 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multi-modal contrastive learning for chemical structure elucidation with VibraCLIP
Pau
Rocabert-Oriols
,
Camilla
Lo Conte
,
Núria
López
 and
Javier
Heras-Domingo†
and
Javier
Heras-Domingo†
 *
*
Institute of Chemical Research of Catalonia (ICIQ-CERCA), Avinguda dels Països Catalans, 16, Tarragona, 43007, Spain
First published on 11th November 2025
Abstract
Identifying molecular structures from vibrational spectra is central to chemical analysis but remains challenging due to spectral ambiguity and the limitations of single-modality methods. While deep learning has advanced various spectroscopic characterization techniques, leveraging the complementary nature of infrared (IR) and Raman spectroscopies remains largely underexplored. We introduce VibraCLIP, a contrastive learning framework that embeds molecular graphs, IR and Raman spectra into a shared latent space. A lightweight fine-tuning protocol ensures generalization from theoretical to experimental datasets. VibraCLIP enables accurate, scalable, and data-efficient molecular identification, linking vibrational spectroscopy with structural interpretation. This tri-modal design captures rich structure–spectra relationships, achieving Top-1 retrieval accuracy of 81.7% and reaching 98.9% Top-25 accuracy with molecular mass integration. By integrating complementary vibrational spectroscopic signals with molecular representations, VibraCLIP provides a practical framework for automated spectral analysis, with potential applications in fields such as synthesis monitoring, drug development, and astrochemical detection.
Introduction
Accelerating the discovery of drugs or decoding complex organic molecules in diverse chemical environments demands rapid, on-the-fly molecular characterization.1–4 Although, during synthetic procedures, organic molecular identification is powered by NMR spectroscopy,5 this technique has limitations when the sample is paramagnetic, has high quadrupoles or cannot be prepared in a liquid or solid form. Alternatively, vibrational spectroscopies like IR and Raman provide information on the patterns within the molecules (functional groups) that correspond to the vibrational modes within the molecule (collective movements). Although both probe molecular vibrations, IR and Raman spectra differ due to distinct selection rules (dipole vs. polarizability6), Fig. 1. These techniques can be employed for remote detection at astronomical distances, such as in the case of the James Webb Space Telescope.7 Despite their widespread use, vibrational spectroscopies often struggle with low throughput, complex interpretation, and their integration with computational tools.Emerging machine learning approaches offer a transformative path forward by enabling the interpretation of complex spectroscopic data and laying the foundation for automated and scalable analysis,8 for instance, in NMR5 spectroscopy. By bridging the gap between qualitative signals and quantitative insights, these tools open new possibilities to understand and design molecular systems with unprecedented speed and precision.9,10 However, in reality human-based experimental identification relies on integrating information from at least two independent techniques. In AI, combining complementary data streams is known as multi-modal learning, but its application to molecular characterization remains a challenge.
Vibrational spectroscopies are often interpreted with Density Functional Theory (DFT), which balances accuracy and cost.11,12 Systematic deviations remain: harmonic frequencies are overestimated requiring empirical scaling.13 Intensities depend on functional and basis; B3LYP generally gives reliable IR intensities, though Raman intensities are more basis sensitive.14–16 Moreover, temperature, intermolecular interactions, and solvation broaden or shift experimental spectra beyond idealized DFT predictions. To address such challenges, recent advancements in AI-driven spectra-to-molecule mapping broadly follow two main strategies: (i) predicting spectral outputs directly from molecular structures, and (ii) interpreting spectroscopic data to infer chemical structures, enabling inverse design. Graph Neural Networks (GNNs) have been developed to predict IR spectra from molecular graphs,17–20 while Convolutional Neural Networks (CNNs) have been applied to classify IR spectra by functional groups.21,22 Other models, such as support vector machines (SVMs), random forest (RF), multilayer perceptrons (MLPs), and deep reinforcement learning have been used to identify functional groups from IR spectra.23–26 SMILES-based representations using the Transformer architecture27 have emerged as an alternative for inferring molecular structures directly from IR spectra.28 Existing approaches focus on individual modalities, such as molecular graphs or single spectral techniques.20,29–31 Unlike, CNNs, RNNs, GNNs, or random forests, which rely on supervised prediction of labels or properties, contrastive learning operates more like a spectroscopic fingerprinting process: it aligns heterogeneous data (Graph, IR, Raman) in a shared latent space by maximizing the similarity of true pairs and minimizing that of mismatches. Just as a chemist identifies a compound by matching experimental spectra against reference patterns, contrastive learning enables unsupervised cross-modal representation learning that is essential for molecular elucidation.
Vibrational spectroscopies like infrared (IR) and Raman are widely used to identify functional groups but remain underutilized when it comes to combining their complementary strengths.32 Integrating both offers a robust foundation for multi-modal analysis, as both spectra originate from the same underlying physics,6 the molecular vibrations expressed as normal modes (Fig. 1). Current methods struggle to unify these modalities, and incorporating spectral data with molecular graphs introduces further challenges in aligning disparate data types within a shared latent space. Overcoming these limitations is essential for advancing multi-modal characterization and extracting deeper molecular insights.
Multi-modal models based on contrastive learning, such as the CLIP architecture,33 have emerged as powerful tools to bridge diverse data modalities. These models are particularly well-suited for characterization techniques, where relationships between different streams of data, such as molecular structures and spectra, must be learned to allow molecular identification. While current applications of contrastive learning focus on dual-modal relationships, they leave room for further exploration in more complex multi-modal tasks (i.e., a figure assigned to its caption). For instance, MolCLR34 leverages self-supervised pre-training on molecular graphs to encode chemically meaningful similarities, improving property prediction tasks with limited labeled data. The CReSS system applies contrastive learning to directly connect 13C NMR spectra with molecular structures, enabling high-recall cross-modal retrieval (i.e., molecular assignment) in large molecular libraries and supporting molecular scaffold determination.35,36 Similarly, the CMSSP framework establishes a shared representation between tandem mass spectrometry (MS/MS) spectra and molecular structures, improving metabolite identification.37 However, these approaches used molecular Morgan fingerprints38,39 together with graph embeddings to better anchor the MS spectra-molecule pair, though combining both may introduce redundant information, underscoring the need for careful feature selection. More recently, the MARASON implementation introduces neural graph matching to retrieval-augmented molecular machine learning, significantly improving mass spectrum simulation accuracy over existing methods.40 Expanding beyond molecular systems, MultiMat introduces a self-supervised multi-modality framework for materials science, leveraging diverse streams of bulk material properties to enhance property prediction, material discovery, and scientific interpretability.41
Despite this rapid progress, existing models remain restricted to dual-modal formulations (e.g., structure–spectrum) alignment. Our benchmarking of recent methods (see SI, S-1) highlights their strengths and trade-offs. For example, Chemprop-IR17 reaches high Spectral Information Similarity (SIS) scores (0.969 theoretical, 0.864 experimental data), while Graphormer-IR19 scales to large datasets but with 139 M trainable parameters. Contrastive approaches like CReSS35 (13C-NMR + SMILES, Top-10 = 91.6%) or CMSSP37 (MS/MS + Graph, Top-10 = 76.3%) demonstrate the potential of cross-modal retrieval but remain constrained by their pairing nature. Among these, the closest approach to our work is SMEN,42 which aligns molecular graphs with IR spectra. While effective (Top-1 = 94.1%, Top-10 = 99.8% in QM9 dataset43), SMEN is limited to two modalities and requires 24 M parameters, more than twice the size of our approach.
VibraCLIP advances this frontier by introducing a tri-modal framework for vibrational spectroscopy that jointly aligns molecular graphs, IR, and Raman spectra in a unified latent space. By exploiting the complementarity of IR and Raman signals, it enables richer and more comprehensive molecular characterization than dual-modal systems currently allow. As demonstrated in the SI (Section S-2), a non-learned baseline further underscores the need for explicit alignment to reliably recover molecular structures from different data streams (i.e., molecular structure and vibrational data). With a maximum of 11 M trainable parameters, VibraCLIP effectively captures complex structure–spectra relationships, enabling molecular elucidation from vibrational data and bridging characterization techniques with molecular interpretation. Its adaptable and scalable design unifies and leverages these complementary modalities, accelerating structural analysis, facilitating knowledge transfer across modalities, and establishing it as a powerful tool for advancing characterization across diverse scientific domains.
Results
VibraCLIP framework
Our model builds on the CLIP architecture,33 adapting cross-modal contrastive learning to vibrational spectroscopy. It aligns IR and Raman spectral data with molecular structures in a shared representation space, enabling accurate identification of organic molecules. The framework operates in two phases: (i) multi-modal contrastive pre-training (learning) and (ii) retrieval with scoring (identification) (Fig. 2). During pre-training on the theoretical QM9S dataset,20 the graph, IR, and Raman encoders process triads of synthetic (calculated) IR and Raman spectra with molecular structures to generate feature embeddings. This objective enables VibraCLIP to learn chemically coherent representations by bringing together IR, Raman, and structural features of the same molecule while separating those from different systems. | ||
| Fig. 2 Overview of VibraCLIP. (A) VibraCLIP pre-training. The SMILES representation is converted into the graph representation (G) and the vibrational spectra are both pre-processed by interpolation and normalization between 0 and 1. Each modality is fed to the VibraCLIP simultaneously in batches of 128 systems, where the graph encoder is based on the DimeNet++ architecture46,47 while the spectral encoders and projection heads are based on fully connected neural networks (FCNN). The contrastive loss is utilized to maximize the agreement between the projected embedding vectors coming from the molecular graph (Gn), IR (In) and Raman (Rn), building a shared latent space. (B) The retrieval strategy, the IR and Raman spectra of an unknown molecule is fed to the spectral encoders and the cross-modality similarity score is then calculated across the database, providing the Top-K-most aligned embeddings vectors, which contains the most likely molecules. | ||
All the employed spectra are standardized and curated following the procedure described in the Vibrational Spectra Pre-processing section. For pre-training, we use the QM9S20 dataset that contains 130![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 optimized organic molecules with synthetic (DFT) spectroscopic data, see Datasets section for details. To generalize, we fine-tuned (realign) the model on an external PubChem dataset,44 which contains 5500 molecules following the same strategy as the QM9S,20,44 expanding the chemical space, together with their corresponding synthetic spectra, and molecular size range encountered by the pre-trained model. This addition, introduces a minimal realignment of the latent space to accommodate unseen molecules. Finally, as it is well-known in the community, experimental and computed IR and Raman spectra differ in peak position and width. Therefore, both experimental realignment and validation are crucial to be predictive under realistic conditions. To this end, 320 gas-phase molecular spectra were used from NIST Webbook45 (IR) and from standard libraries (Raman). The experimental dataset features chemically diverse, real-world compounds with richer spectral complexity, listed in the SI (Section S-5).
000 optimized organic molecules with synthetic (DFT) spectroscopic data, see Datasets section for details. To generalize, we fine-tuned (realign) the model on an external PubChem dataset,44 which contains 5500 molecules following the same strategy as the QM9S,20,44 expanding the chemical space, together with their corresponding synthetic spectra, and molecular size range encountered by the pre-trained model. This addition, introduces a minimal realignment of the latent space to accommodate unseen molecules. Finally, as it is well-known in the community, experimental and computed IR and Raman spectra differ in peak position and width. Therefore, both experimental realignment and validation are crucial to be predictive under realistic conditions. To this end, 320 gas-phase molecular spectra were used from NIST Webbook45 (IR) and from standard libraries (Raman). The experimental dataset features chemically diverse, real-world compounds with richer spectral complexity, listed in the SI (Section S-5).
The representations of organic molecules via the Graph Encoder allows to extract structural patterns from the graph, in our case based on the DimeNet++,46,47 architecture (see Methods section). DimeNet++ is a dedicated graph neural network to learn geometric patterns in molecular structures containing both angular and distance based features thus closely resembling z-matrix molecular representations. DimeNet++ produces a continuous vector space representation of molecular graphs that, in our case, is further enhanced by only concatenating the standardized molecular mass of the molecule (without isotopic considerations) before the projection heads, see Fig. 2. The molecular mass provides additional chemical context crucial to distinguishing between similar structures particularly enhancing the quality of the embeddings in downstream tasks.
Similarly, spectral encoders are needed. In this case, the applied architecture is a multi-layer fully connected neural network (FCNN) designed to transform the IR or Raman spectra data into spectral embeddings representation, see Methods section for details. The encoder first employs an input layer matching the dimensionality of the given spectra, followed by a sequence of two hidden layers with progressively decreasing dimensions ensuring a smooth downsampling in the feature space. The network produces a fixed-length feature vector using a final linear layer.
Next, the projection heads are designed to map embeddings from modality-specific encoders into a shared representation space. It starts by a linear layer reducing the input dimension to the projection dimension, and follows by a GELU activation function48 to introduce non-linearity. Two more layers, one that refines the projection and an optional one for normalization that ensures consistent, well-regularized representations.
The multi-modal training strategy incorporates separate learning rates and optimization schedules for each component to facilitate efficient learning across multiple data types, see Methods for details. The learning ability is then controlled by the contrastive loss functions. In the original CLIP framework,33 two distinct modalities, denoted as A (image) and B (caption), are paired via the cosine similarity metric, see Methods for details. In our adaptation, A = G denotes the molecular graph representation, while the infrared (IR) or Raman spectra correspond to B. Then we can evaluate loss functions, L, for different pairs L(G, IR), L(G, Raman) and L(IR, Raman). The CLIP loss is independently applied between the graph representation and each spectroscopic modality, unifying all three in a single model. The overall loss is the sum of individual CLIP losses involving the graph G (see Contrastive loss functions section).
To evaluate the retrieval accuracy, we implemented a dedicated PyTorch Lightning callback,49–51 executed exclusively on the test dataset in two scenarios: the single and dual spectral modalities. The cosine similarity then scores and ranks all candidate graph embeddings, creating a sorted list of most likely molecules (i.e., Top-K matches) either for single or dual spectra data streams (see Retrieval accuracy section). In the retrieval phase, IR and Raman spectra serve as queries to generate embeddings, which are compared to candidate molecular structures. Candidates are ranked by cosine similarity (eqn (2)), quantifying spectral–structural alignment and prioritizing the most chemically consistent matches. As shown in Fig. 3, retrieval accuracy plots highlight VibraCLIP's effectiveness in matching spectra to molecular structures. Furthermore, the model was also realigned (fine-tuned) using experimental data, demonstrating the effectiveness of the minimal realignment strategy in adapting to real-world spectra.
 | ||
| Fig. 3 VibraCLIP retrieval performance. (A) Performance comparison of VibraCLIP using different contrastive loss strategies without anchoring features. (B) Performance comparison of VibraCLIP using different contrastive loss strategies including the standardized molecular mass as anchoring feature. (C) Latent space realignment on the PubChem dataset44 with the standardized molecular mass as anchoring feature. (D) Latent space realignment on the experimental dataset with the standardized molecular mass as anchoring feature. Training epochs with maximum of 200 epochs with early stopping strategy: (A) IR only 153, IR + Raman 115, IR + Raman (allpairs) 95; (B) IR only 107, IR + Raman (allpairs) 112; (C and D) realignment with 15 epochs. | ||
Performance evaluation
We evaluated VibraCLIP across multiple configurations to assess its performance and adaptability (Fig. 3). These included: (i) IR spectra only, (ii) combined IR and Raman spectra, and (iii) full IR–Raman alignment through contrastive loss (see eqn (5)). To enhance molecular context, we incorporated the standardized molecular mass into the Graph, IR, and Raman embeddings before projection, following approaches like CMSSP.37 To evaluate generalization beyond QM9S,20 we used a PubChem-derived dataset44 constructed by randomly selecting PubChem molecules and computing their IR and Raman spectra, and applying a minimal fine-tuning by updating only the projection heads, leaving the rest of the model frozen. This lightweight realignment enabled adaptation to new chemical distributions with minimal overhead. Additionally, we validated the model on an in-house experimental dataset containing real IR and Raman spectra, again applying fine-tuning only to the final projection layers. Together, these two realignment strategies demonstrate VibraCLIP's ability to generalize across both theoretical and experimental domains.The retrieval accuracies highlight the substantial gains achieved by incorporating the Raman spectra and aligning vibrational modalities within the contrastive learning loss function. Without considering the standardized molecular mass as anchoring feature, adding Raman spectra increases the Top-1 accuracy from 12.4% (IR only) to 55.1%, and further to 62.9% when explicitly aligning IR and Raman embeddings. As shown in Fig. 3A, this improvement underscores the complementary role of Raman spectroscopy in refining molecular identification. For Top-25, performance improves from 63.6% (IR only) to 94.0% and 94.3% with Raman and full IR-Raman alignment, respectively, demonstrating the value of combining vibrational modalities in a unified latent space. Learned alignment is essential, as the non-learning baseline yielded near-random retrieval across modalities (<0.30% Top-K, Section S-2). VibraCLIP surpasses this lower bound by orders of magnitude, confirming that its performance stems directly from the contrastive training objective.
Building on this, the inclusion of the standardized molecular mass as an anchoring feature (i.e., similar to adding the total mass from MS experiment), results in notable improvements across all retrieval thresholds. With mass included, Top-1 accuracy for IR-only models rises from 12.4% to 24.2%, and with the fully aligned IR-Raman loss function, from 62.9% to 81.7%, a remarkable 18.8% absolute gain. As shown in Fig. 3B, the Top-25 accuracy increases to 98.9%, confirming the effectiveness of mass as a chemically grounded global descriptor. This anchoring strategy improves consistency in the latent space, especially in distinguishing structurally similar candidates.
The model was further fined-tuned, realigned, on an external dataset of randomly selected organic molecules from PubChem44 (Fig. 3C) and an experimental dataset (Fig. 3D). These evaluations demonstrate the model's adaptability and generalization to previously unseen chemical and spectral distributions. This realignment was deliberately minimal, updating only the final layer of the projection heads over 15 epochs, while leaving the remaining 8.7 million of the model's 11 million parameters frozen. This lightweight adjustment enabled smooth realignment to new data domains with minimal computational cost.
On the PubChem dataset, realignment yielded strong results: Top-1 accuracy rose to 67.6%, and Top-25 accuracy reached 98.2% when the IR-Raman embeddings alignment is considered in the loss function. While a moderate drop from the QM9S20 benchmarks (98.9%), this performance reflects the increased chemical diversity of PubChem44 and confirms that minimal fine-tuning effectively mitigates dataset shift. Similarly, experimental validation using IR and Raman spectra demonstrated the robustness of the approach, with Top-25 accuracy reaching 100% and Top-1 performance at 34.4% in the fully aligned, mass-anchored setup. These findings confirm that VibraCLIP's realignment strategy is not only data-efficient but also transferable across theoretical and real-world domains.
Discussion
VibraCLIP introduces a new paradigm in molecular characterization by extending contrastive learning beyond dual-modality frameworks to a three-way alignment of molecular Graphs, IR, and Raman spectra. This multi-modal integration not only boosts retrieval performance but also enables direct alignment between spectral modalities, capturing complementary vibrational features that enhance chemical identification. The inclusion of standardized molecular mass as an anchoring feature further enhances embedding consistency, facilitating chemically grounded representations that are crucial for robust retrieval.The necessity of learned alignment is further highlighted when compared to the non-learning baseline (SI, Section S-2), where molecular fingerprints and spectra projected into a shared space yielded near-random retrieval (<0.30%). Similar to comparing an experimental IR spectrum with an uncalibrated reference library, without alignment, meaningful matches cannot be recovered.
The model shows particularly strong performance in Top-K retrieval scenarios. While Top-1 accuracy improves substantially with IR–Raman alignment (from 12.4% to 62.9%) and further to 81.7% with mass anchoring, the real value lies in the Top-25 accuracy of 98.9%. Importantly, this framework is not designed to retrieve a single exact match, but rather to guide molecular identification by narrowing the search space to a small pool of highly similar candidates. In contexts such as drug discovery, high-throughput screening, or the identification of unknown chemical species in extraterrestrial environments,3,4 this level of precision is both meaningful and practically valuable. This highlights that, even when the exact structure is not ranked first, VibraCLIP consistently retrieves chemically similar candidates sharing key scaffolds. As shown in Fig. 4, correct molecules often appear within the top-ranked set, and retrieved structures tend to be structurally coherent, offering actionable insight in practical settings.
To probe the robustness of VibraCLIP under incomplete data scenarios, we performed a missing-data analysis. In the dual-modality setting (Graph, IR with mass anchoring), IR spectra were progressively removed. Interestingly, performance remained stable up to 10–15% missing data, but dropped sharply at 50%, highlighting the model's reliance on complete spectral information in this configuration. In contrast, in the three-modality case (Graph, IR, Raman with the all-pairs loss function), the accuracy decline at 50% missing data, from one of the spectra modalities, was far less severe. This suggest that the model continues to learn effectively from the Graph, IR and the remaining Raman spectra underscoring the complementarity of the two vibrational modalities. When spectra were missing, the spectral encoder part was frozen to prevent weight updates, ensuring stable optimization of the remaining modalities (SI, Section S-6).
VibraCLIP also supports realignment to new data distributions through lightweight fine-tuning. Using only the final projection layers, we adapted the model to a PubChem44 derived dataset and to an internally curated experimental vibrational dataset, confirming transferability across synthetic and real-world domains. Nevertheless, significant limitations remain, largely due to the scarcity of multi-modal, machine learning-ready spectroscopic datasets. The core pre-training relies on QM9S,20 restricted to small organics and element counts of C: 9, N: 7, O: 5, F: 6, leaving larger and chemically richer structures underrepresented. PubChem extends this chemical space (C: 21, N: 6, O: 8, F: 0), and the experimental dataset (C: 30, N: 6, O: 6, F: 6), while limited in size, provides valuable diversity for benchmarking with real-world data, and highlights the opportunity for broader dataset development.
Lastly, while DimeNet++,46,47 provides a strong basis for graph encoding, future VibraCLIP iterations can adopt more expressive GNNs to capture complex molecular features. Its modular design enables expansion to modalities such as NMR,35,36 UV-Vis, and mass spectrometry,37 opening new paths to AI-driven molecular identification.
In summary, VibraCLIP is a scalable, efficient, and generalizable framework for spectral interpretation. By embedding molecular and spectral information in a unified latent space, it provides groundwork for next-generation tools in molecular discovery, structural elucidation, and AI-augmented spectroscopy.
Methods
Datasets
We utilized the QM9S dataset,20 an extension of QM9 (ref. 43) with theoretical spectroscopic data, comprising 130![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 organic molecules with re-optimized geometries. It includes diverse molecular properties, from scalar values (e.g., energies, partial charges) to high-order tensors (e.g., Hessian matrices, quadrupole and octopole moments, and polarizabilities). Spectral data, including IR, Raman, and UV-Vis spectra, were computed via frequency analysis and time-dependent DFT at the B3LYP/def-TZVP level of theory using Gaussian16.52 The inclusion of IR and Raman spectra in QM9S enabled the development of VibraCLIP, a model designed for multi-modal alignment and spectroscopic representation learning.
000 organic molecules with re-optimized geometries. It includes diverse molecular properties, from scalar values (e.g., energies, partial charges) to high-order tensors (e.g., Hessian matrices, quadrupole and octopole moments, and polarizabilities). Spectral data, including IR, Raman, and UV-Vis spectra, were computed via frequency analysis and time-dependent DFT at the B3LYP/def-TZVP level of theory using Gaussian16.52 The inclusion of IR and Raman spectra in QM9S enabled the development of VibraCLIP, a model designed for multi-modal alignment and spectroscopic representation learning.
VibraCLIP was fine-tuned on 5500 molecules from the PubChem-derived subset of QM9S,20,44 expanding the chemical space and molecular size range encountered by the pre-trained model. This subset includes SMILES representations, 3D coordinates, and Hessian matrices. IR and Raman spectra were inferred using the DetaNet model,20 trained and validated on QM9S, which accurately predicts these spectral features. This fine-tuning process improved VibraCLIP's ability to generalize to a broader range of molecular and spectroscopic data.
Experimental realignment and validation were performed using IR spectra from the NIST Webbook45 and Raman spectra from the OMNIC software's standard library for the same molecules. The resulting dataset includes 320 examples, each combining molecular structure, IR, and Raman spectra. Unlike computational benchmarks such as QM9S20 or PubChem,44 which remain biased toward small, synthetically accessible molecules and lack the chemical richness of real systems, our experimental set features diverse compounds with more complex spectra. Although limited in size, it provides a crucial first step toward validating VibraCLIP in real-world scenarios, underscoring both its generalization ability and the need for larger, experimentally grounded benchmarks.
Molecular graph representation
Given a SMILES representation coming from the QM9S dataset, the corresponding molecular graph G is built, in which each node represents an atom and each edge represents a chemical bond between atoms. Initially, the SMILES string is converted into a molecular structure with the RDKit library53 for molecular processing, where the molecule is saturated with hydrogen, and a 3D conformation is generated using the ETKDG embedding algorithm.54 While ETKDG generally produces reasonable geometries, conformer generation may fail for certain strained systems, flexible molecules with many rotatable bonds, or cases requiring higher-level quantum refinement. Such cases are excluded from pre-training. After that, atom features including atomic type, aromaticity and hybridization states (sp, sp2, sp3) are extracted. Bond attributes are encoded by bond type, and pairwise atomic distances are added as additional edge attributes. Furthermore, the molecular mass is standardized by the dataset-wide mean and standard deviation, which is then embedded as meta-data within the graph. The resulting graph comprises node features, edge indices and attributes, 3D positions and standardized molecular mass, making it suitable for a wide range of graph neural networks encoders.Vibrational spectra pre-processing
The processing of vibrational spectra is applied uniformly to both IR and Raman data, involving two key steps to standardize and prepare the spectra for training (80%), validation (10%) and testing (10%). First, both the x-axis (cm−1) and y-axis (intensity) of each spectrum are interpolated, reducing the original resolution from 3501 data points to 1750. This step preserves the spectral shape, ensures consistent dimensionality across all samples, and makes the dataset more manageable computationally. Following interpolation, each spectrum is normalized using the Min–Max scaling strategy,55 which adjusts intensity values to fall within a range of 0 to 1. This normalization step ensures consistency across spectra, enhancing comparability and model performance by eliminating scale variability in the data.Since VibraCLIP is a multi-modal model, the processed IR and Raman spectra are also incorporated within the graph object, following a common strategy in PyTorch Geometric51 for multi-modal data integration. This approach enables seamless access to multiple data types within the model and supporting efficient multi-modal alignment and representation learning. Further implementation details are available in the SI (S-3).
Model architecture
We further enhanced the molecular graph representation by concatenating the standardized molecular mass to the molecular vector generated by DimeNet++. This addition improved the model's overall performance, as the molecular mass provides additional chemical context that proved to be valuable for distinguishing between similar structures. Importantly, this addition occurs before passing the graph embeddings to the projection head, allowing the projection model to fully leverage this SI. We believe that the projection head benefits from the inclusion of the standardize molecular mass by refining the embeddings in a way that better captures molecular distinctions relevant to the downstream tasks.
Each hidden layer consists of a linear transformation followed by an activation function, which introduces non-linearity to enhance the network's expressiveness. Then batch normalization is enabled and applied after each linear layer to stabilize training by normalizing the activations. Finally, the network outputs a fixed-length feature vector through an additional linear layer, which serves as the final layer. This spectral encoder produces embeddings that capture meaningful spectral information, enabling effective interaction with other modalities for alignment within the VibraCLIP model.
Multi-modal training strategy
The multi-modal training strategy for VibraCLIP incorporates separate learning rates and optimization schedules for each component to facilitate efficient learning across multiple data types. Specifically, distinct learning rates are assigned to the graph neural network (GNN) encoder, both the spectral encoders, allowing each modality-specific encoder to adapt at an optimal pace. The projection heads for each modality (Graph, IR, Raman) are grouped under a single learning rate, promoting alignment within the shared representation space. Optimization is carried out through the AdamW optimizer,57 which combines weight decay for regularization with adaptive gradient steps, and a ReduceLROnPlateu scheduler58 that adjusts the learning rates based on validation loss. Additionally, an early stopping mechanism monitors also the validation loss, halting training if no improvement is observed over 15 epochs, thereby preventing overfitting and ensuring efficient convergence. This tailored optimization approach enhances the model's ability to learn complex multi-modal representations while preserving computational efficiency. See details in the SI (S-4).Contrastive loss functions
In the original CLIP framework,33 two distinct modalities, denoted as A and B, are paired with corresponding samples Ai and Bi across a batch of N samples, where i represents the batch index. Following encoding via modality-specific encoders fA and fB, the resulting embeddings are represented by ai = fA(Ai) and bi = fB(Bi). The objective of CLIP is to establish a robust alignment between these modalities through a shared objective function, effectively bridging A and B in the embeddings space, as shown in following equation: | (1) |
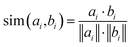 | (2) |
Therefore, the symmetric loss can be represented as:
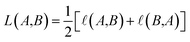 | (3) |
It is worth noting that the CLIP model was originally presented in the context of image-caption pairs, where A represents an image modality and B a text modality.
VibraCLIP for vibrational spectroscopy
An intuitive approach to multi-modal pre-training for smart characterization of organic molecules involves directly adapting the two-modalities CLIP model to spectroscopic-specific data. In our adaptation, G denotes the molecular graph representation analog to the image in the original CLIP model, while the infrared (IR) or Raman spectra take the role of the text caption in CLIP's original implementation. This framework enable us to consider two distinct strategies for multi-modal pre-training in smart characterization for vibrational spectroscopy of organic molecules, paring G with IR or G with Raman. The loss function for such strategies would be represented as L(G, IR), L(G, Raman) and L(IR, Raman), where L is given by eqn (3).Graph-centered VibraCLIP
Alternatively, rather than limiting CLIP to a single vibrational spectroscopy, we extend the original CLIP framework to incorporate multiple modality pairs by positioning the molecular graph representation at the core of the model. This central role leverages the graph's rich structural information and its capacity to interface meaningfully with both IR and Raman spectra. By computing the CLIP loss between the graph and each spectroscopic modality independently, we effectively unify all three modalities through a shared structural epicenter. The resulting loss function for this graph-centered strategy is defined by aggregating the CLIP losses involving the graph G, with L as defined in eqn (3): | (4) |
All pairs VibraCLIP
As a further extension, we can extend the graph-centered strategy by also adding the interaction between vibrational spectra, in our case, IR and Raman. Including such relationship between IR and Raman embeddings, adds the additional loss term that directly encourages alignment between the two spectral modalities of the sample, creating a more cohesive shared latent space by pulling all three modalities closer together. Then, the resulting loss function for the all pairs strategy can be expressed as follows, where L is defined in eqn (3): | (5) |
Retrieval accuracy
For evaluating the retrieval accuracy, we implemented a dedicated PyTorch Lightning callback,49–51 executed exclusively on the test dataset. This callback assesses two retrieval scenarios. In the first, the model aligns the molecular graph representation with a single spectral modality, either IR or Raman, to retrieve the most similar graph embedding and its associated molecular structure. In the second scenario, both IR and Raman spectra are utilized simultaneously, allowing the model to retrieve the molecular entity that best aligns with both spectra. Each of these strategies are used depending on the number of modalities that VibraCLIP is trained on. Top-K accuracy is defined as the percentage of test cases in which the correct molecular structure is ranked within the fist K candidates, ordered by cosine similarity between the spectral and structural embeddings.Single spectra retrieval strategy
In this retrieval accuracy scenario, we evaluate the model's alignment of molecular graph embeddings with a single spectra embedding (either IR or Raman). The callback first generates graph and spectral embeddings for each sample in the test dataset by passing batches through the model's forward pass in inference mode, storing the embeddings alongside SMILES identifiers.The cosine similarity (eqn (2)) scores between spectral and graph embeddings are then calculated. For each target spectrum, the model ranks all candidate graph embeddings based on similarity, creating a sorted list of likely matches. These similarity scores are stored and exported as pickle file for further analysis of the retrieval accuracy and Top-K matches.
Dual spectra retrieval strategy
We assessed the model's ability to align molecular graph embeddings with both IR and Raman spectra simultaneously. This strategy differs from the single-spectrum approach by incorporating both IR and Raman spectral embeddings in the similarity calculations. Here, the cosine similarity scores are computed not only between the graph and each spectral embedding but also between IR and Raman embeddings themselves.To obtain a combined similarity measure, the geometric mean (GM) of the three pairwise similarity scores (Graph-IR, Graph-Raman, IR-Raman) is calculated, providing a comprehensive metric for alignment across three modalities. This combined similarity helps identify the molecular graph that aligns with both spectra most closely. Specifically, a low score in any one pair will drastically reduce the overall geometric mean, reflecting the joint alignment among all three modalities. The results are saved as a pickle file, enabling analysis of retrieval accuracy in multi-modal alignment within the VibraCLIP framework.
 | (6) |
These enhancements in similarity scoring establish a robust and interpretable multi-modal alignment, ensuring that VibraCLIP captures the intricate relationships between molecular structures and vibrational spectra. Employing the geometric mean across three modalities, our approach maximizes retrieval precision while mitigating discrepancies from individual spectral contributions.
Author contributions
P. R.-O. and J. H.-D. contributed equally to the implementation of the VibraCLIP code. C. L. C. contributed to the state-of-the-art benchmark, non-learning baseline and the missing data analysis. N. L. provided overall project guidance. J. H.-D. conceived and supervised the project. All authors contributed to writing the manuscript and approved the final version.Conflicts of interest
There are no conflicts of interest to declare.Data availability
VibraCLIP implementation supporting this study is publicly available on GitHub at (https://github.com/jherasdo/vibraclip). A mirrored version is also maintained at (https://github.com/LopezGroup-ICIQ/vibraclip) to ensure accessibility. Further details on usage can be found in the repository's documentation. A supplementary data repository is available on Zenodo (https://doi.org/10.5281/zenodo.15348391), providing the datasets, training checkpoints, and callback output files used in this study.Supplementary information: Benchmarks with recent models, a non-learning baseline, model implementation details, hyperparameter optimization, retrieval accuracies from experimental data, and a missing-data analysis. These provide additional insights into the performance and intepretability of the proposed approach. See DOI: https://doi.org/10.1039/d5dd00269a.
Acknowledgements
The authors thank the Institute of Chemical Research of Catalonia (ICIQ) Summer Fellow Program for its support. We also acknowledge the Department of Research and Universities of the Generalitat de Catalunya for funding through grant (reference: SGR-01155), and PID2024-157556OB-I00 funded by MICIU/AEI/10.13039/501100011033/FEDER, UE. Additionally, we are grateful to Dr Georgiana Stoica and Mariona Urtasun from the ICIQ Research Support Area (Spectroscopy and Material Characterization Unit) for their valuable assistance. Computational resources were provided by the Barcelona Supercomputing Center (BSC), which we gratefully acknowledge.References
- J. Jiménez-Luna, F. Grisoni and G. Schneider, Nat. Mach. Intell., 2020, 2, 573–584 CrossRef.
- V. Barone, S. Alessandrini, M. Biczysko, J. R. Cheeseman, D. C. Clary, A. B. McCoy, R. J. DiRisio, F. Neese, M. Melosso and C. Puzzarini, Nat. Rev. Methods Primers, 2021, 1, 38 CrossRef.
- R. C. Fortenberry, ACS Phys. Chem. Au, 2023, 4, 31–39 CrossRef PubMed.
- J. Cernicharo, A. M. Heras, A. Tielens, J. R. Pardo, F. Herpin, M. Guélin and L. Waters, Astrophys. J., 2001, 546, L123 CrossRef.
- Y. Luo, W. Chen, Z. Su, X. Shi, J. Luo, X. Qu, Z. Chen and Y. Lin, Nat. Commun., 2025, 16, 2342 CrossRef PubMed.
- J. M. Hollas, Modern spectroscopy, John Wiley & Sons, 4th edn, 2004 Search PubMed.
- J. P. Gardner, J. C. Mather, M. Clampin, R. Doyon, M. A. Greenhouse, H. B. Hammel, J. B. Hutchings, P. Jakobsen, S. J. Lilly and K. S. Long, et al. , Space Sci. Rev., 2006, 123, 485–606 CrossRef.
- M. W. Mullowney, K. R. Duncan, S. S. Elsayed, N. Garg, J. J. van der Hooft, N. I. Martin, D. Meijer, B. R. Terlouw, F. Biermann and K. Blin, et al. , Nat. Rev. Drug Discovery, 2023, 22, 895–916 CrossRef PubMed.
- N. J. Szymanski, C. J. Bartel, Y. Zeng, M. Diallo, H. Kim and G. Ceder, npj Comput. Mater., 2023, 9, 31 CrossRef.
- G. Durant, F. Boyles, K. Birchall and C. M. Deane, Nat. Comput. Sci., 2024, 4, 735–743 CrossRef PubMed.
- E. E. Zvereva, A. R. Shagidullin and S. A. Katsyuba, J. Phys. Chem. A, 2011, 115, 63–69 CrossRef PubMed.
- L. Bastonero and N. Marzari, npj Comput. Mater., 2024, 10, 55 CrossRef.
- J. P. Merrick, D. Moran and L. Radom, J. Phys. Chem. A, 2007, 111, 11683–11700 CrossRef PubMed.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865 CrossRef PubMed.
- E. E. Zvereva, A. R. Shagidullin and S. A. Katsyuba, J. Phys. Chem. A, 2011, 115, 63–69 CrossRef PubMed.
- C. A. Jiménez-Hoyos, B. G. Janesko and G. E. Scuseria, Phys. Chem. Chem. Phys., 2008, 10, 6621–6629 RSC.
- C. McGill, M. Forsuelo, Y. Guan and W. H. Green, J. Chem. Inf. Model., 2021, 61, 2594–2609 CrossRef CAS PubMed.
- N. Saquer, R. Iqbal, J. D. Ellis and K. Yoshimatsu, Digital Discovery, 2024, 3, 602–609 RSC.
- C. M. Stienstra, L. Hebert, P. Thomas, A. Haack, J. Guo and W. S. Hopkins, J. Chem. Inf. Model., 2024, 4613–4629 CrossRef CAS.
- Z. Zou, Y. Zhang, L. Liang, M. Wei, J. Leng, J. Jiang, Y. Luo and W. Hu, Nat. Comput. Sci., 2023, 3, 957–964 CrossRef CAS.
- A. A. Enders, N. M. North, C. M. Fensore, J. Velez-Alvarez and H. C. Allen, Anal. Chem., 2021, 93, 9711–9718 CrossRef CAS PubMed.
- G. Jung, S. G. Jung and J. M. Cole, Chem. Sci., 2023, 14, 3600–3609 RSC.
- J. A. Fine, A. A. Rajasekar, K. P. Jethava and G. Chopra, Chem. Sci., 2020, 11, 4618–4630 RSC.
- K. Judge, C. W. Brown and L. Hamel, Anal. Chem., 2008, 80, 4186–4192 CrossRef CAS PubMed.
- C. Klawun and C. L. Wilkins, J. Chem. Inf. Comput. Sci., 1996, 36, 69–81 CrossRef CAS.
- S. Devata, B. Sridharan, S. Mehta, Y. Pathak, S. Laghuvarapu, G. Varma and U. D. Priyakumar, Digital Discovery, 2024, 3, 818–829 RSC.
- A. Vaswani, Adv. Neural Inf. Process. Syst., 2017, 30 Search PubMed.
- M. Alberts, T. Laino and A. C. Vaucher, Commun. Chem., 2024, 7, 268 CrossRef CAS.
- M. Alberts, O. Schilter, F. Zipoli, N. Hartrampf and T. Laino, Adv. Neural Inf. Process. Syst., 2024, 37, 125780–125808 CrossRef.
- N. J. Williams, L. Kabalan, L. Stojanovic, V. Zólyomi and E. O. Pyzer-Knapp, Sci. Data, 2025, 12, 9 CrossRef PubMed.
- T. Hu, Z. Zou, B. Li, T. Zhu, S. Gu, J. Jiang, Y. Luo and W. Hu, J. Am. Chem. Soc., 2025, 147, 27525–27536 CrossRef PubMed.
- K. Acheson and S. Habershon, J. Chem. Theory Comput., 2024, 21, 307–320 CrossRef.
- A. Radford, J. W. Kim, C. Hallacy, A. Ramesh, G. Goh, S. Agarwal, G. Sastry, A. Askell, P. Mishkin, J. Clark, et al., International conference on machine learning, 2021, pp. 8748–8763 Search PubMed.
- Y. Wang, J. Wang, Z. Cao and A. Barati Farimani, Nat. Mach. Intell., 2022, 4, 279–287 CrossRef.
- Z. Yang, J. Song, M. Yang, L. Yao, J. Zhang, H. Shi, X. Ji, Y. Deng and X. Wang, Anal. Chem., 2021, 93, 16947–16955 CrossRef PubMed.
- F. Xu, W. Guo, F. Wang, L. Yao, H. Wang, F. Tang, Z. Gao, L. Zhang, W. E and Z.-Q. Tian, Nat. Comput. Sci., 2025, 5, 1–9 CrossRef.
- L. Chen, B. Xia, Y. Wang, X. Huang, Y. Gu, W. Wu and Y. Zhou, Anal. Chem., 2024, 96, 16871–16881 CrossRef PubMed.
- N. J. Morehouse, T. N. Clark, E. J. McMann, J. A. van Santen, F. J. Haeckl, C. A. Gray and R. G. Linington, Nat. Commun., 2023, 14, 308 CrossRef PubMed.
- H. L. Morgan, J. Chem. Doc., 1965, 5, 107–113 CrossRef.
- R. Wang, R.-X. Wang, M. Manjrekar and C. W. Coley, arXiv, 2025, preprint, arXiv:2502.17874, DOI:10.48550/arXiv.2502.17874.
- V. Moro, C. Loh, R. Dangovski, A. Ghorashi, A. Ma, Z. Chen, S. Kim, P. Y. Lu, T. Christensen and M. Soljačić, Newton, 2025, 1(1) CrossRef.
- G. C. Kanakala, B. Sridharan and U. D. Priyakumar, Digital Discovery, 2024, 3, 2417–2423 RSC.
- R. Ramakrishnan, P. O. Dral, M. Rupp and O. A. Von Lilienfeld, Sci. Data, 2014, 1, 1–7 CrossRef.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen and B. Yu, et al. , Nucleic Acids Res., 2023, 51, D1373–D1380 CrossRef PubMed.
- P. J. Linstrom and W. G. Mallard, J. Chem. Eng. Data, 2001, 46, 1059–1063 CrossRef.
- J. Gasteiger, S. Giri, J. T. Margraf and S. Günnemann, Machine Learning for Molecules Workshop, NeurIPS, 2020 Search PubMed.
- J. Gasteiger, J. Groß and S. Günnemann, International Conference on Learning Representations (ICLR), 2020 Search PubMed.
- D. Hendrycks and K. Gimpel, arXiv, 2016, preprint, arXiv:1606.08415, DOI:10.48550/arXiv.1606.08415.
- W. Falcon and T. P. L. team, PyTorch Lightning, 2019, https://www.pytorchlightning.ai Search PubMed.
- A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, A. Desmaison, A. Kopf, E. Yang, Z. DeVito, M. Raison, A. Tejani, S. Chilamkurthy, B. Steiner, L. Fang, J. Bai and S. Chintala, arXiv, 2019, preprint, arXiv:1912.01703. DOI:10.48550/arXiv.1912.01703.
- M. Fey and J. E. Lenssen, arXiv, 2019, preprint, arXiv:1903.02428, DOI:10.48550/arXiv.1903.02428.
- M. Frisch, et al., Gaussian 16, Revision B. 01/Gaussian, 2016 Search PubMed.
- G. Landrum, et al., RDKit: Open-source cheminformatics, 2006, https://www.rdkit.org Search PubMed.
- S. Riniker and G. A. Landrum, J. Chem. Inf. Model., 2015, 55, 2562–2574 CrossRef PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- J. Gilmer, S. S. Schoenholz, P. F. Riley, O. Vinyals and G. E. Dahl, International conference on machine learning, 2017, pp. 1263–1272 Search PubMed.
- I. Loshchilov, arXiv, 2017, preprint, arXiv:1711.05101, DOI:10.48550/arXiv.1711.05101.
- A. Al-Kababji, F. Bensaali and S. P. Dakua, International Conference on Intelligent Systems and Pattern Recognition, 2022, pp. 204–212 Search PubMed.
Footnote |
| † Present address: Departament de Química Inorgànica i Orgànica (Secció de Química Inorgànica), Institut de Química Teòrica i Computacional (IQTC), Universitat de Barcelona, C/Martí i Franquès, 1, Barcelona, 08028, Spain. E-mail: E-mail: javier.heras@ub.edu |
| This journal is © The Royal Society of Chemistry 2025 |


