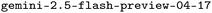Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
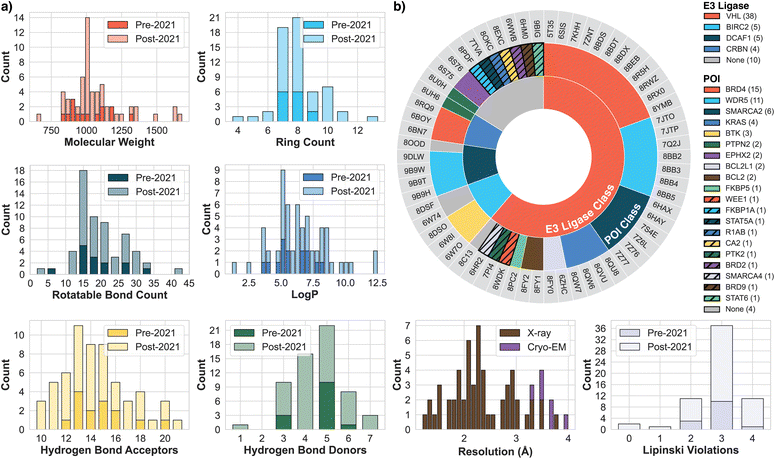
Predicting PROTAC-mediated ternary complexes with AlphaFold3 and Boltz-1
Nils
Dunlop†
 ,
Francisco
Erazo†
,
Francisco
Erazo†
 ,
Farzaneh
Jalalypour
,
Farzaneh
Jalalypour
 and
Rocío
Mercado
and
Rocío
Mercado
 *
*
Department of Computer Science and Engineering, Section for Data Science and AI, Chalmers University of Technology, University of Gothenburg, Chalmersplatsen 4, 412 96 Gothenburg, Sweden. E-mail: rocio.mercado@chalmers.se
First published on 27th October 2025
Abstract
Accurate prediction of protein–ligand and protein–protein interactions is essential for computational drug discovery, yet remains a significant challenge, particularly for complexes involving large, flexible ligands. In this study, we assess the capabilities of AlphaFold 3 (AF3) and Boltz-1 for modeling ligand–mediated ternary complexes, focusing on proteolysis-targeting chimeras (PROTACs). PROTACs facilitate targeted protein degradation by recruiting an E3 ubiquitin ligase to a protein of interest, offering a promising therapeutic strategy for previously undruggable intracellular targets. However, their size, flexibility, and cooperative binding requirements pose significant challenges for computational modeling. To address this, we systematically evaluated AF3 and Boltz-1 on 62 PROTAC complexes from the Protein Data Bank. Both models achieve high structural accuracy by integrating ligand input during inference, as measured by RMSD, pTM, and DockQ scores, even for post-2021 structures absent from AF3 and Boltz-1 training data. AF3 demonstrates superior ligand positioning, producing 33 ternary complexes with RMSD < 1 Å and 46 with RMSD < 4 Å, compared to Boltz-1's 25 and 40, respectively. We explore different input strategies by comparing molecular string representations and explicit ligand atom positions, finding that the latter yields more accurate ligand placement and predictions. By analyzing the relationships between ligand positioning, protein–ligand interactions, and structural accuracy metrics, we provide insights into key factors influencing AF3's and Boltz-1's performance in modeling PROTAC–mediated binary and ternary complexes. To ensure reproducibility, we publicly release our pipeline and results via a GitHub repository and website (https://protacfold.xyz), providing a framework for future PROTAC structure prediction studies.
1 Introduction
Accurate prediction of protein–ligand and protein–protein interactions (PLIs and PPIs) remains a central challenge in structure-based drug discovery,1,2 one made even more pressing by the rise of targeted protein degradation (TPD). TPD modalities such as proteolysis-targeting chimeras (PROTACs) and molecular glues (MGs) move beyond simple functional inhibition of disease drivers to harness the cell's intrinsic protein disposal machinery, the ubiquitin–proteasome system (UPS), offering a different mode-of-action compared to traditional small molecule inhibitors.3–9 By facilitating the complete removal of target proteins, including proteins previously deemed undruggable, TPD has rapidly advanced from concept to dozens of clinical candidates in the past two decades, particularly in oncology.9,10From a computational standpoint, however, modeling PROTAC–mediated ternary complexes is difficult: a large, flexible ligand must cooperatively link two proteins, sampling a vast conformational space.1,11–13 Current computational methods encounter difficulties in accurately predicting complex multi-molecular assemblies, especially when large, flexible ligands mediate the interactions.2,11,14 Traditional docking-based methods have struggled with PROTAC systems due to the dynamic nature of PROTAC–mediated PPIs and the structural variability introduced by flexible linkers.12,15,16 AlphaFold2 (AF2) marked a significant advancement in protein structure prediction, attaining near-native precision for monomeric proteins17,18 and demonstrating some success in modeling transient protein complexes.14 However, it performs poorly on larger multimers and their interfaces,11,19,20 notably when ligand–mediated interactions or conformational changes are involved, as it has not been trained on these.21 Specialized co-folding models have been created to overcome these limitations,22,23 yet significant challenges persist in achieving predictive accuracy for PLIs suitable for drug discovery.
The recent release of AlphaFold3 (AF3) expands AF2's capabilities by incorporating ligand and nucleic acid interactions, learning jointly from protein-small-molecule structures, and thus offers enhanced opportunities for biomolecular complex prediction.2 However, the initial release of the AF3 web server24 does not support PROTACs as of this publication (July 2025), restricting its applicability to model such ternary complexes. Building upon the AF3 framework, Boltz-1 (ref. 25) reportedly achieves comparable accuracy as an open-source alternative, and has been closely followed by the release of Boltz-2 (ref. 26). Recent advances such as Boltz-2 (ref. 26) and Protenix27 have extended the AF3 architecture with larger training sets and new modules for binding affinity prediction and multiple sequence alignment (MSA) processing. However, recent independent benchmarks indicate that while these next-generation generative models offer incremental improvements, they continue to face limitations in physical plausibility and binding site identification, particularly for underrepresented binding modes.28 Given these comparable performance levels, we focused our benchmarking on the most widely adopted and representative frameworks at the time of our study, AF3 and Boltz-1. However, whether powerful deep learning-based structure predictors like AF3 and Boltz-1 can handle PROTAC ternary complexes remains an open challenge due to the structural flexibility and cooperative binding these systems require, and has not been assessed systematically.1,11,13,29
Here we provide that assessment, focusing on modeling PROTAC–mediated ternary complexes due to their growing significance in drug discovery.30 Using the recently released inference code, which accepts explicit ligand coordinates, we benchmark AF3 against Boltz-1 on the 62 crystallographically resolved PROTAC ternary and binary complexes currently in the Protein Data Bank (PDB).31 Our automated pipeline generates inputs, runs three seeds per complex, and extracts accuracy metrics. We show that while both engines achieve near-native structure prediction when ligand information is supplied, AF3 is consistently more accurate on ligand pose. With the best settings, AF3 yields 33 out of 62 structures with an RMSD < 1 Å and 46 with an RMSD < 4 Å, while Boltz-1 produces 25 and 40 for these respective thresholds, indicating near-native accuracy for both methods. Our pipeline and results are publicly available via web (https://protacfold.xyz) and a GitHub repository, providing a framework that other researchers can use for future PROTAC structure predictions.
2 Methods
2.1 Overview of structure prediction pipeline
The structure prediction pipeline, depicted in Fig. 1, integrates several key stages: data preparation, utilization of the PROTACFold automation platform, execution of structural predictions with AF3 and Boltz-1, and comprehensive evaluation of the results. The subsequent sections provide detailed descriptions of each component within the workflow.2.2 Data overview
For this work, we curated a dataset of 62 PROTAC–mediated ternary and binary complexes from the PDB.31 Each structure contains experimentally resolved coordinates for the full PROTAC molecule—including warhead, linker, and E3 ligase binding moiety—as well as at least one of either the POI or the E3 ubiquitin ligase. This collection represents all publicly available, fully crystallized PROTAC ternary and binary complexes as of May 2025. Structures with partially resolved PROTACs, such as those capturing solely warhead, were identified but not included here. The completeness of PROTAC crystallization was validated through careful review of each structure's associated publication, assisted by a Gemini-based prompt.The finalized dataset comprises 48 ternary complexes and 14 binary complexes. Metadata extraction, including chain identifiers, ligand composition, molecular weight, and resolution, was performed using a semi-automated pipeline. Fig. 2 summarizes key dataset characteristics, presenting distributions of PROTAC size, physicochemical properties, and diversity of protein targets.
To ensure extraction accuracy, automated assignments of POIs and E3 ligases were cross-validated against manual labels, with detailed results provided in Appendix F. Structures were additionally confirmed through review of the original publications, and those not definitively identifiable as PROTAC degraders were excluded. An auxiliary set of 62 structures with incomplete or partially resolved PROTACs, including several binary complexes, was compiled but not analyzed further here. All curated structures and preprocessing files utilized in this study are openly available on GitHub at https://github.com/NilsDunlop/PROTACFold.
2.3 Data preparation
Building upon the dataset described previously, we employed our automation platform, https://protacfold.xyz, to systematically extract FASTA sequences, publication details, and ligand annotations, which allowed us to determine the POI, E3 ligase, and whether each structure represented a PROTAC degrader. Our detailed pipeline analysis led to the reclassification of seven structures (5HXB, 6UUO, 6XK9, 7KHL, 7MEU, 7LPS, 8OKB), originally identified as PROTACs in prior work.11,32 Those seven structures were determined to be either MGs or instances of only warhead crystallization. We categorized the final 62-structure dataset based on inclusion in the AF3 and Boltz-1 training data (cut-off: 2021-09-30); of these structures, 16 were included in this training data, while 46 were not.The recent release of AF3 and Boltz-1's inference code33,34 introduced enhanced ligand handling, enabling direct input via either Chemical Compound Dictionary (CCD) codes35 or SMILES strings. CCD codes are unique PDB identifiers, with each entry defining a molecule's connectivity and idealized 3D coordinates, whereas SMILES strings offer a textual line notation for molecules. To determine the optimal input strategy for PROTACs with these new tools, we used our automation platform to systematically prepare model inputs using both representations: canonical isomeric SMILES were generated with OpenEye OEToolkits 2.0.7,36 and CCD codes were retrieved from their respective  files via the PDB GraphQL API.31 Beyond ligand input preparation, the platform also identifies the POI and E3 ligase components using Gemini 2.5 Flash Experimental to process sequences, paper abstracts, and ligand information, all sourced from the PDB (accuracy and prompt details in Appendix E).
files via the PDB GraphQL API.31 Beyond ligand input preparation, the platform also identifies the POI and E3 ligase components using Gemini 2.5 Flash Experimental to process sequences, paper abstracts, and ligand information, all sourced from the PDB (accuracy and prompt details in Appendix E).
We standardized PDB complexes to ensure methodological consistency and focus on the essential components for analysis. This entailed removing accessory proteins and molecules beyond the three main components of binary and ternary structures: POI, E3 ligase, and PROTAC ligand. Components such as elongin-C, elongin-B, extraneous DNA segments, and occasionally solvent molecules (water or ions) were removed. For PDB entries containing multiple ligands, we retained only the PROTACs while excluding all other ligands. For each complex, the required data (POI, E3 ligase, and PROTAC) were compiled into JSON for AF3 and YAML for Boltz-1 according to their input specifications. For AF3, we generated six JSON input files for each PDB entry, corresponding to two ligand representations (CCD or canonical isomeric SMILES) each with three random seeds (24, 37, 42). For Boltz-1, we generated two YAML input files (CCD, SMILES) with the three seeds specified as a runtime argument.
2.4 PROTACFold: PROTAC automation platform
To streamline and support future investigations, we developed the web-based https://protacfold.xyz platform. PROTACFold automates the extraction of key structural and ligand information from the PDB, simplifying the preparation of simulation-ready input files for AF3 and Boltz-1. Users can input a PDB ID and specify the desired number of seeds, and the platform then queries the PDB via its GraphQL API37 to retrieve ligand metadata, FASTA sequences, and publication information. Using this data, PROTACFold determines the POI and E3 ligase components, aided by Gemini 2.5 Flash Experimental. This large language model analyzes the sequence data, abstracts, and protein/ligand annotations to robustly classify components using a detailed prompt (Appendix E).Following PDB information retrieval, the user is redirected to a results page offering options to download all structures (including accessory proteins) or the “cleaned” ternary structures only. The downloaded files include a text document containing the determined POI and E3 ligase names and sequences, the experimental assembly PDB structure, and the AF3 JSON and Boltz-1 YAML input files. Comprehensive guides for then setting up AF3 and Boltz-1 are available on our GitHub repository, aiming to facilitate rapid setup and support future research in this domain.
2.5 Predicting structures with AF3 and Boltz-1
After processing all input files for each of the 62 PROTAC structures (×3 seeds), we initiate biomolecular interaction predictions using AF3 and Boltz-1. The AF3 pipeline33 involves compiling their Docker image, downloading ∼700 GB of public databases for MSA, incorporating model parameters (provided by Google DeepMind following a request for approval), and supplying input JSON files, all configured by our batch script at runtime. Boltz-1 setup,34 by comparison, requires only a Python installation and specification of the cache, input YAML file, and output folder. To ensure a fair comparison between Boltz-1 and AF3, we configured both tools with 10 recycling steps and 25 samples during runtime, mirroring AF3's default parameters. On an A100 GPU, a single prediction with Boltz-1 averages 5–15 minutes, while AF3, with its more extensive MSA calculation, takes 20–30 minutes. Following prediction completion, we aggregate the top predictions (two input strategies, CCD or SMILES, ×3 seeds) by PDB ID and process them through our evaluation script to consolidate all evaluation metrics into a single CSV file.During the initial Boltz-1 prediction runs, we identified that its  file, containing CCD information, was outdated, lacking entries for newer ligands such as A1ANN (PDB ID 9B9W). We thus updated the
file, containing CCD information, was outdated, lacking entries for newer ligands such as A1ANN (PDB ID 9B9W). We thus updated the  file, enabling us to predict newer structures with Boltz-1. Furthermore, three structures (8FY0, 8FY1, and 8FY2) were challenging to predict with SMILES input due to the large ligand, YF8, which triggered a value error due to the default four-character limit for atom names in Boltz-1. This is because Boltz-1 names atoms by combining their chemical symbol (e.g., “CL” for chlorine) with a unique number (e.g., “118”), such that “CL118” exceeds the default four-token limit for atom names in
file, enabling us to predict newer structures with Boltz-1. Furthermore, three structures (8FY0, 8FY1, and 8FY2) were challenging to predict with SMILES input due to the large ligand, YF8, which triggered a value error due to the default four-character limit for atom names in Boltz-1. This is because Boltz-1 names atoms by combining their chemical symbol (e.g., “CL” for chlorine) with a unique number (e.g., “118”), such that “CL118” exceeds the default four-token limit for atom names in  . To predict these structures, we slightly modified the Boltz-1 input parser (Appendix G).
. To predict these structures, we slightly modified the Boltz-1 input parser (Appendix G).
2.6 Molecular dynamics simulations
To evaluate the PROTAC binding stability, we performed atomistic simulations of a select ternary complex in two independent 300 ns replicas. The starting model was launched from the AF3-predicted structure of PDB ID 9B9W, with ligand information provided in CCD format; the structure comprised the target protein (WDR5), an E3 ligase-recruiting protein (DCAF1), and the PROTAC (A1ANN). The AF3-predicted binding sites closely resembled those of the native structure, but exhibited deviations on the POI side, and the PROTAC conformation differed significantly. Our objectives were to examine the flexibility of both the complex and PROTAC, the complex's stability, and the key binding residues.AmberTool's Antechamber 22.0 (ref. 38) was used to assign atom types and generate point charges, while Parmchk2 was used to specify missing parameters. AMBER topology and coordinate files were generated using the tLEaP module of AmberTools24 and converted to GROMACS format via ACPYPE.39 The system was parameterized using the AMBER force field, with AMBER ff14SB for proteins, GAFF for the PROTAC molecule, and TIP3P water. MD simulations were performed using GROMACS 2024.40,41 The system was solvated in a cubic box with at least a 10 Å buffer distance, and ions were added for neutralization. Energy minimization was performed in three steps using the steepest descent algorithm. Equilibration consisted of relaxing the system at constant pressure (1 bar) and temperature (310 K). The production MD simulation ran for 300 ns under NPT conditions at 300 K, employing the particle-mesh Ewald approach to estimate long-range electrostatic interactions. The simulation time step was set to 2 fs, and LINCS42 was used to constrain the length of hydrogen bonds. Post-simulation analysis, including RMSD and distance calculations, was performed using MDAnalysis.43,44
2.7 Evaluation metrics
The accuracy of AF3 and Boltz-1 predictions was assessed by comparing each predicted structure against its experimentally determined reference from the PDB.31 Structural alignment was performed by superimposing predicted and experimental structures based on their Cα atoms using PyMol 3.1.3.45 The quantitative assessment involved computing root mean square deviation (RMSD) and predicted template modeling (pTM) scores.RMSD, representing atomic displacement, is calculated as the square root of the mean squared distances between corresponding Cα atoms following optimal alignment. An RMSD of 0 Å indicates perfect structural alignment, with values below 1 Å considered near-native. Due to the size and inherent flexibility of PROTAC–mediated ternary complexes, a more generous threshold of 4 Å was adopted as an indicator of good structural alignment.46,47 RMSD calculations were performed separately for the entire ternary complex (excluding PROTAC hydrogens), as well as individually for the POI, the E3 ligase, and the PROTAC ligand, though PROTAC ligand RMSDs could not be computed for a few Boltz-1 predictions due to alignment failures against experimentally incomplete ligand structures (e.g., 6HM0, 8OOD).
pTM scores, obtained directly from the AF3 and Boltz-1 prediction outputs, complement RMSD by providing a robust measure less sensitive to outliers. These scores range from 0 to 1, with values greater than 0.5 indicating more accurate structural folds. Additionally, interface-specific pTM scores (ipTM) were analyzed to evaluate protein–protein interfaces.
To assess protein–protein docking accuracy, DockQ v2 (ref. 48) scores were computed, integrating three critical components: fraction of native contacts recovered (fnat), interface RMSD (iRMSD), and ligand RMSD (LRMSD), where the ligand here refers to the secondary protein chain. DockQ scores range from 0 (no similarity to reference) to 1 (perfect agreement), with scores >0.23 indicating acceptable-quality predictions and scores above 0.8 denoting high-quality predictions.15,49 The formula is as follows:
Error bars in figures represent the standard error of the mean (SEM) for each metric. Further visual inspection was performed for select PROTAC complexes involving multiple binding sites (see Fig. 7, 8 and Appendix B). Integrating these complementary metrics enables a comprehensive evaluation of AF3 and Boltz-1 predictions. RMSD provides atomic-level accuracy assessment, pTM measures global fold correctness, and DockQ evaluates the quality of predicted protein–protein interfaces.
2.8 Visualization
Protein structures were visualized using PyMol 3.1.3 (ref. 45) and VMD 1.9.4.50 VMD was also used to process structures for MD simulation. In addition to numerical metrics, we performed a thorough visual analysis to examine ligand placement, structural integrity, and interface quality. We generated side-by-side visualizations of CCD vs. SMILES ligand interactions to better understand the impact of the ligand input on predictions. Further ligand–protein interaction diagrams were generated using the free academic version of LigPlot+ (v2.2.9).51 The visual analysis was essential for evaluating how well AF3 and Boltz-1 captured key interactions.3 Results
3.1 Impact of ligand input strategy on prediction accuracy
 | ||
| Fig. 3 DockQ scores comparing (top) AF3 and (bottom) Boltz-1 predictive capabilities for a subset of 28 PROTAC ternary complexes reported herein with Pereira et al.,11 who used the AF3 web server without ligand inputs (gray bars). Higher DockQ scores indicate better complexes. CCD-based predictions outperform SMILES and no-ligand predictions in DockQ score. The dotted line represents an acceptable threshold value of 0.23. | ||
Furthermore, we compared these results to predictions from Boltz-1 in analogous scenarios (Fig. 3, bottom). Boltz-1 predictions also improve when the PROTAC ligand is provided, achieving higher DockQ scores in 18 of 28 cases, but underperforming relative to AF3 which achieved 26 better predictions. There is a clear difference in performance when comparing the number of acceptable structures (DockQ ≥ 0.23) generated; Pereira et al.,11 for instance, only achieved five acceptable structures, whereas Boltz-1 and AF3 achieve 8 and 21, respectively, when the PROTAC is included in the predictions. These findings highlight that including the PROTAC ligand is beneficial and that AF3 is particularly effective at leveraging the PROTAC–ligand information to predict accurate ternary complexes.
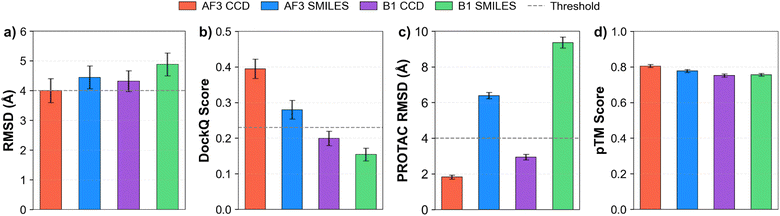
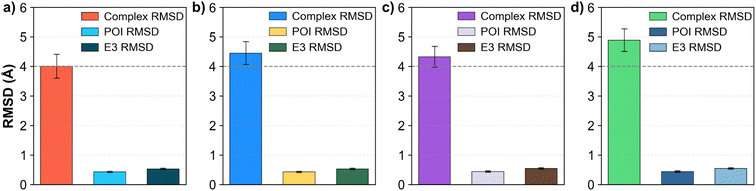
The general trend indicates that using CCD codes yields more accurate ternary complex predictions than using SMILES. AF3 with CCD input achieved a mean complex RMSD of 4.0 Å (Fig. 4a), precisely meeting the acceptable threshold. The other configurations resulted in slightly higher mean RMSD values: 4.45 Å for AF3 with SMILES input, 4.32 Å for Boltz-1 with CCD input, and 4.88 Å for Boltz-1 with SMILES input. When comparing DockQ scores (Fig. 4b), AF3 outperforms Boltz-1. AF3 achieved mean DockQ scores of 0.395 (CCD) and 0.280 (SMILES), both surpassing the acceptable threshold of 0.23. In contrast, Boltz-1's scores of 0.199 (CCD) and 0.154 (SMILES) did not meet this threshold. Analyzing the PROTAC RMSD (Fig. 4c), using CCD codes in AF3 notably reduced the PROTAC RMSD to 1.82 Å. Other configurations resulted in slightly higher PROTAC RMSDs: 6.39 Å for AF3 (SMILES), 2.94 Å for Boltz-1 (CCD), and 9.36 Å for Boltz-1 (SMILES). Lastly, the mean pTM scores (Fig. 4d) were relatively similar across configurations. However, AF3 exhibited slightly higher scores, indicating a stronger prediction confidence: 0.806 (AF3 CCD) and 0.777 (AF3 SMILES), compared to 0.752 (Boltz-1 CCD) and 0.756 (Boltz-1 SMILES).
3.2 Benchmarking AF3 vs. Boltz-1
Focusing on AF3, there is a clear difference between pre-2021 and post-2021 structure predictions. For pre-2021 structures, AF3 achieved low mean RMSD values of 0.92 Å (CCD) and 1.86 Å (SMILES) (Fig. 5a), indicating near-native predictions. In contrast, performance dropped for structures deposited post-2021, with mean RMSD increasing substantially to 4.93 Å (CCD) and 5.20 Å (SMILES), both exceeding the 4 Å acceptability threshold. A similar trend is evident in AF3's DockQ scores (Fig. 5b). Pre-2021 structures yielded high mean DockQ scores of 0.680 (CCD) and 0.467 (SMILES), both comfortably surpassing the 0.23 acceptable threshold. For post-2021 structures, the mean DockQ score with CCD input (0.297) remained above the threshold, while the score obtained with SMILES input (0.218) fell short.
Boltz-1 also shows performance gaps between pre- and post-2021 structure predictions when evaluated by RMSD (Fig. 5e), with some key distinctions to AF3 using CCD inputs. For pre-2021 structures, Boltz-1's mean RMSD values were 2.71 Å (CCD) and 2.86 Å (SMILES); in this scenario, AF3 with CCD input (0.92 Å) outperformed Boltz-1. For post-2021 structures, Boltz-1 yielded mean RMSD values of 4.79 Å (CCD) and 5.47 Å (SMILES). While both models did not meet the acceptability threshold for post-2021 structures with CCD input, Boltz-1 demonstrated a slightly better mean RMSD (4.79 Å) compared to AF3 (4.93 Å). Overall, both models perform comparably on post-2021 structures in terms of RMSD. Boltz-1's DockQ scores (Fig. 5f), however, reveal a considerable performance gap compared to AF3 across both time periods. The pre-2021 mean DockQ scores for Boltz-1 were 0.301 (CCD) and 0.272 (SMILES), while the post-2021 scores dropped to 0.165 (CCD) and 0.115 (SMILES). The most significant DockQ performance discrepancy between the models is observed for pre-2021 structures with CCD input, where Boltz-1 generates predictions with on average 55.7% lower DockQ scores relative to AF3. While Boltz-1's pre-2021 DockQ scores are only marginally above the 0.23 threshold, its post-2021 scores fall well below the benchmark.
PROTAC RMSD scores (Fig. 5c and g) illustrate how the PROTAC RMSD is significantly lower for predictions made with CCD input (pre-2021: 1.78 Å, post-2021: 3.28 Å) compared to SMILES input (pre-2021: 9.22 Å, post-2021: 9.40 Å). Using SMILES input with a given method, there is no difference between predictions made on pre- and post-2021 structures. However, PROTAC RMSD is notably lower for predictions made in otherwise equivalent scenarios when AF3 is than when Boltz-1 is used. pTM scores (Fig. 5d and h) are consistent across pre- and post-2021 structures, both for AF3 and Boltz-1, with only a minor drop in performance for AF3 on unseen structures, and a slightly lower overall performance for Boltz-1 predictions relative to AF3.
AF3 consistently outperformed Boltz-1 in predicting PROTAC–mediated ternary complexes, particularly in capturing the correct interface geometry. It achieved both lower mean RMSD values across POIs (3.65 Å vs. 4.71 Å) and more than twice the average DockQ scores for most systems, indicating stronger modeling of cooperative binding. AF3 produced accurate DockQ predictions (above the 0.23 threshold) for five POIs—including SMARCA4 and BCL2L1, where Boltz-1 failed to achieve acceptable scores. It also showed notably better RMSD performance for challenging systems like WEE1, FKBP5, and BCL2L1. For E3 ligases, AF3 similarly led, achieving acceptable DockQ scores for VHL and CRBN, while Boltz-1 failed to reach the threshold for any E3 ligase.
Despite its overall weaker performance, Boltz-1 demonstrated isolated strengths. It outperformed AF3 on a few difficult POIs, including KRAS and BCL2, achieving lower RMSD values where AF3 predictions were particularly poor. It also yielded a higher DockQ score for PTK2 (0.653 vs. 0.468), suggesting that in some cases, Boltz-1 can more accurately model backbone alignment and protein–protein orientation. However, its difficulty in placing ligands correctly limits its utility for modeling cooperative ternary complexes. Overall, these findings suggest that while both models benefit from explicit ligand input, AF3 is currently more reliable for structure-based PROTAC design, especially when accurate modeling of the ligand–mediated interface is critical.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P), hydrogen bond donors (HBD), and hydrogen bond acceptors (HBA). We observed that increased ligand size and flexibility correlate with higher PROTAC RMSD values. For both size (MW & HAC) and flexibility (RBC), this trend of increasing PROTAC RMSD with increasing size was observed for both models. However, for other properties, such as the number of HBD or HBA, there was less of a correlation with PROTAC RMSD, and more of a correlation with the number of data points: bins with many data points (Fig. 2) generally displayed lower PROTAC RMSDs. The worst performing bins across all properties correlate to where there are few or no structures sampling that property in the training set (pre-2021; Fig. 2). Across all properties, AF3 generally maintains a general advantage; however, specific bins show comparable or better performance for Boltz-1.
P), hydrogen bond donors (HBD), and hydrogen bond acceptors (HBA). We observed that increased ligand size and flexibility correlate with higher PROTAC RMSD values. For both size (MW & HAC) and flexibility (RBC), this trend of increasing PROTAC RMSD with increasing size was observed for both models. However, for other properties, such as the number of HBD or HBA, there was less of a correlation with PROTAC RMSD, and more of a correlation with the number of data points: bins with many data points (Fig. 2) generally displayed lower PROTAC RMSDs. The worst performing bins across all properties correlate to where there are few or no structures sampling that property in the training set (pre-2021; Fig. 2). Across all properties, AF3 generally maintains a general advantage; however, specific bins show comparable or better performance for Boltz-1.
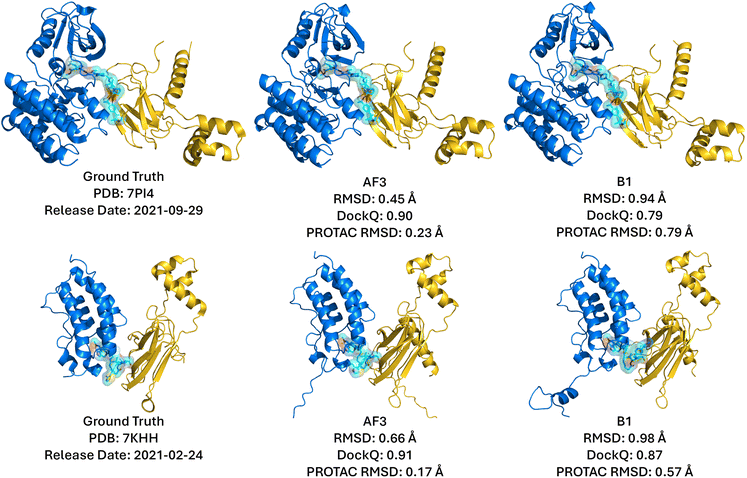
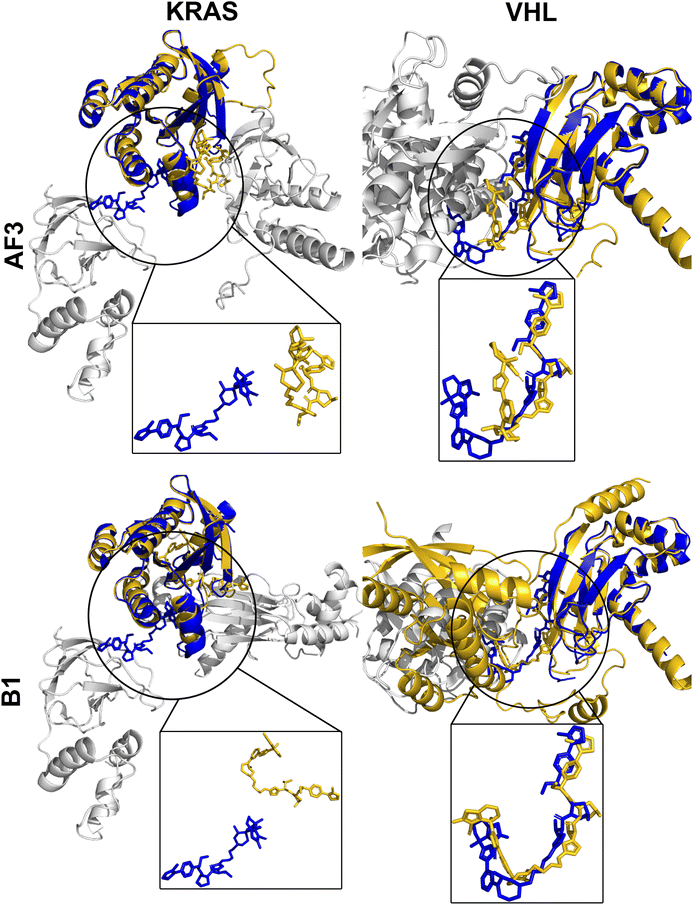
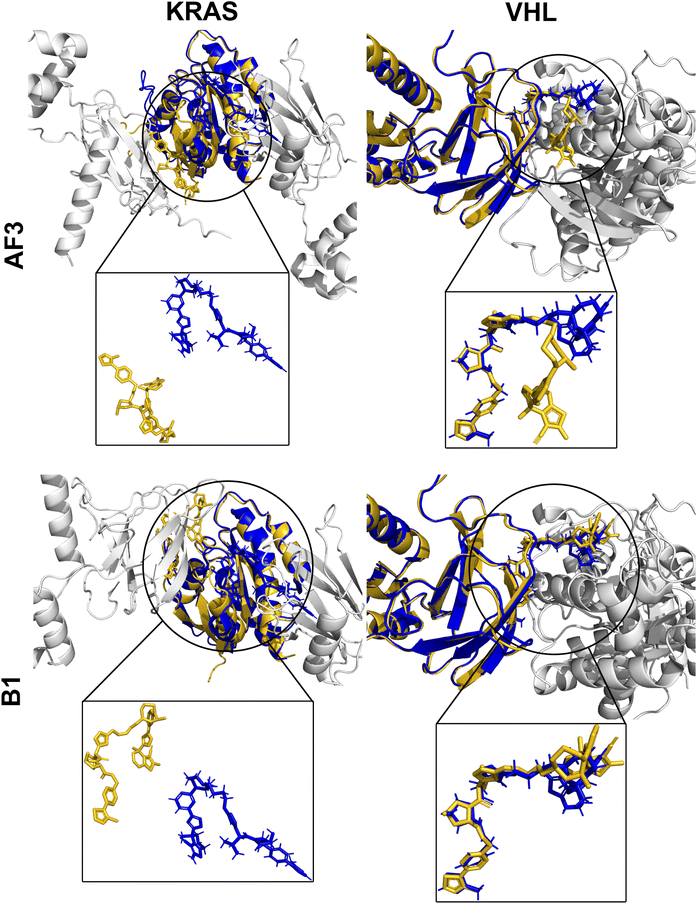
Fig. 7 highlights two well-predicted structures targeting FAK (PDB ID: 7PI4) and BRD4 (PDB ID: 7KHH), both available within the AF3 and Boltz-1 training datasets. In these cases, both models achieved near-native predictions, yielding high DockQ scores (AF3: 0.90, Boltz-1: 0.91) and demonstrating reliable prediction of ligand-binding poses and protein–protein interfaces. Conversely, performance declined significantly for complexes deposited after the training cut-off (Fig. 8), such as those targeting BRD4 (PDB ID: 8BDX) and WDR5 (PDB ID: 9B9W). These recent structures revealed substantial deviations in protein–protein interface predictions, resulting in notably low DockQ scores (AF3: 0.02, Boltz-1: 0.14). Despite this, the individual RMSD values for the POI and E3 ligase remained low (AF3: 0.32, 0.35 Å; B1: 0.32, 0.33 Å), indicating that the primary inaccuracies come from misprediction of the PROTAC and ternary interface rather than errors in protein structure. This likely reflects both limited training data on complex, flexible PROTACs and the inherent constraints of static structure prediction, which cannot fully capture novel PROTAC conformational diversity. Notably, closer inspection of the 9B9W prediction uncovered an intriguing detail: despite misalignment at the protein interface leading to low DockQ and high RMSD, AF3 predicted an alternative, extended PROTAC linker conformation. This predicted conformation proved to be stable in subsequent MD simulations (Fig. 10), suggesting that predictions with poor initial metrics might still reflect biologically relevant conformational states not captured by static crystal structures.
3.3 Low DockQ scores may reveal alternate conformations
While many predicted ternary complexes yielded low DockQ scores and high RMSD values (Fig. 4), a detailed analysis revealed that individual protein components (POIs and E3 ligases) were typically predicted with very high structural accuracy (RMSD often <0.5 Å; Fig. 9). In other words, we observe that poor metrics primarily arise from incorrect relative orientations at protein–protein interfaces rather than inaccuracies in the individual protein folds. Misalignments observed in these predictions suggest that low DockQ scores do not necessarily reflect failed predictions, but may instead indicate alternative binding orientations or twisted conformations at the ternary interface. Such discrepancies could potentially be addressed through refinement techniques, such as energy minimization or MD simulations, which optimize interfaces while preserving individual structures.The inherent conformational flexibility of PROTACs, arising from their long, rotatable-bond-rich linkers (Fig. 2), further complicates accurate prediction of the interfaces. This is because their flexibility allows PROTACs to adopt multiple biologically relevant conformational states that influence their binding modes and functional efficacy, even if only one structure is crystallized and deposited in the PDB. To better understand this flexibility's impact on prediction accuracy, we closely examined several poorly predicted complexes with RMSD > 5.97 Å (AF3: 13 complexes, Boltz-1: 18 complexes), finding that the majority (9 for AF3, 12 for Boltz-1) retained accurate individual protein structures despite large deviations at the ligand interface. Notably, most PROTACs still occupied correct binding sites on both proteins, though their linker conformations and relative orientations differed significantly from experimental observations. In specific cases (e.g., 8DSO and 9DLW; Appendix B), AF3 predictions show slight deviations at the predicted binding sites, while Boltz-1 shows one protein binding site missed from the experimental structure. Furthermore, four complexes (PDB IDs: 8QU8, 8QVU, 8QW6, 8QW7) revealed entirely different predicted binding sites compared to experimental data, suggesting either genuine mispredictions or the possibility of alternative biologically relevant binding modes yet to be experimentally confirmed.
3.4 MD confirms stable DCAF1–PROTAC binding, POI detachment, and linker flexibility
To validate AF3 predictions in a particularly challenging case, we conducted a focused case study using MD simulations on the DCAF1–PROTAC–WDR5 ternary complex (PDB ID: 9B9W). Two independent 300 ns simulations were conducted to evaluate the complex's stability, conformational dynamics, and binding-site interactions (Fig. 10).The predicted complex closely resembled the experimental structure, with minor deviations observed at the DCAF1 binding site (Fig. 10a, left). However, the PROTAC exhibited an overall conformation that differed from the experimental data, particularly in its linker region. Key binding-site residues in the experimental structure were identified using LigPlot+,51 highlighting important interactions, including hydrogen bonds with Ser86 on the POI side and Arg246, Asp304, and His88 on the DCAF1 side (Fig. 10a, right). Our binding analysis primarily focused on the interactions involving Ser86 and Arg246 as key binding sites with the POI and DCAF1, respectively. MD simulations were performed using GROMACS, subjecting the predicted structure to two independent 300 ns simulations. Throughout the simulation, PROTAC binding at the DCAF1 site improved, allowing it to remain stable and maintain its interactions (Fig. 10b, bottom).
Despite an initially well-predicted binding pose that closely matched the experimental structure at the POI site, the PROTAC gradually detached from Ser86 on the POI (Fig. 10b, top). In Fig. 10b, a threshold of 6 Å was established to determine the presence of a hydrogen bond with either one of the key POI residues (Ser86) or an important residue on DCAF1 (Arg246). A distance exceeding this threshold indicates ligand un-binding. For Arg246, the hydrogen bond distance is measured between the NE atom of arginine and the O70 atom of the PROTAC, as seen in the experimental structure, and is generally <4 Å. Similarly, for Ser86, the bond distance is calculated between the OG atom of serine and the N36 atom of the PROTAC, with detachment occurring when this distance surpasses the 6 Å threshold. Although the predicted PROTAC conformation differed from the experimental structure, it exhibited flexibility during the simulation. It was able to move freely within the linker region (Fig. 10c), enabling it to sample multiple conformations and diverse poses (Fig. 10d) while maintaining contacts at the key binding sites. These results further demonstrate that MD refinement enables flexible AF3-predicted PROTAC complexes to access experimentally relevant conformations not captured in the initial prediction.
4 Discussion
4.1 AF3 and Boltz-1 performance
This study demonstrates that both AF3 and Boltz-1 achieve high prediction accuracy for PPIs, reflecting strong capabilities in predicting global folds and interface structures. However, when modeling PROTAC–mediated ternary complexes with explicit ligand information, AF3 clearly outperforms Boltz-1 in accurately capturing ligand binding sites and ligand–mediated interactions. Although Boltz-1 remains competitive with AF3 in overall PPI prediction, it frequently deviates from experimentally resolved ligand binding poses, particularly for novel structures not included in its training set. Among the 62 ternary complexes analyzed, AF3 produced 13 predictions with low DockQ scores, whereas Boltz-1 yielded 18. Further inspection suggests that these low scores often reflect alternative, potentially valid conformations rather than outright prediction failures, as individual protein components typically maintain high structural accuracy. Moreover, the observed prediction difficulties were proportional to ligand size and flexibility, underscoring the challenge posed by the inherent structural plasticity of PROTACs and their highly flexible linkers.To evaluate optimal input strategies for PROTAC modeling, we compared predictions generated using two ligand representations: CCD (explicit 3D atom positions) and SMILES (2D strings with generated 3D conformations via ETKDGv3). We found that CCD inputs produced predictions more closely aligned with experimental structures compared to SMILES, likely due to CCD providing explicit conformational data consistent with AF3 and Boltz-1 training sets and all-atom diffusion models. However, SMILES-based predictions were only marginally less accurate, demonstrating their practical utility for modeling novel PROTACs lacking known atom positions, e.g., in novel PROTAC design and discovery scenarios. Additionally, we assessed model prediction consistency using three random seeds per structure. Both AF3 and Boltz-1 displayed overall consistency, with minor deviations primarily linked to larger, more flexible POIs and E3 ligases.
To further validate model reliability, we conducted MD simulations on a challenging predicted ternary complex (PDB ID: 9B9W). This simulation demonstrated structural stability over 300 ns, reinforcing the validity of AF3 predictions. Across all 62 predicted complexes, we also examined critical residues and molecular properties associated with PROTAC–protein interactions, providing deeper insights into molecular determinants of ternary complex formation. Both AF3 and Boltz-1 performed significantly better on complexes present in their training datasets, particularly for pre-2021 structures predicted with CCD inputs. Interestingly, Boltz-1 slightly outperformed AF3 in RMSD for novel protein–protein interfaces (post-2021), while AF3 consistently surpassed Boltz-1 in ligand binding site accuracy (DockQ). Notably, AF3 achieved comparable DockQ accuracy on novel complexes (0.297) to Boltz-1's average DockQ on familiar structures (0.301), highlighting AF3's strong generalization potential, likely because it has seen more training data. Moreover, confidence metrics (pTM and ipTM scores) effectively predicted RMSD and DockQ outcomes, suggesting their utility for evaluating predictions and guiding experimental validations.
Both AF3 and Boltz-1 notably struggled with four KRAS complexes (PDB IDs: 8QU8, 8QVU, 8QW6, 8QW7), missing binding sites entirely. KRAS's extensive conformational diversity, sensitivity to nucleotide binding-induced rearrangements, and structural sensitivity to mutations make it inherently challenging for static structure-prediction models like AF3 and Boltz-1. Boltz-1 further demonstrated two more mispredictions: a severely mispredicted conformation targeting BTK (8DSO) and another missed binding site targeting DCAF1 (9DLW). Aside from KRAS-related challenges, AF3 consistently delivered more reliable ligand–mediated ternary complex predictions compared to Boltz-1.
Our methodology represents a significant improvement over previous computational approaches by explicitly incorporating ligands into ternary complex predictions. Prior studies frequently overlooked ligand contributions due to limitations of the AF3 server, restricting accurate modeling of ligand–mediated PPIs. Our analysis indicated that individual protein components (POI and E3 ligase) were usually predicted accurately, but their relative orientation often differed from experimental structures, leading to interface misalignment. We hypothesize that refinement through energy minimization or MD simulations may address these discrepancies, particularly when ligand flexibility enables multiple biologically relevant binding conformations.
4.2 Limitations and challenges
Despite AF3 and Boltz-1's promising capabilities in predicting protein complexes, several limitations must be acknowledged, especially regarding PROTAC–mediated ternary complex modeling. While our findings demonstrate that AF3 can achieve high structural accuracy when ligand information is provided, certain challenges remain.First, the limited availability of high-quality experimental data restricts the validation scope for PROTAC predictions. Our study focused on PDB structures where the entire PROTAC ligand was crystallized. We identified and excluded an additional 62 PDB IDs that contained only PROTAC fragments (e.g., warhead, linker, or E3 ligase binder) to concentrate our analysis on complete ternary structures. This necessarily small dataset of complete ternary complexes inherently limits a comprehensive assessment of AF3 and Boltz-1 performance across a diverse range of PROTACs. Consequently, our benchmarking is confined to these existing experimental structures, which may not fully capture the true structural diversity of PROTAC–mediated interactions. Furthermore, we observed that predictive accuracy for both protein and ligand components tends to decline as PROTAC structures increasingly deviate from the AF3 and Boltz-1 pre-2021 training data.
Second, while DockQ is a useful metric for evaluating protein–protein interface accuracy, it has limitations when applied to PROTAC–mediated ternary complexes with flexible or extended interfaces. DockQ calculates the fraction of native contacts (fnat) by defining an interface as any pair of heavy atoms from interacting molecules within 5 Å of each other,52 which is suboptimal for PROTAC complexes where critical interactions may occur at longer distances. Many PROTAC ternary complexes lack inter-protein contacts within this rigid cut-off, leading to DockQ scores that underestimate actual prediction quality. Furthermore, DockQ assumes a rigid-body superposition model, which may not adequately capture the induced-fit effects and alternative conformations characteristic of PROTAC interactions. Consequently, low DockQ scores might reflect valid alternative binding modes rather than outright prediction failures, emphasizing the need for specialized metrics in future work. Future evaluation pipelines should include adapted contact-based metrics to assess PROTAC-relevant binding more accurately. Further, a more nuanced approach incorporating ensemble-based evaluation metrics or energy-based scoring functions could provide deeper insight into ternary complex stability and binding cooperativity. While we initially considered using the updated DockQ v2 for small molecules,48 we decided against it due to its reliance on matching ligand names to compute three-interface DockQ scores, which is incompatible with predictions generated using SMILES.
Third, while AF3 and Boltz-1 provide static structural predictions, they do not capture the dynamics of PROTAC–mediated interactions, which can significantly influence binding affinity and degradation efficacy. MD simulations would provide a complementary validation method by assessing complex stability over time, transient interactions, and induced-fit effects that these models alone cannot model. Although our study included limited MD validation, a comprehensive integration of MD simulations with AF3 and Boltz-1 predictions would further refine these predictions and improve structure-based PROTAC design.
Fourth, analysis of prediction failures reveals that both AF3 and Boltz-1 struggle most with novel PROTACs not represented in the training data, particularly those with large molecular weights and highly flexible linkers. As shown in Fig. 6, prediction accuracy decreases for PROTACs with large molecular weight (MW), heavy atom count (HAC), and rotatable bond count (RBC). AF3 and Boltz-1 achieved near-perfect accuracy on pre-2021 structures (RMSD > 4 Å: 0/16, 3/16), however struggled significantly more with post-2021 structures (RMSD > 4 Å: 19/46, 20/46). A further analysis of success rates by release date is provided in Appendix D. Notably, as shown in Fig. 2, post-2021 structures exhibit higher complexity with increased RBC and ring counts alongside greater diversity of protein targets, reflecting recent advances in crystallization strategies.
Finally, while we propose that AF3 and Boltz-1 can be applied to predict yet-unseen ternary complexes, our study remains purely computational, and the ultimate test of their generalizability would require experimental validation of newly predicted structures. Anecdotally, we observed that these models could generate plausible ternary structures even in cases where experimental crystallization has proven challenging, highlighting the potential risk of overly confident predictions. A definitive accuracy benchmark would require experimentally resolved structures, such as those obtained by cryo-EM. However, given the significant cost and effort associated with these experimental methods, our computational pipeline and publicly available resources aim to reduce the experimental burden by helping researchers prioritize and refine promising PROTAC designs. While our approach is not a replacement for experimental structure determination, it is a valuable tool for guiding experimental efforts, enabling researchers to focus on the most promising PROTAC designs and accelerate the development of degraders with optimized ternary complex formation.
4.3 Reproducibility of study
To ensure the reproducibility of our study and facilitate future research, we have introduced a user-friendly PROTAC prediction pipeline via a publicly accessible web platform https://protacfold.xyz, which integrates PDB and Gemini APIs to automate the generation of AF3 and Boltz-1 input files across various seeds and supports further ligand analysis. Recognizing the challenges in manual POI and E3 ligase determination, especially for researchers new to structural biology, we incorporated a POI and E3 predictive pipeline developed with Gemini 2.5 Flash Experimental. Furthermore, a PyMol script automates the extraction of evaluation metrics from AF3 and Boltz-1 predictions into tabular form.4.4 Future work
One key area for possible future exploration is expanding upon the training data. Recently, Boltz-2 was released,26 with an extended training cut-off date (2023-06-01) and reported improvements in predicting binding affinities. While AF3 performance has been reported to also exceed Boltz-2,26 further research could investigate Boltz-2's performance in predicting PROTAC ternary complexes released between 2021-09-30 and 2023-06-01 in the PDB, providing insights into the importance of not only extending the training data to include additional structures but also including MD trajectories. Such research would pave the way for better training strategies for AF3 (ref. 53) and Boltz models34 to better predict ligand–mediated complex formation. Furthermore, training models on individual PROTAC components (e.g., warheads) may also improve predictions, especially around binding sites.Another important next step will be to include conformational dynamics in the modeling pipeline. AF3 and Boltz-1 only allow for static structural snapshots, not accounting for protein flexibility, ligand-induced conformational changes, or transient binding. MD simulations may provide a better understanding of the stability, cooperativity, and energy involved in forming ternary complexes, especially for PROTACs with flexible linkers or multiple binding possibilities. Improving how we evaluate predictions is also vital. Moving beyond structural superposition methods like DockQ, for instance, by integrating binding free energy calculations, ensemble-based scoring, or MD-derived stability assessments, could refine our ability to distinguish between functional and non-functional PROTAC conformations.
On the experimental front, the scarcity of high-resolution ternary complex structures remains a significant obstacle, restricting the direct benchmarking of AF3 and Boltz-1 predictions. While confidence metrics such as pTM scores from AF2 and AF3 have successfully guided the selection of experimental candidates for snake venom therapeutics54 and tuberculosis vaccine components,55 assessing AF3 and Boltz-1's performance specifically for novel structures not yet represented in the PDB remains an open challenge. Future work combining data-driven computational predictions with systematic experimental validation will be essential to evaluate and enhance the accuracy and generalizability of these models, ultimately expanding their utility in structure-based drug discovery and PROTAC development.
5 Conclusion
To systematically evaluate AF3 and Boltz-1, we developed an automated pipeline to generate input files, predict structures, and quantify predictive accuracy across 62 PROTAC–mediated ternary complexes from the PDB. We explored multiple ligand input strategies, utilizing explicit atom positions (CCD codes) versus SMILES strings, and examined prediction variability using three random seeds. With optimal settings, AF3 achieved near-native accuracy (RMSD < 4 Å) for 46 of 62 complexes, while Boltz-1 achieved this for 40 complexes. AF3 notably outperformed Boltz-1 when explicitly modeling ligand–mediated interactions, highlighting the advantage of including detailed ligand conformational data during predictions. Performance was generally superior for structures present in the training datasets, and prediction accuracy inversely correlated with ligand size and flexibility. Although DockQ scores were useful, they exhibited limitations when evaluating highly flexible complexes, suggesting caution when interpreting these metrics alone.Our findings indicate that both AF3 and Boltz-1 are promising tools for modeling PROTAC–mediated ternary complexes, significantly improving on previous computational approaches. By publicly providing our computational pipeline and prediction results, we offer a reproducible framework for researchers to design, prioritize, and optimize PROTACs and similar therapeutics relying on complex ternary interactions. Nonetheless, AF3 and Boltz-1 are static prediction models and do not fully capture the dynamic conformational landscapes critical to PROTAC efficacy. Future enhancements, such as retraining these models on expanded ligand–protein datasets beyond current PDB cut-offs and integrating extensive MD simulations, will likely further improve prediction accuracy. Experimental validation of computationally predicted complexes also remains essential to assess their real-world applicability in prospective scenarios, ultimately accelerating the development of optimized, ligand-based therapeutic strategies.
Author contributions
ND and FE contributed equally to this work through data curation, formal analysis, investigation, methodology, software, validation, visualization, writing of the original draft, and review and editing of the manuscript. FJ contributed to this work through conceptualization, formal analysis, investigation, methodology, supervision, validation, visualization, and review and editing of the manuscript. RM contributed to this work through conceptualization, formal analysis, funding acquisition, methodology, project administration, resources, supervision, writing of the original draft, and review and editing of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
All data supporting the findings of this study, Predicting PROTAC–mediated ternary complexes with AlphaFold3 and Boltz-1, are openly available:• Predicted and simulated ternary complex structures, along with associated RMSD and DockQ scores, can be accessed via Zenodo at: https://zenodo.org/records/15848838. DOI: https://doi.org/10.5281/zenodo.15848838.
• The full analysis pipeline, code, dataset preparation scripts, and evaluation metrics used in this work are available on GitHub at: https://github.com/NilsDunlop/PROTACFold. DOI: https://doi.org/10.5281/zenodo.15848838.
• An interactive website for PDB ligand analysis and automated AlphaFold3 and Boltz-1 input generation is accessible at: https://protacfold.xyz.
These resources enable full reproduction of the results and support further development and application of PROTAC ternary complex modeling.
Appendix A protein complexes studied in this work
In Table 1, we list the PDB IDs for all 62 PROTAC–mediated protein complexes investigated in this work. Note that while the majority of the complexes listed are confirmed-PROTAC complexes, a few are PROTAC analogues and/or precursors which function via a similar mechanism to PROTACs.| PDB ID | Release date | POI | E3 ligase | Ligand |
|---|---|---|---|---|
| 5T35 | 2017-03-08 | BRD4 | VHL | 759 (MZ1) |
| 6BN7 | 2018-05-30 | BRD4 | CRBN | RN3 (dBET23) |
| 6BOY | 2018-05-30 | BRD4 | CRBN | RN6 (dBET6) |
| 6HM0 | 2019-01-16 | BRD9 | N/A | GBW* (VZ185) |
| 6HAY | 2019-06-12 | SMARCA2 | VHL | FX8 (PROTAC 1) |
| 6HAX | 2019-06-12 | SMARCA2 | VHL | FWZ (PROTAC 2) |
| 6HR2 | 2019-06-12 | SMARCA4 | VHL | FWZ (PROTAC 2) |
| 6SIS | 2019-12-04 | BRD4 | VHL | LFE** (PROTAC 1) |
| 6ZHC | 2020-08-05 | Bcl-xL | VHL | QL8 (Bcl-xL degrader-2) |
| 6W7O | 2020-11-18 | BTK | cIAP1 | TL7 (BCPyr) |
| 6W74 | 2020-11-18 | N/A | BIRC2 | TKY** (BCPyr) |
| 6W8I | 2020-11-18 | BTK | cIAP1 | TKY** (BCPyr) |
| 7KHH | 2021-02-24 | BRD4 | VHL | WEP |
| 7PI4 | 2021-09-29 | FAK1 | VHL | 7QB (GSK215) |
| 7JTO | 2021-10-06 | WDR5 | VHL | VKA (MS33) |
| 7JTP | 2021-10-06 | WDR5 | VHL | X6M (MS67) |
| 6WWB | 2021-11-17 | BRD2 | N/A | YA3 |
| 7Q2J | 2021-11-24 | WDR5 | VHL | 8KH (Homer) |
| 7Z6L | 2022-09-07 | SMARCA2 | VHL | IEI |
| 7Z76 | 2022-09-14 | SMARCA2 | VHL | IEJ |
| 7Z77 | 2022-09-14 | SMARCA2 | VHL | IFF |
| 7ZNT | 2022-09-14 | BRD4 | VHL | IZR |
| 7S4E | 2022-10-05 | SMARCA2 | VHL | 87A (ACBI1) |
| 8BB2 | 2022-11-09 | WDR5 | VHL | Q3X |
| 8BB3 | 2022-11-09 | WDR5 | VHL | Q3X |
| 8BB4 | 2022-11-09 | WDR5 | VHL | Q3R |
| 8BB5 | 2022-11-09 | WDR5 | VHL | Q43 |
| 8C13 | 2022-12-28 | N/A | VHL | SYF (JW48) |
| 7TVA | 2023-02-15 | STAT5A | N/A | KOO (AK-2292) |
| 8BDS | 2023-02-15 | BRD4 | VHL | QIY (PROTAC 48) |
| 8BDT | 2023-02-15 | BRD4 | VHL | QLX (PROTAC 51) |
| 8BDX | 2023-02-15 | BRD4 | VHL | QIY (PROTAC 48) |
| 8BEB | 2023-02-15 | BRD4 | VHL | QIK (PROTAC 49) |
| 8EXC | 2023-02-22 | CA2 | N/A | X2U |
| 8DSF | 2023-03-08 | N/A | BIRC2 | TO0 (BCCov) |
| 8DSO | 2023-03-08 | BTK | cIAP1 | TOO (BCCov) |
| 8OOD | 2023-05-24 | N/A | DCAF1 | VY3 |
| 8PC2 | 2023-11-15 | FKBP5 | VHL | XZW (SelDeg51) |
| 8PDF | 2023-11-15 | FKBP1A | N/A | Y5Q |
| 8QU8 | 2023-12-06 | KRAS | VHL | WYL (ACBI3) |
| 8QVU | 2023-12-06 | KRAS (iso2B) | VHL | WYL (ACBI3) |
| 8QW6 | 2023-12-06 | KRAS | VHL | X4R |
| 8QW7 | 2023-12-06 | KRAS | VHL | X53 |
| 8OKC | 2024-01-17 | SARS-CoV-2 NSP5 | N/A | VQN |
| 8R5H | 2024-02-21 | BRD4 | VHL | 759 (MZ1) |
| 8RWZ | 2024-03-06 | BRD4 | VHL | 759 (MZ1) |
| 8RX0 | 2024-03-06 | BRD4 | VHL | 759 (MZ1) |
| 8FY0 | 2024-04-10 | Bcl-xL | VHL | YF8 (PROTAC 753b) |
| 8FY1 | 2024-04-10 | Bcl-2 | VHL | YF8 (PROTAC 753b) |
| 8FY2 | 2024-04-10 | Bcl-2 | VHL | YFH (PROTAC WH244) |
| 8U0H | 2024-09-04 | PTPN2 | N/A | UB0 (CMDP-2) |
| 8UH6 | 2024-09-11 | PTPN2 | CRBN | WO8 (CMDP-1) |
| 8WDK | 2024-09-18 | WEE1 | VHL | W6U |
| 8RQ9 | 2024-09-25 | BRD4 | CRBN | A1H2F (CFT-1297) |
| 9BIG | 2024-10-02 | STAT6 | N/A | A1AQQ (AK-1690) |
| 9B9H | 2024-11-06 | WDR5 | DCAF1 | A1AM2 (OICR-40333) |
| 9B9T | 2024-11-06 | WDR5 | DCAF1 | A1ANM (OICR-40407) |
| 9B9W | 2024-11-06 | WDR5 | DCAF1 | A1ANN (OICR-40792) |
| 9DLW | 2024-11-06 | WDR5 | DCAF1 | A1BAF (OICR-41114) |
| 8YMB | 2025-02-12 | BRD4 | VHL | A1LY0 (SHD913) |
| 8S75 | 2025-03-12 | EPHX2 | N/A | A1H5L (PROTAC FL412) |
| 8S76 | 2025-03-12 | EPHX2 | N/A | A1H5M (PROTAC JSF67) |
Appendix B interface torsion analysis
Here, we extend upon Fig. 7 and 8 to further investigate a select few of the poorly predicted structures. Fig. 11 and 12 illustrate poorly predicted structures where the extent of misalignment in the protein–protein and protein–ligand interfaces can be visually assessed. We observed that, in most cases, the misalignment can be described as a slight alteration of the torsion at the protein–protein interface. Nevertheless, for some of these poor predictions, we observed how Boltz-1's placed ligands incorrectly on both proteins.Fig. 13–16 instead showcase examples of poorly predicted structures due to inaccurate prediction of the ligand binding site. In all four structures, both AF3 and Boltz-1 missed the KRAS binding site entirely. Notably, these four worst-predicted structures were all from the same study56 analyzing the design of KRAS degraders. KRAS is a significant oncological target that is notoriously difficult to model due to its high structural flexibility.
Appendix C performance breakdown by POI and E3 ligase
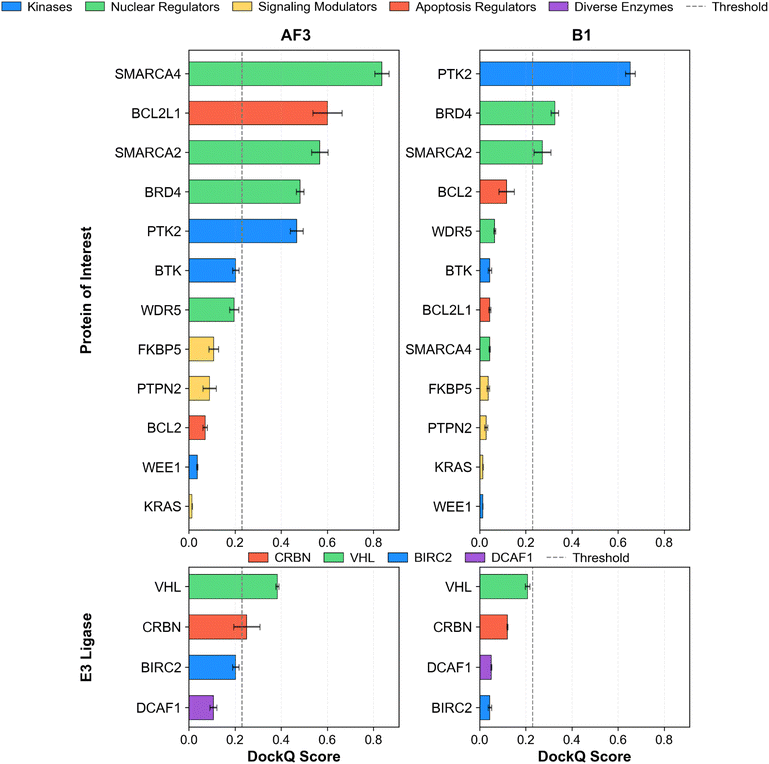
Fig. 17 and 18 assess predictive performance by POI and E3 ligase. The analysis offers insights into the ability of each model to capture specific structural features within various PROTAC ternary complexes.As shown in Fig. 17, while both models struggled with certain POIs, their performance on specific targets varied. Boltz-1 more accurately predicted KRAS (7.91 Å) and BCL2 (8.07 Å), whereas AF3 was more accurate for BCL2L1 (5.20 Å) and WEE1 (10.80 Å). In total, AF3 predicted 13 POIs below the 4 Å threshold, compared to 10 for Boltz-1. For E3 ligases, DCAF1 and VHL exhibited high RMSDs in both models, whereas CRBN and BIRC2 were predicted relatively accurately.
The model's performance gap was more pronounced in terms of DockQ scores (Fig. 18). AF3 had five POIs exceeding the 0.23 threshold, compared to three for Boltz-1. AF3 produced high-quality predictions for SMARCA4 (0.836) and BCL2L1 (0.601), where Boltz-1 failed. For E3 ligases, AF3 achieved acceptable scores for VHL and CRBN, while Boltz-1 failed to do so for any E3 ligase.
Overall, AF3's average POI RMSD was lower than Boltz-1's (3.65 Å vs. 4.71 Å), and its average DockQ score was more than double (0.306 vs. 0.138), indicating its better performance.
Appendix D model performance by release date
Fig. 19 (top) shows model performance relative to the 2021-09-30 training cut-off. Both models perform better on pre-cut-off structures, with AF3 achieving higher success rates overall. Performance peaks near the cut-off date and declines approximately linearly afterward, though AF3 consistently outperforms Boltz-1 except for minor fluctuations in SMILES-based evaluations. Fig. 19 (bottom) presents RMSD success rates (<2.0 Å) by release date, showing near-perfect accuracy for both models at the cut-off, where Boltz-1 matches AF3. AF3 exhibits steady improvements within the training set, whereas Boltz-1 peaks near the cut-off and declines for older structures (2017–2019). The limited number of pre-cut-off PROTAC structures introduces uncertainty in these trends.Appendix E prompt used for the identification of the POI and E3 ligase
In Chart 1, we provide the complete Gemini-2.5 Flash Experimental prompt that is used by https://protacfold.xyz/ to identify the POI and the E3 ligase components from any PDB entry. | ||
| Chart 1 Prompt used with Gemini 2.5 Flash Experimental to annotate the POI and the E3 ubiquitin ligase in a given PDB structure. | ||
The prompt is automatically filled with chain-level metadata retrieved via the PDB GraphQL API. Whenever the crystallographic publication's abstract is available, it is passed to the model as additional context, often supplying explicit functional annotations that further boost classification accuracy. The approach is highly effective, as seen in Appendix F.
Appendix F accuracy of Gemini in POI and E3 ligase classification
As shown in Fig. 20, Gemini correctly detects the presence of a POI in 61 out of 62 test structures (98% accuracy). Meanwhile, its performance for E3 ligase recognition (Fig. 21) is perfect for the same benchmark set (100% accuracy). These results underscore an important point: when a large language model is supplied with the right domain-specific information and given clear, structured instructions, it can help perform tasks in computational drug discovery pipelines that would otherwise require domain expertise.Appendix G Boltz-1 large ligands SMILES parsing
Chart 2 contains the patch applied to lines 928–936 of the function in Boltz-1. The original Boltz-1 parser generated PDB-style atom names by appending the canonical atom index to the element symbol; for large ligands, names such as “CL118” could therefore exceed the four-character limit mandated by the PDB format and crash subsequent file-writing or
function in Boltz-1. The original Boltz-1 parser generated PDB-style atom names by appending the canonical atom index to the element symbol; for large ligands, names such as “CL118” could therefore exceed the four-character limit mandated by the PDB format and crash subsequent file-writing or 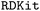 routines. The revised code first retrieves the canonical atom order
routines. The revised code first retrieves the canonical atom order 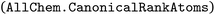 , then computes how many digits can safely follow the element symbol
, then computes how many digits can safely follow the element symbol 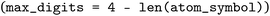 . If no space remains, the index is wrapped with modulo 10 to guarantee at least one trailing digit; otherwise the index is zero-padded up to the available width. The resulting string, always ≤4 characters, is stored via
. If no space remains, the index is wrapped with modulo 10 to guarantee at least one trailing digit; otherwise the index is zero-padded up to the available width. The resulting string, always ≤4 characters, is stored via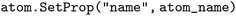 before 3-D conformed generation proceeds. This modification enables Boltz-1 to parse contemporary PDB entries that contain very large ligands without format violations or runtime errors.
before 3-D conformed generation proceeds. This modification enables Boltz-1 to parse contemporary PDB entries that contain very large ligands without format violations or runtime errors.
 | ||
Chart 2 Updated lines 928-936 within the Boltz-1 SMILES  parsing code to handle large ligand inputs by ensuring the atom symbol and index are always less than four characters. parsing code to handle large ligand inputs by ensuring the atom symbol and index are always less than four characters. | ||
Acknowledgements
FJ and RM acknowledge the funding provided by the Wallenberg AI, Autonomous Systems, and Software Program (WASP), supported by the Knut and Alice Wallenberg Foundation. RM would also like to acknowledge Anders Källberg, Christian Tyrchan, and Eva Nittinger for their previous collaboration on AF2 for PROTAC ternary complex prediction, which informed this follow-up study. The computations and data storage were enabled by resources provided by Chalmers e-Commons. The computations and data storage were enabled by resources provided by the National Academic Infrastructure for Supercomputing in Sweden (NAISS), partially funded by the Swedish Research Council through grant agreement no. 2022-06725.Notes and references
- W. Li, J. Zhang, L. Guo and Q. Wang, J. Chem. Inf. Model., 2022, 62, 523–532 CrossRef CAS PubMed.
- J. Abramson, J. Adler, J. Dunger, R. Evans, T. Green, A. Pritzel, O. Ronneberger, L. Willmore, A. J. Ballard and J. Bambrick, et al. , Nature, 2024, 1–3 Search PubMed.
- Z. Hu and C. M. Crews, ChemBioChem, 2022, 23, e202100270 CrossRef CAS.
- M. J. Bond and C. M. Crews, RSC Chem. Biol., 2021, 2, 725–742 RSC.
- M. Békés, D. R. Langley and C. M. Crews, Nat. Rev. Drug Discovery, 2022, 21, 181–200 CrossRef.
- Y.-W. Wang, L. Lan, M. Wang, J.-Y. Zhang, Y.-H. Gao, L. Shi and L.-P. Sun, Eur. J. Med. Chem., 2023, 247, 115037 CrossRef CAS PubMed.
- Z. Liu, M. Hu, Y. Yang, C. Du, H. Zhou, C. Liu, Y. Chen, L. Fan, H. Ma and Y. Gong, et al. , Mol. Biomed., 2022, 3, 46 CrossRef CAS PubMed.
- L. Zhao, J. Zhao and K. Zhong, et al. , Signal Transduction Targeted Ther., 2022, 7, 113 CrossRef CAS.
- J. M. Tsai, R. P. Nowak, B. L. Ebert and E. S. Fischer, Nat. Rev. Mol. Cell Biol., 2024, 25, 740–757 CrossRef CAS PubMed.
- D. Pliatsika, C. Blatter and R. Riedl, Drug Discovery Today, 2024, 29, 104178 CrossRef CAS PubMed.
- G. P. Pereira, C. Gouzien, P. C. T. Souza and J. Martin, Bioinformatics Advances, 2025, 5, vbaf056 CrossRef PubMed.
- M. Ignatov, A. Jindal, S. Kotelnikov, D. Beglov, G. Posternak, X. Tang, P. Maisonneuve, G. Poda, R. A. Batey and F. Sicheri, et al. , J. Am. Chem. Soc., 2023, 145, 7123–7135 CrossRef CAS PubMed.
- E. Rovers and M. Schapira, J. Chem. Inf. Model., 2024, 64, 6162–6173 CrossRef CAS PubMed.
- R. Yin, B. Y. Feng, A. Varshney and B. G. Pierce, Protein Sci., 2022, 31, e4379 CrossRef CAS PubMed.
- G. P. Pereira, B. Jiménez-García, R. Pellarin, G. Launay, S. Wu, J. Martin and P. C. Souza, J. Chem. Inf. Model., 2023, 63, 6823–6833 CrossRef CAS PubMed.
- D. Zaidman, J. Prilusky and N. London, J. Chem. Inf. Model., 2020, 60, 4894–4903 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek and A. Potapenko, et al. , Nature, 2021, 596, 583–589 CrossRef CAS.
- J. Durairaj, A. M. Waterhouse, T. Mets, T. Brodiazhenko, M. Abdullah, G. Studer, G. Tauriello, M. Akdel, A. Andreeva and A. Bateman, et al. , Nature, 2023, 622, 646–653 CrossRef CAS PubMed.
- P. Bryant, G. Pozzati and A. Elofsson, Nat. Commun., 2022, 13, 1265 CrossRef CAS.
- M. van Breugel, I. Rosa e Silva and A. Andreeva, Commun. Biol., 2022, 5, 312 CrossRef CAS.
- Z. Yang, X. Zeng, Y. Zhao and R. Chen, Signal Transduction Targeted Ther., 2023, 8, 115 CrossRef.
- R. Evans, M. O'Neill, A. Pritzel, N. Antropova, A. Senior, T. Green, A. Žídek, R. Bates, S. Blackwell and J. Yimet al., bioRxiv, 2021, preprint, DOI:10.1101/2021.10.04.463034.
- M. Gao, D. Nakajima An, J. M. Parks and J. Skolnick, Nat. Commun., 2022, 13, 1744 CrossRef CAS.
- DeepMind, AlphaFold 3 – Web Server, 2024, https://alphafoldserver.com, accessed: 2025-02-28 Search PubMed.
- J. Wohlwend, G. Corso, S. Passaro, N. Getz, M. Reveiz, K. Leidal, W. Swiderski, L. Atkinson, T. Portnoi, I. Chinn, J. Silterra, T. Jaakkola and R. Barzilay, bioRxiv, 2025, preprint, DOI:10.1101/2024.11.19.624167.
- S. Passaro, G. Corso, J. Wohlwend, M. Reveiz, S. Thaler, V. Ram Somnath, N. Getz, T. Portnoi, J. Roy and H. Starket al., bioRxiv, 2025, preprint, DOI:10.1101/2025.06.14.659707.
- B. A. A. Team, X. Chen, Y. Zhang, C. Lu, W. Ma, J. Guan, C. Gong, J. Yang, H. Zhang, K. Zhang, S. Wu, K. Zhou, Y. Yang, Z. Liu, L. Wang, B. Shi, S. Shi and W. Xiao, bioRxiv, 2025, preprint, DOI:10.1101/2025.01.08.631967.
- E. Nittinger, Ö. Yoluk, A. Tibo, G. Olanders and C. Tyrchan, Artif. Intell. Life Sci., 2025, 100136 CAS.
- S. Zheng, Y. Tan, Z. Wang, C. Li, Z. Zhang, X. Sang, H. Chen and Y. Yang, Nat. Mach. Intell., 2022, 4, 739–748 CrossRef.
- C. Insights, PROTAC Drug Development: Advancing Targeted Protein Degradation, 2025, https://www.cas.org/resources/cas-insights/protac-drug-development, accessed: 2025-02-28 Search PubMed.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed.
- F. Erazo, N. Dunlop, F. Jalalypour and R. Mercado, ChemRxiv, ChemRxiv, 2025, preprint, DOI:10.26434/chemrxiv-2025-gfd6c.
- DeepMind, AlphaFold 3 – GitHub Repository, 2024, https://github.com/google-deepmind/alphafold3, accessed: 2025-02-28 Search PubMed.
- J. Wohlwend, Boltz-1 GitHub Repository, 2024, https://github.com/jwohlwend/boltz, accessed: 2025-04-03 Search PubMed.
- J. D. Westbrook, C. Shao, Z. Feng, M. Zhuravleva, S. Velankar and J. Young, Bioinformatics, 2015, 31, 1274–1278 CrossRef PubMed.
- OpenEye Scientific Software, OpenEye Toolkits, version 2024.2.1, 2024, http://www.eyesopen.com Search PubMed.
- Y. Rose, J. M. Duarte, R. Lowe, J. Segura, C. Bi, C. Bhikadiya, L. Chen, A. S. Rose, S. Bittrich, S. K. Burley and J. D. Westbrook, J. Mol. Biol., 2021, 433, 166704 CrossRef CAS PubMed.
- J. Wang, W. Wang, P. A. Kollman and D. A. Case, J. Chem. Inf. Comput. Sci., 2001, 41, 242–246 CrossRef.
- A. W. Sousa da Silva and W. F. Vranken, BMC Res. Notes, 2012, 5, 1–8 CrossRef PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1, 19–25 CrossRef.
- G. D. Team, GROMACS 2024 Documentation, 2024 Search PubMed.
- B. Hess, H. Bekker, H. J. Berendsen and J. G. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- R. J. Gowers, M. Linke, J. Barnoud, T. J. E. Reddy, M. N. Melo, S. L. Seyler, J. Domanski, D. L. Dotson, S. Buchoux and I. M. Kenney and O. Beckstein, MDAnalysis: a Python package for the rapid analysis of molecular dynamics simulations (No. LA-UR-19-29136), Los Alamos National Laboratory (LANL), Los Alamos, NM, United States, 2019 Search PubMed.
- N. Michaud-Agrawal, E. J. Denning, T. B. Woolf and O. Beckstein, J. Comput. Chem., 2011, 32, 2319–2327 CrossRef CAS PubMed.
- Schrödinger, LLC, The PyMOL molecular graphics system, version 3.1, 2010, https://pymol.org Search PubMed.
- D. Weerakoon, R. J. Carbajo, L. De Maria, C. Tyrchan and H. Zhao, J. Chem. Inf. Model., 2022, 62, 340–349 CrossRef CAS PubMed.
- C. Crowe, M. A. Nakasone, S. Chandler, C. Craigon, G. Sathe, M. H. Tatham, N. Makukhin, R. T. Hay and A. Ciulli, Sci. Adv., 2024, 10, eado6492 CrossRef CAS PubMed.
- C. Mirabello and B. Wallner, Bioinformatics, 2024, 40, btae586 CrossRef CAS PubMed.
- M. Karelina, J. J. Noh and R. O. Dror, eLife, 2023, 12, RP89386 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graph., 1996, 14, 33–38 CrossRef CAS PubMed.
- R. A. Laskowski and M. B. Swindells, J. Chem. Inf. Model., 2011, 51, 2778–2786 CrossRef CAS PubMed.
- S. Basu and B. Wallner, PLoS One, 2016, 11, e0161879 CrossRef.
- lucidrains, alphafold3-pytorch: PyTorch implementation of AlphaFold3 – GitHub Repository, 2024, https://github.com/lucidrains/alphafold3-pytorch, accessed: 2025-05-18 Search PubMed.
- S. Vázquez Torres, M. Benard Valle, S. P. Mackessy, S. K. Menzies, N. R. Casewell, S. Ahmadi, N. J. Burlet, E. Muratspahić, I. Sappington, M. D. Overath, E. Rivera-de Torre, J. Ledergerber, A. H. Laustsen, K. Boddum, A. K. Bera, A. Kang, E. Brackenbrough, I. A. Cardoso, E. P. Crittenden, R. J. Edge, J. Decarreau, R. J. Ragotte, A. S. Pillai, M. Abedi, H. L. Han, S. R. Gerben, A. Murray, R. Skotheim, L. Stuart, L. Stewart, T. J. A. Fryer, T. P. Jenkins and D. Baker, Nature, 2025, 639, 225–231 CrossRef.
- S. Pambudi, A. S. Irsyad, D. I. S. Utomo, A. Nurhasanah and T. Novianti, BIO Web of Conferences, 2024, 127, 04001 CrossRef.
- J. Popow, W. Farnaby, A. Gollner, C. Kofink, G. Fischer, M. Wurm, D. Zollman, A. Wijaya, N. Mischerikow and C. Hasenoehrl, et al. , Science, 2024, 385, 1338–1347 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |