Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Probiotics selection for low-lactose fermented goat milk products
Mariia Antsyperova ,
Tamara Arseneva
,
Tamara Arseneva ,
Aleksei Fedorov
,
Aleksei Fedorov ,
Elena Lemeshonok,
Lyudmila Zabodalova and
Denis Baranenko
,
Elena Lemeshonok,
Lyudmila Zabodalova and
Denis Baranenko *
*
International Research Centre “Biotechnologies of the Third Millennium”, Faculty of Biotechnologies (BioTech), ITMO University, Lomonosova str., 9, Saint-Petersburg, 191002, Russia. E-mail: denis.baranenko@itmo.ru; Tel: +7-9219343335
First published on 3rd December 2025
Abstract
The consumption of fermented goat milk products can reduce the risk of developing various non-communicable diseases, which is critical for people with lactose intolerance living in low- and middle-income countries. The present study aims to determine the most appropriate probiotic microorganism for effective low-lactose goat milk fermentation. Twelve industrial strains were evaluated for their ability to efficiently ferment lactose residues, free galactose, and glucose. The strains were evaluated for technological and kinetic parameters of biomass cultivation in low-lactose goat milk obtained by β-galactosidase processing. The comparative analysis revealed the needed carbohydrate fermentation ability of Bifidobacterium bifidum strains BB01 and AC-1579; they both demonstrated the fastest carbohydrate consumption onset of 6.5 and 5.5 h, respectively, in the following order: lactose, galactose, and glucose. Carbohydrate metabolism in the other studied B. bifidum strains started significantly later, not earlier than 11.5 h, and proceeded more slowly. The highest specific growth rate of 0.81 h−1 was found for B. bifidum BB01. B. bifidum BB01 exhibited the highest fermentation rate to a pH of 4.94 within 7 h and a significant increase in biomass of 2.47 log10CFU ml−1, providing more than 109 viable probiotic cells in 1 ml of the fermented product. Other strains fermented milk to a pH of 4.59–4.97 within 11–14 h. The product was safe after refrigerated storage at 4 ± 2 °C for 28 days with 99.5% of viable probiotic cells remaining (9.17 log10CFU ml−1). The resulting product is widely available and may be highly recommended for people with lactose intolerance.
Sustainability spotlightOur work aligns with the UN's Sustainable Development Goals, specifically targeting good health and well-being through improved nutritional access and sustainable consumption patterns. The detailed analysis of probiotic strains facilitates the creation of novel functional products with significant health benefits, implementing advancements in bioeconomy within the locally available small-scale dairy producers. Furthermore, the identification of efficient starter cultures for low-lactose goat milk fermentation is needed for expanding the availability of functional dairy products to lactose-intolerant populations. The focus on efficient fermentation also aids in maintaining probiotic viability throughout processing and storage, which is critical in developing stable dairy-based probiotic foods. Additionally, increasing process intensiveness and reducing production duration contribute to achieving the tasks of sustainable production goals. |
1 Introduction
Probiotic-enriched food products are known to positively affect the digestive system by restoring intestinal microflora, which is crucial for human health. Probiotics can significantly benefit human health by enhancing resistance to cancer, boosting immune function, offering protection against allergies, and positively influencing cholesterol levels in the blood.1–5 The review of observational cohort studies showed that one of the possible ways to reduce the risk of developing type 2 diabetes mellitus may be the consumption of fermented milk products.6 The consumption of fermented dairy products is associated with the decrease in food intake and increase in satiety, improvement of glycemic and insulin resistance, altered gut hormone response, substitution of less healthy foods, change in gut microbiota, and an increase in body fat reduction.7 Fermented dairy products may be particularly beneficial when using goat milk, as they were shown to improve glucose homeostasis, pancreatic conditions and insulin sensitivity.8,9 Possible mechanisms for the antidiabetic effect of goat milk include activation of pancreatic protein kinase B (AKT) and hepatic and skeletal muscle adenosine monophosphate activated protein kinase (AMPK).10,11 In addition, goat breeding is currently popular worldwide in low-income countries and in dry areas.Although milk is a staple food in many countries12 lactose intolerance affects 68% of the global population, including low- and middle-income countries (LMICs).13 At the same time, self-perceived lactose-intolerant respondents had a significantly higher rate of diagnosed diabetes mellitus.14 Thus, it seems relevant to enrich the human diet with low-lactose fermented products based on goat milk. Producing lactose-free and low-lactose dairy products involves several methods designed to reduce or eliminate lactose content while maintaining the nutritional and sensory properties of the food. The physical removal of lactose from milk is achieved with membrane filtration techniques such as ultrafiltration (UF) and nanofiltration (NF); however, these require high-tech and expensive equipment.15 Enzymatic hydrolysis is a common method involving adding β-galactosidase enzyme to milk or milk products and breaking down lactose into glucose and galactose which are easier to metabolize.16 The use of fermentation can also reduce lactose content by adding lactic acid bacteria and bifidobacteria fermenting lactose into lactic acid. Probiotic lactic acid bacteria and bifidobacteria in fermented milk products improve lactose digestion and eliminate symptoms of intolerance in lactose maldigesters. Active microbial β-galactosidase in fermented milk products survives gastric passage and is released by bile salts into the small intestine, where it supports lactose digestion.17
Despite the developments in the use of probiotics in goat yogurt and fermented dairy drinks, it is difficult to predict which strains will be the most effective.18 Not much research has been conducted on the selection of strains for the production of a fermented drink based on low-lactose goat milk. Pawlos et al. determined a significantly lower amount (about 0.3 log10CFU g−1) of Bifidobacterium animalis ssp. lactis Bb-12 in low-lactose fermented milk in comparison to the control sample. The lactose hydrolysis of goat milk resulted in a higher hardness and more distinct sweet and goaty taste than the control.19 Araújo et al. developed a lactose-free goat milk beverage with bioactive rich jambo pulp using Lacticaseibacillus paracasei BGP-1, Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus.20 Viable probiotic Lacticaseibacillus paracasei counts reduced significantly from 8.23–8.58 to 7.38–7.85 log10CFU ml−1 during 28 days of refrigerated storage. The global acceptance decreased significantly over the storage time and was satisfactory during the 21 days of storage. However, the information on the choice of microorganism strains for the production of probiotic low-lactose fermented goat milk products is still scarce.
Probiotics in fermented dairy products, for example yogurt, can interact with starter cultures, affecting their survival and activeness in the product.21,22 When probiotics are added to fermented foods, there are several factors to consider, as they may influence the ability of probiotics to survive in a product and become active when entering the consumer's gastrointestinal tract due to possible synergetic or antagonistic interactions of probiotics with the starter cultures. The interactions of probiotics with the food matrix may be even more intensive when probiotics are used as a component of the starter culture.23 Co-inoculation with yogurt cultures may suppress the initial growth of bifidobacteria but does not significantly affect their survival during storage.24 It is worth noting that a possible solution is to add a free or preferably encapsulated form of probiotic microorganisms in the required quantity to the product after fermentation.25 However, this method requires more complex technological equipment and also increases the overall requirements for the enterprise level, which may be difficult within the framework of organizing local sustainable production in LMICs. Another approach is to use a single microorganism, which provides a stable and controlled fermentation process, allowing for high levels of active bacteria in the product, as well as predictable high product quality characteristics. Co-cultivation with traditional yogurt starters can indeed create a competitive environment that often suppresses the growth of B. bifidum during fermentation due to the higher acidification rate of yogurt starters and competition for nutrients.26 Monoculture fermentation avoids this antagonistic effect. Furthermore, and most critically for a probiotic product, the viability of B. bifidum during storage remains stable and does not fall below the recommended probiotic threshold.27,28 The development and implementation of sustainable production protocols for probiotic fermented goat milk products will improve the quality of local sources of nutrients and thereby address adverse health outcomes, including in LMICs.
The aim of this work is to determine a probiotic microorganism for the effective fermentation of lactose residues, free galactose and glucose in low-lactose goat milk obtained by hydrolysis with β-galactosidase to ensure that the content of live probiotics is over 109 CFU ml−1 and high quality indicators of the finished product. For this purpose, 12 promising and widely available microorganisms have been evaluated for milk carbohydrate fermentation ability, kinetics of biomass, and organic acid accumulation during milk fermentation. For the selected microorganism, the physicochemical and quality indicators of the fermented beverage have been studied during refrigerated storage for 28 days.
2 Materials and methods
2.1 Materials
Goat milk was obtained from the CJSC “breeding complex Prinevskoe”, Leningrad region, Russia. The milk had the following physico-chemical characteristics: a mass fraction of fat of 3.9 ± 0.1%, protein mass fraction of 3.1 ± 0.1%, nonfat milk solids of 8.6 ± 0.1%, pH of 6.7 ± 0.05, and titratable acidity of 19.5 ± 0.1 °Т. Low-lactose goat milk was obtained by fermentation with 0.08% v/v β-galactosidase from Kluyveromyces lactis (Lactasis 6500K, LLC “Kaprina”, Russia) at a temperature of 40 °C for 4 h to a lactose content of less than 0.1%.This study used well-known and widely available in Russia probiotics Lactobacillus acidophilus strains AT-41, H9, 57S, and 8 (VKPM, Russia), Bifidobacterium bifidum strains BB01 (Alce International s.r.l., Italy), AC-1579 (VKPM, Russia), No. 1 (LLC “ProBioPharm”, Russia), BF3 DSM 29040 (LLC “BioVid”, Russia), and 791 (CJSC “Ecopolis”, Russia). In addition, we studied Streptococcus thermophilus TA 40 (DuPont Nutrition & Health, Denmark), Lactobacillus delbrueckii subsp. bulgaricus 19 (VKPM, Russia) and yogurt starter YF-L811 (Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus from Chr. Hansen Holding A/S, Denmark) commonly used with these probiotics. A yoghurt starter and individual microorganisms included in its composition were selected to evaluate the effect on lactose, and various strains of L. acidophilus and B. bifidum were used as they are promising and beneficial for the gastrointestinal tract when used in the manufacture of functional foods. All microorganisms were obtained in a pure lyophilized form.
For the cultivation of microorganisms, nutrient media produced by HiMedia (India) were used. The ingredient concentrations of the MRS-broth were as follows (g l−1): proteose peptone – 10.0, HM peptone B# – 10.0, yeast extract – 5.0, dextrose (glucose) – 20.0, polysorbate 80 (Tween 80) – 1.0, ammonium citrate – 2.0, sodium acetate – 5.0, magnesium sulphate – 0.1, manganese sulphate – 0.05, and dipotassium hydrogen phosphate – 2.0. The ingredient concentrations of the Bifidobacterium-broth were as follows (g l−1): tryptone – 20.0, yeast extract – 10.0, peptone – 10.0, dextrose (glucose) – 20.0, tomato juice (solids) – 16.65, polysorbate 80 (Tween 80) – 2.0, and agar – 1.0. The ingredient concentrations of the M17 medium were as follows (g l−1): peptone – 2.5, tryptone – 2.5, soya peptone – 5.0, yeast extract – 2.5, HM peptone B# – 5.0, lactose – 5.0, ascorbic acid – 0.5, and disodium-β-glycerophosphate – 19.0.
2.2 Methods
To determine the fermentation ability of milk carbohydrates, the prepared microorganism concentrates at an amount of 5 µL were introduced into 10 ml of the nutrient medium stained with bromocresol purple (20 mg l−1) and incubated for 72 h at the temperature specific to each strain similar to that in Section 2.1. The ability to ferment carbohydrates was determined by the colour change, and the pH level was measured at the same time.29,30
To identify the beginning of carbohydrate intake of B. bifidum strains, the prepared B. bifidum concentrates in the amount of 0.2 µL were introduced into 0.2 ml of the prepared nutrient medium and placed in a 96-well plate. The plates were incubated in the plate reader SPECTROstar Nano (BMG LABTECH, Germany) for 72 h at a temperature of 37 °C. The optical density was determined at 600 nm every 30 min. The beginning of carbohydrate intake was determined by identifying the first time point at which the optical density value is significantly different from the initial value.
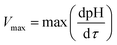 | (1) |
The duration at which the maximum acidification rate was observed (τm) and the duration at which the pH of 5.0 was reached (τe) were considered responses that characterized the process kinetics.34
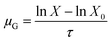 | (2) |
The doubling time (td, h) of the concentration of colony-forming units:
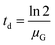 | (3) |
The multiplication rate (MR):
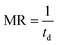 | (4) |
| Attribute | Definition (from none to intensive) |
|---|---|
| Fermented flavour | The taste stimulated by lactic acid |
| Sweet flavour | The taste stimulated by milk carbohydrates |
| Goaty flavour | Animal-like, lingering, associated with a sharp taste; caprylic acid |
| Fermented odour | The intensity of odour associated with sour milk, i.e. lactic acid |
| Goaty odour | Animal-like, lingering, associated with a harsh odour; caprylic acid |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.37,38 The gastric juice model consisted of 0.9% saline with pepsin added to a content of 2000 U ml−1 and hydrochloric acid to pH = 2.5. The sample was maintained at 37 °C and constantly stirred at 170 rpm on an orbital shaker. The survival rate (SR, %) was calculated using the next formula:
9.37,38 The gastric juice model consisted of 0.9% saline with pepsin added to a content of 2000 U ml−1 and hydrochloric acid to pH = 2.5. The sample was maintained at 37 °C and constantly stirred at 170 rpm on an orbital shaker. The survival rate (SR, %) was calculated using the next formula:
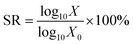 | (5) |
3 Results and discussion
3.1 Milk carbohydrate fermentation by bacteria
To assess the effect of microorganisms on the carbohydrate composition of milk, each studied microorganism was cultured in nutrient media containing one of the studied milk carbohydrates: glucose, galactose, or lactose. The studied microorganism concentrate was added to the stained nutrient medium and incubated for 72 h at the temperature specific to each strain. The milk carbohydrate fermentation ability was determined after 72 h by colour change and pH measurement. All studied microorganisms can process lactose to varying degrees (Fig. 1). This is typical for lactic acid bacteria of the species Lactobacillus and Streptococcus40,41 and also for B. bifidum.42,43 S. thermophilus TA 40, L. delbrueckii subsp.bulgaricus 19 and B. bifidum strains BB01, AC-1579, No. 1, BF3 DSM 29040, and 791 have an obvious ability to ferment glucose. L. acidophilus strains AT-41, H9, 57S, and 8 have not shown the ability to actively metabolize glucose. Some strains of L. acidophilus utilize glucose more actively than other carbohydrates41,44 and at the same time are extremely sensitive to their own acidic products of glucose utilization.45 B. bifidum strains BB01, AC-1579, No. 1, BF3 DSM 29040, and 791 actively utilize galactose, which is consistent with the results of B. bifidum carbohydrate metabolism pathway studies.42,46,47 Galactose-negative Streptococcus and Lactobacillus strains metabolize the glucose moiety and excrete galactose into the extracellular medium.40 | ||
| Fig. 1 Milk carbohydrate fermentation ability of the studied bacteria: (A) – lactose; (B) – glucose; (C) – galactose. Small letters refer to significant differences (p < 0.05). | ||
In low-lactose milk obtained by enzymatic hydrolysis, the main part of carbohydrates is monosaccharides; therefore, milk becomes sweet due to the presence of approximately 25 g l−1 of free glucose.48–50 Low-lactose milk also poses potential health risks to consumers because it contains free galactose that increases neuroinflammatory diseases, contributes to metabolic disorders, and can potentially affect the liver and kidneys.51–53 B. bifidum strains can ferment a range of carbohydrates including glucose, lactose, galactose, mannitol, and xylose, producing acetate, lactate, ethanol, and formate as fermentation products.42 The ability of the studied strains to ferment galactose is confirmed by the presence of the corresponding enzymes for B. bifidum No. 1 and 79154 and AC-1579,55 the available research on B. bifidum BF3 DSM 29040,56–58 and promotion of the growth of B. bifidum BB01 with stachyose and galactooligosaccharides.59–61 Next B. bifidum strains were selected for further study: BB01, AC-1579, No. 1, BF3 DSM 29040, and 791 as capable of fermenting both glucose and galactose in higher quantities. The strain selection allows sweetness suppression and complete fermentation of glucose and galactose in low-lactose milk.
Three of the selected strains of bifidobacteria (No. 1, BF3 DSM 29040, and 791) utilize milk carbohydrates in the following order: glucose, lactose, and galactose (Table 2). This confirms the information about the frequent accumulation of galactose during milk fermentation.62,63 However, lactose and galactose are undesirable carbohydrates in low-lactose dairy products: lactose due to lactase deficiency,62,64 and high levels of galactose lead to a number of serious health and functionality problems in some fermented dairy products.65,66 Thus, for the fermentation of low-lactose milk, it is preferable for microorganisms to utilize primarily the remains of lactose, then free galactose, and then glucose. The B. bifidum strains that consume carbohydrates in this order are BB01 and AC-1579.
| B. bifidum strain | Beginning of carbohydrate intake, h | ||
|---|---|---|---|
| Lactose | Glucose | Galactose | |
| BB01 | 6.5 | 9.0 | 8.0 |
| AC-1579 | 5.5 | 6.5 | 6.0 |
| No. 1 | 13.5 | 12.0 | 15.0 |
| BF3 DSM 29040 | 12.5 | 11.5 | 13.0 |
| 791 | 14.5 | 13.5 | 16.5 |
3.2 Obtaining an active and viable starter culture
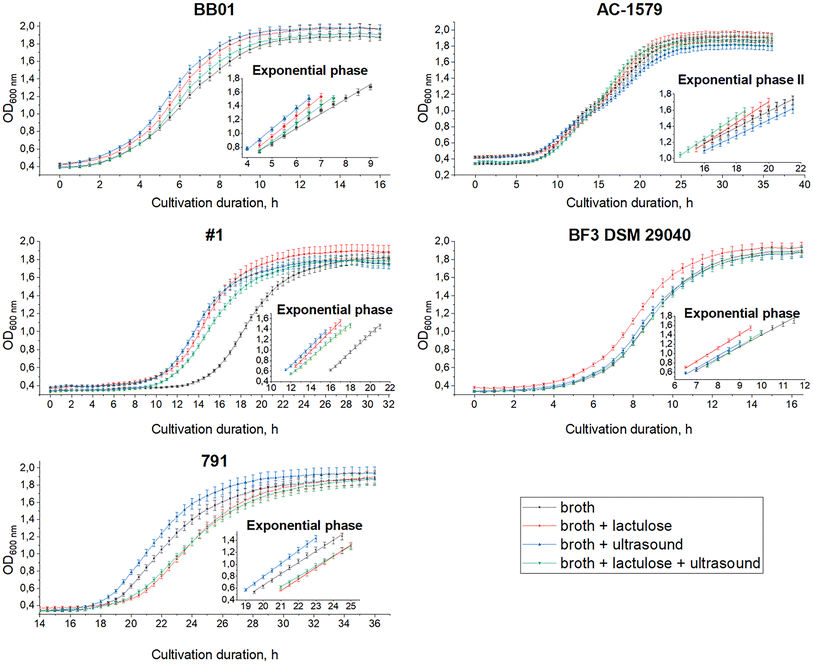
Before introduction into milk, B. bifidum cells underwent submerged cultivation at 37 °C in Bifidobacterium-broth-based media with and without additional ultrasonic treatment and with and without additional lactulose, to obtain the most active and viable starter culture.The composition and method of the nutrient medium processing were selected by comparing the kinetic curves of B. bifidum growth at optical density (OD) at 600 nm in the prepared nutrient media (Fig. 2). The identified exponential growth phases are also shown in Fig. 2. The AC-1579 strain exhibited diauxic growth, so exponential phase II data were used to identify the best conditions. The values for the best culture media (cultivation time to achieve the highest optical density) differ significantly (p < 0.05) from those of the other culture media studied.
The conditions presented in Table 3 were selected for the cultivation of B. bifidum strains. At the end of cultivation under the specified conditions, the strains were washed from the nutrient medium with a physiological solution of 0.9% NaCl and concentrated 10 times and the number of viable cells was determined. The number of viable bifidobacteria in 1 ml of the obtained microorganism concentrate was in the range from 9.75 (B. bifidum BB01) to 11.07 (B. bifidum 791).
| B. bifidum strain | Lactulose, 6.7 g l−1 | Ultrasonic processing | Duration of cultivation, h | Number of viable cells in concentrate, log10CFU ml−1 |
|---|---|---|---|---|
| BB01 | − | + | 6.5 | 9.75 ± 0.25 |
| AC-1579 | + | + | 18.5 | 9.95 ± 0.23 |
| No. 1 | + | − | 16.5 | 9.93 ± 0.24 |
| BF3 DSM 29040 | + | − | 9.5 | 10.92 ± 0.25 |
| 791 | − | + | 23.0 | 11.07 ± 0.22 |
For B. bifidum strains AC-1579, No. 1 and BF3 DSM 29040 Bifidobacterium nutrient medium with the addition of lactulose increased the number of viable cells. Lactulose is available for culture media and has properties that promote the growth of some bacteria and enhance fermentation processes.67–69
Ultrasonic treatment of the nutrient medium promoted accumulation of more biomass for B. bifidum strains BB01, AC-1579 and 791. Low frequency (20–100 kHz) and low-intensity (<1 W cm−2) ultrasonication can enhance microorganism growth and metabolite production, suggesting that it could be very beneficial for the fermentation industry.70,71 Ultrasound generates pores on the cell membrane, which provide and promote the release of intracellular enzymes from the cells through the effect of acoustic cavitation.72,73 When treating microorganisms in nutrient broth, ultrasound can not only induce the permeability of substances through the microbial membrane, but also modify the ingredients of fermentation broth to accelerate the nutrient supply, thereby promoting microbial growth.74,75 Studies suggest that nutrient media ultrasonic treatment can enhance lactic acid bacteria fermentation processes and increase peptide content and viable cell count,76–78 as well as ultrasound treatment of bifidobacteria enhances galactose utilization.72
3.3 Titratable acidity and pH of fermented goat milk
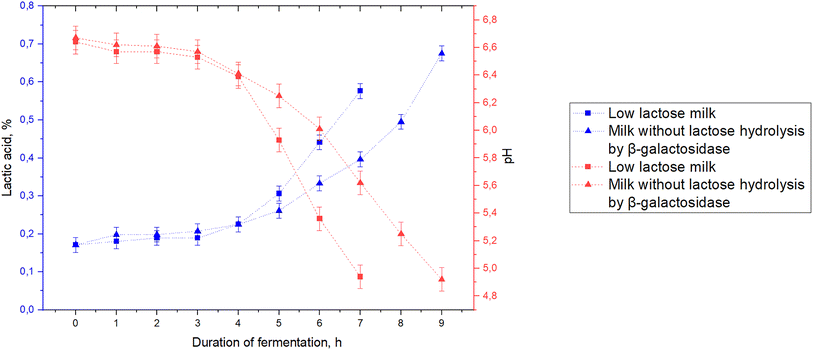
Typically, liquid starters prepared using skim milk in an amount of 5% of the volume of raw materials or lyophilized starters for direct addition are used in the fermented milk product technology.79 A B. bifidum concentrate is used to obtain a fermented milk drink based on low-lactose goat milk, since the use of starters in standard form can increase the total lactose content.80 Concentrates of each selected B. bifidum strain (BB01, AC-1579, No. 1, BF3 DSM 29040, and 791) were added to low-lactose milk in an amount of 0.1% (v/v) at a temperature of 37 ± 2 °C. Fermentation was carried out at 37 ± 1 °C to achieve the organoleptic characteristics corresponding to the decrease of the excessive sweetness of β-galactosidase fermented milk and a pH of 4.6–4.9.The graphs featuring the increase of lactic acid content and pH decrease during low-lactose milk fermentation are shown in Fig. 3. The low-lactose goat milk before the addition of bifidobacteria concentrate had a titratable acidity of 19 ± 1.24 °T (0.17 ± 0.02% lactic acid) and pH of 6.65 ± 0.07.
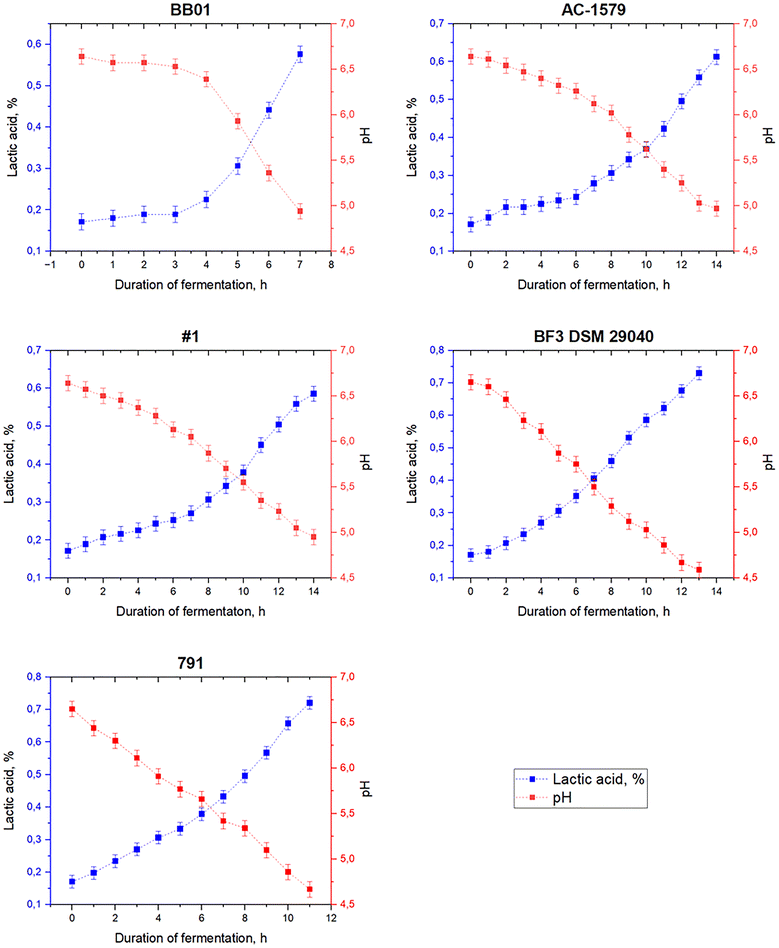 | ||
| Fig. 3 Lactic acid content and pH during low-lactose milk fermentation with B. bifidum concentrates. | ||
When low-lactose milk was fermented with B. bifidum BB01 concentrate, the active acidity reached a value of 4.94 ± 0.07 during fermentation for 7 h, while the titratable acidity was 64 ± 1.24 °T (0.58 ± 0.02% lactic acid). For strains B. bifidum AC-1579 and No. 1, the fermentation duration until the desired organoleptic characteristics were achieved was 14 h. At the end of fermentation, the product with the addition of B. bifidum AC-1579 had a pH of 4.97 ± 0.07 and the product with the addition of B. bifidum No. 1 had a pH of 4.95 ± 0.07, and titratable acidity (TA) values were 68 ± 1.24 °T (0.61 ± 0.02% lactic acid) and 65 ± 1.24 °T (0.59 ± 0.02% lactic acid), respectively. Excessive sweetness of the β-galactosidase fermented milk in the product with B. bifidum BF3 DSM 29040 concentrate was suppressed only at an active acidity of 4.59 ± 0.07 and a TA of 81 ± 1.24 °T (0.73 ± 0.02% lactic acid), which took 13 h. The product with B. bifidum 791 corresponds to the specified organoleptic characteristics after 11 h of fermentation, exhibiting a pH of 4.67 ± 0.07 and TA of 80 ± 1.24 °T (0.72 ± 0.02% lactic acid).
The obtained pH values and duration of fermentation of goat low-lactose milk by the B. bifidum strains BB01, AC-1579, No. 1, BF3 DSM 29040 and 791 are within the range of values previously known for bifidobacteria. The pH value of fermented milk and duration of fermentation vary depending on the specific Bifidobacterium strain and milk used.36,81–83 At the same time, adding the Bifidobacterium strain to the starter culture can reduce the duration of fermentation, compared to fermentation with the yogurt starter alone.82
3.4 Bifidobacterum bifidum strain growth and acidification kinetics parameters in fermented milk
Cooling to a storage temperature (4 ± 2 °C) was performed after fermentation for 1 ± 0.25 h. While cooling, residual fermentation and acid accumulation occur. The number of live B. bifidum in the products was evaluated before and after fermentation and cooling (Fig. 4).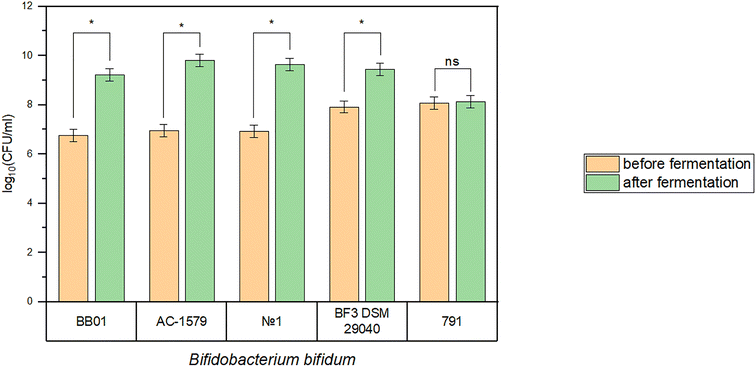 | ||
| Fig. 4 The number of live B. bifidum in the products before and after fermentation and cooling. (*) significant difference; (ns) non-significant difference (p < 0.05). | ||
The greatest increase in biomass was observed for B. bifidum strains AC-1579, No. 1 and BB01 from 6.95 to 9.80 log10CFU ml−1, from 6.93 to 9.65 log10CFU ml−1, and from 6.75 to 9.22 log10CFU ml−1, respectively. B. bifidum BF3 DSM 29040 showed an increase in the number of live bifidobacteria from 7.92 to 9.45 log10CFU ml−1. Changes in the number of B. bifidum 791 during cooling were insignificant (p > 0.05). The calculated values of biomass increase and the values of pH and TA after cooling to the storage temperature are shown in Table 4.
| B. bifidum strain | Fermentation duration, h | Biomass growth, log10CFU ml−1 | Acidity at 4 ± 2 °C | |
|---|---|---|---|---|
| pH | TA, °Т | |||
| BB01 | 7 | 2.47 ± 0.23 | 4.58 ± 0.07 | 75 ± 1.24 |
| AC-1579 | 14 | 2.85 ± 0.24 | 4.84 ± 0.07 | 76 ± 1.24 |
| No. 1 | 14 | 2.72 ± 0.24 | 4.82 ± 0.07 | 70 ± 1.24 |
| BF3 DSM 29040 | 13 | 1.54 ± 0.25 | 4.57 ± 0.07 | 75 ± 1.24 |
| 791 | 11 | Insignificant | 4.67 ± 0.07 | 80 ± 1.24 |
Some authors report that bifidobacteria are more active during goat milk fermentation compared with cow milk fermentation due to the specific composition and structure of goat milk. Namely, goat milk is distinguished by its higher content of some mineral compounds and short-chain fatty acids and better bioavailability of proteins.81,84–87 The viable cell count of bifidobacteria in fermented goat milk can reach up to 10.3 log10CFU ml−1,36,88 showing varying biomass gains.83
The growth kinetics of each B. bifidum strain was characterized by the specific growth rate, doubling time and multiplication rate (Fig. 5).
The highest specific growth rate (h−1) was found for B. bifidum BB01, almost two times lower values were found for B. bifidum strains AC-1579 and No. 1, a three times lower value was found for B. bifidum BF3 DSM 29040, and a 62 times lower value was found for B. bifidum 791. The value of the doubling time of the number of colony-forming units (h) correlates with the same values, but in the opposite direction. The multiplication rate for the studied strains is within the range from 0.019 (B. bifidum 791) to 1.172 (B. bifidum BB01), and the value of 0.676 is reached by B. bifidum AC-1579, 0.646 – by B. bifidum No. 1, and 0.392 – by B. bifidum BF3 DSM 29040.
Acidification kinetics parameters of the studied B. bifidum strains are given in Table 5. The maximum acidification rate was within the range from 2.5 unit pH min−1 by B. bifidum No. 1 to 9.5 unit pH min−1 by B. bifidum BB01. The acidification rate values of 4.0 unit pH min−1 were reached by B. bifidum strains AC-1579 and BF3 DSM 29040; 3.5 unit pH min−1 by B. bifidum 791. The slow pH reduction rate by some B. bifidum strains can be advantageous for the generation of bioactive peptides during milk fermentation.89
| Parameters | Low-lactose milk + B. bifidum | Milk + B. bifidum BB01 | ||||
|---|---|---|---|---|---|---|
| BB01 | AC-1579 | No. 1 | BF3 DSM 29040 | 791 | ||
| Vmax (unit pH min−1) | 9.5 | 4.0 | 2.5 | 4.0 | 3.5 | 6.2 |
| τm (min) | 360 | 540 | 360 | 300 | 420 | 420 |
| τe (min) | 411 | 810 | 810 | 611 | 565 | 525 |
Considering the microbiological and physicochemical parameters described above, as well as organoleptic characteristics, B. bifidum BB01 was selected as the most promising strain for the production of low-lactose beverages. B. bifidum BB01 successfully carries out fermentation of low-lactose goat milk in 7 h at 37 ± 1 °C.
3.5 Fermentation of low-lactose milk and milk without enzymatic hydrolysis
To identify possible differences in the production cycle, a low-lactose fermented milk drink was compared with the one made of milk fermentation without enzymatic hydrolysis, which is more often used to produce fermented milk products from goat milk. The difference in fermentation duration and acid accumulation in both types of milk has been established (Fig. 6).The initial low-lactose and natural milk before adding the bifidobacteria concentrate had the same TA of 19 ± 1.24 °T (0.17 ± 0.02% lactic acid) and pH of 6.65 ± 0.07. During fermentation of low-lactose milk, pH reached a value of 4.94 ± 0.07 during fermentation for 7 h, while TA was 64 ± 1.24 °T (0.58 ± 0.02% lactic acid). Fermentation of natural milk that did not undergo an enzymatic hydrolysis with β-galactosidase took 9 h until the required organoleptic characteristics were achieved with pH equal to 4.92 ± 0.07, and the TA value was significantly higher than that of low-lactose milk – 75 ± 1.24 °T (0.68% ± 0.02% lactic acid). After cooling to the storage temperature of 4 ± 2 °C, TA of the product from low-lactose milk (75 ± 1.24 °T) was significantly lower than that of regular milk (88 ± 1.24 °T). The product from low-lactose milk had a significantly lower pH level of 4.58 ± 0.07 compared to that of the product from goat milk without hydrolysis of lactose – 4.78 ± 0.07. Acids formed by bifidobacteria during fermentation of raw materials with varying carbohydrate compositions dissociate differently;72,90,91 this changes the relationship between pH and TA.90,92–94
Microbial fermentation was found to increase acid production in hydrolysed milk, resulting in accelerated acid-induced coagulation.95,96 Due to the acceleration of fermentation in low-lactose milk, possible changes in its carbohydrate composition are suggested. In this work, high temperature treatment pasteurisation at 90 ± 2 °C was used to inactivate β-galactosidase. This temperature is sufficient to inactivate the enzyme β-galactosidase obtained from Kluyveromyces lactis.97 In milk subjected to high-temperature treatment for inactivation of β-galactosidase, transformation occurs.98,99 The glucose residue of lactose is transformed into fructose, which together with the galactose residue, forms a stereoisomer of lactose–lactulose.98,100 The amount of lactulose formed in hydrolysed-lactose milk samples after heat inactivation can reach 77.9–130 mg l−1.101,102 Lactulose is a prebiotic that can theoretically stimulate the growth and development of lactic acid microflora, including bifidobacteria, and contributes to their viability.67,103 Bifidobacterium strains show different growth rates and acidification patterns in milk with added prebiotics and some strains may perform better.104
The results of the organoleptic evaluation of the fermented products are shown in Table 6. Significant differences between the samples were observed for sweet flavour, goaty flavour and odour (p < 0.05). There were no significant differences in fermented flavour and odour (p > 0.05). Unpleasant goaty taste characteristics were significantly reduced in fermented low-lactose milk and sweet taste characteristics were significantly higher.
| Attribute | Fermented goat milk product | |
|---|---|---|
| Low-lactose milk | Milk without enzymatic hydrolysis | |
| a Significantly different (p < 0.05) in comparison with the control group (fermented goat milk product from milk without enzymatic hydrolysis) | ||
| Fermented flavour | 6.30 ± 0.83 | 6.80 ± 0.78 |
| Sweet flavour | 6.40 ± 0.77a | 2.87 ± 0.80 |
| Goaty flavour | 2.47 ± 0.82a | 4.67 ± 0.77 |
| Fermented odour | 6.77 ± 0.81 | 7.03 ± 0.92 |
| Goaty odour | 3.17 ± 0.88a | 5.43 ± 0.86 |
Using B. bifidum and other starters can improve the sensory and functional properties of fermented milk, such as reducing undesirable flavours and increasing the content of beneficial organic acids.36,105 Using low-lactose milk improved the sensory, physicochemical, and technological properties of fermented goat milk, reducing goaty flavour and odour.
3.6 Viscosity of fermented dairy products
Dynamic viscosity was measured in low-lactose fermented milk drinks and drinks from goat milk without lactose hydrolysis produced by thermostatic and tank methods. These are the two main methods of producing fermented dairy products and the end products differ in consistency and viscosity. In the tank method (stirred fermented dairy), the product is mechanically treated gently in large-volume containers at the end of the fermentation process and then packaged in consumer containers. In the thermostatic method (set fermented dairy), fermentation takes place in sealed consumer containers; thus, the product is not mechanically destroyed until the packaging is opened by the consumer.106Viscosity values of fermented milk products from milk without lactose hydrolysis were significantly higher than those of low-lactose fermented milk products (Fig. 7A). As seen from the figure, the viscosity of the natural dairy product ranged from 2947 to 116 mPa s for the thermostatic method of production and from 1230 to 73 mPa s for the tank method within an increase in the shear rate from 1 to 100 s−1 in 300 s. The viscosity of low-lactose fermented milk products was from 2305 to 104 mPa s for the thermostatic method and from 334 to 24 mPa s for the tank method within the same increase in the shear rate and duration. The dynamic viscosity for a thermostatic product made from natural milk decreased by 25.4 times with the shear rate increase, for a tank product – by 16.9, for a thermostatic low-lactose product – by 13.9, and for a tank low-lactose product – by 22.2.
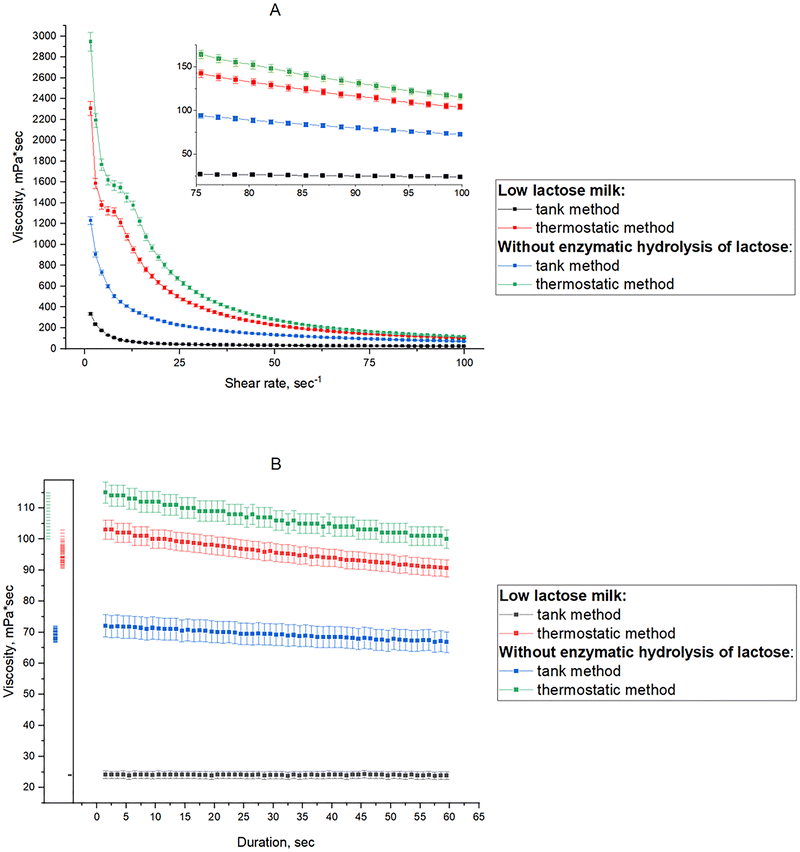 | ||
| Fig. 7 Dynamic viscosity of fermented goat milk products: (A) – dependence of viscosity on the shear rate; (B) – dynamic viscosity at a constant shear rate of 100 s−1. | ||
Previous studies reported that various factors influence the viscosity and the texture profiles of fermented milk products, including acidity, pH, and protein contents.107,108 Lactose-reduced milk was reported to result in a more fluid consistency after fermentation; in addition, the viscosity depends on the composition of the milk, especially the protein/lactose ratio.109,110
The viscosity of the samples produced by the thermostatic method decreased by 12 mPa s from 103 to 91 mPa s for the low-lactose product and by 15 mPa s from 115 to 100 mPa s for the product from milk without lactose hydrolysis at a constant shear rate of 100 s−1 for 60 s after the previous 300 s of shear deformation (Fig. 7B). No significant decrease in viscosity was observed in products produced by the tank method under the same shear deformation conditions. Thus, the viscosity changed insignificantly from 72 to 67 mPa s in the product from milk without lactose hydrolysis. The viscosity also remained at the level of 24 mPa s in the low-lactose product. The fermented low-lactose products showed similar rheological behavior depending on the production method, but their viscosity was lower compared to the fermented products from milk without lactose hydrolysis.
At a constant shear rate, a denser network of protein and fat globules results in higher apparent viscosity, and it takes longer to reduce to a stable value.110,111 When lactose is hydrolysed before or simultaneously with fermentation, a reduction in viscosity and gel strength is observed in the fermented product, despite the fact that lactose increases protein hydration,112 but it also depends on the microbial culture used.96,113 The acidification properties of B. bifidum in milk are linked to its ability to produce exopolysaccharides, which can influence the texture, viscosity and pH of milk-based products.83,114 In general, bifidobacteria are known to synthesize heteropolysaccharides (HePSs) as their primary form of exopolysaccharides. These HePSs are characterized by their complex chemical structure, being built from repeating units that incorporate multiple different monosaccharide residues. Literature data indicate that the most frequently identified constituent HePSs include hexoses, such as glucose and galactose, and deoxy sugars such as rhamnose.115–117 The specific monosaccharide ratio and linkage patterns are strain-dependent and directly influence the rheological properties of the final fermented product. The composition of carbohydrates in milk significantly affects the synthesis and structural properties of EPSs produced by bifidobacteria118 and lactose hydrolysis can increase exopolysaccharide production.113
3.7 Carbohydrate fermentation ability of B. bifidum BB01
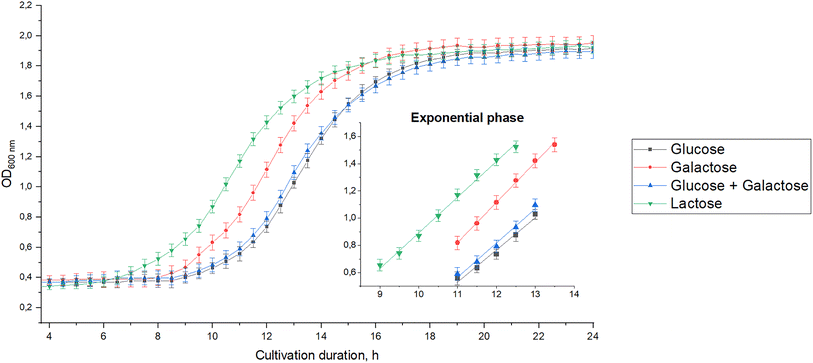
B. bifidum BB01 was cultured in nutrient media containing one of the studied milk carbohydrates: glucose, galactose, or lactose also in a mixture of free glucose and galactose. Kinetic growth curves at optical density (OD) at 600 nm and identified exponential growth phases are shown in Fig. 8. The kinetic growth analysis in defined media revealed a clear hierarchy in carbohydrate preference by B. bifidum BB01. The kinetic data demonstrate that lactose was, in fact, the most efficient substrate, supporting the highest growth rate. Galactose alone supported faster growth than glucose. The kinetics of glucose compared to a glucose–galactose mixture were insignificant (p > 0.05). This quantitative profile provides a mechanistic basis for the strain's performance in low-lactose milk, where the residual lactose and products of its hydrolysis (especially galactose) become the primary drivers of the rapid fermentation which we observed. The concomitant changes in pH values (Fig. 9) at the end of the fermentation period corroborated the optical density data, confirming active consumption of the preferred carbohydrates.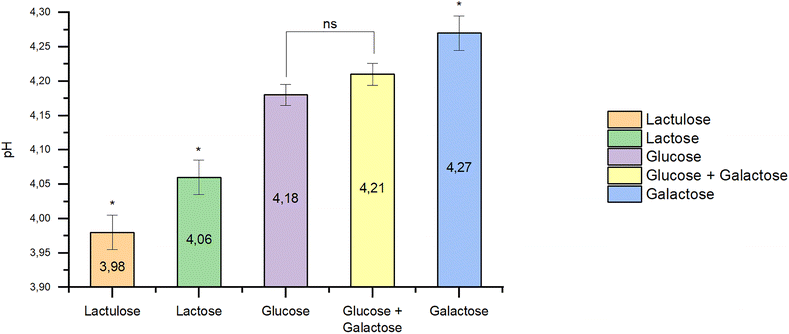 | ||
| Fig. 9 pH changes during carbohydrate fermentation by B. bifidum BB01. (*) significant difference; (ns) non-significant difference (p < 0.05). | ||
Lactulose may appear in low-lactose milk due to the isomerization during high-temperature processing for β-galactosidase inactivation.67,98–100,103 Our studies also revealed differences in acid accumulation, organoleptic and rheological properties of B. bifidum BB01 fermented products obtained from low-lactose milk and milk without enzymatic hydrolysis. Therefore, it was decided to begin testing the hypothesis about the positive effect of the possible lactulose formed on accelerating the fermentation process. Data on pH changes during lactulose fermentation by B. bifidum BB01 were obtained and compared with those of the fermentation of other milk carbohydrates (Fig. 9).
The pH levels after milk carbohydrate fermentation by B. bifidum BB01 were significantly different (p < 0.05). Lactulose showed a higher value of fermentation ability compared to other milk carbohydrates. The pH in the nutrient medium with lactulose was 3.98, with glucose – 4.18, and with galactose – 4.27. The pH value in the nutrient medium with lactose, which simulated the composition of milk without enzymatic hydrolysis of lactose by β-galactosidase, was 4.06.
Previously, a positive effect on the growth of B. bifidum BB01 was found for some prebiotics, namely: fructo-oligosaccharides, xylo-oligosaccharides, inulin, and galacto-oligosaccharides.61,119 Lactulose also significantly increases the growth of various B. bifidum strains.67,68,120–122 The addition of lactulose to infant formulas and fermented milk products enhances the growth and acidification profiles of Bifidobacterium species.68,123
3.8 In vitro gastric survival of B. bifidum BB01
The effect of the fermented milk product matrix on the in vitro gastric survival of B. bifidum BB01 was studied in comparison with the pure strain concentrate (Fig. 10). After 1.5 h of incubation with constant stirring in model gastric juice at 37 °C the B. bifidum BB01 concentrate showed a survival rate of 56% while maintaining a viable cell number of 5.92 ± 0.27 out of 10.64 ± 0.22 log10CFU ml−1. The viability of B. bifidum BB01 cells in the fermented milk product matrix was 89%, which is significantly higher than that of the concentrate. The number of viable cells in the fermented milk product during the test decreased from 9.21 ± 0.25 to 8.16 ± 0.24 log10CFU ml−1. Therefore, the consumption of B. bifidum BB01 can be more effective as part of a fermented milk product.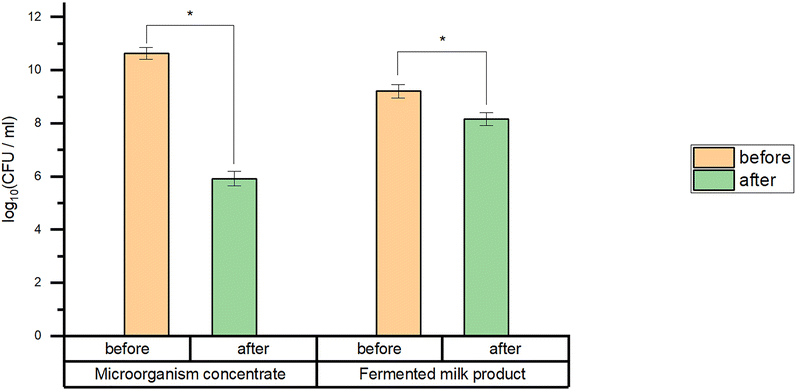 | ||
| Fig. 10 In vitro gastric survival of B. bifidum BB01 in the fermented milk product and in the form of a concentrate. (*) significant difference (p < 0.05). | ||
Bifidobacterium strains behave very differently when exposed to an in vitro simulated gastric environment.124 Some Bifidobacterium strains show high survival rates of over 90% after being treated at pH 2.5 for 3 h.82 Moreover, they can survive in the stomach for up to 90 min, when ingested with fermented milk products, affecting the number of bacteria that enters the small intestine.124 Studies show that in healthy adults, Bifidobacterium strains survive transit through the gastrointestinal tract when consumed as a component of fermented milk products.125
3.9 Post-acidification and characteristics of fermented milk products during storage
Post-acidification allows evaluating the activity of fermenting microorganisms and the product food matrix stability during the shelf life. Measurements of post-acidification (TA and pH) were made during the storage of a low-lactose fermented milk product obtained by the tank method for 28 days at 4 ± 2 °C (Fig. 11). Over 28 days, the TA significantly increased by 10 °T and the pH significantly decreased by 0.19. This is consistent with the results for other Bifidobacterium strains, which demonstrate the TA value increase and the pH values decrease by 0.1–0.4 over fermented goat milk storage time.36,81,82,107,126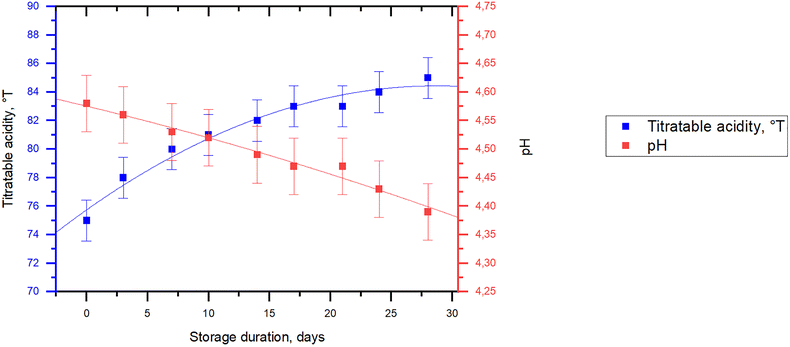 | ||
| Fig. 11 Low-lactose fermented milk product post-acidification characteristics during storage for 28 days at 4 ± 2 °C. | ||
There were no significant changes in the physicochemical properties and microbiological indicators of the product during 28 days (Table 7). Foreign microorganisms, yeasts, molds and coliform bacteria were absent by the end of the storage period, which allows us to assume that a decent shelf life for this product is 22 days with a reserve factor of 1.3 at a storage temperature of 4 ± 2 °C.
| Indicator | Value |
|---|---|
| Protein, g | 3.1 ± 0.1 |
| Fat, g | 3.9 ± 0.1 |
| Carbohydrates, g | 4.7 ± 0.1 |
| Including lactose, no more than, g | 0.1 ± 0.03 |
| pH | 4.39 ± 0.07 |
| TA, °T | 85 ± 1.89 |
| Coliform bacteria, mass of the product in which they are not detected, g | 10 |
| Yeasts and molds | Not found |
| B. bifidum BB01, log10CFU ml−1 | 9.17 |
The appearance of the low-lactose fermented milk product from goat milk is shown in Fig. 12 and its organoleptic characteristics are given in Table 8.
 | ||
| Fig. 12 Low-lactose fermented milk product obtained by the tank method after storage for 28 days at 4 ± 2 °C. | ||
| Indicator | Characteristic |
|---|---|
| Flavour and odour | Clean, fermented milk taste, without foreign tastes and smells, and moderately sweet taste |
| Colour | Milky white |
| Consistency and appearance | Homogeneous, with a broken clot, and moderately viscous |
The amount of bifidobacteria in the product on the 28th day was not less than 9.17 log10CFU ml−1. Thus, 99.5% of B. bifidum BB01 cells remained viable. This corresponds to various known survival rates of bifidobacteria and lactic acid bacteria in fermented milk.81,126–128 Some Bifidobacterium strains may be used in the manufacture of fermented goat milk products that comply with the therapeutic minimum for probiotic bacteria of 6.0 log10CFU g−1, as specified by FAO/WHO, within 28, 21 or 14 days of cold storage.36,81,129 The resulting product from low-lactose goat milk contains quite a high content of bifidobacteria; therefore, with its regular use, it can be considered a probiotic product with the potential to have beneficial effects on human health in a regular diet.
4 Conclusions
The combination of enzymatic lactose hydrolysis and low-lactose milk fermentation with B. bifidum BB01 allows obtaining a low-lactose fermented milk product. This production technology is advantageous as it requires inexpensive equipment and involves simple processes that can be implemented by a wide range of users, including both researchers and small businesses. B. bifidum BB01 and β-galactosidase from Kluyveromyces lactis are produced by industrial companies in a ready-to-use form and require no additional preparation operations before application. This makes this product technology widely available, including for people living in LMICs.The fermented goat milk product produced with B. bifidum BB01 exhibited faster fermentation and a maximum acidification rate and specific growth rate out of 12 studied microorganism strains. The finished product contains more than 9 log10CFU g−1 probiotic cells, which retain their viability for 28 days under refrigerated storage at 4 ± 2 °C. Looking forward, the defined and predictable fermentation kinetics of B. bifidum BB01 in low-lactose milk make it an ideal candidate for its potential integration into functional foods with probiotic consortia. For instance, it could be used in a sequential fermentation strategy, where B. bifidum BB01 is first added to ensure high final probiotic counts without competitive suppression, followed by the use of a traditional yogurt starter for rapid acidification. Alternatively, B. bifidum BB01 could be co-cultured with other well-compatible probiotic strains that share complementary metabolic pathways, creating products with enhanced microbial diversity. The study results also showed that this product is of high quality and safety and can be used as a supplementary source of nutrients for people with lactose intolerance. Our investigations contribute to the development of sustainable low-lactose fermented milk products and thereby address adverse health outcomes, including in LMICs.
Conflicts of interest
The authors confirm that they have no conflicts of interest with respect to the work described in this manuscript.Data availability
All data supporting the findings of this study are available within the article. Raw data, including experimental protocols, analytical results, and characterization data, are available from the corresponding author upon reasonable request.Acknowledgements
This study was financially supported by the Russian Science Foundation (grant no. 23-16-00243, https://rscf.ru/project/23-16-00243/).References
- S. Soleymani, F. Ebrahimi, H. Rezaeizadeh and R. Rahimi, in Nutraceuticals and Cancer Signaling, ed. S. M. Jafari, S. M. Nabavi and A. S. Silva, Springer Nature Switzerland AG, 2021, vol. 18, pp. 467–527 Search PubMed.
- M. Aponte, N. Murru and M. Shoukat, Front. Microbiol., 2020, 11, 562048 CrossRef PubMed.
- H. S. Gill, S. Grover, V. K. Batish and P. Gill, in Prebiotics and Probiotics Science and Technology, Springer New York, New York, NY, 2009, pp. 901–948 Search PubMed.
- A. Mishra, P. P. Dash, A. Usmani, S. P. Singh and A. K. Sirbaiya, in Probiotic Research in Therapeutics, Springer Singapore, Singapore, 2021, pp. 69–94 Search PubMed.
- S. A. Ibrahim, P. J. Yeboah, R. D. Ayivi, A. S. Eddin, N. D. Wijemanna, S. Paidari and R. V. Bakhshayesh, Int. J. Food Sci. Technol., 2023, 58, 4948–4964 CrossRef.
- S. F. Awwad, A. Abdalla, F. C. Howarth, L. Stojanovska, A. Kamal-Eldin and M. M. Ayyash, J. Dairy Sci., 2022, 105, 4722–4733 CrossRef.
- S. Panahi and A. Tremblay, J. Am. Coll. Nutr., 2016, 35, 717–731 CrossRef PubMed.
- S. Sunarti, N. Nurliyani, A. S. A. Tyas, S. D. Kristian and P. Prasetyastuti, Rom. J. Diabetes Nutr. Metab. Dis., 2015, 22, 261–267 Search PubMed.
- B. Han, L. Zhang, Y. Hou, J. Zhong, K. Hettinga and P. Zhou, Food Res. Int., 2022, 157, 111254 CrossRef PubMed.
- X. Chen, Z. Zhang, H. Niu, X. Tian, H. Tian, W. Yao, H. He, H. Shi, C. Li and J. Luo, Mol. Nutr. Food Res., 2023, 68, 2200842 CrossRef PubMed.
- W. Liu, Y. Zhou, H. Sun, R. Li, Y. Qin, L. Yu, Y. Chen, Y. Li, Y. Tan, R. Zhao, W. Zhang, S. Jiang and Y. Xu, Mol. Nutr. Food Res., 2021, 65, 2000888 CrossRef PubMed.
- N. Vissamsetti, M. Simon-Collins, S. Lin, S. Bandyopadhyay, R. Kuriyan, W. Sybesma and D. Tomé, Curr. Dev. Nutr., 2024, 8, 102049 CrossRef PubMed.
- C. L. Storhaug, S. K. Fosse and L. T. Fadnes, Lancet Gastroenterol. Hepatol., 2017, 2, 738–746 Search PubMed.
- T. A. Nicklas, H. Qu, S. O. Hughes, M. He, S. E. Wagner, H. R. Foushee and R. M. Shewchuk, Am. J. Clin. Nutr., 2011, 94, 191–198 Search PubMed.
- H. Zhang, Y. Tao, Y. He, J. Pan, K. Yang, J. Shen and C. Gao, ACS Omega, 2020, 5, 8543–8550 Search PubMed.
- D. Mangan, B. V McCleary, H. Culleton, C. Cornaggia, R. Ivory, V. A. McKie, E. Delaney and T. Kargelis, J. Sci. Food Agric., 2019, 99, 947–956 Search PubMed.
- M. de Vrese, A. Stegelmann, B. Richter, S. Fenselau, C. Laue and J. Schrezenmeir, Am. J. Clin. Nutr., 2001, 73, 421s–429s CrossRef PubMed.
- C. S. Ranadheera, C. A. Evans, S. K. Baines, C. F. Balthazar, A. G. Cruz, E. A. Esmerino, M. Q. Freitas, T. C. Pimentel, A. E. Wittwer, N. Naumovski, J. S. Graça, A. S. Sant'Ana, S. Ajlouni and T. Vasiljevic, Compr. Rev. Food Sci. Food Saf., 2019, 18, 867–882 CrossRef PubMed.
- M. Pawlos, A. Znamirowska, M. Kluz, K. Szajnar and M. Kowalczyk, J. Microbiol., Biotechnol. Food Sci., 2020, 9, 751–755 CrossRef.
- N. G. Araújo, I. M. Barbosa, T. L. S. Lima, R. T. Moreira and H. R. Cardarelli, J. Food Sci. Technol., 2022, 59, 3806–3818 CrossRef PubMed.
- C. G. Vinderola, P. Mocchiutti and J. A. Reinheimer, J. Dairy Sci., 2002, 85, 721–729 CrossRef PubMed.
- E. W. Ng, M. Yeung and P. S. Tong, Int. J. Food Microbiol., 2011, 145, 169–175 CrossRef PubMed.
- K. J. Heller, Am. J. Clin. Nutr., 2001, 73, 374s–379s CrossRef PubMed.
- A. Samona and R. K. Robinson, Int. J. Dairy Technol., 1994, 47, 58–60 CrossRef.
- A. Dantas, S. Verruck, M. H. M. Canella, E. Hernandez and E. S. Prudencio, Food Res. Int., 2021, 150, 110742 CrossRef.
- M. Hedberg, P. Hasslöf, I. Sjöström, S. Twetman and C. Stecksén-Blicks, Oral Microbiol. Immunol., 2008, 23, 482–485 CrossRef PubMed.
- N. P. Shah, W. E. V Lankaputhra, M. L. Britzb and W. S. A. Kyle, Short Commun., 1995, 5, 515–521 Search PubMed.
- C. G. Vinderola, N. Bailo and J. A. Reinheimer, Food Res. Int., 2000, 33, 97–102 CrossRef.
- M. Hedberg, P. Hasslöf, I. Sjöström, S. Twetman and C. Stecksén-Blicks, Oral Microbiol. Immunol., 2008, 23, 482–485 CrossRef CAS PubMed.
- K. Reiner, Carbohydrate Fermentation Protocol, American Society for Microbiology, 2016 Search PubMed.
- A. K. Niamah, Future Food: J. Food Agric. Soc., 2019, 7, 1–8 Search PubMed.
- N. A. Bolívar-Jacobo, R. A. Reyes-Villagrana, G. P. Espino-Solís, A. L. Rentería-Monterrubio, M. M. Arévalos-Sánchez, R. Sánchez-Vega, E. Santellano-Estrada, D. Chávez-Flores and A. Chávez-Martínez, Fermentation, 2023, 9, 356 CrossRef.
- Official Methods of Analysis of AOAC International, ed. G. W. Latimer, Oxford University Press, New York, 2023 Search PubMed.
- M. F. Bezerra, D. F. S. Souza and R. T. P Correira, Int. J. Dairy Technol., 2012, 65, 437–443 CrossRef CAS.
- R. M. Maier and I. L. Pepper, in Environmental Microbiology, Elsevier, 2015, pp. 37–56 Search PubMed.
- S. Guo, M. Chen, T. Wu, K. Liu, H. Zhang and J. Wang, J. Dairy Sci., 2022, 105, 9426–9438 CrossRef CAS.
- H. I. Ko, C. H. Jeong, S. W. Hong, J.-B. Eun and T.-W. Kim, Microbiol. Spectrum, 2022, 10, e01625 Search PubMed.
- M. Minekus, M. Alminger, P. Alvito, S. Ballance, T. Bohn, C. Bourlieu, F. Carrière, R. Boutrou, M. Corredig, D. Dupont, C. Dufour, L. Egger, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. Mackie, S. Marze, D. J. McClements, O. Ménard, I. Recio, C. N. Santos, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and A. Brodkorb, Food Funct., 2014, 5, 1113–1124 RSC.
- Official Methods of Analysis of AOAC International, ed. G. W. Latimer, Oxford University Press, New York, 22nd edn, 2023 Search PubMed.
- C. F. Iskandar, C. Cailliez-Grimal, F. Borges and A.-M. Revol-Junelles, Trends Food Sci. Technol., 2019, 88, 121–132 CrossRef CAS.
- M. W. Hickey, A. J. Hillier and G. R. Jago, Appl. Environ. Microbiol., 1986, 51, 825–831 CrossRef CAS.
- W. de Vries and A. H. Stouthamer, J. Bacteriol., 1968, 96, 472–478 CrossRef CAS PubMed.
- P. L. Møller, F. Jørgensen, O. C. Hansen, S. M. Madsen and P. Stougaard, Appl. Environ. Microbiol., 2001, 67, 2276–2283 CrossRef.
- D. Srinivas, B. K. Mital and S. K. Garg, Int. J. Food Microbiol., 1990, 10, 51–57 CrossRef CAS PubMed.
- R. Suissa, R. Oved, H. Maan, U. Hadad, O. Gilhar, M. M. Meijler, O. Koren and I. Kolodkin-Gal, Front. Microbiol., 2022, 13, 949932 CrossRef.
- V. Füreder, B. Rodriguez-Colinas, F. V. Cervantes, L. Fernandez-Arrojo, A. Poveda, J. Jimenez-Barbero, A. O. Ballesteros and F. J. Plou, J. Agric. Food Chem., 2020, 68, 4930–4938 CrossRef.
- F. De Bruyn, J. Beauprez, J. Maertens, W. Soetaert and M. De Mey, Appl. Environ. Microbiol., 2013, 79, 7028–7035 CrossRef CAS PubMed.
- X. Qi and R. F. Tester, Clin. Nutr., 2019, 33, 18–28 Search PubMed.
- R. A. Clemens, J. M. Jones, M. Kern, S. Lee, E. J. Mayhew, J. L. Slavin and S. Zivanovic, Compr. Rev. Food Sci. Food Saf., 2016, 15, 433–470 CrossRef CAS PubMed.
- A. Li, J. Zheng, X. Han, S. Yang, S. Cheng, J. Zhao, W. Zhou and Y. Lu, Foods, 2023, 12, 2553 CrossRef CAS.
- G. T. Berry, in Rosenberg's Molecular and Genetic Basis of Neurological and Psychiatric Disease, Elsevier, 2015, pp. 615–626 Search PubMed.
- C. Bo-Htay, S. Palee, N. Apaijai, S. C. Chattipakorn and N. Chattipakorn, J. Cell. Mol. Med., 2018, 22, 1392–1410 CrossRef CAS.
- H. Emamat, H. Farhadnejad, H. Poustchi and A. Hekmatdoost, Nutr. Food Sci., 2019, 49, 359–367 CrossRef.
- A. G. Tochilina, I. V. Belova, I. V. Soloveva, T. N. Ilyicheva, D. B. Gelashvili, V. A. Zhirnov and S. B. Molodtsova, Opera Med. Physiol., 2023, 10, 72–105 Search PubMed.
- A. A. Korzhenkov, A. V Tepliuk, K. V Sidoruk, K. E. Voyushin, M. V Patrushev, I. V Kublanov and S. V Toshchakov, Data Brief, 2021, 34, 106710 CrossRef CAS PubMed.
- M. Bazanella, T. V Maier, T. Clavel, I. Lagkouvardos, M. Lucio, M. X. Maldonado-Gòmez, C. Autran, J. Walter, L. Bode, P. Schmitt-Kopplin and D. Haller, Am. J. Clin. Nutr., 2017, 106, 1274–1286 CrossRef CAS PubMed.
- M.-J. Kwak, S.-K. Kwon, J.-K. Yoon, J. Y. Song, J.-G. Seo, M. J. Chung and J. F. Kim, Syst. Appl. Microbiol., 2016, 39, 429–439 CrossRef.
- Cell Biotech Co Ltd, WO2016186246A1, 2015.
- G. Shu, S. Wang, H. Chen, M. Hu, T. Qin and Q. Ma, Acta Univ. Cibiniensis, Ser. E: Food Technol., 2015, 19, 11–18 Search PubMed.
- Q.-L. Tao, Food Sci. Technol. Int., 2011, 6, 6–8 Search PubMed.
- G. Shu, N. Lei, H. Chen, M. Hu and H. Yang, Acta Univ. Cibiniensis, Ser. E: Food Technol., 2016, 20, 41–52 Search PubMed.
- L. Alm, J. Dairy Sci., 1982, 65, 346–352 CrossRef PubMed.
- Q. Wu, C. K. W. Cheung and N. P. Shah, Trends Food Sci. Technol., 2015, 41, 24–36 CrossRef.
- R. Catanzaro, M. Sciuto and F. Marotta, Nutr. Res., 2021, 89, 23–34 CrossRef PubMed.
- Q. Wu and N. P. Shah, Food Microbiol., 2017, 62, 178–187 CrossRef PubMed.
- H. Hu, G. Qimu, J. Nie, N. Wu and T. Dan, J. Dairy Sci., 2024, 107, 6558–6575 CrossRef.
- P. S. Panesar and S. Kumari, Biotechnol. Adv., 2011, 29, 940–948 CrossRef PubMed.
- R. P. De Souza Oliveira, A. C. Rodrigues Florence, P. Perego, M. N. De Oliveira and A. Converti, Int. J. Food Microbiol., 2011, 145, 22–27 CrossRef PubMed.
- M. Nooshkam, A. Babazadeh and H. Jooyandeh, Trends Food Sci. Technol., 2018, 80, 23–34 CrossRef.
- C. Dai, Z. Shu, X. Xu, P. Yan, M. Dabbour, B. Kumah Mintah, L. Huang, R. He and H. Ma, Ultrason. Sonochem., 2023, 100, 106611 Search PubMed.
- J. P. Pagnossa, G. Rocchetti, A. C. Ribeiro, R. H. Piccoli and L. Lucini, Curr. Opin. Food Sci., 2020, 31, 24–30 Search PubMed.
- T. M. P. Nguyen, Y. K. Lee and W. Zhou, Food Chem., 2012, 130, 866–874 CrossRef.
- G. Huang, S. Chen, C. Dai, L. Sun, W. Sun, Y. Tang, F. Xiong, R. He and H. Ma, Ultrason. Sonochem., 2017, 37, 144–149 CrossRef PubMed.
- J. T. Guimarães, C. F. Balthazar, H. Scudino, T. C. Pimentel, E. A. Esmerino, M. Ashokkumar, M. Q. Freitas and A. G. Cruz, Ultrason. Sonochem., 2019, 57, 12–21 CrossRef PubMed.
- C. Dai, F. Xiong, R. He, W. Zhang and H. Ma, Ultrason. Sonochem., 2017, 36, 191–197 CrossRef PubMed.
- G. Huang, S. Chen, Y. Tang, C. Dai, L. Sun, H. Ma and R. He, Ultrason. Sonochem., 2019, 51, 315–324 CrossRef PubMed.
- B. D. Dahroud, R. R. Mokarram, M. S. Khiabani, H. Hamishehkar, A. Z. Bialvaei, M. Yousefi and H. S. Kafil, Int. J. Biol. Macromol., 2016, 86, 462–467 CrossRef PubMed.
- Y. Tao and D.-W. Sun, Crit. Rev. Food Sci. Nutr., 2015, 55, 570–594 CrossRef PubMed.
- P. Sfakianakis and C. Tzia, Foods, 2014, 3, 176–193 CrossRef PubMed.
- S. Cui, M. Hu, Y. Sun, B. Mao, Q. Zhang, J. Zhao, X. Tang and H. Zhang, Microorganisms, 2022, 11, 48 CrossRef PubMed.
- A. Mituniewicz-Małek, M. Ziarno, I. Dmytrów and J. Balejko, J. Dairy Sci., 2017, 100, 6972–6979 CrossRef.
- W. J. Liu, Y. F. Chen, L. Y. Kwok, M. H. Li, T. Sun, C. L. Sun, X. N. Wang, T. Dan, M. Menghebilige, H. P. Zhang and T. S. Sun, J. Dairy Sci., 2013, 96, 6807–6817 CrossRef CAS.
- P. H. P. Prasanna, A. S. Grandison and D. Charalampopoulos, Int. Dairy J., 2012, 23, 36–44 CrossRef CAS.
- C. A. Butts, G. Paturi, D. I. Hedderley, S. Martell, H. Dinnan, H. Stoklosinski and E. A. Carpenter, Food Funct., 2021, 12, 3104–3119 RSC.
- P. H. P. Prasanna and D. Charalampopoulos, Food Biosci., 2018, 21, 72–79 CrossRef CAS.
- C. Kehagias, J. Csapó, S. Konteles, E. Kolokitha, S. Koulouris and Zs. Csapó-Kiss, Int. Dairy J., 2008, 18, 396–402 CrossRef.
- Š. Ročková, V. Rada, J. Havlík, R. Švejstil, E. Vlková, V. Bunešová, K. Janda and I. Profousová, Czech J. Anim. Sci., 2013, 58, 99–105 CrossRef.
- C. Gonzalez-Gonzalez, T. Gibson and P. Jauregi, Int. J. Food Microbiol., 2013, 167, 131–137 CrossRef PubMed.
- C. Gonzalez-Gonzalez, T. Gibson and P. Jauregi, Int. J. Food Microbiol., 2013, 167, 131–137 CrossRef PubMed.
- K. Adhikari, A. Mustapha, I. U. Grün and L. Fernando, J. Dairy Sci., 2000, 83, 1946–1951 CrossRef PubMed.
- E. Lauer and O. Kandler, Arch. Microbiol., 1976, 110, 271–277 CrossRef PubMed.
- H. B. Moonga, S. E. Schoustra, A. R. Linnemann, J. van den Heuvel, J. Shindano and E. J. Smid, LWT--Food Sci. Technol., 2021, 136, 110350 CrossRef.
- H. H. Wilkowske, J. Dairy Sci., 1954, 37, 22–29 CrossRef.
- A. Mituniewicz-Małek, M. Ziarno, I. Dmytrów and J. Balejko, J. Dairy Sci., 2017, 100, 6972–6979 CrossRef.
- M. Manafi, Standardization of Different Levels of Lactose Hydrolysis in the Preparation of Lactose Hydrolyzed Yoghurt, 2009, vol. 10 Search PubMed.
- T. Huppertz, in Yogurt in Health and Disease Prevention, Elsevier, 2017, pp. 387–394 Search PubMed.
- T. L. de Albuquerque, M. de Sousa, N. C. Gomes e Silva, C. A. C. Girão Neto, L. R. B. Gonçalves, R. Fernandez-Lafuente and M. V. P. Rocha, Int. J. Biol. Macromol., 2021, 191, 881–898 CrossRef.
- M. Aider and D. de Halleux, Trends Food Sci. Technol., 2007, 18, 356–364 CrossRef.
- Y.-S. Song, H.-U. Lee, C. Park and S.-W. Kim, Carbohydr. Res., 2013, 369, 1–5 CrossRef.
- F. Zokaee, T. Kaghazchi, A. Zare and M. Soleimani, Process Biochem., 2002, 37, 629–635 CrossRef.
- M. C. Messia, T. Candigliota and E. Marconi, Food Chem., 2007, 104, 910–917 CrossRef.
- L. N. de Oliveira Neves and M. A. Leal de Oliveira, Int. Dairy J., 2020, 106, 104710 CrossRef.
- B. Tungland, in Human Microbiota in Health and Disease, Elsevier, 2018, pp. 289–348 Search PubMed.
- C. Martínez-Villaluenga and R. Gómez, Int. Dairy J., 2007, 17, 116–122 CrossRef.
- M. Czarnowska-Kujawska and B. Paszczyk, Molecules, 2021, 26, 6063 CrossRef PubMed.
- A. Haque, R. K. Richardson and E. R. Morris, Food Hydrocolloids, 2001, 15, 593–602 CrossRef.
- R. Tian, Z. Yu, L. Yu, F. Tian, J. Zhao, H. Zhang, W. Chen and Q. Zhai, Food Biosci., 2022, 50, 102167, DOI:10.1016/j.fbio.2022.102167.
- M. N. Oliveira, I. Sodini, F. Remeuf and G. Corrieu, Int. Dairy J., 2001, 11, 935–942 CrossRef.
- F. Alvarez, M. Arguello, M. Cabero, F. A. Riera, R. Alvarez, J. R. Iglesias and J. Granda, Fermentation of Concentrated Skim-Milk, Effects of Different Protein/Lactose Ratios Obtained by Ultrafiltration-Diafiltration, 1998, vol. 76 Search PubMed.
- N. Lussier, A. Gilbert, D. St-Gelais and S. L. Turgeon, Dairy, 2023, 4, 108–123 CrossRef.
- B. Abu-Jdayil, M. S. Nasser and M. Ghannam, Food Sci. Technol. Res., 2013, 19, 277–286 CrossRef.
- G. H. Meletharayil, H. A. Patel, L. E. Metzger and T. Huppertz, Int. Dairy J., 2016, 61, 107–113 CrossRef.
- C. Schmidt, S. Mende, D. Jaros and H. Rohm, Dairy Sci. Technol., 2016, 96, 199–211 CrossRef.
- F. Aboulfazli, A. S. Baba and M. Misran, Int. J. Food Sci. Technol., 2015, 50, 942–949 CrossRef.
- Y. Xu, Y. Cui, F. Yue, L. Liu, Y. Shan, B. Liu, Y. Zhou and X. Lü, Food Hydrocolloids, 2019, 94, 475–499 CrossRef.
- N. Castro-Bravo, J. M. Wells, A. Margolles and P. Ruas-Madiedo, Front. Microbiol., 2018, 9, 2426 CrossRef.
- C. Hidalgo-Cantabrana, B. Sánchez, C. Milani, M. Ventura, A. Margolles and P. Ruas-Madiedo, Appl. Environ. Microbiol., 2014, 80, 9–18 CrossRef.
- Y. Xu, Y. Cui, F. Yue, L. Liu, Y. Shan, B. Liu, Y. Zhou and X. Lü, Food Hydrocolloids, 2019, 94, 475–499 CrossRef.
- C. He, H. Man, S. Guowei, M. Qi and Q. Tao, in Proceedings 2011 International Conference on Human Health and Biomedical Engineering, IEEE, 2011, pp. 981–984 Search PubMed.
- D. Özer, S. Akin and B. Özer, Food Sci. Technol. Int., 2005, 11, 19–24 CrossRef.
- S. Cui, J. Gu, X. Liu, D. Li, B. Mao, H. Zhang, J. Zhao and W. Chen, J. Sci. Food Agric., 2021, 101, 5721–5729 CrossRef.
- Y. Bouhnik, A. Attar, F. A. Joly, M. Riottot, F. Dyard and B. Flourié, Eur. J. Clin. Nutr., 2004, 58, 462–466 CrossRef.
- U. K. Dubey and V. V. Mistry, J. Dairy Sci., 1996, 79, 1156–1163 CrossRef.
- N. Berrada, J.-F. Lemeland, G. Laroche, P. Thouvenot and M. Piaia, J. Dairy Sci., 1991, 74, 409–413 CrossRef.
- P. Pochart, P. Marteau, Y. Bouhnik, I. Goderel, P. Bourlioux and J. Rambaud, Am. J. Clin. Nutr., 1992, 55, 78–80 CrossRef.
- G. K. Deshwal, S. Tiwari, A. Kumar, R. K. Raman and S. Kadyan, Trends Food Sci. Technol., 2021, 109, 499–512 CrossRef.
- M. Gueimonde, S. Delgado, B. Mayo, P. Ruas-Madiedo, A. Margolles and C. G. de los Reyes-Gavilán, Food Res. Int., 2004, 37, 839–850 CrossRef.
- B. Ebel, F. Martin, L. D. T. Le, P. Gervais and R. Cachon, J. Dairy Sci., 2011, 94, 2185–2191 CrossRef CAS PubMed.
- S. N. Raeisi, L. I. I. Ouoba, N. Farahmand, J. Sutherland and H. B. Ghoddusi, Food Control, 2013, 34, 691–697 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |