactions with the host.18 This approach provides valuable insights into potential interactions that are difficult to detect by characterizing bioactives independently or by using general ecological metrics such as alpha/beta diversity.19 Here, we integrate colostrum cytokine concentrations, microbiota composition and metabolomics in the context of maternal obesity as an approach for identifying distinctive co-occurring networks. By exploring these interconnections, we provide a proof of concept of the systemic nature of colostrum and begin to dimension the complex biological networks at play within this microenvironment.
Materials and methods
Experimental model and study participant details
This is an observational study conducted in accordance with the ethical principles outlined in the Declaration of Helsinki. The protocol was approved by the IRB at Escuela de Medicina y Ciencias de la Salud, Tecnologico de Monterrey, with the ID: P000487; it has been registered at Clinical Trials with the ID: NCT04812847. Every participant was provided with details regarding the study, and written consent was acquired from each of them, ensuring the confidentiality of the personal information.Healthy breastfeeding mothers who had given birth to full-term infants in the Hospital Regional Materno Infantil de Alta Especialidad were recruited between November 2020 and July 2022. The inclusion criteria for participants encompassed mothers between 18 and 35 years, with confirmed residency within the metropolitan area of Monterrey. The exclusion criteria included a history of antibiotic usage in the 3 months prior delivery, a prolonged antibiotic exposure exceeding 3 weeks at any stage of pregnancy, antibiotic requirement for more than 24 hours post-delivery, immunosuppressive, or immunomodulatory corticosteroid therapy, bariatric surgery, experienced delayed lactogenesis or issues with milk supply, pre-term delivery or if required intensive care post-delivery. Women with pregnancy complications such as gestational diabetes mellitus (GDM), hypertensive disorders, thyroid dysfunction, or history of feeding disorders were also excluded, as well as cases involving multiple birth or neonates with congenital anomalies.
Pre-pregnancy BMI was determined by the clinician and reported in the corresponding clinical records. Participants were divided into two groups according to the World Health Organization (WHO) classification of the body mass index (BMI):20 women with obesity (“WO”; BMI ≥ 30 kg m−2; n = 28) and mothers with normal weight (“NW”; BMI ≤ 25 kg m−2; n = 20).
Sample collection and processing
Colostrum samples were collected within the first 48 hours post-partum from a total of 48 mothers by a trained medical team, utilizing sterile gloves for the process. Following a gentle cleansing of the areola with sterile water, when possible, 3 mL of colostrum was obtained by manual expression into a sterile 15 mL polypropylene tube. To ensure the purity of the sample, the initial drops were discarded. After collection, the samples were promptly transported to the laboratory and stored at −20 °C until subsequent processing.Cytokine measurement
Due to limited sample volume availability, cytokine quantification was performed in a subset of 47 colostrum samples (NW = 20 and WO = 27). Samples were centrifuged (3000g, 15 minutes) to remove any cellular structure. Cytokines were quantified in the supernatant using a LEGENDplex® kit (inflammation cytokine markers thirteen, Bio Legend Cat #740![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 808) which includes IL-1β, TNFα, IL-6, IL-8, IL-10 and IL-12p70, according to the manufacturer's instructions with some modifications. Briefly, standard curves with known concentrations of each cytokine were generated through double serial dilutions, resulting in an eight-point curve performed in duplicate. Five microliters of the supernatant were combined with a fluorescent bead's reagent at a 1
808) which includes IL-1β, TNFα, IL-6, IL-8, IL-10 and IL-12p70, according to the manufacturer's instructions with some modifications. Briefly, standard curves with known concentrations of each cytokine were generated through double serial dilutions, resulting in an eight-point curve performed in duplicate. Five microliters of the supernatant were combined with a fluorescent bead's reagent at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio and incubated for two hours at room temperature. Following incubation, the beads were washed by resuspending them in wash buffer and then centrifuged (250g, 5 minutes). The supernatant was carefully discarded, and a mix of secondary antibodies was added and incubated for one hour. Subsequently, streptavidin–phycoerythrin (SA–PE) conjugates were introduced into the same reaction and incubated for 30 minutes at room temperature. After an additional washing step, the samples were analysed using a BD® FACSCelesta flow cytometer fitted with 405 nm, 488 nm, and 633 nm lasers and operated through BD® FACSDiva software v.8. The detection limits for cytokines were IL-1β, 1.5 ± 0.6 pg mL−1; TNF-α, 0.9 ± 0.8 pg mL−1; IL-6, 1.5 ± 0.7 pg mL−1; IL-8, 2.0 ± 0.5 pg mL−1; IL-10, 2.0 ± 0.5 pg mL−1; IL-12p70, 2.0 ± 0.2 pg mL−1. Data analysis was performed using BD software, which is accessible online at the website of LEGENDplex™ cloud-based data analysis software (https://legendplex.qognit.com).
1 volume ratio and incubated for two hours at room temperature. Following incubation, the beads were washed by resuspending them in wash buffer and then centrifuged (250g, 5 minutes). The supernatant was carefully discarded, and a mix of secondary antibodies was added and incubated for one hour. Subsequently, streptavidin–phycoerythrin (SA–PE) conjugates were introduced into the same reaction and incubated for 30 minutes at room temperature. After an additional washing step, the samples were analysed using a BD® FACSCelesta flow cytometer fitted with 405 nm, 488 nm, and 633 nm lasers and operated through BD® FACSDiva software v.8. The detection limits for cytokines were IL-1β, 1.5 ± 0.6 pg mL−1; TNF-α, 0.9 ± 0.8 pg mL−1; IL-6, 1.5 ± 0.7 pg mL−1; IL-8, 2.0 ± 0.5 pg mL−1; IL-10, 2.0 ± 0.5 pg mL−1; IL-12p70, 2.0 ± 0.2 pg mL−1. Data analysis was performed using BD software, which is accessible online at the website of LEGENDplex™ cloud-based data analysis software (https://legendplex.qognit.com).
Genomic DNA extraction and 16S sequencing
Genomic DNA extraction from 48 colostrum samples (NW = 20 and WO = 28) was performed following an optimized phenol–chloroform protocol, as previously described.13 Briefly, 1 mL of colostrum was utilized for DNA extraction whenever feasible. The procedure involved the removal of the fat layer using a sterile hyssop and pelleting bacterial cells with a high-speed centrifugation followed by a sterile PBS wash. The resulting pellet was then treated with a lysis buffer and mechanically lysed using a FastPrep system (MP Biomedicals, Santa Ana, CA). Enzymatic lysis with proteinase K, lysozyme, and RNAse was subsequently performed. DNA extraction was carried out using a phenol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chloroform
chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) isoamyl-alcohol (25
isoamyl-alcohol (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixture, followed by precipitation of DNA with isopropanol. The DNA pellet was washed with 70% ethanol and resuspended in nuclease-free water. As controls, an in-house mock community with two bacterial isolates (Escherichia coli and Pseudomonas putida in a 1
1) mixture, followed by precipitation of DNA with isopropanol. The DNA pellet was washed with 70% ethanol and resuspended in nuclease-free water. As controls, an in-house mock community with two bacterial isolates (Escherichia coli and Pseudomonas putida in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) and a DNA extraction of sterile PBS were included as positive and negative controls, respectively. DNA integrity was assessed by agarose gel electrophoresis, and concentration was determined using a NanoDrop ND-1000 UV spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Unless stated otherwise, all reagents were obtained from Sigma-Aldrich.
1 ratio) and a DNA extraction of sterile PBS were included as positive and negative controls, respectively. DNA integrity was assessed by agarose gel electrophoresis, and concentration was determined using a NanoDrop ND-1000 UV spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Unless stated otherwise, all reagents were obtained from Sigma-Aldrich.
For sequencing, the prepared DNA samples were subjected to a Zymo Research's Quick-16S kit, utilizing phased primers 341F (5′-CCTACGGGDGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) targeting the V3–V4 regions of the 16S rDNA. The sequencing was performed on a V3 MiSeq 622cyc flowcell, generating 2 × 301 bp paired-end reads.
Colostrum taxonomic and differential profiles
The sequencing reads were imported into QIIME2 v.2022.8 and demultiplexed for subsequent analysis.21 DADA2 plugin was used for denoising and quality control with optimized parameters to remove low quality regions of the sequences (--p-trim-left-f 12 --p-trim-left-r 16 --p-trunc-len-f 285 --p-trunc-len-r 250).22 Taxonomic species profiling was accomplished by aligning amplicon sequence variants (ASVs) against the Silva 138.1 database with 99% of identity threshold,23 using the q2-feature-classifier classify-sklearn naive Bayes taxonomy classifier.24 To ensure data quality, potential contaminants identified through batch effect analysis or the positive control (archaea, mitochondria/chloroplast, Cyanobacteria, Streptomyces, Stenotrophomonas, and Pseudomonas) were removed.25 Additionally, rare taxa were identified as ASVs with a total read count of ≤18 in at least three samples and subsequently excluded from the analysis.26Rarefaction curves were generated to assess the sequencing depth and ensure adequate coverage of microbial diversity in colostrum. The rarefaction was conducted at a depth of 7712 sequences. Diversity analyses were performed in R using vegan v2.6-427 and phyloseq v1.44.0 libraries.28 Alpha diversity analyses included the calculation of the Shannon diversity index, observed ASVs, evenness, and phylogenetic distance. Comparisons between groups were conducted using the Mann–Whitney's U test. Beta diversity was assessed through ANOSIM (performed with 999 permutations) using the UniFrac and robust Aitchison distance matrices. A Principal Coordinate Analysis (PCoA) and a Robust Principal Component Analysis (RPCA) were created and visualized using EMPEROR.29 Differential abundance analysis at the taxonomic family level was conducted using DESeq2 v1.40.2 with batch correction applied30 and ANCOMBC v2.6.0.31 Only those families that were significantly different in both tools were graphed.
Colostrum metabolite extraction
Due to limited sample volume availability, 25 colostrum samples (NW = 5 and WO = 20) were used to extract metabolites. 100 μL of colostrum was combined with 600 μL of acetonitrile (ACN). The mixture was vortexed for 1 minute and subjected to 30 minutes of sonication in ice-cold water to enhance the extraction of metabolites. Subsequently, the samples were centrifuged using optimized centrifuge parameters (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 817g, 4 °C for 10 minutes). From the resulting supernatant, 400 μL were carefully collected for vacuum evaporation of ACN. The dried metabolome was then resuspended in 100 μL of an injection solution composed of a water
817g, 4 °C for 10 minutes). From the resulting supernatant, 400 μL were carefully collected for vacuum evaporation of ACN. The dried metabolome was then resuspended in 100 μL of an injection solution composed of a water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile mixture in an 80
acetonitrile mixture in an 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ratio containing 0.1% formic acid (v/v). To ensure effective resuspension, the samples were vortexed for 1 minute and sonicated in ice-cold water for 30 minutes. To obtain a particle-free solution suitable for injection, the resuspended samples underwent centrifugation at the optimized parameters before being transferred to the injection vials. Quality control (QC) samples were generated by pooling all resuspended samples in equal volumes.
20 ratio containing 0.1% formic acid (v/v). To ensure effective resuspension, the samples were vortexed for 1 minute and sonicated in ice-cold water for 30 minutes. To obtain a particle-free solution suitable for injection, the resuspended samples underwent centrifugation at the optimized parameters before being transferred to the injection vials. Quality control (QC) samples were generated by pooling all resuspended samples in equal volumes.
LC-MS2 data acquisition
We employed the instrumentation and followed the equipment's parameters previously described,32 with minor modifications. Four μL of resuspended metabolomes were injected into an LC Agilent 1260 system (Agilent Technologies, Inc., Santa Clara, CA, USA). The separation of metabolites was done using a ProtID-Chip-43 II column (C18, 43 mm, 300 Å, 5 μm particle size, equipped with a 40 nL enrichment column). Mobile phase consisted of water with 0.1% formic acid (solution A) and acetonitrile (ACN) with 0.1% formic acid (solution B). The chromatographic method separation started with a mobile phase composition of 5% B, which was then linearly increased to 40% B over 20 minutes. Subsequently, the gradient was elevated to 100% B within 5 minutes and maintained at this composition for an additional 5 minutes. Next, the system was returned to its initial condition of 5% B within 1 minute and held for 9 minutes to ensure complete column re-equilibration before the next sample analysis. The total process time was 40 min, utilizing a flow rate of 300 nL min−1. To mitigate potential carryover effects, two blank samples of 6 μL each – one comprising the injection solution and the other comprising ACN – were run between every sample injection. The separated metabolites were then introduced into an Agilent 6530A Q-TOF mass spectrometer (Agilent Technologies, Inc., Santa Clara, CA, USA) through a Chip Cube–LC interface using nanospray ionization in positive mode. Data-dependent acquisition was used. For MS1, mass range was 110–2000 m/z with 4 spectra per s velocity. The top 5 most intense precursor ions per cycle reaching 150 cps were selected for fragmentation (MS2 acquisition) in a mass range of 50–2000 m/z, at 3 spectra per s rate. The active exclusion option was on, set to 2 spectra, and released after 0.25 min. Ramped collision energy was used with a slope of 6 and an offset of 4. Calibration was done before sample acquisition and every 24 hours with an ESI-L low mix concentration tuning mix solution (Agilent Technologies, Inc., Santa Clara, CA, USA) to ensure a mass accuracy <5 ppm for MS1 and MS2 data. The samples were randomly allocated in the autosampler for data acquisition.LC-MS2 data processing
Processing of LC-MS2 datasets was performed to reach two main goals: feature or peak (signals with m/z and retention time) extraction (peak-picking) and metabolite annotation at the structure (metabolomics standard initiative (MSI) levels 2 and 3), substructure, and chemical class (MSI, level 3) levels.33 Raw datasets were transformed from commercial .d format to open source .mzXML format using an MSconvert tool within ProteoWizard v3.34 Transformed datasets were processed in MZmine v2.53 for peak-picking using the parameters described in Table S1.†35 For feature annotation by spectral matching (MSI, level 2), open-source raw and processed data were uploaded and analyzed in the Global Natural Products Social Molecular Networking (GNPS) platform36 for classical molecular networking37 and feature-based molecular networking (FBMN),38 respectively. To expand the annotations not achieved by spectral matching, we employed MolDiscovery39 and Dereplicator+40 within the GNPS environment. Additionally, we employed SIRIUS v5.8.141 for database-independent chemical formula annotation and correction by ZODIAC,42 in-silico structural metabolite annotation by CSI:FingerID,43 and chemical class assignment by CANOPUS algorithm (MSI, level 3). The chemical landscape representation was created using the molecular network provided by the FBMN analysis in GNPS, enriched with the chemical superclass annotations of CANOPUS. Substructure (motifs; MSI, level 3) annotation was done using the MS2LDA webpage.44 Molecular structure assignments were filtered by a mass error less than 10 ppm to keep annotations with high-mass accuracy. Additionally, peaks and annotations derived from blank samples were filtered out from the analysis.Quantification and statistical analysis
Unless other specified, all statistical analyses were performed and plotted using GraphPad Prism version 8.0.1, GraphPad Software, San Diego, California, USA (https://www.graphpad.com). The clinical data (maternal age, gestation weeks, delivery mode and neonatal sex) were compared between groups using independent-sample median test or Fisher's exact test to evaluate their influence as confounding factors. The Kruskal–Wallis test and GLM were used to assess the influence of maternal and neonatal demographic and clinical data on the microbiota composition, metabolomic profile, and cytokine quantification. Mann–Whitney's U test was used for evaluating the difference in cytokine concentrations between the NW and WO groups. A p-value ≤ 0.05 was considered statistically significant.For metabolomic data analysis, feature abundance normalization was conducted using the quantile method, and subsequent differential abundance analysis was carried out within the NormalyzerDE online platform.45 Metabolites exhibiting a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-fold change (Log
2-fold change (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC) of ±0.58 and a p-value <0.05 (calculated with the limma package46) were considered differentially abundant between WO and NW groups. Volcano plots were created to facilitate data visualization using EnhancedVolcano R package v1.16.0.47 To assess differences among groups at the chemical class level, the summed abundance (area under the curve) of all the quantified features belonging to each respective class was calculated, followed by a two-group comparison using the Wilcoxon test, assuming unequal variance, implemented with the wilcox.test() function in R. For comparing colostrum chemical diversity between groups, the Shannon and Simpson indexes were calculated as community ecology metrics48 using the vegan package v2.6-4. Statistical significance was determined through a two-group comparison using the Wilcoxon test, as described above.
2FC) of ±0.58 and a p-value <0.05 (calculated with the limma package46) were considered differentially abundant between WO and NW groups. Volcano plots were created to facilitate data visualization using EnhancedVolcano R package v1.16.0.47 To assess differences among groups at the chemical class level, the summed abundance (area under the curve) of all the quantified features belonging to each respective class was calculated, followed by a two-group comparison using the Wilcoxon test, assuming unequal variance, implemented with the wilcox.test() function in R. For comparing colostrum chemical diversity between groups, the Shannon and Simpson indexes were calculated as community ecology metrics48 using the vegan package v2.6-4. Statistical significance was determined through a two-group comparison using the Wilcoxon test, as described above.
Bacterial interactive network was constructed by linking taxonomic families that exhibited a Spearman rank correlation exceeding an absolute value of 0.5 and a p-value < 0.05. Network analysis was performed using MicrobiomeAnalyst.49 Correlations between clinical data, cytokine concentrations, bacterial families, and metabolite abundances were calculated with Spearman's correlation (ρ) using R. Only correlations with a p < 0.10 were kept for visualization in the heatmaps.
Results
Demographic and clinical characteristics of the study cohort
This study cohort consisted of 48 women between 18 and 35 years old. As per the study design, the body mass index (BMI) was significantly higher in the group “with obesity” (WO) compared to the “Normal Weight” (NW) group (medians: 22.45 vs. 32.05, in the NW and WO groups, respectively, p < 0.0001, Table 1). There was no difference in maternal age, duration of gestation, delivery mode or biological sex of the offspring.| Variables | Mothers with normal weight (NW) | Mothers with obesity (WO) | P-Value |
|---|---|---|---|
| BMI = body mass index [kg m−2].a Values expressed as median (interquartile range); statistical analysis by an independent-samples Mann–Whitney U test.b Values expressed as frequency (intra-group percentage); statistical analysis by Fisher's exact test.c Cytokine concentrations are reported as median [pg ml−1] (interquartile range); statistical analysis by the Mann–Whitney U test. NW = 20 and WO = 27.d Metabolite abundances are reported as median (interquartile range); statistical analysis by the Mann–Whitney U test. NW = 5 and WO = 20. | |||
| Number of participants | 20 | 28 | |
| Maternal age (years)a | 23.5 (23.0 to 28.8) | 26.0 (21.0 to 29.0) | 0.60 |
| BMIa | 22.5 (22.0 to 23.7) | 32.1 (31.0 to 36.8) | <0.0001 |
| Gestational age (weeks)a | 39.0 (38.0 to 39.0) | 38.4 (38.0 to 40.0) | 0.28 |
| Way of deliveryb | 0.16 | ||
| Vaginal | 18 (90) | 20 (71.4) | |
| Caesarean | 2 (10) | 8 (28.6) | |
| Neonatal characteristics | |||
| Sex assigned at birthb | 0.14 | ||
| Female | 12 (60) | 10 (35.7) | |
| Male | 8 (40) | 18 (64.3) | |
| Cytokinesc | |||
| IL-12p70 | 0.76 (0.01 to 1.77) | 1.15 (0.10 to 2.33) | 0.30 |
| IL-10 | 4.33 (1.91 to 9.49) | 2.72 (1.23 to 5.75) | 0.34 |
| IL-1β | 16.53 (3.86 to 76.56) | 8.16 (2.96 to 31.34) | 0.31 |
| TNF-α | 7.66 (3.43 to 28.22) | 11.49 (4.62 to 18.50) | 0.94 |
| IL-6 | 46.54 (26.64 to 236.9) | 53.05 (26.74 to 85.60) | 0.40 |
| IL-8 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 498 (3221 to 31 498 (3221 to 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 737) 737) |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 294 (10 294 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 881 to 36 881 to 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 852) 852) |
0.19 |
| Metabolitesd | |||
| (12Z)-9,10-Dihydroxyoctadec-12-enoic acid | 2706 (2011 to 4156) | 2406 (1576 to 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 898) 898) |
0.72 |
| 12(13)Ep-9-KODE | 3164 (2746 to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 827) 827) |
3091 (2046 to 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 327) 327) |
0.41 |
| 15-HEDE | 1070 (741 to 3694) | 244.5 (141.60 to 749.90) | 0.02 |
| 1-O-Elenoloyl-2-O-(9Z-octadecenoyl) glycerol | 4432 (2313 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 431) 431) |
9572 (5217 to 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 373) 373) |
0.11 |
| 3-Acetyl kabiramide D | 180.2 (30.04 to 2515) | 210.9 (11.00 to 1650) | 0.77 |
| 4-Hydroxynonenal | 2290 (974.70 to 4708) | 1361 (912.60 to 3593) | 0.67 |
| 7,26-Epoxy-2,7-dihydro-cycloirid-16-enal | 192.10 (88.44 to 1046) | 366.70 (316.80 to 1169) | 0.37 |
| 8-Hydroxy-9,10-epoxystearic acid | 8762 (7063 to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 353) 353) |
6994 (5829 to 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 920) 920) |
0.49 |
| 8S-Hydroxy-9E,11Z,14Z-eicosatrienoic acid | 8441 (1712 to 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 819) 819) |
1937 (998.60 to 5005) | 0.15 |
| 9(S)-HOTrE | 3370 (1432 to 5267) | 2938 (2053 to 3423) | 0.92 |
| Aminopentol | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 710 (7385 to 26 710 (7385 to 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 755) 755) |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 520 (12 520 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 087 to 45 087 to 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 859) 859) |
0.13 |
| Carveol | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 052 (6423 to 19 052 (6423 to 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 590) 590) |
7979 (4180 to 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 588) 588) |
0.37 |
| Conodutarine A 19′-ketone | 81.16 (2.79 to 8660) | 451.70 (28.10 to 2134) | 0.87 |
| Decanoylcarnitine | 2622 (2023 to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 551) 551) |
1563 (639.30 to 2862) | 0.04 |
| M531 | 1483 (977 to 2144) | 2771 (1946 to 8447) | 0.02 |
| Ervahainamidine B | 3150 (27.33 to 6736) | 1138 (47 to 4016) | 0.76 |
| Isomotuporin D | 102.10 (11.63 to 1986) | 295.70 (25.36 to 967.90) | 0.87 |
| Kaimonolide A | 1683 (1100 to 1786) | 2625 (1532 to 6364) | 0.15 |
| LysoPC(16:0) | 668 (362.50 to 1594) | 671.90 (547.30 to 1152) | 0.72 |
| Linoleic acid | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 852 (1272 to 41 852 (1272 to 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 630) 630) |
4997 (1412 to 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 942) 942) |
0.92 |
| Linolenic acid | 4284 (1943 to 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102) 102) |
2387 (1329 to 9357) | 0.30 |
| Monoelaidin | 1041 (214.50 to 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 263) 263) |
896.80 (319.4 to 5932) | 0.72 |
| 1-Monolinolenoyl-rac-glycerol | 5879 (3722 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 378) 378) |
6145 (2921 to 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 338) 338) |
0.87 |
| Myxochromide S3 | 3208 (2865 to 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 604) 604) |
3883 (2274 to 9778) | 0.53 |
| PA (18:4(6Z,9Z,12Z,15Z)/22:2(13Z,16Z)) | 499.20 (294.90 to 873.60) | 819.70 (528.80 to 1420) | 0.19 |
| Palmitic acid | 1839 (1548 to 3776) | 1462 (963 to 2440) | 0.22 |
| Pargamicin B | 400 (218.70 to 7937) | 406.20 (71.86 to 1409) | 0.41 |
| PC (18:1(9Z)/16:0) | 1040 (93.18 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 347) 347) |
530 (125 to 2864) | 0.57 |
| PE (18:4(6Z,9Z,12Z,15Z)/20:1(11Z)) | 1183 (769.40 to 1521) | 1839 (1001 to 4197) | 0.27 |
| PE (18:4(6Z,9Z,12Z,15Z)/P-18:1(11Z)) | 1736 (1250 to 2028) | 2559 (1583 to 6457) | 0.11 |
| Progesterone | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 698 (3653 to 11 698 (3653 to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 922) 922) |
7071 (3148 to 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 685) 685) |
0.77 |
| Sphingosine | 1547 (942.10 to 2905) | 2675 (1420 to 3388) | 0.37 |
| TG (16:0/18:1(9Z)/20:4(5Z,8Z,11Z,14Z)) | 1603 (625.50 to 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 417) 417) |
3493 (667 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 753) 753) |
0.92 |
| TG (16:1(9Z)/18:2(9Z,12Z)/18:3(6Z,9Z,12Z)) | 393.30 (176.8 to 6832) | 533.60 (186.70 to 2450) | 0.92 |
| Vitamin E | 559.80 (160.60 to 690.60) | 271.80 (105.50 to 1005) | 0.67 |
There were no evident alterations in colostrum pro-inflammatory cytokines in maternal obesity
We quantified colostrum cytokines relevant in inflammatory processes. All cytokines were detected in all samples. However, no significant differences were observed between the groups and correlations with the maternal BMI (Table 1). Although we explored the relationships between the maternal clinical characteristics (maternal age, gestational age, and way of delivery) and colostrum cytokine concentrations, no clear correlation patterns were observed across the groups.Changes in the microbial composition in the colostrum of mothers with obesity
We analysed the colostrum microbial community using 16S ribosomal RNA amplicon sequencing. We obtained 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280
280![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 755 high-quality reads, corresponding to 45
755 high-quality reads, corresponding to 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 615 reads per sample. We identified 1255 amplicon sequence variants (ASVs) annotated within 4 major phyla: Firmicutes (66.5%, NW; 77.5%, WO), Proteobacteria (21.9%, NW; 12.3%, WO), Actinobacteriota (4.5%, NW; 4.8%, WO), and Bacteroidota (2.3%, NW; 1.6%, WO) (Fig. 1A). Overall, the taxonomic families Staphylococcaceae (37.8%, rank 0.17%–97.8%) and Streptococcaceae (28.8%, rank 0.02%–92.7%) exhibited the highest relative abundance. Lower abundances were observed for Gemellaceae (3.7%), Burkholderiaceae (2.2%), Enterobacteriaceae (2%), and Sphingomonadaceae (1.3%) taxonomic families. Additional families (102), including Bacillaceae, Lactobacillaceae and Bifidobacteriaceae, contributed to <1% of the total relative abundance (Fig. 1B and raw data). There was no difference between the groups looking at the main ecological diversity indices (Fig. S1†). The Firmicutes to Bacteroidota (F/B) ratio and the Firmicutes to Proteobacteria (F/P) ratio were higher in samples from women with obesity, although these differences were not statistically significant (mean ratio: 28.7 vs. 47.7, p = 0.30, and 3.0 vs. 6.3, p = 0.14, respectively). In contrast, the Proteobacteria to Bacteroidota (P/B) ratio was significantly decreased in WO (9.4 vs. 7.6, p = 0.046), which agrees with a relative reduction in the abundance of the Proteobacteria phylum, although not significant (21.9% ± 23.5% vs. 12.3% ± 17.2%, p = 0.14; Fig. S2†).
615 reads per sample. We identified 1255 amplicon sequence variants (ASVs) annotated within 4 major phyla: Firmicutes (66.5%, NW; 77.5%, WO), Proteobacteria (21.9%, NW; 12.3%, WO), Actinobacteriota (4.5%, NW; 4.8%, WO), and Bacteroidota (2.3%, NW; 1.6%, WO) (Fig. 1A). Overall, the taxonomic families Staphylococcaceae (37.8%, rank 0.17%–97.8%) and Streptococcaceae (28.8%, rank 0.02%–92.7%) exhibited the highest relative abundance. Lower abundances were observed for Gemellaceae (3.7%), Burkholderiaceae (2.2%), Enterobacteriaceae (2%), and Sphingomonadaceae (1.3%) taxonomic families. Additional families (102), including Bacillaceae, Lactobacillaceae and Bifidobacteriaceae, contributed to <1% of the total relative abundance (Fig. 1B and raw data). There was no difference between the groups looking at the main ecological diversity indices (Fig. S1†). The Firmicutes to Bacteroidota (F/B) ratio and the Firmicutes to Proteobacteria (F/P) ratio were higher in samples from women with obesity, although these differences were not statistically significant (mean ratio: 28.7 vs. 47.7, p = 0.30, and 3.0 vs. 6.3, p = 0.14, respectively). In contrast, the Proteobacteria to Bacteroidota (P/B) ratio was significantly decreased in WO (9.4 vs. 7.6, p = 0.046), which agrees with a relative reduction in the abundance of the Proteobacteria phylum, although not significant (21.9% ± 23.5% vs. 12.3% ± 17.2%, p = 0.14; Fig. S2†).
To our knowledge, this is the first report identifying an increase abundance of the Chitinophagaceae family in the colostrum of women with obesity, with a 0.78 Log2FC relative to samples from the NW group (Fig. 1C). In contrast, 7 taxonomic families were less abundant, including Brevibacteraceae (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC: −0.96), Burkholderiaceae (Log
2FC: −0.96), Burkholderiaceae (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC: −1.04), Lactobacillaceae (Log
2FC: −1.04), Lactobacillaceae (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC: −1.10), Comamonadaceae (Log
2FC: −1.10), Comamonadaceae (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC: −1.16), Rhodospirillales (Log
2FC: −1.16), Rhodospirillales (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC: −1.40), Bacillaceae (Log
2FC: −1.40), Bacillaceae (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC: −1.60), and Planococcaceae (Log
2FC: −1.60), and Planococcaceae (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC: −1.62).
2FC: −1.62).
Maternal obesity correlates with changes in the colostrum metabolite environment
We used untargeted metabolomics to explore discrepancies in the colostrum of women with obesity compared to normal weight subjects. Our pipeline led to the quantification of 2808 features across the samples. We annotated 1027 MS2-containing features across 12 superclasses, leaving 473 features unassigned (Fig. 2A and Table S2†). Metabolite diversity at the class level was assessed using the Shannon and Simpson indexes, revealing a reduction in the colostrum samples from the WO group compared to the NW group (p = 0.05; Fig. S3†). Subsequently, we observed 73 features with decreased abundance and 49 features with increased abundance in the WO group (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC ± 0.58, limma test, p < 0.05; Fig. 2B). Decreased features were mostly linked to carboxylic acids, fatty acyls and prenol lipids, whereas the distribution of the increased features was primarily carboxylic acids, organooxygen compounds, azoles, and organic phosphines (Fig. 2C). To gain insights into the metabolic markers of obesity, we performed correlation analyses between metabolites (identified at the chemical class level) and maternal BMI levels. We identified 57 features distributed across 11 chemical classes that were significantly associated with the maternal BMI (Fig. 2D and Table S3†), mainly comprising carboxylic acids and derivatives (n = 31), followed by fatty acyls (n = 7), and organooxygen compounds (n = 6). Through high-confidence putative structural annotation (mass error ≤ 10 ppm) and differential abundance analysis (limma test), we found that decanoylcarnitine (alongside an unidentified carnitine, as revealed through MS2LDA analysis) and 15-HEDE (fatty acyl) presented reduced abundance in the colostrum of the WO group compared to the NW group (Fig. 2E and Table 1). This is the first time that 15-HEDE is detected in differential abundance in the colostrum of mothers with obesity.
2FC ± 0.58, limma test, p < 0.05; Fig. 2B). Decreased features were mostly linked to carboxylic acids, fatty acyls and prenol lipids, whereas the distribution of the increased features was primarily carboxylic acids, organooxygen compounds, azoles, and organic phosphines (Fig. 2C). To gain insights into the metabolic markers of obesity, we performed correlation analyses between metabolites (identified at the chemical class level) and maternal BMI levels. We identified 57 features distributed across 11 chemical classes that were significantly associated with the maternal BMI (Fig. 2D and Table S3†), mainly comprising carboxylic acids and derivatives (n = 31), followed by fatty acyls (n = 7), and organooxygen compounds (n = 6). Through high-confidence putative structural annotation (mass error ≤ 10 ppm) and differential abundance analysis (limma test), we found that decanoylcarnitine (alongside an unidentified carnitine, as revealed through MS2LDA analysis) and 15-HEDE (fatty acyl) presented reduced abundance in the colostrum of the WO group compared to the NW group (Fig. 2E and Table 1). This is the first time that 15-HEDE is detected in differential abundance in the colostrum of mothers with obesity.
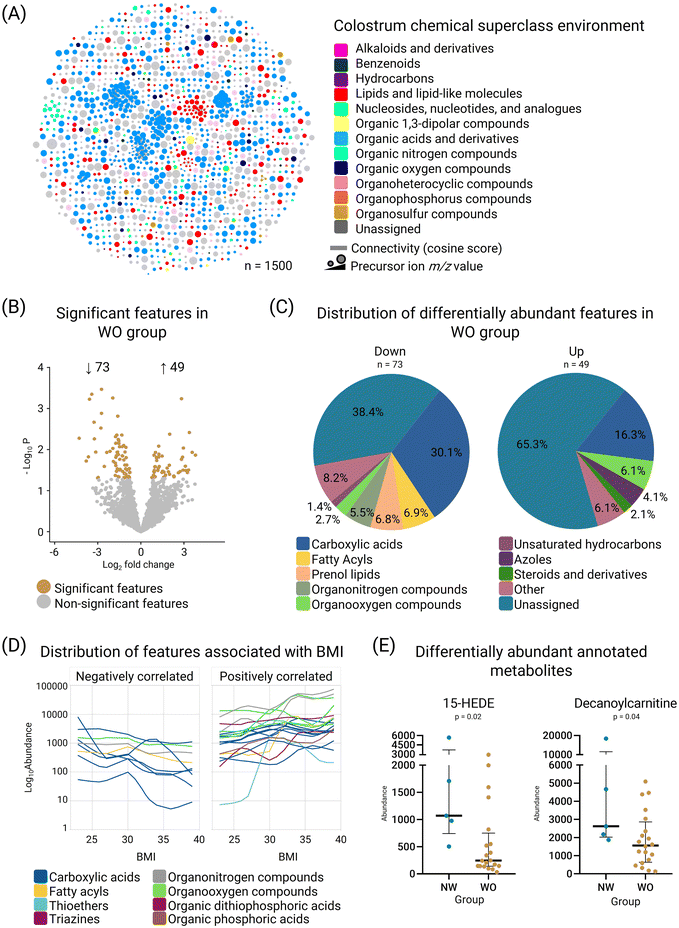 | ||
Fig. 2 Metabolite environment in the colostrum of women with obesity (WO). (A) Molecular network of the identified MS2-containing features in colostrum samples. Each node represents a feature, and the color denotes its assigned chemical superclass. Node size highlights the m/z value. Edge thickness indicates the MS2 similarity (cosine score) among the features and (B) volcano plot of all quantified features highlighting the differentially abundant features between the WO and NW groups. Brown dots represent significantly increased or decreased features in the WO group (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC ± 0.58, limma p < 0.05), while gray dots indicate features without significant differences; (C) pie charts of the chemical class distribution of the differentially increased (n = 49) or decreased (n = 73) features in the colostrum samples from the WO group. “Other” classification corresponds to classes that only contained one feature; (D) LOESS curves of each feature significantly associated with maternal BMI classified by their Spearman's correlation value (ρ). Curves are color-coded based on the feature chemical class assigned as described in the color key. Only features with a correlation value higher than absolute 0.45 were plotted; (E) boxplots of the relative abundance of the dysregulated (limma test, p < 0.05) lipid related identified metabolites, 15-HEDE and decanoylcarnitine; NW, normal weight group (n = 5); WO, with obesity group (n = 20). See also Tables S2 and S3.† 2FC ± 0.58, limma p < 0.05), while gray dots indicate features without significant differences; (C) pie charts of the chemical class distribution of the differentially increased (n = 49) or decreased (n = 73) features in the colostrum samples from the WO group. “Other” classification corresponds to classes that only contained one feature; (D) LOESS curves of each feature significantly associated with maternal BMI classified by their Spearman's correlation value (ρ). Curves are color-coded based on the feature chemical class assigned as described in the color key. Only features with a correlation value higher than absolute 0.45 were plotted; (E) boxplots of the relative abundance of the dysregulated (limma test, p < 0.05) lipid related identified metabolites, 15-HEDE and decanoylcarnitine; NW, normal weight group (n = 5); WO, with obesity group (n = 20). See also Tables S2 and S3.† | ||
Bacterial co-occurrence network in the colostrum microbiota
Expanding upon the abundance analysis of individual elements, we used co-occurrence networks to define distinctive bacterial interaction modules within the samples. This approach provides a comprehensive view of the coexistence of bacterial taxa and their potential influence with each other. Using Spearman's rank coefficients, we defined five distinct modules of taxonomic families according to their degree of interaction. A positive correlation in this context denotes a potential coordinated and mutualistic interaction, either through direct or indirect mechanisms, which implies that the presence of specific taxa positively influences the presence of others within the microbial community. Conversely, a negative correlation indicates a direct or indirect antagonistic interaction, wherein the coexistence of certain taxa is perturbed by the presence of others (Fig. 3 and raw data).In the first module, composed of Firmicutes members, Streptococcaceae appears as a central taxon, negatively correlated to Staphylococcaceae and positively correlated to Gemellaceae. These associations suggest potential competition between dominant breastmilk bacterial families and highlights the role of Streptococcaceae in the microbial structure. The second module includes mostly members of Proteobacteria, including Yersiniaceae, Erwiniaceae, Shewanellaceae, Reyranellaceae and Rhodanobacteraceae. Strong positive correlations were observed between Candidatus Kaiserbacteria and both Reyranellaceae (ρ = 0.81) and Rhodanobacteraceae (ρ = 0.83), suggesting cooperation or shared environmental preferences. Notably, their similar median relative abundance in both groups indicate that the interactions are potentially resilient to the maternal BMI. In the third module, we observed moderate positive correlations (ρ ≈ 0.51–0.53) among Saccharimonadales, Fusobacteriaceae, Neisseriaceae and Carnobacteriaceae. Neisseriaceae and Carnobacteriaceae showed higher median relative abundance in the colostrum of the NW group, indicating potential obesity-mediated disruptions on the mutualistic interactions of this module. The fourth module presented positive correlations between Prevotellaceae, Campylobacteraceae, Veillonellaceae and Peptostreptococcaceae. Veillonellaceae was the most interconnected family within this module, suggesting a key structural role in the bacterial network. Conversely, Prevotellaceae showed a decreased median relative abundance in the WO group, which may imply shifts in the dynamics of the colostrum microbiota. The fifth module was the largest and most diverse, comprising 19 taxonomic families with positive correlations. Potential key taxa in the network included Alcaligenaceae and was identified as a potential key player, participating in 7 interactions, followed by Caulobacteraceae and Nocardiopsaceae, each involved in 6 interactions. We identified families with reductions in relative abundance in the colostrum of women with obesity, including Comamonadaceae, Burkholderiaceae, Xanthobacteraceae, Beijerinckiaceae, Sphingomonadaceae, Caulobacteraceae, Bifidobacteriaceae, Nocardiaceae, Deinococcaceae, and Moraxellaceae, which suggest that maternal obesity may compromise a core mutualistic module essential to colostrum's ecological and functional integrity. Further investigation is required to unravel the complex network of interactions that may hold biological significance in the context of maternal obesity.
Lipid mediators are correlated with pro-inflammatory cytokines and Proteobacteria members in the colostrum
We performed a co-occurrence analysis to explore potential interactions between putatively annotated metabolites and cytokines in the colostrum (Fig. 4A and B). We found that 15-HEDE negatively correlated with IL-12p70 and IL-10, suggesting a potential role of this regulatory lipid in inflammation. Lysophosphatidylcholine (16:00; LysoPC [16:00]), on the other hand, correlated positively with IL-1 and IL-8, suggesting a role in pro-inflammatory signalling pathways (Fig. 4B and Table S4†). | ||
| Fig. 4 Correlation analysis between colostrum bioactive elements. (A) Scheme of the different correlations performed between the bioactive elements. (B) Heatmap of Spearman's correlation between metabolites and cytokines in colostrum; (C) Heatmap of Spearman's correlation analysis of taxonomic families and metabolites in colostrum; (D and E) Heatmap of Spearman's correlation analysis of taxonomic families and cytokines in the colostrum of women with (D) normal weight and (E) obesity. Red represents a positive correlation, white represents a low correlation, and blue represents a negative correlation, as shown in the color key. *p-value < 0.05; #p-value < 0.10. See also Tables S4 and S5.† | ||
In addition, we analysed the interrelationship between metabolites and bacterial groups. For instance, the regulatory lipid 15-HEDE, found to be decreased in the colostrum of women with obesity, exhibited positive correlations with taxonomic families such as Beijerinckiaceae, Burkholderiaceae, and Erwiniaceae, which were also less abundant in the colostrum of women with obesity (Fig. 1C and Fig. 4C). Moreover, despite the absence of a significant difference between the study groups, 12(13) Ep-9-KODE showed a positive correlation with Micrococcaceae, Aeromonadaceae, and Beijerinckiaceae. On the other hand, LysoPC (16:00) showed negative correlations with Xanthobacteraceae, Reyranellaceae, Xanthomonadaceae, Caulobacteraceae, Cellulomonadaceae, Pasteurellaceae, Rhodospirillales, Streptococcaceae, Weeksellaceae and Bacteroidaceae, but exhibited positive correlations with Planococcaceae, Alphaproteobacteria and Burkholderiaceae.
We also identified that the steroid hormone progesterone displayed negative correlations with Caulobacteraceae, Comamonadaceae, Devosiaceae, Nocardioidaceae and Staphylococcaceae, which may imply instances where negatively affects abundance of the bacterial community. Furthermore, fat-soluble vitamin E was negatively correlated with Comamonadaceae, Neisseriaceae, Fusobacteriaceae, Staphylococcaceae, Erwiniaceae and Carnobacteriaceae (Fig. 4C and Table S4†). Further research into the specific metabolic pathways and ecological roles associated with these metabolites could support their significance in the colostrum system and their implications in obesity.
Pro-inflammatory cytokines are negatively correlated to Proteobacteria members in the colostrum of mothers with obesity.
We also delve into the interplay between microbial communities and immune-modulating factors (Table S5†). In samples obtained from normal weight participants, members of the Proteobacteria phylum such as Enterobacteriaceae and Burkholderiaceae displayed negative correlations with IL-10, while Prevotellaceae showed a positive correlation with this anti-inflammatory cytokine. Other members of the Proteobacteria phylum, including Neisseriaceae and Pasteurellaceae, exhibited positive correlations with IL-6, TNF-α, IL-1β, and IL-8, suggesting a potential pro-inflammatory modulation (Fig. 4D). Meanwhile Veillonellaceae, Prevotellaceae, and Gemellaceae presented a positive correlation with IL-1β, and Dermabacteraceae and Brevibacteraceae were positively correlated with IL-12p70. Additionally, Porphyromonadaceae was positively correlated with IL-6.
In contrast, samples from women with obesity showed negative correlations between the taxonomic families Aeromonadaceae, Xanthomonadaceae, and Staphylococcaceae, and the immune-modulating factors IL-6, TNF-α, and IL-12p70. Devosiaceae, Mycobacteriaceae, and Nocardioidaceae were negatively correlated with IL-8, while Corynebacteriaceae, Staphylococcaceae, Leptotrichiaceae and Tannerellaceae showed positive correlations (Fig. 4E). Notably, Peptostreptococcaceae exhibited a positive correlation with IL-8 across the entire cohort independently of the study group.
Discussion
Colostrum is a complex and dynamic fluid that seeds neonatal intestines within the initial hours of life. The impact of colostrum depends on the composition and interactions among its bioactive components.10,14 Integrative research initiatives, such as the BEGIN project, are exploring breastmilk as a biological system, emphasizing the interactions and coordinated functionality between its bioactive components.10,50 In this study, we profiled the colostrum microbiota, cytokines, and metabolites and established networks of co-occurring elements to provide insights into the effect of maternal obesity on interacting bioactive components.Despite extensive research on the associations between breastmilk components and maternal obesity, the findings remain heterogeneous.4,11,13,51–53 We identified subtle differences while analysing individual bioactive components, i.e. cytokines, microbiota, and metabolites. While variability can be attributed to technical and methodological factors,54–56 studying colostrum as a dynamic biological system with interconnected components could provide valuable insights into the relationships within microbial communities and their interactions with bioactive molecules.
In our study, we identified a negative correlation between Staphylococcaceae and Streptococcaceae, indicating a potential competitive relationship. This association could be influenced by ecological factors within the mammary gland microenvironment, such as niche competition, resource availability, or immune responses, which could benefit the dominance of one bacterial group over the other.57,58 Moreover, co-occurrence network analysis revealed bacterial relationships among Proteobacteria members exhibiting reduced relative abundance in the colostrum of women with obesity. Within the same co-occurrence module, these taxa demonstrated positive correlations, indicating mutualistic interactions and functional similarities.57
The observed variations in the colostrum microbiota of the WO group may impact the microbiota development of neonates. Offspring born to mothers with obesity presented a decrease in the abundance of Gammaproteobacteria two weeks after birth.59 This is critical for the establishment of a healthy gut microbiome functioning, since Proteobacteria members are the primary colonizers during the first week of life and play a crucial role in preparing anaerobic conditions in the gut for successive colonization by strict anaerobes.60–62 Additionally, their presence in the postnatal period is essential for promoting a healthy immune system, facilitating host–microbe interactions. A reduction in Proteobacteria supply in the colostrum of mothers with obesity may compromise immune system training during the critical early days of life, potentially leading to impaired immunological tolerance and increased risk of immune-related diseases later in life.63,64
Alterations in specific microbial taxa within breastmilk may potentially be linked to cytokine levels, exerting an impact on breast-fed infants.65 We report that the abundance of Enterobacteriaceae in the colostrum of women with normal weight was negatively correlated with TNF-α, while Proteobacteria such as Neisseriaceae was positively correlated with TNF-α and IL-6. This pattern aligns with previous observations in mice, where the presence of the early colonizer Enterobacteria triggers TNF-α production during the first week of life. This interaction prevents excessive inflammatory and autoimmune gastrointestinal disorders in the future.64 Furthermore, the production of TNF-α by macrophages and monocytes within the first three days of life, stimulated by the microbiota, is a prerequisite for the maturation of pre-conventional dendritic cell 1 (pre-cDC1) into conventional dendritic cell 1 (cDC1).66
The increased levels of Gram-negative bacteria in the gut are known to stimulate the local production of pro-inflammatory cytokines.67 In mothers with obesity (WO group), we observed negative correlations between pro-inflammatory cytokines (TNF-α, IL-6, and IL-12p70) and certain bacterial families such as Aeromonadaceae, Xanthomonadaceae and Staphylococcaceae (Fig. 5). Our unexpected correlative findings align with the observations of lower TNF-α and IL-6 concentrations in the breastmilk of mothers with obesity compared to mothers with normal BMI.68 This suggests a potential local regulatory system within the mammary gland that may modulate systemic inflammation during obesity.
Metabolites are key elements of microbial communities as they reflect interactions within the ecosystem. Different metabolites have been associated with interactions in the breastmilk of women with obesity.5,69,70 We observed a reduction in fatty acyls and prenol lipids, which suggests potential alterations in the lipid profile of colostrum associated with maternal obesity. For instance, we observed that decanoylcarnitine exhibited a negative correlation with BMI and was found to be decreased in the WO group. Our results contrast previous observations where elevated levels of various acylcarnitines were detected in the serum and breastmilk of individuals with obesity.5,71 Acylcarnitines play a crucial role in mediating the beta-oxidation of fatty acids within the mitochondria, a pivotal process in energy production.72
In addition, the breastmilk of women with overweight or obesity exhibits decreased levels of polyunsaturated fatty acids (PUFAs).73 The metabolism of PUFAs processed through the enzymatic action of lipoxygenases and cyclooxygenases can lead to the formation of oxylipins.74 Breastmilk has a diverse array of bioactive lipids, which play significant roles within the neonatal immune system.75,76 The presence of oxylipins in breastmilk may be a marker of maternal health and could influence the health outcomes and inflammatory status of breast-fed infants.74
Some hydroxy fatty acids appear to be linked to inflammatory processes with robust functions, for example, with anti-inflammatory and pro-resolving properties, either directly or through their conversion into lipid mediators.77,78 One notable example is 15-HEDE, a macrophage-oxidized product of the inflammatory modulator 6-PUFA eicosadienoic acid (EDA).79,80 We found that 15-HEDE levels were decreased in the colostrum of women with obesity, showing a negative correlation with IL-10 levels and positive correlations with Proteobacteria members, such as Beijerinckiaceae and Burkholderiaceae (Fig. 5). Despite the fact that 15-HEDE has been described as an inhibitor of 5-lipoxygenase (5-LO),81 it has also been reported to induce vasodilation and vascular hyperpermeability, acting as a pro-inflammatory lipid mediator that exacerbates allergic rhinitis.82 Elevated levels of 15-HEDE have also been observed in allergic airway inflammation in mice after a house dust mite exposure and in psoriasis.79,83 The negative correlation with IL-10 suggests that IL-10 may act as a modulator of inflammation resolution in colostrum. In the chronic inflammatory milieu of obesity, circulating levels of IL-10 are elevated compared to normal weight women.84,85 Therefore, we hypothesized a potential compensatory mechanism to counterbalance chronic inflammatory processes and mitigate the impact of reduced but highly immunogenic bacteria. In normal-weight mothers, the higher abundance of 15-HEDE may enhance the vasodilatory capacity of the mammary vasculature, facilitating efficient nutrient transfer and immunological component delivery to the neonate.86
While progesterone showed no significant difference between the NW and WO groups, its potential role in shaping colostrum microbial ecology warrants further investigation. We observed negative correlations when compared to specific bacterial taxa, including Staphylocococcaceae, Ferrovibrionales, Caulobacteriaceae, and Devosiaceae (Fig. 5). Progesterone has been shown to influence microbial communities in vitro by reducing the growth, adhesion, and virulence of certain Staphylococcus strains.87 While progesterone is dominant during late pregnancy, pregnant women with obesity often exhibit reduced serum progesterone levels.88,89 Moreover, progesterone levels in the breastmilk of women with a high fat and protein diet are diminished90 and negatively correlated with infant weight at 6 months.91
Our study presents some limitations. For instance, detailed information regarding maternal diet and lifestyle was not collected, which could represent potential confounding factors. Although small cohorts are common in colostrum-related studies, we acknowledge that the implementation of larger sample sizes would enhance the statistical power and robustness of findings. Some correlations in our study fell within the moderate range (|ρ| = 0.40–0.59). These values, though statistically significant, should be interpreted with caution, as they may miss nonlinear relationships or interactions mediated by unmeasured variables. In addition, due to limited sample volumes, this work presents high variability in sample size across datasets (16S sequencing, cytokines, and metabolomics), which may negatively impact data integration and interpretation. Finally, while co-occurrence networks offer insights into the microbial community structure and their interactions with bioactive molecules that traditional methods might overlook, they do not establish directional interactions or causal mechanisms.
In summary, using co-occurrence networks, we integrated metataxonomic data, metabolomics, and cytokine profiles to provide novel insights and a proof of concept for colostrum as an interconnected system and demonstrate the influence of maternal obesity on bioactive interactions within this microenvironment. Our analysis identified a negative co-occurrence between the lipid mediator 15-HEDE and IL-10 while also showing a positive association with microbiota members such as Beijerinckiaceae and Burkholderiaceae – findings that have not been previously reported in the colostrum of mothers with obesity. Additionally, we identified that Aeromonadaceae, Xanthomonadaceae, and Staphylococcaceae were negatively correlated with pro-inflammatory cytokines (TNF-α, IL-6, and IL-12p70), suggesting a potential regulatory effect on inflammatory responses in colostrum associated with maternal obesity. Our findings reveal that maternal obesity influences alterations in the ecosystem structure. However, further research is needed to elucidate the neonatal health implications of the disrupted interactions among the bacterial community, metabolites and local inflammatory markers observed in the colostrum of women with obesity.
Author contributions
JSGV: data curation, formal analysis, investigation, methodology, writing – original draft, and writing – review & editing. KCC: data curation, formal analysis, investigation, supervision, and writing – review & editing. ESSS: investigation, methodology, and writing – review & editing. MRAG: investigation and methodology. CNLV: investigation and methodology. RACC: formal analysis, methodology, and writing – review & editing. AMU: supervision, formal analysis, methodology, and writing – review & editing. VJLD: investigation, methodology, supervision, and writing – review & editing. MEGB: conceptualization, investigation, methodology, supervision, funding acquisition, writing – original draft, and writing – review & editing. CLC: conceptualization, data curation, formal analysis, funding acquisition, project administration, supervision, writing – original draft, and writing – review & editing.Data availability
All fastq-format reads analyzed have been deposited and are publicly available at the NCBI Sequence Read Archive (SRA). Metabolite raw datasets are available at the GNPS/MassIVE public repository and are available as of the date of publication. Accession numbers are listed in the ESI (Table S6†). This paper does not report the original code.Conflicts of interest
All authors have read the manuscript and declare no conflicts of interest.Acknowledgements
This work was financially supported by the Centro de Biotecnología FEMSA from Tecnológico de Monterrey and the Institute for Obesity Research from Tecnológico de Monterrey. We thank the National Council for Humanities, Sciences and Technology (CONAHCYT) for providing a scholarship to JSGV, Frontera de la Ciencia grant no. CF/2023/G/990 to MEGB, the BIOCODEX Microbiota Foundation grant to CLC, and grant no. 314964 (Program F0001-2020-02) that supported the functioning of the LC-MS equipment. Finally, we would like to thank Diana Priscila Bonilla Ruelas and Alan G. Hernández Melgar for their technical support.References
- L. F. Stinson, A. S. M. Sindi, A. S. Cheema, C. T. Lai, B. S. Mühlhäusler, M. E. Wlodek, M. S. Payne and D. T. Geddes, The human milk microbiome: who, what, when, where, why, and how?, Nutr. Rev., 2021, 79, 529–543 CrossRef PubMed.
- G. A. G. Lokossou, L. Kouakanou, A. Schumacher and A. C. Zenclussen, Human breast milk: from food to active immune response with disease protection in infants and mothers, Front. Immunol., 2022, 13, 849012 CrossRef CAS PubMed.
- S. Perrella, Z. Gridneva, C. T. Lai, L. Stinson, A. George, S. Bilston-John and D. Geddes, Human milk composition promotes optimal infant growth, development and health, Semin. Perinatol., 2021, 45, 151380 CrossRef PubMed.
- S. Moossavi, S. Sepehri, B. Robertson, L. Bode, S. Goruk, C. J. Field, L. M. Lix, R. J. de Souza, A. B. Becker, P. J. Mandhane, S. E. Turvey, P. Subbarao, T. J. Moraes, D. L. Lefebvre, M. R. Sears, E. Khafipour and M. B. Azad, Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors, Cell Host Microbe, 2019, 25, 324–335 CrossRef CAS PubMed.
- E. Isganaitis, S. Venditti, T. J. Matthews, C. Lerin, E. W. Demerath and D. A. Fields, Maternal obesity and the human milk metabolome: associations with infant body composition and postnatal weight gain, Am. J. Clin. Nutr., 2019, 110, 111–120 CrossRef PubMed.
- M. C. Neville, E. W. Demerath, J. Hahn-Holbrook, R. C. Hovey, J. Martin-Carli, M. A. McGuire, E. R. Newton, K. M. Rasmussen, M. C. Rudolph and D. J. Raiten, Parental factors that impact the ecology of human mammary development, milk secretion, and milk composition-a report from “Breastmilk Ecology: Genesis of Infant Nutrition (BEGIN)” Working Group 1, Am. J. Clin. Nutr., 2023, 117(Suppl 1), S11–S27 CrossRef PubMed.
- Y. Wan, J. Jiang, M. Lu, W. Tong, R. Zhou, J. Li, J. Yuan, F. Wang and D. Li, Human milk microbiota development during lactation and its relation to maternal geographic location and gestational hypertensive status, Gut Microbes, 2020, 11, 1438–1449 CrossRef CAS PubMed.
- O. Koren, L. Konnikova, P. Brodin, I. U. Mysorekar and M. C. Collado, The maternal gut microbiome in pregnancy: implications for the developing immune system, Nat. Rev. Gastroenterol. Hepatol., 2024, 21, 35–45 CrossRef PubMed.
- P. Christian, E. R. Smith, S. E. Lee, A. J. Vargas, A. A. Bremer and D. J. Raiten, The need to study human milk as a biological system, Am. J. Clin. Nutr., 2021, 113, 1063–1072 CrossRef CAS PubMed.
- S. M. Donovan, N. Aghaeepour, A. Andres, M. B. Azad, M. Becker, S. E. Carlson, K. M. Järvinen, W. Lin, B. Lönnerdal, C. M. Slupsky, A. L. Steiber and D. J. Raiten, Evidence for human milk as a biological system and recommendations for study design-a report from “Breastmilk Ecology: Genesis of Infant Nutrition (BEGIN)” Working Group 4, Am. J. Clin. Nutr., 2023, 117(Suppl 1), S61–S86 CrossRef CAS PubMed.
- R. Piñeiro-Salvador, E. Vazquez-Garza, J. A. Cruz-Cardenas, C. Licona-Cassani, G. García-Rivas, J. Moreno-Vásquez, M. R. Alcorta-García, V. J. Lara-Diaz and M. E. G. Brunck, A cross-sectional study evidences regulations of leukocytes in the colostrum of mothers with obesity, BMC Med., 2022, 20, 388 CrossRef PubMed.
- K. Daiy, V. Harries, K. Nyhan and U. M. Marcinkowska, Maternal weight status and the composition of the human milk microbiome: A scoping review, PLoS One, 2022, 17, e0274950 CrossRef CAS PubMed.
- J. S. Gámez-Valdez, J. F. García-Mazcorro, A. H. Montoya-Rincón, D. L. Rodríguez-Reyes, G. Jiménez-Blanco, M. T. A. Rodríguez, R. P.-C. de Vaca, M. R. Alcorta-García, M. Brunck, V. J. Lara-Díaz and C. Licona-Cassani, Differential analysis of the bacterial community in colostrum samples from women with gestational diabetes mellitus and obesity, Sci. Rep., 2021, 11, 24373 CrossRef PubMed.
- J. W. Ruan, Y.-C. Liao, P. C. Chen, Y. J. Chen, Y. H. Tsai, P. J. Tsai, Y. J. Yang, C. C. Shieh, Y. C. Lin and C. Y. Chi, The composition of the maternal breastmilk microbiota influences the microbiota network structure during early infancy, J. Microbiol., Immunol. Infect., 2023, 56, 1084–1097 CrossRef CAS PubMed.
- S. Moossavi, F. Atakora, K. Miliku, S. Sepehri, B. Robertson, Q. L. Duan, A. B. Becker, P. J. Mandhane, S. E. Turvey, T. J. Moraes, D. L. Lefebvre, M. R. Sears, P. Subbarao, C. J. Field, L. Bode, E. Khafipour and M. B. Azad, Integrated analysis of human milk microbiota with oligosaccharides and fatty acids in the CHILD cohort, Front. Nutr., 2019, 6, 58 CrossRef PubMed.
- R. M. Pace, J. E. Williams, B. Robertson, K. A. Lackey, C. L. Meehan, W. J. Price, J. A. Foster, D. W. Sellen, E. W. Kamau-Mbuthia, E. W. Kamundia, S. Mbugua, S. E. Moore, A. M. Prentice, D. G. Kita, L. J. Kvist, G. E. Otoo, L. Ruiz, J. M. Rodríguez, R. G. Pareja, M. A. McGuire, L. Bode and M. K. McGuire, Variation in human milk composition is related to differences in milk and infant fecal microbial communities, Microorganisms, 2021, 9, 1153 CrossRef CAS PubMed.
- J. E. Williams, W. J. Price, B. Shafii, K. M. Yahvah, L. Bode, M. A. McGuire and M. K. McGuire, Relationships among microbial communities, maternal cells, oligosaccharides, and macronutrients in human milk, J. Hum. Lact., 2017, 33, 540–551 CrossRef PubMed.
- J. A. Siles, M. García-Sánchez and M. Gómez-Brandón, Studying Microbial Communities through Co-Occurrence Network Analyses during Processes of Waste Treatment and in Organically Amended Soils: A Review, Microorganisms, 2021, 9, 1165 CrossRef PubMed.
- A. Barberán, S. T. Bates, E. O. Casamayor and N. Fierer, Using network analysis to explore co-occurrence patterns in soil microbial communities, ISME J., 2012, 6, 343–351 CrossRef PubMed.
- World Health Organization , Obesity and overweight, https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight, (accessed May 2, 2024).
- E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, Y. Bai, J. E. Bisanz, K. Bittinger, A. Brejnrod, C. J. Brislawn, C. T. Brown, B. J. Callahan, A. M. Caraballo-Rodríguez, J. Chase, E. K. Cope, R. Da Silva, C. Diener, P. C. Dorrestein, G. M. Douglas, D. M. Durall, C. Duvallet, C. F. Edwardson, M. Ernst, M. Estaki, J. Fouquier, J. M. Gauglitz, S. M. Gibbons, D. L. Gibson, A. Gonzalez, K. Gorlick, J. Guo, B. Hillmann, S. Holmes, H. Holste, C. Huttenhower, G. A. Huttley, S. Janssen, A. K. Jarmusch, L. Jiang, B. D. Kaehler, K. B. Kang, C. R. Keefe, P. Keim, S. T. Kelley, D. Knights, I. Koester, T. Kosciolek, J. Kreps, M. G. I. Langille, J. Lee, R. Ley, Y.-X. Liu, E. Loftfield, C. Lozupone, M. Maher, C. Marotz, B. D. Martin, D. McDonald, L. J. McIver, A. V. Melnik, J. L. Metcalf, S. C. Morgan, J. T. Morton, A. T. Naimey, J. A. Navas-Molina, L. F. Nothias, S. B. Orchanian, T. Pearson, S. L. Peoples, D. Petras, M. L. Preuss, E. Pruesse, L. B. Rasmussen, A. Rivers, M. S. Robeson, P. Rosenthal, N. Segata, M. Shaffer, A. Shiffer, R. Sinha, S. J. Song, J. R. Spear, A. D. Swafford, L. R. Thompson, P. J. Torres, P. Trinh, A. Tripathi, P. J. Turnbaugh, S. Ul-Hasan, J. J. J. van der Hooft, F. Vargas, Y. Vázquez-Baeza, E. Vogtmann and J. G. Caporaso, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, 37, 852–857 CrossRef CAS PubMed.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581–583 CrossRef CAS PubMed.
- E. Pruesse, C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies and F. O. Glöckner, SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB, Nucleic Acids Res., 2007, 35, 7188–7196 CrossRef CAS PubMed.
- N. A. Bokulich, B. D. Kaehler, J. R. Rideout, M. Dillon, E. Bolyen, R. Knight, G. A. Huttley and J. Gregory Caporaso, Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin, Microbiome, 2018, 6, 90 CrossRef PubMed.
- M. C. de Goffau, S. Lager, S. J. Salter, J. Wagner, A. Kronbichler, D. S. Charnock-Jones, S. J. Peacock, G. C. S. Smith and J. Parkhill, Recognizing the reagent microbiome, Nat. Microbiol., 2018, 3, 851–853 CrossRef CAS PubMed.
- Q. Cao, X. Sun, K. Rajesh, N. Chalasani, K. Gelow, B. Katz, V. H. Shah, A. J. Sanyal and E. Smirnova, Effects of rare microbiome taxa filtering on statistical analysis, Front. Microbiol., 2020, 11, 607325 CrossRef PubMed.
- J. Oksanen, F. G. Blanchet, R. Kindt, P. Legendre, P. Minchin, R. O'Hara, G. Simpson, P. Solymos, M. Stevenes and H. Wagner, Vegan: Community Ecology Package, R package, 2022 Search PubMed.
- P. J. McMurdie and S. Holmes, phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data, PLoS One, 2013, 8, e61217 CrossRef CAS PubMed.
- Y. Vázquez-Baeza, M. Pirrung, A. Gonzalez and R. Knight, EMPeror: a tool for visualizing high-throughput microbial community data, Gigascience, 2013, 2, 16 CrossRef PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15, 550 CrossRef PubMed.
- H. Lin and S. D. Peddada, Analysis of compositions of microbiomes with bias correction, Nat. Commun., 2020, 11, 3514 CrossRef CAS PubMed.
- V. M. Flores-Núñez, D. A. Camarena-Pozos, J. D. Chávez-González, R. Alcalde-Vázquez, M. N. Vázquez-Sánchez, A. G. Hernández-Melgar, J. Xool-Tamayo, A. Moreno-Ulloa and L. P. P. Martínez, Synthetic Communities Increase Microbial Diversity and Productivity of Agave tequilana Plants in the Field, Phytobiomes J., 2023, 7, 435–448 CrossRef.
- E. L. Schymanski, J. Jeon, R. Gulde, K. Fenner, M. Ruff, H. P. Singer and J. Hollender, Identifying small molecules via high resolution mass spectrometry: communicating confidence, Environ. Sci. Technol., 2014, 48, 2097–2098 CrossRef CAS PubMed.
- J. D. Holman, D. L. Tabb and P. Mallick, Employing proteowizard to convert raw mass spectrometry data, Curr. Protoc. Bioinf., 2014, 46, 13.24.1–13.24.9 Search PubMed.
- T. Pluskal, S. Castillo, A. Villar-Briones and M. Oresic, MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data, BMC Bioinf., 2010, 11, 395 CrossRef PubMed.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W.-T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C.-C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C.-C. Liaw, Y.-L. Yang, H.-U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. B. P. D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O'Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti and N. Bandeira, Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- A. T. Aron, E. C. Gentry, K. L. McPhail, L.-F. Nothias, M. Nothias-Esposito, A. Bouslimani, D. Petras, J. M. Gauglitz, N. Sikora, F. Vargas, J. J. J. van der Hooft, M. Ernst, K. B. Kang, C. M. Aceves, A. M. Caraballo-Rodríguez, I. Koester, K. C. Weldon, S. Bertrand, C. Roullier, K. Sun, R. M. Tehan, C. A. Boya P, M. H. Christian, M. Gutiérrez, A. M. Ulloa, J. A. Tejeda Mora, R. Mojica-Flores, J. Lakey-Beitia, V. Vásquez-Chaves, Y. Zhang, A. I. Calderón, N. Tayler, R. A. Keyzers, F. Tugizimana, N. Ndlovu, A. A. Aksenov, A. K. Jarmusch, R. Schmid, A. W. Truman, N. Bandeira, M. Wang and P. C. Dorrestein, Reproducible molecular networking of untargeted mass spectrometry data using GNPS, Nat. Protoc., 2020, 15, 1954–1991 CrossRef CAS PubMed.
- L.-F. Nothias, D. Petras, R. Schmid, K. Dührkop, J. Rainer, A. Sarvepalli, I. Protsyuk, M. Ernst, H. Tsugawa, M. Fleischauer, F. Aicheler, A. A. Aksenov, O. Alka, P.-M. Allard, A. Barsch, X. Cachet, A. M. Caraballo-Rodriguez, R. R. Da Silva, T. Dang, N. Garg, J. M. Gauglitz, A. Gurevich, G. Isaac, A. K. Jarmusch, Z. Kameník, K. B. Kang, N. Kessler, I. Koester, A. Korf, A. Le Gouellec, M. Ludwig, C. Martin H, L.-I. McCall, J. McSayles, S. W. Meyer, H. Mohimani, M. Morsy, O. Moyne, S. Neumann, H. Neuweger, N. H. Nguyen, M. Nothias-Esposito, J. Paolini, V. V. Phelan, T. Pluskal, R. A. Quinn, S. Rogers, B. Shrestha, A. Tripathi, J. J. J. van der Hooft, F. Vargas, K. C. Weldon, M. Witting, H. Yang, Z. Zhang, F. Zubeil, O. Kohlbacher, S. Böcker, T. Alexandrov, N. Bandeira, M. Wang and P. C. Dorrestein, Feature-based molecular networking in the GNPS analysis environment, Nat. Methods, 2020, 17, 905–908 CrossRef CAS PubMed.
- L. Cao, M. Guler, A. Tagirdzhanov, Y.-Y. Lee, A. Gurevich and H. Mohimani, MolDiscovery: learning mass spectrometry fragmentation of small molecules, Nat. Commun., 2021, 12, 3718 CrossRef CAS PubMed.
- H. Mohimani, A. Gurevich, A. Shlemov, A. Mikheenko, A. Korobeynikov, L. Cao, E. Shcherbin, L.-F. Nothias, P. C. Dorrestein and P. A. Pevzner, Dereplication of microbial metabolites through database search of mass spectra, Nat. Commun., 2018, 9, 4035 CrossRef PubMed.
- K. Dührkop, M. Fleischauer, M. Ludwig, A. A. Aksenov, A. V. Melnik, M. Meusel, P. C. Dorrestein, J. Rousu and S. Böcker, SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information, Nat. Methods, 2019, 16, 299–302 CrossRef PubMed.
- M. Ludwig, L.-F. Nothias, K. Dührkop, I. Koester, M. Fleischauer, M. A. Hoffmann, D. Petras, F. Vargas, M. Morsy, L. Aluwihare, P. C. Dorrestein and S. Böcker, Database-independent molecular formula annotation using Gibbs sampling through ZODIAC, Nat. Mach. Intell., 2020, 2, 629–641 CrossRef.
- K. Dührkop, H. Shen, M. Meusel, J. Rousu and S. Böcker, Searching molecular structure databases with tandem mass spectra using CSI:FingerID, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12580–12585 CrossRef PubMed.
- J. Wandy, Y. Zhu, J. J. J. van der Hooft, R. Daly, M. P. Barrett and S. Rogers, Ms2lda.org: web-based topic modelling for substructure discovery in mass spectrometry, Bioinformatics, 2018, 34, 317–318 CrossRef CAS PubMed.
- J. Willforss, A. Chawade and F. Levander, NormalyzerDE: Online Tool for Improved Normalization of Omics Expression Data and High-Sensitivity Differential Expression Analysis, J. Proteome Res., 2019, 18, 732–740 CrossRef CAS PubMed.
- M. E. Ritchie, B. Phipson, D. Wu, Y. Hu, C. W. Law, W. Shi and G. K. Smyth, limma powers differential expression analyses for RNA-sequencing and microarray studies, Nucleic Acids Res., 2015, 43, e47 CrossRef PubMed.
- K. Blighe, S. Rana and M. Lewis, EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling, Bioconductor - open source software for bioinformatics, R package version 1.26.0, 2025, DOI:10.18129/b9.bioc.enhancedvolcano.
- F. R. Passos Mansoldo, R. Garrett, V. da Silva Cardoso, M. A. Alves and A. B. Vermelho, Metabology: Analysis of metabolomics data using community ecology tools, Anal. Chim. Acta, 2022, 1232, 340469 CrossRef CAS PubMed.
- J. Chong, P. Liu, G. Zhou and J. Xia, Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data, Nat. Protoc., 2020, 15, 799–821 CrossRef CAS PubMed.
- J. T. Smilowitz, L. H. Allen, D. C. Dallas, J. McManaman, D. J. Raiten, M. Rozga, D. A. Sela, A. Seppo, J. E. Williams, B. E. Young and M. K. McGuire, Ecologies, synergies, and biological systems shaping human milk composition-a report from “Breastmilk Ecology: Genesis of Infant Nutrition (BEGIN)” Working Group 2, Am. J. Clin. Nutr., 2023, 117(Suppl 1), S28–S42 CrossRef PubMed.
- S. Enstad, S. Cheema, R. Thomas, R. N. Fichorova, C. R. Martin, P. O'Tierney-Ginn, C. L. Wagner and S. Sen, The impact of maternal obesity and breast milk inflammation on developmental programming of infant growth, Eur. J. Clin. Nutr., 2021, 75, 180–188 CrossRef CAS PubMed.
- H. Nuss, A. Altazan, J. Zabaleta, M. Sothern and L. Redman, Maternal pre-pregnancy weight status modifies the influence of PUFAs and inflammatory biomarkers in breastmilk on infant growth, PLoS One, 2019, 14, e0217085 CrossRef CAS PubMed.
- C. R. Sims, M. E. Lipsmeyer, D. E. Turner and A. Andres, Human milk composition differs by maternal BMI in the first 9 months postpartum, Am. J. Clin. Nutr., 2020, 112, 548–557 CrossRef PubMed.
- C. Gómez-Gallego, J. M. Morales, D. Monleón, E. du Toit, H. Kumar, K. M. Linderborg, Y. Zhang, B. Yang, E. Isolauri, S. Salminen and M. C. Collado, Human Breast Milk NMR Metabolomic Profile across Specific Geographical Locations and Its Association with the Milk Microbiota, Nutrients, 2018, 10, 1355 CrossRef PubMed.
- K. E. Lyons, C.-A. O’ Shea, G. Grimaud, C. A. Ryan, E. Dempsey, A. L. Kelly, R. P. Ross and C. Stanton, The human milk microbiome aligns with lactation stage and not birth mode, Sci. Rep., 2022, 12, 5598 CrossRef CAS PubMed.
- G. E. Leghi, P. F. Middleton and B. S. Muhlhausler, A methodological approach to identify the most reliable human milk collection method for compositional analysis: a systematic review protocol, Syst. Rev., 2018, 7, 122 CrossRef PubMed.
- M. Loftus, S. A.-D. Hassouneh and S. Yooseph, Bacterial associations in the healthy human gut microbiome across populations, Sci. Rep., 2021, 11, 2828 CrossRef CAS PubMed.
- C. J. Robinson, B. J. M. Bohannan and V. B. Young, From structure to function: the ecology of host-associated microbial communities, Microbiol. Mol. Biol. Rev., 2010, 74, 453–476 CrossRef CAS PubMed.
- D. J. Lemas, B. E. Young, P. R. Baker, A. C. Tomczik, T. K. Soderborg, T. L. Hernandez, B. A. de la Houssaye, C. E. Robertson, M. C. Rudolph, D. Ir, Z. W. Patinkin, N. F. Krebs, S. A. Santorico, T. Weir, L. A. Barbour, D. N. Frank and J. E. Friedman, Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome, Am. J. Clin. Nutr., 2016, 103, 1291–1300 CrossRef CAS PubMed.
- S. Beretta, M. Apparicio, G. H. Toniollo and M. V. Cardozo, The importance of the intestinal microbiota in humans and dogs in the neonatal period, Anim. Reprod., 2023, 20, e20230082 CrossRef PubMed.
- C. D. Moon, W. Young, P. H. Maclean, A. L. Cookson and E. N. Bermingham, Metagenomic insights into the roles of Proteobacteria in the gastrointestinal microbiomes of healthy dogs and cats, Microbiologyopen, 2018, 7, e00677 CrossRef PubMed.
- U. Pandey and P. Aich, Postnatal intestinal mucosa and gut microbial composition develop hand in hand: A mouse study, Biomed. J., 2023, 46, 100519 CrossRef CAS PubMed.
- K. Hou, Z.-X. Wu, X.-Y. Chen, J.-Q. Wang, D. Zhang, C. Xiao, D. Zhu, J. B. Koya, L. Wei, J. Li and Z.-S. Chen, Microbiota in health and diseases, Signal Transduction Targeted Ther., 2022, 7, 135 CrossRef PubMed.
- J. Mirpuri, M. Raetz, C. R. Sturge, C. L. Wilhelm, A. Benson, R. C. Savani, L. V. Hooper and F. Yarovinsky, Proteobacteria-specific IgA regulates maturation of the intestinal microbiota, Gut Microbes, 2014, 5, 28–39 CrossRef PubMed.
- E. Cortés-Macías, M. Selma-Royo, K. Rio-Aige, C. Bäuerl, M. J. Rodríguez-Lagunas, C. Martínez-Costa, F. J. Pérez-Cano and M. C. Collado, Distinct breast milk microbiota, cytokine, and adipokine profiles are associated with infant growth at 12 months: an in vitro host-microbe interaction mechanistic approach, Food Funct., 2023, 14, 148–159 RSC.
- A. Köhler, S. Delbauve, J. Smout, D. Torres and V. Flamand, Very early-life exposure to microbiota-induced TNF drives the maturation of neonatal pre-cDC1, Gut, 2021, 70, 511–521 CrossRef PubMed.
- E. Amabebe and D. O. Anumba, Diabetogenically beneficial gut microbiota alterations in third trimester of pregnancy, Reprod. Fertil., 2021, 2, R1–R12 Search PubMed.
- M. C. Collado, K. Laitinen, S. Salminen and E. Isolauri, Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk, Pediatr. Res., 2012, 72, 77–85 CrossRef CAS PubMed.
- F. Bardanzellu, M. Puddu, D. G. Peroni and V. Fanos, The human breast milk metabolome in overweight and obese mothers, Front. Immunol., 2020, 11, 1533 CrossRef CAS PubMed.
- J. L. Saben, C. R. Sims, B. D. Piccolo and A. Andres, Maternal adiposity alters the human milk metabolome: associations between nonglucose monosaccharides and infant adiposity, Am. J. Clin. Nutr., 2020, 112, 1228–1239 CrossRef PubMed.
- H. H. Chen, Y. J. Tseng, S. Y. Wang, Y. S. Tsai, C. S. Chang, T. C. Kuo, W. J. Yao, C. C. Shieh, C. H. Wu and P. H. Kuo, The metabolome profiling and pathway analysis in metabolic healthy and abnormal obesity, Int. J. Obes., 2015, 39, 1241–1248 CrossRef CAS PubMed.
- T. Purdom, L. Kravitz, K. Dokladny and C. Mermier, Understanding the factors that effect maximal fat oxidation, J. Int. Soc. Sports Nutr., 2018, 15, 3 CrossRef PubMed.
- M. Zhang, L. Simon Sarkadi, M. Üveges, J. Tormási, E. Benes, R. A. Vass and S. G. Vari, Gas chromatographic determination of fatty acid composition in breast milk of mothers with different health conditions, Acta Aliment., 2022, 51, 625–635 CAS.
- R. E. Walker, Oxylipins as potential regulators of inflammatory conditions of human lactation, Metabolites, 2022, 10, 994 CrossRef PubMed.
- G. A. Weiss, H. Troxler, G. Klinke, D. Rogler, C. Braegger and M. Hersberger, High levels of anti-inflammatory and pro-resolving lipid mediators lipoxins and resolvins and declining docosahexaenoic acid levels in human milk during the first month of lactation, Lipids Health Dis., 2013, 12, 89 CrossRef CAS PubMed.
- R. Ottria, M. D. Porta, O. Xynomilakis, S. Casati, R. Cazzola and P. Ciuffreda, Lipids and lipid signaling molecules in Human milk and infant formula, a chemical characterization of relevant biochemical components, J. Nutr. Biochem., 2024, 126, 109580 CrossRef CAS PubMed.
- B. Koletzko and M. Rodriguez-Palmero, Polyunsaturated fatty acids in human milk and their role in early infant development, J. Mammary Gland Biol. Neoplasia, 1999, 4, 269–284 CrossRef CAS PubMed.
- A. D. George, S. Burugupalli, S. Paul, T. Mansell, D. Burgner and P. J. Meikle, The Role of Human Milk Lipids and Lipid Metabolites in Protecting the Infant against Non-Communicable Disease, Int. J. Mol. Sci., 2022, 23, 7490 CrossRef CAS PubMed.
- J. Kolmert, S. Piñeiro-Hermida, M. Hamberg, J. A. Gregory, I. P. López, A. Fauland, C. E. Wheelock, S.-E. Dahlén, J. G. Pichel and M. Adner, Prominent release of lipoxygenase generated mediators in a murine house dust mite-induced asthma model, Prostaglandins Other Lipid Mediators, 2018, 137, 20–29 CrossRef CAS PubMed.
- Y. S. Huang, W. C. Huang, C. W. Li and L. T. Chuang, Eicosadienoic acid differentially modulates production of pro-inflammatory modulators in murine macrophages, Mol. Cell. Biochem., 2011, 358, 85–94 CrossRef CAS PubMed.
- F. Haviv, J. D. Ratajczyk, R. W. DeNet, Y. C. Martin, R. D. Dyer and G. W. Carter, Structural requirements for the inhibition of 5-lipoxygenase by 15-hydroxyeicosa-5,8,11,13-tetraenoic acid analogues, J. Med. Chem., 1987, 30, 254–263 CrossRef CAS PubMed.
- K. Miyata, D. Horikami, Y. Tachibana, T. Yamamoto, T. Nakamura, K. Kobayashi and T. Murata, 15-hydroxy eicosadienoic acid is an exacerbating factor for nasal congestion in mice, FASEB J., 2022, 36, e22085 CrossRef CAS PubMed.
- T. Takeichi, F. Kinoshita, H. Tanaka, S. Fujita, Y. Kobayashi, M. Nakatochi, K. Sugiura and M. Akiyama, The lipoxygenase–hepoxilin pathway is activated in cutaneous plaque lesions of psoriasis, J. Cutan. Immunol. Allergy, 2019, 2, 15–24 CrossRef.
- K. Esposito, A. Pontillo, F. Giugliano, G. Giugliano, R. Marfella, G. Nicoletti and D. Giugliano, Association of low interleukin-10 levels with the metabolic syndrome in obese women, J. Clin. Endocrinol. Metab., 2003, 88, 1055–1058 CrossRef CAS PubMed.
- N. Subramanian, B. Tavira, K. Hofwimmer, B. Gutsmann, L. Massier, J. Abildgaard, A. Juul, M. Rydén, P. Arner and J. Laurencikiene, Sex-specific regulation of IL-10 production in human adipose tissue in obesity, Front. Endocrinol., 2022, 13, 996954 CrossRef PubMed.
- V. J. Taylor, Lactation from the inside out: Maternal homeorhetic gastrointestinal adaptations regulating energy and nutrient flow into milk production, Mol. Cell. Endocrinol., 2023, 559, 111797 CrossRef CAS PubMed.
- F. Kalaycı-Yüksek, D. Gümüş, V. Güler, A. Uyanık-Öcal and M. Anğ-Küçüker, Progesterone and Estradiol alter the growth, virulence and antibiotic susceptibilities of Staphylococcus aureus, New Microbiol., 2023, 46, 43–51 Search PubMed.
- M. Nuriel-Ohayon, H. Neuman, O. Ziv, A. Belogolovski, Y. Barsheshet, N. Bloch, A. Uzan, R. Lahav, A. Peretz, S. Frishman, M. Hod, E. Hadar, Y. Louzoun, O. Avni and O. Koren, Progesterone Increases Bifidobacterium Relative Abundance during Late Pregnancy, Cell Rep., 2019, 27, 730–736 CrossRef CAS PubMed.
- I. Bartha, I. Joumady, M. Cuerva and J. L. Bartha, The effect of maternal obesity and lipid profile on first-trimester serum progesterone levels, Am. J. Obstet. Gynecol. MFM, 2023, 5, 100959 CrossRef CAS PubMed.
- M. Lu, H. Xiao, K. Li, J. Jiang, K. Wu and D. Li, Concentrations of estrogen and progesterone in breast milk and their relationship with the mother's diet, Food Funct., 2017, 8, 3306–3310 RSC.
- M. Messripour, A. Forooghi-Abary and F. Dashti, Association between sex hormones in human breast milk and infant growth and development, Arch. Iran. Med., 2002, 5, 166–169 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo05637j |
| This journal is © The Royal Society of Chemistry 2025 |



