Open Access Article
Open Access ArticleArtificial intelligence-driven innovation in Ganoderma spp.: potentialities of their bioactive compounds as functional foods
Sonali
Khanal
a,
Aman
Sharma
a,
Manjusha
Pillai
a,
Pratibha
Thakur
b,
Ashwani
Tapwal
b,
Vinod
Kumar
 c,
Rachna
Verma
de and
Dinesh
Kumar
c,
Rachna
Verma
de and
Dinesh
Kumar
 *ae
*ae
aSchool of Bioengineering and Food Technology, Shoolini University of Biotechnology and Management Sciences, Solan 173229, India. E-mail: chatantadk1973@gmail.com
bICFRE-Himalayan Forest Research Institute, Shimla, 171013, India
cSchool of Water, Energy and Environment, Cranfield University, Cranfield MK43 0AL, UK
dSchool of Biological and Environmental Science, Shoolini University of Biotechnology and Management Sciences, Solan 173229, India
eCentre of Advanced Innovation Technologies, VSB-Technical University of Ostrava, 70800, Ostrava-Poruba, Czech Republic
First published on 7th April 2025
Abstract
Ganoderma spp., which are essential decomposers of lignified plant materials, can affect trees in both wild and cultivated settings. These fungi have garnered significant global interest owing to their potential to combat several chronic, complicated, and infectious diseases. As technology progresses, researchers are progressively employing artificial intelligence (AI) for studying various fungal strains. This novel approach has the potential to accelerate the knowledge and application of Ganoderma spp. in the food industry. The development of extensive Ganoderma databases has markedly expedited research on them by enhancing access to information on bioactive components of Ganoderma and promoting collaboration with the food sector. Progress in AI techniques and enhanced database quality have further advanced AI applications in Ganoderma research. Techniques such as machine learning (ML) and deep learning employing various methods, including support vector machines (SVMs), Bayesian networks, artificial neural networks (ANNs), random forests (RFs), and convolutional neural networks (CNNs), are propelling these advancements. Although AI possesses the capacity to transform Ganoderma research by tackling significant difficulties, continuous investment in research, data dissemination, and interdisciplinary collaboration are necessary. AI could facilitate the development of customized functional food products by discerning patterns and correlations in customer data, resulting in more specific and accurate solutions. Thus, the future of AI in Ganoderma research looks auspicious, presenting prospects for ongoing advancement and innovation in this domain.
Sustainability spotlightThe study “Artificial intelligence-driven innovation in Ganoderma spp.: potentialities of their bioactive compounds as functional foods” makes significant contributions to a number of important areas that are in line with the Sustainable Development Goals (SDGs) of the UN: this study addresses chronic diseases and improves preventive healthcare through sustainable, natural remedies by investigating the potential of bioactive substances from Ganoderma spp. as functional foods. The incorporation of artificial intelligence into natural product research stimulates creativity and propels the creation of cutting-edge instruments and techniques for environmentally friendly manufacturing in the nutraceutical and functional food sectors. Using AI to effectively extract and use Ganoderma spp. components promotes sustainable and resource-efficient procedures, cutting down waste and promoting circular economy principles. |
1. Introduction
Ganoderma P. Karst. (Ganodermataceae and Basidiomycota) is a diverse genus of wood-decaying fungi, encompassing species that induce white rot in the roots and lower trunks of trees from several plant groups. These fungi are crucial in the decomposition of lignified plant matter, frequently impacting trees in both natural and managed ecosystems. The laccate Ganoderma species, characterized by their glossy or varnished appearance, are significant fungi that facilitate the disintegration of living trees and decomposition of woody detritus.1 It is a commonly utilized medicinal fungus recognized for its potential anticancer and immunotherapeutic attributes, owing to its low toxicity and efficacy in combination therapy. Nonetheless, the precise molecular pathways remain inadequately delineated, and numerous findings are derived from in vitro research.2 Hence, future studies should concentrate on the interaction between Ganoderma and clinical chemotherapeutic agents to mitigate their adverse effects. Examining the principal bioactive components, conducting in vivo pharmacokinetic studies, and investigating the processes of immune regulation and interactions are crucial for enhancing Ganoderma's clinical uses.3 Over 1000 healthy food products containing Ganoderma have received certification from the Chinese government, indicating its extensive utilization and acknowledged advantages. In ancient recipes, G. lucidum is sometimes used with ginseng (Panax ginseng) to prepare soups that are said to calm the nerves, ease asthma, and improve the immune system. Furthermore, it is combined with Sanqi (Panax notoginseng) to create herbal Sanqi wine, purported to enhance blood circulation and facilitate relaxation.4 Herbal wines containing G. lucidum, whether alone or in combination with other medicinal herbs, are recognized for their possible anti-aging properties and capacity to promote bodily equilibrium. Tea prepared with G. lucidum, whether used independently or in conjunction with other herbs, is esteemed for its contribution to immune support and the preservation of overall health equilibrium.5As illustrated in Table 1, Ganoderma spp. play various roles, extending beyond medicinal uses to include important industrial applications. Notwithstanding its acknowledged pharmacological benefits and widespread use as a functional food, the specific mechanisms of Ganoderma's active components and the regulations governing their compatibility with other ingredients remain little understood. Nevertheless, a contemporary scientific foundation for understanding the mechanisms of its active components and the rule of compatibility remains inadequate.
| Field | Key area | Description | Methods/techniques | Commercial importance | Challenges/limitations | References |
|---|---|---|---|---|---|---|
| Taxonomy & identification | Morphological and molecular classification | Specified according to fruiting body form, spore morphology, and DNA markers (ITS, LSU rDNA) | Light microscopy, SEM, ITS sequencing | Ensures accurate species identification for pharmaceutical and industrial uses | Morphological resemblance leads to taxonomic confusion; molecular methods are expensive | 6 and 7 |
| Phytochemistry | Bioactive compound profiling | Polymeric in nature, contains polysaccharides, triterpenoids, proteins, phenolics, sterols, and flavonoids | HPLC, GC-MS, FTIR, NMR spectroscopy | Pharmaceutical, nutraceutical, and cosmetic formulation basis | The problem of standardization, which is based on differences in cultivation conditions | 8 |
| Biochemical activity analysis | Ganoderic acids, β-glucans, and proteins possess antioxidant, anti-inflammatory, and immunomodulatory activities | LC-MS, ELISA, UV-vis spectrophotometry | Facilitates functional food and drug development | Species and cultivation method variation in bioactive content | 9 and 10 | |
| Cultivation & processing | Growth, & production | Raised on solid (logs, sawdust) or liquid (submerged) culture to increase bioactive compound yield | Spawn inoculation, bioreactors | Industrial-scale commercial growth for mushroom and extract manufacture | Requires controlled environment, risks of contamination | 11 |
| Extraction, & purification | Hot water, ethanol, or supercritical CO2 are used for the extraction of bioactive compounds | Soxhlet extraction, ultrasonic-assisted extraction (UAE) | Pharmaceuticals and nutraceutical-grade extracts | Extraction efficiency is variable; bioavailability issues | 12 and 13 | |
| Standardization and formulation | Standardized extracts are produced as capsules, tablets, and functional foods | Pharmacopoeia standards, HPLC fingerprinting | Ensures constancy and efficiency in medicinal usage | Regulatory challenges; bioactive composition variance | 14 and 15 | |
| Biomedical & therapeutic applications | Pharmaceutical use | Applied in immunomodulation, anticancer, hepatoprotection, and neuroprotection | Preclinical and clinical trials | Possible alternative medicine; incorporated into contemporary drug formulations | Few large-scale clinical trials; FDA/EMA approval requirements | 16 and 17 |
| Functional foods and nutraceuticals | Added to teas, coffee, and wellness dietary supplements | Spray drying, encapsulation | Growing market in the functional food business | Consumer cynicism; flavor and solubility concerns | 18 and 19 | |
| Biotechnology & environmental applications | Enzyme production, & bioremediation | Yields laccase, and peroxidase, contributes to bioremediation of contaminants | Fermentation, solid-state cultivation | Used for waste treatment and green industrial operations | High production cost; requires optimization for industrial application | 20–22 |
| Nanotechnology | Nano formulations | Ganoderma-derived nanoparticles used for drug delivery, antimicrobial coatings, and functional materials | Green synthesis of nanoparticles, surface modification techniques | Emerging applications in medicine, food safety and material science | Need for large-scale validation; stability and regulatory concerns | 23 and 24 |
| Biosensor technology | Ganoderma-based biosensors | Bioactive compounds from Ganoderma used in fluorescence-based biosensor for detecting toxins, heavy metals, and pathogens | DNA based biosensor, fluorescence-based detection | Potential for real-time monitoring in healthcare, and environmental analysis | Stability, reproducibility, and integration into commercial devices remain challenges | 25 and 26 |
| Role of AI in Ganoderma | AI-driven identification & drug discovery | Machine learning (ML) and deep learning (DL) used to predict bioactive compounds, optimize cultivation, and automate species identification | AI-based molecular docking, image recognition, neural networks | Accelerates drug discovery, improves yield prediction, and ensures precise quality control | Requires large datasets, computational resources, and validation of AI-generated results | 27–29 |
The extraction of polysaccharides and other active components from the woody and intricately textured fruiting bodies of Ganoderma spp. continues to be a significant challenge, despite the high efficiency achieved with other edible and therapeutic mushrooms.30 Several state-of-the-art techniques including subcritical water extraction, ultrasound-assisted extraction, microwave-assisted extraction, and enzyme-assisted extraction have been employed to increase the effectiveness of extracting bioactive components. These methods aim to improve agricultural precision and efficiency in conjunction with cross-industry technologies such as the IoT.31 Maximizing therapeutic benefits is fundamentally contingent upon the bioaccessibility of Ganoderma spp., the percentage of compounds that stay intact during gastrointestinal transit, the fraction absorbed by epithelial cells, and their stability following processing and storage.32 Innovative delivery methods such as nanoencapsulation have emerged as an effective approach to enhance the therapeutic efficacy of polyphenols in functional food. It may significantly enhance the stability of these biomolecules in gastrointestinal environments and promote their absorption by protecting them from degradation.33 Moreover, the integration of bioinformatics with AI-driven predictive models has the potential to improve the design of these delivery systems, thereby ensuring that bioactive molecules effectively reach their intended targets.34 In addition to predicting their metabolic pathways and potential interactions in vivo, AI may assist in the discovery of novel bioactives with optimal physicochemical properties for absorption. This capability aligns with the evolving healthcare paradigm that prioritizes AI-based preventive nutrition. Finding potential functional food ingredients with enhanced bioactivity and safety features has been made easier by the quick screening of huge chemical libraries using AI algorithms.35 This synergy enhances efficiency, accuracy, and automation across several processes including food production, distribution, and waste management. Blockchain technology ensures food safety and authenticity through unparalleled transparency in supply chains, hence guaranteeing these attributes; AI-driven predictive analytics facilitate resource efficiency and waste reduction.36 Incorporating AI in the exploration and development of functional food components would facilitate the creation of tailored nutritional solutions that enhance bioavailability while addressing unmet consumer demands in a safe, sustainable, and cost-effective manner.37
The application of AI has emerged since AI technology has markedly enhanced the reliability and accuracy of diagnosis, target screening, and novel food product development. AI is changing the food business by tackling important issues like lowering food waste, making the supply chain more efficient, and improving the delivery, logistics, and safety of food. To make sure that food safety rules are followed, AI makes the supply chain more open by keeping an eye on processes and making things better, like predicting prices, production, inventory control, and transportation. It also improves the customer experience by making suggestions that are more relevant to them, answering questions with chatbots, and making pricing methods more efficient. AI also helps with predicting demands, finding the best delivery routes, and judging the success of suppliers. Tracking shelf life and analyzing consumption trends help cut down on waste. These new ideas make sure that processes run smoothly, products are of high quality, and customers are satisfied.38
In the realm of food biotechnology, artificial intelligence is rapidly emerging as an essential tool for understanding the various applications of AI within the food industry. Artificial intelligence significantly aids in the development of new skills, products, and services while enhancing the processing speed and accuracy. AI uses a variety of techniques for machine learning and deep learning, including support vector machines (SVMs), Bayesian networks, artificial neural networks (ANNs), random forests (RFs), and convolutional neural networks (CNNs).39 Furthermore, AI has significantly contributed to the development of innovative methodologies for discovering and producing industrial enzymes, including those utilized in food applications.40 Screening and detection of the natural bioactives present in Ganoderma spp. have not yet been established, creating challenges in maintaining consistency and reliability. This review highlights how these technologies might improve the accuracy, efficiency, and scalability of bioactive chemical discovery, specifically for applications in the food business. It also assesses advanced AI techniques such as predictive analytics and simulation models to enhance fermentation processes, enzyme synthesis, and product standardization. This article reconciles established methods with growing AI capabilities, addressing existing limits and fostering a more sustainable and innovative application of Ganoderma in the food industry.
2. Predicting bioactivity of Ganoderma spp. by leveraging AI for the food industry
2.1. Classical bioinformatics-driven methods with integrated AI
Bioinformatics-based approaches play a central role in screening the applications of bioactive compounds from fungi (S. Singh et al., 2024).41 They serve important applications for food safety, preservation, and functional properties, with the screening of bioactive compounds extracted from fungi through computational approaches such as genome mining, molecular docking, and machine learning algorithms, leading to the identification, prediction, and validation of bioactive compounds with potential therapeutic or functional properties.42,43Table 2 provides a comprehensive illustration of the computational algorithms used in Ganoderma research, including detailed descriptions and their specific applications.| Algorithm type | Algorithm name | Description | Specific application in Ganoderma research | Suggested visualization | References |
|---|---|---|---|---|---|
| Network pharmacology | Random walk with restart (RWR) | A probabilistic approach that assists in the identification of potential drug–target interactions by spreading information in a biological network | Applied to rank bioactive compounds in Ganoderma spp. for therapy | Network graph (Cytoscape) | 44 and 45 |
| Topological analysis (degree, betweenness, closeness, etc.) | Analyses the significance of genes/proteins considering connectivity in a biological network | Distinguishes the major hub genes in Ganoderma-mediated disease pathways (e.g., inflammation, cancer) | Protein–protein interaction (PPI) network visualization (STRING, Cytoscape) | 46 and 47 | |
| Machine learning | Support vector machine (SVM) | A supervised machine learning model classifying compounds based on molecular and pharmacological features | Utilized for the prediction of drug–target interactions and the classification of Ganoderma triterpenoids and polysaccharides | Classification plots (Python, R) | 46 and 48 |
| Random forest (RF) | An ensemble learning algorithm based on decision trees for classification and regression tasks | Screening candidate bioactive compounds from Ganoderma species for certain diseases | Decision tree diagram | 49 and 50 | |
| Molecular docking | AutoDock, AutoDock Vina | Computational programs that forecast how small molecules interact with target proteins, their binding affinity | Screens binding interactions of Ganoderma bioactive with disease proteins such as NF-κB and VEGFR | Docking pose visualization (PyMOL, Chimera) | 51 and 52 |
| Molecular dynamics (MD) | GROMACS, AMBER | Represents the physical motions of molecules to predict binding stability in a biological setting | Examines stability of Ganoderma bioactive in receptor binding sites at physiological conditions | RMSD, RMSF, and hydrogen bonding plots | 53 and 54 |
| Pharmacokinetics & ADME | SwissADME, pkCSM | Forecasts drug absorption, distribution, metabolism, and excretion (ADME) characteristics, as well as toxicity | Assesses the bioavailability and drug-likeness of Ganoderma metabolites for oral drugs | Radar plots, box plots | 55 and 56 |
| Pathway enrichment analysis | Gene ontology (GO), KEGG pathway | Identifies biological processes and pathways associated with specific genes and compounds | Enables comprehension of mechanisms of Ganoderma bioactive compounds in cancer, neuroprotection, and immunomodulation | KEGG pathway maps, functional enrichment charts | 57 and 58 |
Grienke et al.59 used computational approaches to predict bioactive compounds from G. lucidum for functional foods and nutraceuticals. From 279 Ganoderma constituents, the researchers established a database holding chemical structures and biological activities and performed in silico screening to make a simulation of the interaction between the above-mentioned compounds and biological targets related to viral infections and metabolic syndrome. They assessed how such compounds would bind to specific targets using 3D pharmacophore modeling, identifying key molecular interaction features. The researchers also applied 3D molecular docking in the prediction of the binding affinity between bioactive compounds and their targets; this is one of the most critical tools in bioinformatics for the prediction of molecular interaction using genome mining techniques, encompassing the screening of fungal genomes for BGCs or biosynthetic gene clusters that encode the manufacturing of bioactive secondary metabolites.60 Low-yielding NRPs (nonribosomal peptides) were heterologously expressed using systems to transfer them into other microbial hosts and enhance the production of useful compounds for food preservation and health-promoting ingredients. Another study by Cao et al.61 used bioinformatics enzyme modeling to predict enzymes responsible for the biosynthesis of the terpenic compound γ-cadinene. The focus was on the identification and modeling of terpene synthases, particularly γ-cadinene enzymes, of G. lucidum and G. sinensis. The researchers used bioinformatics tools to predict and experimentally validate three γ-cadinene enzymes. They generated an initial enzyme model, model 1, based on conserved amino motifs and further utilized this model to screen 67 homologous sequences from the NCBI database to fine-tune the model to model 2. However, the bioinformatics analysis confirmed that conserved regions in both models were highly similar, and five sequences were experimentally verified as γ-cadinene enzymes. This approach was also applied to other enzymes, such as Δ6-protoilludene from fungi and (−)-α-bisabolol from plants.62
Similarly, Schuller et al.63 utilized genome mining in combination with CRISPR-Cas9 genome editing to activate silent biosynthetic gene clusters in fungi. They used CRISPR-Cas9 to manipulate fungal genomes for the production of bioactive compounds that were not previously expressed, thus discovering new metabolites with practical relevance in food safety. Additionally, Yilmaz et al.64 developed the FunARTS tool, a bioinformatics tool that uses gene-guided screening for identifying fungal secondary metabolite gene clusters (BGCs) with bioactive potential. The FunARTS tool employs machine learning algorithms to predict and prioritize bioactive compounds, helping researchers focus on the most promising compounds for food industry applications, with an accuracy rate of 88%. Hautbergue et al.65 have integrated multi-omics approaches combining genome mining with metabolomics and CRISPR-Cas9 technologies to explore the full metabolic potential of fungi. Through integration, the novel bioactive compounds can be profiled by the entire set of metabolites produced under certain conditions, and this is important for discovering substances that are relevant to the process of enhancing food quality.
Shah et al.66 used mass spectrometry and molecular docking in the study of the bioactive compounds of Aspergillus ficuum for their different anti-bacterial, anti-inflammatory, and antioxidant activities. For the mass spectrometry analysis, detailed molecular structures emerged; hence, molecular docking was conducted for the prediction of the binding affinity of the compounds to various biological targets, thus validating their therapeutic potential in food safety applications. Kaliaperumal et al.67 utilized HPLC and NMR spectroscopy for the identification and structural characterization of secondary metabolites produced by Penicillium verruculosum. By using these techniques, such as detailed determination of the chemical structure of the metabolites, their interaction with biological targets was predicted by molecular docking. The authors confirmed that the anticancer activity of the metabolites opens potential avenues for functional food applications. Together, these studies underscore a wide range of applications for bioinformatics-based approaches in the food sector and especially in quality evaluation. Applied mass spectrometry combined with bioinformatics spectral library searches to screen and characterize fungal extracts for bioactive peptides. High-resolution mass spectrometry can analyze the structure of peprides in details, while bioinformatics tools can facilitate predictions of potential biological activities such as antimicrobial properties, hence greatly used in food preservation.68 Genome mining, molecular docking, and machine learning tools enable the identification of bioactive compounds that can be used in food preservation, flavor enhancement, and functional food development. These methods also have considerable significance in the discovery of natural preservatives, antimicrobial agents, antioxidants, and other bioactive compounds that advance improving nutritional value, safety, and food quality overall.69 For instance, several bioactive chemicals with potential uses in food preservation and health promotion have been identified as a result of genome mining of Ganoderma spp. which is illustrated in Fig. 1. Furthermore, these computational approaches provide an efficient and scalable way to evaluate fungal metabolites for application in food development, supporting the creation of safer, healthier, and more sustainable food products. It has been demonstrated that these substances, which include polysaccharides and triterpenoids, have anti-inflammatory, anti-cancer, and antioxidant qualities. Researchers can create focused interventions to enhance food safety and human health by comprehending the molecular mechanisms underlying these impacts.
2.2. AI-driven methods for the food industry
In recent years, the food industry has experienced a revolution toward using advanced technologies, especially artificial intelligence (AI), for screening and detection of natural components.70 The integration of AI with tools of genomics, proteomics, and metabolomics has revolutionized the identification and characterization of molecules responsible for the quality and authenticity of natural products.71 The production of biosynthetic gene clusters (BGCs) and data mining processes have emerged as critical requirements to identify genes that are related to secondary metabolism, while SVM and ANN, pattern recognition strategies driven by AI, play significant roles in discovering metabolic pathways and modules.72 New methodologies for predicting the production of metabolites have led to innovations that further open the fields of medicine and industry to more applications. The tools based on AI, which include deep learning and next-generation sequencing, come in handy in processing large datasets, like mass spectrometry spectra, to improve the prediction and classification of natural products. These innovations are improving quality control in the food industry and help ensure the safety of consumers, besides leading to better production processes.73 In recent years, food quality and safety have garnered increased attention due to rising food consumption and heightened consumer knowledge of food quality assurances. Detection and analysis techniques for volatile organic compounds (VOCs) are effective instruments for evaluating food product quality, owing to their non-destructive, environmentally friendly, continuous, and real-time monitoring advantages.74,75 ML-supported electronic nose, colorimetric sensor array (CSA), and gas chromatography (GC) hyphened techniques (e.g., GC-MS and GC-IMS) are becoming a hot research area in food sciences.76Machine learning and an electronic nose, or “e-nose,” can be used to anticipate the scents of beer. Following the detection of volatile compounds by nine gas sensors, two ANN models were created: one to forecast the intensity of ten sensory descriptors from trained panellists, and another to forecast the concentration of seventeen volatile compounds from GC-MS. By providing the beer industry with a quick, easy, and affordable way to predict aromas and perform quality control in real time, this approach increases the production efficiency and consistency.77 Moreover, some other researchers have developed a tool for rapid flavor characterization and quality evaluation of fermented bean curd by combining traditional analysis methods with sensor technology and algorithms. Employing HS-SPME-GC/MS and quantitative descriptive analysis (QDA), researchers discerned 63 volatile chemicals, with 13 principal components distinctly differentiating the samples. A cost-effective CSA was subsequently constructed using these chemicals. Integrating this system with powerful machine learning methods, including linear discriminant analysis (LDA), achieved an astounding accuracy of 97.22% in differentiating flavor profiles. The results indicate that integrating CSA with AI methodologies offers an effective and cost-efficient approach for food flavor profiling and quality assurance.78 Integrating AI with conventional quantitative approaches will facilitate the accurate identification of essential chemicals in Ganoderma spp. samples, hence increasing comprehensive profiling and quality assurance processes (Fig. 2). This integration guarantees enhanced accuracy, efficiency, and scalability in the analysis of intricate biological samples.
2.3. Leveraging machine learning in Ganoderma spp.
ML algorithms have become vastly important in predicting compound bioactivity, especially within those of natural origin. More or less, they are categorized into the forms of supervised and unsupervised methods.72 This kind uses labeled datasets for outcome prediction. In unsupervised learning, patterns and structures are identified in unlabelled data. Both are crucial tools when used to gauge bioactivity by chemical properties and biological effects.79 Zongur et al.80 proved the efficiency of support vector machines (SVM) and random forests (RF) in predicting the antifungal activity of extracts of Viburnum opulus L. against Fusarium strains. The SVM is a supervised learning algorithm that classifies data, finding the optimum hyperplane that separates the different classes, whereas RF utilizes an ensemble method to create multiple decision trees and aggregate those for more robust predictions. The given algorithms managed to score accuracy levels of 92.6%, thus proving the potential for the identification of bioactive compounds via ML. This study showed how ML can predict bioactivity at a low error rate, which is so important for the development of natural antifungal agents. This was complemented by Rielding et al.81 for predicting the bioactivity of fungal secondary metabolites (SMs) obtained from BGCs using ML. They showed that the accuracy of the model was between 51% and 68%, thereby showing some difficulties in working with smaller datasets. This accuracy level emphasizes the necessity of further developing datasets to better predict bioactivity screening.Building on the results above, Yang et al.82 utilized molecular docking in conjunction with ML for the prediction of anti-tumor activity of compounds derived from G. lucidum. Molecular docking is essentially a simulation of interactions between molecules: drug candidates and their biological target proteins and can be used to predict binding affinity and activity. They established an accuracy of 86% in the study; clearly, the integration of molecular docking with machine learning facilitates the streamlined identification of anti-tumor compounds, prioritizing them for experimental validation. In addition, Ni et al.83 prepared a model for the quantitative determination of polysaccharides and moisture in G. lucidum samples using NIR direct scanning with no destruction of the products and no crushing need of the samples. It utilizes the LSSVM algorithm as a machine learning approach to augment the model accuracy; the latter was further optimized using the ALO algorithm. The models of polysaccharides and moisture have good prediction efficiency, with Rp values of 0.9218 and 0.9581, respectively, indicating strong reliability to be used in the preliminary screening and process control. This methodology can conduct a speedy, non-destructive on-site quality test on natural products, which would provide important convenience for producers and consumers.
Riahi84 used supervised machine-learning techniques to optimize the hot water extraction process of polysaccharides from G. lucidum. This study applied the supervised learning technique to fine-tune the extraction parameters, resulting in a 10% increase in yield and a 20% improvement in the bioactivity of the extracted polysaccharides, hence bringing to view how ML can enhance traditional bioprocesses. Abrantes-Coutinho et al.85 pursued another route to couple electrochemical data with machine learning for glucose biosensing using G. applanatum lectin. Electrochemical sensors measure changes in current or voltage resulting from chemical reactions, and when integrated with ML models, they can detect specific analytes such as glucose with high accuracy. Their investigation led to a detection rate of 94%, proving that ML-based improvement in sensor designs boosts the accuracy of detecting glucose to be applied towards medical diagnosis.
Similarly, Zhu et al.86 utilized Fourier-transform infrared (FTIR) spectroscopy in combination with machine learning to distinguish between G. lucidum and G. sinense. FTIR spectroscopy determines the absorption of infrared light by molecular vibrations, giving each compound a unique spectral signature. They obtained an excellent classification accuracy of 99%, which therefore further indicates that the use of ML can be feasible in species identification and quality control in the pharmaceutical industry. Qiao et al.87 used an electronic nose (E-nose), combining it with FTIR spectroscopy and SVM, to distinguish pure from adulterated G. lucidum spore powder. SVM is an algorithm for training machine learning classifiers between classes. The authors were able to establish 95% accuracy in pure versus adulterated samples through such authentication, offering a fast yet reliable method for the determination of product authenticity. Chen et al.88 showed how GA and ANN are used to optimize extraction procedures in biotechnological applications. It employed GA, an optimization technique based on natural selection, in combination with ANN, a machine learning technique that simulates biological neural networks in terms of processing information, for the prediction and optimization of bioactive polysaccharides extracted from Bletilla striata. The optimization process resulted in a 16% increase in extraction efficiency, highlighting the ability of ML techniques to fine-tune extraction protocols. Similarly, Jandoust et al.89 used ANN to predict the production of bioactive compounds in Momordica charantia in response to elicitors. Their model achieved 87% accuracy in predicting metabolite production, demonstrating how ANN can help optimize biotechnological processes for the production of valuable bioactive compounds. Another application of ANN was done by a team of researchers with ANN and SVM, which were used to predict alginate lyase yield from Cunninghamella echinulata. The optimized production conditions through ANN and SVM led to an increase in enzyme yield by 14%, thereby demonstrating the ability of these machine learning models to optimize processes in biotechnology.90 The authors further demonstrated ML's potential in predicting antimicrobial properties using ANN to predict the extract of mushrooms. Their model identified key compounds that effectively inhibited bacterial biofilm formation with 76% accuracy and highlighted how ML can guide the discovery of new antimicrobial agents.91
The application of new advanced computational techniques and ML methods to the identification and screening of bioactive compounds in the food industries has substantially progressed over the last few years. This process is not only faster but also better for the reliability and efficiency of identifying potentially bioactive molecules.
For example, Zhang et al.92 developed an all-inclusive in silico protocol for screening antifungal peptides that enabled the identification of peptides active against Candida albicans, Candida krusei, Cryptococcus neoformans, and Candida parapsilosis. Their strong classification models yielded clear accuracy, with AUCs reaching 0.99, and allowed screening of more than three million peptide library sequences to identify three candidates with high antifungal indices. This computational approach provides a fast and efficient alternative to the traditional in vitro screening methods, hence minimizing laboratory work. Correspondingly, Walker et al.93 developed an ML bioinformatics approach to predict the biological activities (antibacterial, antifungal, or antitumor) of natural products isolated from BGCs. Their classifiers, which achieved 57–79% accuracy, performed especially well in predicting antibacterial activity, although antifungal predictions were less efficient due to sparse data. Structural features of natural products associated with biological activity, as identified in this study, have proved valuable in the analysis of the key molecular features critical to drug discovery.
Kim et al.94 investigated Natura Predicta™, a tool that uses natural language processing (NLP) for predicting and analyzing the bioactivity of botanical ingredients in Health Functioning Foods (HFF). They indicated similarities in strong bioactivity between several traditional herbal ingredients such as Camellia sinensis, G. lucidum, and Citrus sinensis, opening them to further research with HFF. However, the study highlighted some challenges, such as the dependency on PubMed for the data and polymorphism in nomenclature in botanical terms. Sun et al.95 developed QSAR models to predict nephrotoxicity in TCMs, using ANN and SVM. The natural product models showed the best results and high accuracy in predicting nephrotoxic ingredients by ANN (96.7%) and SVM (93.3%), meaning that it holds great promise in screening the toxic ingredients in TCMs.
Finally, Liu et al.96 used SERS combined with machine learning algorithms for the differentiation of wild and cultivated medicinal fungus Ophiocordyceps sinensis. Through optimized SVM models to analyze the SERS spectra, they obtained an impressive accuracy of 98.95%, thereby confirming the usability of this method for quality control and origin identification. This approach offers a high-speed and efficient tool for the authentication of medicinal raw materials such as O. sinensis. These studies collectively demonstrate the potential power of computationally based techniques, machine learning, and spectroscopy in the screening of active compounds for their accelerating roles in the discovery and development of functional bioactive ingredients in food and pharmaceutical industries.
These techniques would optimize bioactive compound identification, extraction, and production from natural sources, enhancing food quality, safety, and nutritional value. This method, which combines SERS with ML, has a huge amount of promise for identifying and verifying different Ganoderma spp. Researchers will be able to properly tell the difference between different species of these fungi by looking at their unique spectral signatures. This method could change the way quality control and standardization are done for Ganoderma-based goods, making sure that people get real, effective supplements. ML algorithms predict bioactivity, enhance yield efficiency, and ensure raw material authenticity. Thus, by streamlining production processes, eliminating waste, and improving quality control, these technological developments provide more sustainable cost-effective solutions for the food industry and the opportunity to create healthier and innovative foods.
2.4. Deep learning approaches in Ganoderma spp.
Deep learning techniques have transformed the detection of bioactive compounds, particularly in fungi and mushrooms, with wide-ranging applications in food safety, quality control, and identification of bioactive compounds. These AI-based models, especially CNN-based ones, have gained maximum usage in terms of improving identification, classification, and prediction accuracy in the food industry, presenting an application that aims to prevent poisoning from toxic fungi using artificial intelligence technology, more particularly CNNs and transfer learning.97 CNNs are composed of stacks, developed to handle visual data, meaning that they typically contain convolutional layers, pooling layers, and fully connected layers, while transfer learning is a technique wherein a pre-trained model on one task is fine-tuned for a new, related task. This method uses the knowledge gained on one dataset to apply it to another; it's particularly useful when the amount of data for training a model from scratch is limited. It is widely used in applications such as image recognition and drug discovery. The application, developed using Flutter, allows the photographing of fungi, which are then processed by a deep learning model built upon the Efficient NetV2 CNN architecture. With a classification accuracy of 97%, the model allows users to identify whether fungi are poisonous or non-poisonous. This AI-driven approach significantly reduces the risk of poisoning due to misidentification, demonstrating the potential for deep learning in enhancing public safety in food consumption. Similarly, AI is used for classifying fungal–fungal interactions, particularly for biocontrol fungi. Using deep learning networks, like DenseNet121, a type of CNN whereby each layer is connected to every other layer in a dense block. The study achieved 95% accuracy in classifying interactions between plant pathogens, such as Fusarium graminearum, and fungal strains. This enhanced the consistency and efficiency of the models that analyzed fungal interactions, eradicating subjective visual observations characteristic of the more traditional methods.98Additionally, this technology can be used for sustainable agriculture, as it could be applied in screening the biocontrol agent. Another promising development was mentioned by Korshunova et al.,99 in which molecular generation was optimized using generative neural networks and reinforcement learning. Generative neural networks are models that can be trained to generate new instances of data that closely mimic a certain training dataset, and reinforcement learning is a style of machine learning, where an agent learns to make decisions by interacting with an environment. This study focuses on the optimization of the bioactivity of protein targets through the use of transfer learning, experience replay, and real-time shaping rewards for AI-generated molecules. The developed method has been validated experimentally, which shows a hit rate of 27% for compounds with activity against the pharmacological target. This approach describes how AI can mimic the decision-making process of medicinal chemists, thus putting forward new chemical entities for drug discovery. Posansee et al.100 discovered AI-assisted methodologies for determining mTOR inhibitors from fungi. By applying the deep learning models based on PCA and structural methods, the researchers detected a trihydroxy sterol from Lentinus polychrous Lev. as a potential mTOR inhibitor. By integrating virtual screening with molecular docking, the study verified the role of steroid cores in the inhibition of mTOR, therefore bestowing importance to the AI-surveyed method in drug discovery, especially within the food and pharmaceutical sectors.
Dong et al.101 used deep learning to determine the content of polysaccharides in Lentinula edodes, using NIR spectroscopy. Based on comparisons between models, such as siPLS and 1D-CNN, this work demonstrated how 1D-CNN models surpassed others with an accuracy of 95.50%. This development opens new avenues for the food industry, mainly for the quality control and nutritional grading of edible mushrooms, with products consistently meeting market standards. Authors also utilized FT-NIR spectroscopy and deep learning, specifically a particular type of deep neural network known as residual neural networks (ResNet) to identify various species of bolete mushrooms. ResNet improves training deep networks by using residual connections, through which networks learn more quickly and faster. The results show a 100% accuracy with the potential of improving species identification through the application of deep learning techniques. Wang et al.102 developed a deep learning-based method to determine the shelf life of Phlebopus portentosus, a type of porcini mushroom, using Fourier Transform Near-Infrared (FT-NIR) spectroscopy. The method, combined with machine learning and two-dimensional correlation spectroscopy (2DCOS), demonstrated high accuracy and non-destructive rapid results in determining freshness levels. This approach could significantly enhance food safety and reduce food waste by providing fast and reliable freshness analysis for mushrooms in the food industry.103 Authors harnessed deep learning approaches, especially CNNs, for the identification of edible mushroom species and assessment of their quality. Exploiting spectral data from FTIR or MIR spectra and 2DCOS, it obtained an accuracy of up to 99.76% in species identification and geographic origin determination. These methods including ANN help in the detection of physical damage via NIR spectra and are non-destructive and fast and reliable tools for mushroom industry quality control and traceability.104
Fungal contamination was assessed in rapeseed by implementing CNN, MLPN, and RBFN for mold identification. The results showed that the best performance, with only 14% classification error, was obtained using CNN; its accuracy was higher than other networks. This deep learning method might prove to be a good way of providing efficient detection of crop's fungal contamination that may lead to enhanced food safety.105 Li et al.106 proposed a deep learning model that integrated CNN and Bi-LSTM (Bi-LSTM is a variant of RNN that reads input in both forward and backward directions. Therefore, it tries to capture the context from information that can be provided by both past and future knowledge for tasks like sequential data, such as peptide classification or bioactivity prediction from protein sequences) for bioactive peptide classification from protein sequences. Using stacked architecture enhanced the model's predictive performance and revealed the distribution of amino acids among different types of peptides. Such a method is useful in finding new bioactive peptides, which have food industry applications specifically in generating functional foods with health benefits. Collectively, these studies point out the revolutionary capabilities of AI in the food industry primarily in food safety, quality control, and bioactive compound discovery. These AI models, making use of techniques such as CNNs, reinforcement learning, and transfer learning, would be able to analyze huge volumes of data autonomously, discover compounds, and provide precision and efficiency in the evaluation of food quality. The future of Ganoderma research and manufacturing holds enormous promise as it embraces AI-driven technology. These advancements will not only improve the quality and efficacy of Ganoderma-based products but also pave the way for the development of novel medicines and functional foods. Food processing and product development are integrated with these technologies to enhance more sustainable, safer, and higher-quality food products for the benefit of consumers and industries all over the world. Other AI-driven approaches in various fungal species with their bio-functional activities are illustrated in Table 3.
| Fungal species | Bioactive compound | AI technique used | Bio-functional activities | References |
|---|---|---|---|---|
| Ganoderma lucidum | Polysaccharides, triterpenes | Metabolomics analysis | ACE inhibitory antitumor and antioxidant properties | 107 |
| Cordyceps militaris | Cordycepin, polysaccharides | Machine learning models | Immunomodulation, antioxidant, and ACE inhibitory | 108 and 109 |
| Lentinula edodes | Eritadenine, polysaccharides | QSAR models | Anti-inflammatory and antidiabetic | 110 |
| Hericium erinaceus | Erinacines, hericenones | AI-driven pathway analysis | Identification of neuroprotective and antioxidant bioactive pathway | 111 |
| Agaricus bisporus | β-Glucan, phenolic compound | Chemometric analysis | Antitumor, antioxidant, and immunomodulatory | 112 |
| Inonotus obliquus | Betulinic acid, polysaccharides | Spectrometric analysis with AI | Prediction of anti-inflammatory potential | 113 |
| Coprinus comatus | Polysaccharide, protein | Deep learning and machine learning for metabolite prediction | Optimizing compounds for antidiabetic effects | 114 |
| Aspergillus oryzae | Polyphenols, kojic acid | AI-driven quality control system | Antidiabetic and anticancer | 115 and 116 |
| Rhizopus oryzae | Fumaric acid | AI-driven process optimization | Antioxidant | 117 |
| Neurospora crassa | Carotenoids, polyketides | Machine learning for pathway prediction | Antifungal | 118 and 119 |
3. Integrating AI to revolutionize Ganoderma food production
Food has been the most direct source of life support and energy accumulation for humans since the beginning of time when body tissues were formed. Technology, engineering, microbiology, physics, chemistry, biochemistry, and sensory evaluation are all included in the multidisciplinary subject of food science.120 In today's world, AI has an impact on food production by copying how humans think, learn, and store knowledge.121,122 AI helps a lot in the current food processing cycle, making it easier to sort, analyze, and package food. When people eat the food, AI works well to check how happy customers, are and to handle issues with getting food where it needs to go, farmers in all areas of farming have started to use AI-based tools to analyze their work.123 Numerous researchers use neural networks to predict agricultural production. AI, ML, and DL significantly influence various contemporary agricultural challenges, particularly those associated with climate change. These sophisticated computer algorithms facilitate the selection and categorization of agricultural goods depending on variables such as soil composition. They enhance agriculture by monitoring crops, analyzing soil, and managing water resources.124 In modern times, AI contributes to ensuring food safety and quality from production to consumption, addressing concerns related to climate change. AI utilizes predictive models to identify prospective dietary dangers, rendering it an invaluable instrument for managing future uncertainties.125The implementation of AI, data analytics, and intelligent processes in manufacturing, production, and operations has several challenges and managerial implications. Various impediments may emerge, including the identification of requisite skills and abilities for people, the adoption of applications, and the management of diverse productivity and performance challenges. Numerous potential and associated obstacles exist for the supported supply management duties. Consequently, the study needs to bolster operations by advocating for AI methodologies that enhance intelligent processes throughout diverse industrial sectors, while also anticipating potential hazards and vulnerabilities.126,127 The dynamic conditions that currently exist are pressuring food production enterprises to consistently innovate new goods. As a result, significant efforts have been made over the past ten years to pinpoint the variables that influence the effectiveness of new product development initiatives.128 The research indicates that a significant portion of the food industry's prior efforts in the creation of new products have been lost due to product failure. According to reports, new food products fail 70–80% of the time.129 AI is integrated into every stage of the food production cycle, encompassing sorting, analysis, and packing. Furthermore, it is advantageous to examine consumer happiness with challenges within the food supply chain and distribution during the consuming phase.130
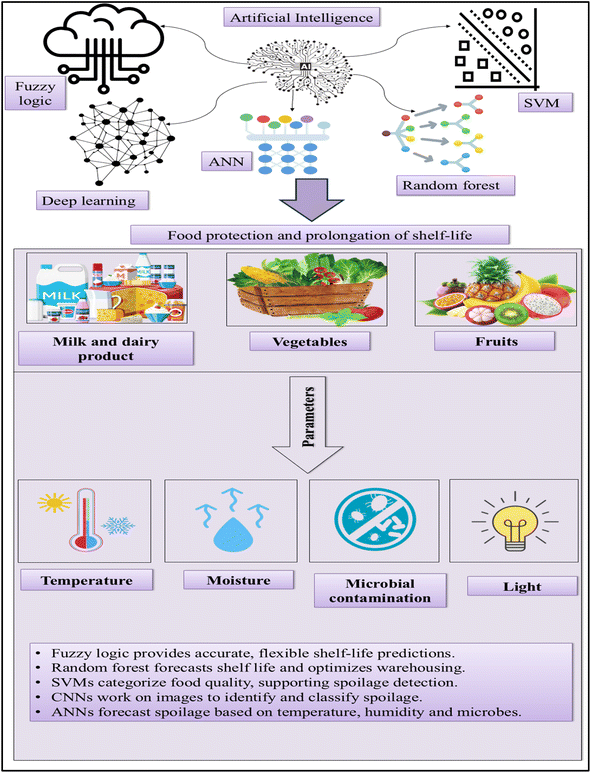
Food processing facilities employ environmental monitoring methods to identify contamination and confirm the efficacy of control measures. To simulate sampling tactics and corrective measures, recent studies have created agent-based models of Listeria contamination in food facilities.131 Model performance and decision support can be enhanced by combining machine learning with agent-based modeling.132 In order to extend food shelf life with AI models, Fig. 3 demonstrates how ML algorithms and predictive modelling optimize storage conditions, detect spoilage, and enhance food quality monitoring.
Popular approaches that have been used in the food sectors include expert systems, fuzzy logic, ANN, adaptive neuro-fuzzy inference systems (ANFIS), and ML. The application of AI in the food business has been growing over the past few decades due to its many benefits. AI is transforming the way food is produced from Ganoderma spp., making the process of cultivation and extraction more efficient and optimizing product development. AI controls the environment in such a way that the Ganoderma is cultured uniformly and efficiently, enhancing the production of bioactive triterpenoids. Additionally, AI enhances sustainability by facilitating improved agricultural practices with efficient, optimized resource utilization, and minimized environmental impact.133 It is also very important in enhancing yield, nutritional absorption, and synthesis of beneficial substances during the cultivation of Ganoderma. It ensures quality control and process optimization through machine learning and data analytics in the growth of fungi and manufacturing bio-composites during maintenance, which are consistent and have a lower ecological footprint. In addition, AI facilitates the development of mycelium-bound bio-composites made from lignocellulosic substrates by optimizing various stages of the production process.134 Ongoing research continues to explore AI's potential in scaling up and advancing Ganoderma food production, offering a promising future for both sustainability and innovation in this field. Ongoing research continues to highlight AI's potential in scaling and advancing Ganoderma-based food production.
While the implementation of AI in Ganoderma spp. research and other biotechnology domains may be expensive, certain methods can be employed to effectively manage these expenditures. One approach is to employ cloud-based AI solutions offered by companies such as Google Cloud AI, AWS SageMaker, and Microsoft Azure AI, which provide scalable, pay-as-you-go alternatives, thereby eliminating the necessity for substantial upfront infrastructure investments. Additionally, open-source AI frameworks such as TensorFLow, PyTorch, and Scikit-learn may significantly reduce software license costs by providing robust tools for bioinformatics analysis.135 Moreover, pre-trained AI models and transfer learning are suitable for tasks such as species identification, since they mitigate computing demands. Efficient data management techniques such as dimensionality reduction and feature selection minimize redundant computations, thereby saving resources.136 Distributed computing and crowdsourcing offer economical solutions for data processing and annotation; also, obtaining government grants and funding from organizations such as the National Institutes of Health (NIH) and National Science Foundation (NSF) helps alleviate budgetary burdens. Furthermore, employing energy-efficient algorithms and enhancing AI models would contribute to reducing costs and energy consumption. Integrating these technologies will allow resource-constrained universities to more effectively and sustainably incorporate AI into Ganoderma research.
4. Challenges, limitations and future prospects
Predictive modeling in species distribution modeling (SDM) has improved greatly with AI, using improved machine learning techniques in the context of G. lucidum. This technique incorporates comprehensive environmental predictions and uses multiple algorithms to give a holistic output analysis of the species distribution. Through this, scientists can best identify the ecological niche of G. lucidum and predict its potential distribution in varying climates.137 In addition, AI is also important in genomic and transcriptomic studies, which analyze large data sets to determine key genes that play important roles in critical biological pathways. This makes it possible to gain deeper insights into the pathogenesis, biosynthesis of bioactive compounds, and cellular development of Ganoderma spp.138 Moreover, AI-powered detection and identification methods such as sensors based on sub-20 nm gold electrodes have shown excellent sensitivity in identifying specific Ganoderma species. These sensors allow accurate discrimination of biomolecules, a key capability for identifying the most virulent strains. In biotechnology, AI speeds up the identification of potential uses of Ganoderma, such as enzyme production, bioremediation, and bioactive compound extraction, thus promoting advancement in both the industrial and pharmaceutical industries. Despite these advantages, AI-based approaches present several significant challenges.139 One of the main limitations is the data-hungry and complex nature of AI models, especially in genomic analysis and ecological niche modeling. Models are highly dependent on large, high-quality datasets to make reliable predictions, and the lack of complete data can hinder their development and validation.140 Furthermore, hybrid AI systems that combine mechanical and cybernetic models use heuristic designs, i.e., their optimal setups are not always clearly defined, which can hinder their effectiveness. Another concern is the interpretability of AI models, especially deep learning and neural networks, which tend to be “black boxes.” Their lack of transparency makes model predictions difficult to interpret, potentially restricting their use in biological studies.141 Additionally, the use of AI technologies requires a significant amount of computational capacity and technical skills, serving as a tremendous barrier for smaller research teams or less endowed institutions. These may discourage the global spread and acceptance of AI among Ganoderma studies, despite its revolutionary role.The regulatory concerns of Ganoderma spp. products primarily focus on ensuring their quality, safety, and efficacy. Primary issues are the standardization of bioactive compounds, since discrepancies in manufacturing and processing may result in variations in active component concentrations, therefore affecting consumer safety and product reliability.142 Moreover, precise labelling is crucial, since mislabelling or insufficient transparency about ingredients and strength may deceive consumers and jeopardize their health. Ensuring product quality relies on compliance with Good Manufacturing Practices (GMP), prompting regulatory agencies to emphasize adherence to these standards.143
Although GMP is necessary but pre-market is not, manufacturers are nonetheless responsible for product integrity since Ganoderma products are regulated by the Food and Drug Administration (FDA) in the US as dietary supplements. However, regulations in the European Union and other Asian countries, such as China, are occasionally more stringent and require clinical data to support health claims and establish technical specifications for Ganoderma products.144 The incorporation of Ganoderma spp. into mainstream healthcare is made possible by translational efforts to bridge traditional uses and modern therapeutic applications, standardizing extraction techniques to guarantee consistent levels of bioactive compounds, conducting clinical trials, and using evidence-based research to validate health claims.145
The integration of artificial intelligence in the food industry brings forth both challenges and future directions. AI has the potential to completely transform the food sector, particularly in terms of maximizing the sustainability, safety, and quality of Ganoderma species utilized in functional foods and pharmaceuticals. Complex data from electronic noses and GC-MS may be precisely analyzed thanks to advanced ML techniques such as SVM and deep learning. This opens up new health advantages and treatment possibilities by enabling quick, high-accuracy screening of bioactive substances, such as terpenoids and polysaccharides, in Ganoderma spp., namely G. lucidum and G. boninense. To increase the yields and improve the synthesis of bioactive compounds, AI-driven prediction models may optimize growth conditions by examining variables such as temperature, humidity, and soil health. AI is also used in precision farming to track crop health and predict the best times to harvest, guaranteeing optimal quality for food and medicine applications. AI can also improve food safety by putting in place real-time quality control systems that identify contamination or rotting threats early on, guaranteeing the lifespan and safety of goods made from Ganoderma. Forward-thinking enterprises, such as Swiss Agrotech company Gayama, best represent the future of AI in agriculture. Gayama uses hyperspectral imagery and AI analysis to advance resource management, insect identification, and agricultural yield prediction, giving farmers the insights they need to boost productivity and sustainability.146,147 As AI continues to evolve, its integration into food systems, especially in the context of Ganoderma, promises not only improved health outcomes but also a more efficient, resilient, and sustainable food industry.
A key challenge for AI in food systems is ensuring reliable and robust data, as AI relies on accurate datasets for effective predictions and recommendations. Data security, privacy, and addressing biases in AI models are critical issues that must be addressed to prevent discriminatory outcomes. Promoting fairness and inclusivity, alongside transparency, accountability, and responsible AI usage, is essential to build consumer and stakeholder trust. Regulatory frameworks should be developed to uphold ethical standards.148 Looking ahead, AI's potential in the food sector is promising, with advancements in machine learning, natural language processing, and computer vision, as well as increased IoT and sensor integration, enabling real-time monitoring and addressing global challenges like sustainability and food security.149,150
5. Conclusion
The potential of food biotechnology and pharmaceutical applications is particularly promising, especially concerning functional foods and therapeutic treatments. Ganoderma P. Karst., a medicinal mushroom, has been utilized for centuries as an anticancer and immunopotentiating agent, although the molecular mechanisms underlying its bioactivity remain unclear. Future research would maximize potential by identifying such pathways and examining the interaction of Ganoderma with other pharmaceuticals. AI significantly transforms the food business by optimizing production processes and improving the quality and safety control of food products. Utilizing advanced ML, it can interpret complex data sets from electronic noses or gas chromatography sources, hence facilitating the discovery of bioactive compounds inside Ganoderma. AI models can forecast growth circumstances, thus facilitating the production of more efficient quantities and superior yields of Ganoderma useful substances. Notwithstanding these advancements, significant obstacles persist in integrating AI into food systems. Challenges include data accuracy and quality, security, and the potential for bias in AI model development. To cultivate trust between consumers and AI, industry stakeholders must address regulatory concerns that are essential for fostering transparency and accountability through ethical AI usage rules. These holds promise for the future as it may enhance sustainability and efficiency in food production, especially for the cultivation and utilization of Ganoderma. Advancements in technology suggest that AI could significantly influence global challenges, including food security and environmental sustainability, by fostering a more resilient and inventive agricultural business.Data availability
Data are available from the authors upon request.Author contributions
Sonali Khanal, Aman Sharma, Manjusha Pillai and Pratibha Thakur drafted the manuscript. Ashwani Tapwal, Rachna Verma, Vinod Kumar and Dinesh helped in manuscript revision. All authors read and approved the final manuscript for publication.Conflicts of interest
The authors do not possess any financial or other competing interests that require disclosure.Acknowledgements
This research work has been funded by DST-SERB, Govt. of India under CRG project vide sanction order number CRG/2021/001815 and this article has been produced with the financial support of the European Union under the REFRESH – Research Excellence for region Sustainability and High-tech industries project number CZ.10.03.01/00/22_003/0000048 via the operational Programme Just Transition.References
- A. L. Loyd, B. W. Held, E. R. Linder, J. A. Smith and R. A. Blanchette, Fungal Biol., 2018, 122, 254–263 Search PubMed.
- S. C. Karunarathna, A. Ediriweera, K. Prasannath, Y. Mingfei and K. K. Hapuarachchi, N. Z. J. Bot., 2024, 1–85 Search PubMed.
- Y. Cao, X. Xu, S. Liu, L. Huang and J. Gu, Front. Pharmacol, 2018, 9, 1217 CrossRef CAS PubMed.
- N. Yadav, R. Deshmukh and R. Majumder, Pharmacological Research - Modern Chinese Medicine, 2024, 100423 Search PubMed.
- K. K. Hapuarachchi, W. A. Elkhateeb, S. C. Karunarathna, C. R. Cheng, A. R. Bandara, P. Kakumyan, K. D. Hyde, G. M. Daba and T. C. Wen, Mycosphere, 2018, 9, 1025–1052 CrossRef.
- A. Senapati, S. Basak and L. Rangan, Drug Saf., 2021, 45, 193–213 CrossRef PubMed.
- M. Ghobad-Nejhad, R. H. Nilsson and A. Ordynets, Microbial Genomics: Clinical, Pharmaceutical, and Industrial Applications, 2024, pp. 173–203 Search PubMed.
- O. F. Nwabor, H. Onyeaka and O. Nwaiwu, Forest Fungi: Biodiversity, Conservation, Mycoforestry and Biotechnology, 2025, pp. 125–145 Search PubMed.
- M. Reste, K. Ajazi, A. Sayi-Yazgan, R. Jankovic, B. Bufan, S. Brandau, E. S. Bækkevold, F. Petitprez, M. Lindstedt, G. J. Adema and C. R. Almeida, Front. Immunol., 2024, 1439413, DOI:10.3389/FIMMU.2024.1439413.
- S. C. Karunarathna, A. Ediriweera, K. Prasannath, Y. Mingfei and K. K. Hapuarachchi, N. Z. J. Bot., 2024, 1–85, DOI:10.1080/0028825X.2024.2375996.
- M. Berovic and J. J. Zhong, Adv. Biochem. Eng./Biotechnol., 2022, 184, 125–161 CrossRef PubMed.
- A. Carreira-Casais, P. Otero, P. Garcia-Perez, P. Garcia-Oliveira, A. G. Pereira, M. Carpena, A. Soria-Lopez, J. Simal-Gandara and M. A. Prieto, Int. J. Environ. Res. Public Health, 2021, 18, 9153 CAS.
- Y. A. Bhadange, J. Carpenter and V. K. Saharan, ACS Omega, 2024, 9, 31274–31297 CrossRef CAS PubMed.
- P. K. Mukherjee, S. Banerjee, B. Das Gupta and A. Kar, Evidence-Based Validation of Herbal Medicine: Translational Research on Botanicals, 2022, pp. 1–41 Search PubMed.
- S. Bagade, D. D. Patil and A. Shirkhedkar, Herbal Bioactive-Based Drug Delivery Systems: Challenges and Opportunities, 2022, pp. 393–407 Search PubMed.
- S. S. Gaikwad, Y. Y. Morade, A. M. Kothule, S. J. Kshirsagar, U. D. Laddha and K. S. Salunkhe, Heliyon, 2023, e16561, DOI:10.1016/j.heliyon.2023.e16561.
- K. Chaudhary and A. Rajora, Phytochem. Rev., 2024, 24, 165–195 CrossRef.
- C. Caleja, A. Ribeiro, M. F. Barreiro and I. C. F. R. Ferreira, Curr. Pharm. Des., 2017, 23, 2787–2806 CAS.
- L. Sammugam and V. R. Pasupuleti, Crit. Rev. Food Sci. Nutr., 2019, 59, 2746–2759 Search PubMed.
- H. Forootanfar and M. A. Faramarzi, Biotechnol. Prog., 2015, 31, 1443–1463 CrossRef CAS PubMed.
- M. Mahuri, M. Paul and H. Thatoi, Syst. Microbiol. Biomanuf., 2023, 3, 533–551 CrossRef CAS.
- K. Sellami, A. Couvert, N. Nasrallah, R. Maachi, M. Abouseoud and A. Amrane, Sci. Total Environ., 2022, 806, 150500 CrossRef CAS PubMed.
- C. Della Torre, D. Maggioni, A. Ghilardi, M. Parolini, N. Santo, C. Landi, L. Madaschi, S. Magni, S. Tasselli, M. Ascagni, L. Bini, C. La Porta, L. Del Giacco and A. Binelli, Environ. Pollut., 2018, 241, 999–1008 CrossRef CAS PubMed.
- N. J. Rowan and C. M. Galanakis, Sci. Total Environ., 2020, 141362, DOI:10.1016/J.SCITOTENV.2020.141362.
- N. M. Bakhori, N. A. Yusof, A. H. Abdullah and M. Z. Hussein, Biosensors, 2013, 3, 419–428 CAS.
- S. W. Dutse, N. A. Yusof, H. Ahmad, M. Z. Hussein, Z. Zainal and R. Hushiarian, Int. J. Electrochem. Sci., 2013, 8, 11048–11057 CAS.
- I. Kalita, S. Bhattacharjee and M. Saharia, Biotechnology, Multiple Omics, and Precision Breeding in Medicinal Plants, 2025, pp. 135–145 Search PubMed.
- R. Gupta, D. Srivastava, M. Sahu, S. Tiwari, R. K. Ambasta and P. Kumar, Mol. Diversity, 2021, 25, 1315–1360 CAS.
- C. F. Shi, H. T. Yang, T. T. Chen, L. P. Guo, X. Y. Leng, P. B. Deng, J. Bi, J. G. Pan and Y. M. Wang, Bioresour. Technol., 2022, 357, 127248 CrossRef CAS PubMed.
- C. Pascale, R. Sirbu and E. Cadar, Eur. J. Nat. Sci. Med., 2022, 5, 40–48 Search PubMed.
- Megha and N. Singh, Transforming Agriculture Residues for Sustainable Development: from Waste to Wealth, 2024, pp. 109–129 Search PubMed.
- A. C. S. Pais, E. R. Coscueta, M. M. Pintado, A. J. D. Silvestre and S. A. O. Santos, Heliyon, 2024, 10(7), 27 Search PubMed.
- H. Cao, O. Saroglu, A. Karadag, Z. Diaconeasa, G. Zoccatelli, C. A. Conte-Junior, G. A. Gonzalez-Aguilar, J. Ou, W. Bai, C. M. Zamarioli and others, Food Front., 2021, 2, 109–139 CAS.
- L. K. Vora, A. D. Gholap, K. Jetha, R. R. S. Thakur, H. K. Solanki and V. P. Chavda, Pharmaceutics, 2023, 15, 1916 CrossRef CAS PubMed.
- Y. Zhang, X. Bao, Y. Zhu, Z. Dai, Q. Shen and Y. Xue, Trends Food Sci. Technol., 2024, 104578 CrossRef CAS.
- H. Onyeaka, P. Tamasiga, U. M. Nwauzoma, T. Miri, U. C. Juliet, O. Nwaiwu and A. A. Akinsemolu, Sustainability, 2023, 15, 10482 Search PubMed.
- R. Roy, S. Marakkar, M. P. Vayalil, A. Shahanaz, A. P. Anil, S. Kunnathpeedikayil, I. Rawal, K. Shetty, Z. Shameer, S. Sathees and others, Recent Adv. Food, Nutr. Agric., 2022, 13, 27–50 Search PubMed.
- E. Ramirez-Asis, J. Vilchez-Carcamo, C. M. Thakar, K. Phasinam, T. Kassanuk and M. Naved, Mater. Today: Proc., 2022, 51, 2462–2465 Search PubMed.
- C. Martinez-Castillo, G. Astray, J. C. Mejuto and J. Simal-Gandara, eFood, 2020, 1, 69–76 Search PubMed.
- A. Amore and S. Philip, Frontiers in Industrial Microbiology, 2023, 1, 1–4 Search PubMed.
- S. Singh, K. Kumar, A. Rao and V. K. Prajapati, in Endophytic Fungi: the Hidden Sustainable Jewels for the Pharmaceutical and Agricultural Industries, ed. B. P. Singh, A. M. Abdel-Azeem, V. Gautam, G. Singh and S. K. Singh, Springer International Publishing, Cham, 2024, pp. 191–208 Search PubMed.
- T. A. Holton, V. Vijayakumar and N. Khaldi, Trends Food Sci. Technol., 2013, 34, 5–17 CAS.
- V. Sharma and I. N. Sarkar, Briefings Bioinf., 2013, 14, 238–250 Search PubMed.
- D. H. Le, 2017 4th NAFOSTED Conference on Information and Computer Science, NICS 2017 – Proceedings, 2017, pp. 242–247 Search PubMed.
- X. Wu, S. Cao, Y. Zou and F. Wu, PLoS One, 2023, 18, e0294772 CAS.
- N. Chen, G. Wan and X. Zeng, Front. Oncol., 2022, 12, 833375 CAS.
- T. T. Nguyen, T. K. Dao, D. T. Pham and T. H. Duong, Symmetry, 2024, 16, 462 Search PubMed.
- R. Vyas, S. Bapat, E. Jain, S. S. Tambe, M. Karthikeyan and B. D. Kulkarni, Comb. Chem. High Throughput Screening, 2015, 18, 658–672 CAS.
- Y. Li, J. Y. Zhang and Y. Z. Wang, Anal. Bioanal. Chem., 2018, 410, 91–103 CAS.
- T. Shen, H. Yu and Y. Z. Wang, Molecules, 2020, 25, 1442 CAS.
- Z. Wu, X. Chen, W. Ni, D. Zhou, S. Chai, W. Ye, Z. Zhang, Y. Guo, L. Ren and Y. Zeng, RSC Adv., 2021, 11, 11821–11843 RSC.
- Y. Xie and S. Wang, Quality Control of Chinese Medicines, 2024, pp. 187–228 Search PubMed.
- Y. Yu, S. Xu, R. He and G. Liang, J. Agric. Food Chem., 2023, 71, 2684–2703 CrossRef CAS PubMed.
- S. Z. Gheshlaghi, A. Ebrahimi and Z. Faghih, J. Biomol. Struct. Dyn., 2022, 40, 10835–10851 CrossRef CAS PubMed.
- Y. O. Ayipo, I. Ahmad, Y. S. Najib, S. K. Sheu, H. Patel and M. N. Mordi, J. Biomol. Struct. Dyn., 2023, 41, 1959–1977 CAS.
- J. Shi, J. Chen, C. Cheng, W. Li, M. Li, S. Ye, Z. Liu and Y. Ding, Curr. Pharm. Des., 2025 DOI:10.2174/0113816128365423250126035306.
- J. Zhang, W. Zhang, H. Zhang, W. Zhang, C. He, H. Yu and G. Xin, Food Biosci., 2024, 62, 105249 CAS.
- X. Ma, Z. Huo, M. Shi, P. Zhang, T. Yang, J. Xiao and N. Gong, Liaoning Acad. Agric. Sci., 2024 DOI:10.21203/RS.3.RS-4380809/V1.
- U. Grienke, T. Kaserer, F. Pfluger, C. E. Mair, T. Langer, D. Schuster and J. M. Rollinger, Phytochemistry, 2015, 114, 114–124 CAS.
- A. Vassaux, L. Meunier, M. Vandenbol, D. Baurain, P. Fickers, P. Jacques and V. Leclère, Biotechnol. Adv., 2019, 37, 107449 CAS.
- R. Cao, X. Wu, Q. Wang, P. Qi, Y. Zhang, L. Wang and C. Sun, ACS Omega, 2022, 7, 7229–7239 CrossRef CAS PubMed.
- R. Cao, X. Wu, Q. Wang, P. Qi, Y. Zhang, L. Wang and C. Sun, ACS Omega, 2022, 7, 7229–7239 CAS.
- A. Schüller, L. Studt-Reinhold and J. Strauss, Pharmaceutics, 2022, 14, 1837 Search PubMed.
- T. M. Yilmaz, M. D. Mungan, A. Berasategui and N. Ziemert, Nucleic Acids Res., 2023, 51, W191–W197 CAS.
- T. Hautbergue, E. L. Jamin, L. Debrauwer, O. Puel and I. P. Oswald, Nat. Prod. Rep., 2018, 35, 147–173 CAS.
- Z. A. Shah, K. Khan, H. U. Rashid, T. Shah, M. Jaremko and Z. Iqbal, BMC Microbiol., 2022, 22, 295 CAS.
- K. Kaliaperumal, L. Salendra, Y. Liu, Z. Ju, S. K. Sahu, S. Elumalai, K. Subramanian, N. M. Alotaibi, N. Alshammari, M. Saeed and R. Karunakaran, Front. Microbiol., 2023, 1216928, DOI:10.3389/fmicb.2023.1216928.
- T. Ahmed, A. Juhász, U. Bose, N. Shiferaw Terefe and M. L. Colgrave, J. Funct. Foods, 2024, 119, 106343 CAS.
- V. Bisht, B. Das, A. Hussain, V. Kumar and N. K. Navani, npj Science of Food, 2024, 8, 67 Search PubMed.
- T. Bidyalakshmi, B. Jyoti, S. M. Mansuri, A. Srivastava, D. Mohapatra, Y. B. Kalnar, K. Narsaiah and N. Indore, Food Eng. Rev., 2024, 1–28 Search PubMed.
- A. Ikram, H. Mehmood, M. T. Arshad, A. Rasheed, S. Noreen and K. T. Gnedeka, CyTA--Journal of Food, 2024, 22, 2393287 Search PubMed.
- Y. Yuan, C. Shi and H. Zhao, ACS Synth. Biol., 2023, 12, 2650–2662 Search PubMed.
- A. Shankar and K. K. Sharma, Appl. Microbiol. Biotechnol., 2022, 106, 3465–3488 CAS.
- E. Kiran, K. Kaur and P. Aggarwal, Qual. Assur. Saf. Crop Foods, 2022, 14, 9–18 CAS.
- F. Y. H. Kutsanedzie, L. Hao, S. Yan, Q. Ouyang and Q. Chen, Sens. Actuators, B, 2018, 254, 597–602 CAS.
- Y. Feng, A. Soni, G. Brightwell, M. M. Reis, Z. Wang, J. Wang, Q. Wu and Y. Ding, Trends Food Sci. Technol., 2024, 104555 CAS.
- C. Gonzalez Viejo and S. Fuentes, Fermentation, 2020, 6, 104 Search PubMed.
- X. Zhang, X. Huang, J. H. Aheto, Y. Ren, L. Wang, S. Yu and Y. Wang, Food Biosci., 2024, 60, 104291 CAS.
- M. K. Goshisht, ACS Omega, 2024, 9, 9921–9945 Search PubMed.
- A. Zongur, H. Kavuncuoglu, E. Kavuncuoglu, T. D. Capar, H. Yalcin and M. A. Buzpinar, J. Microbiol. Methods, 2022, 192, 106379 Search PubMed.
- O. Riedling, A. S. Walker and A. Rokas, Microbiol. Spectrum, 2024, 12, e0340023 Search PubMed.
- Q. Yang, L. Yao, F. Jia, G. Pang, M. Huang, C. Liu, H. Luo and L. Fan, Chemom. Intell. Lab. Syst., 2024, 255, 105271 CAS.
- H. Ni, W. Fu, J. Wei, Y. Zhang, D. Chen, J. Tong, Y. Chen, X. Liu, Y. Luo and T. Xu, LWT--Food Sci. Technol., 2023, 184, 115001 CrossRef CAS.
- A. Riahi, PhD thesis, University College London, 2022.
- V. E. Abrantes-Coutinho, A. O. Santos, B. E. B. Holanda, H. R. A. Costa and T. M. B. F. Oliveira, Bioelectrochemistry, 2023, 151, 108392 CrossRef CAS PubMed.
- Y. Zhu, T. L. Tan and W. K. Cheang, Chemom. Intell. Lab. Syst., 2017, 171, 55–64 Search PubMed.
- J. Qiao, J. Li, C. Liu, Y. Zhang, S. Liu, X. Weng and Z. Chang, Efficient Detection technology of Ganoderma lucidum Spore Powder Quality Based on Electronic Nose, available at SSRN 4990847, DOI:10.2139/ssrn.4990847.
- H. Chen, B. Wang, J. Li, J. Xu, J. Zeng, W. Gao and K. Chen, Int. J. Biol. Macromol., 2023, 226, 982–995 CrossRef CAS PubMed.
- S. Jandoust, A. Shojaeiyan, M. Ayyari, M. Tohidfar, H. Ahmadi and S. N. Ebrahimi, Ind. Crops Prod., 2023, 192, 115984 CrossRef CAS.
- C. E. De Farias Silva, G. Y. S. C. M. Costa, J. V. Ferro, F. de Oliveira Carvalho, B. M. V. da Gama, L. Meili, M. C. dos Santos Silva, R. M. R. G. Almeida and J. Tonholo, React. Kinet., Mech. Catal., 2022, 135, 3155–3171 Search PubMed.
- J. Vunduk, A. Klaus, V. Lazić, M. Kozarski, D. Radić, O. Šovljanski and L. Pezo, Antibiotics, 2023, 12, 627 Search PubMed.
- J. Zhang, L. Yang, Z. Tian, W. Zhao, C. Sun, L. Zhu, M. Huang, G. Guo and G. Liang, ACS Med. Chem. Lett., 2022, 13, 99–104 CAS.
- A. S. Walker and J. Clardy, J. Chem. Inf. Model., 2021, 61, 2560–2571 CAS.
- S. H. Kim and D. Choi, Food Supplements and Biomaterials for Health, 2023, e17, DOI:10.52361/fsbh.2023.3.e17.
- Y. Sun, S. Shi, Y. Li and Q. Wang, Food Chem. Toxicol., 2019, 128, 163–170 CAS.
- Q.-H. Liu, J.-W. Tang, Z.-W. Ma, Y.-X. Hong, Q. Yuan, J. Chen, X.-R. Wen, Y.-R. Tang and L. Wang, Curr. Res. Food Sci., 2024, 9, 100820 CAS.
- S. Oral, I. Ökten and U. Yüzgeç, Bitlis Eren Üniversitesi Fen Bilimleri Dergisi, 2023, 12, 226–241 Search PubMed.
- M. Mansourvar, S. D. Petersen, S. Tavakoli, J. B. Hoof, D. L. Corcoles, S. M. Pittroff, L. Jelsbak, N. B. Jensen, L. Ding and R. J. N. Frandsen, Comput. Struct. Biotechnol. J., 2024 DOI:10.1016/j.csbj.2024.11.027.
- M. Korshunova, N. Huang, S. Capuzzi, D. S. Radchenko, O. Savych, Y. S. Moroz, C. I. Wells, T. M. Willson, A. Tropsha and O. Isayev, Commun. Chem., 2022, 5, 129 Search PubMed.
- K. Posansee, M. Liangruksa, T. Termsaithong, P. Saparpakorn, S. Hannongbua, T. Laomettachit and T. Sutthibutpong, ACS Omega, 2023, 8, 38373–38385 CAS.
- X. Dong, X. Gao, R. Wang, C. Liu, J. Wu and Q. Huang, Int. J. Med. Mushrooms, 2023, 25, 13–28 Search PubMed.
- L. Wang, J. Li, T. Li, H. Liu and Y. Wang, ACS Omega, 2021, 6, 19665–19674 CAS.
- X. Chen, H. Liu, J. Li and Y. Wang, Vib. Spectrosc., 2022, 121, 103404 CAS.
- J.-E. Dong, J. Zhang, Z.-T. Zuo and Y.-Z. Wang, Spectrochim. Acta, Part A, 2021, 249, 119211 CAS.
- K. Przybył, J. Wawrzyniak, K. Koszela, F. Adamski and M. Gawrysiak-Witulska, Sensors, 2020, 7305, DOI:10.3390/s20247305.
- Y. Li, X. Li, Y. Liu, Y. Yao and G. Huang, Pharmaceuticals, 2022, 15, 707 CAS.
- Q. Wu, Y. Li, K. Peng, X.-L. Wang, Z. Ding, L. Liu, P. Xu and G.-Q. Liu, J. Agric. Food Chem., 2019, 67, 8149–8159 CAS.
- Y. Xiao, M. Sun, Q. Zhang, Y. Chen, J. Miao, X. Rui and M. Dong, J. Food Sci. Technol., 2018, 55, 1244–1255 CAS.
- H. Liu, H. Liu, J. Li and Y. Wang, ACS Omega, 2023, 8, 19663–19673 CAS.
- U. Ejaz, M. Afzal, M. Naveed, Z. S. Amin, A. Atta, T. Aziz, G. Kainat, N. Mehmood, M. Alharbi and A. F. Alasmari, Sci. Rep., 2024, 14, 5751 CrossRef CAS PubMed.
- J. Raman, H. Lakshmanan, P. A. John, C. Zhijian, V. Periasamy, P. David, M. Naidu and V. Sabaratnam, Int. J. Nanomed., 2015, 5853–5863 CAS.
- M. Idrees, S. Yaqoob, S. F. Leo, A. A. Khan, L. Sun, Y. Yang, D. Li, Y. Fu and Y. Li, Int. J. Med. Mushrooms, 2021, 79–90 CrossRef PubMed.
- C. W. Wold, C. Kjeldsen, A. Corthay, F. Rise, B. E. Christensen, J. Ø. Duus and K. T. Inngjerdingen, Carbohydr. Polym., 2018, 185, 27–40 CrossRef CAS PubMed.
- S. Pilafidis, P. Diamantopoulou, K. Gkatzionis and D. Sarris, Appl. Sci., 2022, 12, 11426 CrossRef CAS.
- A. E. A. Magro, L. C. Silva, G. B. Rasera and R. J. S. De Castro, Bioresources and Bioprocessing, 2019, 6, 1–9 CrossRef.
- T. Williams, D. Parker and B. Taubman, J. Am. Soc. Brew. Chem., 2022, 80, 427–434 CAS.
- D. L. Abd Razak, N. Y. Abd Rashid, A. Jamaluddin, S. A. Sharifudin, A. Abd Kahar and K. Long, J. Saudi Soc. Agric. Sci., 2017, 16, 127–134 CrossRef.
- D. C. Gomes, in Fungi Bioactive Metabolites: Integration of Pharmaceutical Applications, Springer, 2024, pp. 651–681 Search PubMed.
- N. P. Keller, Nat. Rev. Microbiol., 2019, 17, 167–180 CrossRef CAS PubMed.
- R. Esmaeily, M. A. Razavi and S. H. Razavi, Trends Food Sci. Technol., 2024, 143, 104286 Search PubMed.
- P. Hamet and J. Tremblay, Metabolism, 2017, 69, S36–S40 Search PubMed.
- C. Krittanawong, M. Aydar, H. U. H. Virk, A. Kumar, S. Kaplin, L. Guimaraes, Z. Wang and J. L. Halperin, Can. J. Cardiol., 2022, 38, 185–195 Search PubMed.
- M. Javaid, A. Haleem, I. H. Khan and R. Suman, Adv. Agrochem, 2023, 2, 15–30 Search PubMed.
- Y. Akkem, S. K. Biswas and A. Varanasi, Eng. Appl. Artif. Intell., 2023, 120, 105899 Search PubMed.
- S. Karanth, E. O. Benefo, D. Patra and A. K. Pradhan, J. Agric. Food Res., 2023, 11, 100485 Search PubMed.
- A. Bin Rashid and M. D. A. K. Kausik, Hybrid Advances, 2024, 7, 100277 Search PubMed.
- T. Papadopoulos, U. Sivarajah, K. Spanaki, S. Despoudi and A. Gunasekaran, Int. J. Prod. Res., 2022, 60, 4361–4364 Search PubMed.
- V. Zatsu, A. E. Shine, J. M. Tharakan, D. Peter, T. V. Ranganathan, S. S. Alotaibi, R. Mugabi, A. Bin Muhsinah, M. Waseem and G. A. Nayik, Food Chem.:X, 2024, 24, 101867 Search PubMed.
- G. Dijksterhuis, R. de Wijk and M. Onwezen, Foods, 2022, 11, 767 Search PubMed.
- I. Kutyauripo, M. Rushambwa and L. Chiwazi, J. Agric. Food Res., 2023, 11, 100502 Search PubMed.
- Y. Jung, C. Qian, C. Barnett-Neefs, R. Ivanek and M. Wiedmann, J. Food Prot., 2024, 87, 100337 Search PubMed.
- M. G. Juan, J. Ram, J. Suckling, M. Leming, Y. Zhang, J. Ram, P. Bonomini, F. E. Casado, D. Charte, F. Charte, R. Contreras, R. J. Duro, A. Fern and R. Mart, Neurocomputing, 2020, 410, 237–270 Search PubMed.
- D. R. Kale, J. Nalvade, P. S. Randive and S. Hirve, Artif. Intell., 2024 Search PubMed.
- E. Soh, N. Saeidi, A. Javadian, D. E. Hebel and H. Le Ferrand, PLoS One, 2021, 16, e0260170 CAS.
- K. Anbalagan, International Journal of Computer Engineering and Technology, 2024, 15, 622–635 Search PubMed.
- F. Abdullakutty, Y. Akbari, S. Al-Maadeed, A. Bouridane, I. M. Talaat and R. Hamoudi, Comput. Struct. Biotechnol. Rep., 2024, 1, 100019 Search PubMed.
- M. Mathur and P. Mathur, Trop. Ecology, 2025 DOI:10.1007/s42965-025-00374-z.
- T. Wu, M. Cai, H. Hu, C. Jiao, Z. Zhang, Y. Liu, J. Chen, C. Xiao, X. Li, X. Gao, S. Chen, Q. Wu and Y. Xie, J. Fungi, 2022, 1081, DOI:10.3390/jof8101081.
- J. Ran, H. Xu, Z. Wang and X. Bai, Front. Chem., 2025, 13, 1534216 Search PubMed.
- Y. Fang, D. Wu, N. Gao, M. Lv, M. Zhou, C. Ma, Y. Sun and B. Cui, G3:Genes, Genomes, Genet., 2024, 14, jkae005 CAS.
- R. Zbinden, N. M. A. Van Tiel, B. A. Kellenberger, L. Hughes and D. Tuia, in 11th International Conference on Learning Representations (ICLR) Workshops, 2023 Search PubMed.
- F. Zhao and S. Iacono, in Encyclopedia of Big Data, Springer, 2022, pp. 89–97 Search PubMed.
- M. K. Gupta, V. Balodi, A. D. Tripathi, A. Agarwal, N. P. Nirmal and others, in Unleashing the Power of Functional Foods and Novel Bioactives, Elsevier, 2025, pp. 449–465 Search PubMed.
- J. Intrasook, T. W. Tsusaka and A. K. Anal, J. Food Drug Anal., 2024, 32, 112 Search PubMed.
- A. Awoke and B. N. Cosendey, International Journal of Ethnoscience and Technology in Education, 2025, 2, 64–89 Search PubMed.
- P. Kataria, Food Sci. Nutr. Case, 2024, foodsciencecases20240003 Search PubMed.
- G. A. Abraham and A. K. Singh, Journal of Krishi Vigyan, 2024, 12, 424–428 CrossRef.
- A. Rejeb, K. Rejeb, S. Zailani, J. G. Keogh and A. Appolloni, Artificial Intelligence in Agriculture, 2022, 6, 111–128 CrossRef.
- K. Spanaki, U. Sivarajah, M. Fakhimi, S. Despoudi and Z. Irani, Ann. Oper. Res., 2022, 308, 491–524 CrossRef.
- I. Vågsholm, N. S. Arzoomand and S. Boqvist, Front. Sustain. Food Syst., 2020, 4, 16 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |