Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Investigation of arene and heteroarene nitration supported by high-throughput experimentation and machine learning†
Taline
Kerackian
*ab,
Clément
Wespiser
 b,
Matthieu
Daniel
b,
Eric
Pasquinet
b,
Matthieu
Daniel
b,
Eric
Pasquinet
 b and
Eugénie
Romero
b and
Eugénie
Romero
 *a
*a
aDépartement Médicaments et Technologies pour la Santé (DMTS), SCBM, Université Paris Saclay, CEA, INRAE, 91191 Gif-sur-Yvette, France. E-mail: eugenie.romero@cea.fr
bCEA, DAM, Le Ripault, F-37260 Monts, France
First published on 24th June 2025
Abstract
Access to the nitro functional group is a widespread and longstanding transformation of interest in many fields of chemistry. However, the robustness and specificity of this transformation can remain challenging, particularly in the case of heteroarene nitration. Based on this observation, a comprehensive investigation was initiated to screen nitration conditions on various arenes and heteroarenes. A systematic and diverse study of both nitrating agents and activating reagents was conducted using high-throughput experimentation to afford high-quantity and high-quality data generation. General trends were identified and correlated with the electronic properties of the heteroarenes; notably, the difficult nitration of electron-poor heteroarenes was highlighted. Original combinations of reagents were found to perform well in nitration reactions. The obtained data were also used to design a predictive tool relying on machine learning in order to provide the best nitration reaction conditions depending on the targeted substrate. The limited predictive efficiency obtained pointed out the importance of diversification and chemically relevant encoding of the data set.
Introduction
Nitro heteroarenes are highly versatile compounds with diverse applications due to their multifaceted properties.1–7 Their broad utility highlights the need for efficient nitration methods. Two main synthetic routes exist to obtain the nitro functionality: (i) oxidation of amines8 or azides;9 (ii) direct nitration using a nitrating agent.1,3 The first method is limited when applied to heteroarenes. For the latter, the most common nitration methodology is the electrophilic substitution of electron-rich or electron-neutral arenes10–12 using a mixture of nitric and sulfuric acid. This approach turns out to be ineffective when applied to some electron-poor, nitrogen-containing heteroarenes and suffers from limited selectivity control. Direct nitration can be carried out from various substrates, namely via direct C–H functionalization13–16 or ipso-functionalization17 from the corresponding carboxylic acid or halogenated heteroarene, offering broader chemical exploration opportunities and enhanced selectivity control (Fig. 1a). Nevertheless, C–H and ipso-functionalization methodologies to access nitro compounds remain scarce when applied to heteroarenes,18,19 with only a limited number of examples and generally poor yields. Nitration methodologies have extended to the use of metallic salts as nitrating agents, allowing access to safer reaction conditions by replacing nitric acid.20–22 They also provide alternatives to electrophilic substitution reactions. Numerous reactions involve radical transformations, in which nitro radicals can be intermediates.23,24 This strategy may offer improved compatibility with electron-poor arenes. Transition metal-catalyzed nitration reactions have also been developed,25 with a predominance of palladium and copper catalysis. Recent reports of N-nitro compounds used as nitrating agents26,27 have further expanded the scope of applicable molecules. Altogether, these methodologies show a wide range of nitrating agents and activating reagents. As reaction intermediates can be difficult to characterize, proposed mechanisms often display inconsistencies.28–30 Thus, we considered it relevant to systematically study the outcome of nitration reactions by reacting diverse nitrating agents with activating reagents. To do so, we relied on high-throughput experimentation (HTE).31–33 HTE has proven to be an efficient and resource-economical approach for screening reaction parameters. It allows systematic variation of conditions and enables a high number of experiments to be performed in parallel. The standardization of reactions, from preparation to analysis, makes the results highly comparable and enables efficient identification of general reactivity trends.
can be intermediates.23,24 This strategy may offer improved compatibility with electron-poor arenes. Transition metal-catalyzed nitration reactions have also been developed,25 with a predominance of palladium and copper catalysis. Recent reports of N-nitro compounds used as nitrating agents26,27 have further expanded the scope of applicable molecules. Altogether, these methodologies show a wide range of nitrating agents and activating reagents. As reaction intermediates can be difficult to characterize, proposed mechanisms often display inconsistencies.28–30 Thus, we considered it relevant to systematically study the outcome of nitration reactions by reacting diverse nitrating agents with activating reagents. To do so, we relied on high-throughput experimentation (HTE).31–33 HTE has proven to be an efficient and resource-economical approach for screening reaction parameters. It allows systematic variation of conditions and enables a high number of experiments to be performed in parallel. The standardization of reactions, from preparation to analysis, makes the results highly comparable and enables efficient identification of general reactivity trends.
Together, these advantages support the generation of qualitative experimental data, which serves as excellent input for machine learning processes, in contrast to classical bench-scale data typically described in the literature.34–36 This observation was recently emphasized, as predictive tools developed using standard experimental data from the literature have shown limited efficiency, largely due to a lack of data standardization and the absence of negative results, issues that HTE can address. Machine learning and artificial intelligence have been applied in chemistry, notably to generate predictive tools.37 Most reported HTE campaigns study well-known reactions and generally high-yielding transformations.38–45 During our exploration of heteroarene nitration, we initially observed the limited amount of available literature and also faced significant challenges reproducing reported reaction conditions. Hence, we chose to study the challenging nitration reaction, which is especially low-yielding on electron-poor, nitrogen-containing heteroarenes. Our goal was to test combinations of various substrates and reagents in order to identify optimal reaction conditions and to uncover reactivity trends depending on selected scaffolds.
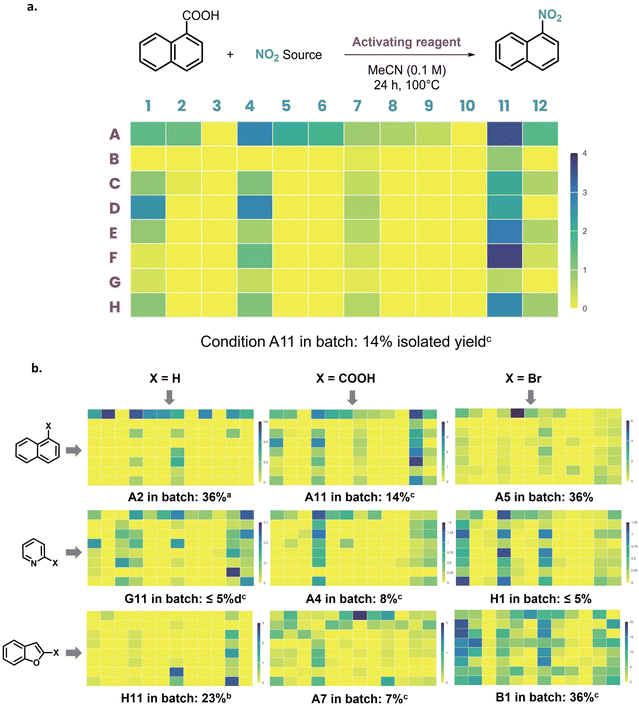
Based on the existing literature,3,23 we designed a 96-well HTE plate to test 12 different nitrating agents and 8 different activating reagents. This plate was systematically evaluated on: (i) arenes, (ii) electron-rich heteroarenes, and (iii) electron-poor heteroarenes. Each class of substrate was evaluated under three reactivity modes: (i) direct C–H functionalization of the non-functionalized substrate, (ii) ipso-functionalization from the corresponding carboxylic acid, and (iii) ipso-functionalization from the corresponding halogenated substrate. The overall HTE campaign led to the performance of 864 different reactions. The data collected will be used to develop a predictive model based on machine learning. Different types of molecular encoding will be tested, and the ability of the model to accurately predict nitration outcomes on new substrates will be evaluated.
Experimental results and discussions
This investigation aims to test various substrates' ability to undergo nitration reactions. The first parameter to select was the set of scaffolds to be examined. Naphthalene was selected as an electron-neutral aryl moiety; benzofuran was chosen as an electron-rich heteroarene, as a lone pair is involved in the overall aromaticity of the molecule; and pyridine was picked as a nitrogen-containing heteroarene displaying electron-poor character, as the lone pair on nitrogen is not delocalized (Fig. 1a). All these substrates were directly exposed to the designed nitration plate to perform direct C–H functionalization.Then, ipso-functionalization was evaluated using 1-naphthoic acid, picolinic acid, benzofuran-2-carboxylic acid, 1-bromonaphthalene, 2-bromopyridine, and 2-bromobenzofuran. Each of these nine different scaffolds was submitted to a 96-well plate designed to study nitration reaction parameters. Most nitration reactions can be regarded as involving a combination of a nitrating agent and an activating reagent. Our goal was to test various combinations of these two species (Fig. 1b). Such variation, based on literature precedent, would allow us to reproduce reported conditions but also permit original combinations of reagents giving the opportunity for a fortunate discovery.
Since the variety of reported nitrating agents is tremendous, we chose to select it as the parameter with the highest number of screened candidates (Fig. 1c). Twelve nitrating agents were selected. Nitric acid and tert-butyl nitrite, two of the main nitration reagents, were picked. Since nitronium tetrafluoroborate provides a solid and stable source of the reactive nitronium ion, many modern methodologies employ it as a nitrating agent.46,47 We naturally picked it as a nitrating agent of interest to screen. Then, both nitrate and nitrite alkali metal salts were selected with sodium as the cationic species. To screen a different cationic counterpart, potassium nitrite was also picked. A soluble nitrate salt, tetrabutylammonium nitrate, was selected. Bismuth(III), silver(I), and iron(III) were chosen, as they are the most commonly used metallic nitrate salts. It has to be noted that both bismuth and iron nitrate salts come as hydrated metal complexes: bismuth(III) nitrate pentahydrate and iron(III) nitrate nonahydrate. Finally, the two most reported N-nitro compounds were selected, namely N-nitrosuccinimide (Succ-NO2) and N-nitrosaccharin (Sacc-NO2).19,48–50 The number of equivalents of nitrating agent was set at two equivalents across the plate.
On the other hand, seven different activating reagents were selected (Fig. 1c). In addition, one line was set to be free from any activating reagent, allowing evaluation of nitrating agents on their own. Persulfates, generally activated thermally, are the most common activating reagents used in nitration reactions. They are readily available radical precursors; here, potassium persulfate was selected. Another common radical precursor is 2,2′-azobis(2-methylpropionitrile) (AIBN). Notably, it was used in catalytic amounts with nitric acid as a nitrating agent to perform nitration under mild conditions.51 Silver species have been used in nitration reactions involving a carboxylic acid species to assist decarboxylation.47,52 Following the same activation pathway, a Lewis acid magnesium salt (magnesium perchlorate hydrate) was also picked. Indeed, Lewis acid species were reported to suit nitration reactions.53 Consequently, two different copper sources were selected: copper(II) trifluoromethanesulfonate54 and copper(I) iodide. N,N′-Dimethylethylenediamine (DMEDA) was also added as a ligand with copper(I) iodide.55 Tris(dibenzylideneacetone)dipalladium(0) together with tBuBrettPhos was screened as a potent catalyst for nitration reactions.56,57 The number of equivalents of activating reagents followed the closest paper of reference.
Acetonitrile was chosen as the most commonly used solvent in nitration reactions. An average concentration of 0.1 M was selected at a reaction scale of 10 μmol, and the reaction was run at 100 °C for 24 hours under an air atmosphere. Each plate was prepared and worked up after reaction by addition of an internal standard, and the crude reactions were analyzed by UHPLC-UV-MS (see details in the ESI†). The general HTE workflow applied in this study, including home-made software for design and visualization, was assessed in a previously reported publication and is detailed in the ESI.†58
Results are presented in Fig. 2 and detailed for the nitration of 1-naphthoic acid in Fig. 2a Quantification of product formation was done by calculating the ratio between the nitration product Area Under Peak (AUP) and the internal standard (biphenyl) Area Under Peak (AUP). The results can only be compared within a single plate using heat maps (Fig. 2b) due to substrate-dependent UV response, but trends can be observed between plates. A total of nine HTE plates and 864 reactions were performed. A large number of unsuccessful results were obtained: 487 reactions gave no quantifiable product formation, representing 56% of the overall 864 reactions conducted. These unfruitful results are still of major importance. As previously mentioned, predictive algorithms developed with experimental data from the literature have recently shown limited efficiency, partly due to the lack of negative results reported in the literature to train machine learning models.59–61
In the context of this project, the negative data will address this drawback and hopefully help produce more accurate predictions of the chances of success for nitration reactions. The large percentage of negative results obtained during this HTE campaign also confirms the challenging nature of nitration reactions. Thanks to this study, clear trends in the activity of nitrating agents or activating reagents can be observed across the plate, depending on the substrate (Fig. 2b and 3). Notably, only potassium persulfate is a versatile activating reagent, usually giving rise to better results than in the absence of an activating reagent, except for benzofuran, where no reactivity enhancement is observed. Nonetheless, less common activities were observed from other agents. For example, AIBN displayed significant activation performance on several scaffolds (picolinic acid, 2-bromopyridine, and 2-bromobenzofuran). All other activating reagents failed to show clear activity. Notably, silver carbonate and magnesium perchlorate hydrate did not allow better performance with carboxylic acid derivatives, thus showing no specific decarboxylation enhancement. Additionally, copper and palladium catalysts did not perform better with bromide derivatives, displaying no specific catalytic activity.
On the part of nitrating agents, as expected, nitric acid is a recurring adequate nitro source. Tert-butyl nitrite (tBuONO) also demonstrated good activity across the plates, although diminished compared to nitric acid. Interestingly, both iron and bismuth nitrate metallic salts showed significant activities on several substrates. Finally, N-nitrosaccharin (Sacc-NO2) exhibited high activity with most of the substrates, confirming the strong interest in newly developed N-nitro reagents.
As expected, N-nitrosuccinimide (Succ-NO2) showed no significant activity under thermal activation, since this reagent generally requires light activation.62 Other nitrate and nitrite sources (silver nitrate, sodium nitrite, sodium nitrate, potassium nitrite, and tetrabutylammonium nitrate), as well as nitronium tetrafluoroborate, showed no significant reactivity in the global results. Interestingly, in the absence of an activating reagent, a significant number of nitrating sources do not show diminished activity, thus indicating no requirement for activation. The overall profiles confirmed the high difference in reactivity between arenes and heteroarenes (Fig. 2b and 3b). Pyridine and benzofuran moieties display a disparity in reactivity toward the nitration reaction. This confirms the difficulty of designing versatile nitration conditions across different arenes and heteroarenes.
To confirm the obtained results, batch reactions were performed by selecting high-yielding entries for each compound (yields are displayed below each corresponding heat map in Fig. 2b). The obtained yields corroborate the challenge that comes with nitration reactions. The generally lower yields observed with pyridine derivatives confirm the reactivity trend in the nitration of aromatic rings: arenes > electron-rich heteroarenes ≫ electron-poor heteroarenes. Significantly lower yields are obtained from the ipso-functionalization of carboxylic acids with naphthyl and benzofuran moieties. For benzofuran, the ipso-functionalization of the bromo derivative also gave the best result on the overall plate. Ipso-functionalization of carboxylic acid, when applied to the pyridine moiety, gave the highest ratios. For arenes, electrophilic substitution seems to be the preferred transformation. However, it should be noted that for both naphthalene and benzofuran,63 regioisomers were observed. In the case of naphthalene, several dinitronaphthalene compounds were also formed,64 pointing out the lack of selectivity of this methodology.65 The selected high-yielding entries present a large variety of nitrating agents, highlighting the value of a broad HTE campaign studying nitration conditions depending on the reacting scaffolds. However, activating reagents are less diverse, with potassium persulfate being overrepresented. Notably, the high-yielding entries selected for compound isolation were mostly original reaction conditions (marked with a “c” in Fig. 2). Remarkably, the best-yielding entry for the benzofuran moiety—obtained from the reaction of 2-bromobenzofuran with bismuth nitrate pentahydrate in the presence of AIBN—was, to the best of our knowledge, never reported. Additionally, the reactions selected for isolation were all different, emphasizing the relevance of the conducted study. This large HTE campaign thus allowed for the identification of an unusual mixture of nitrating and activating reagents for the nitration of arenes and heteroarenes.
Evaluation of predictive algorithms
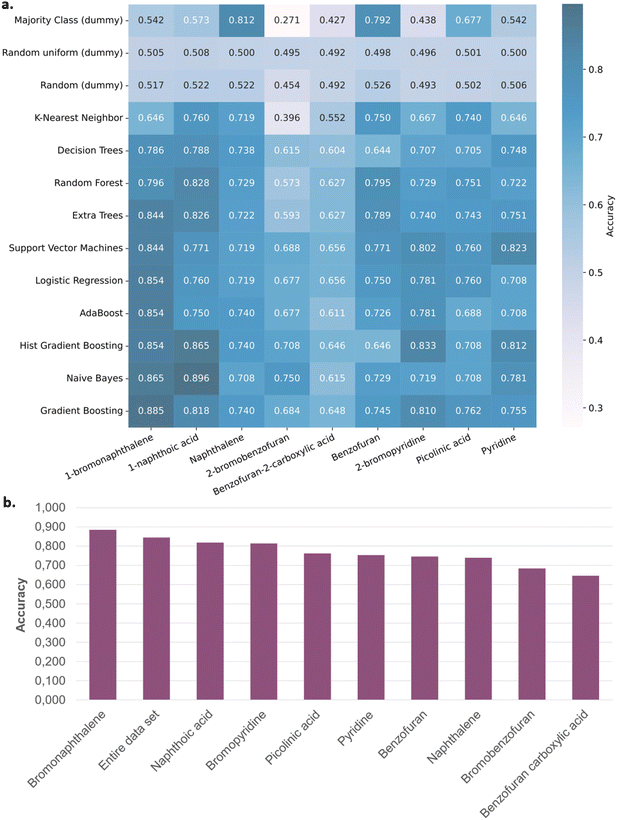
From the results of those 864 reactions, producing high-quality and high-quantity data, we then sought to valorize them through machine learning. As demonstrated by other groups in recent years, a predictive algorithm could be developed to generate the best conditions depending on the targeted substrate.34,38,40,66 In our case, we envisioned the possibility of predicting suitable nitrating and activating reagents for a specific molecule. The pool of data generated presents an uncommon repartition of results, with 56% of the reactions not leading to any product formation (see ESI†). From this observation, a binary classifier was envisioned to predict whether a nitration reaction is likely to succeed given a substrate, nitrating agent, and activating agent. Among these three independent variables, the two latter were numerically encoded as categorical variables through one-hot encoding, whereas two types of chemically relevant encoding67,68 were investigated for the substrate. First, the substrates' molecular structures were encoded as Morgan fingerprints69 with a radius of 2 and fingerprint sizes of 512, 1024, and 2048, respectively referred to as MorganFP-2-512, MorganFP-2-1024, and MorganFP-2-2048 in the remainder. The open-source RDKit package was used for this purpose. Its built-in descriptors module was used as well to feature the substrate as numerical vectors mainly containing electrotopological descriptors, with (rdkitDescr-210) and without (rdkitDescr-125) descriptors related to the occurrence of predefined molecular fragments.70,71 Vectors resulting from the concatenation of the chemically-relevant encoding of the substrates and categorical one-hot encoding of the nitrating and activation agents were then randomly split into train set (80%) and test set (20%) and used as inputs to several classification algorithms implemented in the open source scikit-learn library (AdaBoost, Decision Trees, Extra Trees, Gradient Boosting, Hist Gradient Boosting, K-Nearest Neighbor, Logistic Regression, Naive Bayes, Random Forest, Support Vector Machines). Dummy classifiers (Majority Class, Random, Random uniform) were systematically tested as well as baselines, allowing for the evaluation of the ability of the other models to perform better than basic prediction models. For each combination of descriptors and algorithms, the whole process of train/test splitting, model training and model evaluation was repeated fifty times to mitigate the potential bias due to the unrepresentative splitting of the dataset and average the results of stochastic algorithms. The average accuracies on the test set are computed across these fifty repeats and reported in Fig. 4 (see ESI for details on the classification metrics†). The average accuracies of all machine learning models are significantly higher than the ones of the dummy models, indicating that the formers indeed learned some input–output relationships (Fig. 4a and b). Overall, Gradient Boosting and Hist Gradient Boosting, both relying on ensemble learning, gave the best results and look especially well appropriate for this data set (Fig. 4b). The overall study has been evaluated in terms of balanced accuracy and no major variations have been reported. All related graphs are reported in the ESI.†Next, selecting MorganFP-2-1024 as the substrates' featurization method, the evaluation of the ability of the classification models to accurately guess the reaction success on an unseen substrate was conducted, to mimic a real-world scenario where the reaction outcome on a new substrate would be sought to be determined before experiment. The leave-one-out strategy was used for this purpose: every experiment related to one specific substrate was taken out of the dataset, and classification models were trained on the remaining experiments. The left-out experiments were then used as the test set and classification accuracies were calculated for each model. This operation was repeated for each substrate.
Unfortunately, the accuracy of the predicting model varies significantly depending on the unseen substrate evaluated (Fig. 5a). As an example, when Gradient Boosting is used, the variations of the model are enormous depending on the left-out substrate (Fig. 5b). In addition, accuracies tend to diminish compared to the result obtained with the entire data set. Only bromonaphthalene, when not present in the training set and evaluated by the trained model, gives better accuracy than the one obtained with the entire data set. Benzofuran moieties give especially reduced accuracies. This result could outline the difficult prediction of heteroarene reactivity. From this hypothesis, we decided to only select one class of substrates to train the model, to hopefully obtain more accurate predictions. Electron-poor pyridine moieties displayed a significantly different reactivity than the other two studied scaffolds. They were thus selected and used to train an “electron-poor” model (Fig. 6). However, when compared to the results obtained with the entire data set, no significant improvement is observed.
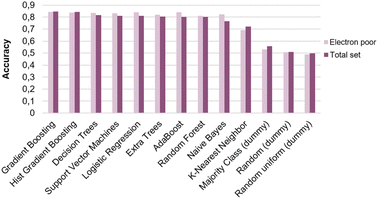 | ||
| Fig. 6 Graph of the results expressed as the calculated accuracies depending the model and the data set examined. | ||
Overall, the capacity of the model to accurately predict product formation on an unseen molecule is limited. We hypothesize that this limitation comes from the limited set of evaluated molecules62,72 and incomplete feature-engineering of the reactive system. Indeed, nitration and activation agents were only categorically encoded in this study, providing no chemically-relevant information about these reagents, whereas some of their physico-chemical properties are likely to be important for reactivity prediction. The same goes for the substrates, although to a lesser extent, for which no descriptors stemming from electronic structure calculations were used. These machine-learning considerations and insufficient chemical diversity of substrates could both contribute to restraining the model from accurately classifying the reactivity of substrates in nitration reaction, thus explaining the diminished accuracy of the models on unseen molecules.
To further explore how negative results contribute to the performances of machine learning models, the experimental dataset produced in this study was split into different trainsets with varying proportions of successful/unsuccessful reactions. These proportions varied between 10% and 95% of successful experiments, and the accuracy of the best model is reported accordingly in Fig. 7. The same results in terms of balanced accuracy are reported in the ESI.†
For highly unbalanced trainsets, it is clear that the best models are obtained when the test set is split in the same way. Otherwise, the classification accuracy dramatically decreases. On the other hand, balanced trainsets, containing around 40 to 60% of successful reactions, always give reasonable accuracy which is much less dependent on the testset split. Because the testset repartition is typically unknown in real-world scenarios, this provides further evidence that reactions typically considered unworthy of publication actually are precious to developing robust data-driven models. A complementary study was performed using only two out of the three types of descriptors to train the algorithm (see Fig. 27 in the ESI†). The balanced accuracies obtained for each split revealed that the nitration agent is the most important descriptor to take into account to correctly predict the outcome of a reaction. On the other hand, ignoring the activation agent or the substrate itself does not significantly affect the model's performance. This might originate from strong similarities between the left-out substrate's reactivity and the training set's reactivity towards the same pairs of activation/nitration agents. Further exploration of these questions will be the object of a following study.
Conclusions
From the identified heteroarene nitration challenge, we carried out a large HTE campaign for the evaluation of nitration conditions depending on various scaffolds. The designed model HTE plate allowed for the screening of 12 nitrating reagents and 7 activating agents. This plate was applied to three different classes of scaffolds: (i) an arene (naphthalene core), (ii) an electron-rich heteroarene (benzofuran core), and (iii) an electron-poor heteroarene (pyridine core). Three different substrates were selected for each moiety, giving rise to 9 different compounds screened and 864 different reactions. The results confirmed the lower reactivity of electron-poor, nitrogen-containing heteroarenes.The high diversity of nitrating agents occurring in the best-yielding results proved the interest in performing such a large-spectrum HTE campaign. It also highlighted original reaction conditions. Over the 9 HTE plates, 5 high-yielding reaction conditions were previously unreported mixtures of nitrating and activating reagents. Finally, the high-quality and high-quantity data were used to develop a predictive tool relying on machine learning to evaluate the best nitration conditions depending on the targeted substrate. Although the model gave satisfying metrics when trained on the overall dataset, it revealed limited generalization capability on unseen substrates. A higher chemical diversity of targeted substrates and a more thorough featurization of the whole reactive system could allow for improved accuracy. Together, HTE and machine learning allowed for an extensive exploration of the nitration reaction, paving the way for a new methodology to address this challenging transformation.
Data availability
The data supporting this article (HTE data set and ML codes/outputs) have been included as part of the ESI† and are available on GitHub at https://github.com/DAM-LDMM/2025_HTE-ML-nitration and on Zenodo with the doi: https://doi.org/10.5281/zenodo.15691760.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We warmly thank the GIPSI team (C. Petat and P. Drevet) for the development of HTDesign® (CEA Paris-Saclay, DRF/JOLIOT). We thank S. Lebrequier, T. D'Anfray and D. Buisson for the development of analytical methods and fruitful discussions. We also warmly thank the technical support of the SCBM.Notes and references
- S. S. Patel, D. B. Patel and H. D. Patel, ChemistrySelect, 2021, 6, 1337–1356 CrossRef CAS.
- L. F. Albright, R. V. C. Carr and R. J. Schmitt, Nitration, 1996, 1, 1–9 Search PubMed.
- Y.-E. Qian, L. Zheng, H.-Y. Xiang and H. Yang, Org. Biomol. Chem., 2021, 19, 4835–4851 RSC.
- G. Yan and M. Yang, Org. Biomol. Chem., 2013, 11, 2554–2566 RSC.
- G. A. Olah, S. C. Narang, J. A. Olah and K. Lammertsma, Proc. Natl. Acad. Sci. U. S. A., 1982, 79, 4487–4494 CrossRef CAS.
- H. Sepehrmansourie and M. Zarei, J. Org. Chem. Res., 2023, 9, 1–5 Search PubMed.
- S. Patterson and S. Wyllie, Trends Parasitol., 2014, 30, 289–298 CrossRef CAS PubMed.
- A. Capperucci and D. Tanini, Chemistry, 2022, 4, 77–97 CrossRef CAS.
- G. K. S. Prakash and M. Etzkorn, Angew. Chem., Int. Ed., 2004, 43, 26–28 CrossRef PubMed.
- G. A. Olah, Industrial and Laboratory Nitrations, 1976, vol. 1, pp. 1–47 Search PubMed.
- J. I. Murray, M. V. S. Elipe, K. D. Baucom, D. B. Brown, K. Quasdorf and S. Caille, J. Org. Chem., 2022, 87, 1977–1985 CrossRef CAS PubMed.
- R. G. Coombes, R. B. Moodie and K. Schofield, J. Chem. Soc. B, 1968, 800–804 RSC.
- Y.-X. Li, L.-H. Li, Y.-F. Yang, H.-L. Hua, X.-B. Yan, L.-B. Zhao, J.-B. Zhang, F.-J. Jic and Y.-M. Liang, Chem. Commun., 2014, 50, 9936–9938 RSC.
- B. Kilpatrick, M. Hellera and S. Arns, Chem. Commun., 2013, 49, 514–516 RSC.
- D. Koley, O. C. Colón and S. N. Savinov, Org. Lett., 2009, 11, 4172–4175 CrossRef CAS PubMed.
- J. Moon, H. K. Ji, N. Ko, H. Oh, M. S. Park, S. Kim, P. Ghosh, N. K. Mishra and I. S. Kim, Arch. Pharmacal Res., 2021, 44, 1012–1023 CrossRef CAS PubMed.
- K. Bozorov, J.-Y. Zhao and H. A. Aisa, Arkivoc, 2017, 41–66 Search PubMed.
- R. Calvo, K. Zhang, A. Passera and D. Katayev, Nat. Commun., 2019, 10, 3410 Search PubMed.
- K. Zhang, A. Budinská, A. Passera and D. Katayev, Org. Lett., 2020, 22, 2714–2719 CrossRef CAS PubMed.
- D. M. Badgujar, M. B. Talawar and P. P. Mahulikar, Propellants, Explos., Pyrotech., 2016, 41, 24–34 CrossRef CAS.
- G. K. S. Prakash, C. Panja, T. Mathew, V. Surampudi, N. A. Petasis and G. A. Olah, Org. Lett., 2004, 6, 2205–2207 CrossRef CAS PubMed.
- S. Mukhopadhyay and S. Batra, Eur. J. Org Chem., 2019, 2019, 6424–6451 CrossRef CAS.
- J. Huang, F. Ding, P. Rojsitthisak, F.-S. He and J. Wu, Org. Chem. Front., 2020, 7, 2873–2898 RSC.
- S.-Z. Song, Y. Dong, G.-P. Ge, Q. Li and W.-T. Wei, Synthesis, 2020, 52, 796–806 CrossRef CAS.
- L.-R. Song, Z. Fanab and A. Zhang, Org. Biomol. Chem., 2019, 17, 1351–1361 RSC.
- S. Patra, V. Valsamidou and D. Katayev, Chimia, 2024, 78, 32–39 CrossRef CAS PubMed.
- T. Yang, X. Li, S. Deng, X. Qi, H. Cong, H.-G. Cheng, L. Shi, Q. Zhou and L. Zhuang, JACS Au, 2022, 2, 2152–2161 CrossRef CAS PubMed.
- L. Eberson and F. Radner, Acc. Chem. Res., 1987, 20, 53–59 CrossRef CAS.
- G. Bontempelli, G.-A. Mazzocchin, F. Magno and R. Seeber, J. Electroanal. Chem. Interfacial Electrochem., 1974, 55, 101–107 CrossRef CAS.
- T. L. Broder, D. S. Silvester, L. Aldous, C. Hardacre and R. G. Compton, J. Phys. Chem. B, 2007, 111, 7778–7785 CrossRef CAS PubMed.
- M. Shevlin, ACS Med. Chem. Lett., 2017, 8, 601–607 CrossRef CAS PubMed.
- S. M. Mennen, C. Alhambra, C. L. Allen, M. Barberis, S. Berritt, T. A. Brandt, A. D. Campbell, J. Castañón, A. H. Cherney, M. Christensen, D. B. Damon, J. E. de Diego, S. García-Cerrada, P. García-Losada, R. Haro, J. M. Janey, D. C. Leitch, L. Li, F. Liu, P. C. Lobben, D. W. C. MacMillan, J. Magano, E. McInturff, S. Monfette, R. J. Post, D. Schultz, B. J. Sitter, J. M. Stevens, I. I. Strambeanu, J. Twilton, K. Wang and M. A. Zajac, Org. Process Res. Dev., 2019, 23, 1213–1242 CrossRef CAS.
- X. Caldentey and E. Romero, Chem.:Methods, 2023, e202200059 CAS.
- B. Mahjour, R. Zhang, Y. Shen, A. McGrath, R. Zhao, O. G. Mohamed, Y. Lin, Z. Zhang, J. L. Douthwaite, A. Tripathi and T. Cernak, Nat. Commun., 2023, 14, 3924 CrossRef CAS PubMed.
- K. McCullough, T. Williams, K. Mingle, P. Jamshidi and J. Lauterbach, Phys. Chem. Chem. Phys., 2020, 22, 11174–11196 RSC.
- X. Li, P. M. Maffettone, Y. Che, T. Liu, L. Chen and A. I. Cooper, Chem. Sci., 2021, 12, 10742–10754 RSC.
- Z. Tu, T. Stuyver and C. W. Coley, Chem. Sci., 2023, 14, 226–244 RSC.
- E. King-Smith, S. Berritt, L. Bernier, X. Hou, J. L. Klug-McLeod, J. Mustakis, N. W. Sach, J. W. Tucker, Q. Yang, R. M. Howard and A. A. Lee, Nat. Chem., 2024, 16, 633–643 CrossRef CAS PubMed.
- D. F. Nippa, K. Atz, A. T. Müller, J. Wolfard, C. Isert, M. Binder, O. Scheidegger, D. B. Konrad, U. Grether, R. E. Martin and G. Schneider, Commun. Chem., 2023, 6, 256 CrossRef CAS PubMed.
- A. V. Kalikadien, C. Valsecchi, R. van Putten, T. Maes, M. Muuronen, N. Dyubankova, L. Lefort and E. A. Pidko, Chem. Sci., 2024, 15, 13618–13630 RSC.
- J. Y. Wang, J. M. Stevens, S. K. Kariofillis, M.-J. Tom, D. L. Golden, J. Li, J. E. Tabora, M. Parasram, B. J. Shields, D. N. Primer, B. Hao, D. Del Valle, S. DiSomma, A. Furman, G. G. Zipp, S. Melnikov, J. Paulson and A. G. Doyle, Nature, 2024, 626, 1025–1033 CrossRef CAS PubMed.
- S. Tcyrulnikov, A. K. Hubbell, D. Pedro, G. P. Reyes, S. Monfette, D. J. Weix and E. C. Hansen, J. Am. Chem. Soc., 2024, 146, 6947–6954 CrossRef CAS PubMed.
- N. P. Romer, D. S. Min, J. Y. Wang, R. C. Walroth, K. A. Mack, L. E. Sirois, F. Gosselin, D. Zell, A. G. Doyle and M. S. Sigman, ACS Catal., 2024, 14, 4699–4708 CrossRef CAS.
- M. Christensen, Y. Xu, E. E. Kwan, M. J. Di Maso, Y. Ji, M. Reibarkh, A. C. Sun, A. Liaw, P. S. Fier, S. Grosser and J. E. Hein, Chem. Sci., 2024, 15, 7160–7169 RSC.
- P. Raghavan, A. J. Rago, P. Verma, M. M. Hassan, G. M. Goshu, A. W. Dombrowski, A. Pandey, C. W. Coley and Y. Wang, J. Am. Chem. Soc., 2024, 146, 15070–15084 CrossRef CAS PubMed.
- Y. V. Guk, M. A. Ilyushin, E. L. Golod and B. V. Gidaspov, Russ. Chem. Rev., 1983, 52, 284 CrossRef.
- P. Natarajan, R. Chaudhary and P. Venugopalan, Org. Chem., 2015, 80, 10498–10504 CrossRef CAS PubMed.
- I. Mosiagin, A. J. Fernandes, A. Budinská, L. Hayriyan, K. E. O. Ylijoki and D. Katayev, Angew. Chem., Int. Ed., 2023, 62, e202310851 CrossRef CAS PubMed.
- S. Patra, R. Giri and D. Katayev, ACS Catal., 2023, 13, 16136–16147 CrossRef CAS.
- Both Succ-NO2 and Sacc-NO2 were prepared accordingly to the procedures described in the ESI†.
- J. P. Das, P. Sinha and S. Roy, Org. Lett., 2002, 4, 3055–3058 CrossRef CAS PubMed.
- H. Yan, J. Mao, G. Rong, D. Liu, Y. Zhenga and Y. He, Green Chem., 2015, 17, 2723–2726 RSC.
- J. Sun, J.-K. Qiu, Y.-N. Wu, W.-J. Hao, C. Guo, G. Li, S.-J. Tu and B. Jiang, Org. Lett., 2017, 19, 754–757 CrossRef CAS PubMed.
- P. J. A. Joseph, S. Priyadarshini, M. L. Kantam and H. Maheswaran, Tetrahedron Lett., 2012, 53, 1511–1513 CrossRef.
- S. Saito and Y. Koizumi, Tetrahedron Lett., 2005, 46, 4715–4717 CrossRef CAS.
- B. P. Fors and S. L. Buchwald, J. Am. Chem. Soc., 2009, 131, 12898–12899 CrossRef CAS PubMed.
- G. K. S. Prakash and T. Mathew, Angew. Chem., Int. Ed., 2010, 49, 1726–1728 CrossRef CAS PubMed.
- T. Kerackian, G. Chacktas, D. Durand, E. Romero and J. Flow, Chem, 2024, 14, 367–375 Search PubMed.
- P. Raccuglia, K. C. Elbert, P. D. F. Adler, C. Falk, M. B. Wenny, A. Mollo, M. Zeller, S. A. Friedler, J. Schrier and A. J. Norquist, Nature, 2016, 533, 73–76 CrossRef CAS PubMed.
- P. M. Pflüger and F. Glorius, Angew. Chem., Int. Ed., 2020, 59, 18860–18865 CrossRef PubMed.
- J. Schleinitz, M. Langevin, Y. Smail, B. Wehnert, L. Grimaud and R. Vuilleumier, J. Am. Chem. Soc., 2022, 144, 14722–14730 CrossRef CAS PubMed.
- K. Zhang, B. Jelier, A. Passera, G. Jeschke and D. Katayev, Chem.–Eur. J., 2019, 25, 12929–12939 CrossRef CAS PubMed.
- In the case of benzofuran the identification of regioisomers was impossible since all C-H bond have been substituted.
- Impossible identification of regioisomers of dinitronaphthalene.
- No other identifiable significant side products were observed in any other reactions.
- D. T. Ahneman, J. G. Estrada, S. Lin, S. D. Dreher and A. G. Doyle, Science, 2018, 360, 186–190 CrossRef CAS PubMed.
- P. Carracedo-Reboredo, J. Liñares-Blanco, N. Rodríguez-Fernández, F. Cedrón, F. J. Novoa, A. Carballal, V. Maojo, A. Pazos and C. Fernandez-Lozano, Comput. Struct. Biotechnol. J., 2021, 19, 4538–455 CrossRef CAS PubMed.
- J. Deng, Z. Yang, I. Ojima, D. Samaras and F. Wang, Briefings Bioinf., 2022, 23, 1–19 CAS.
- H. L. Morgan, J. Chem. Doc., 1965, 5, 107–113 CrossRef CAS.
- https://www.rdkit.org/docs/GettingStartedInPython.html#list-of-available-descriptors, consulted on 27/01/2025.
- https://rdkit.org/docs/source/rdkit.Chem.Fragments.html, consulted on 27/01/2025.
- M. Wen, S. M. Blau, X. Xie, S. Dwaraknathd and K. A. Persson, Chem. Sci., 2022, 13, 1446–1458 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5dd00086f |
| This journal is © The Royal Society of Chemistry 2025 |