Complexation of starch and konjac glucomannan during screw extrusion exhibits obesity-reducing effects by modulating the intestinal microbiome and its metabolites†
Fanrui
Liu‡
a,
Hao
Wan‡
c,
Honghao
Fan
*d,
Zhihong
Zhang
a,
Hua
Dai
a and
Hai
He
 *ab
*ab
aHeinz Mehlhorn Academician Workstation, Key Laboratory of Tropical Translational Medicine of Ministry of Education, School of Public Health, Hainan Medical University, Haikou 571199, Hainan Province, China
bDepartment of Endocrinology and Metabolism, Shunde Hospital, Southern Medical University (The First People's Hospital of Shunde), Foshan 528300, Guangdong Province, China. E-mail: h.hai@hainmc.edu.cn; h.hai@muhn.edu.cn
cDepartment of Laboratory, Qianjiang City Center Hospital, Qianjiang 433100, Hubei Province, China
dNJUST-YX Artificial Intelligence Biomedical Technology Innovation Center, Nanjing University of Science and Technology, Nanjing 210094, Jiangsu Province, China. E-mail: fhh201308@163.com
First published on 9th December 2024
Abstract
Dietary interventions have been shown to improve gut health by altering the gut flora, preventing obesity, and mitigating inflammatory disorders. This study investigated the benefits of a rice starch–konjac glucomannan (ERS–KGM) complex, produced via screw extrusion, for gut health and obesity prevention. Analyzed through in vitro starch digestion, scanning electron microscopy, and structural analysis, the ERS–KGM complex exhibited a notable increase in resistant starch content due to its well-ordered structure. When administered to mice on a high-fat diet for 8 weeks, the ERS–KGM complex significantly reduced body weight, white adipose tissue mass, adipocyte size, and food intake while increasing water consumption. It also improved glucose metabolism, insulin sensitivity, and lipid profiles by lowering serum triglycerides and total glycerol content. Enhanced metabolic biomarkers and enzyme activities were observed, specifically involving glycerophospholipid metabolism. It decreased the activities of aldehyde dehydrogenase, lactate dehydrogenase, and amino acid transaminase while increasing antioxidant enzymes like glutathione peroxidase and superoxide dismutase. Additionally, it elevated glycogen and positively altered gut microbiota by enriching Firmicutes, Desulfobacterota, and Bifidobacterium. This change enhanced the ability to degrade specific compounds and elevated the concentrations of short-chain fatty acids in feces. These findings suggest that the ERS–KGM complex could serve as a dietary supplement for obesity prevention.
1. Introduction
The rapid spread of obesity has brought great challenges to global human health and social economy. Obesity has a profound impact on quality of life, and its effects can range from mild to severe, causing varying degrees of dysfunction in the metabolic processes of different bodily systems.1 Obesity disrupts normal metabolism by affecting liver function, adipose tissue metabolism, gut microbiome balance, and other key physiological pathways. As a result, the body becomes chronically metabolically dysfunctional over an extended period, substantially increasing the risk of developing cardiovascular disease, non-alcoholic fatty liver disease, hypertension, type 2 diabetes, etc.2 Internationally, dietary intervention is widely recognized as a potent approach to preventing and managing obesity. The strategic optimization of nutritional components in the diet, particularly the scientifically informed balance of carbohydrates, significantly influences obesity management and prevention.3Starch, the primary component of staple foods, can be categorized into three fractions based on their digestive resistance: rapidly digestible starch (RDS), slowly digestible starch (SDS), and resistant starch (RS).4 RS can evade digestion by human enzymes and reach the colon, where the gut microbiota can break it down through fermentation. Subsequently, RS has been shown to treat several chronic illnesses by modifying gut microbiota, including obesity and colon cancer. Notably, Roseburia, Blautia, Ruminococcus bromine, and other beneficial species are favorably promoted by RS, a strong and plentiful fiber that has the ability to alter the gut microbiota, resulting in an increased short-chain fatty acids (SCFAs) production.1 Consequently, various technologies, such as chemical, genetic modification, and enzymatic, are being employed in the production of starchy foods to enhance the content of RS.5,6
Screw extrusion technology has become a promising new processing technology in the food manufacturing industry due to its advantages of easy operation, green environmental protection, and high safety. Due to the mixing, stirring, crushing, shearing, and thermal effects during extrusion, the original multi-scale structure of starch will be destroyed, and at the same time, the starch molecules will be induced to reorganize and assemble to form new multi-scale aggregates, which will change the digestibility of starch.7,8 In terms of starch, screw extrusion can alter its gelatinization and retrogradation properties, thereby influencing its digestibility and the glycemic index of the final product—a factor that is increasingly important in the development of foods targeting health-conscious consumers.7 Moreover, screw extrusion is utilized in the creation of novel ingredients, including prebiotics and encapsulated nutrients, which are incorporated into functional foods and supplements.9 This broadens the scope of how extrusion technology is applied in meeting the demand for innovative and health-promoting food products. In addition, the digestibility of starch is not only affected by the internal multi-scale structure of starch and the processing method but also by the molecular interactions of other components in the food system. The starch multi-scale structure evolves through interactions between macromolecular compounds like non-starch polysaccharides, mediated by intermolecular forces such as hydrogen bonding, hydrophobic attractions, and van der Waals forces. This leads to the formation of physical barriers that prevent starch from coming into contact with amylase, thereby reducing the digestibility of starch.10,11
However, there are limited reports on starch–non starch polysaccharides complex that can act as prebiotics to prevent metabolic diseases. In this study, we produced a complex interaction between rice starch, water, and konjac glucomannan (KGM) during screw extrusion (ERS), and identified a molecular mechanism of multi-scale structural characteristics regulating the digestibility of starch. In animal experiments, we conducted an insulin tolerance test (ITT), oral glucose tolerance test (OGTT), and serum glucose test to assess the impact of the starch–KGM on high-fat diet (HFD)-induced obesity. Additionally, adipose tissue and body weight were recorded, and measurements were taken for liver enzyme activities, antioxidant enzyme activities, and serum lipids. To get a comprehensive understanding of the mechanism of action, the impact of the ERS–KGM complex treatment on serum metabolomic and gut microbiota composition and functions was also investigated. The study lies in the specific exploration of the molecular mechanisms through which KGM and rice starch interact during screw extrusion to regulate starch digestibility and its consequent impact on obesity. While previous studies have demonstrated that KGM and RS can alleviate obesity,12,13 this research leverages the screw extrusion process to alter the multi-scale structural characteristics of starch. This technique not only enhances the resistant starch content but also explores the previously underreported molecular interactions between starch and non-starch polysaccharides, such as KGM.
2. Materials and methods
2.1 Production of starch–KGM complex using screw extrusion
The preparation process was conducted according to our prior investigation.11 8% KGM, a food-grade substance with a molecular weight of 1.37 × 106 Da, was incorporated into rice starch (Remy-DR, Beneo-orafti, Oreye, Belgium) by Yuanye Biotech Co., Ltd in Shanghai, China. The moisture content of the mixtures was precisely set at 40% using a mixing agitator operating at 100 rpm for 5 min, before being extruded into an extruder (co-rotating twin-screw, LTD HK-36, L/D ratio 52, 90 rpm, 85 °C) manufactured by Nanjing KY Chemical Machinery Co., Ltd in Nanjing, China. The high shear stress involved in screw extrusion has previously been shown to cause the mixtures to gelatinize.14 Following a 48-hour drying process at 45 °C (moisture content <8%), the extrudates were subsequently crushed at room temperature using a laboratory-scale shredder (DM-6, Asone, Osaka, Japan). Subsequently, the powder was sifted through a 100-mesh screen and stored at room temperature in vacuum-sealed bags in the dark. The protocol named the resulting preparation the extruded rice starch (ERS)–KGM complex, with ERS reserved for the starch alone.2.2 Structure characterization
The circular metal tube containing the extruded starch samples was wrapped with a double-sided carbon tape sealed with bonding, and a sputter coater was used to apply gold plating. With 300× magnification and an accelerating voltage of 20 kV, starch particles were imaged using an EM-30 Plus scanning electron microscope (SEM, COXEM, Korea).The protocol outlined by He et al.15 was followed while performing small-angle X-ray scattering (SAXS) observations on a NanoSTAR SAXS (Bruker AXS GmbH, Karlsruhe, Germany).
An ATR SLR reflection cell was used with a Tensor 37 (Bruker Inc., Germany) to measure the Fourier transform infrared (FTIR) spectrum. With air as the background, measurements were made in 64 scans with a scanning range of 4000–4 cm−1 and a resolution of 4 cm−1. Using OMNIC 8.0 (Nicolet, Madison, USA), an area spanning from 1200–900 cm−1 was chosen for analysis. The enhancement factor and half-height full width were set at 19 and 1.9 cm−1, respectively. The ratio (R1047/1022) of peak intensities at 1047 cm−1 and 1022 cm−1 was calculated by PeakFit 4.12 (Systat Software Inc., San Jose, USA).
A Bruker Avance III 400 MHz WB spectrometer was employed to record the solid-state 13C cross-polarization and magic angle spinning nuclear magnetic resonance (CP/MAS NMR) spectra at 100.62 MHz. Using the previously published procedure, PeakFit 4.2 (Systat Software Inc.) was used to prepare amorphous samples and perform a quantitative study of 13C CP/MAS NMR spectra.11
The X-ray diffractometer (XRD, D/tex Ultra, Brand Rigaku Ultima IV, Texas, USA) was employed to determine the XRD patterns of the extruded starch samples. Measurements were taken with 0.01° increments and a rate of 5° per minute, spanning the angular range of 2θ from 5° to 40°, utilizing a diffractometer set at 40 kV and 40 mA. The relative degree of crystallinity (RC) was calculated by analyzing the diffractograms using MDI Jade 6.0 software.
2.3 In vitro starch digestion
1 g of dry extruded starch samples was added to 20 mL of 0.1 M sodium acetate buffer, pH 5.2, as previously described.11 The samples were then hydrolyzed with an enzyme mixture (0.5 mL, 37 °C, 190 rpm) that included amyloglucosidase (20 U mL−1, A3306, activity 318 U mL−1, Sigma-Aldrich Co., Ltd, St Louis, MO, USA) and porcine pancreatic α-amylase (300 U mL−1, P7545, activity 8 × USP, Sigma-Aldrich Co., Ltd). The in vitro digestion process involved sampling 0.5 mL of digestible fluid at intervals of 0, 20, and 120 min. To inactivate the enzyme, the digestible fluid was thoroughly mixed with 25 mL of 70% ethanol. Centrifugation at 4000 rpm for 5 min was followed by incubation in a water bath at 45 °C for 20 min, with the supernatant (0.1 mL) combined with 3 mL of glucose oxide-peroxidase (K-GLUC, Wicklow, Ireland). The digestibility of extruded starch samples was evaluated by determining the contents of hydrolyzed glucose from the eqn (1)–(3):16| RDS (%) = [(G20 − G0) × 0.9/TS] × 100% | (1) |
| SDS (%) = [(G120 − G20) × 0.9/TS] × 100% | (2) |
| RS (%) = [(TS − RDS − SDS)/TS] × 100% | (3) |
2.4 Experiments on animals
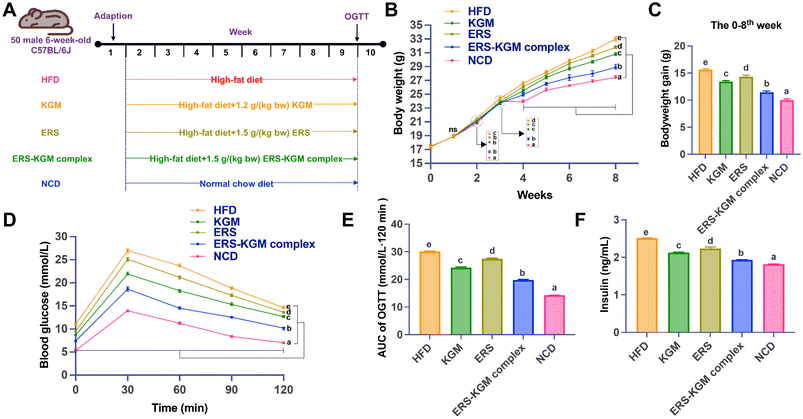
This study was performed in strict accordance with the Guidelines for Care and Use of Laboratory Animals of Hainan Medical University (ethics number: HYLL-2023-101) and approved by the Animal Ethics Committee of Hainan Medical University (Haikou, China). The Hainan Medical University Laboratory Animal Center [no. SYXK(Qiong) 2022-0013, Haikou, China] supplied 50 six-week-old male C57BL/6J mice at the acquisition time. After being acclimated for a week, ten mice each were randomly assigned to five separate groups: (1) HFD group: high-fat diet; (2) KGM group: HFD with 1.2 g per [kg body weight (bw)] KGM; (3) ERS group: HFD with 1.5 g per (kg bw) ERS group; (4) ERS–KGM complex group: HFD with 1.5 g per (kg bw) ERS–KGM complex; (5) NCD group: normal chow diet. The dosage of KGM, ERS, and the ERS–KGM complex was carefully determined based on the outcomes of preclinical animal experiments and previous scientific studies.12,13 Intragastric samples, comprising KGM, ERS, and ERS–KGM complex, were administered orally via gavage daily from week two to week nine (Fig. 1A). The animals had ad libitum access to food and water, with feeding sessions scheduled twice daily at 8:00 AM and 6:00 PM. They were kept in a carefully controlled environment, maintained at a temperature range of 22–24 °C with a humidity level of 50–60%. The lighting conditions followed a 12-hour light/12-hour dark cycle. For housing, the animals were grouped together in cages, facilitating social interaction while adhering to welfare standards.Throughout the trial, the body weight, food intake, and water intake of each mouse were monitored and recorded weekly. At the end of the ninth week, a sample of each mouse's feces was taken and kept at −80 °C. Mice fasted for twelve hours and were put to sleep by inhaling ether. After obtaining blood samples by heart puncture, serum was extracted and stored at 3000g for 10 min at 4 °C. The liver and epididymal white adipose tissues (WATs) were taken immediately and placed in a cold 0.85% saline solution before being dried with filter paper and weighed. A portion of the liver tissues and WATs were preserved in a 10% formalin solution to prepare them for histopathologic experiments. After being processed into 20% liver homogenate, the residual liver tissue samples were stored at −80 °C.
2.5 Blood biochemical analysis
The OGTT and ITT offer essential information on insulin sensitivity and the functionality of pancreatic beta-cells. These tests were carried out according to the methods outlined in a previous experimental protocol.17 At week 9, an OGTT was conducted after an overnight fast of 12 h. About three days before the experiment ended, an ITT was conducted after a 5-hour fasting period.High- and low-density lipoprotein cholesterol (HDL-c and LDL-c), triacylglycerols (TG), and total cholesterol (TC) kits (Nanjing Jiancheng Biology Engineering Institute, Nanjing, China) were used to test blood serum. Superoxide dismutase (SOD), glutathione peroxidase (GSH-PX), aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (AKP), and malondialdehyde (MDA) kits (Nanjing Jiancheng Biology Engineering Institute) were used to assess the liver homogenate.
2.6 Histological analysis
WATs and preserved liver tissues were quickly embedded in paraffin. The thickness of adipose tissues in embedded paraffin blocks was measured at 8 μm, whereas the thickness of liver tissues was measured at 5 μm. The sections were stained using the conventional technique for oil red O and hematoxylin and eosin (H&E) staining. Panoramic MIDI (3D HISTECH, Jinan, China) was used to take histological pictures.2.7 Analysis of SCFAs
After thorough mixing, 900 μL of filtered water was soaked with 100 milligrams of cecum contents. The mixes underwent a 15-minute centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. The filtered supernatant, obtained using a 0.22 μm filter membrane, was analyzed by high-performance liquid chromatography (HPLC) to detect and quantify the SCFAs present in the sample. The HPLC conditions employed were as follows: a model 1260 instrument with UV detection at 210 nm, coupled to an Agilent C18 column (150 mm × 4.6 mm, 5 μm particle size) operated at 40 °C. The mobile phases comprised a 30% methanol–water mixture (A) and 0.05 mol L−1 potassium dihydrogen phosphate (B), with a 70% B composition. The system used a flow rate of 1.0 mL min−1, and injections were made at a volume of 10 μL.
000g. The filtered supernatant, obtained using a 0.22 μm filter membrane, was analyzed by high-performance liquid chromatography (HPLC) to detect and quantify the SCFAs present in the sample. The HPLC conditions employed were as follows: a model 1260 instrument with UV detection at 210 nm, coupled to an Agilent C18 column (150 mm × 4.6 mm, 5 μm particle size) operated at 40 °C. The mobile phases comprised a 30% methanol–water mixture (A) and 0.05 mol L−1 potassium dihydrogen phosphate (B), with a 70% B composition. The system used a flow rate of 1.0 mL min−1, and injections were made at a volume of 10 μL.
2.8 Serum metabolomic analysis
The methodology for extracting serum lipid metabolites was based on the protocols established in prior research.18 Following overnight storage at −20 °C for protein precipitation, the lower solution was collected and centrifuged (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 15 min) for lipidomics analysis. The metabolome was analyzed using the supernatant. Afterward, the UPLC-Q exactive HF-X (Accucore C30, 2.6 μm, 100 mm × 2.1 mm i.d.; Thermo, USA) was used to identify the serum samples by lipid metabolomics analysis. Mobile phase A: 50% aqueous acetonitrile solution containing formic acid (0.1%) and ammonium acetate (10 mmol L−1). Mobile phase B: acetonitrile, isopropanol, and water were mixed in the ratio of 10
000g, 15 min) for lipidomics analysis. The metabolome was analyzed using the supernatant. Afterward, the UPLC-Q exactive HF-X (Accucore C30, 2.6 μm, 100 mm × 2.1 mm i.d.; Thermo, USA) was used to identify the serum samples by lipid metabolomics analysis. Mobile phase A: 50% aqueous acetonitrile solution containing formic acid (0.1%) and ammonium acetate (10 mmol L−1). Mobile phase B: acetonitrile, isopropanol, and water were mixed in the ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 88
88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 with formic acid (0.1%) and ammonium acetate (10 mmol L−1). The temperature of the column was adjusted to 40 °C, and the injection volume was 5 μL.
2 with formic acid (0.1%) and ammonium acetate (10 mmol L−1). The temperature of the column was adjusted to 40 °C, and the injection volume was 5 μL.
2.9 Analysis of gut microbiota
Guangzhou IGE Biotechnology Ltd (Guangzhou, China) conducted a study in which ten mice per group were analyzed to determine their composition of intestinal flora, specifically focusing on the gut microbiota. Guangzhou IGE Biotechnology Ltd prepared and tested 100 mg of fecal material from each mouse to amplify the 16s rRNA V4 region (forward: GTGCCAGCMGCCGCGGTAA, reverse: GTACHVGGGTWTCTAAT). The study was based on species annotations and amplicon sequence variants (ASVs) generated from high-throughput sequencing.2.10 Analytical statistics
Statistical analyses were conducted with Prism 10 software (GraphPad, USA), and the data are shown as the mean ± standard deviation (SD). One-way analysis of variance (ANOVA) followed by Tukey's test was used to investigate any significant differences between the groups. A statistically significant P-value was defined as being less than 0.05.Lipid metabolomics data were analyzed by considering metabolites with a variable importance in the projection (VIP) value ≥1 and a P-value <0.05 as differentially expressed metabolites (DEMs).19 The identification of key metabolic pathways was performed using the ChiPlot platform (https://www.chiplot.online).
For gut microbiota analysis, raw sequences through quality primary screening were divided into libraries and samples according to index and barcode information, and barcode sequences were removed and primers were removed. Sequence denoising was performed according to the Qiime2 DADA2 analysis process to obtain the final ASVs.20 The specific composition of each group at different taxonomic levels of the species was demonstrated for an overall overview. Based on the distribution of ASVs in different samples, the Alpha diversity level of each sample was assessed and the sparse curve reflected whether the sequencing depth was appropriate. The distance from each sample to the distance matrix is calculated to measure Beta diversity differences and the significance of differences between different groups. Statistically significant biomarkers between different groups were identified through the analysis of the significance of differences between groups. The gene function or phenotype of a sample is predicted by functional predictive analysis.
3. Results
3.1 Multi-scale structures
The SEM of the ERS and ERS–KGM complex are presented in Fig. S1A.† Following the screw extrusion process, the ERS and ERS–KGM complex underwent a profound transformation, surrendering their initial elliptical and spherical granular structures to adopt an irregular, flaky morphology, characterized by layered striations on the surface of the particles. The surface of the ERS–KGM complex showed significantly fewer stripped layers and was significantly smoother than that of ERS, featuring a distinct structure that profoundly influenced the attachment ability of amylase, affecting their digestibility.The SAXS maps of the ERS and ERS–KGM complex exemplify the effectiveness of SAXS in visualizing the fractal nature of starch structures (Fig. S1B†). After the screw extrusion treatment, the scattering peak [ca. 0.6 nm−1 (q)] disappeared, indicating that the semicrystalline layered structure of ERS was disturbed. A semicrystalline layer structure was absent from the ERS–KGM complex, and its SAXS diagram resembled that of the ERS. The fractal dimension of the ERS or ERS–KGM complex was 1 < α < 3 (Dm = α), and the SAXS scattering signal power-law equation was computed by matching I(q) ∼ q−α for the α value, as shown in Table 1. The Dm value of the ERS–KGM complex dramatically exceeded that of ERS, with the addition of KGM leading a significant contribution to this enhancement.
| Samples | RDS (%) | SDS (%) | RS (%) | Amorphous | Single helix | Double helix | RC (%) | R 1047/1022 | α = Dm |
|---|---|---|---|---|---|---|---|---|---|
| a The value is derived from three repeated experiments with a mean ± SD (n = 3). One-way ANOVA followed by Tukey's test was used to investigate significant differences between the groups. P < 0.05 is shown by different small-case letters within each column. RS: resistance starch; SDS: slowly digested starch; RDS: rapidly digested starch; ERS–KGM complex: extruded rice starch–konjac glucomannan complex; RC: relative crystallinity; R1047/1022: peak intensities ratio at 1047 and 1022 cm−1; α = Dm: fractal dimension; ERS: extruded rice starch; ANOVA: analysis of variance; SD: standard deviation. | |||||||||
| ERS | 62.13 ± 0.83b | 14.67 ± 0.93a | 23.20 ± 0.99a | 73.10 ± 0.30b | 8.00 ± 0.53a | 18.90 ± 0.41a | 21.90 ± 0.13a | 0.570 ± 0.006a | 1.610 ± 0.047a |
| ERS–KGM complex | 46.47 ± 1.04a | 22.39 ± 1.01b | 31.14 ± 1.14b | 59.11 ± 0.72a | 17.83 ± 0.63b | 23.06 ± 0.45b | 25.60 ± 0.14b | 0.593 ± 0.001b | 1.718 ± 0.046b |
The infrared spectrum of the ERS and ERS–KGM complex is exhibited in Fig. S1C.† The intensity of the spectral band at 1022 cm−1 indicates the presence of amorphous regions in the starch crystal, whereas the intensity of the spectral band at 1047 cm−1 is related to the crystallinity of starch.21 Thus, inherent differences in double-helical structure and molecular order within the ERS and ERS–KGM complex may be quantified and characterized by R1047/1022. The value of R1047/1022, as shown in Table 1, exhibits significant differences when compared to ERS, with the ERS–KGM complex having a greater proportion of crystalline area, accompanied by a more ordered alignment of the double helix.
Fig. S1D† presents the 13C CP/MAS NMR spectra of the ERS and ERS–KGM complex. Four prominent signal peaks can be observed in the spectrum; specifically, the anomeric C1 peak falls within the range of 90–110 ppm, while C2, C3, and C5 peaks appear as a superposition resonance between 68–78 ppm, the C4 peak is found between 79–84 ppm, and the C6 peak falls within the range of 58–65 ppm. The presence of multiple peaks in the C1 region is often correlated with the double-helical structure of starch, providing valuable insights into the crystalline polymorphic form of these biopolymers.22 As presented in Table 1, the following order applied to the double and single helix contents: ERS–KGM complex, followed closely by ERS. The increased content of single helix structures could be attributed to the presence of the V-type structure in the ERS–KGM complex, which is predominantly localized in the region that interfaces with lipids and amylopectin in starch.
The X-ray diffractograms of the ERS and ERS–KGM complex are presented in Fig. S1E.† A distinct X-ray absorption peak was observed at 2θ values of 15.4°, 17.5°, 18.4°, 20.1°, and 23.4° for the ERS and ERS–KGM complex, which were shown to be crystalline structures of the A and V types.23 It is likely that the V-type crystal structure derived from the molecular structure of the amylose-fatty acid complex, which formed as amylose underwent conformational changes during screw extrusion. As demonstrated in Table 1, the ERS–KGM complex exhibits higher RC than ERS, which can be attributed to the enhanced fractal structure, short-range order, single helix, and double helix structures mentioned above.
3.2 In vitro starch digestibility
The multi-scale structure of starch significantly affects its digestibility. As presented in Table 1, the ERS contained 62.13% RDS, 14.67% SDS, and 23.20% RS. Following screw extrusion treatment, the ERS–KGM complex showed a notable increase, while the RDS content exhibited a significant decline of 46.47%. Additionally, SDS and RS contents surged by 22.39% and 31.14%, respectively. The interaction between KGM and starch molecules during screw extrusion results in the formation of a dense, hydrogen-bonded structure, featuring both single and double helices along with localized molecular chain arrangements.3.3 Intake of food and water, WATs mass
Table 2 reveals the consumption patterns differed significantly (P < 0.05) between the HFD group and the NCD group, with the HFD group exhibiting higher values for food intake (2.55 g per d per mouse), water intake (2.65 mL per d per mouse), and WATs (1.22 g). In comparison to the HFD group, treatment with the ERS–KGM complex led to a highly statistically significant increase in water consumption and a reduction in food intake (P < 0.05). Interestingly, there were no significant differences in water consumption among the KGM, ERS, and ERS–KGM complex groups (P > 0.05) or in food intake between the KGM and ERS groups (P > 0.05). As a result, the ERS–KGM complex displayed a significant preference for reducing food consumption by modulating satiety levels. Under the HFD condition, where mice intervened with the ERS–KGM complex exhibited the lowest WATs mass.| Parameter | HFD | KGM | ERS | ERS–KGM complex | NCD |
|---|---|---|---|---|---|
| a The value is derived from ten repeated experiments with a mean ± SD (n = 10). One-way ANOVA followed by Tukey's test was used to investigate significant differences between the groups. P < 0.05 is shown by different small-case letters within each column. HFD: high-fat diet; KGM: konjac glucomannan; ERS–KGM complex: extruded rice starch–konjac glucomannan complex; NCD: normal chow diet; ERS: extruded rice starch; WATs: white adipose tissues; ANOVA: analysis of variance; SD: standard deviation. | |||||
| Food intake | 2.55 ± 0.03c | 2.49 ± 0.03abc | 2.53 ± 0.03b | 2.47 ± 0.02a | 2.73 ± 0.05d |
| Water intake | 2.65 ± 0.05a | 2.86 ± 0.06c | 2.77 ± 0.04bc | 2.88 ± 0.04c | 3.01 ± 0.04d |
| WATs weight | 1.22 ± 0.14d | 0.80 ± 0.03c | 0.85 ± 0.04c | 0.72 ± 0.03b | 0.65 ± 0.02a |
3.4 Body weight and serum glucose-related parameters
Fig. 1B and C depict the body weight and weight gain of mice, respectively. It seems that the KGM, ERS, and ERS–KGM complex did not cause any pathological problems in the mice, as Fig. 1B illustrates, since the body weight of the mice increased over time in all groups. Additionally, mice in the HFD group exhibited significantly higher body weights than those in the NCD group, confirming that HFD exacerbates weight gain and obesity. After two weeks, the HFD group experienced a substantial gain in body weight, exceeding that of the NCD group (P < 0.05). After four weeks, mice feeding KGM, ERS, or the ERS–KGM complex showed significantly lower body weights and reduced weight gains compared to the HFD group (all P < 0.05). Following four weeks of feeding, the ERS–KGM complex intervention reduced body weight and weight gain compared to the KGM or ERS group alone (P < 0.05, Fig. 1C). Notably, the ERS–KGM complex revealed greater efficacy in reducing weight compared to the KGM or ERS group alone. Next, the changes in serum glucose-related parameters treated by the KGM, ERS, or ERS–KGM complex were explored. The KGM, ERS, and ERS–KGM complex groups had higher OGTT values at 0, 30, 60, 90, and 120 min than the HFD group (Fig. 1D). Fig. 1E shows that the AUC values of glucose were considerably lower in the ERS–KGM complex group compared to the KGM or ERS group alone (P < 0.05). In addition, the insulin values of the ERS–KGM complex group were considerably lower than those of the KGM or ERS group alone (Fig. 1F, P < 0.05).3.5 Serum lipoprotein metabolism
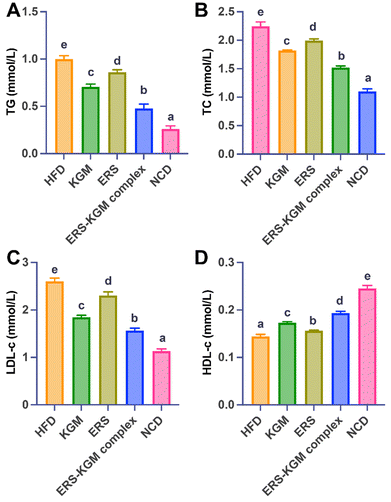
Fig. 2A and B show that KGM, ERS, or ERS–KGM complex reduced the TG and TC contents in mice compared with the HFD group (P < 0.05). Additionally, the ERS–KGM complex showed obvious effects on TG and TC reduction in mice compared with the KGM or ERS group alone. Compared to the HFD group, the KGM, ERS, or the ERS–KGM complex group demonstrated a significant elevation of HDL-c levels and a corresponding decrease in LDL-c levels in mice, as seen in Fig. 2C and D (P < 0.05). The anti-digestibility of the ERS–KGM complex may reduce TC levels in mice by decreasing LDL-c and increasing HDL-c content.3.6 Morphology of liver and WATs
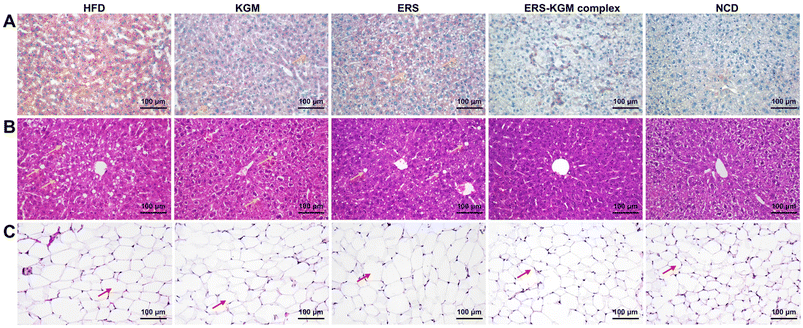
The tissues were stained with H&E and oil red O findings for the WATs and liver tissues are shown in Fig. 3. Fig. 3A and B reveal that cells in the HFD group displayed pronounced, spherical lesions resulting from the accumulation of lipid-filled vesicles. This led to compaction and deformation of the cells, with some exhibiting broken cytoplasms and compromised membranes. Following ERS treatment, there was a significant reduction in fat accumulation and a notable improvement in liver cell integrity. In contrast, the KGM group exhibited minimal fat droplets and cell damage. Furthermore, the ERS–KGM complex group demonstrated even fewer fat droplets and cell breakages than treatment with either the ERS or KGM alone. The hepatic steatosis brought on by the HFD could be dramatically reversed by the ERS–KGM complex, which also had a liver-protecting effect. According to Fig. 3C, compared to the HFD group, the NCD group exhibited smaller cells with more uniform spacing. The cells in the HFD group were almost 3–4 times larger than those in the NCD group. Notably, mice treated with ERS exhibited smaller cell sizes. On the other hand, compared to the HFD group, the cells from the mice treated with KGM or the ERS–KGM complex were much smaller. Additionally, the ERS–KGM complex exhibited the most pronounced effects in reducing lipid accumulation and protecting cells from damage. The ERS–KGM complex consistently exhibited the most pronounced improvement in cell integrity, fat vesicle reduction, uniform cell size, and compact arrangement, in line with its effects on lowering WATs, serum LDL-c, TC, and TG levels.3.7 Serum liver enzyme and antioxidant enzyme activities
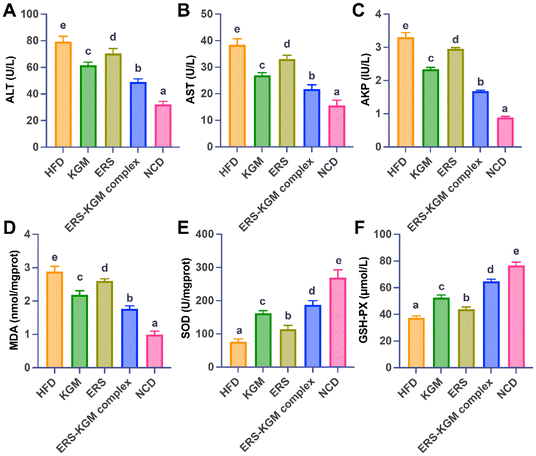
As seen by Fig. 4A–C, mice treated with ERS–KGM complex had substantially lower blood levels of ALT, AST, and AKP (P < 0.05) than mice treated with KGM or ERS group alone. The ERS–KGM complex significantly enhanced endurance and liver protection in mice, while also reducing fatigue, thereby outperforming the KGM or ERS group alone. Fig. 4D indicates that significantly higher MDA concentrations were observed in the HFD group compared to the NCD group. Consequently, the KGM, ERS, or ERS–KGM complex group significantly decreased MDA levels in mice (P < 0.05), with the ERS–KGM complex exhibiting the most pronounced effect. As shown in Fig. 4E and F, the HFD, KGM, ERS, and ERS–KGM complex groups had significantly lower levels of SOD and GSH-PX than the NCD group (P < 0.05). Furthermore, compared to the KGM or ERS group alone, SOD and GSH-PX activities were considerably increased after ERS–KGM complex treatment (P < 0.05). Notably, the ERS–KGM complex displayed a significant enhancement of antioxidant capacity in the organism, outperforming the effects of either the KGM or ERS group alone. Furthermore, the ERS–KGM complex was presumed to facilitate the promotion of SOD and GSH-PX activities by concurrently suppressing MDA activity, which may rely on its ability to optimize SCFAs production during digestion.3.8 Short-chain fatty acids
As illustrated in Table 3, acetate, propionate, and butyrate emerged as the primary SCFAs present in the five groups. Compared to the NCD group, butyrate, acetate, propionate, and total SCFAs in the feces of the HFD group undergo a significant reduction. In contrast, these molecules significantly increased in the KGM, ERS, or ERS–KGM complex group compared with the HFD group. In particular, compared to the KGM or ERS group alone, the ERS–KGM complex group resulted in a significant enhancement of total SCFAs production, with propionate and butyrate exhibiting the most notable increase. This suggests that the ERS–KGM complex can more effectively counteract the negative effects of HFD on the levels of SCFAs in feces. Notably, this result corroborated the above finding that administering the ERS–KGM complex to obese mice led to a significant reduction in liver damage.| Parameter | Acetate (μg g−1) | Propionate (μg g−1) | Butyrate (μg g−1) | Total SCFAs (μg g−1) |
|---|---|---|---|---|
| a The value is derived from five repeated experiments with a mean ± SD (n = 5). One-way ANOVA followed by Tukey's test was used to investigate significant differences between the groups. P < 0.05 is shown by different small-case letters within each column. HFD: high-fat diet; KGM: konjac glucomannan; ERS–KGM complex: extruded rice starch–konjac glucomannan complex; NCD: normal chow diet; ERS: extruded rice starch; SCFAs: short-chain fatty acids; ANOVA: analysis of variance; SD: standard deviation. | ||||
| HFD | 1232.62 ± 26.56a | 128.26 ± 8.03a | 146.74 ± 3.92a | 1542.21 ± 33.81a |
| KGM | 1542.35 ± 24.18c | 185.07 ± 6.64bc | 167.10 ± 2.83ab | 1925.35 ± 23.24c |
| ERS | 1439.62 ± 38.17b | 176.94 ± 5.83b | 158.10 ± 1.73ab | 1818.66 ± 41.09b |
| ERS–KGM complex | 1542.76 ± 47.03c | 197.39 ± 4.98bc | 186.11 ± 6.87b | 1978.27 ± 41.90c |
| NCD | 2452.53 ± 26.38d | 611.28 ± 29.93d | 511.46 ± 43.35c | 3631.59 ± 73.62d |
3.9 Serum metabolomic
In this study, we exhaustively counted the number of lipids grouped by subclass. Specifically, the lipid metabolic pathways classification system categorizes lipid metabolites into eight major classes: polyketides (PK), glycolipids (SL), isoprenoid lipids (PR), steroids (ST), sphingolipids (SP), fatty acyls (FA), glycerophospholipids (GP), and glycerol esters (GL). The lipids within these classes are categorized into 96 distinct subclasses (Fig. 5A). The results revealed that most classes were GL and GP. Based on the degree of unsaturation of lipids, they can be categorized into four distinct types: odd-numbered fatty acyls (ODD), polyunsaturated fatty acyls (PUFA), monounsaturated fatty acyls (MUFA), and saturated fatty acyls (SAF). This study utilizes pie charts to effectively visualize the distribution of various saturated lipids, showcasing the relative proportions of PUFA (39.23%), SFA (22.87%), ODD (21.2%), and MUFA (16.69%) (Fig. 5B). To better understand the metabolic differences in HFD and ERS–KGM complex groups, the differential metabolites were screened with P < 0.05 and a fold change (FC) > 2.0 as the basis. We identified 154 species with distinct groups in the HFD and ERS–KGM complex groups, as illustrated in Fig. 5C, which revealed 87 metabolites to be upregulated and 67 to be downregulated. 238 metabolites were significantly altered in the ERS–KGM complex group relative to the HFD group (Fig. 5D). A total of 85 metabolites are identified as common to both HFD vs. NCD and ERS–KGM complex vs. HFD comparisons, indicating their coexistence between the three groups. Subsequent analysis revealed the presence of 49 TG, 14 DG, and 4 MGDG in the GL of both HFD and ERS–KGM complex groups. The heatmap revealed that under HFD stimulation, certain GL molecular species exhibited increased levels, while others demonstrated a decrease (Fig. 5E). The results indicate that the KGM, ERS, or ERS–KGM complex group regulates biomarkers and related pathways to enhance host health. Notably, the gut flora of the host is also affected by these modifications in small molecule metabolites.3.10 Gut microbiota
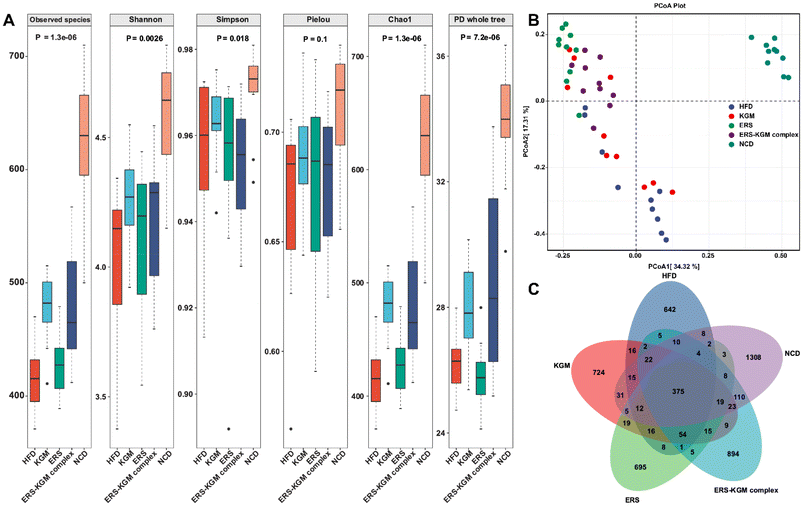
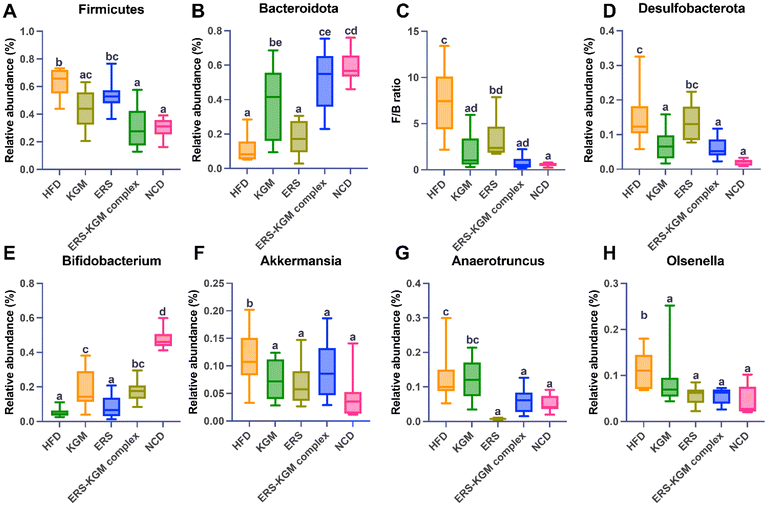
The species accumulation curves, rarefaction curves, and rank-abundance curves are illustrated in Fig. S2A–S2C.† The results showed that the sequencing data was trustworthy, and most of the intestinal microbiota was comprehensively captured at the current sequencing depth in each sample. Significant variations were found between the five groups according to the α-diversity analysis (Fig. 6A). Compared to the HFD group, the NCD group exhibited significantly greater bacterial diversity, relative abundance, and evenness. The KGM, ERS, or ERS–KGM complex group showed a slight and beneficial effect on gut bacterial community diversity and richness, which was comparable to the NCD group while still discernible from the HFD group. Samples from various groups were well clustered and divided into three groups (Fig. S3A†) according to the hierarchical clustering analysis based on the Bray–Curtis distance algorithm. This suggests that KGM, ERS, or the ERS–KGM complex group caused notable changes in the composition of gut microbiota. Furthermore, the ERS and ERS–KGM complex groups showed little variation, suggesting a high degree of similarity in the gut microbiome composition between the two groups. The HFD group was clearly separated from the NCD group, as demonstrated by the results of the PCA, PCoA, and NMDS analyses. Similarly, the KGM, ERS, or ERS–KGM complex group was clearly separated from the HFD and NCD groups (Fig. 6B and S3B, S3C†), indicating that these factors impacted the composition of the gut microbiota. According to the PCoA analysis, β-diversity identified a substantial difference in the structures of the gut microbial community between the HFD and NCD groups along the PC1 axis (Fig. 6B). In contrast, the KGM, ERS, or ERS–KGM complex group reversed the changes in bacterial populations induced by the HFD, displaying distinct patterns that differed from those seen in the HFD group. By employing a Venn diagram, the species shared by several microbial communities were identified (Fig. 6C). Our analysis revealed a shared species pool of 375 across the five groups, with 724, 695, and 894 species unique to the KGM, ERS, and ERS–KGM complex groups, respectively. This suggests that the distinct fine structures of the KGM, ERS, or ERS–KGM complex group were responsible for mediating their interventional effects on the intestinal flora environment by altering its variability.The phylum-level microbial alterations for each of the experimental groups were determined. According to Fig. S4A,† the three most common phyla in all groups were Firmicutes, Bacteroidetes, and Desulfobacterota, comprising more than 90% of the total phyla detected at this level. Firmicutes and Desulfobacterota were much more abundant in mice with HFD-induced obesity, but Bacteroidetes were less common in the HFD group (Fig. 7A, B, and D). Furthermore, the HFD group exhibited a noticeably higher Firmicutes/Bacteroidetes (F/B) ratio than the NCD group. Interestingly, mice fed with the KGM, ERS, or ERS–KGM complex group showed a significant reduction in their F/B ratio (Fig. 7C). Fig. S4B† shows the genus levels of the top 10 dominant bacteria in the five groups. The HFD group had reduced relative Bifidobacterium levels compared to the NCD group (Fig. 7E). The KGM, ERS, or ERS–KGM complex group significantly raised the Bifidobacterium levels. On the other hand, mice treated with the KGM, ERS, or ERS–KGM complex group showed a reduction in many genera, including Olsenella, Anaerotruncus, and Akkermansia (Fig. 7F–H). However, a significant abundance of the conditionally pathogenic bacteria Anaerotruncus was discovered in the HFD group.
Fig. S4C† are the distributions of LDA values >3 consistently identified by LEfSe analysis in all groups. Alloprevotella, Blautia, Acholeplasmatales, Anaeronlasma, and Acholeplasmataceae were characteristic of the KGM group. Firmicutes, Clostridia, Lachnospiraceae, Lachnospirales, Oscillospirales, Desulfovibrionaceae, Desulfovibrionales, Desullobaclerole, Desulfovibrionia, Oscillospiraceae, Lachnospiraceae, Colidextribacter, Faecalibaculum, Ruminococcaceae, Curvibacter, Acetatifactor, Tuzzerella, and Peptococcus were characteristic of the ERS group. Erysipelatoclostridium, Erysipelatoclostridiaceae, Lactobacillales, Lactobacillaceae, Lactobacillus, Olsenella, Lachnoclostridium, Eggerthellaceae, Christensenellaceae, Christensenellales, Peptostreptococcaceae, Romboutsia, and Enterococcaceae were characteristic of the ERS–KGM complex group. The HFD specifically promoted Bacteroidaceae, Bacteroides, Rikenellaceae, Alistipes, Tannerellaceae, Parabacteroides, Butyricimonas, Paraprevotella, and Bacteroidales. Conversely, NCD mice were dominated by Muribaculaceae, Muribaculaceae, Prevotellaceae, Prevotellaceae, Prevotellaceae, Clostridia, Clostridia, Spirochaetia, Spirochaetales, Treponema, Oscillospira, Burkholderiales, Gammaproteobacteria, Proteobacteria, Corinharteriales Incertas Sedic, Muribaculum, Rikenella, Catenibactenum, Paludicola, and Anaerovoracaceae. Therefore, the HFD significantly altered the gut microbiota of mice, potentially compromising their host health. Notably, the adverse effects of HFD were mitigated by treating the KGM, ERS, or the ERS–KGM complex. The unique bacterial profiles present in each group were elucidated by LEfSe, demonstrating statistically significant differences between them. Specifically, the HFD group exhibited a notable enrichment of Bacteroidaceae, while the NCD group was characterized by a prominent presence of Coriobacteria, as shown in Fig. S4D.† The gut microbiota of the KGM and ERS groups was markedly enhanced in Acholeplasmataceae and DesuIfovibrionaceae, while the ERS–KGM complex group was rich in Lachnospiraceae. The ERS–KGM complex treatment altered the dominant microbial types in the gut microbiome based on the results.
4. Discussion
The findings from this study provide valuable insights into the potential of ERS–KGM complex as a dietary intervention for improving gut health and preventing obesity. The ERS–KGM complex exhibits unique structural properties, derived from the screw extrusion process, which have led to a substantial increase in RS content. A plausible explanation could arise from the underlying mechanics of screw extrusion procedures, where previous research has shown that increased glucan chain disaggregation and rearrangement occur upon the interaction between KGM and starch molecules. This interaction yields a dense, hydrogen-bonded structure consisting of both single and double helices, as well as localized molecular chain arrangements.11,24 Most of the crystalline area of starch polymers comprises amylopectin side clusters associated with ordered double-helical and single-helical amylose-lipid complex.25 This suggests that the physical modifications induced by screw extrusion helped to create a well-ordered structure that is more resistant to enzymatic digestion, an important attribute for promoting gut health and metabolic benefits.4.1 Impact on metabolic health and obesity markers
The ERS–KGM complex demonstrated notable improvements in various metabolic health markers in mice fed an HFD. Specifically, the ERS–KGM complex reduced WATs mass, body weight, and adipocyte size, which are critical indicators of obesity. Similar to another research, supplementation with RS was shown to decrease ghrelin production in human participants while increasing the release of satiety hormones such as tyropeptide 3–36, cholecystokinin octapeptide, and glucagon-like peptide 1 (GLP-1).26 The potent anti-digestibility of the ERS–KGM complex, a key factor behind this phenomenon, induces the conversion of white fat in mice to brown fat, thereby hindering weight gain.24 This response is accompanied by the inhibition of ghrelin release and reduction in fat accumulation and adiposity. The observed reduction in food intake, along with an increase in water consumption, could suggest that the ERS–KGM complex promotes satiety and hydration, contributing to its weight management effects.In addition to these effects on body composition, the ERS–KGM complex significantly improved glucose metabolism. It enhanced glucose tolerance, lowered insulin levels, and alleviated insulin resistance in mice. The addition of the ERS–KGM complex to a regimen was found to have a substantial impact on glucose homeostasis and insulin resistance, yielding outcomes comparable to those reported for RS, including increased insulin sensitivity and reduced liver damage in mice fed an HFD.27,28 The ability of the ERS–KGM complex to lower serum triglyceride and total glycerol content further reinforces its potential role in reducing metabolic disorders associated with obesity, such as dyslipidemia.
4.2 Molecular and enzymatic modulation
The study also highlights the ERS–KGM complex's ability to modulate critical enzymes and biomarkers associated with oxidative stress and inflammation. For instance, reducing ALT, AST, AKP, and MDA activities suggests decreased cellular stress and damage. Key indicators of liver function, including AKP, ALT, and AST, suggest liver damage when their levels are elevated in serum, indicating cellular injury and compromised structural integrity of the liver membrane.29 When polyunsaturated fatty acids in membranes are attacked by free radicals, MDA is the breakdown product. Therefore, MDA levels may serve as an indicator of the degree of oxidative damage inside cells.30 Conversely, the enhanced activities of GSH-PX and SOD indicate a strengthened antioxidant defense system. This shift in enzyme activity profiles may contribute to the overall health benefits observed in the mice, as oxidative stress and inflammation are well-known contributors to metabolic diseases, including obesity. Elevated levels of ROS, which promote protein oxidation, are a major cause of muscle fatigue. The protective effect of the dietary intervention was assessed by measuring the activities of SOD and GSH-PX enzymes.31 Its superiority was shown by its capability to upregulate the activities of GSH-PX and SOD, which enhanced the antioxidant and hepatoprotective properties of the liver.32Furthermore, the ERS–KGM complex increased HDL-c and decreased TG, TC, and LDL-c levels, suggesting that it improved energy storage and metabolic efficiency. This enhancement in energy metabolism could be another factor behind the improved glucose handling and reduced adiposity in the mice. The anti-digestibility of the ERS–KGM complex may contribute to the reduction in TC levels in mice by decreasing LDL-c content and increasing HDL-c content, which is comparable to the outcomes of the Mediterranean and hypocaloric diets in mitigating metabolic syndrome risk.33 As previously noted, mice administered RS exhibited significantly lower levels of TG, TC, and LDL-c.34 An in-depth analysis of our study suggests that the anti-digestibility of the ERS–KGM complex may be due to the preservation of the high-viscosity properties of KGM, as well as the enzyme-resistant structure of the ERS–KGM complex.
4.3 Gut microbiota and serum metabolites
Obesity often disrupts lipid profiles, increasing glycerolipid levels. However, dietary interventions, such as the introduction of arabinoxylan, show promise in reversing these effects by impacting key metabolic pathways like glycerolipid and glycerophospholipid metabolism.35 Our findings reveal that the ERS–KGM complex significantly impacted lipid metabolism, especially within the GL and GP classes. Through serum metaboiltes analysis, we observed that dietary interventions can lead to changes in the unsaturation levels of fatty acyls, with noticeable shifts in the PUFA, SFA, MUFA, and ODD. This suggests that dietary interventions could modulate lipid profiles towards a more balanced and potentially health-favorable lipidomic state. The significant differential expression of metabolites in the ERS–KGM complex group compared to the HFD group underscores the potential of diet-based regulation of lipid metabolism. With 154 distinct metabolites altered, many of which are involved in key metabolic pathways, these changes point toward potential biomarkers for metabolic health improvement. Additionally, dietary fibers have been shown to reduce body weight and dyslipidemia, partly by modulating key lipid metabolic pathways.36 Moreover, changes in bile acid metabolism and increases in the hormone GLP-1, driven by dietary fibers like glucomannan and arabinoxylan, are linked to improved responses in obesity and type 2 diabetes.37 The alterations in small molecule metabolites induced by the ERS–KGM complex suggests a cascading effect wherein changes in the gut environment and microflora contribute significantly to lipid homeostasis and metabolic regulation. Gut microbiota is known to produce and modulate systemic bioactive metabolites, such as SCFAs and bile acids, which can further influence host metabolism by affecting insulin sensitivity, lipid oxidation, and immune responses.4.4 Gut microbiota modulation
The ERS–KGM complex enriched the population of beneficial microbes such as Bifidobacterium at the genus level and Firmicutes and Desulfobacterota at the phylum level. These microbial shifts are particularly important because gut microbiota is critical in energy metabolism, fat storage, and overall metabolic health. A noteworthy earlier finding revealed a favorable relationship between blood leptin levels and the Bifidobacterium population.38 The peptide hormone leptin is well-established for its ability to reduce body weight and food intake in mice, as it promotes increased energy expenditure and appetite regulation. In contrast, leptin-deficient animals are more susceptible to obesity due to decreased energy expenditure and hyperphagia.39 Probiotic bacteria, particularly from the well-known Bifidobacterium species, produce lactate and acetate, which help alleviate the effects of toxins and inhibit the growth of harmful microorganisms. Notably, its abundance in the HFD group may also contribute to an abnormal colonic state and compromise the function of the intestinal mucus barrier.40,41The increase in SCFAs detected in the feces of mice suggests that the ERS–KGM complex promotes fermentation in the gut, which is a key mechanism through which resistant starch exerts its health benefits.42,43 SCFAs, such as butyrate, acetate, and propionate, have been shown to enhance gut barrier integrity, reduce inflammation, and regulate energy homeostasis, all crucial for preventing obesity and improving metabolic health. The enrichment of gut microbiota that can degrade fluorobenzoate further supports the idea that the ERS–KGM complex enhances metabolic capacity and functional diversity in the gut. Research has consistently demonstrated that acetate can inhibit adipocyte lipolysis, which lowers the flow of free fatty acids to the liver.44 On the other hand, butyrate plays a crucial role in inhibiting tumorigenesis by preventing the formation of tumor cells and restricting the division of colonic mucosal cells.45 Notably, Clostridiales are characterized by numerous butyrate-producing strains. The augmented production of butyrate can be attributed to the substantial increase in Clostridiales abundance, primarily induced by RS.46 Furthermore, the ability of acetate and propionate production to decrease cholesterol levels was consistent with the fact that obese mice fed the RS diet had lower plasma TC levels compared to those fed the HFD.30 Paolella et al.47 reports that the liver absorbs around 90% of the propionate produced by intestinal microbiota in mammary animals, which lowers hepatic lipid levels and lessens liver damage brought on by an HFD. Obesity is typically associated with abnormal lipid profiles, including elevated GL levels.48 Notably, the ERS–KGM complex group has been shown to ameliorate metabolic disorders induced by obesity, with beneficial effects on glucose metabolism, GP metabolism, phosphatidylinositol signaling pathways, and primary bile acid biosynthesis. One important route that has a greater influence on obesity is GP metabolism.49 The metabolism of bile acids plays a crucial role in regulating RS through subtle alterations in bile acid profiles.50 According to a previous study, circulating bile acid concentrations and high levels of the incretin hormone GLP-1 are strongly linked to obesity.49
A PICRUSt2 study utilized the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to gain insight into the metabolic pathways of the microbiota, anticipating that changes in the composition of the gut microbiota would reflect changes in microbiota function (Fig. 8A).51 Significant upregulation was observed in several KEGG pathways, including calcium signaling (P < 0.01), protein digestion and absorption (P < 0.001), steroid hormone biosynthesis (P < 0.01), biosynthesis of siderophore group nonribosomal peptides (P < 0.001), glycosaminoglycan degradation (P < 0.01), apoptosis (P < 0.001), glycan degradation (P < 0.01), and penicillin and cephalosporin biosynthesis (P < 0.05) in the ERS group compared to the HFD group, suggesting enhanced fecal bacterial activity. The administration of the ERS–KGM complex significantly enhances the fermentation activity in the gut, characterized by a notable promotion of fluorobenzoate degradation (P < 0.05) (Fig. 8B–D). This finding is consistent with the observation of higher levels of SCFAs in the feces of mice fed the ERS–KGM complex. Given their established role as ligands for SCFAs and their status as a recognized therapeutic target for metabolic disorders, the ERS–KGM complex boosted the synthesis of SCFAs, particularly propionate, and butyrate, and modified the composition of the gut microbiota. Our in vivo mouse model results further supported the idea that the positive effects of the ERS–KGM complex on metabolism were largely mediated by SCFAs, which is in line with our above findings.
4.5 Implications for human health and future research
This study provides a promising foundation for potentially using extruded starchy foods as nutritional supplements for obesity prevention. The combination of starch and KGM appears to offer a multifaceted approach to improving metabolic health through both direct physiological effects and modulation of the gut microbiota. However, while the findings in mice are encouraging, further research in human populations is necessary to validate these effects and determine optimal dosages and formulations. Future studies should also explore the long-term impact of the ERS–KGM complex supplementation on metabolic health, especially in diverse populations with varying dietary habits and microbiota profiles. Additionally, it would be beneficial to investigate whether other types of RS to enhance the range of dietary interventions available for metabolic health.5. Conclusion
In conclusion, this work investigated the possible relationship between the physiological activities of the ERS–KGM complex produced via screw extrusion and its multi-scale structural characteristics. In obese mice, the ERS–KGM complex showed exceptional efficacy in mitigating the metabolic abnormalities induced by an HFD. Notably, the ERS–KGM complex exhibited superior performance over its individual components (ERS or KGM) in regulating body and WATs gain, while effectively promoting lipid metabolism and inhibiting lipogenesis in obese mice, which correlated with distinct patterns of expression of genes related to lipid metabolism in the liver. In addition, the ERS–KGM complex significantly boosted the production of total SCFAs, with propionate and butyrate exhibiting the most pronounced elevations compared to the ERS or KGM alone. According to high-throughput 16S rRNA gene sequencing, the ERS–KGM complex may modulate the diversity of gut microbiota by virtue of its unique structure, which in turn may influence fatty acid metabolism and alter the production of microbial metabolites. Therefore, we hypothesized that the protective effects of the ERS–KGM complex on obese mice might be mediated by its involvement in the gut microbiota-SCFAs-liver metabolism axis. These findings strongly suggest that the ERS–KGM complex may serve as a promising and safe candidate for the prevention of obesity.Author contributions
Fanrui Liu: methodology, validation, formal analysis, investigation, data curation, writing – original draft, visualization. Hao Wan: methodology, validation, formal analysis, investigation, data curation, visualization. Honghao Fan, methodology, resources, writing – review & editing, visualization, supervision. Zhihong Zhang: methodology, investigation. Hua Dai: methodology, investigation. Hai He: methodology, resources, writing – review & editing, visualization, supervision, funding acquisition.Data availability
All relevant data are within the manuscript and its additional files.Conflicts of interest
The authors declare that there is no conflict of interest regarding the publication of this article.Acknowledgements
The study was supported by the Hainan Provincial Natural Science Foundation of China (no. 824MS066), Guangdong Basic and Applied Basic Research Foundation (no. 2022A1515110478), National Natural Science Foundation of China (no. 82304137), and Talent Project of Hainan Medical University (no. RZ2300005977 and RZ2300002106).References
- H. Liu, M. Zhang, Q. Ma, B. Tian, C. Nie, Z. Chen and J. Li, Health beneficial effects of resistant starch on diabetes and obesity via regulation of gut microbiota: a review, Food Funct., 2020, 11, 5749–5767 RSC
.
- J. Geng, Q. Ni, W. Sun, L. Li and X. Feng, The links between gut microbiota and obesity and obesity related diseases, Biomed. Pharmacother., 2022, 147, 112678 CrossRef CAS PubMed
.
- N. Hwalla and Z. Jaafar, Dietary management of obesity: a review of the evidence, Diagnostics, 2020, 11, 24 CrossRef
.
- L. A. Bello-Perez, P. C. Flores-Silva, E. Agama-Acevedo and J. Tovar, Starch digestibility: past, present, and future, J. Sci. Food Agric., 2020, 100, 5009–5016 CrossRef CAS PubMed
.
- S. P. Bangar, A. O. Ashogbon, A. Singh, V. Chaudhary and W. S. Whiteside, Enzymatic modification of starch: A green approach for starch applications, Carbohydr. Polym., 2022, 287, 119265 CrossRef
.
- Y. Zhong, J. Xu, X. Liu, L. Ding, B. Svensson, K. Herburger, K. Guo, C. Pang and A. Blennow, Recent advances in enzyme biotechnology on modifying gelatinized and granular starch, Trends Food Sci. Technol., 2022, 123, 343–354 CrossRef CAS
.
- X. Huang, H. Liu, Y. Ma, S. Mai and C. Li, Effects of extrusion on starch molecular degradation, order–disorder structural transition and digestibility—A review, Foods, 2022, 11, 2538 CrossRef CAS
.
- B. Wang, Y. Dong, Y. Fang, W. Gao, X. Kang, P. Liu, S. Yan, B. Cui and A. M. Abd El-Aty, Effects of different moisture contents on the structure and properties of corn starch during extrusion, Food Chem., 2022, 368, 130804 CrossRef CAS PubMed
.
- A. Homayouni-Rad, A. M. Mortazavian, H. Pourjafar and S. K. Moghadam, Extrusion and Co-extrusion: A Technology in Probiotic Encapsulation with Alternative Materials, Curr. Pharm. Biotechnol., 2024, 25, 1986–2000 CAS
.
- Y. Fourati, A. Magnin, J.-L. Putaux and S. Boufi, One-step processing of plasticized starch/cellulose nanofibrils nanocomposites via twin-screw extrusion of starch and cellulose fibers, Carbohydr. Polym., 2020, 229, 115554 CrossRef CAS PubMed
.
- H. He, X. Zhang, W. Liao and J. Shen, Characterization and in vitro digestion of rice starch/konjac glucomannan complex prepared by screw extrusion and its impact on gut microbiota, Food Hydrocolloids, 2023, 135, 108156 CrossRef CAS
.
- Y. Wen, D. Zhu, J. Sun, Q. Yan and Z. Jiang, Anti-obesity effect and mechanism of konjac mannooligosaccharides, Food Sci., 2020, 41, 115–121 Search PubMed
.
- J. Wu, M. Qiu, C. Zhang, C. Zhang, N. Wang, F. Zhao, L. V. Liqiao, J. Li, A. G. A. Lyu-Bu and T. Wang, Type 3 resistant starch from Canna edulis modulates obesity and obesity-related low-grade systemic inflammation in mice by regulating gut microbiota composition and metabolism, Food Funct., 2021, 12, 12098–12114 RSC
.
- T. Jiang, F. Chen, Q. Duan, X. Bao, S. Jiang, H. Liu, L. Chen and L. Yu, Designing and application of reactive extrusion with twice initiations for graft copolymerization of acrylamide on starch, Eur. Polym. J., 2022, 165, 111008 CrossRef CAS
.
- H. He, B. Zheng, H. Wang, X. Li and L. Chen, Insights into the multi-scale structure and in vitro digestibility changes of rice starch-oleic acid/linoleic acid complex induced by heat-moisture treatment, Food Res. Int., 2020, 137, 109612 CrossRef CAS
.
- H. N. Englyst, S. M. Kingman, G. J. Hudson and J. H. Cummings, Measurement of resistant starch in vitro and in vivo, Br. J. Nutr., 1996, 75, 749–755 CrossRef CAS PubMed
.
- T. Wang, J. Han, H. Dai, J. Sun, J. Ren, W. Wang, S. Qiao, C. Liu, L. Sun and S. Liu, Polysaccharides from Lyophyllum decastes reduce obesity by altering gut microbiota and increasing energy expenditure, Carbohydr. Polym., 2022, 295, 119862 CrossRef CAS PubMed
.
- J. Wen, M. Li, J. Hu, J. Wang, Z. Wang, C. Chen, J. Yang, X. Huang, M. Xie and S. Nie, Different dietary fibers unequally remodel gut microbiota and charge up anti-obesity effects, Food Hydrocolloids, 2023, 140, 108617 CrossRef CAS
.
- S. Fan, M. Shahid, P. Jin, A. Asher and J. Kim, Identification of metabolic alterations in breast cancer using mass spectrometry-based metabolomic analysis, Metabolites, 2020, 10, 170 CrossRef CAS PubMed
.
- N. A. Bokulich, B. D. Kaehler, J. R. Rideout, M. Dillon, E. Bolyen, R. Knight, G. A. Huttley and J. G. Caporaso, Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin, Microbiome, 2018, 6, 1–17 CrossRef
.
- I. Capron, P. Robert, P. Colonna, M. Brogly and V. Planchot, Starch in rubbery and glassy states by FTIR spectroscopy, Carbohydr. Polym., 2007, 68, 249–259 CrossRef CAS
.
- H. Tang and B. P. Hills, Use of 13C MAS NMR to study domain structure and dynamics of polysaccharides in the native starch granules, Biomacromolecules, 2003, 4, 1269–1276 CrossRef CAS
.
- M. E. Rodriguez-Garcia, M. A. Hernandez-Landaverde, J. M. Delgado, C. F. Ramirez-Gutierrez, M. Ramirez-Cardona, B. M. Millan-Malo and S. M. Londoño-Restrepo, Crystalline structures of the main components of starch, Curr. Opin. Food Sci., 2021, 37, 107–111 CrossRef CAS
.
- Q. Wang, L. Li, T. Wang and X. Zheng, A review of extrusion-modified underutilized cereal flour: Chemical composition, functionality, and its modulation on starchy food quality, Food Chem., 2022, 370, 131361 CrossRef CAS PubMed
.
- S. Pérez and E. Bertoft, The molecular structures of starch components and their contribution to the architecture of starch granules: A comprehensive review, Starch/Staerke, 2010, 62, 389–420 CrossRef
.
- J. Zhou, R. J. Martin, R. T. Tulley, A. M. Raggio, K. L. McCutcheon, L. Shen, S. C. Danna, S. Tripathy, M. Hegsted and M. J. Keenan, Dietary resistant starch upregulates total GLP-1 and PYY in a sustained day-long manner through fermentation in rodents, Am. J. Physiol.: Endocrinol. Metab., 2008, 295, E1160–E1166 CrossRef CAS
.
- Q. Chen, D. Wang, Y. Gu, Z. Jiang and Z. Zhou, Tangeretin prevents obesity by modulating systemic inflammation, fat browning, and gut microbiota in high-fat diet-induced obese C57BL/6 mice, J. Nutr. Biochem., 2022, 101, 108943 CrossRef CAS
.
- F. Hu, Y. Niu, X. Xu, Q. Hu, Q. Su and H. Zhang, Resistant dextrin improves high-fat-high-fructose diet induced insulin resistance, Nutr. Metab., 2020, 17, 1–11 CrossRef
.
- Q. Yao, X. Jiang, Y. Zhai, L. Luo, H. Xu, J. Xiao, L. Kou and Y. Zhao, Protective effects and mechanisms of bilirubin nanomedicine against acute pancreatitis, J. Controlled Release, 2020, 322, 312–325 CrossRef CAS PubMed
.
- T. Ilyés, C. N. Silaghi and A. M. Crăciun, Diet-related changes of short-chain fatty acids in blood and feces in obesity and metabolic syndrome, Biology, 2022, 11, 1556 CrossRef PubMed
.
- Y. Chen, J. Wang, Z. Jing, J. M. Ordovas, J. Wang and L. Shen, Anti-fatigue and anti-oxidant effects of curcumin supplementation in exhaustive swimming mice via Nrf2/Keap1
signal pathway, Curr. Res. Food Sci., 2022, 5, 1148–1157 CrossRef CAS
.
- X. Meng, Z. Wang, S. Liang, Z. Tang, J. Liu, Y. Xin, H. Kuang and Q. Wang, Hepatoprotective effect of a polysaccharide from Radix Cyathulae officinalis Kuan against CCl4-induced acute liver injury in rat, Int. J. Biol. Macromol., 2019, 132, 1057–1067 CrossRef CAS PubMed
.
- S. Pisanu, V. Palmas, V. Madau, E. Casula, A. Deledda, R. Cusano, P. Uva, S. Vascellari, F. Boi and A. Loviselli, Impact of a moderately hypocaloric Mediterranean diet on the gut microbiota composition of Italian obese patients, Nutrients, 2020, 12, 2707 CrossRef CAS PubMed
.
- S. He, Z. Xiong, L. Li, Y. Wang, C. Wang, B. Zheng, H. Zeng and Y. Zhang, Lotus seed resistant starch ameliorates high-fat diet induced hyperlipidemia by fatty acid degradation and glycerolipid metabolism pathways in mouse liver, Int. J. Biol. Macromol., 2022, 215, 79–91 CrossRef CAS PubMed
.
- X. Fu, Z. Liu, R. Li, J. Yin, H. Sun, C. Zhu, Q. Kong, H. Mou and S. Nie, Amelioration of hydrolyzed guar gum on high-fat diet-induced obesity: Integrated hepatic transcriptome and metabolome, Carbohydr. Polym., 2022, 297, 120051 CrossRef CAS PubMed
.
- Z. Yu, N. Wang, F. Geng and M. Ma, High-density lipoproteins from egg yolk's effect on hyperlipidemia in a high-fat-diet obese mouse using lipidomic analysis, Food Biosci., 2020, 33, 100492 CrossRef CAS
.
- S. N. Chaudhari, D. A. Harris, H. Aliakbarian, J. N. Luo, M. T. Henke, R. Subramaniam, A. H. Vernon, A. Tavakkoli, E. G. Sheu and A. S. Devlin, Bariatric surgery reveals a gut-restricted TGR5 agonist with anti-diabetic effects, Nat. Chem. Biol., 2021, 17, 20–29 CrossRef CAS
.
- S. Delgado, E. O'sullivan, G. Fitzgerald and B. Mayo, In vitro evaluation of the probiotic properties of human intestinal Bifidobacterium species and selection of new probiotic candidates, J. Appl. Microbiol., 2008, 104, 1119–1127 CrossRef CAS PubMed
.
- S. Arora, Role of neuropeptides in appetite regulation and obesity–a review, Neuropeptides, 2006, 40, 375–401 CrossRef CAS PubMed
.
- A. I. Álvarez-Mercado, M. Navarro-Oliveros, C. Robles-Sánchez, J. Plaza-Díaz, M. J. Sáez-Lara, S. Muñoz-Quezada, L. Fontana and F. Abadía-Molina, Microbial population changes and their relationship with human health and disease, Microorganisms, 2019, 7, 68 CrossRef PubMed
.
- J. Jin, J. Wang, R. Cheng, Y. Ren, Z. Miao, Y. Luo, Q. Zhou, Y. Xue, X. Shen and F. He, Orlistat and ezetimibe could differently alleviate the high-fat diet-induced obesity phenotype by modulating the gut microbiota, Front. Microbiol., 2022, 13, 908327 CrossRef
.
- D. L. Topping and P. M. Clifton, Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides, Physiol. Rev., 2001, 81, 1031–1064 CrossRef CAS
.
- E. E. Blaak, E. E. Canfora, S. Theis, G. Frost, A. K. Groen, G. Mithieux, A. Nauta, K. Scott, B. Stahl and J. Van Harsselaar, Short chain fatty acids in human gut and metabolic health, Benefic. Microbes, 2020, 11, 411–455 CAS
.
- H. Ge, X. Li, J. Weiszmann, P. Wang, H. Baribault, J. Chen, H. Tian and Y. Li, Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids, Endocrinology, 2008, 149, 4519–4526 CrossRef CAS PubMed
.
- J. C. Encarnação, A. M. Abrantes, A. S. Pires and M. F. Botelho, Revisit dietary fiber on colorectal cancer: butyrate and its role on prevention and treatment, Cancer Metastasis Rev., 2015, 34, 465–478 CrossRef
.
- J. Wu, J. Wang, Z. Lin, C. Liu, Y. Zhang, S. Zhang, M. Zhou, J. Zhao, H. Liu and X. Ma, Clostridium butyricum alleviates weaned stress of piglets by improving intestinal immune function and gut microbiota, Food Chem., 2023, 405, 135014 CrossRef CAS PubMed
.
- G. Paolella, C. Mandato, L. Pierri, M. Poeta, M. Di Stasi and P. Vajro, Gut-liver axis and probiotics: their role in non-alcoholic fatty liver disease, World J. Gastroenterol., 2014, 20, 15518 CrossRef CAS PubMed
.
- O. Friedland, D. Nemet, N. Gorodnitsky, B. Wolach and A. Eliakim, Obesity and lipid profiles in children and adolescents, J. Pediatr. Endocrinol. Metab., 2002, 15, 1011–1016 CAS
.
- S. Fiorucci, E. Distrutti, A. Carino, A. Zampella and M. Biagioli, Bile acids and their receptors in metabolic disorders, Prog. Lipid Res., 2021, 82, 101094 CrossRef CAS PubMed
.
- P. Lefebvre, B. Cariou, F. Lien, F. Kuipers and B. Staels, Role of bile acids and bile acid receptors in metabolic regulation, Physiol. Rev., 2009, 89, 147–191 CrossRef CAS PubMed
.
- X. Yu, W. Jiang, R. O. Kosik, Y. Song, Q. Luo, T. Qiao, J. Tong, S. Liu, C. Deng and S. Qin, Gut microbiota changes and its potential relations with thyroid carcinoma, J. Adv. Res., 2022, 35, 61–70 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo04275a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |