Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A property graph schema for automated metadata capture, reproducibility and knowledge discovery in high-throughput bioprocess development†
Federico M.
Mione
 a,
Martin F.
Luna
a,
Lucas
Kaspersetz
a,
Martin F.
Luna
a,
Lucas
Kaspersetz
 b,
Peter
Neubauer
b,
Peter
Neubauer
 b,
Ernesto C.
Martinez
ab and
M. Nicolas
Cruz Bournazou
b,
Ernesto C.
Martinez
ab and
M. Nicolas
Cruz Bournazou
 *b
*b
aINGAR (CONICET – UTN), Avellaneda 3657, Santa Fe, Argentina
bTechnische Universität Berlin, Institute of Biotechnology, Chair of Bioprocess Engineering, Berlin, Germany. E-mail: mariano.n.cruzbournazou@tu-berlin.de
First published on 12th June 2025
Abstract
Recent advances in autonomous experimentation and self-driving laboratories have drastically increased the complexity of orchestrating robotic experiments and of recording the different computational processes involved including all related metadata. Addressing this challenge requires a flexible and scalable information storage system that prioritizes the relationships between data and metadata, surpassing the limitations of traditional relational databases. To foster knowledge discovery in high-throughput bioprocess development, the computational control of the experimentation must be fully automated, with the capability to efficiently collect and manage experimental data and their integration into a knowledge base. This work proposes the adoption of graph databases integrated with a semantic structure to enable knowledge transfer between humans and machines. To this end, a property graph schema (PG-schema) has been specifically designed for high-throughput experiments in robotic platforms, focused mainly on the automation of the computational workflow used to ensure the reproducibility, reusability, and credibility of learned bioprocess models. A prototype implementation of the PG-schema and its integration with the workflow management system using simulated experiments is presented to highlight the advantages of the proposed approach in the generation of FAIR data.
1. Introduction
In the era of autonomous discovery,1 high-throughput robotics platforms integrated with liquid handling stations,2,3 have established the foundation for applying artificial intelligence (AI) methodologies in bioprocess development. By increasing significantly the amount of data generated, and the informative content gained through automated parallel experimentation,4 these platforms have revolutionized the discovery process in different areas, presenting new opportunities in biotechnology, pharmaceuticals and bioengineering.5,6Unfortunately, manual metadata annotation methods are error-prone for these complex dynamic experiments, highlighting the need for an automated extraction system to scalable feed machine-readable7 metadata from the workflow management system (WMS) into a database schema which must be specifically designed to enable knowledge discovery and semantic understanding of data generation.8
As robotics facilities evolve over time,9 experimental reproducibility has lagged behind and remains a challenge.10,11 The increasing complexity of cloud12 and autonomous laboratories,13,14 coupled with experimental control using model-based computational methods,15–18 exacerbates the difficulty of ensuring consistent and reproducible results. Moreover, these experiments are dynamic, meaning that in order to maximize the information content of the generated data need to be redesigned online. Without proper metadata collection and format,19 it is impossible to reach experimental reproducibility, even in self-driving laboratories (SDLs), thereby undermining trust in the conclusions drawn from such studies.20,21
The key to ensuring findability, accessibility, interoperability, and reusability (FAIR)22 of experimental data lies in the automatic capture of all metadata, which is essential for answering elaborated queries to a knowledge base. Reproducible experiments and adherence to FAIR principles are crucial to effectively accumulate, share and reuse knowledge and expertise in high-throughput bioprocess development (HTBD).23,24 This paper addresses these challenges by means of a property graph schema (PG-schema) integrated with a WMS that enables comprehensive and automated metadata capture throughout the experimental process, thereby facilitating knowledge discovery and enhancing reproducibility.
As highlighted by Reder et al.,25 many laboratories still lack robust database platforms designed to support the advanced capabilities of AI systems. These platforms are fundamental for storing the knowledge generated and fostering SDLs, facilitating transfer learning, and enabling the automated generation of AI-ready data that is essential for autonomous discovery.26 Providing metadata about the experimental dataset in the form of linked data graph is a key step towards enabling FAIR data. The ability to represent not only data but knowledge with focus on semantic relationships and contextual metadata is increasingly gaining attention27 and is the very aim of knowledge graphs (KGs). A KG uses graph-based data structure to represent entities of a specific domain and the relationships between them.
Although several knowledge representation models exist (Section 2.1), labeled property graphs (LPGs) have been chosen since they provide better support for highly interconnected datasets compared to the relational model. On the other hand, while a flexible storage of information in LPGs without predefined formats or structures, could be considered a beneficial feature for software developers, it may become challenging to navigate and understand when the amount of data scales. Therefore, a conceptualization is needed that defines and delimits what these knowledge bases can represent and how constraints are formalized.28 These constraints are enforced through schemas, providing a formal structure to the data29 and establishing a common vocabulary for entities involved in the domain of analysis. Formalization of a KG as a set of premises grants machines inherent deductive capability for knowledge discovery, enabling them to perform inference reasoning with a level of precision, efficiency, and scale beyond human capabilities.30 The contribution of schemas to efficiently navigate the graph following a predefined set of rules, improve the answer to user queries.31 For this reason, the integration of the PG-schema with the workflow execution timeline, merging entities from both platforms as a single source of knowledge, is a key enabler for generating FAIR data in SDLs.
Computational workflows designed to control and monitor the experimentation provide a modular framework for defining dependencies between tasks, specifying inputs and outputs for each of them. Through the integration of a WMS, the precise documentation of how data has been generated is ensured, facilitating data provenance capture32,33 and adherence to FAIR data principles.34,35
Furthermore, computational methods are not entirely reproducible just by code sharing, as they also depend on the hardware used for execution, along with specific frameworks, dependencies, libraries, and operating systems, including their respective versions.36 Detailed information of executed processes and the computational environment used, promotes experimental and computational reproducibility. Combining provenance data from the WMS, experimental information from the laboratory, and computational methods used into a timeline-based KG structured by a PG-schema, constitutes a significant contribution to the field.
Several approaches exist for integrating a WMS with the storage of structured data and metadata. In ChemOS,37 a platform for orchestrating laboratory software and hardware is introduced, but relies on diverse files formats and an SQL database to store the generated information. ESCALATE38 provides an ontological framework for describing experiments and managing data lake files across various Google Drive folders, with a primary focus on material discovery field, which differs significantly from the dynamic nature of experiments in HTBD. Additionally, The World Avatar,39 also oriented towards material science, incorporates an ontological approach with simpler workflows, presenting a promising method for distributed SDLs. Still, none of the above mentioned methods can provide a seamless integration between a WMS and a schema-based relationship-oriented storage system that facilitates knowledge discovery and ensures the reproducibility of experiments in HTBD.
In this work, a methodology for modeling and prototyping a PG-schema for automatic metadata capture in bioprocess development is presented. The computational workflow for online redesigning parallel experiments is implemented using Apache Airflow® and represented as directed acyclic graphs (DAGs). The WMS is dynamically linked to a Neo4j database employing an LPG data model, with the defined schema serving as the core of the proposed approach. Each task executed within Airflow automatically saves its results and associated metadata into the graph database, aiming to centralize all experimental data in a unified knowledge base. To facilitate interaction with the LPG, a web interface was developed, enabling users to create the experimental design file for Airflow execution, monitor experiments in real time, manage entities for metadata related to laboratory devices, and query historical data. As a case study, a simulated experiment involving 24 parallel E. coli fed-batch cultivations was performed, replacing robotic devices with a local emulator to replicate the cultivation dynamics.
The proposed approach demonstrates the pivotal role of a PG-schema and the use of graph databases for integrating diverse information sources from the laboratory into a KG in order to share a semantic vocabulary and lay down foundations for trust, reproducibility, knowledge discovery and reuse of costly experimental data in HTBD.
The remainder of this article is structured as follows: Section 2 describes the relevant background for the study, Section 3 elaborates on the methodology adopted to model the representation schema and the implementation of the prototype, Section 4 introduces the simulation case study, in Section 5 the significance of results are summarized, and finally, Section 6 presents the conclusions and final remarks of the presented work.
2. Background
2.1 Knowledge graphs
KGs have emerged as an alternative of choice for representing and managing common knowledge about the real world in a formal and structured manner.40 Despite the growing adoption of KG, its definition is still a subject of debate, with several interpretations ranging from specific technical definition to broader perspectives that consider it a field of study on its own.41Every graph, denoted as G = (N, E), is composed of a set of nodes N and edges E, where each edge connects a pair—or more—nodes, either in a directed or undirected relationship. Depending on the specific domain, the nodes and edges can be enriched with attributes and labels. Accordingly, the formal definition of a KG is a graph-based data structure where the entities of interest are represented as nodes and influence or causal relationships between them as edges (Fig. 1).
The concept of KGs gained significant attention in 2012 when Google introduced it to enhance search results. Since then, KGs have become increasingly important in fields such as machine learning, natural language processing, and semantic web technologies, where they serve as a backbone for organizing large volumes of data.28 One of the key strengths of KGs lies in their ability to encode complex relationships and semantic descriptions in a structured format, which facilitates inference and automates reasoning.
There are multiple data representations in the semantic web field, each offering different approaches to data formalization and modeling. Two of the most commonly used are the resource description framework (RDF) and the LPG. While RDF provides a formal structure that is well suited for automated knowledge inference, LPG has gained popularity for its scalability and flexibility in performing graph analytic tasks. Several studies have attempted to link both models, either to leverage their respective strengths42 or to enable interoperability from one to the other.43,44 However, this integration is still not considered a well-established standard. A summary of these two representations is presented in the next subsections.
The main goal of RDF is to create statements about resources to express information with semantic meaning. An RDF statement can be expressed by a uniform structure via triples, consisting of three linked data pieces: subject, predicate and object. Subject is the resource being described by the triple, object is another resource related to the subject, and predicate describes the relationship between them. A collection of RDF triples can be seen as a directed graph where subjects and objects are nodes, and predicates are represented as edges. Due to its structure, one of the key features of RDF is to support a complete atomic decomposition. In other words, detailed information about a resource is expressed through additional sets of triples.
To uniquely identify each component of the triple, RDF uses internationalized resource identifiers (IRIs). IRIs are defined as a superset of the uniform resource locator (URL) and have the same structure with a scheme, path and fragment. Generally, they are used for most popular ontologies in order to reuse predefined vocabularies.
RDF enables the expression of statements about resources through named properties and values. To further enrich these statements, the RDF schema (RDFS) provides a set of reserved words for defining classes and properties, which adds another layer of semantics to the vocabulary used. For querying RDF data, SPARQL46 serves as the standardized query language, also developed by the W3C.
Many graph databases that support RDF also have means for reasoning over the stored knowledge. Several reasoning strategies exist, all of these with the general purpose of generating new statements (implicit knowledge) based on predefined RDF statements (explicit knowledge). This inference process is considered a pivotal feature of the RDF data model.
LPGs lack a formal knowledge representation such as the entity-relationship model for relational databases. While several schema representations exist,47,48 most fail to provide the required means to perform automated schema validation and knowledge inference.
There exists a high number of graph database alternatives that implement LPG data models. Neo4j is one of the most popular49 open source system written in Java with native graph storage capability. Native graph databases are designed with specialized engines highly optimized to support graph workloads and built-in graph functions.50,51
In contrast to RDF, LPG do not have a unified query language. However, there is an official standard known as graph query language (GQL),52 which provides guidelines for data manipulation and basic operations on property graphs. In the case of Neo4j, the formal query language is Cypher,53 a declarative language based on pattern matching. A comparative summary of the two data models is provided in the Table 1.
| Aspect | RDF | LPG |
|---|---|---|
| Data model representation | Subject, predicate, object triples | Nodes, edges with properties and labels |
| Schema definition | RDF schema (RDFS) and OWL | No standardized schema (schema-less or schema-flexible) |
| Data format | Uses standardized data formats (XML, Turtle, JSON-LD) | Custom serialization based on the specific database implementation |
| Data integration | Easy integration with external data sources using IRIs | No native data integration. A linked data approach is viable |
| Graph storage and processing | Does not support native graph storage or processing | Supports native graph storage and native graph processing |
| Reasoning | Enables ontology-based reasoning, allowing for inference to derive implicit knowledge | No native reasoning capabilities. Extra tools needed |
| Scalability | Vertical, focused on enhancing the hardware capacity of a single server | Horizontal, oriented to increase the number of servers through a distributed approach with load balancing |
| Database implementation | Allegro Graph, Blazegraph, Dgraph, Apache Jena | Memgraph, TigerGraph, Neo4j |
| Query language | SPARQL | Gremlin, Cypher |
| Use cases/applications | Semantic web, linked data, integration with ontologies | Big data analysis, social networks, complex graph traversal queries |
2.2 Graph versus relational databases
The increasing complexity and volume of generated information demand new approaches for its organization and processing, especially in contexts where the relationships between data are as important as the data themselves.This challenge underscores the need to develop alternative storage models to traditional databases, whose tabular structure presents limitations in representing complex and dynamic relationships.54 Although the term relational database suggests that this type of system is inherently suited to handling relationships, in practice, this is often not the case, especially when queries involve multiple relationships (or hops) across tables, where both performance and semantic clarity tend to degrade significantly as more metadata are involved in the answer.55
One of the main reasons for this limitation lies in the way relationships are represented, which reduces their semantic richness and can only be reconstructed through an additional application layer. However, when relying solely on the database structure, it becomes difficult for an AI agent to process and interpret these connections effectively.
On the other hand, a graph database with native processing capabilities using index-free adjacency can directly reference its adjacent (neighboring) nodes, meaning that accessing relationships and related data is essentially a memory pointer lookup.56 As a result, native graph processing time becomes proportional to the amount of data processed, rather than increasing exponentially with the number of relationships traversed or hops navigated.
Consider a simple example in the specific domain of interest where an experiment has only one responsible person. Fig. 2A presents the two tables corresponding to the relational model, in which the association is established through a foreign key (FK). In this schema, the only way to infer the relationship between Person and Experiment entities is through the property name (IdResponsible) chosen arbitrarily by the database designer or developer. However, such representations are insufficient for AI agents, as they lack the contextual richness required for reasoning or inference. Extracting meaning from a fragment of an attribute name provides little clarity about the underlying relationship between entities.
To mitigate this, it is possible to explicitly design the relationship—typically in the case of many-to-many associations —by creating a dedicated table to store the links between entities (Fig. 2B). In such a case, the semantics of the relationship is preserved; however, retrieving the related data requires two JOIN operations—links between tables—, which become increasingly inefficient when navigating three or more relationship hops.
In contrast, Fig. 2C illustrates the same scenario modeled in a graph database. In this approach, the Person and Experiment entities are represented as nodes, and their relationship is explicitly defined through a labeled edge that indicates the type of connection. The use of these structures, like subject–predicate–object in RDF triples, significantly enhances the ability of AI agents to reason over data. This representation not only preserves the semantics of the relationships but also enables more expressive and efficient queries, facilitating data analysis, inference processes and contextual interpretation by both humans and AI agents.
3. Methodology
The main objective of the proposed approach is to drastically increase experimental reproducibility, knowledge discovery, and to foster the generation of FAIR data in SDLs. Achieving reproducibility demands a comprehensive understanding of the underlying experiment and its provenance. KGs are essential for understanding the context of the experiment and through the WMS, traceability of events and actions are also part of the represented knowledge.The foundation of the proposed data model lies in ensuring data traceability at each stage of the experimentation process and storing rich metadata, including hyperparameters in the computational workflow established to control the experiment execution. Through the definition of a PG-schema, a common structure is introduced for the storage of HTBD data, enabling standardized data organization and supporting experiment data interoperability.
By employing an LPG as a knowledge storage base with the formal specification of its structure, the obtained data can be enriched with semantic and descriptive metadata. This contextual information provides a better understanding of the experimental design and execution, thereby facilitating subsequent analysis and supporting the transfer of acquired knowledge. As a result, the experimental data become AI-actionable, which is an essential building block for explainable machine learning models.
The use of a WMS to orchestrate the computational tasks involved in execution control is imminent to achieve traceability and enable reproducibility of SDL experiments. Furthermore, it is not possible to ensure the identical conclusions between repeated executions of the same experiment without storing the specific input/output of each task as a timeline related to the execution of the different steps including all the metadata related to the decisions made by software agents.
Apache Airflow is employed as a WMS57 to handle the execution of the tasks necessary to achieve a specific goal throughout the experiment, creating the computational environments required for each step by means of the instantiation of docker containers.58,59 As shown in Fig. 3, the interaction with robotic devices is carried out through the relational database associated with the manufacturer's software for these devices.
This work addresses the formalization of a common schema or vocabulary for the experiments in a robotic platform and its control component, associated with the computational workflow implemented in parallel and directly integrated into the graph database, more specifically an LPG. To interact with the database, a web interface has been implemented to facilitate the access of different types of users to the knowledge stored, to monitor the online progress of an experiment execution or to query historical data.
The following subsections begins with a resume of the PG-schema capabilities (Section 3.1), the data modeling process with its different stages (Section 3.2), the formalization of the PG-schema (Section 3.3), its subsequent prototyping in Python, and the integration with the Airflow orchestrator and the web interface (Section 3.4).
3.1 PG-schema
One of the most promising approaches for specifying a well-structured LPG is the PG-schema formalism29 developed by the Property Graph Schema Working Group of the Linked Data Benchmark Council. This section provides a concise overview of the formalism's capabilities.In the context of relational and semi-structured data, the definition of a schema typically involves two main components: types and constraints. This structure is also applicable to graph databases, and specially LPGs. Types define the structure of the data and the datatype for each element, including nodes and relationships. Complementary, constraints, specifies a set of rules to maintain data consistency and integrity.
In order to define a schema, two alternatives are available:  and
and  . In the first option, a graph instance is considered valid with respect to a schema if it is possible to assign at least one type from that schema to each node and relationship within the instance. In contrast, the
. In the first option, a graph instance is considered valid with respect to a schema if it is possible to assign at least one type from that schema to each node and relationship within the instance. In contrast, the  keyword, allows for creating nodes and relationship in the graph instance without a formal type definition. Accordingly, a statement for graph definition has the following structure:
keyword, allows for creating nodes and relationship in the graph instance without a formal type definition. Accordingly, a statement for graph definition has the following structure:
To define nodes and relationships, the ASCII-art formatting is adopted:  notation is used for node types specification, and
notation is used for node types specification, and 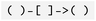 for edge types. Available datatypes for properties are aligned with GQL standards, including
for edge types. Available datatypes for properties are aligned with GQL standards, including  ,
,  ,
,  ,
,  and
and  . An example for defining two nodes and one relationship has the following structure:
. An example for defining two nodes and one relationship has the following structure:
The  clause indicates that a property is not mandatory. Besides, the
clause indicates that a property is not mandatory. Besides, the  keyword allows nodes to have additional properties. Notice that the
keyword allows nodes to have additional properties. Notice that the 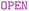 modifier also applies to labels, enabling an additional and arbitrary label to be assigned to a specific node. Multiple labels can be designated using the &-operator. Extra features include abstract types, which define types that cannot be instantiated, and inheritance, to reuse previously defined types.
modifier also applies to labels, enabling an additional and arbitrary label to be assigned to a specific node. Multiple labels can be designated using the &-operator. Extra features include abstract types, which define types that cannot be instantiated, and inheritance, to reuse previously defined types.
The second component, although not always included in definition languages, is nevertheless crucial: the specification of constraints. Formally, a constraint is defined by the statement:  p(x)
p(x)  q(x, ȳ). Here,
q(x, ȳ). Here,  outlines the expressed constraint using combinations of
outlines the expressed constraint using combinations of  ,
,  , and
, and 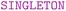 keywords. Both p(x) and q(x, ȳ) represent queries, with ȳ denoting the tuple (y1, y2, …, yn). The keywords definitions are:
keywords. Both p(x) and q(x, ȳ) represent queries, with ȳ denoting the tuple (y1, y2, …, yn). The keywords definitions are:
•  : it is not possible to share one tuple ȳ by two different values of x.
: it is not possible to share one tuple ȳ by two different values of x.
•  : for every output x of p(x) there must be at least one tuple ȳ that satisfies q(x, ȳ).
: for every output x of p(x) there must be at least one tuple ȳ that satisfies q(x, ȳ).
•  : for each x there should be at most one ȳ that satisfies q(x, ȳ).
: for each x there should be at most one ȳ that satisfies q(x, ȳ).
It is possible to use the keyword  inside a query to describe what its output is about (Section 3.3). To simplify the definition, an
inside a query to describe what its output is about (Section 3.3). To simplify the definition, an  keyword is also available:
keyword is also available:
The following constraint statement exemplifies the definition of a mandatory place of birth for a person, allowing at most one city per person while emphasizing that a city is not exclusive, as multiple individuals can be born in the same city:
Using these specifications it is possible to define participation constraints, denial constraints, key constraints, SQL-style CHECK constraints, range constraints and other custom constraints. For a deeper understanding of the formalism, please refer to PG-schema29 and PG-keys.60
3.2 Data modeling process
Despite the absence of a unified schema for designing graph databases,61 the data modeling process is generally standardized and closely related to conceptual modeling in information systems development.29,48 Both representations focus on establishing entities, their characteristics, and relationships between them. The main difference among these processes lies in the iterative nature of graph database modeling, which is essential for refining its structure to align it with the specific domain. The set of queries intended to be answered based on the database content defines how knowledge is going to be structured using the data model.21,56As show in Fig. 4, the first step for the data modeling process begins with a thorough understanding of the domain. This stage encompasses requirements gathering from different sources through activities such as interviews with stakeholders directly involved in the experimentation process, reviewing documentation of the experimental protocols, technical reports and publications, and finally extracting data from existing subsystems, storage, repositories and devices.62 The aim is to capture and define a common vocabulary for the most relevant objects within the domain, thereby creating a knowledge base that represents the data and its associated context (metadata).45
With knowledge acquisition and collaboration of domain experts, it is possible to define the competency questions,63 also referred to in the literature as uses cases. The specification of these queries is a key enabler for identifying the main entities (nodes) and establishing connections between them. These queries serve as a description of what the knowledge base is expected to answer, therefore, constituting a testing indicator for the generated data model as the main result of each iteration.
The next step focuses on modeling the entities and relationships with their properties, generating an initial whiteboard sketch of the data model. A graph data model by itself contains no data, however, it is crucial as it defines the names for labels, relationship types, and properties to be used when the graph is created and instantiated by an external application.
The iterative cycle involves testing the generated model against the defined questions. This requires creating an instance model for the proposed nodes with dummy data, and performing the corresponding Cypher queries to evaluate effectiveness and performance. From this point, the iteration resumes with new entities, relationships or properties added to the data model to account for missing responses or partially answered queries. Additionally, as the data scales, it may be necessary to carry out graph (re)factorization to achieve optimal performance for the defined use cases.
The Neo4j database offers indexing capabilities to ensure performance at query filtering. Therefore, it is important to consider this potential, to reformat the graph for the next iteration. When no more iterations are needed and a concrete data model is achieved, the graph formalism with the correct datatypes for properties and constraints explicitly indicated becomes the expected outcome of the complete data modeling process.
The domain of this study has been extensively addressed and discussed in Mione et al.,57 and is further elaborated here in the case study presented in Section 4. The competency questions derived from the domain analysis process are shown in Table 2.
| In reference to a single experiment: | |
| #1 | Which were the computational methods implemented to control the last experiment performed? |
| #2 | Who were responsible for the last experiment where the objective was strain screening? |
| #3 | Which values were used for the model parameters to calculate the feeding profile in the fifth iteration of the workflow for the experiment with run ID 623? |
| #4 | Which devices were used to analyze the samplings taken in the last experiment performed? |
| #5 | Which computational environment was used for the initial parameter estimation executed in the experiment having the run ID 724? |
| #6 | Which protocol was followed to obtain the acetate concentrations from samples in the last experiment? |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| In reference to multiple experiments: | |
| #7 | From the last 5 experiments aimed at maximizing biomass, in which bioreactors did the DOT measurements reach values below 20% for 5 consecutive samplings at any given time throughout the cultivation? |
| #8 | Which are the bioreactor and the experiment ID where the cell dry weight (CDW) had reached the highest value in gram per liter using the” E. coli BL21(DE3)” strain? |
| #9 | Which experiments were controlled using the macro-kinetic growth model published in the paper Anane et al.?64 |
| #10 | How many experiments did the person “John doe” perform as responsible in the role “laboratory_experimentation”? |
| #11 | How many bioreactors used strains containing the plasmid “pET28-NMB2-mEFGFP-TEVrec-(V2y)15-His?” |
| #12 | Which were the model-based state predictions variables at induction time in the last two experiments preformed aiming to maximize the product Fluo-RFP? |
These questions are categorized into two main types: there are queries that can be answered based on data and metadata from a single experiment, whereas other queries are designed to extract knowledge from multiple experiments. It is considered relevant to differentiate these groups, because the second group of queries is considered transversal (as they traverse the entire database) and provides a strong justification for the use of graph databases over relational ones. This is based on the fact that graph databases allow for filtering operations to be performed directly with the database engine rather than through data processing using a programming language as would be necessary when using a traditional database.
The following entities could be identified from the list: Experiment, Person, Objective, Bioreactor, Strain, Plasmid, WorkflowNode, ComputationalMethod, ComputationalEnvironment, FeedingSetpoint, Measurement, ModelParameters, ModelState, Model, Device and ProtocolTask. Furthermore, based on the protocols of the experimentation process, two more entities were identified: FeedingConfig and InductionConfig. The outcome of the data modeling process which is needed to provide a formal definition of the graph is presented in the next section.
3.3 Knowledge representation and formalization
The proposed schema for storing knowledge using an LPG is defined through the data model presented in Fig. 5, by following the guidelines established for a PG-schema and its predecessor PG-keys, with the complete definition provided in the schema.pgs file of the public repository.It is essential to highlight that the main objective of the presented model is to encompass the computational processes carried out for controlling and monitoring automated experiments. In the future, it should be extended to account for all tasks and protocols involved in the experimental workflow execution,65 with special reference to the different microorganisms and robotic devices available in a laboratory.
To begin with the formal definition, it is mandatory to specify the name of the schema and the type of graph. In this context, the 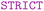 type is selected to ensure that an instance of this graph remains valid only if all its entities and relationships can be associated to at least one entity type or relationship type defined within the schema.
type is selected to ensure that an instance of this graph remains valid only if all its entities and relationships can be associated to at least one entity type or relationship type defined within the schema.
The Experiment node constitutes the first definition, incorporating properties like the identifier, start time, and duration horizon with their units. In accordance with PG-schema guidelines, the possibility of adding extra properties to this node is indicated by the  clause. Therefore, the initial definition is structured as shown in Table 3.
clause. Therefore, the initial definition is structured as shown in Table 3.
Subsequently, six relationships are established to link other entities with the Experiment. The first one connects with the Objective node and includes information about the name and description of the overall objective of the experiment. The second one, describes the relationship with a Person responsible for its execution. Several relationships between Experiment and Person may exist with different roles (property of the relationship), such as supervisor, modeler, planner, or others. The schema formalization for these two relationships can be seen in Table 4.
The third relationship corresponds to the general FeedingConfig profiles in all the bioreactors that made up the platform, specifying parameters like feeding pulse frequency, minimum and maximum volume, substrate concentration, and their units. Additionally, the InductionConfig is stored in a separate node where its configuration with the necessary details is provided, including induction time, concentration value, units, and stock used.
The fifth relationship links the Experiment node to the Bioreactor node, detailing metadata about the Strain and the associated Plasmid used in each of them, both represented as linked nodes. Several bioreactors may be included in a single experiment as the robotic platform allows for parallel experimental processes. Finally, there is the “has_computational_workflow” relationship, which must initialize a WorkflowNode named “start” to begin the execution of an experiment. This relationship may include several parameters for workflow definition, such as the number of iterations to execute, start time of the first iteration, time elapsed between iterations and others. As execution progresses, nodes related to computational tasks are dynamically attached to the graph database, based on task dependencies specified in the computational DAG defined in Airflow. To achieve reproducibility, each WorkflowNode is optionally linked with the corresponding ComputationalMethod and ComputationalEnvironment. The association of these nodes is not mandatory, allowing for the definition of Airflow empty operators, which perform no task, or time sensor operators, which pause the execution for a specified time window. As a result, these special Airflow operators do not require a computational setup to be designated.
In the specific case of closed loop experimental re-design operation, four special nodes are identified as outcomes of each applied computational method and subsequently connected to the workflow node executed. The first node, called ModelParameter, indicates the results obtained from the model parameter adjustment process as new measurements are received from sample processing; the second node, ModelState, stores the states predicted at different times by the model for each bioreactor until the end of the experiment; the third node, FeedingSetpoint, is obtained for each bioreactor during the optimization task which is part of the experiment redesign pipeline; and lastly, new Measurement nodes for each bioreactor are processed and dynamically added to the graph, specifying the ProtocolTask followed and the Device used. The ModelState and ModelParameters nodes are associated with the corresponding Model node, which contains the properties name, description, and optionally the digital object identifier (DOI).
For this type of graphs, all relationships are unidirectional. If a bidirectional representation is required, two separate relationships with opposite directions between the same pair of nodes must be created.
Several formalized constraints according to the adopted guidelines are presented in Table 5. These encompass keys, cardinalities and other specific requirements including the mandatory assignment of the name “start” to the task_id property of the first node in the workflow.
As mentioned before, the main contribution of this scheme focuses around the computational workflow used to control the execution of an experiment, predominantly expressed through the recursive relationship “dependency” on WorkflowNode entity. For a more comprehensive understanding of the data model, Fig. 6 presents the instance model generated with limited test data for the aforementioned use case, covering three iterations of the same control process. The image depicts a detailed view of the primary computational tasks executed by the workflow nodes, illustrating the corresponding nodes for each task type (extensively detailed in Section 3.4.3).
The complete definition of the proposed model can be adapted, as its formulation is open. Other researchers may also extend this model using the  clause, which allows them to inherit the existing formalism while also enabling the rewriting of current formulation in the data model or the incorporation of new entities and relationships associated with the laboratory facilities involved and its specific experimental domain.
clause, which allows them to inherit the existing formalism while also enabling the rewriting of current formulation in the data model or the incorporation of new entities and relationships associated with the laboratory facilities involved and its specific experimental domain.
3.4 Implementation and prototyping
To validate the designed scheme, a Python prototype was experimentally implemented for an specific HTBD scenario at the KIWI-biolab, Chair of Bioprocess Engineering, TU-Berlin. The implementation, the subsequent integration with Airflow, and the web interface created to interact with the corresponding knowledge base are detailed in the following subsections.The use of the OGM facilitates the interaction between the programming language used and the graph database, making it more accessible to developers.66 The queries can be executed using Python's language style, resulting in data structures known as Python objects. When CRUD (create, read, update and delete) operations are executed, the OGM ensures data consistency based on the defined model, allowing the action to proceed without requiring any additional methods to be implemented. These features introduce an abstraction layer that provides a database-agnostic API. However, there are instances where queries become highly complex and cannot be performed through this Python-based interface.54 In such cases, the library incorporates the capability to run Cypher queries for large volumes of data, using the database's native language to handle these requests more efficiently compared to the OGM alternative.
It is important to note that not all properties defined in the PG-schema are present in the current implementation, as some attributes were designated as optional. Additional attributes can be added to certain classes that have room for their inclusion. This also applies to the defined constraints, which cannot be fully implemented explicitly due to limitations of the adopted library. For instance, the definition of the first WorkflowNode with the task_id attribute labeled “start”, or the relationship of the same node to a single resulting type (Measurement, FeedingSetpoint, ModelState, or ModelParameter). These limitations are present at the model definition layer, but to fully enforce the PG-schema, the safeguard implementation was included in the code.
Furthermore, with the aim of simplifying the implementation, the violation of constraints is not managed on the software side. Only unique identifiers violation are considered, and in cases where duplicates exist in the database, a random number is concatenated, and the event is logged into the WMS. This log can then be reviewed and managed by an administrator through the web interface.
To achieve portable and reproducible development, docker is used to create the instance of the Neo4j database with the official image, and an extra container to execute the Neomodel library. Both environments are defined as services in the docker-compose.yml file of the repository.
The definition of the  class and the
class and the  relationship are presented as an example of the implementation. To define a new node class, the Neomodel
relationship are presented as an example of the implementation. To define a new node class, the Neomodel  class —or
class —or  if it allows for additional unmodeled attributes— should be extended. Subsequently, its properties are defined according to the datatypes supported by the library, including
if it allows for additional unmodeled attributes— should be extended. Subsequently, its properties are defined according to the datatypes supported by the library, including  ,
,  , and
, and  . Within the definition of each property, certain parameters can be specified to denote constraints defined in the schema, such as uniqueness (unique_index), mandatory status (required), or enumeration (choices).
. Within the definition of each property, certain parameters can be specified to denote constraints defined in the schema, such as uniqueness (unique_index), mandatory status (required), or enumeration (choices).
Edges can be declared within the node using the  keyword, specifying the destination node class (remembering that relationships are directed) and the relationship name. Afterwards, it is possible to establish the cardinality constraints as a parameter of the relationship, including
keyword, specifying the destination node class (remembering that relationships are directed) and the relationship name. Afterwards, it is possible to establish the cardinality constraints as a parameter of the relationship, including  ,
,  ,
,  , or
, or  . If the relationship has its own properties, a new object must be defined by extending the
. If the relationship has its own properties, a new object must be defined by extending the  class and specifying the attributes and datatypes, similarly to node properties. In Table 6,
class and specifying the attributes and datatypes, similarly to node properties. In Table 6, 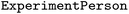 relationship includes the role attribute, and it is added as a model parameter of the defined relationship.
relationship includes the role attribute, and it is added as a model parameter of the defined relationship.
With a focus on automation and controlled execution through the computational workflow, different methods are defined for storing data and metadata in a graph database. The main structure is implemented through specific Airflow callbacks, invoked upon different events during the execution lifecycle of a node (Fig. 7). The four main events that trigger these callbacks are detailed below:
• on_execute: represents the creation event of a node. The function defined in this callback will be executed whenever a given node's virtual environment is created, prior to the execution of its specific task. It receives the node instance context as a parameter, including predecessor tasks, initial state, node type, task name, start execution time, and other details.
• on_success: represents the successfully finished event of a node. The callback defined for this event will be triggered only if there were no errors during the entire execution of the node's specific task. Similarly to the other defined events, it receives the node instance context with all its information. Additional entities are created within the graph database as a consequence of the successful execution of the task (detailed on Section 3.4.3).
• on_failure: represents the event of an interruption of the node execution due to an unsolved error detection. It receives as parameters the node instance context and details of the associated error. No additional entities are generated in the KG due to inconsistencies in the computational method.
• on_retry: represents the event of a new execution call for a given node due to a detected failure. This event will be triggered if and only if this option is enabled for such node.
Each callback invocation demands some execution time within the node. Consequently, if this task takes an unreasonable time, it could significantly affect the metadata collected regarding the node's execution time. However, any failure in these callback functions will not impact the normal processing of the node's specific task.
The execution of the task begins with the dispatch of the Airflow executor, initiated by the scheduler. An instance of the corresponding node (operator) is created, usually a docker container or a Python virtual environment for executing simple computational tasks. At this point, the first callback, on_execute, is invoked. This callback is responsible for adding initial information of the node into Neo4j, including start time, execution status, task name, and other details. Additionally, it assigns the precedence relationships of tasks defined in the corresponding DAG for the computational workflow, based on the information received in the callback context. Subsequently, execution is returned to the node instance to perform the designated task. If an error occurs, an on_failure callback is invoked to record the end time, the failure status, and save an error message in the node's properties. On the other hand, if no errors occur, these parameters are recorded in the on_success callback, along with the corresponding ”successful” status. Whenever the node has a retry parameter defined, this value is recorded in the graph database, and the retry count is incremented by one for each attempted execution. The partial start time of each attempt is stored in a dictionary to compute the total execution time of the node across its successive trials.
This approach is particularly suited for the type of experimentation addressed in this work, where experiment control is executed at intervals of several minutes (10, 30, 60 minutes, etc.), rather than in real time. If adaptation for real-time operation is required, it could similarly be implemented in the WMS using Apache Airflow and the PG-schema with Neomodel. However, an additional communication layer with IoT (internet of things) devices would be needed, using Redis, MQTT, or another message broker specifically designed for such connections, while maintaining parallel communication with the WMS and Neo4j. The PG-schema and Neomodel implementation can be extended to incorporate these new data structures based on the data model presented in Section 3.3.
• Start node: this is an empty node, meaning it does not perform any task; rather, it serves as an execution trigger for each defined workflow. At the end of the dummy execution, the on_success callback loads the metadata.yaml file, which contains information about the DAG definition and experimental metadata. Data related to the DAG is stored as properties of the relationship between the experiment and the initial workflow node instance in Neo4j. The remaining data includes details about the duration of the experiment, feeding and induction configurations, people who are responsible for planning and executing the experiment, the objective, the definition of the bioreactor groups with the corresponding strains and plasmid used as well as the specifications for the computational methods implemented to control them. All of these metadata is stored at the beginning of the Airflow execution to associate the generated dynamic data with these prior metadata.
• Get measurements: this method queries the relational database linked to the robotic devices software in the laboratory, where information is stored upon completion of analytical sample processing. Therefore, it is responsible for retrieving these new measurements for some state variables and creating the corresponding nodes for each of them.
• Parameter re-estimation: as a result of completing this specific task, different nodes for all model parameter types are created, with properties indicating the type of parameter and the assigned value. These nodes are associated with the workflow node entity in the corresponding iteration to achieve traceability of model changes.
• Online redesign: several nodes descriptive of the feeding profile used for each bioreactor are obtained by solving an optimization problem implemented through this method. Each node includes properties for the corresponding time and feeding concentration volume for each pulse of the fed-batch step until the end of the experiment.
• Model state predictions: at the end of this task, nodes corresponding to model state predictions for each bioreactor are generated. Each node includes the type of measurement, the future time for the prediction, and the predicted value until the end of the experiment.
The implementation of the class to define all the available callbacks can be found in the helper.py file, and its corresponding invocation is included in the definition of each Airflow node.
The current implementation considers a specific set of Airflow nodes; however, the architecture is designed to be extensible. Future expansions may include additional tasks, such as a data preprocessing module. Given the heterogeneity of data sources, arising from diverse devices, sampling frequencies, and analytical techniques, a preprocessing layer within the WMS could serve to standardize data structures. Furthermore, an AI-driven agent could be employed to learn from available measurement types and calibration settings, enabling functionalities such as outlier detection, value normalization, and automated inference of measurement units.
This interface allows users not only to query and retrieve the necessary data for their analysis but also to add extra metadata to certain entities. By employing a linked-data approach, users can add references that enrich the information characterizing an entity in the graph database. For instance, it is possible to add the DOI of a reference article where a mathematical model used for process simulation was defined, or the URL of providing the technical specifications for a particular robotic device.
The interface manages several entities related to experimental and computational workflows, including Objective, Person, Strain, Plasmid, Device, ProtocolTask, ComputationalMethod, and Model. By properly managing all these entities, the execution of a specific computational workflow is linked to the corresponding metadata. The configuration of the experiment can be predefined through a semantically structured section that allows users to select and define groups of bioreactors, each one involving a particular strain, or controlling them using a specific computational algorithm, or specifying the different people responsible for the experiment in their respective roles. The result of the process is the metadata.yaml file needed by Airflow to start the execution.
By incorporating information provided by human sources, such as experimental design, configurations, conclusions, and other manually added metadata, knowledge transfer from humans to AI agents is enabled. Conversely, through the use of graph visualization tools, combined with the presented schema, knowledge transfer in the opposite direction is also facilitated.
Several commercial and freeware data analysis tools allow Neo4j to be integrated either as an embedded web interface or as an external application. Examples include Neo4j Bloom,69 ThornViz,70 and KeyLines.71 These tools enable users to navigate and explore the knowledge base, identify relationships between data points, and perform visual comparisons over the schema within the graph, without requiring the implementation of Cypher queries. This type of analysis is not feasible with relational databases, where information can only be retrieved through explicit SQL queries in a tabular format.
In this work, the integration of an external visualization tool into the web interface is left for future development. Nevertheless, as shown in the Results (Section 5), a visual interface included by default in the standard installation of Neo4j is used.
The web component was developed using the Python Flask framework,72 and the Datta Able Flask template to enhance visual implementation and user experience. Information gathered from the web sessions and authentication were fully implemented within Neo4j, thus integrating FlaskLogin with the scheme created in Neomodel, without the need for any additional database. For a complete description of the functionalities and associated source code, please refer to the web folder in the public repository.
4. Case study
This section discusses the instantiation of the PG-schema and the prototype developed through an in silico case study that emulates an experimental setup used at the KIWI-biolab at TU-Berlin.3 The study involves the simulation of robotic devices and their connection to the SQL database for an initial testing phase of the PG-schema. Notably, the WMS has already been tested in real fully automated experiments with three different algorithms running in parallel,57 a groundbreaking achievement that provided crucial insights. Based on this experience and learning, the concept of LPGs has been developed and tailored to overcome the previous limitations.Furthermore, while the current study relies on in silico data, it closely mirrors real experimental conditions, incorporating noise in both the measurements and the sample values of variables of interest. The use of an emulator ensures that the developed system can be seamlessly tested in a real experimental setup, validating its applicability in a real world scenario. To replicate the dynamics of a fed-batch experiment, a bioreactor model for E. coli cultivations is employed (based on the ones presented in the literature73). The simulation framework is designed to support parallel experimentation with 24 mini-bioreactors (MBRs), arranged in 3 columns and 8 rows. This framework includes a MySQL database replica from the robotics device software, enabling the WMS and the graph database to interact with the experimental platform in a standardized manner, whether in simulation mode or during a real HTBD experiment. The emulator reads the profile setpoints for each MBR from the SQL database and using the current estimated states of the bioreactors, simulates the subsequent steps. The simulated data are then stored in the SQL database to ensure readiness for WMS processing.
The parameters of the model used in the emulator are chosen based on data from previous experiments, and the overall experiment duration is set to 16 hours. Different initial glucose concentrations are assigned to the MBRs, four groups of six MBRs are then defined, each one controlled in a closed-loop configuration.
In the emulator's experimental setup, dissolved oxygen tension (DOT) measurements are sampled online every 2 minutes, while biomass, glucose, acetate, and the red fluorescent protein (Fluo-RFP) product, are obtained at-line every hour. This setup simulates the real behavior of the robotic platform, in terms of sampling constraints and analytical processing delays. Each MBR column is sampled every 20 minutes due to task scheduling constraints of the robotic facility, requiring a total of 60 minutes to complete processing of all samples taken from different MBRs. The subsequent at-line analytical processing takes an additional 60 minutes, resulting in the first measurement being written in the database 2 hours after the experiment has begun. Feeding pulses are scheduled every 10 minutes, with a concentration of 200 g L−1 and a minimum and maximum addition volume of 5 μL and 150 μL respectively, starting upon the event of total glucose consumption is triggered, depending on the initial glucose level in each specific MBR. Finally, the induction time is set to occur after completion of the batch and fed-batch phases at 10 hours for all MBRs with a concentration of 5 mM.
The computational workflow begins with the first feeding profile calculation at time zero. Iterations are initiated after 2 hours when measurements become available, with subsequent iterations occurring at one-hour intervals. Each iteration starts by querying the SQL database to retrieve new measurements. This new data is used to update the model parameter distributions for each MBR group using the Variational Bayesian Analysis toolbox.74 The model controlling the operation is the same as the one used by the emulator, but the exact parameter values are considered unknown. Thus, wide prior distribution are used for the model parameters and the initial states. As new data become available during each iteration, the model parameters are updated to better describe the bioreactors operation. Based on the posterior distributions of these model parameters, a new feeding profile is computed by solving an optimization problem, and it is immediately stored in the SQL database. Finally, the remaining part of the experiment is predicted by propagating the current states using the model and the newly computed feeding strategy for each MBR.
In order to obtain the feeding profiles, an objective function has to be defined. During the early stages of process development, the information content of the experiment is deemed very important. Generating different biomass profiles (which involves having different growth rates) for the MBR is a good proxy for information content and is an interesting goal for the experiment. However, important constraints have to be considered to ensure experimental conditions that are relevant for industrial scale. For example, a DOT value above 20% during throughout the complete experiment is typically considered necessary in aerobic cultivation processes. Thus, the optimization problem to be solved is the maximization of the distance between biomass profiles while maintaining the concentration of DOT above the established threshold. By these a comprehensive exploratory experiment over the viable domain space of physiological conditions is designed.
5. Results
Results obtained from a single experiment simulation are illustrated in Fig. 9. The execution scheduled through Airflow (Fig. 9A) is stored in the Neo4j KG. The timeline of the executed computational workflow, the time spent on each task, and the specific outcomes are available for inspection. This enables complete traceability of the computational control implemented for the experiment, ensuring full reproducibility. Monitoring the experiment execution through the web platform and a tab for generating the experimental design are shown in Fig. 9B, where real-time filters can also be applied to focus on the measurements related to certain bioreactors.The plots illustrating the experimental results (Fig. 9C) highlights how the different feeding profiles for the first group of six MBRs explore the action space of the domain, maximizing the distances among state trajectories. Due to the delay caused by the analytical methods, the model and the optimizer are always using data from one or two hours before, which may result in a mismatch between the predictions and the real values. This is particularly challenging for the optimizer, leading to occasional violations of the DOT constraint in certain MBRs. Once new data is available, the feeding profiles are corrected and the constraints are hopefully fulfilled for the rest of the experiment.
In Fig. 9D, the evolution of the Neo4j KG is illustrated as successive executions of the computational workflow advances over time. New nodes are generated to store information gathered from the samples taken, indicating properties and relationships for each of them, thereby ensuring FAIR data generation. This specific case study depicts the sequence of measurements obtained, excluding DOT values due to high-frequency sampling, involving a total of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 520 nodes for this variable type. Once the last iteration has been completed, the 24 MBRs have each accumulated 80 samples of the corresponding state-related measurements. One single experimental run of 24 MBRs over 16 hours cultivation time gives rise to a total of 62
520 nodes for this variable type. Once the last iteration has been completed, the 24 MBRs have each accumulated 80 samples of the corresponding state-related measurements. One single experimental run of 24 MBRs over 16 hours cultivation time gives rise to a total of 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 127 nodes and 140
127 nodes and 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128 relationships, classified as shown in Table 7.
128 relationships, classified as shown in Table 7.
| Node | Instances |
|---|---|
| FeedingSetpoint | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
| ModelState | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
| Measurement | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440 440 |
| ModelParameter | 1288 |
| WorkflowNode | 443 |
| ComputationalEnvironment | 116 |
| Bioreactor | 24 |
| ComputationalMethod | 4 |
| Person | 3 |
| Others | 9 |
| TOTAL |
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 127 127 |
| Relationship | Instances |
|---|---|
| Calculates | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
| Feeds | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
| Part_of | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 488 488 |
| Predicts | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
| Prediction_for | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
| Sample_from | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440 440 |
| Gets | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440 440 |
| Dependency | 442 |
| Executes | 232 |
| Others | 1486 |
| TOTAL |
140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128 128 |
These results underscore the critical role of relationships in data, effectively doubling the number of nodes instances required to represent knowledge. Metadata in the form of relationships are not clearly represented within a relational database approach, as used by platforms like ChemOS or ESCALATE. This limitation prevents the generation of AI-ready data and reduces the potential for applying inference processes in knowledge discovery.
To demonstrate the exploratory analysis capabilities of the proposed PG-schema and the power of graph databases, a Cypher query was executed to analyze the knowledge captured regarding DOT constraint violations (a measurement going below 20%). Initial plot-based results (Fig. 9C) indicate that two MBRs have breached this constraint at least once, including the MBR identified as #19441. Additionally, a deeper analysis provides detailed insights about the timing (WorkflowNode) when the controller detected the violation and the corrective actions taken to adjust the feeding profile for the affected MBR. These findings are shown in Fig. 10, where the violation was detected after 11 hours of experimentation. The corresponding node, identified as “get_measurements_10”, marks the detection event, and the adjusted feeding profile is represented as a time vector in seconds along with the cumulative feed volume.
Neo4j provides several engine-level functions, one of the most notable being vector similarity, which is widely used in recommendation systems. The comparison relies on the Euclidean distance function, which outputs a floating-point value between 0 and 1: values close to 1 indicate strong similarity, while those near to 0 represent significant differences. In this work, this feature is particularly valuable for comparing a feeding profile applied to a MBR against a reference feeding profile, specifically the mean one between all observed profiles for that group.
The primary objective of this similarity-based analysis is to identify a feeding profile that maximizes product yield (Fluo-RFP) at the end of the experiment, near the mean values of all applied profiles for that group. The evaluation metric is computed as the product of the similarity score and the final Fluo-RFP concentration. Fig. 11A presents the corresponding Cypher query and Fig. 11B shows the table results, displaying the MBR ID, the achieved product yield, and the computed comparison values, sorted in descending order. In particular, the MBR with ID #19425 achieves the highest comparative score. However, it is important to highlight that while the MBR with ID #19441 attains the highest product yield, its feeding profile significantly deviates from the reference profile (Fig. 11C).
A comparison between a relational database (MySQL) and a graph database (Neo4j), including a brief analysis focusing on performance and query syntax can be found in the ESI† file, which relies on a database populated with 400 experiments, each containing 24 MBRs.
6. Concluding remarks
The use of graph databases as flexible storage for both data and metadata has proven to be fundamental in enabling autonomous knowledge discovery and data analysis across various domains. In the context of HTBD, the generation of a KG to link experimental data with the corresponding metadata based on the proposed PG-schema is considered a cornerstone for deploying powerful and scalable SDLs.The proposed PG-schema has been designed based on the integration of an experimental-computational workflow choreographer (Apache Airflow) with a graph-based database engine (Neo4j) that use property graphs as semantic models. This allows for comprehensive digital encoding of all relevant metadata about devices, protocols, computational methods, strains, process conditions, models, software versions and hardware configurations. The case study presented demonstrates that the redesign of feeding rates is aligned with the experimental objective established, with all metadata captured in the KG, thus fostering AI-Ready data.
The decision of resorting to Airflow and Neo4j platforms were driven by their open-source nature, scalability, and the robust support and comprehensive documentation provided by their development teams. Still, no generality or flexibility has been compromised through the use of both Airflow and Neo4j, since the framework leverages the use of Docker containers to define all tasks involved in the computational workflow, enabling migration with minimal modifications to the DAG definition. Similarly, for Neo4j storage, it is possible to integrate callbacks with a new framework or reuse the PG-schema with another graph database implementation.
This study underscores the importance of having a well-structured PG-schema for gathering metadata and representing real-world constraints, as exemplified by its application to autonomous robotic facilities. The schema's versatility, however, extends beyond this context; it is open and can be adapted for use in different domains or laboratory environments such as drug, chemistry and materials discovery as well as autonomous hypothesis testing. The computational workflows can also be modified to collect metadata from various types of experimentation or executed using different computational environments, such as cloud-based or high-performance distributed computing. An interesting avenue for future development would be the incorporation of dynamic agents capable of evolving the schema over time.
The integration of the proposed platforms, presents a robust foundation for FAIR data generation in bioprocess development, contributing to achieve SDLs and enhancing machine actionability for autonomous knowledge discovery. Furthermore, with the rise of large language models and their capability to retrieve information from diverse knowledge sources, this PG-schema facilitates the adoption of such technologies, enabling natural language queries to the graph database. By semantically labeling entities and relationships, future projects can readily incorporate this functionality, which opens up a promising avenue for further developments in autonomous experimentation.
Data availability
In the GitHub repository https://github.com/fmione/Property-Graph-Schema (DOI: 10.5281/zenodo.15610711) a detailed step by step guide to deploy the project and reproduce the results and analysis is presented. In addition, screenshots of the web interface and queries for the graph database are included. This comprehensive documentation aims to facilitate the reproducibility of the methodology presented for the PG-schema in a HTBD scenario.Author contributions
Conceptualization: FM, ML, EM; methodology: FM, EM; software: FM, ML, LK; validation: PN, EM, MNCB; formal analysis: FM, ML, EM; investigation: FM, ML, EM; data curation: FM; writing – original draft preparation: FM, EM; writing – review and editing: ML, LK, PN, MNCB; visualization: FM; supervision: ML, EM, MNCB; project administration: MNCB; funding acquisition: PN, MNCB.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the financial support of the German Federal Ministry of Education and Research (01DD20002A – KIWI biolab) and the Open Access Publication Fund of TU-Berlin.References
- P. M. Maffettone, P. Friederich, S. G. Baird, B. Blaiszik, K. A. Brown, S. I. Campbell, O. A. Cohen, R. L. Davis, I. T. Foster, N. Haghmoradi, M. Hereld, H. Joress, N. Jung, H.-K. Kwon, G. Pizzuto, J. Rintamaki, C. Steinmann, L. Torresi and S. Sun, Digital Discovery, 2023, 2, 1644–1659 RSC.
- D. B. Nickel, M. N. Cruz-Bournazou, T. Wilms, P. Neubauer and A. Knepper, Eng. Life Sci., 2017, 17, 1195–1201 CrossRef CAS.
- B. Haby, S. Hans, E. Anane, A. Sawatzki, N. Krausch, P. Neubauer and M. N. Cruz Bournazou, SLAS Technol., 2019, 24, 569–582 CrossRef CAS PubMed.
- M. N. Cruz Bournazou, T. Barz, D. B. Nickel, D. C. Lopez Cárdenas, F. Glauche, A. Knepper and P. Neubauer, Biotechnol. Bioeng., 2017, 114, 610–619 CrossRef CAS PubMed.
- H. G. Martin, T. Radivojevic, J. Zucker, K. Bouchard, J. Sustarich, S. Peisert, D. Arnold, N. Hillson, G. Babnigg, J. M. Marti, C. J. Mungall, G. T. Beckham, L. Waldburger, J. Carothers, S. Sundaram, D. Agarwal, B. A. Simmons, T. Backman, D. Banerjee, D. Tanjore, L. Ramakrishnan and A. Singh, Curr. Opin. Biotechnol., 2023, 79, 102881 CrossRef CAS PubMed.
- J. T. Rapp, B. J. Bremer and P. A. Romero, Nat. Chem. Eng., 2024, 1, 97–107 CrossRef.
- E. Nuñez-Andrade, I. Vidal-Daza, J. W. Ryan, R. Gómez-Bombarelli and F. J. Martin-Martinez, Digital Discovery, 2025, 4(3), 776–789 RSC.
- E. Pignotti, P. Edwards, N. Gotts and G. Polhill, J. Web Semantics, 2011, 9, 222–244 CrossRef.
- L. Kaspersetz, S. Waldburger, M.-T. Schermeyer, S. L. Riedel, S. Groß, P. Neubauer and M.-N. Cruz-Bournazou, Front. Chem. Eng., 2022, 4, 812140 CrossRef.
- M. Baker, Nature, 2016, 533, 452–454 CrossRef CAS.
- B. Miles and P. L. Lee, SLAS Technol., 2018, 23, 432–439 CrossRef.
- C. Arnold, Nature, 2022, 606, 612–613 CrossRef CAS.
- L. Hung, J. A. Yager, D. Monteverde, D. Baiocchi, H.-K. Kwon, S. Sun and S. Suram, Digital Discovery, 2024, 3, 1273–1279 RSC.
- N. Ishizuki, R. Shimizu and T. Hitosugi, Sci. Technol. Adv. Mater. Methods, 2023, 3, 2197519 Search PubMed.
- T. Barz, A. Sommer, T. Wilms, P. Neubauer and M. N. Cruz Bournazou, IFAC-PapersOnLine, 2018, 51, 765–770 CrossRef.
- M. F. Luna, M. N. Cruz Bournazou and E. C. Martínez, Computer Aided Chemical Engineering, Elsevier, 2022, vol. 51, pp. 1111–1116 Search PubMed.
- J. W. Kim, N. Krausch, J. Aizpuru, T. Barz, S. Lucia, E. C. Martínez, P. Neubauer and M. N. Cruz Bournazou, IFAC-PapersOnLine, 2022, 55, 934–939 CrossRef.
- M. C. Barbet, J. Lee, C. E. LaGrotta, R. E. Cornell and M. P. Burke, Combust. Flame, 2024, 267, 113562 CrossRef CAS.
- M. Akhtar, O. Benjelloun, C. Conforti, P. Gijsbers, J. Giner-Miguelez, N. Jain, M. Kuchnik, Q. Lhoest, P. Marcenac, M. Maskey, P. Mattson, L. Oala, P. Ruyssen, R. Shinde, E. Simperl, G. Thomas, S. Tykhonov, J. Vanschoren, J. Van Der Velde, S. Vogler and C.-J. Wu, Proceedings of the Eighth Workshop on Data Management for End-to-End Machine Learning, 2024, pp. 1–6 Search PubMed.
- O. E. Gundersen, Philos. Trans. R. Soc. A, 2021, 379, 20200210 CrossRef PubMed.
- K. Hose, Advances in Databases and Information Systems, Springer Nature, Switzerland, 2023, vol. 13985, pp. 3–15 Search PubMed.
- M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. G. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. C. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, Sci. Data, 2016, 3, 160018 CrossRef PubMed.
- H. Narayanan, M. F. Luna, M. Von Stosch, M. N. Cruz Bournazou, G. Polotti, M. Morbidelli, A. Butté and M. Sokolov, Biotechnol. J., 2020, 15, 1900172 CrossRef CAS PubMed.
- K. Isoko, J. L. Cordiner, Z. Kis and P. Z. Moghadam, Digital Discovery, 2024, 3, 1662–1681 RSC.
- G. K. Reder, A. H. Gower, F. Kronström, R. Halle, V. Mahamuni, A. Patel, H. Hayatnagarkar, L. N. Soldatova and R. D. King, Bioinf. Adv., 2023, 3(1), vbad102 Search PubMed.
- A. A. Volk, R. W. Epps, D. T. Yonemoto, B. S. Masters, F. N. Castellano, K. G. Reyes and M. Abolhasani, Nat. Commun., 2023, 14, 1403 CrossRef CAS.
- S. Sakr, A. Bonifati, H. Voigt, A. Iosup, K. Ammar, R. Angles, W. Aref, M. Arenas, M. Besta, P. A. Boncz, K. Daudjee, E. D. Valle, S. Dumbrava, O. Hartig, B. Haslhofer, T. Hegeman, J. Hidders, K. Hose, A. Iamnitchi, V. Kalavri, H. Kapp, W. Martens, M. T. Özsu, E. Peukert, S. Plantikow, M. Ragab, M. R. Ripeanu, S. Salihoglu, C. Schulz, P. Selmer, J. F. Sequeda, J. Shinavier, G. Szárnyas, R. Tommasini, A. Tumeo, A. Uta, A. L. Varbanescu, H.-Y. Wu, N. Yakovets, D. Yan and E. Yoneki, Commun. ACM, 2021, 64, 62–71 CrossRef.
- D. Di Pierro, S. Ferilli and D. Redavid, Information, 2023, 14, 154 CrossRef.
- R. Angles, A. Bonifati, S. Dumbrava, G. Fletcher, A. Green, J. Hidders, B. Li, L. Libkin, V. Marsault, W. Martens, F. Murlak, S. Plantikow, O. Savkovic, M. Schmidt, J. Sequeda, S. Staworko, D. Tomaszuk, H. Voigt, D. Vrgoc, M. Wu and D. Zivkovic, Proceedings of the ACM on Management of Data, 2023 Search PubMed.
- A. Hogan, E. Blomqvist, M. Cochez, C. D’amato, G. D. Melo, C. Gutierrez, S. Kirrane, J. E. L. Gayo, R. Navigli, S. Neumaier, A.-C. N. Ngomo, A. Polleres, S. M. Rashid, A. Rula, L. Schmelzeisen, J. Sequeda, S. Staab and A. Zimmermann, ACM Comput. Surv., 2021, 54, 1–37 CrossRef.
- H. Abu-Rasheed, C. Weber, J. Zenkert, M. Dornhöfer and M. Fathi, Informatics, 2022, 9, 6 CrossRef.
- E. Deelman, T. Peterka, I. Altintas, C. D. Carothers, K. K. Van Dam, K. Moreland, M. Parashar, L. Ramakrishnan, M. Taufer and J. Vetter, Int. J. High Perform. Comput. Appl., 2018, 32, 159–175 CrossRef.
- S. B. Davidson and J. Freire, Proceedings of the 2008 ACM SIGMOD International Conference on Management of Data, 2008, pp. 1345–1350 Search PubMed.
- C. Goble, S. Cohen-Boulakia, S. Soiland-Reyes, D. Garijo, Y. Gil, M. R. Crusoe, K. Peters and D. Schober, Data Intell., 2020, 2, 108–121 CrossRef.
- S. N. Mitchell, A. Lahiff, N. Cummings, J. Hollocombe, B. Boskamp, R. Field, D. Reddyhoff, K. Zarebski, A. Wilson, B. Viola, M. Burke, B. Archibald, P. Bessell, R. Blackwell, L. A. Boden, A. Brett, S. Brett, R. Dundas, J. Enright, A. N. Gonzalez-Beltran, C. Harris, I. Hinder, C. David Hughes, M. Knight, V. Mano, C. McMonagle, D. Mellor, S. Mohr, G. Marion, L. Matthews, I. J. McKendrick, C. Mark Pooley, T. Porphyre, A. Reeves, E. Townsend, R. Turner, J. Walton and R. Reeve, Philos. Trans. R. Soc., A, 2022, 380, 20210300 CrossRef PubMed.
- O. E. Gundersen, Improving Reproducibility of Artificial Intelligence Research to Increase Trust and Productivity, Oecd Technical Report, 2023 Search PubMed.
- M. Sim, M. G. Vakili, F. Strieth-Kalthoff, H. Hao, R. J. Hickman, S. Miret, S. Pablo-García and A. Aspuru-Guzik, Matter, 2024, 7(9), 2959–2977 CrossRef CAS.
- I. M. Pendleton, G. Cattabriga, Z. Li, M. A. Najeeb, S. A. Friedler, A. J. Norquist, E. M. Chan and J. Schrier, MRS Commun., 2019, 9, 846–859 CrossRef CAS.
- J. Bai, S. Mosbach, C. J. Taylor, D. Karan, K. F. Lee, S. D. Rihm, J. Akroyd, A. A. Lapkin and M. Kraft, Nat. Commun., 2024, 15, 462 CrossRef CAS PubMed.
- S. Purohit, N. Van and G. Chin, in 2021 IEEE International Conference on Big Data (Big Data), 2021, pp. 2672–2677 Search PubMed.
- J. Lin, Y. Zhao, W. Huang, C. Liu and H. Pu, Neural Comput. Appl., 2021, 33, 681–690 CrossRef.
- S. Ferilli, D. Redavid and D. Di Pierro, in Proceedings of the 30th Italian Symposium on Advanced Database Systems, 2022, pp. 256–267 Search PubMed.
- J. Bruyat, P.-A. Champin, L. Médini and F. Laforest, PRSC: from PG to RDF and Back, Using Schemas, 2024 Search PubMed.
- R. Angles, H. Thakkar and D. Tomaszuk, IEEE Access, 2020, 8, 86091–86110 Search PubMed.
- C. Barba-González, J. García-Nieto, M. D. M. Roldán-García, I. Navas-Delgado, A. J. Nebro and J. F. Aldana-Montes, Expert Syst. Appl., 2019, 115, 543–556 CrossRef.
- S. Harris, A. Seaborne and E. Prud’hommeaux, W3C Recommendation, 2013, 21, 778 Search PubMed.
- A. H. Chillón, M. Klettke, D. S. Ruiz and J. G. Molina, IEEE Trans. Knowl. Data Eng., 2024, 36, 2774–2789 Search PubMed.
- N. Beeren, MSc thesis, Eindhoven University of Technology, 2022.
- Redgate Software, DB Engines Ranking, n.d., https://db-engines.com/en/ranking/graph+dbms Search PubMed.
- Y. Tian, ACM SIGMOD Record, 2023, 51, 60–67 CrossRef.
- R. Angles, M. Arenas, P. Barceló, A. Hogan, J. Reutter and D. Vrgoč, ACM Comput. Surv., 2018, 50, 1–40 CrossRef.
- Information technology — Database languages — GQL, 2024, https://www.iso.org/standard/76120.html Search PubMed.
- N. Francis, A. Green, P. Guagliardo, L. Libkin, T. Lindaaker, V. Marsault, S. Plantikow, M. Rydberg, P. Selmer and A. Taylor, in Proceedings of the 2018 International Conference on Management of Data, 2018, pp. 1433–1445 Search PubMed.
- M. Dreger, M. J. Eslamibidgoli, M. H. Eikerling and K. Malek, J. Mater. Inform., 2023, 3(2), 1–14 Search PubMed.
- I. Robinson, J. Webber and E. Eifrem, Graph Databases: New Opportunities for Connected Data, O'Reilly, Beijing, Boston, Farnham, Sebastopol, Tokyo, 2nd edn, 2015 Search PubMed.
- A. Vaisman and E. Zimányi, Data Warehouse Systems: Design and Implementation, Springer Berlin Heidelberg, 2022 Search PubMed.
- F. M. Mione, L. Kaspersetz, M. F. Luna, J. Aizpuru, R. Scholz, M. Borisyak, A. Kemmer, M. T. Schermeyer, E. C. Martinez, P. Neubauer and M. N. Cruz Bournazou, Comput. Chem. Eng., 2024, 187, 108720 CrossRef CAS.
- D. Merkel, Linux J., 2014, 2 Search PubMed.
- C. Boettiger, ACM SIGOPS Operat. Syst. Rev., 2015, 49, 71–79 CrossRef.
- R. Angles, A. Bonifati, S. Dumbrava, G. Fletcher, K. W. Hare, J. Hidders, V. E. Lee, B. Li, L. Libkin, W. Martens, F. Murlak, J. Perryman, O. Savković, M. Schmidt, J. Sequeda, S. Staworko and D. Tomaszuk, in Proceedings of the 2021 International Conference on Management of Data, 2021, pp. 2423–2436 Search PubMed.
- S. Sahu, A. Mhedhbi, S. Salihoglu, J. Lin and M. T. Özsu, VLDB J., 2020, 29, 595–618 CrossRef.
- K. S. Aggour, A. Detor, A. Gabaldon, V. Mulwad, A. Moitra, P. Cuddihy and V. S. Kumar, Integrat. Mater. Manuf. Innovat., 2022, 11, 467–478 CrossRef.
- C. Yang, Y. Zheng, X. Tu, R. Ala-Laurinaho, J. Autiosalo, O. Seppänen and K. Tammi, Adv. Eng. Inform., 2023, 58, 102185 CrossRef.
- E. Anane, P. Neubauer and M. N. C. Bournazou, et al. , Biochem. Eng. J., 2017, 125, 23–30 CrossRef CAS.
- L. Kaspersetz, B. Englert, F. Krah, E. C. Martinez, P. Neubauer and M. N. Cruz Bournazou, SLAS Technol., 2024, 29, 100214 CrossRef.
- L. Costa, N. Freitas and J. R. da Silva, J. Comput. Cult. Heritage, 2022, 15, 44:1–44:18 Search PubMed.
- B. Harenslak and J. d. Ruiter, Data pipelines with Apache Airflow, Manning Publications Co, 2021 Search PubMed.
- C. Sharma and R. Sinha, in Proceedings of the 6th IEEE/ACM International Conference on Big Data Computing, Applications and Technologies, 2019, pp. 71–80 Search PubMed.
- Massive graph Analytics, ed. D. A. Bader, Chapman and Hall/CRC, 1st edn, 2022 Search PubMed.
- V. Lavigne and A. Bergeron-Guyard, in 2023 International Conference on Military Communications and Information Systems (ICMCIS), 2023, pp. 1–7 Search PubMed.
- Cambridge International, KeyLines White Paper, 2024, https://cambridge-intelligence.com/ Search PubMed.
- M. Copperwaite and C. Leifer, Learning Flask Framework: Build Dynamic, Data-Driven Websites and Modern Web Applications with Flask, Packt Publishing, 2015 Search PubMed.
- E. Anane, A. Sawatzki, P. Neubauer and M. N. Cruz-Bournazou, J. Chem. Technol. Biotechnol., 2019, 94, 516–526 CrossRef CAS.
- J. Daunizeau, V. Adam and L. Rigoux, PLoS Comput. Biol., 2014, 10(1), 1–16 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5dd00070j |
| This journal is © The Royal Society of Chemistry 2025 |