Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Improving structural plausibility in diffusion-based 3D molecule generation via property-conditioned training with distorted molecules†
Lucy
Vost
 a,
Vijil
Chenthamarakshan
a,
Vijil
Chenthamarakshan
 b,
Payel
Das
b and
Charlotte M.
Deane
b,
Payel
Das
b and
Charlotte M.
Deane
 *a
*a
aDepartment of Statistics, University of Oxford, Oxford, UK. E-mail: deane@stats.ox.ac.uk
bIBM Research, Yorktown Heights, New York, USA
First published on 24th March 2025
Abstract
Traditional drug design methods are costly and time-consuming due to their reliance on trial-and-error processes. As a result, computational methods, including diffusion models, designed for molecule generation tasks have gained significant traction. Despite their potential, they have faced criticism for producing physically implausible outputs. As a solution to this problem, we propose a conditional training framework resulting in a model capable of generating molecules of varying and controllable levels of structural plausibility. This framework consists of adding distorted molecules to training datasets, and then annotating each molecule with a label representing the extent of its distortion, and hence its quality. By training the model to distinguish between favourable and unfavourable molecular conformations alongside the standard molecule generation training process, we can selectively sample molecules from the high-quality region of learned space, resulting in improvements in the validity of generated molecules. In addition to the standard two datasets used by molecule generation methods (QM9 and GEOM), we also test our method on a druglike dataset derived from ZINC. We use our conditional method with EDM, the first E(3) equivariant diffusion model for molecule generation, as well as two further models—a more recent diffusion model and a flow matching model—which were built off EDM. We demonstrate improvements in validity as assessed by RDKit parsability and the PoseBusters test suite; more broadly, though, our findings highlight the effectiveness of conditioning methods on low-quality data to improve the sampling of high-quality data.
1 Introduction
Drug design involves complex optimisation steps to obtain molecules that achieve desired biological responses. Traditional methods rely on trial-and-error, leading to high costs and limited productivity.1 Computational approaches, especially deep learning models, aim to reduce costs and expedite processes by reducing failures. One way that such models aim to do this is by generating molecules with desirable properties, particularly in terms of binding to their target. To achieve this, a model must first master the fundamental task of generating structurally viable molecules.While many models historically operated in 1D or 2D space,2–4 focus has recently shifted towards developing models capable of directly outputting both atom types and coordinates in 3D. Autoregressive models were once prominent in this domain, generating 3D molecules by adding atoms and bonds iteratively.5–7 However, such models suffer from an accumulation of errors during the generation process and do not fully capture the complexities of real-world scenarios due to their sequential nature, potentially losing global context.8,9 To address these limitations, recent studies have turned to diffusion models, which iteratively denoise data points sampled from a prior distribution to generate samples. Unlike autoregressive models, diffusion-based methods can simultaneously model local and global interactions between atoms. Nevertheless, diffusion in molecule generation has faced criticism for yielding implausible outputs.10,11 There have been ongoing efforts to improve the performance of models trained on small molecules such as those found in the QM9 dataset, and as such the models currently available are capable of reliably generating molecules of this size.12–16 However, achieving success in generating larger molecules, as encountered in datasets like GEOM,17 remains challenging without incorporating additional techniques such as energy minimisation or docking.18
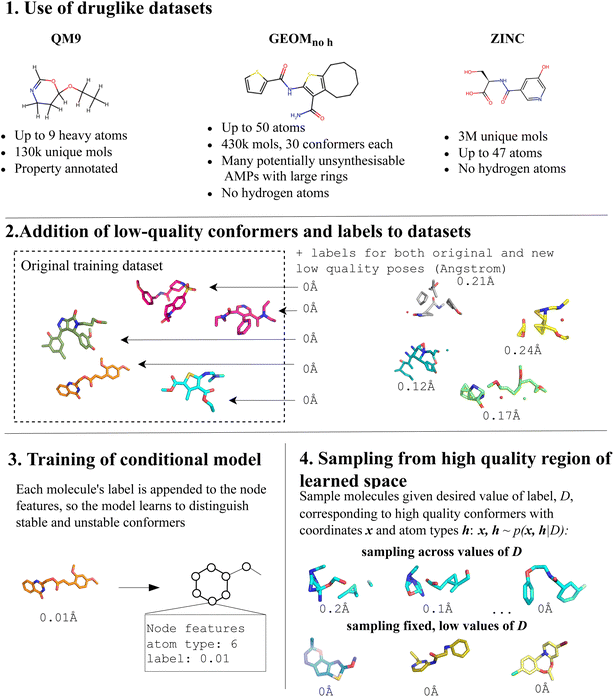
In this paper, we focus on enhancing the ability of a diffusion model to generate plausible 3D druglike molecules. To achieve this, we use the property-conditioning method developed by Hoogeboom et al.13 Instead of conditioning a model on pre-existing properties, we condition on conformer quality, training the model to not only generate molecules, but also to distinguish high- and low-quality chemical structures (Fig. 1).
To achieve this, we generate distorted versions of each of the three datasets we evaluate the method on: QM9, GEOM, and a subset of ZINC. We sample molecules from each dataset and apply random offsets to their original coordinates, based on a maximum distortion value. Each distorted molecule is assigned a label representing the degree of warping applied and is added back to the dataset. Non-distorted molecules are also labeled, identifying them as high-quality conformers. Using these datasets of molecules with varying levels of quality, we train property-conditioned models, encouraging the model to learn to label molecule validity while simultaneously training it to generate molecules.
First, we evaluate our conditioning method with EDM, the first E(3) equivariant diffusion model for molecule generation.13 We then test it on two additional models: a geometry-complete diffusion model14 and a flow matching method,19 both designed to enhance the structural plausibility of generated molecules. Since existing models already achieve strong performance on QM9, leaving little room or need for improvement, we focus on evaluating our approach on slightly larger, more chemically complex molecules. To this end, we employ two datasets of druglike molecules: the GEOM dataset, and another derived from the ZINC database.
Our findings demonstrate that across the models tested, conditioning a model with low-quality conformers enables it to discern between favourable and unfavourable molecular conformations. This allows us to target the area of the learned space corresponding to high-quality molecules, resulting in an improvement of the validity of generated molecules. More broadly, this demonstrates the potential of supplementing molecule generation methodologies not solely with examples of desired molecules but also with instances exemplifying undesired outcomes.
2 Methods
2.1 Generation of 3D molecules
Hoogeboom et al.13 introduced the first E(3)-equivariant diffusion model (EDM) for generating 3D small molecules. Since then, significant efforts have been made to modify the original EDM, whether to adapt the method for structure-based drug design8,20,21 or to enhance the validity of the generated molecules.22 Notable examples of the latter include GCDM (Geometry-Complete Diffusion Model)14 and MolFM.19 GCDM addresses the limitations of diffusion models that rely on molecule-agnostic and non-geometric graph neural networks (GNNs) for 3D graph denoising by introducing a geometry-complete approach. In contrast, MolFM focuses on the issue of unstable probability dynamics in existing diffusion models by incorporating geometric flow matching, merging the advantages of equivariant modeling with stabilised probability dynamics.2.2 Conditioning on conformer quality
The authors of EDM developed an extension to their method to carry out conditional molecule generation. In this instance, property annotations are included alongside each of the molecules in the training dataset, and at inference, molecules can be generated with a desired value of this property. We use this property-conditioning method to train models conditioned on conformer quality. To implement this, we first generated datasets with 3D conformers of molecules of variable quality levels, and corresponding annotations. We generated distorted versions of a subset of molecules from each of the datasets we used (Fig. 2). For each molecule, its 3D coordinates, represented as C = {(xi, yi, zi)} where i denotes the atom index, were obtained. Subsequently, a random number D within the range of 0 to Dmax angstroms, labelled as the maximum distortion, was sampled:| D ∼ U(0,Dmax) |
This value represents the maximum distance in angstrom that could be added to atoms in that molecule: in other words, the sampled distortion value determines the maximum extent of perturbation to be applied to the molecule's structure. Following this, random offsets were generated within the range of −D to D, for each dimension of every atom's coordinates:
| offsetx, offsety, offsetz ∼ U(−D,D) |
These offsets were then applied to the original coordinates:
| sxi = xi + offsetx; syi = yi + offsety; szi = zi + offsetz |
Resulting in a ‘distorted’ version of the molecule. This distorted molecule, along with its corresponding sampled distortion value D, was subsequently added to the training set. Following the generation of the distorted datasets, we use the property-conditioning training protocol outlined by Hoogeboom et al.13 to train on them, using the distortion factor D as the property of interest, and follow the sampling protocol to generate molecules corresponding to D = 0 Å.
2.3 Assessment metrics
For each model architecture, we follow their respective guidelines for training and sampling models, with the exception of cases for which pretrained models have been provided (EDM – QM9, GCDM – GEOMno h, and MolFM – GEOMno h). We then use each trained model to generate 1000 molecules, with the exception of models in the ablation tests, where we sampled 100 molecules per model to efficiently screen the effects of varying both distortion magnitudes and the ratio of distorted to non-distorted molecules. Since these models output only atom types and coordinates, we adhered to the standard practice13,23 of using OpenBabel24 to assign bonds based on interatomic distances. These post-processed molecules were first passed through RDKit's sanitisation checks, giving an RDKit sanitisation pass rate. All molecules were then evaluated using the PoseBusters test suite, which begins with its own sanitisation step – molecules failing this initial check automatically fail all subsequent tests. PoseBusters then tests the physical validity of the molecule by bond pattern matching, correct tetrahedral chirality specification, and appropriate double bond stereochemistry. The suite also assesses intramolecular validity through multiple geometric checks: bond lengths must fall within 0.75–1.25 times the expected bounds from distance geometry, bond angles must similarly be within 0.75–1.25 times their expected ranges, aromatic rings (5 or 6-membered) must maintain planarity within 0.25 Å, and aliphatic carbon–carbon double bonds must show appropriate planarity with their neighbouring atoms (within 0.25 Å). Additionally, PoseBusters checks for internal steric clashes by ensuring interatomic distances between non-covalently bound atoms exceed 0.7 times their lower geometric bounds. Finally, it evaluates energetic feasibility by comparing the molecule's calculated energy (using RDKit's UFF) against an ensemble of 50 conformations generated via ETKDGV3 and relaxed using force field optimisation. We report both the initial RDKit sanitisation pass rate and the number of molecules that successfully pass all seven non-sanitisation PoseBusters tests. Additionally, we assess structural diversity among the generated molecules using the MOSES framework,25 and check for presence of undesirable functional groups in each molecule using the REOS (Rapid Elimination of Swill26,27) functionality in the toolkit28 (see ESI†). For every statistic, we provide 95% confidence intervals computed using SciPy's
toolkit28 (see ESI†). For every statistic, we provide 95% confidence intervals computed using SciPy's  with 1000 samples and the “percentile” method.29
with 1000 samples and the “percentile” method.29
2.4 Datasets
To ensure consistency in our comparisons with pretrained baseline models, we use the same dataset splits and versions as those in the EDM paper. For QM9, this corresponds to the train/validation/test split introduced by Anderson et al.,30 based on the 2014 version of QM9.12 For GEOM, we follow the split introduced by the EDM authors,13 which uses the 2022 version of the dataset.17 To maintain consistency and allow for comparison with the models trained on these datasets, we follow the same splitting regime as proposed by Anderson et al.30 for our ZINC subset.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 small organic molecules with an average of 17.5 atoms (8.2 heavy atoms).
000 small organic molecules with an average of 17.5 atoms (8.2 heavy atoms).
The QM9 dataset has been extensively used to develop and validate machine learning models for molecular property prediction. However, it has also recently become the central benchmark for de novo molecule generation, particularly in the development of diffusion models,13,14 and as such, new diffusion models are capable of reliably generating molecules similar to those found in QM9.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules, of which 317
000 molecules, of which 317![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 928 are mid-sized organic molecules from AICures and MoleculeNet,31 and 133
928 are mid-sized organic molecules from AICures and MoleculeNet,31 and 133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 molecules are from QM9, resulting in an average molecule size of 44.4 atoms (20.1 heavy atoms). For each molecule, a variable number of conformers are given along with their approximate internal energy as calculated with XTB.32 From this dataset, Hoogeboom et al.13 retain the 30 lowest energy conformations for each molecule in their work.
258 molecules are from QM9, resulting in an average molecule size of 44.4 atoms (20.1 heavy atoms). For each molecule, a variable number of conformers are given along with their approximate internal energy as calculated with XTB.32 From this dataset, Hoogeboom et al.13 retain the 30 lowest energy conformations for each molecule in their work.
Similar to Peng et al.,22 we use a version of GEOM from which hydrogen have been removed (GEOMno h), as the positions of hydrogen atoms can often be inferred with a high level of confidence.33 This not only reduces the computational demand of training, but also facilitates more effective learning of heavy atom placements. This leads to the GEOMno h dataset becoming the quickest to train on among the three druglike datasets. We therefore use the GEOMno h dataset for conducting ablation tests.
We generate a training set by selecting a subset of 660![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules from the druglike catalog of the ZINC database. Unlike GEOM, this subset is curated without repeat conformers. Hydrogen atoms are not included, and the average molecule comprises 26.8 heavy atoms.
000 molecules from the druglike catalog of the ZINC database. Unlike GEOM, this subset is curated without repeat conformers. Hydrogen atoms are not included, and the average molecule comprises 26.8 heavy atoms.
3 Results and discussion
In this section, we evaluate the performance of EDM, both conditional and non-conditional, on QM9, GEOMno h and ZINC. We then present a series of ablation tests on the GEOMno h dataset. These tests were used to identify a sensible ratio and distortion level of the distorted molecules. Finally, we assess the broader applicability of our quality conditioning method by assessing it with GCDM and MolFM.3.1 Conditioning on distortion factor
We generated conditional versions of each dataset using parameters Dmax = 0.25 Å and a distorted![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) non-distorted molecule ratio of 1
non-distorted molecule ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50. After training conditional models on these modified datasets, we sampled 1000 molecules from each and evaluated their quality using RDKit and PoseBusters. The results of this evaluation are presented in Table 1.
50. After training conditional models on these modified datasets, we sampled 1000 molecules from each and evaluated their quality using RDKit and PoseBusters. The results of this evaluation are presented in Table 1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 distorted molecules, and sampled with D = 0 Å. Pass rates are shown for RDKit sanitisation, five PoseBusters subtasks, and overall PoseBusters pass rates. The two omitted subtasks, which had identical results across all six models, are provided in the ESI. 95% confidence intervals are shown in brackets
50 distorted molecules, and sampled with D = 0 Å. Pass rates are shown for RDKit sanitisation, five PoseBusters subtasks, and overall PoseBusters pass rates. The two omitted subtasks, which had identical results across all six models, are provided in the ESI. 95% confidence intervals are shown in brackets
| Dataset | RDKit sanitisation, % | Posebusters pass rate, % | |||||
|---|---|---|---|---|---|---|---|
| All atoms connected | Bond lengths | Bond angles | Internal steric clash | Internal energy | All tests passed | ||
| Baseline | |||||||
| QM9 | 92.2 (90.5–93.8) | 100.0 (100.0–100.0) | 100.0 (100.0–100.0) | 99.9 (99.7–100.0) | 100.0 (100.0–100.0) | 88.1 (85.9–90.1) | 81.1 (78.7–83.5) |
| GEOMno h | 84.7 (82.5–86.9) | 74.4 (71.7–77.1) | 65.6 (62.5–68.7) | 73.8 (70.8–76.7) | 97.8 (96.7–98.7) | 75.2 (72.3–78.0) | 62.2 (58.3–66.1) |
| ZINC | 70.6 (67.8–73.3) | 62.4 (59.4–65.3) | 65.0 (61.5–68.4) | 75.3 (72.2–78.5) | 79.5 (76.6–82.4) | 78.5 (75.5–81.4) | 40.0 (37.0–43.0) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Distortion factor conditioning | |||||||
| QM9 | 65.0 (62.0–68.0) | 84.0 (81.7–86.2) | 96.6 (95.2–98.0) | 95.8 (94.3–97.2) | 98.9 (98.0–99.7) | 91.1 (88.8–93.2) | 46.9 (43.9–50.0) |
| GEOMno h | 92.4 (90.7–94.0) | 89.4 (87.5–91.3) | 96.6 (95.5–97.8) | 93.7 (92.1–95.2) | 100.0 (100.0–100.0) | 87.7 (85.5–89.7) | 68.8 (65.9–71.1) |
| ZINC | 95.3 (93.8–96.6) | 95.7 (94.3–96.9) | 94.1 (92.5–95.6) | 93.7 (92.1–95.2) | 99.0 (98.4–99.6) | 89.4 (87.4–91.4) | 74.5 (71.7–77.3) |
Our baseline analysis reveals a clear relationship between molecule size and model performance. The QM9 dataset, comprising molecules smaller than 9 heavy atoms had the highest baseline performance with RDKit and PoseBusters pass rates of 92.2% and 81.1%, respectively. The non-conditioned EDM model performed exceptionally well with QM9, surpassing all conditional variants across both evaluation metrics. This superior performance may be attributed to EDM's specific development for QM9, coupled with the dataset's smaller molecular size, which appears to enable better discrimination between high-quality and low-quality conformers without requiring examples of the latter.
The GEOMno h dataset also showed strong baseline performance as assessed with RDKit, with generated molecules achieving a pass rate of 84.7%. However, the more stringent PoseBusters tests presented more of a challange, with generated molecules presenting a pass rate of 62.2% for PoseBusters. This can particularly be attributed to the tests concerning bond lengths (65.6% pass rate), bond angles (73.8%), connectivity (74.4%) and internal energy (75.2%). Conditional training improved these metrics substantially, reaching pass rates of 96.6% (bond lengths), 93.7% (bond angles), 89.4% (connectivity) and 87.7% (internal energy). Minor improvements were also seen in the steric clash tests (97.8% to 100%).
The baseline model trained on the ZINC subset exhibited markedly lower performance than the other two datasets, with RDKit sanitisation pass rates of 70.6% and PoseBusters pass rates of 40.0%. The most prevalent failures occurred in bond lengths and atom connectivity, with pass rates of only 65.0% and 62.4%, respectively. This performance decline relative to GEOMno h may be attributed to two factors. Firstly, the ZINC dataset's increased diversity, featuring unique conformers rather than multiple conformers per molecule (as found in GEOMno h), may cause the model to prioritise learning atom types over optimising 3D conformer generation. Secondly, and perhaps more significantly, the compositional differences between datasets may play a crucial role. While the ZINC subset exclusively contains medium-sized compounds, GEOMno h incorporates the entire QM9 dataset, resulting in a smaller average molecule size.
Conditional training on the ZINC dataset yielded improved RDKit sanitisation rates and PoseBusters scores. The most notable improvement was observed in the ZINC model's atom connectivity pass rate, which increased from 62.4% to 95.7%, but improvements were also seen in the pass rates of the tests assessing bond lengths (65.0% to 94.1%), bond angles (75.3% to 93.7%), internal steric clash (79.5% to 99.0%), and internal energy (78.5% to 89.4%).
These findings show that while the baseline, non-conditional EDM model excels at generating small compounds, its performance declines when restricted to medium-sized molecules, often producing physically implausible structures. In the next section, we present ablation tests exploring the impact of varying distortion magnitudes and the ratio of distorted to non-distorted molecules when applying our conditioning method.
3.2 Ablation tests
To identify the optimal proportion of distorted molecules and the required degree of distortion for effective conditional training, we performed ablation studies using the GEOMno h dataset (Table 2). This dataset was selected due to its inclusion of drug-like molecule sizes, unlike QM9, whilst being a more computationally tractable set to train than the ZINC dataset. For these ablation studies, we sampled 100 molecules per model to efficiently screen the effects of varying both distortion magnitudes and the ratio of distorted to non-distorted molecules.| D max (Å) | Ratio of distorted:non-distorted molecules | |||||
|---|---|---|---|---|---|---|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 50 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
||||
| RDKit sanitisation, % | PoseBusters pass rate, % | RDKit sanitisation, % | PoseBusters pass rate, % | RDKit sanitisation, % | PoseBusters pass rate, % | |
| 0.1 | 96 (92–99) | 73 (64–81) | 96 (92–99) | 77 (69–85) | 96 (92–99) | 77 (68–85) |
| 0.25 | 95 (90–99) | 52 (42–62) | 97 (92–99) | 81 (73–88) | 96 (92–99) | 77 (68–85) |
| 0.5 | 97 (93–100) | 75 (66–83) | 97 (93–100) | 78 (70–86) | 95 (90–99) | 68 (59–77) |
| 1 | 93 (88–97) | 57 (47–67) | 89 (78–97) | 54 (38–70) | 62 (52–71) | 8 (3–14) |
We introduced varying numbers of distorted molecules at different distortion levels (ranging from 0 Å, indicating no distortion, to the maximum distortion, Dmax Å) into the original GEOMno h dataset. We defined dataset ratios based on the number of distorted and original molecules: for example, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 ratio indicates one distorted molecule was added for every fifty original molecules. We evaluated each model's performance by training conditioned models and sampling 100 molecules, ensuring that the samples were from the low-distortion-factor region of the learned space (formally, enforcing D = 0 Å).
50 ratio indicates one distorted molecule was added for every fifty original molecules. We evaluated each model's performance by training conditioned models and sampling 100 molecules, ensuring that the samples were from the low-distortion-factor region of the learned space (formally, enforcing D = 0 Å).
The model trained on a dataset with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 distorted molecules and a maximum distortion of 0.25 Å exhibited the joint highest RDKit parsability rate of 97%, and the highest PoseBusters pass rate at 81%. While several models reached 97% RDKit sanitisation rates (namely 1
50 distorted molecules and a maximum distortion of 0.25 Å exhibited the joint highest RDKit parsability rate of 97%, and the highest PoseBusters pass rate at 81%. While several models reached 97% RDKit sanitisation rates (namely 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, Dmax = 0.5 Å and 1
20, Dmax = 0.5 Å and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, Dmax = 0.5 Å), these models exhibited slightly lower PoseBusters pass rates (75% and 78%, respectively). Increasing or decreasing Dmax further resulted in PoseBusters performance decreasing across all ratios, primarily due to failures in the internal energy test.
50, Dmax = 0.5 Å), these models exhibited slightly lower PoseBusters pass rates (75% and 78%, respectively). Increasing or decreasing Dmax further resulted in PoseBusters performance decreasing across all ratios, primarily due to failures in the internal energy test.
This observation suggests that if the training includes molecules that are too distorted, the model does not effectively learn to distinguish between subtly flawed and acceptable molecular structures. Distorted molecules should therefore still bear some resemblance to realistic conformers, albeit with deliberately infeasible bond lengths and angles. On the other hand, insufficient distortion compromises the effectiveness of the conditioning classifier, and the models struggle to distinguish between high-quality and low-quality conformations, leading to poor performance in generating desirable molecules.
These results demonstrate the concept of conditioned training on negative data, and give an idea of the extent of distortion and frequency of distorted molecules to add. We used a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, and Dmax = 0.25 Å for all subsequent tests, but note that any dataset would likely benefit from different exact values of these parameters.
50, and Dmax = 0.25 Å for all subsequent tests, but note that any dataset would likely benefit from different exact values of these parameters.
We also examined the quality of molecules generated when sampling from the low-quality region of the learned space (formally, D = Dmax Å). The results of this are shown in the ESI.† The molecules sampled using D = Dmax Å are, as expected, worse than both the conditioned models and the baseline model in terms of PoseBusters pass rates, with the highest reaching only 53%. This poor performance is mainly attributed to failures in the internal energy test.
Having established the parameters for conditional training datasets in terms of quantity of distorted molecules and extent of distortion, and demonstrated that our conditioning method enhances the structural plausibility of generated molecules when EDM is trained on ZINC or GEOMno h, we now move on to testing this approach on other models.
3.3 Testing the conditioning method on additional models
To evaluate the broader applicability of our method, we apply it to two other models: GCDM14 and MolFM.19 The performance of these models when trained on GEOMno h and ZINC is presented in Table 3.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 distorted molecules, and sampled with D = 0 Å. 95% confidence intervals are shown in brackets
50 distorted molecules, and sampled with D = 0 Å. 95% confidence intervals are shown in brackets
| RDKit sanitisation, % | PoseBusters pass rate, % | ||
|---|---|---|---|
| (a) GCDM | |||
| Baseline | GEOMno h | 100.0 (100.0–100.0) | 77.8 (75.2–80.4) |
| ZINC | 56.3 (53.3–59.3) | 40.8 (38–43.6) | |
| Conditional | GEOMno h | 99.9 (99.7–100.0) | 79.7 (77.2–82.1) |
| ZINC | 97.2 (95.8–98.4) | 66.5 (62.5–70.4) | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| (b) MolFM | |||
| Baseline | GEOMno h | 98.6 (97.5–99.6) | 80.8 (77.3–84.1) |
| ZINC | 72.0 (69.2–74.8) | 42.3 (39.2–45.4) | |
| Conditional | GEOMno h | 94.5 (93.0–95.9) | 46.8 (43.7–49.8) |
| ZINC | 93.3 (91.6–94.9) | 45.9 (42.6–49.1) | |
For the GEOM dataset, the GCDM conditional model shows very marginal improvements in PoseBusters performance over the baseline (mostly due to the internal energy test, for which the pass rate increases from 86.2% to 88.6%). However, since baseline performance is already high, conditioning has limited overall impact. GCDM trained on the ZINC subset, on the other hand, shows a much more substantial improvement with conditioning. The baseline model struggles with the connectivity of molecules, which increases from 63.7% in the baseline model to 94.4% when our conditioning method is applied, resulting in an overall boost in both RDKit sanitisation and PoseBusters pass rate.
Training MolFM using the conditional method does not improve the plausibility of generated molecules when using GEOMno h, in which many molecules suffer from connectivity issues. It does, however, improve the plausibility of generated molecules when using the ZINC dataset, by a margin similar to that shown by EDM.
In conclusion, our conditioning method that was developed and tested with EDM is able to, without modification, enhance molecular plausibility across different models when looking at ZINC. These results suggest that the conditioning approach is broadly applicable.
4 Conclusions
In this work, we have demonstrated the effectiveness of including low-quality conformers in a training set and conditioning a diffusion model on a label representing conformer quality to enhance the generation of high-quality druglike molecules. By leveraging datasets derived from GEOM and ZINC, alongside a conditioning method proposed by Hoogeboom et al., we have successfully improved the validity of generated molecules. Our approach, which focuses on sampling molecules with labels corresponding to low distortion factors, leads to enhancements in RDKit parsability and validity as assessed by PoseBusters for the original EDM, as well as for a subsequent diffusion model, GCDM, and a flow-matching model, MolFM.The method shows strongest improvements for diffusion-based models, particularly those built on the EDM framework, which comprise a significant portion of current 3D molecule generation approaches. The approach is also more effective for datasets containing larger, drug-like molecules, as demonstrated by our results with GEOMno h and ZINC. This can be explained mechanistically; larger molecules have more complex conformational spaces where explicit examples of invalid states help define the boundary between high-quality and low-quality conformers. Conversely, the method provides limited benefits for datasets like QM9 where molecules are small (fewer than 9 heavy atoms) and have constrained conformational spaces. In these cases, it appears models can effectively learn to distinguish valid from invalid conformations from the training data alone.
Our findings underscore the importance of considering the quality of conformers in molecule generation processes. The results show that by training models to discern between favorable and unfavorable molecular conformations, we can selectively sample from the high-quality region of learned space, resulting in significant improvements in the validity of generated molecules.
Moving forward, further research could explore additional conditioning methods and datasets to continue improving the quality and diversity of generated molecules. Additionally, investigating the applicability of our approach to other areas of molecular design and exploration could yield valuable insights for drug discovery and beyond. Overall, our study provides a promising avenue for generating valid drug-sized molecules efficiently and effectively.
Data availability
The data, checkpoints, and scripts for training, testing, and evaluation used in this paper are available at https://github.com/lucyvost/distorted_diffusion/tree/main (DOI: https://doi.org/10.5281/zenodo.15010217). Preprocessed versions of the larger two datasets the method was tested on, GEOM and ZINC, are hosted on Zenodo (DOI: https://doi.org/10.5281/zenodo.14825439).Conflicts of interest
There are no conflicts to declare.References
- O. J. Wouters, M. McKee and J. Luyten, Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009-2018, JAMA, 2020, 323(9), 844 Search PubMed . https://jamanetwork.com/journals/jama/fullarticle/2762311.
- J. Ross, B. Belgodere, S. C. Hoffman, V. Chenthamarakshan, Y. Mroueh and P. Das, GP-MoLFormer: A Foundation Model For Molecular Generation, arXiv, 2024, preprint, arXiv:2405.04912, DOI:10.48550/arXiv.2405.04912.
- V. Chenthamarakshan, P. Das, S. Hoffman, H. Strobelt, I. Padhi, K. W. Lim, et al., CogMol: Target-Specific and Selective Drug Design for COVID-19 Using Deep Generative Models, in Advances in Neural Information Processing Systems, Curran Associates, Inc., 2020, vol. 33, pp. 4320–4332, https://proceedings.neurips.cc/paper/2020/hash/2d16ad1968844a4300e9a490588ff9f8-Abstract.html Search PubMed.
- O. Dollar, N. Joshi, D. A. C Beck and J. Pfaendtner, Attention-based generative models for de novo molecular design, Chem. Sci., 2021, 12(24), 8362–8372 RSC . https://pubs.rsc.org/en/content/articlelanding/2021/sc/d1sc01050f.
- Y. Luo and S. Ji, An Autoregressive Flow Model for 3D Molecular Geometry Generation from Scratch, 2022 Search PubMed.
- J. O. Spiegel and J. D. Durrant, AutoGrow4: an open-source genetic algorithm for de novo drug design and lead optimization, J. Cheminf., 2020, 12(1), 25, DOI:10.1186/s13321-020-00429-4.
- X. Peng, S. Luo, J. Guan, Q. Xie, J. Peng and J. Ma, Pocket2Mol: Efficient Molecular Sampling Based on 3D Protein Pockets, arXiv, 2022, preprint, arXiv:2205.07249, DOI:10.48550/arXiv.2205.07249.
- H. Lin, Y. Huang, M. Liu, X. Li, S. Ji and S. Z. Li, DiffBP: Generative Diffusion of 3D Molecules for Target Protein Binding, arXiv, 2022, preprint, arXiv:2211.11214, DOI:10.48550/arXiv.2211.11214.
- Q. Liu, M. Allamanis, M. Brockschmidt and A. L. Gaunt, Constrained Graph Variational Autoencoders for Molecule Design, arXiv, 2019, preprint, arXiv:1805.09076, DOI:10.48550/arXiv.1805.09076.
- C. Harris, K. Didi, A. R. Jamasb, C. K. Joshi, S. V. Mathis, P. Lio, et al., Benchmarking Generated Poses: How Rational is Structure-based Drug Design with Generative Models?, arXiv, 2023, preprint, arXiv:2308.07413, DOI:10.48550/arXiv.2308.07413.
- M. Buttenschoen, G. M. Morris and C. M. Deane, PoseBusters: AI-based docking methods fail to generate physically valid poses or generalise to novel sequences, Chem. Sci., 2024, 15(9), 3130–3139 RSC . http://xlink.rsc.org/?DOI=D3SC04185A.
- R. Ramakrishnan, P. O. Dral, M. Rupp and O. A. von Lilienfeld, Quantum chemistry structures and properties of 134 kilo molecules, Sci. Data, 2014, 1(1), 140022 CrossRef CAS PubMed . https://www.nature.com/articles/sdata201422.
- E. Hoogeboom, V. G. Satorras, C. Vignac and M. Welling, Equivariant Diffusion for Molecule Generation in 3D, arXiv, 2022, preprint, arXiv:2203.17003, DOI:10.48550/arXiv.2203.17003.
- A. Morehead and J. Cheng, Geometry-Complete Diffusion for 3D Molecule Generation and Optimization, arXiv, 2023, preprint, arXiv:2302.04313, DOI:10.48550/arXiv.2302.04313.
- C. Vignac, N. Osman, L. Toni and P. Frossard, MiDi: Mixed Graph and 3D Denoising Diffusion for Molecule Generation, in Machine Learning and Knowledge Discovery in Databases: Research Track, ed. D. Koutra, C. Plant, M. Gomez Rodriguez, E. Baralis and F. Bonchi, Springer Nature Switzerland, Cham, 2023, pp. 560–576 Search PubMed.
- L. Huang, H. Zhang, T. Xu and K. C. Wong, MDM: Molecular Diffusion Model for 3D Molecule Generation, arXiv, 2022, preprint, arXiv:2209.05710, DOI:10.48550/arXiv.2209.05710.
- S. Axelrod and R. Gómez-Bombarelli, GEOM, energy-annotated molecular conformations for property prediction and molecular generation, Sci. Data, 2022, 9(1), 185 CrossRef CAS PubMed . https://www.nature.com/articles/s41597-022-01288-4.
- Y. Ziv, B. Marsden, C. M. Deane, MolSnapper: Conditioning Diffusion for Structure Based Drug Design, bioRxiv, 2024, preprint, DOI:10.1101/2024.03.28.586278.
- Y. Song, J. Gong, M. Xu, Z. Cao, Y. Lan, S. Ermon, et al., Equivariant Flow Matching with Hybrid Probability Transport, arXiv, 2023, preprint, arXiv:2312.07168, DOI:10.48550/arXiv.2312.07168.
- Z. Chen, B. Peng, S. Parthasarathy and X. Ning, Shape-conditioned 3D Molecule Generation via Equivariant Diffusion Models, arXiv, 2023, preprint, arXiv:2308.11890, DOI:10.48550/arXiv.2308.11890.
- J. Guan, X. Zhou, Y. Yang, Y. Bao, J. Peng, J. Ma, et al., DecompDiff: Diffusion Models with Decomposed Priors for Structure-Based Drug Design, in Proceedings of the 40th International Conference on Machine Learning, PMLR, 2023, pp. 11827–11846, ISSN: 2640-3498, https://proceedings.mlr.press/v202/guan23a.html Search PubMed.
- X. Peng, J. Guan, Q. Liu and J. Ma, MolDiff: Addressing the Atom-Bond Inconsistency Problem in 3D Molecule Diffusion Generation, arXiv, 2023, preprint, arXiv:2305.07508, DOI:10.48550/arXiv.2305.07508.
- N. Gebauer, M. Gastegger and K. Schütt, Symmetry-adapted generation of 3d point sets for the targeted discovery of molecules, in Advances in Neural Information Processing Systems, Curran Associates, Inc., 2019, vol. 32, https://papers.nips.cc/paper_files/paper/2019/hash/a4d8e2a7e0d0c102339f97716d2fdfb6-Abstract.html Search PubMed.
- N. M. O'Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch and G. R. Hutchison, Open Babel: An open chemical toolbox, J. Cheminf., 2011, 3(1), 33, DOI:10.1186/1758-2946-3-33.
- D. Polykovskiy, A. Zhebrak, B. Sanchez-Lengeling, S. Golovanov, O. Tatanov and S. Belyaev, et al., Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models, Front. Pharmacol, 2020, 11, 1–13 CrossRef PubMed . https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2020.565644/full.
- W. P. Walters, A. A. Murcko and M. A. Murcko, Recognizing molecules with drug-like properties, Curr. Opin. Chem. Biol., 1999, 3(4), 384–387 CrossRef CAS PubMed . https://www.sciencedirect.com/science/article/pii/S1367593199800581.
- W. P. Walters, M. T. Stahl and M. A. Murcko, Virtual screening—an overview, Drug Discovery Today, 1998, 3(4), 160–178 CAS . https://www.sciencedirect.com/science/article/pii/S135964469701163X.
- P. Walters, PatWalters/useful_rdkit_utils, 2025, Original-date: 2021-12-31T00:24:33Z, https://github.com/PatWalters/useful_rdkit_utils Search PubMed.
- P. Virtanen, R. Gommers, T. E. Oliphant, M. Haberland, T. Reddy and D. Cournapeau, et al., SciPy 1.0: fundamental algorithms for scientific computing in Python, Nat. Methods, 2020, 17(3), 261–272 CrossRef CAS PubMed . https://www.nature.com/articles/s41592-019-0686-2.
- B. Anderson, T. S. Hy and R. Kondor, Cormorant: Covariant Molecular Neural Networks, in Advances in Neural Information Processing Systems, Curran Associates, Inc., 2019, vol. 32, https://proceedings.neurips.cc/paper_files/paper/2019/hash/03573b32b2746e6e8ca98b9123f2249b-Abstract.html Search PubMed.
- Z. Wu, B. Ramsundar, E. Feinberg, J. Gomes, C. Geniesse and A. S. Pappu, et al., MoleculeNet: a benchmark for molecular machine learning, Chem. Sci., 2018, 9(2), 513–530 CAS . https://xlink.rsc.org/?DOI=C7SC02664A.
- C. Bannwarth, E. Caldeweyher, S. Ehlert, A. Hansen, P. Pracht and J. Seibert, et al., Extended tight-binding quantum chemistry methods, Wiley Interdiscip. Rev. Comput. Mol. Sci., 2021, 11(2), e1493 CAS . https://onlinelibrary.wiley.com/doi/pdf/10.1002/wcms.1493.
- X. He, E. Hatcher, L. Eriksson, G. Widmalm and A. D. MacKerell, Bifurcated Hydrogen Bonding and Asymmetric Fluctuations in a Carbohydrate Crystal Studied via X-ray Crystallography and Computational analysis, J. Phys. Chem. B, 2013, 117(25), 7546–7553 CAS . https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3771504/.
- T. Sterling and J. J. Irwin, ZINC 15 – Ligand Discovery for Everyone, J. Chem. Inf. Model., 2015, 55(11), 2324–2337, DOI:10.1021/acs.jcim.5b00559.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00331d |
| This journal is © The Royal Society of Chemistry 2025 |