Open Access Article
Open Access ArticleCryo-EM for atomic characterization of supramolecular gels†
Ravi R.
Sonani
 a,
Simona
Bianco
a,
Simona
Bianco
 b,
Mark A. B.
Kreutzberger
b,
Mark A. B.
Kreutzberger
 a,
Dave J.
Adams
a,
Dave J.
Adams
 b and
Edward H.
Egelman
b and
Edward H.
Egelman
 *a
*a
aDepartment of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA 22903, USA. E-mail: egelman@virginia.edu
bSchool of Chemistry, University of Glasgow, Glasgow, G12 8QQ, UK
First published on 17th December 2024
Abstract
While there have been great advances in the design and synthesis of supramolecular gels, their characterization methods have largely stayed the same, with electron microscopy of dried samples, or small-angle scattering and spectroscopy dominating the approaches used. Although these methods provide valuable insights into structural properties, they are unable to unambiguously generate reliable atomic models that can further guide the site-specific modification of supramolecular gelators. Cryogenic electron microscopy (cryo-EM), allowing the high-resolution imaging of the sample in a hydrated state, has emerged as the dominant technique in structural biology, but has yet to become a routine method in materials science. Here, we describe the use of cryo-EM to determine the atomic structure of the tubular micelle formed by the dipeptide CarbIF, revealing the mechanism of assembly and gelation. Using the CarbIF micelle as an example, we highlight some of the challenges in using cryo-EM to study such materials, and how determination of the helical symmetry can be the most difficult aspect of such a project.
Introduction
Supramolecular gels, formed by low-molecular-weight gelators through non-covalent interactions, have gained significant attention for their wide range of applications in fields such as drug delivery, tissue engineering, catalysis, and sensors.1–6 These gels form three-dimensional networks that are responsive to environmental stimuli, making them ideal candidates for use in various biomedical and industrial applications.7,8 Structural characterization of supramolecular gels is essential for understanding their physical properties, functionality, and self-assembly mechanisms, while also guiding the design of modified gelators with tailored properties.9 Numerous techniques, including small-angle scattering, electron microscopy of dried samples, nuclear magnetic resonance (NMR), X-ray fiber diffraction, spectroscopy, atomic force microscopy, and computational approaches, are commonly used for studying the assembly, molecular packing, and structural features of these gels.9–15 While these techniques provide valuable information about the nature of molecular interactions and overall morphology, they are unable to provide reliable, unambiguous atomic models.Cryo-electron microscopy (cryo-EM) has emerged as a powerful and dominant tool in structural biology, enabling high-resolution visualization of specimens in their native states without the need for crystallization or drying.16–20 Unlike traditional electron microscopy, cryo-EM preserves the hydrated state of a sample, offering insights into molecular structures at the atomic level.21 This technique has proven particularly effective for studying helical polymers, which are often found in biological systems such as virus tails,22 cytoskeletal filaments,23–26 extracellular filaments,27 filamentous enzymes28–32 and amyloid fibrils.33 Structure determination of helical polymers via cryo-EM requires knowledge about the helical symmetry present in them, and often presents ambiguities.34 Helical symmetry, in X-ray crystallographic terms, can be described as a screw symmetry with two parameters: twist, describing rotation around the helical axis, and axial rise, describing translation along the axis, together defining the repetition of identical units. Accurately determining these parameters is critical for solving the structures of helical polymers using cryo-EM.
Determination of helical parameters usually involves examining the power spectrum, which is the averaged intensities of the Fourier transform of the segments of the helical polymer. The power spectrum is only non-zero along layer lines in reciprocal space, each corresponding to a certain periodicity of the helical structure. Once initial estimates of the helical parameters are obtained, the 3D structure of the helical object can be reconstructed via Iterative Helical Real-Space Reconstruction (IHRSR) or conceptually similar methods.35–37 While cryo-EM has been widely used to resolve the structures of biological helical polymers, its application to materials science remains at an early stage, particularly in the study of helical structures in synthetic materials like supramolecular gels.38,39 Nonetheless, cryo-EM offers great potential for understanding the atomic structures and packing of helical polymers in materials science, as exemplified in recent papers,40–43 which could help design advanced materials with customized morphology and properties.
Functionalized dipeptides have emerged as a promising class of supramolecular gelators due to their modular nature, enabling control over their gelation properties.6,44–47 These peptides offer inherent flexibility and functionality, which can be tuned through chemical modifications such as the introduction of non-natural amino acids or functional groups. By adjusting these modifications, researchers can enhance the morphology, solubility, gelation strength, and structural integrity of dipeptide-based supramolecular gels. One such modified dipeptide, CarbIF, has been studied for its gelation properties,48 which occur through a two-step process. In the first step, CarbIF assembles into micelles at alkaline pH, and in the second step, these micelles aggregate to form a supramolecular gel at neutral-to-acidic pH.
Here, we describe the application of cryo-EM to determine the atomic structure of the tubular micelle formed by CarbIF, offering new insights into the dipeptide's mechanism of assembly and gelation. Using CarbIF as a model system, we highlight some of the challenges in employing cryo-EM to study helical polymers, particularly the complexities involved in accurately determining helical symmetry.
Results and discussion
Self-assembly of CarbIF micelles
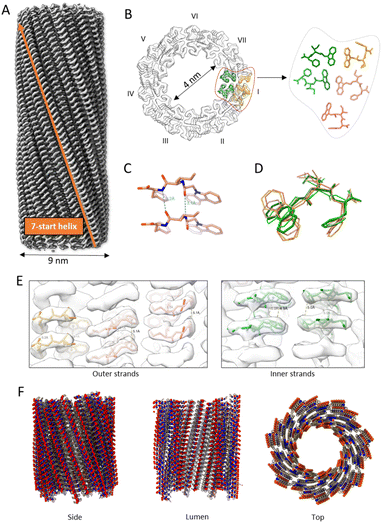
CarbIF is a dipeptide consisting of isoleucine–phenylalanine (IF) modified at its N-terminus (Fig. 1A).48 Under continuous stirring at room temperature, CarbIF self-assembles at pH 11 into long tubular micelles with a diameter of ∼90 Å (Fig. 1B). These CarbIF micelles further transition into a supramolecular gel at lower pH, as previously characterized.48 We aimed to investigate the atomic structure and packing within the CarbIF micelle to gain insights into both the assembly and gelation mechanisms.Cryo-EM structure of the CarbIF micelle reveals the mechanism of assembly and gelation
A total of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 742
742![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 371 overlapping segments of the CarbIF micelle were extracted from motion-corrected cryo-EM micrographs and subjected to 2D classification to remove poor quality segments, resulting in a stack of 1
371 overlapping segments of the CarbIF micelle were extracted from motion-corrected cryo-EM micrographs and subjected to 2D classification to remove poor quality segments, resulting in a stack of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 516
516![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 good CarbIF micelle segments. A representative 2D class average derived from these good segments is shown in Fig. 1C. Unlike lipid micelles, which exhibit an amorphous structure, CarbIF micelles display a highly ordered arrangement, as apparent from high-resolution features in their 2D class average (Fig. 1C). This is further supported by the high-resolution layer line at a spacing of 1/(4.8 Å) in the averaged power spectrum generated from these segments (Fig. 1D). The information from the power spectrum was used to derive potential symmetries49–52 and associated parameters, the axial rise and twist, as outlined in the following section.
102 good CarbIF micelle segments. A representative 2D class average derived from these good segments is shown in Fig. 1C. Unlike lipid micelles, which exhibit an amorphous structure, CarbIF micelles display a highly ordered arrangement, as apparent from high-resolution features in their 2D class average (Fig. 1C). This is further supported by the high-resolution layer line at a spacing of 1/(4.8 Å) in the averaged power spectrum generated from these segments (Fig. 1D). The information from the power spectrum was used to derive potential symmetries49–52 and associated parameters, the axial rise and twist, as outlined in the following section.
The 3D reconstruction of the structure from these 2D segments was determined using the Helical Refinement tool in cryoSPARC.37 This tool is conceptually similar to the Iterative Helical Real Space Reconstruction (IHRSR) algorithm,53 while also integrating a maximum likelihood framework and an accelerated branch-and-bound alignment algorithm. Due to ambiguities in determining the correct symmetry, as described below, all possible symmetries were tested. Applying the correct symmetry (0.692 Å rise and 153.94° twist) resulted in a high-resolution map of the CarbIF micelle, revealing well-defined density for both the peptide backbone and side chains (Fig. 2A). The resultant map allowed atomic model building for the CarbIF micelle. Since the hand determination of the map was ambiguous, as described below, we arbitrarily selected the left-handed map for model building. The CarbIF micelle is a hollow tube with a lumen diameter of ∼4 nm and an outer diameter of ∼9 nm, with a double-layered wall ∼2.5 nm thick. The tube has dipeptides arranged along seven strands (Fig. 2A and B). The asymmetric unit of five CarbIF peptides stacks to form each strand, with two forming the inner layer and three forming the outer layer of the wall. Notably, all five peptides exhibit distinct conformations, consistent with the expected flexibility in the CarbIF geometry (Fig. 2D). Structural analysis suggests that the packing of the micelle is primarily driven by hydrophobic interactions between the aromatic rings of the CarbIF subunits. These aromatic rings within the strands show well-ordered stacking (5.0 ± 0.2 Å) in both the inner and outer strands (Fig. 2E). The association of these strands is further aided by hydrophobic interactions, where the aromatic rings are buried between the two layers of the wall, exposing the polar atoms on the inner and outer surfaces of the micelle (Fig. 2F). Since the hydrophobic interactions are inherently non-specific, their role in the assembly of CarbIF raises the question of what contributes to the micelle's ordered structure. Structural analysis suggests that, while the assembly is predominantly driven by hydrophobic forces, each dipeptide makes two hydrogen bonds with a neighboring dipeptide (Fig. 2C) with their amide nitrogens and carbonyl oxygens. These interactions are likely crucial for maintaining the order within the micelle.
We then sought to understand how the CarbIF micelle adopts the straight, rigid morphology and remains in an unaggregated state at high pH, but becomes flexible and aggregates at lower pH to form a supramolecular gel. The atomic structure of CarbIF provides insights into this behavior. The C-terminal carbonyl group of the dipeptide is exposed on both the outer and inner surfaces of the micelle (Fig. 2F). At high pH, this carbonyl group tends to be negatively charged due to deprotonation, which renders the micelle surface highly negative. The charge repulsion between carbonyl groups likely favors the straight, rigid morphology of CarbIF as observed for other polymers54–56 and further inhibits aggregation. However, at low pH, the increased proton concentration would make the carbonyl neutral, promoting aggregation and formation of the supramolecular gel. This gelation model aligns well with previous observations for a related system38,48 that, upon a decrease in pH, CabIF micelles transform into non-hollow flexible cylinders with a radius of approximately ∼4.9 nm. At lower pH, the carbonyl groups in the lumen of the CabIF micelle form strong ion–dipole interactions between RCOO− and RCOOH, causing the collapse of the hollow lumen.
Challenges in Cryo-EM of CarbIF micelles and similar systems
While the Cryo-EM helical reconstruction for most protein polymers, like bacterial flagella, pilins, and bacteriophage tails, is relatively simple, it is more challenging for helical structures made up of small peptides, like supramolecular gel-forming structures. The main challenge is due to ambiguities in helical symmetry57 and the increased number of symmetries that may appear to be correct at high resolution.51 The power spectrum of a protein polymer such as F-actin or a cytochrome nanowire can be completely unambiguous and have only a single possible symmetry.50,58 This is not the case for short peptides, which typically generate power spectra with many possible symmetries.59 The CarbIF micelle can have up to 24 potential symmetries from analysis of its power spectrum (Fig. 1D). By measuring the radius (r) of the CarbIF micelle from the 2D class average (Fig. 1C) and calculating the ‘R’ value (the distance from the meridian to the first peak of the layer line in the power spectrum, shown in Fig. 1D), the argument x for a Bessel function Jn(x) can be determined using the formula x = 2πRr for layer lines ll1 (at 1/(110 Å)) and ll7 (at 1/(4.8 Å)) (Fig. 1D). It is important to note that the radius, r, is not fixed due to the finite thickness of the tube walls. Considering a range of r values reveals that ll1 may correspond to Bessel orders 4, 5, 6, or 7, while ll7 may correspond to orders 1, 2, or 3, leading to at least 12 distinct combinations (4 × 3). When accounting for the sign of the Bessel orders, where each order can be either positive or negative (corresponding to a right- or left-handed helix, respectively), the total number of possibilities increases to 48. Since the absolute handedness cannot be determined from the power spectrum, there is no need to calculate, for example, both 7/7 and −7/−7 for ll1/ll7, as they represent mirror images. Assuming the Bessel order for ll1 is positive, this results in 24 possible helical symmetries, including combinations such as Bessel orders 7/3, 7/−3, 7/2, 7/−2, etc. for ll1/ll7. CarbIF is not the most complex case, as its smaller diameter limits the number of possible symmetries. Larger structures, like the LD-tube reported previously,59 can have even more potential symmetries. Although many of these potential symmetries have closely related rise and twist values (Fig. 3A), determining the correct symmetry requires testing all possibilities discretely, as typically the algorithm becomes locked into a starting symmetry and will never evolve to a different symmetry. Furthermore, the reconstructed map requires careful manual inspection, as incorrect symmetries can yield maps that appear correct at first glance. Fig. 3 illustrates two CarbIF micelle maps, reconstructed with the correct symmetry and an incorrect (but closely related) symmetry. The map with the incorrect symmetry still results in an estimated resolution of 3.5 Å by cryoSPARC. At first glance, this incorrect symmetry appears correct due to the well-separated strands visible from the top view. However, closer inspection of the structure reveals that the peptide backbone is poorly resolved (Fig. 3C). Additionally, there are vertical connections visible in a side view of the structure that should be resolved at a resolution better than 4 Å. In contrast, the map generated with the correct symmetry shows well-defined separation of the peptide backbone and sidechains (Fig. 3B). Part of the problem is that the measure of resolution used in cryo-EM, the Fourier Shell Correlation (FSC), is not really a measure of resolution, but rather of reproducibility.60 The measure of reproducibility comes from dividing the data set into two halves, independently reconstructing the two half sets, and comparing these two reconstructions in Fourier space to look at their correlation as a function of resolution. Thus, one can impose the same wrong symmetry on the two half maps, and they will show a strong correlation at some high resolution, but the maps themselves are useless. An alternative, independent method for assessing resolution is using a map:model FSC (Table 1), rather than the map:map FSC, which provides more of a reality-based metric for actually estimating the true resolution. | ||
| Fig. 3 (A) Possible helical symmetry parameters for the CarbIF micelle calculated from the power spectrum shown in Fig. 1(D). Cn in parentheses indicates the relevant point group symmetry if one is present. (B and C) Cryo-EM maps of CarbIF generated by imposing the correct (B) and wrong (C) symmetry parameters. Strands are separated in both maps, while the peptide backbone within a strand is resolved well only in the correct map. | ||
| EMBD entry | EMD-45610 |
| PDB file | ESI file |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 50 |
| Pixel size (Å per pixel) | 1.08 |
| Particle images (no.) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 516 516![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 102 |
| Helical parameters (axial rise/twist) | 0.692 Å/153.94° |
| Map:map FSC (0.143) resolution | 3.2 Å |
| Model:map FSC (0.5) resolution | 3.4 Å |
| Model vs. map CC (mask) | 0.90 |
The initial symmetry parameters derived from the power spectrum require further refinement. This is typically achieved through local symmetry searches during each iteration, using the volume generated from the previous iteration of the reconstruction. Starting the local symmetry search after the map reaches ∼6 Å is effective for many biological structures, as they typically contain medium-resolution information due to the presence of protein secondary structures such as α-helices and β-sheets. To illustrate this, we performed a helical reconstruction of the type 1 pilus by imposing an initial helical symmetry of −114.894° twist/8.557 Å rise, which deviated significantly from the correct values of −115.01° twist/7.848 Å rise. With the local symmetry search initiated at 6 Å, the search converged to the correct symmetry, leading to a map with 3.5 Å resolution (Fig. 4A and B). In contrast, such a test for the CarbIF micelle failed. The symmetry parameters did not converge from 153.80° twist/0.685 Å rise to 153.94° twist/0.692 Å rise, despite the relatively small distance from the starting parameters (Fig. 4C and D). This is likely due to the lack of medium-resolution details at 6 Å in the CarbIF micelle, which are essential for guiding the local symmetry search. Therefore, the optimal point at which to initiate the local symmetry search is also a critical factor to test during the helical reconstruction of helical structures formed from small peptides.
Can prior knowledge of the subunit size be used to aid in determining the correct symmetry? The observation of five conformationally different peptides in the asymmetric unit of CarbIF micelles suggests that this approach may not be feasible. CarbIF is not the only case. It was assumed from high-resolution X-ray fiber diffraction that the Lanreotide octapeptide assembled into tubes with two peptides per asymmetric unit.61 A cryo-EM study showed that the actual asymmetric unit in these tubes contained eight peptides, in eight different conformations.62 A dipeptide (L,L)-2NapFF was shown to form an LL-tube with three peptides in the asymmetric unit,59 while it might have been imagined that the tube would contain two peptides, one forming the inner wall and the other forming the outer wall. Additionally, small α-helical peptides have been shown to assemble into nanotubes with multiple peptides per asymmetric unit.63 Such cases are not limited to the peptide polymers, but also extend to protein polymers: archaeal pili have been found with three subunits in the helical asymmetric unit,64,65 archaeal flagellar filaments with six subunits,66 and bacterial flagellar filaments with two or four subunits.67 Assuming in each of these cases that the asymmetric unit would contain a single subunit would lead one to the wrong symmetry.
It is important to note that cryo-EM images do not retain information about the handedness of the structure, resulting in a 50% probability that the Cryo-EM map will exhibit the incorrect hand.51 Additional information is therefore needed to determine the correct hand of the map. For protein polymers, the correct hand can typically be determined by looking at an α-helix, which we know is right-handed. However, this approach may be problematic for structures lacking α-helices, such as cross-β structures and supramolecular gel-forming assemblies built from small peptides. One possible method for identifying the correct hand in these cases involves model building in both possible maps and evaluating which map best accommodates the model, as previously described.62,68 However, this approach typically requires high resolution (better than 2.8 Å) and may not yield definitive results for very short peptides or other small chemical building blocks commonly used in supramolecular gelation, as they can fit equally well in both the correct map and its mirror image. Atomic force microscopy (AFM), which looks at the surface of a structure, offers a means for determining the correct hand.69 Other techniques that visualize surfaces, such as scanning EM (SEM) or metal-shadowed EM, may also be useful but are problematic if the resolution is not sufficient to determine the hand of helical features.
Experimental
Synthesis of CarbIF
The synthesis of CarbIF was carried out as described elsewhere.70Preparation of CarbIF micelles
CarbIF (500 mg) was dispersed in deionized water. A 0.1 M sodium hydroxide solution (NaOH, Sigma Aldrich) was then added in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio to the CarbIF suspension, resulting in a final volume of 50 mL. The mixture was stirred at 1000 rpm overnight to facilitate the complete dissolution of the gelator. Once all the solid material had dissolved, a viscous, free-flowing solution was formed. The pH of the solution was measured and adjusted to 11 ± 0.1 using either 1 M NaOH or 1 M HCl as required.
1 molar ratio to the CarbIF suspension, resulting in a final volume of 50 mL. The mixture was stirred at 1000 rpm overnight to facilitate the complete dissolution of the gelator. Once all the solid material had dissolved, a viscous, free-flowing solution was formed. The pH of the solution was measured and adjusted to 11 ± 0.1 using either 1 M NaOH or 1 M HCl as required.
Vitrification of sample on a copper grid for cryo-electron microscopy
A 3 μL aliquot of the CarbIF micelle suspension was applied to glow-discharged lacey carbon grids. Excess sample was removed by blotting from both sides of the grid, leaving behind a thin film. The grid was then plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). The frozen grids were stored in liquid nitrogen until they were used for cryo-EM imaging.Cryo-electron microscopy
Conclusions
This study demonstrates the potential of cryo-EM as a powerful tool for determining the atomic structure of supramolecular gels, as exemplified by the cryo-EM structure of CarbIF micelles, revealing the mechanism of assembly and gelation. However, challenges remain, particularly in determining helical symmetry, which can be a complex aspect of the cryo-EM-based structural analysis of such polymers. By overcoming these hurdles, cryo-EM could play a transformative role understanding the atomic structure and packing of the supramolecular gel-forming materials and guide the rational design of novel gelators with tailored properties.Data availability
The Cryo-EM map of CarbIF is available at the electron microscopy database (EMD-45610). The atomic model for the CarbIF micelle is provided as ESI.†Author contributions
DJA and EHE conceived the idea. SB prepared the specimens. RRS imaged the samples, performed the analysis and reconstruction, and built the atomic model. EHE and DJA obtained the funding. RRS, MABK and EHE wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
EHE was funded by NIH grant GM122510. SB thanks the University of Glasgow for funding. Cryo-EM was performed at the University of Virginia's Molecular Electron Microscopy Core, supported by NIH grants G20-RR31199, SIG S10-RR025067 and U24-GM116790.Notes and references
- N. M. Sangeetha and U. Maitra, Chem. Soc. Rev., 2005, 34, 821–836 RSC.
- W. Fang, Y. Zhang, J. Wu, C. Liu, H. Zhu and T. Tu, Chem.–Asian J., 2018, 13, 712–729 CrossRef CAS PubMed.
- S. Panja, A. Panja and K. Ghosh, Mater. Chem. Front., 2021, 5, 584–602 RSC.
- X. Cao, A. Gao, J.-t. Hou and T. Yi, Coord. Chem. Rev., 2021, 434, 213792 CrossRef CAS.
- D. K. Smith, Soft Matter, 2024, 20, 10–70 RSC.
- K. Tao, H. Wu, L. Adler-Abramovich, J. Zhang, X. Fan, Y. Wang, Y. Zhang, S. A. Tofail, D. Mei and J. Li, Prog. Mater. Sci., 2024, 142, 101240 CrossRef CAS.
- D. Tripathy, A. S. Gadtya and S. Moharana, Polym.–Plast. Technol. Mater., 2023, 62, 306–326 CAS.
- C. D. Jones and J. W. Steed, Chem. Soc. Rev., 2016, 45, 6546–6596 RSC.
- G. Yu, X. Yan, C. Han and F. Huang, Chem. Soc. Rev., 2013, 42, 6697–6722 RSC.
- D. A. Middleton, J. Madine, V. Castelletto and I. W. Hamley, Angew. Chem., Int. Ed., 2013, 52, 10537–10540 CrossRef CAS PubMed.
- L. Majumder, K. Bera, K. Khamaru, U. Pal, N. C. Maiti and B. Banerji, J. Mol. Struct., 2022, 1266, 133455 CrossRef CAS.
- H. Jiang, M. O. Guler and S. I. Stupp, Soft Matter, 2007, 3, 454–462 RSC.
- M. Cao, S. Lu, W. Zhao, L. Deng, M. Wang, J. Wang, P. Zhou, D. Wang, H. Xu and J. R. Lu, ACS Appl. Mater. Interfaces, 2017, 9, 39174–39184 CrossRef CAS PubMed.
- C. R. MacDonald and E. R. Draper, Beilstein J. Org. Chem., 2024, 20, 2608–2634 CrossRef CAS PubMed.
- T. Allam, D. E. Balderston, M. K. Chahal, K. L. F. Hilton, C. K. Hind, O. B. Keers, R. J. Lilley, C. Manwani, A. Overton, P. I. A. Popoola, L. R. Thompson, L. J. White and J. R. Hiscock, Chem. Soc. Rev., 2023, 52, 6892–6917 RSC.
- X.-C. Bai, G. McMullan and S. H. Scheres, Trends Biochem. Sci., 2015, 40, 49–57 CrossRef CAS PubMed.
- E. H. Egelman, Biophys. J., 2016, 110, 1008–1012 CrossRef CAS PubMed.
- W. Chiu and K. H. Downing, Curr. Opin. Struct. Biol., 2017, 46, iv CrossRef PubMed.
- E. Nogales and S. H. Scheres, Mol. Cell, 2015, 58, 677–689 CrossRef CAS PubMed.
- W. Kühlbrandt, Science, 2014, 343, 1443–1444 CrossRef PubMed.
- J. Dubochet, M. Adrian, J.-J. Chang, J.-C. Homo, J. Lepault, A. W. McDowall and P. Schultz, Q. Rev. Biophys., 1988, 21, 129–228 CrossRef CAS PubMed.
- J. M. Hardy, R. A. Dunstan, T. Lithgow and F. Coulibaly, Biochem. Soc. Trans., 2022, 50, 459–422W CrossRef CAS PubMed.
- T. Fujii, A. H. Iwane, T. Yanagida and K. Namba, Nature, 2010, 467, 724–728 CrossRef CAS PubMed.
- F. Merino, S. Pospich, J. Funk, T. Wagner, F. Kullmer, H. D. Arndt, P. Bieling and S. Raunser, Nat. Struct. Mol. Biol., 2018, 25, 528–537 CrossRef CAS PubMed.
- R. Zhang, G. M. Alushin, A. Brown and E. Nogales, Cell, 2015, 162, 849–859 CrossRef CAS PubMed.
- G. M. Alushin, G. C. Lander, E. H. Kellogg, R. Zhang, D. Baker and E. Nogales, Cell, 2014, 157, 1117–1129 CrossRef CAS PubMed.
- E. H. Egelman, Curr. Opin. Struct. Biol., 2017, 46, 31–37 CrossRef CAS PubMed.
- C. K. Park and N. C. Horton, Nat. Rev. Mol. Cell Biol., 2020, 21, 1–2 CrossRef CAS PubMed.
- C. K. Park and N. C. Horton, Biophys. Rev., 2019, 11, 927–994 CrossRef PubMed.
- K. L. Hvorecny and J. M. Kollman, Curr. Opin. Struct. Biol., 2023, 79, 102530 CrossRef CAS PubMed.
- E. M. Lynch, J. M. Kollman and B. A. Webb, Curr. Opin. Cell Biol., 2020, 66, 28–33 CrossRef CAS PubMed.
- N. Pillay, L. Mariotti, M. Zaleska, O. Inian, M. Jessop, S. Hibbs, A. Desfosses, P. C. R. Hopkins, C. M. Templeton, F. Beuron, E. P. Morris and S. Guettler, Nature, 2022, 612, 162–169 CrossRef CAS PubMed.
- A. W. Fitzpatrick and H. R. Saibil, Curr. Opin. Struct. Biol., 2019, 58, 34–42 CrossRef CAS PubMed.
- E. H. Egelman, eLife, 2014, 3, e04969 CrossRef PubMed.
- E. H. Egelman, Ultramicroscopy, 2000, 85, 225–234 CrossRef CAS PubMed.
- S. He and S. H. Scheres, J. Struct. Biol., 2017, 198, 163–176 CrossRef CAS PubMed.
- A. Punjani, J. L. Rubinstein, D. J. Fleet and M. A. Brubaker, Nat. Methods, 2017, 14, 290–296 CrossRef CAS PubMed.
- R. R. Sonani, S. Bianco, B. Dietrich, J. Doutch, E. R. Draper, D. J. Adams and E. H. Egelman, Cell Rep. Phys. Sci., 2024, 5, 101812 CrossRef CAS PubMed.
- A. Bigo-Simon, L. F. Estrozi, A. Chaumont, R. Schurhammer, G. Schoehn, J. Combet, M. Schmutz, P. Schaaf and L. Jierry, ACS Nano, 2024, 18, 30448–30462 CrossRef CAS PubMed.
- L. Pieri, F. Wang, A.-A. Arteni, M. Vos, J.-M. Winter, M.-H. Le Du, F. Artzner, F. Gobeaux, P. Legrand and Y. Boulard, Proc. Natl. Acad. Sci., 2022, 119, e2120346119 CrossRef PubMed.
- F. Wang, O. Gnewou, S. Wang, T. Osinski, X. Zuo, E. H. Egelman and V. P. Conticello, Matter, 2021, 4, 3217–3231 CrossRef CAS PubMed.
- M. Tirrell, Proc. Natl. Acad. Sci., 2022, 119, e2123197119 CrossRef CAS PubMed.
- F. Wang, O. Gnewou, C. Modlin, L. C. Beltran, C. Xu, Z. Su, P. Juneja, G. Grigoryan, E. H. Egelman and V. P. Conticello, Nat. Commun., 2021, 12, 407 CrossRef CAS PubMed.
- E. Gazit, Annu. Rev. Biochem., 2018, 87, 533–553 CrossRef CAS PubMed.
- X. Liu, Q. Jiang, Y. Yin and G. Liang, Chem & Bio Engineering, 2024, 1, 664–677 Search PubMed.
- D. M. Ryan and B. L. Nilsson, Polym. Chem., 2012, 3, 18–33 RSC.
- S. Fleming and R. V. Ulijn, Chem. Soc. Rev., 2014, 43, 8150–8177 RSC.
- L. J. Marshall, S. Bianco, R. E. Ginesi, J. Doutch, E. R. Draper and D. J. Adams, Soft Matter, 2023, 19, 4972–4981 RSC.
- E. H. Egelman, Curr. Opin. Struct. Biol., 2024, 85, 102788 CrossRef CAS PubMed.
- M. A. B. Kreutzberger, R. R. Sonani and E. H. Egelman, Q. Rev. Biophys., 2024, 57, e16 CrossRef CAS PubMed.
- F. Wang, O. Gnewou, A. Solemanifar, V. P. Conticello and E. H. Egelman, Chem. Rev., 2022, 122, 14055–14065 CrossRef CAS PubMed.
- E. H. Egelman, Methods Enzymol., 2010, 482, 167–183 CAS.
- E. H. Egelman, in Methods in enzymology, Elsevier, 2010, vol. 482, pp. 167–183 Search PubMed.
- J. Skolnick and M. Fixman, Macromolecules, 1977, 10, 944–948 CrossRef CAS.
- T. Odijk, J. Polym. Sci., Part B:Polym. Phys., 1977, 15, 477–483 CAS.
- R. R. Sonani, L. K. Palmer, N. C. Esteves, A. A. Horton, A. L. Sebastian, R. J. Kelly, F. Wang, M. A. Kreutzberger, W. K. Russell and P. G. Leiman, Nat. Commun., 2024, 15, 756 CrossRef CAS PubMed.
- E. H. Egelman, eLife, 2014, 3, e04969 CrossRef PubMed.
- D. P. Baquero, V. Cvirkaite-Krupovic, S. S. Hu, J. L. Fields, X. Liu, C. Rensing, E. H. Egelman, M. Krupovic and F. Wang, Cell, 2023, 186, 2853–2864 CrossRef CAS PubMed.
- R. R. Sonani, S. Bianco, B. Dietrich, J. Doutch, E. R. Draper, D. J. Adams and E. H. Egelman, Cell Rep. Phys. Sci., 2024, 5, 101812 CrossRef CAS PubMed.
- S. Subramaniam, L. A. Earl, V. Falconieri, J. L. Milne and E. H. Egelman, Curr. Opin. Struct. Biol., 2016, 41, 194–202 CrossRef CAS PubMed.
- C. Valery, M. Paternostre, B. Robert, T. Gulik-Krzywicki, T. Narayanan, J. C. Dedieu, G. Keller, M. L. Torres, R. Cherif-Cheikh, P. Calvo and F. Artzner, Proc. Natl. Acad. Sci. U.S.A., 2003, 100, 10258–10262 CrossRef CAS PubMed.
- L. Pieri, F. Wang, A. A. Arteni, M. Vos, J. M. Winter, M. H. Le Du, F. Artzner, F. Gobeaux, P. Legrand, Y. Boulard, S. Bressanelli, E. H. Egelman and M. Paternostre, Proc. Natl. Acad. Sci. U.S.A., 2022, 119, e2120346119 CrossRef PubMed.
- M. A. B. Kreutzberger, S. Wang, L. C. Beltran, A. Tuachi, X. Zuo, E. H. Egelman and V. P. Conticello, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2121586119 CrossRef CAS PubMed.
- J. Liu, G. N. Eastep, V. Cvirkaite-Krupovic, S. T. Rich-New, M. A. B. Kreutzberger, E. H. Egelman, M. Krupovic and F. Wang, Nat. Commun., 2024, 15, 5049 CrossRef CAS PubMed.
- M. C. Gaines, S. Sivabalasarma, M. N. Isupov, R. U. Haque, M. McLaren, C. Hanus, V. A. M. Gold, S. V. Albers and B. Daum, Nat. Commun., 2024, 15, 5050 CrossRef CAS PubMed.
- L. Gambelli, M. N. Isupov, R. Conners, M. McLaren, A. Bellack, V. Gold, R. Rachel and B. Daum, Nat. Commun., 2022, 13, 710 CrossRef CAS PubMed.
- M. A. B. Kreutzberger, R. C. Sobe, A. B. Sauder, S. Chatterjee, A. Pena, F. Wang, J. A. Giron, V. Kiessling, T. R. D. Costa, V. P. Conticello, G. Frankel, M. M. Kendall, B. E. Scharf and E. H. Egelman, Nat. Commun., 2022, 13, 1422 CrossRef CAS PubMed.
- J. Guo, F. Wang, Y. Huang, H. He, W. Tan, M. Yi, E. H. Egelman and B. Xu, Nat. Nanotechnol., 2023, 18, 1094–1104 CrossRef CAS PubMed.
- D. M. Marini, W. Hwang, D. A. Lauffenburger, S. Zhang and R. D. Kamm, Nano Lett., 2002, 2, 295–299 CrossRef CAS.
- J. K. Gupta, D. J. Adams and N. G. Berry, Chem. Sci., 2016, 7, 4713–4719 RSC.
- J. He, T. Li and S.-Y. Huang, Nat. Commun., 2023, 14, 3217 CrossRef CAS PubMed.
- P. V. Afonine, B. K. Poon, R. J. Read, O. V. Sobolev, T. C. Terwilliger, A. Urzhumtsev and P. D. Adams, Acta Crystallogr., Sect. D:Struct. Biol., 2018, 74, 531–544 CrossRef CAS PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D:Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- T. D. Goddard, C. C. Huang, E. C. Meng, E. F. Pettersen, G. S. Couch, J. H. Morris and T. E. Ferrin, Protein Sci., 2018, 27, 14–25 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fd00181h |
| This journal is © The Royal Society of Chemistry 2025 |