Open Access Article
Open Access ArticleAI agents in chemical research: GVIM – an intelligent research assistant system†
Kangyong
Ma
 *
*
College of Physics and Electronic Information Engineering, Zhejiang Normal University, Jinhua City 321000, China. E-mail: kangyongma@outlook.com; kangyongma@gmail.com
First published on 10th January 2025
Abstract
This work utilizes collected and organized instructional data from the field of chemical science to fine-tune mainstream open-source large language models. To objectively evaluate the performance of the fine-tuned models, we have developed an automated scoring system specifically for the chemistry domain, ensuring the accuracy and reliability of the evaluation results. Building on this foundation, we have designed an innovative chemical intelligent assistant system. This system employs the fine-tuned Mistral NeMo model as one of its primary models and features a mechanism for flexibly invoking various advanced models. This design fully considers the rapid iteration characteristics of large language models, ensuring that the system can continuously leverage the latest and most powerful AI capabilities. A major highlight of this system is its deep integration of professional knowledge and requirements from the chemistry field. By incorporating specialized functions such as molecular visualization, SMILES string processing, and chemical literature retrieval, the system significantly enhances its practical value in chemical research and applications. More notably, through carefully designed mechanisms for knowledge accumulation, skill acquisition, performance evaluation, and group collaboration, the system can optimize its professional abilities and interaction quality to a certain extent.
1 Introduction
Large Language Models (LLMs) stand out as one of the most noteworthy achievements in the field of artificial intelligence in recent years and represent a crucial direction for the development of Artificial General Intelligence (AGI).1,2 Since the introduction of ChatGPT and GPT-4o, Large Language Models (LLMs) and Multimodal Large Language Models (MLLMs) have attracted significant interest due to their versatile abilities in understanding, reasoning, and generating content.3 However, the current state of this technology still presents significant deficiencies and imbalances, including persistent illusions, misaligned values, weak specialization, and the black box effect.2 In this scenario, how to apply Large Language Models (LLMs) to different professional fields has become a current research hotspot.Fine-tuning has a significant effect on improving the performance of LLMs in specific application scenarios, which lays the foundation for LLMs to further promote scientific progress in various fields.4,5 For example, research by Ouyang et al. (2022), Wei et al. (2021), and Sanh et al. (2021) demonstrates that fine-tuning language models on a specific set of tasks significantly enhances their ability to understand and execute instructions.6–8 This method not only reduces the reliance on large datasets but also improves the generalization capabilities of the models. Given the scale of LLMs, a common fine-tuning strategy currently involves adjusting a limited number of parameters while keeping the rest fixed.9 This technique, known as Parameter-Efficient Fine-Tuning (PEFT), selectively tunes a small subset of parameters. PEFT has also gained interest beyond NLP, particularly in the CV community, for fine-tuning large-parameter visual models such as Vision Transformers (ViTs), diffusion models, and visual-language models.4
However, fine-tuning large models still has some drawbacks. For example, this method requires substantial computational resources and data. Fine-tuning large models is also prone to overfitting on small-scale datasets and cannot accurately reflect potential risks (e.g., “hallucinations”), which may introduce latent hazards. Additionally, it cannot update its knowledge base in real time.10 The primary reasons for these drawbacks are that both pre-trained large models and fine-tuned large models use parameter memory to construct a parameterized implicit knowledge base.11 Hybrid models that combine parametric memory and non-parametric (i.e., retrieval-based) memory can address some of these issues.12–14 The Retrieval-Augmented Generation (RAG) technique improves the accuracy and reliability of hybrid model generation by integrating knowledge from external databases (non-parametric memory), especially for knowledge-intensive tasks. This approach also allows for continuous knowledge updates and the integration of domain-specific information. RAG synergizes the intrinsic knowledge of large language models with the extensive dynamic repositories of external databases.15
Furthermore, with the continuous development of LLMs, they are seen as potential sparks for Artificial General Intelligence (AGI), providing hope for the construction of general AI agents.16 Currently, AI agents are considered a crucial step towards achieving AGI, encompassing the potential for a wide range of intelligent activities.17–19 In many real-world tasks, the capabilities of agents can be enhanced by constructing multiple cooperative agents.20 Studies have shown that multi-agent systems help encourage divergent thinking21 (Liang et al., 2023), improve factuality and reasoning abilities22 (Du et al., 2023), and provide verification23 (Wu et al., 2023). These features have garnered widespread attention. Currently, the general frameworks for constructing LLM applications with multiple agents include AutoGen,20 crewAI,38 Langchain39 and others. Intelligent agents based on large language models (LLMs) are increasingly permeating various aspects of human production and daily life. However, designing artificial intelligence agents with self-evolution capabilities has become a current research hotspot. For example, Li et al.24 proposed an evolutionary framework for agent evolution and arrangement called EvoluaryAgent. Qian et al.25 proposed a general strategy for inter-task agent self-evolution based on Investigation-Consolidation-Exploitation (ICE).
These artificial intelligence technologies will provide a new paradigm for scientific research and open new avenues for scientific innovation, thereby significantly accelerating the pace of scientific discoveries. The close collaboration between artificial intelligence technologies and scientists heralds the advent of a new era of scientific exploration and technological breakthroughs.26,27
In recent years, despite the rapid development of artificial intelligence technology, especially the emergence of large language models, its application in the field of chemistry has not yet been widely popularized. As an important productivity tool, artificial intelligence not only improves work efficiency but also provides a new paradigm for scientific research. For chemistry, a discipline with a long history, how to combine with this advanced productivity tool to breathe new life into the field has become an important topic facing the new generation of chemists. This research aims to address this challenge by developing a dedicated intelligent assistance system for the field of chemistry through the integration of cutting-edge AI technologies. Specifically, we first collected and organized a large amount of data from the field of chemical science to fine-tune mainstream open-source large language models. Secondly, we designed a set of evaluation systems specifically for the chemistry field to detect the performance of the fine-tuned models and select the best-performing model from them. On this basis, we developed an AI assistant for the chemistry field. This system integrates multi-agent architecture, retrieval-augmented generation (RAG) technology, online search functionality, and an interactive user interface. It not only provides an innovative platform for chemical research and education but also offers valuable research opportunities for exploring multi-agent collaboration in complex systems. By fusing traditional chemical knowledge with cutting-edge AI technology, this system is expected to promote innovative development in the field of chemistry and provide new ideas and tools for solving current scientific and engineering challenges. Fig. 1 illustrates the overall process of this study.
2 Related work
2.1 Fine-tuning LLMs for applications in the field of chemistry
In recent years, with the rapid development of artificial intelligence technology, Large Language Models (LLMs) have been increasingly applied in the field of chemical sciences. Through fine-tuning for specific chemical tasks, these models have demonstrated remarkable potential, bringing new perspectives and methods to chemical research. Currently, significant progress has been made in chemical science research using fine-tuned large language models, covering various aspects from material design to drug discovery. These studies not only showcase the exceptional ability of LLMs in handling complex chemical problems but also provide innovative approaches to addressing long-standing chemical challenges.For example, Kevin Maik Jablonka et al.45 fine-tuned the large language model GPT-3 to perform various tasks in chemistry and materials science, including properties of molecules and materials, as well as chemical reaction outcomes. Zikai Xie et al.46 demonstrated the effectiveness of fine-tuned GPT-3 in predicting electronic and functional properties of organic molecules. Shifa Zhong et al.47 developed quantitative structure–activity relationship (QSAR) models for water pollutant activity/properties by fine-tuning GPT-3 models. Seongmin Kim et al.48 evaluated the effectiveness of pre-trained and fine-tuned large language models (LLMs) in predicting the synthesizability of inorganic compounds and selecting synthetic precursors. Results showed that fine-tuned LLMs performed comparably, and sometimes superiorly, to recent custom machine learning models in these tasks, while requiring less user expertise, cost, and time to develop.
These research findings conclusively demonstrate that fine-tuning LLMs can significantly enhance their application breadth and effectiveness in the field of chemical sciences. This approach not only provides powerful tools for chemical research but also promises to accelerate innovation in chemical sciences, offering new ideas and methods for solving complex chemical problems. As technology continues to advance, we can anticipate that fine-tuned LLMs will play an increasingly important role in the field of chemical sciences, driving chemical research towards deeper and more precise directions.
2.2 AI agents in the field of chemistry
Although large language models (LLMs) have demonstrated excellent performance in tasks across multiple domains, they face challenges in chemistry-related problems and lack the ability to access external knowledge sources, limiting their practicality in scientific applications. To address these deficiencies, researchers have conducted relevant explorations.For example, Kevin Maik Jablonka et al.49 developed ChemCrow, an LLM chemical agent designed to complete chemistry tasks such as organic synthesis, drug discovery, and materials design. By integrating multiple expert-designed chemical tools and using GPT-4 as the LLM, they enhanced the performance of LLMs in the field of chemistry and demonstrated new capabilities. Daniil A. Boiko et al.50 reported Coscientist, a GPT-4-powered artificial intelligence system capable of autonomously designing, planning, and executing complex scientific experiments. Coscientist leverages large language models combined with tools such as internet searches, document retrieval, code execution, and experimental automation. Andrew D. McNaughton et al.51 introduced a system called CACTUS (Chemistry Agent Connecting Tool-Usage to Science), which is an intelligent agent based on large language models (LLMs) designed to enhance advanced reasoning and problem-solving capabilities in the fields of chemistry and molecular discovery by integrating cheminformatics tools.
These research findings demonstrate that AI agents, by expanding the functionality of large language models, enable their more extensive application in the field of chemistry.
3 Materials and methods
3.1 Dataset-related work
This work collects and organizes datasets sourced from the studies of Yin Fang et al.44 and Tong Xie et al.43 For example, Yin Fang et al.44 constructed a dataset called Mol-Instructions, which includes three key components: molecule-oriented instructions, protein-oriented instructions, and biomolecular text instructions. Each component aims to enhance the performance of LLMs in related fields. The data are sourced from multiple authorized biochemical databases, extracted and selected through data mining and AI-assisted generation techniques. Strict quality assurance measures were implemented, including chemical validity verification. Its focus in the chemistry domain is to enhance large language models' (LLMs) understanding and application capabilities in chemistry by providing instruction data specifically targeted at chemical molecules. Here are several key aspects of this dataset's focus on chemistry.For instance, Tong Xie et al.43 constructed a dataset by integrating resources from multiple scientific domains to support natural science research, especially in the fields of physics, chemistry, and materials science. It includes:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 679 science exam questions covering subjects such as physics, chemistry, and biology.
679 science exam questions covering subjects such as physics, chemistry, and biology.
This study collected and organized datasets related to the field of chemical science from the above research for use in this study. Specific details can be found in the ESI†.
3.2 Datasets
During the adjustment of the LLM, various datasets related to chemical sciences were utilized. The fine-tuning data for this work come from the datasets listed in Table 1, with a total of 1.72 million fine-tuning instructions collected and organized. Based on this, two different types of instructions were divided for the fine-tuning training of different large models. Fig. 2 illustrates a representative example of fine-tuned data.| Dataset | Url link | Data format |
|---|---|---|
| ESOL43 | https://github.com/MasterAIEAM/Darwin/blob/main/dataset/ESOL/ESOL.json | Json |
| MoosaviCp43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/MoosaviCp/MoosaviCp.json | Json |
| MoosaviDiversity43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/MoosaviDiversity/MoosaviDiversity.json | Json |
| NagasawaOPV43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/NagasawaOPV/NagasawaOPV.json | Json |
| Chembl43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/chembl/chembl.json | Json |
| matbench_expt_gap43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/matbench_expt_gap/matbench_expt_gap.json | Json |
| matbench_glass43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/matbench_glass/matbench_glass.json | Json |
| matbench_is_metal43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/matbench_is_metal/matbench_is_metal.json | Json |
| matbench_steels43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/matbench_steels/matbench_steels.json | Json |
| Pei43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/Pei/pei.json | Json |
| waterStability43 | https://github.com/MasterAI-EAM/Darwin/blob/main/dataset/waterStability/waterStability.json | Json |
| description_guided_molecule_design44 | https://huggingface.co/datasets/zjunlp/Mol-Instructions/tree/main/data | Json |
| forward_reaction_prediction44 | https://huggingface.co/datasets/zjunlp/Mol-Instructions/tree/main/data | Json |
| molecular_description_generation44 | https://huggingface.co/datasets/zjunlp/Mol-Instructions/tree/main/data | Json |
| reagent_prediction44 | https://huggingface.co/datasets/zjunlp/Mol-Instructions/tree/main/data | Json |
| property_prediction44 | https://huggingface.co/datasets/zjunlp/Mol-Instructions/tree/main/data | Json |
| Retrosynthesis44 | https://huggingface.co/datasets/zjunlp/Mol-Instructions/tree/main/data | Json |
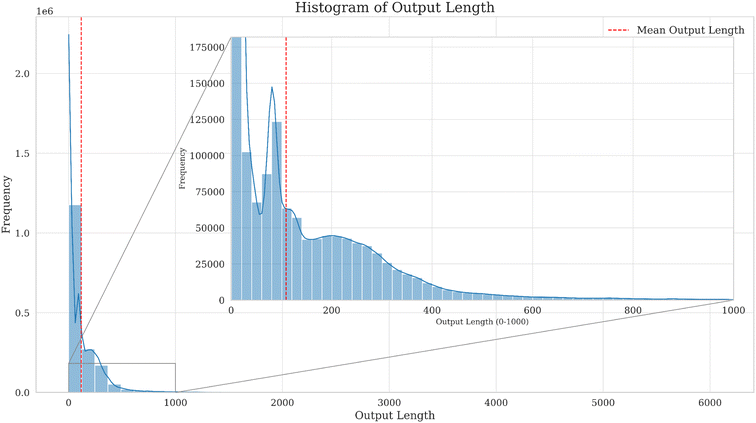
Fig. 3 and 4 show the distribution of output character lengths for the instruction dataset and the usage frequency and types of the 20 most commonly used instructions in this work.
Fig. 3 illustrates the character count (output length) of the output text in the dataset, which exhibits a wide distribution range, covering both short and long texts. The distribution is concentrated in the 0 to 1000 character range. Short texts (texts with fewer characters) appear more frequently, and as the output length increases, the frequency decreases. Kernel Density Estimation (KDE), also known as Parzen's window,28 is one of the most renowned methods for estimating the underlying probability density function of a dataset. The KDE curve provides a smooth estimate of the distribution within this range, aiding in a more intuitive understanding of the text distribution pattern.
The bar chart (Fig. 4) shows the frequency of the 20 most common instructions in the dataset for this study. Among these, “Provide a brief overview of this molecule” and “Provide a description of this molecule” appear significantly more often than other instructions, indicating their prominent role in the dataset. Nonetheless, other types of instructions also appear, demonstrating the diversity of instruction types within the dataset.
3.3 Fine-tuning
In this work, we collected and curated 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 720
720![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313 fine-tuning instructions from the field of chemical science. Using the unsloth29 tool, we fine-tuned open-source large language models including llama-3-8B-Instruct-bnb-4bit, mistral-7B-instruct-v0.3-bnb-4bit, gemma-7B-bnb-4bit, gemma-2-9b-bnb-4bit, Phi-3-mini-4k-instruct, Mistral-Nemo-Instruct-2407-bnb-4bit and Llama-3.1-8B-Instruct-bnb-4bit. We employed the PEFT (Parameter-Efficient Fine-Tuning) method to apply the LoRA (Low-Rank Adaptation) technique for fine-tuning the pre-trained models. The training parameters were configured using SFTTrainer and TrainingArguments. By combining quantization techniques, LoRA technology, and optimized training configurations, we aimed to enhance performance and optimize resource utilization. Table 2 shows parameter settings for the fine-tuning process for LLMs.
313 fine-tuning instructions from the field of chemical science. Using the unsloth29 tool, we fine-tuned open-source large language models including llama-3-8B-Instruct-bnb-4bit, mistral-7B-instruct-v0.3-bnb-4bit, gemma-7B-bnb-4bit, gemma-2-9b-bnb-4bit, Phi-3-mini-4k-instruct, Mistral-Nemo-Instruct-2407-bnb-4bit and Llama-3.1-8B-Instruct-bnb-4bit. We employed the PEFT (Parameter-Efficient Fine-Tuning) method to apply the LoRA (Low-Rank Adaptation) technique for fine-tuning the pre-trained models. The training parameters were configured using SFTTrainer and TrainingArguments. By combining quantization techniques, LoRA technology, and optimized training configurations, we aimed to enhance performance and optimize resource utilization. Table 2 shows parameter settings for the fine-tuning process for LLMs.
| Parameter | Value | Description |
|---|---|---|
| Lora_alpha | 16 | LoRA alpha parameter |
| Max_steps | 60 | Maximum training steps |
| Learning_rate | 2 × 10−4 | Learning rate |
| Weight_decay | 0.01 | Weight decay parameter |
| Seed | 3407 | Random seed |
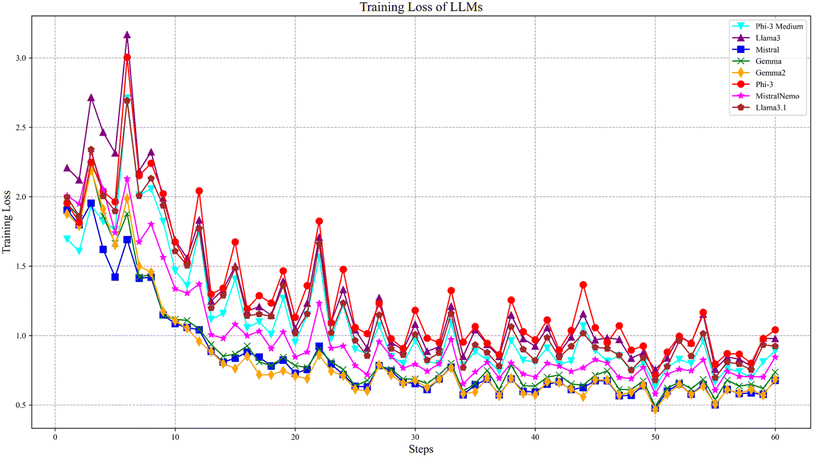
Fig. 5 presents the training loss curve during the training process of LLMs. In the initial phase of training, the loss value is relatively high because the model parameters have not yet been optimized, leading to a significant gap between the predicted results and the actual values. As the training progresses, the model gradually learns and continuously adjusts the parameters, making the predicted results increasingly closer to the actual values. Consequently, the error decreases, and the loss value gradually declines and tends to stabilize.
3.4 Deployment of LLMs (large language models)
After the fine-tuning step in Section 3.3 of the large language model, we employed Ollama for the local deployment and testing of fine-tuned LLMs. Model parameters were set using the Modelfile configuration file. Specifically, the model's temperature was set to 0.8 and the context window size was configured to 8192 tokens. Additionally, three stop markers were defined to control the boundaries of the generated text. The detailed configuration is shown in Fig. 6. After fine-tuning, the four large language models were deployed on a local computer for testing. The four fine-tuned large language models (Llama3-8B, Phi-3-mini, Gemma-7B, and Mistral-7B) were deployed on a local computer with an Intel(R) Core(TM) i5-10210U CPU @ 1.60 GHz (up to 2.11 GHz) and an NVIDIA GeForce MX250 GPU for testing. The two fine-tuned models are tested using Google Colab, with Gemma2-9B tested on a T4 GPU, Phi-3Medium tested on an L4 GPU, Llama3.1-8B tested on a Colab CPU and Mistral NeMo tested on an L4 GPU.3.5 Methods for evaluating the quality of LLM responses
Based on previous research, evaluation after fine-tuning large language models is crucial, as it serves as a key tool for identifying current system limitations and informing the design of more powerful models.30 Therefore, in this work, to assess the performance of different large models after fine-tuning, 100 questions were randomly selected from the dataset for model testing. To evaluate the performance of different models after fine-tuning more objectively, this study specifically designed OptimizedModelEvaluator, an automatic scoring program to evaluate the performance of different models.Different scoring criteria were designed for different questions. Additionally, the evaluator considered some special cases in the field of chemical science, assigning higher weights to key words such as ‘reaction’, ‘mechanism’, 'synthesis', and ‘catalyst’. It also recognizes specific chemical terms (e.g., ‘alkane’, ‘alkene’, and ‘alkyne’), considers conversions between different units when making numerical comparisons (such as kJ to kcal), and applies special processing for questions involving specific concepts such as the LUMO, the HOMO, and orbital energies (comparing the signs (positive or negative) of the extracted answer value and the correct answer value; LUMO and HOMO energies are typically negative, so the correctness of the sign is important). For questions involving MOFs, it pays special attention to key concepts such as ‘linker’, ‘node’, and ‘topology’.
The system employs various methods to evaluate the quality of answers. For numerical problems, it calculates relative errors and assigns corresponding scores. It uses Levenshtein distance31 or simple word set intersections to compute the similarity between answers and standard solutions. BLEU scores32 and ROUGE scores33 are used to assess the quality of generated text and summaries, respectively. The Flesch34 Reading Ease Index is utilized to evaluate text readability. In addition to these methods, the system also incorporates evaluation criteria such as keyword relevance, coherence, conciseness, factual accuracy, and creativity. Fig. 7 presents the scoring criteria for various types of questions.
Through these detailed settings, the evaluator can better assess the model's understanding of concepts related to molecular orbital theory, rather than just simple numerical matching. This enables a comprehensive evaluation of AI models' performance in answering chemistry-related questions, covering multiple dimensions including accuracy, relevance, readability, and creativity. Fig. 8 illustrates the scoring process. (See the ESI† for details).
Based on this classification, we developed a highly customized scoring analysis system, implemented through the OptimizedModelEvaluator class. This system evaluated eight different models, including Llama3, Mistral, Phi-3, Gemma, Gemma2, Phi-3 Medium, MistralNemo, and Llama3.1. Specific scoring criteria and weights were designed for the three main question types: numeric, descriptive, and generate. For numeric type questions, the system focuses on numeric_accuracy (60%), keyword_relevance (20%), and conciseness (20%). The descriptive type considers bleu_score (20%), rouge_scores (20%), keyword_relevance (20%), readability (20%), and coherence (20%). The generate type emphasizes creativity (40%), coherence (30%), and keyword_relevance (30%).
The system also introduced chemistry-specific keyword weights and terminology, assigning different weights to various chemical concepts. For example, reaction, mechanism, and synthesis each account for 0.5 points, while bond, electron, and orbital each account for 0.3 points. Additionally, the system pays special attention to key chemical terms such as alkane, alkene, alkyne, aromatic, nucleophile, and electrophile. To ensure the accuracy of numerical evaluations, the system integrated conversion factors between common units. For instance, 1 kJ = 0.239006 kcal and 1 eV = 96.485 kJ. This carefully designed configuration ensures that the scoring system can accurately capture the characteristics and challenges of different types of questions.
For numeric type questions, the system identifies and extracts values and units, supports the aforementioned unit conversions, and calculates accuracy scores based on relative errors. In terms of keyword relevance scoring, the system uses a predefined keyword_importance dictionary to assign weights to different keywords, while also considering specific chemical terminology.
For descriptive and generate type questions, the system integrates various advanced natural language processing techniques. Text similarity scoring primarily uses Levenshtein distance, with the word set overlap rate as a fallback when unavailable. The system also applies BLEU and ROUGE algorithms to evaluate generated text quality, uses the textstat library to calculate readability, and assesses text coherence based on the word overlap rate between adjacent sentences.
For domain-specific knowledge such as HOMO/LUMO energy levels or MOF structures, the system applies special scoring rules to accurately evaluate these highly specialized chemical concepts. We implemented a complex and refined set of rules in the scoring system. These rules not only consider numerical accuracy but also include unit consistency, relative energy relationships, structural composition, functional properties, and more.
For HOMO/LUMO energy levels, the system first evaluates numerical accuracy, allowing an error range of ±0.1 eV. We also consider unit consistency, prioritizing electron volts (eV) as the standard unit and slightly penalizing answers using non-standard units. Furthermore, the system checks if the relative positions of HOMO and LUMO levels are correct and rewards answers that correctly mention the energy gap.
When evaluating MOF structures, our rules are more comprehensive. The system checks if the answer correctly identifies the metal center, organic linkers, and their connectivity. We also assess descriptions of porosity and specific surface area, as well as the identification and explanation of the MOF's main functions. To encourage more in-depth answers, we provide extra points for mentioning synthesis methods and characterization techniques.
These rules are implemented through the calculate_factual_accuracy method in the OptimizedModelEvaluator class. This method uses regular expressions to extract values and units and combines them with a chemistry knowledge base to provide reference values and expected ranges. The scoring system can dynamically adjust weights based on the depth and accuracy of the provided information.
By implementing these special rules, our scoring system can more accurately evaluate the model's performance in handling complex chemical concepts. This not only improves the accuracy and professionalism of the evaluation but also provides valuable feedback to model developers regarding the model's mastery of specific chemical domain knowledge. This approach allows us to gain a more comprehensive understanding of the capabilities and limitations of large language models in specialized chemical problems, providing important guidance for further improvement and application of these models. Creativity scoring combines uniqueness (degree of difference from standard answers) and coherence, mainly used to evaluate generate type questions.
This comprehensive scoring system is not just a simple word count or hard-coded decision tree, but a complex evaluation tool that integrates multiple techniques and domain knowledge. By preliminarily classifying questions and designing specific scoring criteria and weights for each category, we can more accurately evaluate the model's performance in different types of tasks. This approach enables us to comprehensively and deeply analyze the performance of large language models on complex and diverse chemical problems.
3.6 LLM fine-tuning test results and discussion
This study conducts a comprehensive evaluation of eight fine-tuned large language models: Llama3-8B, Mistral-7B, Phi-3 Mini, Gemma-7B, Gemma2-9B, Phi-3 Medium, Llama3.1 and MistralNemo. Through testing across multiple dimensions, we aim to gain a deep understanding of the performance differences of these models under various tasks and criteria, providing insights for model selection and future optimization directions. Using the automated scoring program introduced in Section 3.5, the fine-tuned models were evaluated with four main metrics: overall score, average performance, multi-dimensional criteria evaluation, and question type classification assessment. Each model was fine-tuned using the same strategy and tested on the same test set (details in the ESI†), ensuring the comparability of the results.The results from Fig. 9, model performance evaluation, show that Mistral NeMo demonstrated the strongest overall performance with an average score of 4.39. The model excelled particularly in descriptive tasks (3.60) while maintaining strong performance in numeric tasks (4.25) and generative tasks (6.24). Mistral and Llama3 follow closely behind, scoring 4.07 and 4.00 respectively, with both performing very similarly. Phi-3 follows with a score of 3.84, showing balanced capabilities across different question types.
Both Mistral and Llama3 performed notably better in generative tasks (Mistral: 6.36 and Llama3: 6.26) compared to descriptive ones (Mistral: 2.25 and Llama3: 2.28). Phi-3 showed particular strength in generative tasks (6.09) and comparative weakness in descriptive ones (2.61).
Notably, Gemma2-9B (3.70 points) shows significant improvement compared to its predecessor Gemma-7B (3.02 points). According to the technical reports,35,37 these gains can be attributed to several key architectural enhancements: First, Gemma2-9B adopts a deeper architecture with 42 transformer layers compared to Gemma-7B's 28 layers, along with an increased model dimension (d_model: 3584 vs. 3072). Second, it introduces novel components including interleaving local-global attentions (with a 4096-token local window and 8192-token global span) and the group-query attention (GQA) mechanism with num_groups = 2. Third, Gemma2 models employ knowledge distillation for training instead of traditional next-token prediction, learning from a larger teacher model on 8 trillion tokens. However, both models still face common challenges in keyword relevance, BLEU score, and ROUGE scores (<0.2), suggesting that while architectural and training advances boost overall capabilities, some fundamental limitations in text generation quality and precision remain.
The iteration from Mistral 7B to Mistral NeMo demonstrates significant architectural advances, scaling up from 7B to 12B parameters while introducing innovations such as the Tekken tokenizer for improved multilingual handling and expanding context length to 128 k tokens. These improvements enhance the model's capabilities across reasoning, instruction following, and multilingual tasks.52,53
We observe that Phi-3-medium (14B parameters), despite its larger capacity with 40 attention heads and 40 layers (embedding dimension 5120), shows more modest improvements on certain benchmarks compared to Phi-3-mini (3.8B parameters, 32 heads, 32 layers, and embedding dimension 3072). This suggests that our current data mixture, while effective for the smaller model architecture, may need further optimization to fully leverage the increased representational capacity of the 14B parameter scale.54
While Llama 3.1 8B incorporated multilingual capabilities and extended context length to 128 K tokens, it scored lower than Llama 3 8B which utilized a comprehensive post-training approach combining supervised fine-tuning, rejection sampling, PPO and DPO. This suggests that the diversity in fine-tuning strategies may play a more crucial role in model performance than expanded linguistic coverage and context length at the 8B parameter scale.36,55,56
Based on the comprehensive evaluation data, the analysis reveals a clear hierarchy in model performance, with Mistral NeMo leading at an average score of 4.39, followed by Mistral (4.07) and Llama3 (4.00). The models demonstrate distinct strengths across different question types, with generative tasks yielding the highest performance scores ranging from 6.2 to 6.4 for top performers. In numeric tasks, models showed moderate capability with scores between 4.02 and 4.25 for the leading models, while descriptive tasks proved most challenging with significantly lower scores, though Mistral NeMo maintained a notable advantage at 3.60 compared to others ranging from 1.97 to 3.01. Looking at specific evaluation criteria, most models exhibited strong creativity (above 0.77) and coherence, with Mistral NeMo particularly excelling in coherence at 0.962. However, all models struggled with keyword relevance, with scores varying across models but generally remaining low. The correlation analysis indicates that numeric accuracy operates largely independently from other metrics, while keyword relevance shows a moderate negative correlation with conciseness (−0.29). These findings suggest that while current models excel at creative and generative tasks, there remains significant room for improvement in precise information extraction and keyword relevance, particularly in descriptive tasks. The substantial variation in performance across different question types also indicates that optimal model selection should be task-dependent rather than assuming that one model will excel universally.
Research findings reveal the significant impact of model iterations on performance improvement, particularly evident in the evolution from Gemma-7B to Gemma2-9B35 and from Mistral-7B to Mistral-Nemo. However, the iteration from Llama3-8B to Llama3.1-8B failed to achieve the expected performance leap, possibly due to different iteration priorities.36 Notably, all tested models face common challenges, especially in keyword relevance and task scoring, highlighting the necessity of introducing additional technologies to address these shortcomings.
Nevertheless, the outstanding performance of these models in creative and generative tasks continues to demonstrate the inherent advantages of large language models in these domains. The test results indicate that fine-tuned large language models can meet researchers' needs to some extent, but still have many limitations, including the inability to update data in real-time, lack of online search capabilities, poor compatibility with specific domains, insufficient response accuracy, and limitations in decision-making for single large models.
Given these limitations exhibited by fine-tuned large language models, this study developed an artificial intelligence assistant for the chemical domain. This system cleverly integrates multi-agent architecture, Retrieval-Augmented Generation (RAG) technology, online search functionality, and a user-friendly interactive interface, aiming to comprehensively address the aforementioned shortcomings and provide researchers with a more intelligent, precise, and practical auxiliary tool.
4 AI agent system for chemistry
This work builds upon the fine-tuning of the aforementioned large language models to design an AI assistant platform specifically tailored for the field of chemistry. The platform integrates multi-agent systems, retrieval-augmented generation, real-time web search, and chemical structure visualization. The system incorporates AI agents with diverse professional backgrounds (such as laboratory directors, senior chemists, safety officers, etc.), simulating a virtual chemistry research team environment. These agents can collaborate and continuously learn to provide researchers with comprehensive and professional support in chemical knowledge, experimental design suggestions, safety guidance, and data analysis. Additionally, the system has the capability to convert chemical structure formulae (SMILES) into visualized images, greatly enhancing the efficiency and intuitiveness of chemical research, education, and team collaboration. The system primarily consists of the following components: a multi-agent system, retrieval-augmented generation (RAG), real-time web search, chemical structure visualization, an agent improvement system and user-friendly interface design.4.1 Multi-agent system
This system is the core architecture of the project, simulating a real chemical team. The system contains five specialized agents, each with a specific role and expertise, together forming a comprehensive and efficient virtual chemical research team. The Lab_Director is responsible for overall task allocation and research direction guidance, ensuring that the team's research direction aligns with the overall goals and coordinating work between agents. The Senior_Chemist provides in-depth chemical knowledge and solutions to complex problems, possessing rich chemical theory and practical experience to handle challenging chemical issues and propose innovative research ideas. The Lab_Manager is responsible for experiment planning and resource management, ensuring the feasibility of experimental plans, managing laboratory resources, optimizing experimental processes, and improving research efficiency. The Safety_Officer ensures that all discussions and suggestions comply with safety standards, focusing on experimental safety, reviewing potential risks of all experimental protocols, and providing safety operation guidance. The Analytical_Chemist focuses on data analysis and instrument use, responsible for interpreting experimental data, providing instrument operation advice, and ensuring data accuracy and reliability. This design allows each agent to have its specific area of expertise, providing in-depth professional knowledge. Agents can complement each other to solve complex problems collaboratively. For example, when the Senior_Chemist proposes an experimental protocol, the Safety_Officer reviews its safety, while the Lab_Manager considers its feasibility. This multi-perspective analysis allows agents with different backgrounds to analyze problems from various angles, providing comprehensive insights. The structure simulates the team dynamics of a real chemistry research group, closely mimicking real team decision-making processes. Each agent in the system is based on a large language model but has specific system prompts to define its role and expertise, and different language models can be substituted to meet the needs of different tasks. AutoGen is used to manage interactions and dialogue flow between agents, adopting a round-robin approach to select speakers, ensuring that each agent has the opportunity to contribute. The above multi-agent design allows the system to analyze and solve chemical problems from multiple perspectives, providing comprehensive insights.AutoGen is an open-source framework for building LLM applications through multi-agent dialogue. In AutoGen, a conversable agent is an entity with a specific role that can send and receive messages to and from other conversable agents, such as starting or continuing a conversation. It maintains its internal context based on the messages sent and received and can be configured to have a range of functionalities, such as being supported by LLMs, tools, or human input. These agents can be implemented through AutoGen's built-in AssistantAgent (powered by GPT-4 for general problem-solving) and UserProxyAgent (configured to gather human input and execute tools).20
(This research can utilize models that have undergone fine-tuning and comprehensive performance testing as the system's response model. All fine-tuned large language models have been uploaded to the Hugging Face platform, allowing researchers to flexibly invoke different models from KANGYONGMA/Chemistry based on specific application scenarios. Additionally, the system supports the use of original base models without fine-tuning to execute tasks, providing greater flexibility and diverse options for research).
4.2 Retrieval-augmented generation (RAG)
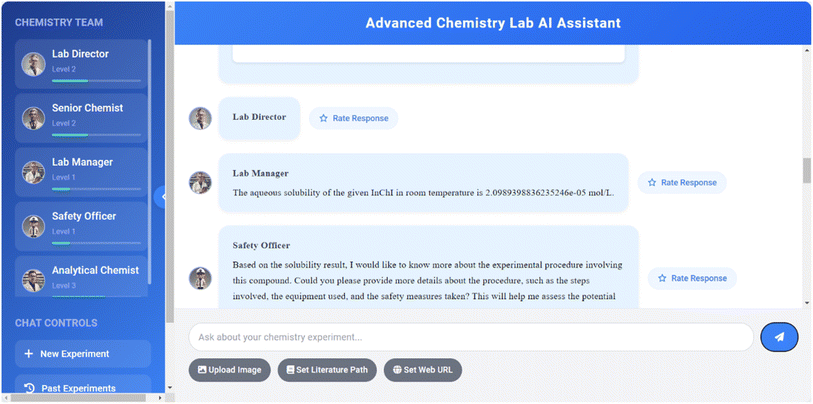
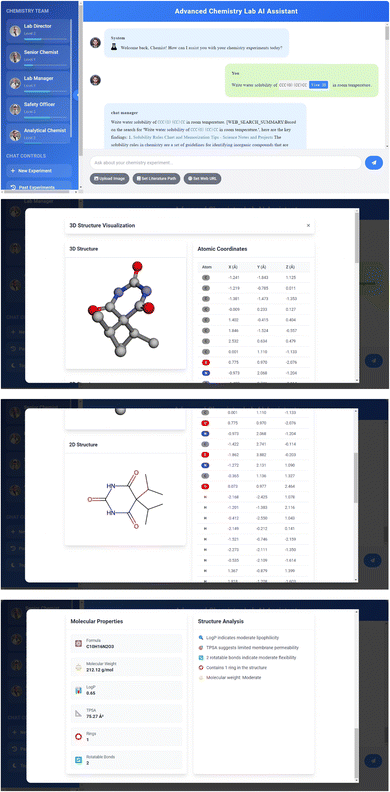
RAG is a core functionality of the system, extending the knowledge base of agents by integrating preloaded chemical literature and experimental data. The RAG workflow includes document loading, text splitting, vector embedding, vector storage, similarity search, context enhancement, and answer generation. This process is implemented using the LangChain library and RetrievalQA chain, significantly improving the accuracy and relevance of answers while reducing the possibility of AI generating false information. RAG technology enables agents to provide answers based on the latest chemical research, cite relevant literature to support views, and associate user queries with existing knowledge bases, thereby greatly enhancing the system's ability to handle complex chemical problems and provide more precise and relevant information. Fig. 10 and 11 demonstrate the AI Agent system's RAG-based responses using local documents.The above examples demonstrate the capability of a Retrieval-Augmented Generation (RAG) based intelligent agent system in accurately answering questions about the water solubility of chemical compounds. The system precisely answered the water solubility of two compounds at room temperature: CCC(O)(CC)CC at 0.14125375446227545 mol L−1, and a compound with a complex InChI representation at 2.0989398836235246 × 10−5 mol L−1. This high-precision response highlights the advantages of RAG technology in cheminformatics applications, especially in tasks requiring precise numerical outputs, where it outperforms traditional fine-tuned large language model approaches. The application of RAG functionality enables the system to retrieve and provide accurate numerical information. Fig. 12 illustrates the Q&A test results based on the RAG.
4.3 Real-time web search
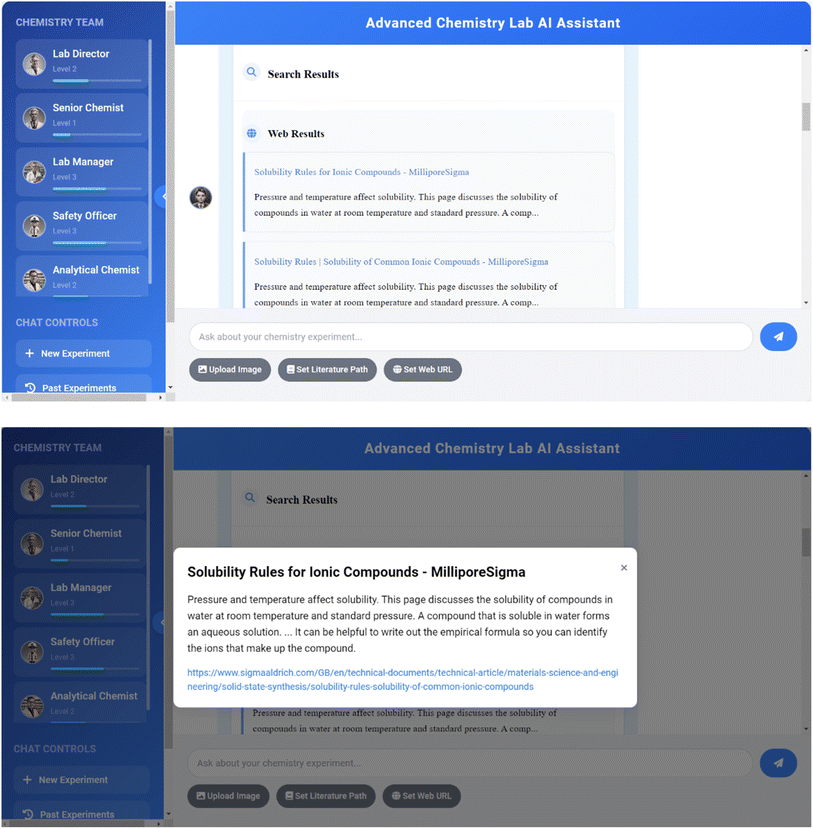
Another important feature of the system is its ability to perform real-time web searches by integrating the Tavily search API40 to supplement the preloaded knowledge base. The workflow of this feature includes query analysis, API calls, result processing, and information integration. The system uses the requests41 library to send API requests and implements error handling and retry mechanisms to ensure stability. This feature allows agents to access the latest chemical research and discoveries, supplement information that may be missing from the preloaded database, and significantly improve the system's ability to answer current affairs questions. By combining preloaded data and real-time search, the system can provide users with comprehensive, up-to-date, and accurate chemical information, excelling particularly in handling emerging research, the latest discoveries, or real-time data-related issues. Fig. 13 showcases the system's real-time web search summarization.The project not only integrates advanced online search functionality but is also equipped with an intelligent summarization system that significantly enhances information retrieval capabilities. The project employs a multi-layered processing architecture that intelligently merges and refines web search results with knowledge base data to present users with precise and concise information summaries. Notably, the search results go beyond simple text summaries by incorporating interactive design elements. Specifically, key content within the summaries includes corresponding hyperlinks, allowing users to trace back to original information sources with just a click. This design enables researchers to conveniently access primary sources and quickly verify the accuracy of search content.
4.4 Chemical structure visualization
This feature greatly enhances the system's interactivity and intuitiveness when discussing chemical structures through comprehensive molecular visualization and analysis. The system converts SMILES strings into both 2D and interactive 3D molecular structures, while also providing detailed molecular property analysis. The visualization process leverages RDKit42 for 2D representation and basic molecular structure display, while utilizing py3Dmol57 for interactive 3D visualization that allows users to manipulate and examine molecular structures from any angle. The system automatically generates optimized 3D conformers using MMFF force field58–64 optimization with explicit hydrogen atoms, enabling accurate structural representation. Beyond visualization, the system calculates and displays a comprehensive set of molecular properties including core molecular descriptors (molecular formula, exact molecular weight, atom and bond counts), physicochemical properties (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for lipophilicity assessment and TPSA for polar surface area), and structural features (rotatable bonds, ring count, and formal charge). The interactive interface provides real-time 3D structure manipulation with zoom, rotation and pan capabilities, along with atom-by-atom positional data and element-specific coloring. The system automatically validates and processes SMILES strings within conversations, offering integrated “View 3D” buttons for instant structure visualization through a performance-optimized viewer with anti-aliasing and customizable display styles. This implementation significantly enhances the visual understanding of chemical concepts and improves the efficiency of discussing complex molecular structures through the integration of both structural visualization and quantitative property analysis. The system's ability to automatically detect and process SMILES strings while providing instant access to both 2D and 3D structural representations, along with detailed molecular properties, creates a seamless and informative environment particularly valuable for chemical education, drug discovery research, and molecular design applications. This enhanced functionality provides chemistry researchers with a comprehensive molecular interaction experience, combining visual inspection capabilities with quantitative chemical property analysis in a single, integrated interface, Fig. 14.
P for lipophilicity assessment and TPSA for polar surface area), and structural features (rotatable bonds, ring count, and formal charge). The interactive interface provides real-time 3D structure manipulation with zoom, rotation and pan capabilities, along with atom-by-atom positional data and element-specific coloring. The system automatically validates and processes SMILES strings within conversations, offering integrated “View 3D” buttons for instant structure visualization through a performance-optimized viewer with anti-aliasing and customizable display styles. This implementation significantly enhances the visual understanding of chemical concepts and improves the efficiency of discussing complex molecular structures through the integration of both structural visualization and quantitative property analysis. The system's ability to automatically detect and process SMILES strings while providing instant access to both 2D and 3D structural representations, along with detailed molecular properties, creates a seamless and informative environment particularly valuable for chemical education, drug discovery research, and molecular design applications. This enhanced functionality provides chemistry researchers with a comprehensive molecular interaction experience, combining visual inspection capabilities with quantitative chemical property analysis in a single, integrated interface, Fig. 14.
4.5 Agent improvement system
The adaptive learning system implements a basic framework with four interconnected components: knowledge enrichment, capability enhancement, performance assessment, and refinement mechanisms. While still in its early stages, the system takes initial steps toward knowledge expansion and skill development, allowing agents to gradually build upon their fundamental capabilities through user interactions and feedback.The framework consists of two primary classes: ChemistryAgent and ChemistryLab. The ChemistryAgent class maintains a developing knowledge repository and growing skill set through the knowledge_base and skills attributes, working to expand its capabilities via the learn() and acquire_skill() methods. A preliminary performance tracking system has been implemented through history-based assessment, with the evaluate_performance() method beginning to analyze effectiveness based on user feedback.
The refinement process, managed by the improve() and refine_skills() methods, represents early efforts toward developing new capabilities and refining existing ones. The system makes initial attempts to identify potential areas for enhancement by examining interaction patterns and user responses. At the group level, the ChemistryLab class introduces basic knowledge sharing among agents and implements foundational assessment cycles.
This architecture takes preliminary steps toward enabling incremental adjustments based on interactions and feedback, aiming to gradually enhance its domain expertise and interaction quality in chemistry-related discussions. While the current design creates a basic responsive framework that shows potential for adapting to user needs, it acknowledges substantial room for improvement across all aspects. The user feedback interface, shown in Fig. 15, provides initial support for ongoing refinement of the system's developing capabilities.
Through basic mechanisms including knowledge base expansion, skill development, and feedback incorporation, agents work toward building their understanding of chemical concepts and problem-solving approaches. This measured approach to capability enhancement represents early progress while acknowledging the significant work still needed to achieve more sophisticated and comprehensive functionality. The system remains in its nascent stages, with considerable opportunities for advancement in areas such as response accuracy, contextual understanding, and adaptive learning mechanisms.
4.6 User-friendly interface design
The project includes an intuitive web interface that can display real-time conversations between agents, agent status, and feedback mechanisms, providing a better interactive experience.4.7 Functionality expansion
During the system design phase, the team fully considered the potential impact of model updates and iteration and therefore reserved corresponding upgrade and development space. Fig. 16 demonstrates the image recognition capabilities after the integration of multi-modal large models, which provides an important foundation for expanding more functionalities in the future. This extensible architecture also creates opportunities for incorporating advanced inference-time computation strategies, such as chain-of-thought prompting and Monte Carlo tree search, which could enhance the system's reasoning capabilities in complex chemical analysis tasks. While balancing real-time performance with computational resources remains a challenge, our multi-agent framework is well-positioned to accommodate such future optimizations, particularly in tasks requiring sophisticated chemical reasoning and analysis.The system's architecture leverages the capabilities of large language models through a flexible model-calling mechanism that can integrate different advanced models as they become available. The implementation incorporates specialized chemistry domain functions, including molecular visualization and SMILES string processing, to address specific requirements in chemical research. Fig. 17 illustrates the structure of the AI agents within the chemistry system. The agents' prompts can be found in the ESI.†
The system's effectiveness stems from its integrated design combining language model capabilities with domain-specific chemical tools. Through structured knowledge organization, targeted skill implementation, performance monitoring, and coordinated agent interactions, it aims to provide reliable support for chemical research tasks. This modular approach allows for systematic updates and refinements as underlying technologies advance, helping to maintain consistent and efficient assistance for complex chemical problems.
5 Conclusion
This study utilized 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 720
720![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313 instruction data points from the field of chemical science to fine-tune 8 mainstream open-source large language models, including Llama3-8B, Mistral-7B, Phi-3 Mini, Gemma-7B, Gemma2-9B, Phi-3 Medium, Llama3.1, and MistralNemo. Through an automatic scoring program specifically designed to evaluate the quality of responses from large language models in the chemistry domain, the Mistral NeMo model demonstrated the most outstanding performance, achieving a score of 4.39 points, surpassing other models. Based on these research results, an innovative chemical intelligent assistant system was designed. This system can utilize fine-tuned models as its primary models and call upon different large models according to task requirements. Furthermore, the system deeply integrates professional knowledge and requirements from the chemistry field, featuring specialized functionalities such as molecular visualization, SMILES string processing, and chemical literature retrieval. Through the integration of knowledge bases, continuous performance monitoring, and interactive feedback mechanisms, the system shows potential for gradual improvement in its professional capabilities and response quality. While still in its early stages with considerable room for enhancement, these initial steps suggest promising directions for developing more effective AI assistance tools in chemistry applications. The current implementation, though requiring further refinement, represents an exploratory effort toward better supporting chemical research and analysis.
313 instruction data points from the field of chemical science to fine-tune 8 mainstream open-source large language models, including Llama3-8B, Mistral-7B, Phi-3 Mini, Gemma-7B, Gemma2-9B, Phi-3 Medium, Llama3.1, and MistralNemo. Through an automatic scoring program specifically designed to evaluate the quality of responses from large language models in the chemistry domain, the Mistral NeMo model demonstrated the most outstanding performance, achieving a score of 4.39 points, surpassing other models. Based on these research results, an innovative chemical intelligent assistant system was designed. This system can utilize fine-tuned models as its primary models and call upon different large models according to task requirements. Furthermore, the system deeply integrates professional knowledge and requirements from the chemistry field, featuring specialized functionalities such as molecular visualization, SMILES string processing, and chemical literature retrieval. Through the integration of knowledge bases, continuous performance monitoring, and interactive feedback mechanisms, the system shows potential for gradual improvement in its professional capabilities and response quality. While still in its early stages with considerable room for enhancement, these initial steps suggest promising directions for developing more effective AI assistance tools in chemistry applications. The current implementation, though requiring further refinement, represents an exploratory effort toward better supporting chemical research and analysis.
Data availability
The complete implementation code for this work is publicly accessible through our GitHub repository (https://github.com/KangyongMa/GVIM). The trained models and associated datasets are hosted on Hugging Face (https://huggingface.co/KANGYONGMA). For long-term preservation and reproducibility, all models, code, and datasets have been archived on Zenodo (https://doi.org/10.5281/zenodo.14609584).Author contributions
Kangyong Ma was responsible for the conception and design of this study. He conducted the data analysis and interpretation. He wrote the original draft of the manuscript and created the visualizations.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
This work involves the following AI technologies: Llama3-8B, Mistral-7B, Phi-3 Mini, Gemma-7B, Gemma2-9B, Phi-3 Medium, Llama3.1, and MistralNemo. The aforementioned open-source large language models were used for fine-tuning tests in this work. Tavily Search AI was used for online searches, and sentence-transformers/all-mpnet-base-v2 was used for RAG (Retrieval-Augmented Generation). Additionally, Claude 3.5 Sonnet was used to address code issues encountered in this research, assist in developing the web UI interface, optimize the multi-agent framework, and expand multi-agent tools. The agent avatar in this work was generated by Stable Diffusion 3. The manuscript was polished using Claude 3.5 Sonnet and ChatGPT-4o. In this study, Claude 3.5 Sonnet and ChatGPT-4o were utilized to alleviate the repetitive labor in research and writing, thereby reducing the workload of human scientists. We are grateful for the assistance of these AI technologies in completing this work. I would like to express my gratitude to the REPLICATE team for their support in providing a $100 voucher. We would like to express our special gratitude to Groq for providing free API access. This work utilized the LLaMA-3 70B and Mixtral-8x7B models on the Groq platform, which significantly advanced our research progress. This work has no financial support.References
- E. Pavlick, Symbols and grounding in large language models, Philos. Trans.: Math., Phys. Eng. Sci., 2023, 381(2251), 20220041 Search PubMed.
- W. Chen, L. Yan-Yi and G. Tie-Zheng, et al., Systems engineering issues for industry applications of large language model, Appl. Soft Comput., 2024, 151, 111165 CrossRef.
- H. Xiao, F. Zhou, X. Liu, et al., A comprehensive survey of large language models and multimodal large language models in medicine, arXiv, 2024, preprint, arXiv:2405.08603, DOI:10.48550/arXiv.2405.08603.
- Z. Han, C. Gao, J. Liu, et al., Parameter-efficient fine-tuning for large models: a comprehensive survey, arXiv, 2024, preprint, arXiv:2403.14608, DOI:10.48550/arXiv.2403.14608.
- M. Livne, Z. Miftahutdinov and E. Tutubalina, et al., Nach0: multimodal natural and chemical languages foundation model, Chem. Sci., 2024, 15, 8380–8389 RSC.
- L. Ouyang, J. Wu, J. Xu, et al., Training language models to follow instructions with human feedback, arXiv, 2022, preprint, arXiv:2203.02155, DOI:10.48550/arXiv.2203.02155.
- J. Wei, M. Bosma, V. Y. Zhao, et al., Finetuned language models are zero-shot learners, arXiv, 2022, preprint, arXiv:2109.01652, DOI:10.48550/arXiv.2109.01652.
- V. Sanh, A. Webson, C. Raffel, et al., Multitask prompted training enables zero-shot task generalization, arXiv, 2022, preprint, arXiv:2110.08207, DOI:10.48550/arXiv.2110.08207.
- B. Lester, R. Al-Rfou, N. Constant, The power of scale for parameter-efficient prompt tuning, arXiv, 2021, preprint, arXiv:2104.08691, DOI:10.48550/arXiv.2104.08691.
- H. Nori, N. King, S. M. Mckinney, et al., Capabilities of gpt-4 on medical challenge problems, arXiv, 2023, preprint, arXiv:2303.13375, DOI:10.48550/arXiv.2303.13375.
- P. Lewis, E. Perez, A. Piktus, et al., Retrieval-augmented generation for knowledge-intensive nlp tasks, arXiv, 2021, preprint, arXiv:2005.11401, DOI:10.48550/arXiv.2005.11401.
- K. Guu, K. Lee, Z. Tung, et al., Realm: retrieval-augmented language model pre-training, arXiv, 2020, preprint, arXiv:2002.08909, DOI:10.48550/arXiv.2002.08909.
- S. D. Vladimir Karpukhin BOG, S. E. C. Ledell Wu, Dense passage retrieval for open-domain question answering, arXiv, 2020, preprint, arXiv:2004.04906, DOI:10.48550/arXiv.2004.04906.
- F. Petroni, P. Lewis, A. Piktus, et al., How context affects language models' factual predictions, arXiv, 2020, preprint, arXiv:2005.04611, DOI:10.48550/arXiv.2005.04611.
- Y. Gao, Y. Xiong, X. Gao, et al., Retrieval-augmented generation for large language models: a survey, arXiv, 2024, preprint, arXiv:2312.10997, DOI:10.48550/arXiv.2312.10997.
- Z. Xi, W. Chen, X. Guo, et al., The rise and potential of large language model based agents: a survey, arXiv, 2023, preprint, arXiv:2309.07864, DOI:10.48550/arXiv.2309.07864.
- M. J. Wooldridge and N. R. Jennings, Intelligent agents: theory and practice, Knowl. Eng. Rev., 1995, 10(2), 115–152 CrossRef.
- Y. Shoham, Agent oriented programming, in Knowledge Representation and Reasoning Under Uncertainty, Logic at Work [International Conference Logic at Work, Amsterdam, The Netherlands, December 17-19, 1992]. Vol 808 of Lecture Notes in Computer Science, ed. Masuch M., Pólos L.Springer, 1992, pp. 123–129 Search PubMed.
- M. Hutter, Universal artificial intelligence: Sequential decisions based on algorithmic probability, Springer Science & Business Media, 2004 Search PubMed.
- Q. Wu, G. Bansal, J. Zhang, et al., Autogen: enabling next-gen llm applications via multi-agent conversation, arXiv, 2023, preprint, arXiv:2308.08155, DOI:10.48550/arXiv.2308.08155.
- L. Tian, Z. He, W. Jiao, et al., Encouraging divergent thinking in large language models through multi-agent debate. arXiv, 2023, preprint, arXiv:2305.19118, DOI:10.48550/arXiv.2305.19118.
- Y. Du, S. Li, A. Torralba, et al., Improving factuality and reasoning in language models through multiagent debate, arXiv, 2023, preprint, arXiv:2305.14325, DOI:10.48550/arXiv.2305.14325.
- Y. Wu, F. Jia, S. Zhang, et al., An empirical study on challenging math problem solving with gpt-4.arXiv, 2023, preprint, arXiv:2306.01337, DOI:10.48550/arXiv.2306.01337.
- Li T. S. Q. C. Shimin, Agent alignment in evolving social norms, arXiv, 2024, preprint, arXiv:2401.04620, DOI:10.48550/arXiv.2401.04620.
- S. L. Y. Q. Cheng Qian, M. Sun, Investigate-consolidate-exploit: a general strategy for inter-task agent self-evolution, arXiv, 2024, preprint, arXiv:2401.13996, DOI:10.48550/arXiv.2401.13996.
- K. M. Merz, G. Wei and F. Zhu, Editorial: harnessing the power of large language model-based chatbots for scientific discovery, J. Chem. Inf. Model., 2023, 63(17), 5395 CrossRef CAS PubMed.
- C. Stokel-Walker and R. Van Noorden, What ChatGPT and generative AI mean for science, Nature, 2023, 614(7947), 214–216 CrossRef CAS PubMed.
- Y. C. Chen, A tutorial on kernel density estimation and recent advances, Biostat. Epidemiol., 2017, 1(1), 161–187 CrossRef.
- D. Han, M. Han and Unsloth Team, Unsloth, 2023, http://github.com/unslothai/unsloth Search PubMed.
- Y. Chang, X. Wang, J. Wang, Y. Wu, L. Yang and K. Zhu, et al., A Survey on Evaluation of Large Language Models, ACM Trans. Intell. Syst. Technol., 2024, 15(3), 1–45, DOI:10.1145/3641289.
- P. Coates, F. Breitinger, Identifying document similarity using a fast estimation of the levenshtein distance based on compression and signatures, arXiv, 2023, preprint, arXiv:2307.11496, DOI:10.48550/arXiv.2307.11496.
- E. Reiter, A Structured Review of the Validity of BLEU, J. Comput. Linguist., 2018, 44(3), 393–401, DOI:10.1162/coli_a_00322.
- M. Zhang, C. Li and M. Wan, et al., Rouge-sem: better evaluation of summarization using rouge combined with semantics, Expert Syst. Appl., 2024, 237, 121364 CrossRef.
- J. S. Hershenhouse, D. Mokhtar and M. B. Eppler, et al., Accuracy, readability, and understandability of large language models for prostate cancer information to the public, Prostate Cancer Prostatic Dis., 2024 DOI:10.1038/s41391-024-00826-y.
- Gemma Team, Gemma 2: Improving Open Language Models at a Practical Size, in International Conference on Learning Representations (ICLR), OpenReview.net, 2024 Search PubMed.
- Llama Team, AI @ Meta. The Llama 3 Herd of Models, 2024, available from: https://llama.meta.com/.
- Gemma Team, Gemma: Open Models Based on Gemini Research and Technology, arXiv, 2024, preprint, arXiv:2403.08295v4, DOI:10.48550/arXiv.2403.08295.
- CrewAI from crewAI - the platform for Multi AI agent systems.
- H. Chase, LangChain LLM App Development Framework. 2023, available from: https://langchain.com/.
- Tavily Search API, available from: https://tavily.com/.
- K. Reitz. Requests: HTTP for Humans, available from: https://requests.readthedocs.io/en/.
- V. F. Scalfani, V. D. Patel and A. M. Fernandez, Visualizing chemical space networks with RDKit and NetworkX, J. Cheminf., 2022, 14, 87 Search PubMed.
- T. Xie, Y. Wan, W. Huang, et al., DARWIN Series: Domain Specific Large Language Models for Natural Science, arXiv, 2023, preprint, arXiv:2308.13565, DOI:10.48550/arXiv.2308.13565.
- Y. Fang, X. Liang, N. Zhang, et al., Mol-Instructions: A Large-Scale Biomolecular Instruction Dataset for Large Language Models, in ICLR, OpenReview.net, 2024 Search PubMed.
- K. M. Jablonka, P. Schwaller and A. Ortega-Guerrero, et al., Leveraging large language models for predictive chemistry, Nat. Mach. Intell., 2024, 6, 161–169, DOI:10.1038/s42256-023-00788-1.
- Z. Xie, X. Evangelopoulos, Ö. H. Omar, A. Troisi, A. I. Cooper and L. Chen, Fine-tuning GPT-3 for machine learning electronic and functional properties of organic molecules, Chem. Sci., 2024, 15, 500–510, 10.1039/d3sc04610a.
- S. Zhong and X. Guan, Developing Quantitative Structure–Activity Relationship (QSAR) Models for Water Contaminants’ Activities/Properties by Fine-Tuning GPT-3 Models, Environ. Sci. Technol. Lett., 2023, 10, 872–877, DOI:10.1021/acs.estlett.3c00599.
- S. Kim, Y. Jung and J. Schrier, Large Language Models for Inorganic Synthesis Predictions, J. Am. Chem. Soc., 2024, 146(29), 19654–19659, DOI:10.1021/jacs.4c05840.
- A. M. Bran, S. Cox, O. Schilter, C. Baldassari, A. D. White and P. Schwaller, Augmenting large language models with chemistry tools, Nat. Mach. Intell., 2024, 6, 525–535, DOI:10.1038/s42256-024-00832-8.
- D. A. Boiko, R. MacKnight and B. Kline, et al., Autonomous chemical research with large language models, Nature, 2023, 624, 570–578, DOI:10.1038/s41586-023-06792-0.
- A. D. McNaughton, G. Ramalaxmi, A. Kruel, C. R. Knutson, R. A. Varikoti, N. Kumar, CACTUS: Chemistry Agent Connecting Tool-Usage to Science, arXiv, 2024, preprint, arXiv:2405.00972, DOI:10.48550/arXiv.2405.00972.
- Mistral AI team. Mistral nemo, 2024, available from: https://mistral.ai/news/mistral-nemo/.
- A. Q. Jiang, A. Sablayrolles, A. Mensch, et al., Mistral 7B, arXiv, 2023, preprint, arXiv:2310.06825, DOI:10.48550/arXiv.2310.06825.
- M. Abdin, J. Aneja, A. Salim, et al., Phi-3 Technical Report: A Highly Capable Language Model Locally on Your Phone, arXiv, 2024, preprint, arXiv:2404.14219v4, DOI:10.48550/arXiv.2404.14219.
- Meta Large Language Model. Introducing Llama 3.1: Our most capable models to date, available from: https://ai.meta.com/blog/meta-llama-3-1/.
- Meta Large Language Model. Introducing Meta Llama 3: The most capable openly available LLM to date, available from: https://ai.meta.com/blog/meta-llama-3/.
- N. Rego and D. Koes, 3Dmol.js: molecular visualization with WebGL, Bioinformatics, 2015, 31(8), 1322–1324, DOI:10.1093/bioinformatics/btu829.
- T. A. Halgren, Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94, J. Comput. Chem., 1996, 17, 490–519 CrossRef CAS.
- T. A. Halgren, Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions, J. Comput. Chem., 1996, 17, 520–552 CrossRef CAS.
- T. A. Halgren, Merck molecular force field. III. Molecular geometries and vibrational frequencies for MMFF94, J. Comput. Chem., 1996, 17, 553–586 CrossRef CAS.
- T. A. Halgren and R. B. Nachbar, Merck molecular force field. IV. Conformational energies and geometries for MMFF94, J. Comput. Chem., 1996, 17, 587–615 CAS.
- T. A. Halgren, Merck molecular force field. V. Extension of MMFF94 using experimental data, additional computational data, and empirical rules, J. Comput. Chem., 1996, 17, 616–641 CrossRef CAS.
- T. A. Halgren, MMFF VI. MMFF94s option for energy minimization studies, J. Comput. Chem., 1999, 20, 720–729 CrossRef CAS PubMed.
- T. A. Halgren and V. I. I. MMFF, Characterization of MMFF94, MMFF94s, and other widely available force fields for conformational energies and for intermolecular-interaction energies and geometries, J. Comput. Chem., 1999, 20, 730–748 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00398e |
| This journal is © The Royal Society of Chemistry 2025 |