 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Towards greener-by-design fine chemicals. Part 2: technological frontiers
Theodore A.
Gazis
a,
Rodolfo I.
Teixeira
 b,
Giulio
Volpin
b,
Giulio
Volpin
 c,
Ashish
Yewale
b,
Mert Can
Ince
a,
Mark J.
Ford
c,
Jan
Harmsen
d,
Marco
Uboldi
e,
Alice
Melocchi
c,
Ashish
Yewale
b,
Mert Can
Ince
a,
Mark J.
Ford
c,
Jan
Harmsen
d,
Marco
Uboldi
e,
Alice
Melocchi
 e,
Mattia
Sponchioni
e,
Mattia
Sponchioni
 a,
Andrea
Aramini
f,
Renzo
Luisi
a,
Andrea
Aramini
f,
Renzo
Luisi
 g,
Brahim
Benyahia
b and
Gianvito
Vilé
g,
Brahim
Benyahia
b and
Gianvito
Vilé
 *a
*a
aDepartment of Chemistry, Materials, and Chemical Engineering “Giulio Natta”, Politecnico di Milano, Piazza Leonardo da Vinci 32, 20133 Milano, Italy. E-mail: gianvito.vile@polimi.it
bChemical Engineering Department, Loughborough University, Epinal Way, LE11 3TU Loughborough, Leicestershire, UK
cBayer AG, Crop Science Division, Alfred-Nobel-Straße 50, 40789 Monheim, Germany
dHarmsen Consultancy BV, Hoofdweg Zuid 18, 2912 ED Zuidplas, The Netherlands
eSezione di Tecnologia e Legislazione Farmaceutiche “Maria Edvige Sangalli”, Dipartimento di Scienze Farmaceutiche, Università degli Studi di Milano, via Giuseppe Colombo 71, 20133 Milano, Italy
fResearch & Early Development, Dompé Farmaceutici S.p.A., 67100 L'Aquila, Italy
gFlow Chemistry and Microreactor Technology (FLAME-Lab), Department of Pharmacy – Drug Sciences, University of Bari “A. Moro”, 70125 Bari, Italy
First published on 20th November 2025
Abstract
Over the past three decades, the pharmaceutical and agrochemical sectors have embarked on a transformative journey towards greener-by-design processes, firmly rooted in the principles of green chemistry. Building on this foundation, green engineering frameworks have expanded the focus beyond environmental concerns to encompass product quality, economic viability, and the evolving demands of modern healthcare. At the heart of this transformation is continuous and smart manufacturing due to its capacity to reduce raw material use, waste, and energy consumption. While attention has understandably centered on replacing or refining conventional batch operations, the breadth of progress is far wider. Advanced analytics and digitization, as exemplified by AI-driven modeling, are nurturing the rise of “smart factories” that autonomously optimize performance in real time. A prime illustration lies in the purification of fine chemicals, where real-time analytics and advanced process control slash solvent requirements, an acute pollution hotspot, while ensuring consistent product quality. Meanwhile, 3D printing has introduced a genuinely disruptive dimension, challenging traditional notions of scale and location through on-demand, flexible production. In this piece, we explore how these converging technological frontiers lay the groundwork for the patient-centered, eco-conscious pharmaceutical and agrochemical facilities of the future.
1. Introduction
Nearly three decades ago, the fine chemicals industry experienced a watershed moment with the advent of green chemistry. This framework redefined reaction efficiency by shifting emphasis from mere product yield to a holistic strategy prioritizing resource conservation and waste reduction. The introduction of the E(nvironmental)-Factor, defined as the ratio of waste mass to product mass, highlighted the sectors heavy reliance on stoichiometric protocols, high solvent volumes, and inefficient equipment, resulting in E-factors that were significantly higher than those typical in bulk chemical manufacturing (Table 1).1 Over time, it became clear that solvent inefficiency had been seriously underestimated, with more recent figures placing pharmaceutical E-factors near 182 kg of waste per kg of product.2 This realization has galvanized the rise of a “greener-by-design” philosophy in fine chemical production, particularly in the pharmaceutical and agrochemical sectors.| Industry segment | Tons per annum | E-factor (kg of waste per kg product) |
|---|---|---|
| Oil refining | 106–108 | <1 |
| Bulk chemicals | 104–106 | 1–50 |
| Fine chemicals | 10–104 | >50 |
A major pollution hotspot lies in the synthesis of active ingredients, which typically accounts for 75–90% of the carbon footprint of small molecules.3,4 Major contributors to this footprint include organic solvents, fossil-fuel feedstocks, inefficient catalytic protocols, and the intrinsic constraints of reaction and processing conditions. Recent breakthroughs targeting these core synthetic pillars have been detailed in Part 1 of this two-volume review. While greener active ingredient synthesis is key, green engineering plays an equally important role, as sustainability can only truly arise from the synergy between chemistry and engineering. In this context, the 12 principles of green engineering,5 and the Sandestin 9 principles of green engineering6 provide foundational frameworks that guide the design of inherently sustainable processes and technologies. Importantly, whereas green chemistry is primarily concerned with waste prevention and hazard reduction, green engineering embodies a broader outlook by encompassing aspects of profitability and product quality within the framework of green process design.7 This is reflected in the relationship between process mass intensity (PMI), which quantifies the total mass of all materials used per unit product, and E-factor (waste only), captured succinctly by PMI = E-factor + 1.1
At first glance, sustainability may appear incompatible with profitability; however, in the manufacture of fine chemicals, the two often go hand in hand. Mounting evidence supports this view,1,8 as reductions in raw material consumption, waste-disposal costs, energy usage, and improvements in capacity utilization can collectively offset initial investments. Only in specific cases, such as when reagents like phosphorus oxychloride (POCl3) or phosphorus pentachloride (PCl5) cannot be replaced with greener alternatives, a divergence may arise. Nevertheless, even in such instances, the overall process can be designed to remain green, with high solvent recovery, efficient throughput, and minimized waste generation. A case in point is Pfizer's 2021 Environmental, Social, Governance Report,9 which documented a 27% reduction in hazardous waste from 114![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 metric tons in 2020 to 82
000 metric tons in 2020 to 82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 in 2021, largely attributed to process intensification. Concurrently an annual revenue growth of 95% was recorded.10
700 in 2021, largely attributed to process intensification. Concurrently an annual revenue growth of 95% was recorded.10
A key force driving the evolution of modern process development is the industry's commitment to “continuous improvement”, an imperative shaped by growing demands for sustainability, efficiency, and cost-effectiveness. Many companies have formalized this pursuit through structured “continuous excellence” programs, which blend lean manufacturing (requiring maximal productivity with minimal waste), six Sigma (requiring minimal process variability and defects), and digital innovation to systematically enhance operations.11 These initiatives aim not only to reduce waste and variability but also to accelerate innovation cycles, improve product quality, and optimize resource utilization.
However, the implementation of continuous improvement varies widely across regions, shaped by regulatory environments and cultural attitudes towards operational excellence.12 Japanese firms often operate within rigid hierarchies that can hinder innovation; European companies typically emphasize regulatory compliance alongside collaborative improvement; U.S. firms favor agile, market-driven approaches supported by proactive regulation. Even within the same region, company-specific factors, such as leadership and organizational culture, can significantly influence adoption. In Ireland's pharmaceutical sector, for instance, 97% of surveyed professionals reported using continuous improvement strategies to boost productivity and quality, yet 45% cited strict regulations as a barrier for further implementation, particularly due to added validation and regulatory requirements.13
In this context, process development is no longer seen as a one-time effort but as an iterative, data-driven strategy for sustained advancement. In fact, traditional linear models of innovation,14 once characterized by sequential steps like research, development, engineering, and commercial start-up, have been gradually replaced by more adaptive and systematically organized frameworks. The linear approach of improvement lacked the responsiveness needed to navigate the increasing complexity of technical, economic, and regulatory environments, resulting in inefficiencies, delays, or missed opportunities. Over the past two decades, stage-gate models have become the industry standard, providing structured pathways for chemical innovation through well-defined stages and decision-making checkpoints, or “gates”. As outlined by Harmsen,15,16 the most widely adopted stage-gate model for both process and product innovation comprises six phases: discovery (idea generation and proof of principle), concept (initial design and validation through experiments), feasibility (pilot-scale design and risk assessment), development (pilot plant execution and Front End Engineering Design (FEED) preparation), Engineering, Procurement, and Construction (EPC), and implementation (start-up and commercial manufacturing, with flexibility for multi-product production, especially in the pharmaceutical and agrochemical sectors).
The second force behind both sustainability and economic growth has been the standardization of international quality standards, spearheaded by the International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use (ICH). Fundamental manufacturing concepts have been formalized in ICH guidelines (particularly Q7,17 Q8,18 Q9,19 and Q1020) including process intensification (streamlining manufacturing steps for efficiency), quality by design (ensuring product quality from the outset, rather than relying on end-point checks), and good manufacturing practice (consistently meeting high quality and safety standards). Building on these principles, the ICH has dedicated its latest guideline, ICH Q13 (2023)21 to continuous manufacturing, an approach already endorsed by the Food and Drug Administration (FDA)22 and European Medicines Agency (EMA).23 The United States Pharmacopeia (USP) has reinforced this direction in its 2022 Annual Report, projecting that continuous manufacturing will become an industry staple alongside traditional batch processing within the next decade.24
To contextualize the projected benefits of continuous manufacturing, it is instructive to consider the conventional industrial practice for the production of active ingredients. Typically, manufacturing involves a series of sequential batch unit operations, involving reaction, purification, blending, and packaging (Fig. 1). Historically, these stages have been spatially and temporally segregated, resulting in prolonged processing, repeated start-stop cycles, storage and transportation costs between stages, and substantial facility footprints.25 In this context, however, it must be mentioned that crop protection manufacturing frequently employs telescoped batch reactions with minimal or no isolation of intermediates, as repeated interruptions would be highly inefficient. Here, the focus is on robust, scalable, and integrated processes that enable streamlined, high-throughput production, different from pharmaceutical models where final formulation and patient-specific customization may play a greater role.26 Continuous manufacturing continuously feeds materials through multiple integrated operations, minimizing processing times and lowering contamination risk by closed-loop design. Its reconfigurable, modular design adapts readily to manufacturing and supply chain requirements at a fraction of the space requirement.27 Moreover, unlike batch processes, which can operate with periodic, offline inspections, more sophisticated real-time control systems are required to dynamically regulate process parameters during long-term operation. Thus, two additional components are essential: process analytical technologies (PAT), such as inline infrared spectroscopy, for continuous detection of impurities and product quality assurance; and (2) process control models to adjust operating conditions in real time.
 | ||
| Fig. 1 Comparison of conventional batch manufacturing and integrated continuous manufacturing. Top: traditional batch pharmaceutical process, in which synthesis, purification, blending, granulation, and tableting occur in separate unit operations, often at different locations, with intermediate storage, testing, and shipping steps extending production timelines to several months. Bottom: continuous manufacturing process integrating all stages, from synthesis to final tablet coating, into a streamlined, single-site workflow supported by PAT and real-time process control, reducing production times from months to days. Reproduced from ref. 28, with permission from Springer Nature, copyright 2015. | ||
With clear regulatory momentum and documented benefits, numerous pharmaceutical companies have begun implementing continuous manufacturing processes.29 Janssen's continuous process for the antiretroviral darunavir exemplifies the substantial reductions in operational time and environmental that impact continuous operation confers.30 By shifting from a conventional batch process to continuous mode, production was compressed from two weeks to a single day, and the required space was scaled back from seven rooms to only two.31
Concurrently, a surge of research over the past 20 years has explored various continuous operations like continuous blending,32 continuous crystallization33 and continuous drying.34 Moreover real-time monitoring with PAT35–38 and advanced process control strategies39–41 have been reviewed extensively. Others have explored interconnected units for end-to-end manufacturing and continuous plant operation.27 All of the above developments in continuous manufacturing have been expertly reviewed elsewhere, and readers are directed there for a more detailed discussion.
Nevertheless, continuing down the road to greener-by-design pharmaceuticals and agrochemicals require more than merely developing and integrating continuous versions of existing unit operations. It demands embracing the autonomy and digital transformation envisioned by Industry 4.0, in tandem with the sustainability ethos of Industry 5.0.42 Specifically, Industry 4.0 shifts the focus from manual supervision of automated systems to autonomous manufacturing powered by real-time data acquisition and analysis, adaptive process optimization and innovative manufacturing methods.43,44 Emerging technological frontiers are poised to transform pharmaceutical and agrochemical manufacturing by unlocking new capabilities that extend beyond conventional process intensification. In particular, smart manufacturing (Section 2) and innovative fabrication methods (Section 3) stand out as key enablers, offering unprecedented opportunities to enhance efficiency, precision, and sustainability across the chemical and pharmaceutical industries.
Smart manufacturing is redefining the production of pharmaceuticals and fine chemicals by integrating real-time data acquisition, digital modeling, and advanced purification strategies. Since the FDAs 2004 endorsement of PATs,45 continuous monitoring has become instrumental during the later stages of drug manufacturing to ensure consistent product quality. However, integrating PAT into earlier active pharmaceutical ingredient (API) synthesis has been impeded by the complexity of chemical reactions being conducted. The recent miniaturization of spectroscopic instruments, including benchtop systems, has broadened the arsenal available to process chemists to monitor them. Moreover, the integration of PATs with emerging technologies, such as photo- and electrochemistry, is expanding the capabilities of fine chemical manufacturing. Finally, the fusion of PAT with flow reactors and artificial intelligence (AI)-driven process control is establishing automated reaction platforms enhancing real-time decision making and operational efficiency. These aspects are described in Section 2.1.
Traditional process models are essential in designing and controlling continuous manufacturing systems. Often, these models are resource intensive and rely on arbitrary parameter estimation procedures which may result in several limitations such as poor prediction capabilities and lack of adaptability. The adoption of digital twins and AI-driven models (presented in Section 2.2) supports real-time decision-making, allowing manufacturing systems to dynamically adjust to process variations and enhance efficiency and sustainability.
Beyond reaction monitoring, smart manufacturing also extends to downstream processes, where solvent-intensive separation operations remain a major environmental and economic concern. While solvent minimization initiatives for active ingredient synthesis have received considerable attention (see the dedicated sections in Part 1: Synthetic Frontiers), purification strategies have often lagged behind in innovation. Section 2.3 will highlight continuous purification as a testbed for advanced inline analytics and smart manufacturing, illustrating how these methods can cut solvent use and deliver broader environmental and economic benefits.
Technological innovation is reshaping pharmaceutical manufacturing, moving beyond traditional mass production towards more flexible, patient-centered, and data-driven approaches. Among these advancements, 3D printing is a disruptive technology poised to transform mass production into flexible, on-demand, and intrinsically “smart” systems (Section 3). Beyond rapid prototyping, it enables the fabrication of complex, customized reactor components (such as novel static mixers or multi-inlet geometries) that are difficult or impossible to manufacture using conventional methods. This is particularly impactful in flow chemistry, where 3D printing allows the integration of tailored mixing units directly into reaction channels, enhancing mass and heat transfer, improving reaction control, and accelerating process intensification. Moreover, it is a critical step towards the realization of personalized medicine and point-of-care manufacturing models, where treatments can be adapted to individual patient needs and produced closer to the point of use.
The aim of this review is to assess how emerging disruptive technologies in these critical research areas can enable greener-by-design pharmaceutical and agrochemical manufacturing. We will explore the current state of these innovations, pinpoint existing hurdles, and propose strategic pathways for their seamless implementation. Importantly, we will showcase relevant case studies where these technological frontiers can shape how synthetic routes are designed from the outset, and actively guide and accelerate the adoption of the synthetic strategies discussed in Part 1: Synthetic Frontiers. To conclude, we will provide industry-informed perspectives on the transformative potential of these technologies and the steps needed for successful adoption across their respective sectors.
2. Smart manufacturing
2.1. Real-time data acquisition
In a recent perspective, Qian and co-workers highlighted that information integration, dynamic risk assessment, decision-making support, and the lack of early warning systems are major barriers to green manufacturing.46 To address these, inline or online analytical tools, collectively termed as process analytical technology (PAT), provide comprehensive, real-time process insights, enabling swift, evidence-based decisions.37 As emphasized elsewhere, the integration of in situ analytics through PAT is essential for advancing both the quality and sustainability of chemical manufacturing at any scale (Fig. 2).47 | ||
| Fig. 2 Overview on how PAT enhances sustainability throughout the value chain, from small-scale research to process development and large-scale manufacturing. Reproduced from ref. 47, with permission from the American Chemical Society, copyright 2022. | ||
At the industrial level, PAT keeps critical process parameters within predefined optimal conditions. It directly leads to reduced raw material waste (i.e., through less product quality failure), minimized energy consumption, and enhanced overall process efficiency and safety. As the sector transitions from batch to continuous manufacturing, PAT facilitates self-optimization, scale-up, and fault detection. This enhances safety, improves operational flexibility, and ensures consistent product quality throughout production.48 Consequently, PAT has gained broad endorsement from both regulatory agencies and the fine chemical industry.45,49–52
At this large scale, PAT enables real-time monitoring and precise control of both individual unit operations and entire continuous plants in data-rich environments.35 A milestone was the first end-to-end continuous manufacturing process developed by the Novartis-MIT Center for Continuous Manufacturing. Multiple PATs (e.g., density meters, UV-vis) were employed to tightly regulate critical material and reaction parameters, minimizing disturbances across interconnected unit operations.53,54 Albeit sustainability was not explicitly discussed, this flow-based, real-time approach allowed the mixing, granulation, drying, and tablet compression unit operations to be merged into a single continuous screw extrusion. As a result, the 21-unit operations were reduced to 14, the residence times were shortened from 300 to 47 h and solvent consumption was minimized all while ensuring consistent product quality. In follow-up work, Benyahia et al. used a plant-wide dynamic model of a continuous pharmaceutical pilot plant to refine process design, decrease impurities and lower E-factor.55 In agrochemical settings, probes and sensors are also permanently integrated into production infrastructure at predefined control points, enabling continuous monitoring of critical process parameters and product attributes. The Bayer Production Network provides a striking example, with over 4000 PAT installations and more than 40 dedicated Near InfraRed (NIR) spectroscopy systems embedded across its manufacturing sites.
On the lab and pilot scale, the primary focus of this section, PAT creates a data-rich environment that supports modelling and automation. Amongst numerous benefits, it shortens optimisation cycles (thus cutting waste), minimizes human error and provides detailed insights on reaction mechanisms (i.e., real-time kinetics or the detection of unstable intermediates). Its use in this small-scale context is by no means mandatory, yet the sustainability gains are tangible, especially as smart-manufacturing strategies take hold. PAT is particularly well suited to continuous-flow synthetic protocols, a topic explored in greater detail in Part 1: Synthetic Frontiers. It is crucial to emphasize, however, that PAT operates within the chemical constraints of a given process. It does not alter the intrinsic nature of the chemistry involved. For instance, if process optimization is conducted exclusively using inherently toxic solvents or petrochemical feedstocks (Part 1: Synthetic Frontiers), PAT may improve control, efficiency, and data transparency, but it cannot overcome the fundamental sustainability limitations of these choices. The technology is only as green as the synthetic chemistry it monitors. Classic analytical equipment readily available to offline analysis can be adapted for real-time monitoring in continuous-flow setups. Among these techniques, infrared spectroscopy, particularly Near-Infrared Spectroscopy (NIR), is widely applied for real-time analysis.56 Unlike FTIR, which probes fundamental molecular vibrations in the mid-infrared region, NIR analyzes overtone and combination bands, making it especially effective for tracking bulk properties such as moisture content, concentration, and blend uniformity. Its non-destructive nature, rapid acquisition, and ease of automation make NIR a robust tool for continuous process control. Complementing IR-based methods, Raman spectroscopy offers distinct advantages, especially in aqueous or alcoholic media where –OH groups interfere strongly with IR but scatter weakly in Raman.57 Raman also operates reliably over a broad range of temperatures and pressures and has acquisition times comparable to FTIR, making it suitable for a wide range of reaction conditions.
UV-vis absorption spectroscopy, while more limited in structural resolution and prone to signal overlap, remains useful for reactions involving chromophoric species.58 Its versatility and simplicity continue to support its use in kinetic profiling and reaction tracking. Benchtop nuclear magnetic resonance (NMR) provides structural information with minimal sample preparation.59 While lower in sensitivity and resolution than high-field NMR, benchtop systems remain attractive for real-time monitoring of well-resolved species, especially when complemented by other techniques.
In contrast to spectroscopic methods, HPLC/UPLC physically separates components before detection (e.g., via UV, ELSD, CAD, or MS), enabling accurate quantification even in complex mixtures.60 Though slower (5–30 min acquisition time), it is well suited for reactions with longer residence times or where immediate feedback is not critical. Importantly, HPLC/UPLC is increasingly integrated into PAT frameworks. For example, in 2024, Cai et al., through the real-time application of HPLC-MS, investigated how palladium(II) species suppress epimerization during Buchwald–Hartwig C–N couplings used to synthesize a RORγ inhibitor.61 Crucially, HPLC allowed the authors to resolve temporal trends in substrate consumption and product formation, revealing that epimerization correlates with Pd speciation.
As seen in Table 2, each analytical technique presents limitations, and the lack of a universal analytical tool poses a significant challenge in monitoring multistep syntheses and complex reaction mixtures. Nonetheless, using multiple methods in tandem has significantly advanced the sustainable development of drugs and agrochemicals.37,62–82 Below we showcase recent examples where PAT facilitated greener flow synthesis of pharmaceutically and agriculturally relevant compounds.
| FTIR | Raman | UV-vis | NMR | HPLC/UPLC | |
|---|---|---|---|---|---|
| a Average analysis time observed in literature. b Steadily improving. c Multicomponent regression analysis might be necessary. d Depending on method and column selection. e Depending on nuclei, structure, and analysis time. f Automation and inline integration is possible. | |||||
| Sensitivity | Good | Good | Good | Fairb | Excellent |
| Time efficiencya | ∼15 s | ∼15 s | 30 s–1 min | >2 min | >5 min |
| Quantification method | Calibration | Calibration | Calibration | Direct Integration | Calibration |
| Mixture resolution | Fairc | Fairc | Fairc | Faire | Goodd |
| Impurity detection | Poor | Poor | Poor | Faire | Goodd |
| Sampling | Flow through | Flow through | Flow through | Flow through | Sample loopf |
| Structural information | Good | Good | Poor | Good | None |
Zhang and co-workers introduced a compact, reconfigurable continuous-flow platform for end-to-end API synthesis and formulation employing advanced PATs, such as inline FTIR, to regulate temperature, pressure, and flow rates thereby reducing waste (Scheme 1).83 For instance, real-time monitoring during diphenhydramine hydrochloride synthesis improved API yield to 82% while curbing solvent use. With reconfigurable modules operating in continuous mode, the platform accommodated APIs with diverse chemical profiles (e.g., lidocaine hydrochloride, diazepam, fluoxetine hydrochloride), reducing reaction times from hours to minutes. Its modular configuration facilitated cross contamination-free transitions between APIs, ensuring product quality in adherence to U.S. Pharmacopeia standards. Another related study was reported by Gilmore and co-workers and featured online FTIR and NMR analytics for continuous rufinamide synthesis.84
 | ||
| Scheme 1 Reconfigurable continuous-flow platform using online FTIR for multistep API synthesis. The system enables rapid switching between synthetic routes, illustrated here for the preparation of Lidocaine hydrochloride (top) and Diazepam (bottom). Each workflow combines controlled heating stages, inline phase separations, and real-time FTIR monitoring to track key functional-group transformations and ensure consistent product quality. Adapted from ref. 83. | ||
Kappe and co-workers also developed a modular, reconfigurable continuous-flow platform integrating FTIR, NMR and UPLC, to optimize an organometallic multistep reaction.85 The process involved deprotonating tert-butyl propionate with lithium diisopropylamide (LDA), reacting the resultant enolate with 4-fluorobenzaldehyde, and quenching with water to form the desired aldol product. FTIR recorded the ester peaks rapid disappearance at 1730 cm−1, confirming >90% deprotonation within 3.9 seconds of residence time. A 43 MHz benchtop NMR tracked 4-fluorobenzaldehyde conversion every 43 seconds, capturing intermediate formation and product concentration, while online UPLC, equipped with automated subsampling and dilution, distinguished the desired product from diastereomers. This approach afforded 70% yield and 4.2 g h−1 productivity upon scale-up, showcasing how PAT integration drives data-informed optimization, real-time parameter tuning, and reduced impurities.
However, it is important to distinguish between such reaction optimization, typically performed at the laboratory or pilot scale, and the sustained optimal operation required in a manufacturing environment. The two phases differ significantly in goals and timeframes: optimization seeks to identify the best reaction conditions, usually through iterative adjustments under tightly controlled settings. In contrast, once the process is transferred to production, the role of PAT evolves. Rather than guiding discovery, PAT in manufacturing serves as a real-time feedback system to ensure that the pre-established optimal conditions are consistently maintained. It enables process control, rapid fault detection, and assurance of product quality at scale, where robustness and reproducibility become critical. Recognizing this distinction is essential to properly frame the role of PAT across the development-to-production continuum.
A major obstacle to broader PAT adoption in multistep processes is the sheer complexity of implementing and managing it across each reaction stage. Bourne and co-workers tackled this by devising a telescoped continuous-flow platform to yield an aryl ketone hydroisoquinoline derivative (C-5), using a single online HPLC instrument for in-depth monitoring.86 Their process combined a Heck coupling, intramolecular cyclization, and acid-catalyzed hydrolysis. Daisy-chained multipoint sampling valves at each reactor outlet facilitated sequential analyses of individual reaction profiles, including aryl bromide conversion and vinyl ether and ketal intermediates. HPLC data guided adjustments to residence times, reagent stoichiometries, and temperatures. For instance, PAT revealed vinyl ether 3 formation was optimal at 125–140 °C with less than 14-min residence times, while controlling TsOH equivalents was critical for high yields in the final hydrolysis. After 14 h of optimization, the telescoped process furnished an 81% overall yield. Most importantly, relying on a single HPLC instrument significantly reduced setup complexity and costs without sacrificing detailed process insights.
Another hurdle for PAT to overcome is monitoring highly viscous fluids in continuous-flow mode, as stagnant boundary layers near sensors can slow response times. Veser and co-workers addressed this by testing the performance of inline real time FTIR in five flow configurations: horizontal flow, vertical flow, static mixer, horizontal nozzle, and vertical nozzle.87 The static mixer and nozzle setups improved response times by disrupting boundary layers and assisting fluid exchange at the expense of higher pressure drops. Among these, the horizontal nozzle yielded the fastest response. Computational fluid dynamics (CFD) simulations corroborated these observations, highlighting how flow dynamics and sensor placement affect data accuracy and system efficiency in high-viscosity processes.
Kappe and co-workers developed a multistep continuous-flow synthesis of Mesalazine, deploying different PATs in each reaction stage management (Scheme 2).88 The sequence comprised of three telescoped steps: nitration, hydrolysis and hydrogenation. In the nitration step, inline NMR spectroscopy quantified the ratios between 5-nitro-2-chlorobenzoic (5N-2ClBA) to its undesired 3-nitro-2-chlorobenzoic (3N-2ClBA) isomer at different stoichiometries and temperatures, following a published procedure.89 An NMR signal was recorded every 12 s while an indirect hard model (IHM) addressed signal overlap with less than 5% concentration uncertainty, ensuring optimal regioselectivity. For the hydrolysis of 5N-2ClBA to 5-nitrosalicylic acid (5-NSA), UV-vis spectroscopy acquired 2 s spectra, tracking reaction progress in real time. A data driven neural network (NN) model was trained on ∼35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 spectra obtained by systematically ramping the reactor temperature from 20 to 210 °C, spanning 0–100% starting material conversion. This enabled rapid, accurate optimization of alkaline hydrolysis conditions with minimal errors, an impressive feat given that NN-based concentration predictions required only 1.4 ms per spectrum. For the final hydrogenation step, inline FTIR tracked the loss of nitro stretching bands (∼1530–1350 cm−1) and the rise of amino bands (∼3300–3400 cm−1), allowing swift detection of deviations. A partial least squares (PLS) regression model delivered concentration estimates with <5% error for all reaction components. A final UHPLC analysis at the process endpoint confirmed a 79% overall assay yield and a throughput of 1.6 g h−1. Collectively, these PAT tools, combined with data analytics and advanced modelling improved process efficiency, flagged impurities, illustrating their synergistic role in modern flow chemistry.
000 spectra obtained by systematically ramping the reactor temperature from 20 to 210 °C, spanning 0–100% starting material conversion. This enabled rapid, accurate optimization of alkaline hydrolysis conditions with minimal errors, an impressive feat given that NN-based concentration predictions required only 1.4 ms per spectrum. For the final hydrogenation step, inline FTIR tracked the loss of nitro stretching bands (∼1530–1350 cm−1) and the rise of amino bands (∼3300–3400 cm−1), allowing swift detection of deviations. A partial least squares (PLS) regression model delivered concentration estimates with <5% error for all reaction components. A final UHPLC analysis at the process endpoint confirmed a 79% overall assay yield and a throughput of 1.6 g h−1. Collectively, these PAT tools, combined with data analytics and advanced modelling improved process efficiency, flagged impurities, illustrating their synergistic role in modern flow chemistry.
 | ||
| Scheme 2 Integrated continuous-flow route to mesalazine (5-ASA). The process converts 2-chlorobenzoic acid (2ClBA) into 5-nitro-2-chlorobenzoic acid (5N-2ClBA) via nitration monitored by inline NMR. Subsequent alkaline hydrolysis produces 5-nitrosalicylic acid (5-NSA), tracked by inline UV-Vis spectroscopy. The final catalytic hydrogenation step yields mesalazine (5-ASA), and inline IR is used to follow the reduction. Product quality and conversion are verified through online UHPLC analysis. Adapted from ref. 88. | ||
PATs are also broadly compatible with emerging technologies such as photochemistry, electrochemistry and biocatalysis. For instance, Ley and co-workers employed inline FTIR spectroscopy for the photochemical continuous-flow conversion of oxadiazolines to non-stabilized diazo compounds under a 310 nm UV lamp.62 FTIR spectroscopy played a critical role in detecting the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch of methyl acetate at 1746 cm−1 and the product N
O stretch of methyl acetate at 1746 cm−1 and the product N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretch at 2040 cm−1, allowing precise assessment of conversion efficiency. This approach enabled safe and efficient generation of volatile diazo species with an 80-min residence time at 10 °C, but also facilitated their immediate application in metal-free C(sp2)–C(sp3) cross-coupling reactions with boronic acids to prepare muscle-relaxant Baclofen in good yields.
N stretch at 2040 cm−1, allowing precise assessment of conversion efficiency. This approach enabled safe and efficient generation of volatile diazo species with an 80-min residence time at 10 °C, but also facilitated their immediate application in metal-free C(sp2)–C(sp3) cross-coupling reactions with boronic acids to prepare muscle-relaxant Baclofen in good yields.
More recently, George and co-workers progressed a continuous-flow metallaphotoredox C–O coupling from a small tubular reactor to a Taylor Vortex Reactor with the aid of inline FTIR real-time monitoring.90 The substrate scope included the antidepressant fluoxetine and an intermediate of the tuberculosis drug Delamanid with throughputs as high as 11 kg day−1.
Beyond photocatalysis, electrochemistry can also benefit from PAT, particularly the electrochemical Birch reduction, which is a commonly employed protocol (Fig. 3).91 George and colleagues applied a battery of real-time spectroscopy techniques including FTIR, Raman, and absorbance-transmittance-excitation Emission Matrix (A-TEEM) spectroscopy in the continuous-flow Birch reductions of naphthalene derivatives.92 Herein, FTIR tracked the consumption of reactants and product formation, while Raman and A-TEEM helped identify elusive reaction intermediates and product fingerprints. These PATs assisted in optimizing electrolyte and proton source concentrations ultimately achieving 98% selectivity for the single ring reduced product. Notably, the authors scaled up the protocol for a Ropinirole intermediate, leading to 95% yield and 114 g day−1 productivity. Other electrochemical examples such as silane oxidation93 and C(sp3)–H amidation94 have employed inline FTIR under continuous-flow conditions, while Cantillo and colleagues95 described a spinning electrode reactor equipped with online UV-vis for scalable electrochemistry.
 | ||
| Fig. 3 Electrochemical Birch reduction monitored by PAT. (a) Schematic of the electrochemical Birch reduction of an aniline, leading to an intermediate relevant to the synthesis of Ropinirole hydrochloride. (b) Representative inline Raman spectra collected during the reaction, with characteristic vibrational bands of reactants and intermediates. (c) Excitation-Emission Matrix fluorescence map acquired during the process, showing the evolution of fluorescent species as a function of excitation and emission wavelengths. Reproduced from ref. 92, with permission from the American Chemical Society, copyright 2022. | ||
Perhaps PAT’s most significant impact on sustainable manufacturing lies in its ability to generate data-rich environments that support modelling, digitization, and, critically, reactor automation. In this context, Jensen's Group fabricated an autonomous robotic flow platform for synthesizing Sonidegib, combining multiple PATs (LC-MS, FTIR, and UV-vis) with Bayesian optimization to telescope three reaction steps: nucleophilic aromatic substitution (SNAr), nitro reduction, and amide coupling (Fig. 4).96 For the SNAr step, researchers conducted 25 initial experiments to broadly map the parameter space, with FTIR monitoring the disappearance of the C–F bond signal and LC-MS tracking product yields and impurities. Subsequently, a Bayesian Gaussian Process Regression (GPR) model steered the optimization by iteratively refining temperature (70–120 °C), residence time (2–10 min), and base concentration (1–3 equiv). This positive feedback loop resulted in a SNAr yield of 93% with <0.1% impurities. In the following step, FTIR measured the nitro-to-amine conversion at a <5% validation error, allowing dynamic adjustments to the Pd/C catalyst loading and hydrogen pressure, which ensured steady conversion and high selectivity. Finally, LC-MS helped suppress guanidinium impurities during the amide coupling, achieving an overall purity of >99%. The fully autonomous system reached a throughput of 1.8 g h−1 and 89% overall yield, illustrating the synergy of advanced PATs, machine learning, and automation in efficient API production.
 | ||
| Fig. 4 Autonomous reactors featuring PAT feedback for self-optimization. (a) Schematic of the integrated flow system combining inline FT-IR, LC-MS, and NMR for real-time analysis and closed-loop decision making. (b) Optimization workflow showing the reaction parameters explored and the multi-objective Bayesian algorithm used to maximize yield and productivity while minimizing reagent consumption. (c) Photograph of the full experimental setup, illustrating automated sampling, photochemical reactor modules, and the PAT-driven feedback loop. Panels (a) and (b) are reproduced from ref. 96, with permission from the American Chemical Society, copyright 2022. Panel (c) is reproduced from ref. 97, with permission from Science, copyright 2024. | ||
More recently, Slattery et al. unveiled RoboChem, an automated system designed to optimize, intensify, and scale-up photochemical processes.97 Integrating inline NMR with a capillary photoreactor equipped with tunable LEDs, RoboChem employs Bayesian optimization to iteratively refine reaction conditions, achieving over 95% yields and boosting space-time yields by up to 500% across various photocatalytic transformations. Beyond these instances, numerous additional PAT-driven autonomous reactors have been documented, as recently reviewed.98,99
Despite ongoing challenges such as sensor reliability in viscous flows, complex data handling, and the absence of a universal PAT, these technologies remain influential in advancing sustainable production. Future endeavors will likely center on integrating PAT more closely with digital innovations, thereby enhancing sensitivity, resilience, and speed while moving closer to fully automated systems.100 In such systems, PAT will supply the real-time feedback essential for process self-optimization and control thus laying the groundwork for the next wave of modelling and AI-based process management.
2.2. Next-generation digital models
Based on their reliance on physical principles, models can be categorized into three classes: first-principle, data-driven, and hybrid models.102 First principles models are derived from the fundamental underlying physical, chemical, thermodynamic, and flow phenomena which typically integrate into the mass, energy, momentum, and population balance equations. Data-driven models are built based on the input-output behavior of a process whereas hybrid models can combine features both from first principles and data-driven models. A mathematical model can be represented by a single or a set of equations, typically algebraic, ordinary differential, or partial differential equations. Additionally, depending on their intended scope, mathematical models can be built to capture the dynamics or steady state behavior of a process.
Mathematical models play a crucial role in decision-making and process optimization, supporting the design, operation, and control of chemical and industrial processes.103 One of their key functions is evaluation, allowing for the prediction of process behavior under various conditions to assess feasibility and performance. They also enable optimal design by facilitating the development of cost-effective and sustainable processes and plants. Another important application is scale-up, where models assess the scalability of a process and optimize its performance for industrial implementation. Additionally, they contribute to process and plant-wide control, ensuring safe, resilient, and sustainable real-time operation. Beyond these operational aspects, mathematical models assist in calculating inventories, predicting and optimizing environmental impacts to support sustainability assessments. Furthermore, they enhance fault-free operation by detecting process faults and predicting maintenance needs, ultimately improving reliability and efficiency.
A recent review by Destro and Barolo104 has highlighted the essential role of mathematical models in fostering innovation and ensuring consistent quality throughout the manufacturing lifecycle of pharmaceuticals. Notably, these models increasingly incorporate environmental indicators (e.g., process mass intensity, E-factor) to evaluate sustainability across various unit operations from synthesis, dosage to packaging and logistics. Indeed, mathematical models have been an integral component of various ICH guidelines (Q8, Q9, Q10).18–20 This regulatory momentum, coupled with projected economic benefits, has amplified interest in mathematical models for pharmaceutical research.105–107 In this context, Gani et al. introduced a framework for selecting optimal reaction pathways in pharmaceutical manufacturing, featuring a series of modeling workflows to identify possible reaction routes, monitor processes, simulate operations, evaluate outcomes, optimize strategies, and inform real-time decisions.108
The advent of industry 4.0 and the inherent digital transformation consolidated even further the role of mathematical models in real-time decision making and operation and unveiled a new class of high-fidelity mathematical models referred to as digital twins (DT) (Fig. 5). A DT is a virtual replica of a process or a plant able to leverage PAT data and deliver real-time insights and foresights allowing more effective and resilient operation and most importantly enhanced quality and emissions control.109,110 DT consist of three core components: the virtual component utilizes real-time process simulation and optimization models (e.g., MATLAB, COMSOL, gPROMS, AspenTech) to conduct comprehensive system analyses, the physical component conducts data collection through human–machine interfaces (HMI) and machine–machine interfaces (MMI) (i.e., OPC, Modbus, MQTT) facilitating seamless information flow between equipment and control systems, and finally, the data management component relies on internet-of-things (IoT) platforms (i.e., Predix, Mindsphere, SEEQ) for secured connectivity, analytics, and scalable storage. The ability to access real time data, pertaining to the inputs (e.g., feed concentrations, flow rates, temperatures) and outputs (e.g., product concentration, purity, side products) along with their dynamic variations, allows the DT to predict more accurately and reliably key performance indicators and critical quality attributes (e.g., productivity). This ultimately improves performance compared to conventional modelling approaches, while minimizing material use and reducing waste.
For instance, DTs employed in continuous direct compression lines accurately predicted critical product attributes such as tablet composition, weight, thickness, and hardness supporting steady-state analysis, dynamic response prediction, and process risk minimisation.111 Furthermore, DTs can detect equipment faults, such as contamination in a bioreactor, thereby minimizing downtime and enhancing reliability. Incorporating integrated flowsheet models further enhance DT capabilities by simulating interactions between material properties, equipment status, and operating conditions, thus enabling sensitivity analysis,112,113 design-space identification,114 and process optimization.115,116 Currently, the vast majority of DT models lack real-time adaptability to changes in the physical system but advancements in their data management components promise to unlock their full potential. Indeed, forecasts suggest DT implementation could elevate pharmaceutical productivity by 150–200%. As a result, the use of DT technologies in pharmaceutical manufacturing industry is growing rapidly with an expected compound annual growth rate of 31.3% by 2034.117
In both the pharmaceutical and agrochemical sectors, embracing AI has potential to fuel efficiency, accuracy, and scalability (Fig. 6a).124 Although AI has been primarily used in drug discovery and clinical studies,125–132 its role extends to critical manufacturing processes. A recent FDA whitepaper (FDA 2023)133 identifies four key AI applications in drug production: process design optimization, advanced process control (APC), smart monitoring and maintenance, and trend analysis. These align with the five process approaches (structure, energy, synergy, time, and data) outlined by Van Gerven134 and Lopez-Guajardo.135 In particular, the “data” approach stipulates the integration of information and expertise to refine process control (Fig. 6b), a role AI can aptly fulfil across the entire manufacturing lifecycle (Fig. 6c). Consequently, AI-driven innovations form the backbone of Industry 4.0.
 | ||
| Fig. 6 AI-enabled strategies for modern process development. (a) Hierarchical overview of synthetic API process development, from route mapping and narrowing to scouting, selection, and subsequent process optimization. Green highlights indicate opportunities where AI-driven tools can enhance decision making, accelerate route design, and improve starting points for future projects. (b) Illustration of a digitalized process-development workflow, showing how sensor data, automated control, and cloud-based AI systems integrate to optimize operations and support manufacturing decisions. (c) Conceptual data and knowledge framework for Process Intensification 4.0, linking structural, operational, and digital domains to energy, space, data, and time efficiency. Reproduced from ref. 124, with permission from the American Chemical Society, copyright 2009. | ||
AI capabilities have led to the development of hybrid models that blend parametric and nonparametric methodologies.136 For instance, ML algorithms (XGB, kNN, NN, RF, DT, LR, and PLS) and hybrid physical ML models have been used to predict drug solubility in binary solvent mixtures (Fig. 7).137–140 In tableting, artificial neural networks (ANNs) can model multiple outputs more effectively than traditional multivariate analysis.141 ANNs have also demonstrated a predilection for correlating material specifications with in vivo drug performance, as recently demonstrated by a single layer model with five hidden nodes.142 Finally, ML models were developed to predict the life-cycle environmental impact of chemicals,143,144 laying the ground for more effective greener-by-design processes, particularly in the pharmaceutical and agrochemical sector where new chemical entities are continuously discovered and introduced.
However, reliable predictions require large, high-quality datasets.145,146 For instance in computer-aided tetrosynthesis (CAR), tools such as Synthia and IBM RXN, have shown considerable promise in proposing plausible synthetic routes.147,148 However, these tools often inherit historical biases and overlook modern green chemistry principles. A telling example is the CAR-designed synthesis of IM-204, a herpes (HSV) antiviral.118 Although the AI-generated route achieved a threefold yield increase over literature reports, it recommended >1 mol% Pd(OAc)2 for Suzuki coupling, and HATU for amide, conditions that are suboptimal from both a green and scale-up perspective. These reflect the limitations of current CAR datasets and scoring functions, which prioritize feasibility over sustainability. Consequently, while CAR tools mark a significant step forward, their practical use in pharmaceutical manufacturing relies, at least for the present, on expert oversight to ensure safe and sustainable outcomes.
To continue advancing towards reliable and autonomous machine learning models in synthesis planning and beyond, the incorporation of domain-specific knowledge is essential to ensure process stability, efficiency, and regulatory compliance.149–152 Combining advanced PATs with physical processes can address challenges like data scarcity, scalability, and interpretability.153–155 For instance, a hybrid method improved control in continuous dry granulation by coupling a first-principles roll compaction model with an NN-based ribbon milling model with partial least square regression for real-time PAT measurements.156 Other advantages of these hybrid models are a broader knowledge base, greater transparency in the modelling process, and cost-effective model development.157,158 However, successful dataset integration to conventional ML models requires thorough data pre-processing, including noise reduction, normalization, and feature selection.
To address this limitation, deep learning (DL), a transformative subset of ML, can be deployed. DL enables hierarchical learning from complex, high-dimensional datasets by automatically extracting features from raw data. Consequently, it can uncover intricate patterns standard ML might overlook.159 For instance, Krumme et al. applied a DL neural network to monitor a wet granulation line in solid dosage production.160 The model approach managed seven critical process parameters (e.g., API mass flow, liquid feed rate, dryer conditions) and tracked eight quality attributes, such as API content, particle size distribution, and loss on drying. Utilizing near-infrared spectroscopy, the model achieved a calibration error of less than 10%, demonstrating precise monitoring and control of complex and noisy pharmaceutical formulation datasets.64,161
Furthermore, due to DL's capacity to automatically extract features without explicit engineering, it outperforms other ML algorithms in prediction accuracy.162 For instance, convolutional neural networks (CNNs) excel in image analysis (segmentation, classification, and object detection) by extracting both high- and low-level features from modern in situ imaging probes.163–166 These architectures comprise of specialized layers for (a) feature extraction, (b) pooling for dimensionality reduction and (c) fully connected layers for classification or prediction. However, training CNNs is resource-intensive so models like AlexNet, GoogLeNet, and ResNet with 8, 22, and 152 layers, respectively are based on existing knowledge to minimize data and computational demands.
Recurrent neural networks (RNN) represent another powerful class of DL networks designed for sequential or time-series tasks including optimization167 and process control.168,169 In contrast with the traditional feedforward NNs, RNNs feature feedback loops that provide memory-like behavior, effectively modelling temporal dependencies.
Likewise, Farkas and colleagues employed advanced RNN variants (NARX and LRNN) to build a digital twin for low-dosage continuous powder blending.137 The digital twin comprised of three key components: NN models to represent the blending process, PAT monitoring for data collection and PLS regression for data processing. By exploiting this digital twin, they thoroughly defined the blending process design space, enhancing process understanding and optimization. Notably, the NARX model was comparable to residence time distribution models in terms of accuracy and reliability, illustrating how deep learning techniques can match or outperform traditional soft sensing models when handling complex, high-dimensional data.170,171
Recent ML advances have introduced statistical approaches like Bayesian inference.172 These frameworks quantify how failure scenarios affect critical product attributes by: (a) assessing failure probability and severity, (b) modelling probabilistic parameter distributions, (c) accounting for mean values, and (d) addressing uncertainties arising from limited sampling.141,173 Consequently, the models adeptly balance critical product quality attributes with manufacturing cost, yield, and throughput objectives. For example, Treitler and colleagues applied Bayesian modelling to predict failure rates in drug manufacturing elucidating probabilistic impurity distributions with respect to pre-defined acceptance criteria.150
Similarly, Bayesian Neural Networks (BNNs) extend ML and DL capabilities by addressing uncertainty quantification.152 By representing probability distributions over weights, BNNs capture both aleatoric (data-driven) and epistemic (model-driven) uncertainties, enabling more robust decision-making.154 Building on this concept, Bayesian Recurrent Neural Networks (BRNNs) fuse the modelling abilities of RNNs with Bayesian inference enabling a probabilistic approach to input–output mapping. Unlike traditional RNNs, BRNNs assign probability distributions to parameters, thereby quantifying both types of uncertainty. This probabilistic modelling improves decision accuracy, mitigates risks, and increases confidence in predictions.153
Importantly, “confidence”, or more precisely the lack of transparency, is a major issue with AI and ML models as their decision-making process is often opaque. Explainable Artificial Intelligence (XAI) seeks to address this shortcoming by employing surrogate models, sensitivity analyses, and visualization tools, that clarify how input variables influence outcomes,155,156,174 and these methods have been also widely applied in industry.130,175–177 Though XAI is still an emerging field,178 its synergy with advanced AI techniques promises to make ML approaches both trustworthy and compliant with industry standards.
Beyond industrial settings, AI has also fueled substantial automation of academic laboratory workflows. The Ley group, for instance, created a cloud-based system that interconnects any laboratory setup via TCP/IP protocols, delivering real-time data collection and storage that streamline experimentation.64,179 Security challenges associated with the online nature of the platform are mitigated by strictly controlled departmental network access, user authentication, and IT safeguards. LabMate.ML, developed by Rodrigues and co-workers, exemplifies a self-evolving AI engine.180 Employing adaptive random forest models, LabMate.ML identifies optimal reaction parameters with minimal initial data requirements. OpenFlowChem161 and Rxn Rover181 are alternative open-source platforms.
While LabMate.ML was an important early demonstration of a self-evolving AI engine using adaptive random forests, it did not gain widespread adoption. Similarly, platforms like OpenFlowChem and Rxn Rover laid the groundwork for networked lab automation, but the field has since evolved towards more democratized, robust systems under the banner of “self-driving laboratories”. In this context, the work by Doyle and co-workers represents a significant advance in data-driven reaction optimization by introducing a robust, generalizable framework for Bayesian optimization in chemical synthesis.182 Unlike earlier tools such as LabMate.ML, which focused on model-free learning with limited uptake in the community, this work builds on widely accepted Gaussian process models and open-source, user-friendly software (EDBO), enabling broad applicability across both batch and flow systems. The study also emphasizes Active Learning, now an established paradigm in iterative chemical optimization, by dynamically balancing exploration and exploitation across high-dimensional reaction spaces.183 This methodological rigor and real-world applicability go beyond earlier demonstrations, positioning Bayesian optimization as a cornerstone of modern computer-assisted synthesis.184 As a follow up study, Panayides et al. presents a significant advance in the integration of open-source digital infrastructures for autonomous reaction optimization in flow chemistry.185 Using Node-RED, the authors developed a cost-effective and highly customizable dashboard for control and automation of flow reactors, integrated with Summit, a Python-based machine learning library, to execute closed-loop Bayesian optimization of an allylation reaction. This work importantly lowered the barrier to entry by decoupling from proprietary software and demonstrating seamless interconnection between hardware, analytics (HPLC), and ML-based decision-making. Overall, it is now well acknowledged that the shift towards Gaussian process-based approaches is a more standardized framework in the field. Today's “self-driving labs” combine integrated robotics, inline analytics, and active-learning optimization strategies with community-supported tools. These developments signal a transition from isolated academic demonstrators to robust, transferable technologies capable of transforming chemical discovery across academia and industry.185 Recent reviews have provided comprehensive overviews of the rapidly evolving field of machine learning-guided chemical synthesis and optimization. For readers seeking a broader context and deeper insight into the integration of Bayesian optimization, active learning, and self-driving laboratories, we refer to these more extensive discussions.98,186–188
It is worth noting that despite the tremendous advantages of mathematical models, building predictive and reliable models for both single processes and plants in the pharmaceutical and agrochemical sectors is often a laborious and challenging endeavor. Typically, large sets of experimental data are required for parameter estimations or model training. This translates into large sets of experiments or repeated experimental campaigns at different scales based on trial-and-error methodologies or complex design of experiments such as factorial and Box-Behnken designs. To address these challenges, high-throughput screening (HTS) methodologies have been increasingly employed. Despite their advantages, conventional HTS approaches frequently encounter limitations, including operational costs, substantial labor demands and occasionally low success rates, restricting broader adoption, not to mention consideration of green principles.189 Consequently, building reliable models may be cost prohibitive in terms of resources, labor, and environmental footprint. This highlights the urgent need for greener and more effective design of experiments. Model-Based Design of Experiments (MBDoE) may overcome some of these challenges by identifying reduced sets of optimal experimental campaigns that specifically minimize uncertainties of a selected mathematical model or DT (Fig. 8).190 MBDOE has notably curtailed experimental effort in automated flow reactors,191,192 crystallization,193,194 freeze-drying195 and other unit operations. For example, Van Hauwermeiren et al. reported a 72% reduction in both experimental time and material use, which are crucial factor for scarce and costly APIs.196 Similarly, MBDoE decreased experimental efforts by 70% during tablet lubrication for direct compression.197 However, the benefits of MBDoE may be significantly affected and often limited by the quality of the selected model, parameter correlations, and poor data information content. Recently, a new method was proposed to address these shortcomings by combining (a) systematic model selection and discrimination, (b) evaluation of the data information content based on the estimability approach, also known as practical identifiability, and (c) MBDoE.194 This approach successfully built the most reliable model amongst a set of possible candidates, reduced model uncertainties and parameter correlations, and, most importantly, reduced the number of required experiments from 8 (or more) to 2 based on factorial designs. As a side note, MBDoE can be designed to address multiple objectives and identify the most realistic trade-offs or compromises, be it cost, model quality, or environmental emissions, a process known as multi-objective optimization (MOO).198
2.3. Towards autonomous production
With the increased need for smart plug-and-play processes that require minimum supervision, a new class of real time optimization strategies known as self-optimization has emerged. This strategy leverages real-time data and advanced algorithms to dynamically adjust and improve processes autonomously (without operator intervention).
As practices shift towards continuous manufacturing, automation is a subject of considerable interest. Automated optimization platforms typically consist of flow reactors, PAT instruments (Section 2.1), and self-optimization algorithms that iteratively propose and test alternative reaction conditions (Fig. 10). These adaptive algorithms provide real-time, multi-variate, and multi-objective optimizations, at a fraction of the cost and time.82,212,213 By eliminating the need for repeated intervention and reactive corrections, they enable more resilient operation and self-adaptive manufacturing.
 | ||
| Fig. 10 Combinatorial library synthesis using SNOBFIT for self-optimization. (a) Conceptual overview of the stopped-flow reaction system used for rapid combinatorial screening, enabling controlled mixing of multiple reagents and integration with inline analytical tools (NIR, HPLC-MS, Prep-LC). (b) Schematic of the automated platform, including reagent preparation, sequential injection, reaction under high-pressure/temperature conditions, and real-time analysis within a closed-loop SNOBFIT optimization workflow. (c) Heatmap representation of the resulting amide library, generated from the combinatorial reaction of five carboxylic acids with five amines using four coupling agents across nine reaction conditions (900 total reactions), illustrating reactivity patterns and optimal conditions discovered by the self-optimization algorithm. Reproduced from ref. 223, with permission from the Royal Society of Chemistry, copyright 2022. | ||
It is worth noting that some of the traditional optimization algorithms can be adapted to self-optimization environments by incorporating shifting response surfaces. As an illustrative example, Lapkin and co-workers proposed a methodology that combines MBDoE with self-optimization for the C–H activated continuous-flow synthesis of aziridines, rapidly generating reliable process models under minimal experimental workload to meet cost, productivity and sustainability goals.214 Earlier, Reizman and Jensen used feedback DoE to optimize the alkylation of 1,2-diaminocyclohexane, iteratively refining a linear response surface model for multiple solvents and ultimately narrowing them from 10 to 1 in just 67 experiments.215
Beyond improving synthetic processes, the effective identification of steady-state conditions via real-time optimization and self-optimization strategies enables shorter yet data-rich sequential experiments. This approach significantly enhances data collection efficiency and accelerates process development. Girard and co-workers exemplified this advantage in their study on the self-optimization of continuous crystallization of Nirmatrelvir, the antiviral component of a COVID-19 treatment (Paxlovid). Employing mixed-integer self-optimization algorithms coupled in conjunction with iterative DoE, the experiments were strategically grouped based on similar temperature conditions. This strategy effectively minimized equilibration times between achievable steady state conditions, ensuring lower energy consumption, reduced material waste, and improved process efficiency.216
Two classes of algorithms are popular in the context of self-optimization, namely: local optimizers applied within a confined experimental region (e.g., Nelder-Mead simplex217,218) and global optimizers across the entire reaction space (i.e., Stable Noisy Optimization by Branch and Fit (SNOBFIT),219 Bayesian optimization182).
Global optimizers manage experimental noise more effectively than local methods. For example, SNOBFIT autonomously optimized 4 parameters in a telescoped amide coupling followed by an elimination reaction for the synthesis of Osimertinib, a lung cancer treatment.65 The optimization was complete in just 42 experiments over 26 h. The algorithm can also achieve purity and yield improvements in multi-step reactions.220–222 SNOBFIT was recently employed in high-throughput design-make-test-analyse (DMTA) platforms to complete approximately ∼900 reactions in 192 h for combinatorial library synthesis and at-line reaction analysis of bioactive compounds. Notably, reactant and solvent usage was reduced by 90% compared to fully continuous alternatives (Fig. 10).223 Moreover, the algorithms capabilities extend beyond reaction optimization, enabling the refinement of manufacturing aspects such as the reaction-extraction phase in multistep processes.224
BO offers an alternative global strategy by iteratively building a surrogate model and acquisition function from existing data, then refining predictions through targeted experimental tests.96,97,225,226 This excels at tackling complex problems with limited function evaluations and can manage experiment parallelization. A recent example applied BO to the continuous-flow optimization of the Buchwald–Hartwig synthesis of pyridinylbenzamide, a pharmaceutical intermediate. Two separate optimizations were carried out: one seeded with 14 prior data points and another with only 5. Both converged to the same optimal conditions (79% yield), but the smaller prior dataset required 27.6% fewer experiments overall (21 vs. 29 experiments). This counterintuitive result highlights a critical point: more prior data does not necessarily translate into greater efficiency, and careful selection of initial data points can be as important as the optimization algorithm itself.227
To enhance the initial sampling process, Latin hypercube sampling (LHS) is often applied, generating a diverse and representative set of starting points, that comprehensively covers the parameter space with fewer experiments. For example, BO with an adaptive expected improvement (BOAEI) algorithm optimized the oxidation of four organic sulfides to sulfoxides using hydrogen peroxide and phosphotungstic acid as a catalyst, across four continuous parameters.207 Process evaluation identified optimal conditions in under 25 experiments and 24 h, yielding up to 25% more yield than conventional one-point screening, for each substrate. Similarly, the BOAEI algorithm optimized the telescoped synthesis of an aryl ketone precursor for 1-Methyltetrahydroisoquinoline, reaching an 81% yield in just 13 experiments within 14 h.86 The integration of multipoint sampling provided detailed reaction profiles, ensuring a comprehensive understanding of each step and streamlining global optimization for multistep syntheses.
BO can also be connected to other optimizers in order to expedite response-surface mapping and pinpoint global optima228 and by using Kernel Density Estimation for categorical data, further reduce experimental needs.229,230 Similarly, Experimental Design via BO techniques like density functional theory encoding, Gaussian process surrogate modelling, and expected improvement acquisition are combined to reduce experimental load.182 Finally, incorporating Kernel Density Estimation for data smoothing enables BO to optimize categorical variables without converting them into continuous forms.230,231
Other classes of ML techniques have been effectively employed in self-optimization, leveraging advanced algorithms to further reduce experimental effort and enhance decision-making in dynamic environments. For example, Zare and co-workers combined a long short-term memory model with deep reinforcement learning (DRL) to self-optimize four two-component microdroplet reactions by iteratively identifying optimal flow rates, voltage, and pressure, achieving optimal results in fewer steps than customary optimization routines.167 Additionally, the deployment of DRL agents based on deep deterministic policy gradient proved effective as a self-optimization strategy for flow chemistry.232
Regardless of whether they consider the partial or entire reaction space, self-optimization protocols can refine a single performance metric at a time (e.g., single objective optimization). For example, Felpin's group refined multiple single-objective reactions through tailored experiments,233 while deMello's group employed constrained optimization to balance trade-offs and identify a global optimum.234 However, these methods often struggle to address conflicting criteria such as cost minimization and environmental performance.235,236 Multi-Objective Optimization (MOO) algorithms are designed to find the Pareto front, which captures a set of possible compromises between conflicting or competing criteria. Established methods like NSGA-II can accurately approximate the front, but often require substantial computation and expertise to operate, making them unsuitable for self-optimizing platforms.
In contrast, Thomson sampling efficient multi-objective (TS-EMO), a machine learning based approach is purpose-built for global MOO scenarios, where function evaluations are inherently costly. It was reported that TS-EMO can cut experimental requirements by 50% relative to standard single-objective algorithms, fostering dynamic data re-evaluation and active learning.224 Moreover, this method was successfully applied to optimize a Sonogashira synthesis of Lanabecestat (AZD3293), achieving optimal conditions within just 13 h.224 This eliminated the need for additional experiments when downstream work-up specifications changed, significantly reducing the consumption of high-value catalytic reagents during process development. In the same study, TS-EMO identified 18 Pareto-optimal solutions in a 65-h, 109-experiment multi-step Claisen-Schmidt condensation and separation process, optimizing key objectives such as purity, space–time yield, and reaction mass efficiency.224 Moreover, it converged on the Pareto front within 68 and 78 experiments, respectively, when optimizing space-time yield and E-factor of SNAr and N-benzylation reactions.237 TS-EMO also adeptly tackled both discrete (solvent, ligand) and continuous (temperature, residence time, concentration) variables, autonomously optimizing SNAr and Sonogashira reactions in 18 and 22 h, despite the challenges posed by localized low space-time yields.238
To predict and gauge the performance of these autonomous approaches, in-silico benchmarking tools like SUMMIT and OLYMPUS are commonly utilised. SUMMIT assesses an array of algorithms to streamline industrial reaction steps, substantially cutting down on experimental overheads.239 OLYMPUS furthers this by considering experimental noise, constructing response surfaces for different scenarios and examining multiple algorithms in real-time (Fig. 11).240 Applying Olympus, the Bourne group benchmarked multiple MOO algorithms, including TS-EMO, ParEGO, EIM-EGO, NSGA-II, and BS-TESMO to pinpoint optimal Sonogashira conditions with fewer experiments. Such tools augment and reduce resource consumption, yielding more sustainable manufacturing.241
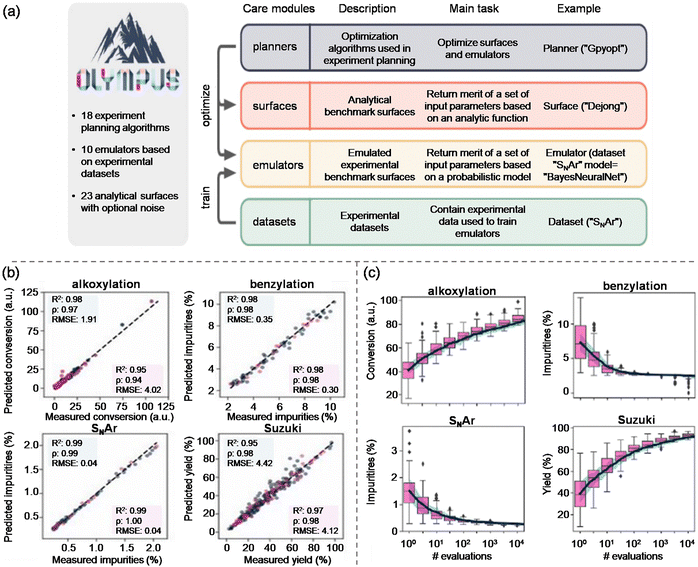 | ||
| Fig. 11 Overview and benchmarking of the OLYMPUS platform for experiment planning. (a) Structure of the OLYMPUS framework, highlighting its four core modules, i.e., planners, surfaces, emulators, and datasets. Each module supports experiment planning through optimization tools, analytical benchmark surfaces, probabilistic emulators, and curated experimental datasets. (b) Parity plots comparing experimentally measured versus emulator-predicted outcomes for representative reactions, including alkoxylation, benzylation, SNAr, and Suzuki couplings. High correlation coefficients demonstrate the accuracy of OLYMPUS emulators in reproducing experimental behavior. (c) Example performance curves for random-search optimization across different reaction surfaces (alkoxylation, benzylation, SNAr, and Suzuki coupling). The plots show how impurity or yield metrics evolve with increasing numbers of evaluations, illustrating the benchmarking capabilities of OLYMPUS for assessing optimization strategies. Reproduced from ref. 240, with permission from IoP Science, copyright 2021. | ||
Achieving environmental and operational excellence demands a holistic and hierarchical plant-wide control strategy which can effectively reconcile optimization and stabilization requirements at different segments of the plant (Fig. 12a).53,249 This may include combining reaction-purification and upstream and downstream operations. By integrating such an advanced control and optimization framework, operational disturbances like product quality deviations can be promptly resolved250 and environmental footprints, production times, and operational costs can be reduced relative to batch processes in real-time.251,252
 | ||
| Fig. 12 (a) Flowsheet of a fully integrated continuous pharmaceutical pilot plant, illustrating upstream synthesis, liquid-liquid extraction, crystallization, solid handling, extrusion, and tablet coating operations. The diagram highlights the interconnected unit operations and sequential processing required to convert raw materials into coated tablets. (b) Plant-wide mass flow diagram showing the major input and output streams, including solvents, reagents, excipients, product, and waste. This schematic emphasizes how material flows are distributed across the pilot plant and supports holistic process monitoring and optimization. Reproduced from ref. 249, with permission from the American Chemical Society, copyright 2012. | ||
A common approach for plant-wide control involves PID controllers.53,244,253 These feedforward networks perform adequately, but often struggle with prolonged dead times, necessitating extensive detuning to maintain stability. Model Predictive Control (MPC)252,254–258 and hybrid MPC-PID controllers256,259,260 offer stronger performance and flexibility.203–205 For example, MPC surpasses PID in controlling the outlet mass flow of feeders and blenders grappling with mass flow disturbances.261,262
Recent dynamic plant-wide models incorporate mass and energy balances, population balance equations, thermodynamics, and unit correlations (Fig. 12b). Some archetypical systems contain over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 differential and 2000 algebraic equations.55,263,264 At the Novartis-MIT Center for Continuous Manufacturing, Barton and co-workers developed a hierarchical plant-wide control structure, that uses dynamic models for parametric sensitivities in control loops, accurately predicting system behavior during start-up and shutdown.100,249 In this same end-to-end system, an MPC strategy with the quadratic dynamic matrix control (QDMC) algorithm efficiently managed plant-wide dynamics.258,265 QDMC's computational efficiency and independence from system dimensionality make it suitable for real-time oversight of high-dimensional processes. To overcome this challenge, Stochastic MPC factors in the probabilistic distribution of uncertain parameters, offering greater flexibility than traditional worst-case MPC. A novel fast SMPC algorithm unites the input-output framework of QDMC with generalized Polynomial Chaos (gPC) theory for efficient uncertainty propagation and improved control.265 Meanwhile, Multivariate Statistical Process Control (MSPC) enables simultaneous monitoring and optimization of interconnected variables, though it cannot directly regulate processes.266,267
000 differential and 2000 algebraic equations.55,263,264 At the Novartis-MIT Center for Continuous Manufacturing, Barton and co-workers developed a hierarchical plant-wide control structure, that uses dynamic models for parametric sensitivities in control loops, accurately predicting system behavior during start-up and shutdown.100,249 In this same end-to-end system, an MPC strategy with the quadratic dynamic matrix control (QDMC) algorithm efficiently managed plant-wide dynamics.258,265 QDMC's computational efficiency and independence from system dimensionality make it suitable for real-time oversight of high-dimensional processes. To overcome this challenge, Stochastic MPC factors in the probabilistic distribution of uncertain parameters, offering greater flexibility than traditional worst-case MPC. A novel fast SMPC algorithm unites the input-output framework of QDMC with generalized Polynomial Chaos (gPC) theory for efficient uncertainty propagation and improved control.265 Meanwhile, Multivariate Statistical Process Control (MSPC) enables simultaneous monitoring and optimization of interconnected variables, though it cannot directly regulate processes.266,267
Importantly, successful plant-wide control hinges on real-time process data to identify trends, pinpoint correlations, and detect faults, transforming raw information into actionable insights. Thus, integration of in-process control tests (e.g., weight, hardness), PAT tools (e.g., Infrared Spectroscopy) and univariate/multivariate data analytics are essential to reinforce control strategies in continuous manufacturing.160
As a final point to consider, startup and shutdown operations in continuous manufacturing plants can generate substantial waste due to dynamic transitions and inherent deviations from steady state performance and targeted quality attributes.55 Startup may require several residence times before steady-state conditions (e.g., flowrate, temperature) are achieved leading to significant waste. Similarly, shutdown operations often leave behind considerable residual material, further contributing to environmental burdens. Recently, several model-based optimal control alternatives of a multistage continuous crystallizer were developed and benchmarked showing a reduction of startup time and waste by up to 80%.268 Similarly, novel optimal shutdown strategies have been effective in converting nearly 80% of the residual material into on spec product.269
2.4. A case study on sustainability: the role of continuous purification
Purification is among the most resource- and waste-intensive stages in the downstream processing of pharmaceuticals and agrochemicals given its high solvent consumption and environmental impact. The most employed purification techniques include crystallization, chromatography, membrane separation and liquid–liquid extractions. Despite the broader move towards continuous processes, batch separations remain the norm, largely due to relatively low initial investment and operational versatility.112,270 The latter is especially important in the pharmaceutical sector, where a diverse drug portfolio demands an equally varied range of purification techniques. However, batch methods often entail prolonged processing times, batch-to-batch variability,27 and inefficient impurity removal, which collectively drive up solvent usage. The extent of resource consumption as well as their process efficiency depends on the specific batch method, as reflected in their PMI values (Fig. 13a). Batch crystallization (PMI 100-210) provides precise purity and particle-size control and resists clogging, but suffers from batch-to-batch variability.33,271–273 Batch chromatography (PMI 170-310) excels in enantiomeric purification but is affected by limited throughput and high resin costs stemming from inefficient utilization of the stationary phase.274–277 Membrane-based batch purification (PMI 240-660) effectively processes larger molecules (>500 g mol−1), but requires high solvent volumes and frequent membrane replacement.278–280 Lastly, liquid–liquid extraction (LLE) benefits from mild conditions, but has substantial space and energy requirements alongside disproportionately large PMI values (up to 3200).281–283Over the past two decades, continuous purification has gained traction, driven by the desire for better product consistency, shorter lead times, and lower PMI7,282 with much research and development now focused on chiral pharmaceutical separations.284 Beyond higher throughput and reduced contamination risk, continuous purification methods fit smoothly into end-to-end manufacturing, eliminating offline bottlenecks and human intervention, features particularly beneficial for reactive or unstable intermediates.285 They are particularly suited for multistep continuous-flow synthesis, a topic explored in greater detail at Part 1: Synthetic Frontiers, where inline purification techniques (i.e., phase separation, immobilized reagents, and nanofiltration) not only reduce solvent use and manual handling but also enable seamless integration of complex transformations.286,287 This positions continuous purification as a key enabler of autonomous manufacturing and, by extension, Industry 4.0. This section thus explores the operational and environmental benefits of continuous purification, demonstrating how smart manufacturing principles of real-time analytics and advanced process control can transform the purification process.
A prominent continuous crystallization technique is the Mixed Suspension Mixed Product Removal (MSMPR) crystallizer, where a well-mixed crystal slurry is continuously withdrawn while fresh supersaturated feed is introduced, maintaining near-constant operating conditions.292 It is important to note, that although referred to as continuous, these units often feature prolonged residence times under controlled stirring rather than a strictly continuous feed and discharge.293 The technique is scalable and offers reduced operational costs, while inherent limitations, such as low per-stage yield or clogging, can be mitigated through process modeling and cascade configurations.294
In a recent study, Mazzotti and co-workers investigated the continuous crystallization of L-glutamic acid (LGA), an amino acid precursor of neurotransmitter glutamate.295 LGA exists in two polymorphic forms, the metastable αLGA and the thermodynamically stable βLGA, each with distinct solubility and handling characteristics. Conventional batch approaches struggle with polymorphic control with αLGA dissolving and reprecipitating as βLGA. Additionally, the strong tendency of αLGA to agglomerate causes poor suspension stability and clogging.
To address these issues, single and two-stage (cascade) MSMPR crystallizers were tested. The cascade configuration outperformed the batch operation, yielding up to 73% βLGA at 4.4 kg m−3 h−1 with a PMI of 200. When solvent recovery was implemented, it was further reduced to 5.7. The cascade sequence also displayed 55% higher yields compared to the single MSMPR. To effectively monitor the continuous process, various PAT tools were considered. Their suitability was determined by using tp/ta as an assessment metric, where tp is process duration and ta is analyzer time. When tp/ta < 1, the PAT tool is unsuitable as the analysis time exceeds the process time, preventing timely intervention. Both Raman (tp/ta ≈ 10) and UV-vis spectroscopy (tp/ta > 10) accurately distinguished between αLGA and βLGA thus facilitating stable continuous operation.296
Seidel-Morgenstern and colleagues demonstrated similar advantages by implementing enantiomeric preferential crystallization (PC) for L-threonine, an essential amino acid common in dietary supplements. Batch crystallization of a mixture of D-/L-threonine in water tends to co-crystallize the undesired counter-enantiomer. Two alternative setups were explored to alleviate this issue: a single gram-scale MSMPR unit and a coupled pair featuring liquid-phase exchange.297 After excluding startup phases, the study tested four scenarios at various temperatures and residence times. At 38 °C and τ = 46.5 min, the coupled MSMPR system doubled productivity to 6.3 g h−1 L−1 while maintaining >99% enantiomeric purity. Additionally, its PMI fell to 4.1 from a batch benchmark of 7.3. This drop was largely attributed to the lower solvent consumption enabled by continuously exchanging liquid phases between the two MSMPR chambers. Notably, the coupled configuration reduced process interruptions, bolstering steady-state operation.
As an illustrative example, Pais et al. implemented a pilot-scale inline SMB system to separate four stereoisomers of nadolol, a nonselective β-blocker for hypertension and angina pectoris.302 Optimizing solvent composition through advanced simulation tools proved crucial in balancing separation efficiency, throughput, and solvent use. Switching from pure methanol which yielded 0.33 g L−1 h−1 at a 26.1 L g−1 solvent consumption to a mixture of methanol/acetonitrile/diethylamine (25/75/0.1, v/v/v), increased productivity by 133% and cut solvent consumption by 63%. This improvement was reflected in a PMI of 9.62, in stark contrast to typical batch values of 100–210. Importantly, the target stereoisomer was recovered with 100% purity.
Crucial to optimizing the separation efficiency of the system was the integration of inline and real-time PAT. UV-vis and NIR spectroscopy (estimated tp/ta ≈ 103) monitored concentration profiles, ensuring high purity without offline HPLC delays. Mass spectrometry (estimated tp/ta ≈ 102–103) tracked solvent composition to prevent adsorption equilibrium shifts. This data on critical process parameters was integrated into a feedback control system to maintain optimal separation efficiency and system performance.
Beyond PAT, process control techniques such as MPC are integral to continuous purification, and especially SMB chromatography, due to the system's inherent nonlinearity and multi-zone complexity. The advent of AI and ML is bound to increase MPC efficiency as highlighted in Section 2.2. For instance, Chang-Ha et al. developed a ML dynamic model to optimize the commercial-scale SMB separation of C10–C14n-paraffins from kerosene, which are vital feedstocks for detergents, plasticizers, and solvents.303
The existing industrial process used zeolite 5A with n-pentane at 177 °C and 24 bar to ensure sufficient diffusion, optimized using a conventional dynamic model. However, limited data and the nonlinear behavior of adsorption, combined with complex flow and feed interactions, challenged the model. Its reliance on simplified assumptions (ideal plug flow, constant velocity, uniform particles) failed to capture real fluid dynamics, pressure changes, and mass transfer limits.
To overcome these limitations, an ML approach was implemented using real industrial data to enhance predictive accuracy and optimize impurity removal. Initially, several ML algorithms, including random forest, ANN and DNN were evaluated to identify the most suitable for the process in question. Among these, random forest emerged as the most effective, offering the highest predictive accuracy when handling high-variance data, particularly for forecasting aromatic content in n-paraffin products. Despite its high accuracy, it is worth noting that the random forest decision-making process is non-transparent (black box), a major limitation in the highly regulated pharmaceutical sector. Explainable Artificial Intelligence analysis promises to address this issue as discussed in Section 2.2. While no dedicated PAT instrument was present, key parameters such as p-xylene concentration in the flush zone were tracked to guide impurity removal. As a result, overall process performance improved significantly, achieving 99.6% purity with recovery rates of 95.7% (nC10), 88.9% (nC12), and 81.6% (nC14). The SMB process was also sustainable with a PMI value of 13.6. These findings underscore the growing synergy between machine learning algorithms and continuous purification strategies.
A key membrane-based technique is nanofiltration, which selectively separates solutes by molecular size, charge, and hydrophobicity.306 Nanofiltrations are well suited for removing organic impurities, endotoxins, and multivalent salts during drug purification. Nevertheless, fouling, suboptimal chemical resistance, and limited membrane lifespans pose significant hurdles. In addition, although long-term operating savings may offset initial investment costs, the high cost of this technique is a challenge for small-scale applications.307,308
In response, Livingston and colleagues developed a two-stage nanomembrane cascade to remove genotoxic 4-dimethylaminopyridine (DMAP) from roxithromycin, a broad spectrum antibiotic.309 The separation system was based on the molecular weight differences between DMAP and roxithromycin (837 g mol−1vs. 122 g mol−1). The system was optimized under varying flow rates and transmembrane pressures (5–30 bar). At pressures above 6 bar, roxithromycin rejection approached 99.2% with only 22% DMAP rejection. Under optimized conditions, 1 m3 of industrial stream was purified in 40 h at an estimated 99.4% yield, although the outlet flow still contained 0.5 kg m−3 DMAP.
From a sustainability standpoint, the process was benchmarked against five batch and continuous purification scenarios. Single-stage batch diafiltration exhibited PMI of roughly 1400, mainly due to excessive solvent use, whereas optimized two-stage batch processes, despite full solvent recovery, remained between PMI 200 and 300. By contrast, a continuous two-stage cascade without solvent recovery reduced PMI below 200, and the same system with 100% solvent recovery achieved a PMI of 21, reflecting higher product yields and a smaller environmental footprint.
For instance, Alcázar and colleagues developed a membrane-based continuous LLE process for a fully autonomous, end-to-end process.311 Their streamlined approach involved generating organozinc reagents from readily available alkyl halides, prior to performing Negishi coupling for broad (hetero)aryl halide functionalization. Consequently, 54 target molecules were produced comprising of a 30-member combinatorial list of indazole derivatives and a 24-compound medicinal chemistry library focused on mGluR2 NAMs, a promising class of antidepressant compounds.
Conductivity-based membrane LLE was employed to detect the interface between the organic (non-conductive) and aqueous (highly conductive) phases, simplifying separation and avoiding emulsions. A Tecan liquid handler automated the phase separation, eliminating manual intervention, while mass-triggered preparative HPLC and LC-MS enabled automated purification, compound identification, and fraction collection. Additionally, an automated evaporator removed solvents during post-purification, and a liquid handling system reformatted the purified compounds for high-throughput screening. This fully automated workflow minimized reagent loss, enhanced reproducibility, and eliminated pre-evaporation steps, significantly improving overall process performance and environmental sustainability.
3. Innovative fabrication through 3D printing
The previous section examined how smart manufacturing transforms continuous processes through real-time data acquisition, advanced analytics, and AI control systems to minimize waste and ensure high product quality. All of the examples focused on mass production. Innovative fabrication techniques like 3D printing offer a paradigm shift, one that emphasizes customization, rapid prototyping, and personalized small-scale manufacturing. Indeed, 3D printing is widely hailed as one of the 20th century's most transformative technologies, reshaping numerous manufacturing sectors. For example, 3D printing enables the fabrication of lightweight components to improve fuel efficiency,312 supports product customization without major cost increases,313 and allows for decentralized production due to its compact size,314–320 reducing dependence on global supply chains.In contrast to conventional subtractive manufacturing, where cutting, drilling or milling removes excess material from a bulk to attain the desired product, 3D printing follows an additive paradigm, layering materials in sequence along the x, y, and z axes.321–327 This rich and diverse field, sometimes termed as additive manufacturing, encompasses several techniques that differ in starting materials, deposition methods, and resultant product characteristics (see Box 1).321,322
Box 1. Standard steps involved during 3D printing.All 3D printing approaches share key production stages:328–331• Conceptualization: creating the initial design using computer-aided software (CAD) • Conversion: exporting the design to a printer-readable format (commonly .stl), which encodes dimensions, placement and orientation. This step often involves detecting and correcting errors or resolution loss using specialized software. • Equipment setup: configuring parameters (movement, speed, temperature) in G-code, the command language of 3D-printers. Although many devices auto-generate G-code, operators often refine it manually, making this a semi-automated step with significant user input. • 3D printing: sequentially depositing layers by precise head and build-plate in the xy and z directions. The total print time depends on the chosen technique, machine capabilities, object size, operational parameters, and resolution. • Object finishing: removing surplus material or support structures and performing any required post-processing (cleaning, polishing, sanding). This standardized workflow of 3D printing, from digital design to post-processing, not only ensures reproducibility and customization but also aligns closely with the principles of greener-by-design process development. In pharmaceutical applications, these advantages translate into smaller footprints, reduced solvent use, and intensified synthesis or purification steps, paving the way for sustainable, modular, and agile manufacturing systems. |
Each method offers distinct advantages in resolution, material compatibility, and cost, topics thoroughly explored elsewhere and beyond the scope of this review. Nonetheless, Fig. 14 briefly outlines the 3D printing methods most pertinent to pharmaceutical manufacturing.
 | ||
| Fig. 14 Additive manufacturing techniques employed in pharmaceutical production. The schemes illustrate four commonly used additive manufacturing modalities: (i) material extrusion, in which molten material is deposited layer-by-layer; (ii) Vat photopolymerization, where a photo-curable resin is selectively hardened using light; (iii) binder jetting, involving selective deposition of a binding liquid to join powder particles; and (iv) powder bed fusion, where powder layers are fused by localized thermal energy to build the desired structure. Reproduced from ref. 332, with permission from Wiley, copyright 2019. | ||
3.1. 3D printing trends
The fine chemical sector is emerging as one of the most dynamic and swift adopters of 3D printing.333–337 In the context of chemical production, 3D printing has enabled the design and fabrication of bespoke components that are difficult or impossible to achieve through conventional manufacturing techniques, and that can be used to enhance both batch and continuous processes.338 These include 3D-printed flow reactors with precisely engineered geometries for tailored residence time distribution, heat and mass transfer efficiency, and integrated sensing. Additionally, additive manufacturing has been applied to create structured catalyst supports, static mixers, and column packing materials with fine-tuned porosity and surface area. For example, Cronin and Burke's work on 3D-printed reactionware has enabled compact, modular setups for multi-step synthesis, facilitating process intensification, automation, and integration with analytical tools.339 Similarly, Pérez-Ramírez and co-workers340 and Kappe and co-workers341 have demonstrated how metal additive manufacturing could be harnessed for robust flow hydrogenations involving highly exothermic reactions, offering superior heat transfer and structural stability. Van Summeren342 and Zogg and co-workers343 have brought these innovations into the industrial field, using custom 3D-printed reactors and inserts to solve scale-up challenges for highly exothermic reductions and heterogeneous hydrogenations. Another notable example by CSIRO's FloWorks underscores the potential of metal additive manufacturing in catalysis. They fabricate static mixer scaffolds from robust metals such as titanium and stainless steel followed by 3D coating with catalytic metals including Pd and Ni, to enable efficient hydrogenations in flow.344,345 Compared to polymer 3D prints used in early development, metal-based prints like titanium or Inconel better withstand industrial conditions through greater thermal conductivity, solvent resistance, and mechanical durability. These attributes facilitate strong catalyst adhesion and reactor–catalyst integration, enabling levels of process intensification that are generally unattainable with polymeric systems.A very recent and interesting direction is the 3D printing of catalysts themselves for fine chemical manufacturing. Here, Vilé and co-workers integrated atomically dispersed catalytic layers onto 3D-printed static mixers, combining the advantages of single-atom catalysis with structured flow design for efficient and selective hydrogenations.346 In a follow-up study, the same authors developed self-standing, photocatalytically active structured catalysts via high-resolution photopolymerization, directly embedding single-atom catalytic functionality into the printed material.347 This eliminated the need for washcoating, improved adhesion, and offered a scalable, one-step manufacturing route for advanced structured catalysts. Thus, 3D printing is redefining reactor design by blurring the line between reactor and catalyst to enable compact, high-efficiency catalytic systems.
In terms of applications to formulated products, early research predominantly used off-the-shelf 3D printing materials to fabricate simple monolithic drug products with rather basic shapes, often overlooking pharmaceutical quality standards.348–354 However, the field has swiftly progressed to (a) engineering complex constructs (e.g., hollow, multi-layer, multi-material), (b) developing sophisticated release profiles (e.g., prolonged or pulsatile), (c) targeting diverse administration routes (oral, ocular, auricular, vaginal)355–362 and (d) targeting the development of temporally evolving structures (4D printing).357,363 At present, much industrial and academic interest regarding the use of 3D printing focuses on personalized medicine.348–351 Personalized medicine is a medical approach that tailors every treatments phase – from diagnosis to therapy – to the specific genetic, physiological, or pathological profiles of individual patients or patient subgroups.364–366 Specifically, 3D printing is a critical enabler of personalized medicine through:367–371
• Customized design and composition: this improves patient adherence through appealing shapes and sizes, plus tailor-made colors, flavors, and textures.
• Enhanced treatment safety: this enables features like embedded Braille, aiding identification for patients with disabilities.
• Precise and personalized drug dosing: this optimizes therapeutic efficacy by adjusting dosages and drug combinations to individual patient needs.
• Co-formulation of multiple APIs: this incorporates multiple APIs with distinct release kinetics into a single dosage form.
• Complex release patterns: this prevents burst effects, enables zero-order release in prolonged matrices, and promotes rapid dissolution of poorly soluble drugs.
Beyond personal medicine, numerous studies have highlighted the versatility of 3D printing. A single printer can fabricate a wide spectrum of dosage forms, including capsules, tablets, suppositories, vaginal rings, implants and inserts, accommodating various administration routes.333,370 Moreover, 3D printing extends beyond drug production by enabling the creation of custom testing devices (e.g., Franz diffusion cells) and organ-on-a-chip platforms, which are particularly useful in research settings.372–374 Scientists have also paired 3D printing with conventional production processes to yield innovative hybrid methodologies. One example is the production of bilayer tablets featuring a standard injection-molded prolonged-release layer and a customizable Fused Deposition Modeling (FDM)-printed immediate-release layer.375 This hybrid setup permits the administration of a second customized drug dose or even a different API, each with its own release kinetics. Another approach merges 2D inkjet and 3D printing to combat substandard or counterfeit medicines.376,377 Recent developments further broaden the scope of 3D printing. 3D Bioprinting combines cells, growth factors, and other bioactive substances that evolve into tissues or organs over time,378,379 while 4D printing furnishes objects capable of changing shape in response to external non mechanical stimuli.354,357,363,380–382 Concurrently, artificial intelligence is increasingly being investigated to expedite development by reducing trial-and-error screening.383 The transformative scope and versatility of 3D printing have drawn major investment across sectors, with industrial use falling into two main areas: large-scale production and small-scale, on-demand manufacturing (personalized treatment).
3.2. 3D printing for large-scale manufacturing
The first application of 3D printing in industrial settings has followed the traditional one-size-fits-all approach mass production model (Fig. 15, top).370 As 3D printing is inherently designed for precision and complexity rather than mass replication, the technology has undergone significant evolution to meet the particular needs of mass production. | ||
| Fig. 15 3D-printing equipment used across different production scales. The top row shows systems designed for large-scale industrial pharmaceutical manufacturing, while the bottom row highlights compact or benchtop 3D-printing units suitable for decentralized or on-demand drug production. Reproduced from ref. 384–386, with permission from Elsevier, copyright 2025. | ||
One notable example is ZipDose, a formulation platform pioneered by Aprecia using MIT-licensed technology.387,388 ZipDose yields “powder-like” formulations of well-established APIs and excipients, featuring a highly porous architecture that rapidly disintegrates in the oral cavity using minimal liquid or saliva. Even at high API loads, these rapidly dispersing tablets retain sufficient strength for packaging in crush-resistant cavities possessing a distinct advantage over both freeze-dried and conventional orally disintegrating tablets. Aprecia's manufacturing process uses standard powder blending and liquid binder preparations in a specialized 3D printing process. A key technological leap was separating and immobilizing the powder and liquid stations while moving the build modules underneath them in a circular pattern which enabled near-continuous production in a single facility. Later generations of the equipment transitioned from a linear design to a circular platform to further minimize space requirements, while clinically-certified construction materials, easy cleanability, and integrated process controls ensured regulatory standards were met. As proof of concept, Aprecia produced high-dose systems containing 1g of a cardiometabolic drug. These disintegrated swiftly both in vitro and in vivo. Their first commercial offering was Spritam, a high dose levetiracetam product for epilepsy. Albeit Spritam comprised of an established API and common excipients, the approval process was complex as it was classified as a new dosage form. Nonetheless, it was quickly vetted through clinical studies, confirming bioequivalence to immediate release levetiracetam in 33 healthy volunteers.
Building on ZipDose, Aprecia has since unveiled ZipCupTM, an orodispersible system.389 The outer sections are shaped like nested cups, into which the API formulation is enclosed and sealed. This design accommodates diverse payloads and forms, including granules and enables various release profiles.
Other companies have also ventured into large scale 3D printing for pharmaceuticals. Triastek Inc. launched the MED formulation platform, capable of producing 150–200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units daily for clinical trials or high-volume manufacturing.389–392 MED is a hybrid system, that fuses elements of hot melt extrusion with 3D printing to continuously convert powder feeds into a softened or molten state, bypassing the need for pre-prepared filaments. With parallel modules for feeding, mixing, and printing MED can craft multi-compartment, multi-drug structures. Over 70 products have undergone in vitro, and several have advanced to human trials under the brand 3DμS (3D Microstructure for Modified Release) targeting oral, gastric and colonic release. Examples include several open-source products like T19 (rheumatoid arthritis), T20 (cardiovascular and clotting disorders), T21 (ulcerative colitis), T22 (pulmonary arterial hypertension) and D23 (primary immunoglobulin A nephropathy).
000 units daily for clinical trials or high-volume manufacturing.389–392 MED is a hybrid system, that fuses elements of hot melt extrusion with 3D printing to continuously convert powder feeds into a softened or molten state, bypassing the need for pre-prepared filaments. With parallel modules for feeding, mixing, and printing MED can craft multi-compartment, multi-drug structures. Over 70 products have undergone in vitro, and several have advanced to human trials under the brand 3DμS (3D Microstructure for Modified Release) targeting oral, gastric and colonic release. Examples include several open-source products like T19 (rheumatoid arthritis), T20 (cardiovascular and clotting disorders), T21 (ulcerative colitis), T22 (pulmonary arterial hypertension) and D23 (primary immunoglobulin A nephropathy).
Similarly, Laxxon Medical introduced SPID (Screen Printed Innovation Drug), a manufacturing platform inspired by flatbed screen printing methods common in textiles and electronics.376,393–395 Here, a screen mesh deposits semi-solid paste onto a flat substrate, with drug shapes dictated by a blocking stencil. The platform has successfully produced various acetaminophen tablet forms with hardness comparable to commercial equivalents. Moreover, SPID can finely control drug internal structure, facilitating heterogeneous distribution of APIs and excipients and thus sophisticated release profiles. SPID is touted as highly scalable, potentially producing 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–40
000–40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units at once without significant process modifications. Laxxon aims to extend drug patents and facilitate the relaunch of generic APIs in a manner akin to Spritam with potential disease targets including epilepsy, diabetes, rheumatoid arthritis, and prostate carcinoma. Beyond SPID, Laxxon Medical is focusing its R&D endeavors on 3D printing additional medications such as levodopa and dapsone, with elaborate shapes, and embedded anti-counterfeiting features.
000 units at once without significant process modifications. Laxxon aims to extend drug patents and facilitate the relaunch of generic APIs in a manner akin to Spritam with potential disease targets including epilepsy, diabetes, rheumatoid arthritis, and prostate carcinoma. Beyond SPID, Laxxon Medical is focusing its R&D endeavors on 3D printing additional medications such as levodopa and dapsone, with elaborate shapes, and embedded anti-counterfeiting features.
Notably, 3D printing advances are not confined to therapeutics. The nutraceutical company Nourished employs a proprietary large-scale printer with seven simultaneous printheads to construct layered vegan supplements enriched with active ingredients like probiotics, vitamin B12, and calcium. Customers are able to customize their 3D-printed supplements by choosing from 28 constituents or completing an online questionnaire.396–398
3.3. 3D printing for personalized small-scale manufacturing
Undoubtedly, 3D printing's greatest promise is in relocating production hubs from large industrial sites to hospitals and neighborhood pharmacies (Fig. 15, bottom). This paradigm shift replaces the traditional mass-manufacturing model with customization, yielding several advantages. For instance, pharmaceuticals worth around US$283 billion currently require cold-chain storage and transport, a burden that decentralized production could notably lessen. Central to this concept is the simplification of pharmaceutical compounding, a cornerstone of personalized medicine.399–401 In compounding, pharmacists prepare tailor-made medications when off-the-shelf products are inadequate. Traditionally, compounding entails: (a) assessing patient-specific dosage and ingredient sensitivities amongst other factors (b) weighing and mixing APIs with excipients, (c) adjusting characteristics like flavor or texture for greater compliance, (d) meeting quality standards, and (e) packaging and labeling the final product. By automating many of these steps (e.g., weighing, mixing, and packaging), 3D printing can reduce human error and production time, while in principle enabling performances not attainable through conventional compounding techniques. Furthermore, a single 3D printer can customize the type, quantity, and strength of multiple APIs in one dosage form; modify excipients to avoid allergens or improve palatability; and tailor product design, physical properties, and release kinetics.Several companies have begun capitalizing on this burgeoning market. FabRx Ltd., for example, offers M3DIMAKERTM, a modular 3D printer compatible with FDM, Drop-on-demand Piezoelectric Ejection (DPE), and Single-Sided Extrusion (SSE) printheads.375 A key feature is its suite of Process Analysis Technologies (PAT), which enable real-time quality monitoring through weighing and imaging systems, Raman confocal microscopy, and near-infrared spectroscopy.358,402–404 Such nondestructive PAT are economically essential given the small-scale nature of personalized medicine. In tandem, FabRx's M3DISEEN software applies AI-driven modeling to predict printability and product quality.405 Through, academic collaborations, FabRx has demonstrated the M3DIMAKERTM's versatility in furnishing a multitude of dosage forms, shapes and flavors, with a particular emphasis on SSE-manufactured chewable medications for pediatric use.406–408 These include hydrocortisone and isoleucine formulations for adrenal insufficiency and maple syrup urine disease respectively as well as chewable forms containing citrulline, isoleucine and valine to treat short-chain enoyl-CoA hydratase deficiency and ornithine transcarbamylase deficiency. These 3D-printed drugs have demonstrated efficacy comparable to customary compounded treatments, while improving patient adherence.
As an alternative to M3DIMAKERTM, CurifyLabs has introduced Pharma Printer another SSE 3D printer likely due to the inherent stability of this technique.409–413 Pharma Printer has already been operated in two Nordic university hospitals by personnel with no prior 3D printing experience.414 In a recent multi-site study spanning 30 pharmacies in eight European countries, Pharma Printer manufactured propranolol hydrochloride tablets of varying strengths using CuraBlend, a GMP-certified excipient base.413 The manufacturing process followed a standardized 4-phase protocol, certifying each batch through non-destructive PAT quality checks (weighing, NIR, and HPLC).415,416 Additionally, the printer integrates built-in mixing and direct blister-pack filling, while CurifyLabs supplies predefined device settings, quality dossiers, and automated digital batch records for a more automated and streamlined operation.
Other noteworthy players include CELLINK, which renewed its partnership with AstraZeneca for drug discovery;417 Vitae Industries and Craft Health which are developing pharmaceutical-grade printers;418,419 DiHeSys is collaborating with Harro Höfliger on FlexDosePrinter for oral-dispersible films;420 and 3DForMe, a DPE-based printer from the University of Bari and PharmaLabor.421–423 All of the aforementioned companies supply “inks” or formulation development services to ensure uniform product quality. Although such standardization contradicts the core tenet of 3D-printing, namely end-user customization, it is essential for pharmacists, as extensive on-site product testing cannot be pursued within pharmacies. Indeed, concerns over pharmacist liability, the absence of established guidelines, and quality control are commonly cited reasons for the hesitant adoption of 3D printing in community pharmacies.424 In response, recent research focuses on validating the 3D printing process and implementing non-destructive quality checks to reassure all stakeholders.425,426
In conclusion, the compact footprint, flexibility, and versatility of 3D printing present an enticing path to automating the production of small, highly personalized batches of therapeutics at the point of care, be it hospitals or pharmacies. Yet, lingering questions regarding the regulatory environment, operational logistics, and cost-efficiency persist. Ultimately, whether this approach will fully meet stakeholder expectations, including financial viability for industry participants, remains to be seen.
4. Outlook and future perspectives
The convergence of continuous manufacturing, smart analytics, digital modeling, and advanced fabrication techniques is unlocking new opportunities for innovation in pharmaceutical and agrochemical production. As these technologies are increasingly deployed in tandem, their individual strengths are not merely additive but synergistic, accelerating development timelines, enhancing process control, and contributing to more sustainable manufacturing practices across the value chain.Central to this transformation is the role of data. While the current discourse often emphasizes the abundance of data, in practice, the success of AI-driven optimization and process control strategies hinges far more on data quality. Today, predictive models are typically introduced late in development, once a process has already been optimized through conventional means. This practice diminishes their potential to meaningfully impact decision-making during critical stages. However, by integrating PAT and advanced experimental design methodologies earlier in development, more relevant and actionable data can be captured from the outset. This shift enables richer insights and more effective process optimization strategies throughout the development cycle.427,428
Nonetheless, this transition is not without its challenges. Many industrial environments remain encumbered by data silos, fragmented knowledge management systems, and concerns over data confidentiality. These factors hinder collaboration and limit the generalizability of models trained on academic or public datasets when applied to the far more complex, variable, and specialized conditions of real-world manufacturing. Bridging this gap requires a multifaceted approach. One key strategy is to better expose academic researchers to industrially relevant, data-rich experimentation during their training, either through research exchanges or through collaborative programs. In parallel, the pharmaceutical and agrochemical industries can support this evolution by openly sharing non-competitive data, including negative results that are typically underreported but highly informative for algorithm training and model generalization.429 Initiatives such as the Open Reaction Database exemplify how structured community efforts can contribute to high-quality, standardized datasets that are critical for the development of robust, generalizable predictive models.430
Technologies like federated learning have already shown promise in drug discovery contexts.431 While not yet widespread in manufacturing, these frameworks offer a path forward to unlock cross-sector collaboration without compromising proprietary data.432–434
Even with improved datasets and model architectures, hybrid approaches that integrate algorithmic predictions with expert intuition will remain essential, and, especially in tightly regulated environments, human oversight is crucial, not only for compliance but also for interpreting the outputs of complex models in context.
Nevertheless, the need for flexible, adaptive manufacturing platforms is growing rapidly, particularly as pharmaceutical pipelines diversify. With increasing investment in complex molecules such as peptides, oligonucleotides, and antibody-drug conjugates, manufacturing strategies must evolve to accommodate distinct production requirements while maintaining agility. Modular, “modality-agnostic” systems are particularly promising in this regard, allowing for the rapid reconfiguration of production lines and seamless integration of Quality by Design principles. Each module can be independently validated, scaled, and optimized, reducing both development time and regulatory risk.435
In parallel, the optimization of the chemistry underlying the production of high-value fine chemicals has become increasingly important. The strategies articulated here contribute to this effort, but several challenges continue to constrain progress. Continuous manufacturing, for instance, often suffers from sensitivity to recipe variations, mixing inefficiencies, and heat transfer limitations, factors that can compromise the control of critical quality attributes such as crystal size, morphology, polymorphism, and purity. These attributes, in turn, have direct implications for drug efficacy, safety, and downstream processing characteristics. To address these limitations, future research must focus on the simultaneous optimization of product characteristics, process configurations, and operational parameters. Such a comprehensive approach will ensure that economic, regulatory, and environmental objectives are met. This includes fine-tuning crystallization recipes and outcomes (e.g., solvent selection, particle size distribution), identifying optimal technologies (e.g., MSMPR, fluidized beds, tubular or segmented reactors), and optimizing dynamic operations such as startup and shutdown conditions to ensure consistent, on-spec production trajectories across the phase diagram.
The ultimate objective is the realization of fully autonomous, integrated digital manufacturing platforms. These would be capable of executing continuous screening and optimization workflows through robotic systems, automating tasks like sample extraction and injection into analytical platforms (e.g., HPLC), and integrating all hardware, software, data processing, and visualization tools within a centralized supervisory framework. Such platforms will represent a major leap towards the concept of digital chemistry at scale.
In this evolving technological landscape, 3D printing is emerging as a transformative tool. Once confined to rapid prototyping, additive manufacturing now enables the fabrication of complex reactor geometries that enhance mixing, heat transfer, and mass transport, particularly relevant in photochemical and electrochemical processes where conventional equipment may be inadequate.436 These advantages extend beyond efficiency to scalability and decentralization. By enabling just-in-time production of custom parts or even active ingredients near the point of use, 3D printing has the potential to make supply chains more resilient, lower transportation costs, and improve access in underserved regions. In a context where contract development and manufacturing organizations (CDMOs) are playing an increasingly central role, additive manufacturing also offers a competitive edge in demonstrating responsiveness and technical sophistication.
The future of pharmaceutical and agrochemical production will undoubtedly be shaped by the rise of connected and intelligent plant operations. As digital infrastructure matures, manufacturers are beginning to integrate real-time PAT systems, digital twins, and advanced control algorithms into cohesive production environments. These “smart” plants can monitor operations continuously, detect deviations early, and in some cases, self-correct to maintain optimal performance. Importantly, such connectivity enables not just unit-level control but end-to-end optimization across entire production lines and networks. Yet, smart manufacturing of APIs and finished pharmaceutical products still plays only a modest role in commercial-scale production. Most manufacturing remains rooted in conventional processes and equipment, often operated in a partially batchwise manner. Nevertheless, significant progress has been made in developing new methods for process design, control, and intensification. Techniques such as PAT and digital twins are central to this shift, enabling both high-fidelity monitoring and real-time correction, thus ensuring tighter quality control and process efficiency.
Looking ahead, several developments are anticipated to redefine the manufacturing landscape: first, the implementation of newly designed, intensified processes at commercial scale; second, the widespread adoption of PAT and digital twins for superior quality control and online optimization; third, the integration of 3D printing for patient-specific formulations; and finally, the contribution of modular design and engineering to accelerate and scale continuous manufacturing. These connected operations offer advantages that extend far beyond productivity. They serve as critical enablers that can bring the synthetic innovations discussed in Part 1: Synthetic Frontiers into real-world production. A prime example is the multigeneration development of belzutifan (Fig. 16), where PAT and modelling were not merely supportive tools but instrumental in driving sustainable process transformations.75,128,437–447
 | ||
| Fig. 16 Evolution of the synthetic route to belzutifan, highlighting the progressive redesign from long medicinal chemistry sequences to increasingly efficient manufacturing processes. Generation 1 introduces a streamlined 5-step route and a fully continuous key transformation, while Generation 2 exploits biocatalysis, telescoped operations, and water-based SNAr chemistry to further intensify the process. These advances were enabled by modern development methodologies, including digitalized workflows, PAT-guided optimization, DoE-driven understanding, and data-centric decision making, which together allowed rapid identification of superior conditions and robust scale-up strategies. The resulting improvements in yield, PMI, organic solvent use, and reduction of chlorinated/amide/ether solvents demonstrate how integrated innovation in chemistry, engineering, and digital tools can transform the sustainability and manufacturability of an API. Adapted from ref. 446 and 447. | ||
In the first-generation commercial synthesis, a notable implementation of PAT and modelling tools was the flow photochemical benzylic bromination step.443 Inline NMR, UV sensors, and LED actinometry enabled real-time conversion tracking, while photon flux modelling guided photon dose control and reactor scaling. This allowed the elimination of overbromination impurities and reduced reaction time from hours to minutes. These tools collectively eliminated overbromination impurities and reduced reaction times from hours to minutes. However, a downstream Ru-catalyzed dynamic kinetic resolution (DKR) introduced a disproportionate burden in terms of cost, precious metal usage, and E-factor, compounded by separate solvent handling steps.445 These and other inefficiencies ultimately prompted Merck to redesign the synthesis.
In the second-generation route, PAT and modelling were deeply integrated to support a transition to biocatalysis, earth-abundant metals, aqueous media, and process intensification.447 A key transformation was the dioxygenase-catalyzed benzylic hydroxylation that replaced Pd/C oxidation.438,441 This step was enabled by a suite of PAT tools (e.g., dissolved oxygen probes, inline FTIR, and gas-flow sensors) that controlled oxygen levels, while kLa and Njs modelling informed scale-up and optimized mass transfer within a water/1-octanol biphasic system. This reduced enzyme loading, minimized process volumes, and eliminated hazardous solvents. Further downstream, the Ru-catalyzed DKR was replaced with a Keto Reductase (KRED) process through enzyme evolution and PAT-enabled process development.439In situ FTIR, dissolved oxygen probes, and real-time imaging revealed that oxygen mass transfer and indanone dissolution are sequentially rate-limiting, and that 1-octanol plays a critical role in preventing indanone aggregation, maintaining particle size, and improving reaction conversion. Beyond real-time reaction tracking PAT also provided mechanistic insights which facilitated the process development of an efficient and robust biocatalytic transformation.
In the final SNAr step, PAT (UPLC with EasySampler) was used to map hydrolysis impurity formation and support the switch from formamide to water as solvent.440 Detailed kinetic profiling revealed sensitivity to CO2 evolution, prompting headspace pressure and nitrogen sweep adjustments. While no formal mechanistic model was used, PAT-guided empirical optimization, controlled phenol deprotonation and minimized hydrolysis. The aqueous SNAr was coupled with a reactive crystallization that allowed direct isolation of the API without extraction or solvent exchange. This eliminated multiple purification steps and exemplified solvent minimisation using water as the reaction and isolation medium.
Across both generations of development, PAT and modeling were not passive monitoring tools but fundamental enablers of deep process innovation. The second generation process shortened the linear sequence from 16 to 9 steps (a 44% reduction), increased overall yield from 4% to 26% (a >6-fold improvement), and cut PMI by over 75%, from 2039 to 508.447 This includes a 34% PMI reduction compared to the generation one commercial route. The impact on solvent use was even more striking: total organic solvent consumption was reduced from ∼1400 to ∼261 liters (81% reduction), while toxic and genotoxic solvents were nearly eliminated. Consequently, by minimizing waste, optimizing resource use, and facilitating sustainability assessments smart manufacturing lays the foundation for a more responsible and resilient chemical manufacturing sector.
To fully realize these benefits, however, several systemic challenges must still be overcome. Regulatory frameworks need to evolve to accommodate emerging technologies while preserving safety and efficacy standards. The introduction of ICH Q13 guidelines on continuous manufacturing is an encouraging development, but further clarity and standardization will be required for technologies like AI-based control and additive manufacturing.
Talent development is equally critical. The sector will increasingly rely on professionals with T-shaped skillsets: individuals who possess deep expertise in one area, combined with a broad understanding of related fields. These individuals must also exhibit strong learning agility and openness to technological change. Academic institutions and industry alike will need to collaborate closely to foster such profiles through curricula that balance specialization with interdisciplinary fluency.
Lastly, economic factors will play a decisive role in determining the pace of technological adoption. Although many innovations promise long-term cost savings and performance gains, their implementation often entails significant upfront investment. As such, companies must weigh the return on investment carefully, particularly given the uncertainties inherent in drug development. Strategic partnerships can play a key role in reducing risk, sharing knowledge, and accelerating implementation through access to best-in-class technologies.
In summary, the scientific and technological frontiers explored in this review present a compelling vision for the future of pharmaceutical and agrochemical manufacturing. Embracing these innovations is not merely a strategic advantage: it is a necessity. In an era of growing scrutiny and complex societal demands, the industry must commit to continuous innovation, deep collaboration, and bold integration of emerging tools. Only through such efforts can it deliver safer, more sustainable, and more accessible products for the patients and communities it serves.
Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
The authors gratefully acknowledge financial support from the European Commission through a Marie Skłodowska-Curie Fellowship under the “SOLCAT” project (T.A.G., grant agreement No. 101152890); the Horizon Europe's “Global Challenges and European Industrial Competitiveness” programme for the “SusPharma” project (R.I.T., A.A., R.L., B.B., grant agreement No. 101057430); the UKRI Horizon Europe Guarantee Scheme supporting the “SusPharma” project at Loughborough University (R.I.T., B.B., grant agreement 10038378); the Marie Skłodowska-Curie Doctoral Network “GreenDigiPharma” (M.C.I., grant agreement No. 101073089); and the European Research Council for the ERC Starting Grant “SAC_2.0” (G.V., grant agreement No. 101075832).References
- R. A. Sheldon, Green Chem., 2023, 25, 1704–1728 RSC.
- F. Roschangar, J. Li, Y. Zhou, W. Aelterman, A. Borovika, J. Colberg, D. P. Dickson, F. Gallou, J. D. Hayler, S. G. Koenig, M. E. Kopach, B. Kosjek, D. K. Leahy, E. O’Brien, A. G. Smith, M. Henry, J. Cook and R. A. Sheldon, ACS Sustainable Chem. Eng., 2022, 10, 5148–5162 CrossRef CAS.
- L. J. Diorazio and A. Mullen, Curr. Res. Green Sustain. Chem., 2022, 5, 100247 CrossRef CAS.
- C. Jimenez-Gonzalez, C. S. Ponder, Q. B. Broxterman and J. B. Manley, Org. Process Res. Dev., 2011, 15, 912–917 CrossRef CAS.
- P. T. Anastas and J. B. Zimmerman, Environ. Sci. Technol., 2003, 37, 94A–101A CrossRef PubMed.
- M. A. Abraham and N. Nguyen, Environ. Prog., 2003, 22, 233–236 CrossRef CAS.
- D. Dallinger and C. O. Kappe, Curr. Opin. Green Sustain. Chem., 2017, 7, 6–12 CrossRef.
- R. A. Sheldon, Green Chem., 2017, 19, 18–43 RSC.
- An overview of Pfizer's approach to Environment, Social, and Governance commitments and reporting, https://www.pfizer.com/sites/default/files/investors/financial_reports/annual_reports/2021/esg/?linkId=158814291, (accessed March 3, 2025).
- Pfizer's year in review | Pfizer 2021 Annual Report, https://www.pfizer.com/sites/default/files/investors/financial_reports/annual_reports/2021/, (accessed March 3, 2025).
- B. M. S. Ramos Da Silva, V. A. Nepomuceno De Oliveira and J. Magalhães, in Advances in Information Quality and Management, ed. M. Khosrow-Pour, IGI Global, 2024, pp. 1–21 Search PubMed.
- L. Sanchez and B. Blanco, Total Qual. Manag. Bus. Excell., 2014, 25, 986–1001 CrossRef.
- O. McDermott, J. Antony, M. Sony and S. Daly, Processes, 2021, 10, 73 CrossRef.
- G. H. Vogel, Process Development: From the Initial Idea to the Chemical Production Plant, Wiley-VCH, Weinheim, 2006 Search PubMed.
- J. Harmsen, A. B. De Haan and P. L. J. Swinkels, Product and Process Design: Driving Sustainable Innovation, De Gruyter, 2024 Search PubMed.
- J. Harmsen, Kirk-Othmer Encyclopedia of Chemical Technology, ed. Kirk-Othmer, Wiley, 1st edn, 2024, pp. 1–19 Search PubMed.
- ICH Q7 Good manufacturing practice for active pharmaceutical ingredients - Scientific guideline | European Medicines Agency (EMA), https://www.ema.europa.eu/en/ich-q7-good-manufacturing-practice-active-pharmaceutical-ingredients-scientific-guideline, (accessed March 3, 2025).
- ICH Q8 (R2) Pharmaceutical development - Scientific guideline | European Medicines Agency (EMA), https://www.ema.europa.eu/en/ich-q8-r2-pharmaceutical-development-scientific-guideline, (accessed March 3, 2025).
- ICH Q9 Quality risk management - Scientific guideline | European Medicines Agency (EMA), https://www.ema.europa.eu/en/ich-q9-quality-risk-management-scientific-guideline, (accessed March 3, 2025).
- ICH Q10 Pharmaceutical quality system - Scientific guideline | European Medicines Agency (EMA), https://www.ema.europa.eu/en/ich-q10-pharmaceutical-quality-system-scientific-guideline, (accessed March 3, 2025).
- ICH Q13 Continuous Manufacturing of Drug Substances and Drug Products, https://www.fda.gov/regulatory-information/search-fda-guidance-documents/q13-continuous-manufacturing-drug-substances-and-drug-products, (accessed February 25, 2025).
- FDA Guidance for Industry: Quality Considerations for Continuous Manufacturing - ECA Academy, https://www.gmp-compliance.org/guidelines/gmp-guideline/fda-guidance-for-industry-quality-considerations-for-continuous-manufacturing, (accessed March 3, 2025).
- N. Mihokovic, Continuous manufacturing - EMA perspective and experience, https://dc.engconfintl.org/biomanufact_iii/69, (accessed January 26, 2025) Search PubMed.
- USP 2022 Annual report, https://www.usp.org/about/annual-report, (accessed August 23, 2025).
- K. P. Cole and M. D. Johnson, Expert Rev. Clin. Pharmacol., 2018, 11, 5–13 CrossRef CAS PubMed.
- M. Sarkis, A. Bernardi, N. Shah and M. M. Papathanasiou, Processes, 2021, 9, 457 CrossRef.
- A. Domokos, B. Nagy, B. Szilágyi, G. Marosi and Z. K. Nagy, Org. Process Res. Dev., 2021, 25, 721–739 CrossRef CAS.
- S. L. Lee, T. F. O’Connor, X. Yang, C. N. Cruz, S. Chatterjee, R. D. Madurawe, C. M. V. Moore, L. X. Yu and J. Woodcock, J. Pharm. Innov., 2015, 10, 191–199 CrossRef.
- M. M. Nasr, M. Krumme, Y. Matsuda, B. L. Trout, C. Badman, S. Mascia, C. L. Cooney, K. D. Jensen, A. Florence, C. Johnston, K. Konstantinov and S. L. Lee, J. Pharm. Sci., 2017, 106, 3199–3206 CrossRef CAS PubMed.
- EMA approves Janssen's Prezista continuous manufacturing line, https://www.europeanpharmaceuticalreview.com/news/62587/ema-continuous-manufacturing/, (accessed March 3, 2025).
- J. Wahlich, Pharmaceutics, 2021, 13, 1311 CrossRef CAS.
- L. Pernenkil and C. L. Cooney, Chem. Eng. Sci., 2006, 61, 720–742 CrossRef CAS.
- M. Jiang and R. D. Braatz, CrystEngComm, 2019, 21, 3534–3551 RSC.
- H. Chen, E. Diep, T. A. G. Langrish and B. J. Glasser, AIChE J., 2020, 66, e16902 CrossRef CAS.
- E. J. Kim, J. H. Kim, M.-S. Kim, S. H. Jeong and D. H. Choi, Pharmaceutics, 2021, 13, 919 CrossRef PubMed.
- L. L. Simon, H. Pataki, G. Marosi, F. Meemken, K. Hungerbühler, A. Baiker, S. Tummala, B. Glennon, M. Kuentz, G. Steele, H. J. M. Kramer, J. W. Rydzak, Z. Chen, J. Morris, F. Kjell, R. Singh, R. Gani, K. V. Gernaey, M. Louhi-Kultanen, J. O’Reilly, N. Sandler, O. Antikainen, J. Yliruusi, P. Frohberg, J. Ulrich, R. D. Braatz, T. Leyssens, M. von Stosch, R. Oliveira, R. B. H. Tan, H. Wu, M. Khan, D. O’Grady, A. Pandey, R. Westra, E. Delle-Case, D. Pape, D. Angelosante, Y. Maret, O. Steiger, M. Lenner, K. Abbou-Oucherif, Z. K. Nagy, J. D. Litster, V. K. Kamaraju and M.-S. Chiu, Org. Process Res. Dev., 2015, 19, 3–62 CrossRef CAS.
- A. M. Kearney, S. G. Collins and A. R. Maguire, React. Chem. Eng., 2024, 9, 990–1013 RSC.
- P. Neugebauer, M. Zettl, D. Moser, J. Poms, L. Kuchler and S. Sacher, Int. J. Pharm., 2024, 661, 124412 CrossRef CAS.
- F. Destro, P. K. Inguva, P. Srisuma and R. D. Braatz, Curr. Opin. Chem. Eng., 2024, 45, 101035 CrossRef.
- M. Jelsch, Y. Roggo, P. Kleinebudde and M. Krumme, Eur. J. Pharm. Biopharm., 2021, 159, 137–142 CrossRef CAS PubMed.
- M. Schwenzer, M. Ay, T. Bergs and D. Abel, Int. J. Adv. Manuf. Technol., 2021, 117, 1327–1349 CrossRef.
- X. Xu, Y. Lu, B. Vogel-Heuser and L. Wang, J. Manuf. Syst., 2021, 61, 530–535 CrossRef.
- V. Malheiro, J. Duarte, F. Veiga and F. Mascarenhas-Melo, Pharmaceutics, 2023, 15, 2545 CrossRef CAS PubMed.
- H. M. Inuwa, A. Ravi Raja, A. Kumar, B. Singh and S. Singh, Mater. Today Proc., 2022, 62, 3593–3598 CrossRef.
- PAT—A Framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance, https://www.fda.gov/regulatory-information/search-fda-guidance-documents/pat-framework-innovative-pharmaceutical-development-manufacturing-and-quality-assurance, (accessed March 3, 2025).
- S. Mao, B. Wang, Y. Tang and F. Qian, Engineering, 2019, 5, 995–1002 CrossRef CAS.
- D. Hebrault, A. J. Rein and B. Wittkamp, ACS Sustainable Chem. Eng., 2022, 10, 5072–5077 CrossRef CAS.
- M. A. Morin, W. (Peter) Zhang, D. Mallik and M. G. Organ, Angew. Chem., Int. Ed., 2021, 60, 20606–20626 CrossRef CAS.
- S. Bordawekar, A. Chanda, A. M. Daly, A. W. Garrett, J. P. Higgins, M. A. LaPack, T. D. Maloney, J. Morgado, S. Mukherjee, J. D. Orr, G. L. I. Reid, B.-S. Yang and H. W. I. Ward, Org. Process Res. Dev., 2015, 19, 1174–1185 CrossRef CAS.
- C. H. Pérez-Beltrán, A. M. Jiménez-Carvelo, A. Torrente-López, N. A. Navas and L. Cuadros-Rodríguez, Food Eng. Rev., 2023, 15, 24–40 CrossRef.
- S.-H. Lee, J.-K. Kim, J.-P. Jee, D.-J. Jang, Y.-J. Park and J.-E. Kim, J. Pharm. Investig., 2022, 52, 649–682 CrossRef.
- P. D. Rege, A. Schuster, J. Lamerz, C. Moessner, W. Göhring, P. Hidber, H. Stahr, O. M. Andrei, J. Burren, A. Moesching, D. Coleman and S. Hildbrand, Org. Process Res. Dev., 2024, 28, 1003–1017 CrossRef CAS.
- S. Mascia, P. L. Heider, H. Zhang, R. Lakerveld, B. Benyahia, P. I. Barton, R. D. Braatz, C. L. Cooney, J. M. B. Evans, T. F. Jamison, K. F. Jensen, A. S. Myerson and B. L. Trout, Angew. Chem., Int. Ed., 2013, 52, 12359–12363 CrossRef CAS PubMed.
- P. L. Heider, S. C. Born, S. Basak, B. Benyahia, R. Lakerveld, H. Zhang, R. Hogan, L. Buchbinder, A. Wolfe, S. Mascia, J. M. B. Evans, T. F. Jamison and K. F. Jensen, Org. Process Res. Dev., 2014, 18, 402–409 CrossRef CAS.
- B. Benyahia, Computer Aided Chemical Engineering, Elsevier, 2018, vol. 41, pp. 141–157 Search PubMed.
- J. P. Coates, Process Analytical Technology, John Wiley & Sons, Ltd, 2010, pp. 157–194 Search PubMed.
- N. L. Jestel, Process Analytical Technology, John Wiley & Sons, Ltd, 2010, pp. 195–243 Search PubMed.
- M. A. Liauw, L. C. Baylor and P. E. O’Rourke, Process Analytical Technology, John Wiley & Sons, Ltd, 2010, pp. 81–106 Search PubMed.
- J. C. Edwards and P. J. Giammatteo, Process Analytical Technology, John Wiley & Sons, Ltd, 2010, pp. 303–335 Search PubMed.
- T. Graf, L. Naumann, L. Bonnington, J. Heckel, B. Spensberger, S. Klein, C. Brey, R. Nachtigall, M. Mroz, T. V. Hogg, C. McHardy, A. Martinez, R. Braaz and M. Leiss, J. Chromatogr. A, 2024, 1729, 465013 CrossRef CAS.
- I. Cai, T. C. Malig, K. L. Kurita, J. S. Derasp, L. E. Sirois and J. E. Hein, ACS Catal., 2024, 14, 12331–12341 CrossRef CAS PubMed.
- A. Greb, J.-S. Poh, S. Greed, C. Battilocchio, P. Pasau, D. C. Blakemore and S. V. Ley, Angew. Chem., Int. Ed., 2017, 56, 16602–16605 CrossRef CAS.
- C. Battilocchio, B. J. Deadman, N. Nikbin, M. O. Kitching, I. R. Baxendale and S. V. Ley, Chem. – Eur. J., 2013, 19, 7917–7930 CrossRef CAS PubMed.
- D. E. Fitzpatrick, T. Maujean, A. C. Evans and S. V. Ley, Angew. Chem., Int. Ed., 2018, 57, 15128–15132 CrossRef CAS.
- N. Holmes, G. R. Akien, A. John Blacker, R. L. Woodward, R. E. Meadows and R. A. Bourne, React. Chem. Eng., 2016, 1, 366–371 RSC.
- E. C. Aka, E. Wimmer, E. Barré, D. Cortés-Borda, T. Ekou, L. Ekou, M. Rodriguez-Zubiri and F.-X. Felpin, Org. Process Res. Dev., 2020, 24, 745–751 CrossRef CAS.
- N. Vasudevan, E. Wimmer, E. Barré, D. Cortés-Borda, M. Rodriguez-Zubiri and F. Felpin, Adv. Synth. Catal., 2021, 363, 791–799 CrossRef CAS.
- V. Mancino, B. Cerra, A. Piccinno and A. Gioiello, Org. Process Res. Dev., 2018, 22, 600–607 CrossRef CAS.
- E. Içten, A. J. Maloney, M. G. Beaver, D. E. Shen, X. Zhu, L. R. Graham, J. A. Robinson, S. Huggins, A. Allian, R. Hart, S. D. Walker, P. Rolandi and R. D. Braatz, Org. Process Res. Dev., 2020, 24, 1861–1875 CrossRef.
- F. Guan, A. J. Blacker, B. Hall, N. Kapur, J. Wen and X. Zhang, J. Flow Chem., 2021, 11, 763–772 CrossRef CAS.
- V. K. Sthalam, A. K. Singh and S. Pabbaraja, Org. Process Res. Dev., 2019, 23, 1892–1899 CrossRef CAS.
- K. E. Konan, A. Abollé, E. Barré, E. C. Aka, V. Coeffard and F.-X. Felpin, React. Chem. Eng., 2022, 7, 1346–1357 RSC.
- H.-W. Hsieh, D. J. Griffin, A. M. K. Nambiar, N. Sarkar, H. Youssef Ismail, K. Saigal, D. E. Shen, N. Goudas-Salomon, R. Wimalasinghe, A. Zeng, O. R. Thiel and M. G. Beaver, Org. Process Res. Dev., 2024, 28, 2844–2853 CrossRef CAS.
- P. Sagmeister, F. F. Ort, C. E. Jusner, D. Hebrault, T. Tampone, F. G. Buono, J. D. Williams and C. O. Kappe, Adv. Sci., 2022, 9, 2105547 CrossRef.
- N. Salehi Marzijarani, A. J. Fine, S. M. Dalby, R. Gangam, S. Poudyal, T. Behre, A. R. Ekkati, B. M. Armstrong, C. S. Shultz, Z. E. X. Dance and K. Stone, Org. Process Res. Dev., 2022, 26, 533–542 CrossRef CAS.
- K. P. Cole, J. M. Groh, M. D. Johnson, C. L. Burcham, B. M. Campbell, W. D. Diseroad, M. R. Heller, J. R. Howell, N. J. Kallman, T. M. Koenig, S. A. May, R. D. Miller, D. Mitchell, D. P. Myers, S. S. Myers, J. L. Phillips, C. S. Polster, T. D. White, J. Cashman, D. Hurley, R. Moylan, P. Sheehan, R. D. Spencer, K. Desmond, P. Desmond and O. Gowran, Science, 2017, 356, 1144–1150 CrossRef CAS PubMed.
- T. A. Hamlin and N. E. Leadbeater, Beilstein J. Org. Chem., 2013, 9, 1843–1852 CrossRef.
- H. Lange, C. F. Carter, M. D. Hopkin, A. Burke, J. G. Goode, I. R. Baxendale and S. V. Ley, Chem. Sci., 2011, 2, 765–769 RSC.
- L. Capaldo, Z. Wen and T. Noël, Chem. Sci., 2023, 14, 4230–4247 RSC.
- P. Bianchi and J. M. Monbaliu, Angew. Chem., Int. Ed., 2024, 63, e202311526 CrossRef CAS.
- N. M. Ralbovsky and J. P. Smith, Talanta, 2023, 252, 123787 CrossRef CAS.
- P. Liu, H. Jin, Y. Chen, D. Wang, H. Yan, M. Wu, F. Zhao and W. Zhu, Chin. Chem. Lett., 2024, 35, 108877 CrossRef CAS.
- A. Adamo, R. L. Beingessner, M. Behnam, J. Chen, T. F. Jamison, K. F. Jensen, J.-C. M. Monbaliu, A. S. Myerson, E. M. Revalor, D. R. Snead, T. Stelzer, N. Weeranoppanant, S. Y. Wong and P. Zhang, Science, 2016, 352, 61–67 CrossRef CAS.
- S. Chatterjee, M. Guidi, P. H. Seeberger and K. Gilmore, Nature, 2020, 579, 379–384 CrossRef CAS PubMed.
- P. Sagmeister, J. D. Williams, C. A. Hone and C. O. Kappe, React. Chem. Eng., 2019, 4, 1571–1578 Search PubMed.
- A. D. Clayton, E. O. Pyzer-Knapp, M. Purdie, M. F. Jones, A. Barthelme, J. Pavey, N. Kapur, T. W. Chamberlain, A. J. Blacker and R. A. Bourne, Angew. Chem., Int. Ed., 2023, 62, e202214511 Search PubMed.
- N. Al Azri, C. Clifford, R. M. Enick and G. Veser, Org. Process Res. Dev., 2024, 28, 1657–1667 Search PubMed.
- P. Sagmeister, R. Lebl, I. Castillo, J. Rehrl, J. Kruisz, M. Sipek, M. Horn, S. Sacher, D. Cantillo, J. D. Williams and C. O. Kappe, Angew. Chem., Int. Ed., 2021, 60, 8139–8148 CrossRef CAS PubMed.
- P. Sagmeister, J. Poms, J. D. Williams and C. O. Kappe, React. Chem. Eng., 2020, 5, 677–684 RSC.
- R. I. Teixeira, T. H. Waldron Clarke, A. Love, X.-Z. Sun, S. Kayal and M. W. George, Org. Process Res. Dev., 2025, 29, 34–47 CrossRef CAS.
- B. K. Peters, K. X. Rodriguez, S. H. Reisberg, S. B. Beil, D. P. Hickey, Y. Kawamata, M. Collins, J. Starr, L. Chen, S. Udyavara, K. Klunder, T. J. Gorey, S. L. Anderson, M. Neurock, S. D. Minteer and P. S. Baran, Science, 2019, 363, 838–845 CrossRef CAS PubMed.
- D. S. Lee, A. Love, Z. Mansouri, T. H. Waldron Clarke, D. C. Harrowven, R. Jefferson-Loveday, S. J. Pickering, M. Poliakoff and M. W. George, Org. Process Res. Dev., 2022, 26, 2674–2684 CrossRef CAS PubMed.
- M. E. Avanthay, O. H. Goodrich, D. Tiemessen, C. M. Alder, M. W. George and A. J. J. Lennox, JACS Au, 2024, 4, 2220–2227 CrossRef CAS.
- A. Choi, O. H. Goodrich, A. P. Atkins, M. D. Edwards, D. Tiemessen, M. W. George and A. J. J. Lennox, Org. Lett., 2024, 26, 653–657 CrossRef CAS.
- N. Petrović, B. K. Malviya, C. O. Kappe and D. Cantillo, Org. Process Res. Dev., 2023, 27, 2072–2081 CrossRef.
- A. M. K. Nambiar, C. P. Breen, T. Hart, T. Kulesza, T. F. Jamison and K. F. Jensen, ACS Cent. Sci., 2022, 8, 825–836 CrossRef CAS.
- A. Slattery, Z. Wen, P. Tenblad, J. Sanjosé-Orduna, D. Pintossi, T. Den Hartog and T. Noël, Science, 2024, 383, eadj1817 CrossRef CAS PubMed.
- G. Tom, S. P. Schmid, S. G. Baird, Y. Cao, K. Darvish, H. Hao, S. Lo, S. Pablo-García, E. M. Rajaonson, M. Skreta, N. Yoshikawa, S. Corapi, G. D. Akkoc, F. Strieth-Kalthoff, M. Seifrid and A. Aspuru-Guzik, Chem. Rev., 2024, 124, 9633–9732 CrossRef CAS.
- A. D. Clayton, Chem. Methods, 2023, 3, e202300021 CrossRef CAS.
- R. Lakerveld, B. Benyahia, P. L. Heider, H. Zhang, A. Wolfe, C. J. Testa, S. Ogden, D. R. Hersey, S. Mascia, J. M. B. Evans, R. D. Braatz and P. I. Barton, Org. Process Res. Dev., 2015, 19, 1088–1100 CrossRef CAS.
- M. Ierapetritou, M. Sebastian Escotet-Espinoza and R. Singh, Continuous Manufacturing of Pharmaceuticals, John Wiley & Sons, Ltd, 2017, pp. 33–105 Search PubMed.
- T. F. O’Connor, S. Chatterjee, J. Lam, D. H. P. de la Ossa, L. Martinez-Peyrat, M. H. N. Hoefnagel and A. C. Fisher, Int. J. Pharm. X, 2024, 8, 100274 Search PubMed.
- S. Chatterjee, C. M. V. Moore and M. M. Nasr, Comprehensive Quality by Design for Pharmaceutical Product Development and Manufacture, John Wiley & Sons, Ltd, 2017, pp. 9–24 Search PubMed.
- D. Bonvin, C. Georgakis, C. C. Pantelides, M. Barolo, M. A. Grover, D. Rodrigues, R. Schneider and D. Dochain, Ind. Eng. Chem. Res., 2016, 55, 6891–6903 CrossRef CAS.
- H. G. Jolliffe and D. I. Gerogiorgis, Chem. Eng. Res. Des., 2016, 112, 310–325 CrossRef CAS.
- K. C. Aroh and K. F. Jensen, React. Chem. Eng., 2018, 3, 94–101 RSC.
- S. Diab and D. I. Gerogiorgis, Comput. Chem. Eng., 2018, 111, 102–114 CrossRef CAS.
- E. Papadakis, J. M. Woodley and R. Gani, Computer Aided Chemical Engineering, Elsevier, 2018, vol. 41, pp. 597–656 Search PubMed.
- R. V. Barenji, Y. Akdag, B. Yet and L. Oner, Int. J. Pharm., 2019, 567, 118445 CrossRef CAS PubMed.
- Y. Chen, O. Yang, C. Sampat, P. Bhalode, R. Ramachandran and M. Ierapetritou, Processes, 2020, 8, 1088 CrossRef.
- M. Moreno-Benito, K. T. Lee, D. Kaydanov, H. M. Verrier, D. O. Blackwood and P. Doshi, Int. J. Pharm., 2022, 628, 122336 CrossRef CAS PubMed.
- F. Boukouvala and M. G. Ierapetritou, J. Pharm. Innov., 2013, 8, 131–145 CrossRef.
- B. Nagy, B. Szilágyi, A. Domokos, B. Vészi, K. Tacsi, Z. Rapi, H. Pataki, G. Marosi, Z. K. Nagy and Z. K. Nagy, Chem. Eng. J., 2021, 419, 129947 CrossRef CAS.
- A. Rogers and M. Ierapetritou, Comput. Chem. Eng., 2015, 81, 32–39 CrossRef CAS.
- M. Dosta, J. D. Litster and S. Heinrich, Adv. Powder Technol., 2020, 31, 947–953 CrossRef.
- R. C. Dias, O. Korhonen, J. Ketolainen, J. A. Lopes and T. Ervasti, Int. J. Pharm., 2023, 639, 122969 CrossRef CAS.
- Digital Twin Technology in Pharmaceutical Manufacturing Market, https://www.visiongain.com/digital-twin-technology-in-pharmaceutical-manufacturing-market/, (accessed March 26, 2025).
- R. I. Teixeira, M. Andresini, R. Luisi and B. Benyahia, JACS Au, 2024, 4, 4263–4272 CrossRef CAS PubMed.
- R. I. Teixeira and B. Benyahia, Chem. Eng. Res. Des., 2025, 216, 367–375 CrossRef CAS.
- C. Castiello, P. Junghanns, A. Mergel, C. Jacob, C. Ducho, S. Valente, D. Rotili, R. Fioravanti, C. Zwergel and A. Mai, Green Chem., 2023, 25, 2109–2169 RSC.
- A. Wołos, D. Koszelewski, R. Roszak, S. Szymkuć, M. Moskal, R. Ostaszewski, B. T. Herrera, J. M. Maier, G. Brezicki, J. Samuel, J. A. M. Lummiss, D. T. McQuade, L. Rogers and B. A. Grzybowski, Nature, 2022, 604, 668–676 CrossRef.
- K. Huanbutta, K. Burapapadh, P. Kraisit, P. Sriamornsak, T. Ganokratanaa, K. Suwanpitak and T. Sangnim, Eur. J. Pharm. Sci., 2024, 203, 106938 CrossRef CAS PubMed.
- B. Van Snick, J. Dhondt, K. Pandelaere, J. Bertels, R. Mertens, D. Klingeleers, G. Di Pretoro, J. P. Remon, C. Vervaet, T. De Beer and V. Vanhoorne, Int. J. Pharm., 2018, 549, 415–435 CrossRef CAS.
- D. J. Griffin, C. W. Coley, S. A. Frank, J. M. Hawkins and K. F. Jensen, Org. Process Res. Dev., 2023, 27, 1868–1879 CrossRef CAS.
- K.-K. Mak and M. R. Pichika, Drug Discov. Today, 2019, 24, 773–780 CrossRef.
- C. Selvaraj, I. Chandra and S. K. Singh, Mol. Divers., 2022, 26, 1893–1913 CrossRef CAS.
- J. Jiang, X. Ma, D. Ouyang and R. O. Williams, Pharmaceutics, 2022, 14, 2257 CrossRef CAS.
- R. Han, H. Yoon, G. Kim, H. Lee and Y. Lee, Pharmaceuticals, 2023, 16, 1259 CrossRef CAS PubMed.
- L. K. Vora, A. D. Gholap, K. Jetha, R. R. S. Thakur, H. K. Solanki and V. P. Chavda, Pharmaceutics, 2023, 15, 1916 CrossRef CAS PubMed.
- Y. Djoumbou-Feunang, J. Wilmot, J. Kinney, P. Chanda, P. Yu, A. Sader, M. Sharifi, S. Smith, J. Ou, J. Hu, E. Shipp, D. Tomandl and S. P. Kumpatla, Front. Chem., 2023, 11, 1292027 CrossRef CAS PubMed.
- C. Sarkar, B. Das, V. S. Rawat, J. B. Wahlang, A. Nongpiur, I. Tiewsoh, N. M. Lyngdoh, D. Das, M. Bidarolli and H. T. Sony, Int. J. Mol. Sci., 2023, 24, 2026 CrossRef PubMed.
- V. P. Ananikov, Artif. Intell. Chem., 2024, 2, 100075 CrossRef.
- FDA Releases Two Discussion Papers to Spur Conversation about Artificial Intelligence and Machine Learning in Drug Development and Manufacturing, https://www.fda.gov/news-events/fda-voices/fda-releases-two-discussion-papers-spur-conversation-about-artificial-intelligence-and-machine, (accessed March 4, 2025).
- T. Van Gerven and A. Stankiewicz, Ind. Eng. Chem. Res., 2009, 48, 2465–2474 CrossRef CAS.
- E. A. López-Guajardo, F. Delgado-Licona, A. J. Álvarez, K. D. P. Nigam, A. Montesinos-Castellanos and R. Morales-Menendez, Chem. Eng. Process. - Process Intensif., 2022, 180, 108671 CrossRef.
- Z. Wu, J. Luo, D. Rincon and P. D. Christofides, Chem. Eng. Res. Des., 2021, 168, 275–287 CrossRef CAS.
- Á. K. Beke, M. Gyürkés, Z. K. Nagy, G. Marosi and A. Farkas, Eur. J. Pharm. Biopharm., 2021, 169, 64–77 CrossRef.
- M. S. Alhajeri, J. Luo, Z. Wu, F. Albalawi and P. D. Christofides, Chem. Eng. Res. Des., 2022, 179, 77–89 CrossRef CAS.
- J. Wollschläger and F. Montanari, Digit. Discov., 2024, 3, 1749–1760 RSC.
- A. D. Vassileiou, M. N. Robertson, B. G. Wareham, M. Soundaranathan, S. Ottoboni, A. J. Florence, T. Hartwig and B. F. Johnston, Digit. Discov., 2023, 2, 356–367 RSC.
- R. Schenkendorf, X. Xie, M. Rehbein, S. Scholl and U. Krewer, Processes, 2018, 6, 27 CrossRef.
- M. F. Simões, G. Silva, A. C. Pinto, M. Fonseca, N. E. Silva, R. M. A. Pinto and S. Simões, Eur. J. Pharm. Biopharm., 2020, 152, 282–295 CrossRef PubMed.
- Y. Sun, X. Wang, N. Ren, Y. Liu and S. You, Environ. Sci. Technol., 2023, 57, 3434–3444 CrossRef CAS PubMed.
- D. Zhang, Z. Wang, C. Oberschelp, E. Bradford and S. Hellweg, ACS Sustainable Chem. Eng., 2024, 12, 2700–2708 CrossRef CAS.
- D. L. Galata, L. A. Mészáros, N. Kállai-Szabó, E. Szabó, H. Pataki, G. Marosi and Z. K. Nagy, Eur. J. Pharm. Sci., 2021, 159, 105717 CrossRef CAS.
- Y. Hayashi, Y. Nakano, Y. Marumo, S. Kumada, K. Okada and Y. Onuki, Int. J. Pharm., 2021, 609, 121158 CrossRef CAS PubMed.
- M. A. Hardy, B. Nan, O. Wiest and R. Sarpong, Tetrahedron, 2022, 104, 132584 CrossRef CAS PubMed.
- Science of Synthesis on IBM RXN for Chemistry, https://science-of-synthesis-datasets.thieme.com/science-of-synthesis-ibm-rxn-chemistry/, (accessed July 6, 2025).
- M. Sen and R. Ramachandran, Adv. Powder Technol., 2013, 24, 51–59 CrossRef.
- A. J. Freitag, A. J. Rogers, J. E. Tabora and D. S. Treitler, Org. Process Res. Dev., 2024, 28, 511–523 CrossRef CAS.
- K. P. Kusumo, L. Gomoescu, R. Paulen, S. García Muñoz, C. C. Pantelides, N. Shah and B. Chachuat, Ind. Eng. Chem. Res., 2020, 59, 2396–2408 CrossRef CAS.
- M. Magris and A. Iosifidis, Artif. Intell. Rev., 2023, 56, 11773–11823 CrossRef.
- W. Sun, A. R. C. Paiva, P. Xu, A. Sundaram and R. D. Braatz, Comput. Chem. Eng., 2020, 141, 106991 CrossRef CAS.
- J. E. Tabora, F. Lora Gonzalez and J. W. Tom, AIChE J., 2019, 65, e16744 CrossRef CAS.
- B. Nagy, Á. Szabados-Nacsa, G. Fülöp, A. Turák Nagyné, D. L. Galata, A. Farkas, L. A. Mészáros, Z. K. Nagy and G. Marosi, Int. J. Pharm., 2023, 633, 122620 CrossRef CAS PubMed.
- L. Gentiluomo, D. Roessner, D. Augustijn, H. Svilenov, A. Kulakova, S. Mahapatra, G. Winter, W. Streicher, Å. Rinnan, G. H. J. Peters, P. Harris and W. Frieß, Eur. J. Pharm. Biopharm., 2019, 141, 81–89 CrossRef CAS.
- M. Von Stosch, S. Davy, K. Francois, V. Galvanauskas, J. Hamelink, A. Luebbert, M. Mayer, R. Oliveira, R. O’Kennedy, P. Rice and J. Glassey, Biotechnol. J., 2014, 9, 719–726 CrossRef CAS PubMed.
- L. Rajulapati, S. Chinta, B. Shyamala and R. Rengaswamy, AIChE J., 2022, 68, e17715 CrossRef CAS.
- A. Chakraborty, A. Sivaram and V. Venkatasubramanian, Comput. Chem. Eng., 2021, 154, 107470 CrossRef CAS.
- Y. Roggo, V. Pauli, M. Jelsch, L. Pellegatti, F. Elbaz, S. Ensslin, P. Kleinebudde and M. Krumme, J. Pharm. Biomed. Anal., 2020, 179, 112971 CrossRef CAS.
- N. Cherkasov, Y. Bai, A. J. Expósito and E. V. Rebrov, React. Chem. Eng., 2018, 3, 769–780 RSC.
- L. Madarász, L. A. Mészáros, Á. Köte, A. Farkas and Z. K. Nagy, Int. J. Pharm., 2023, 641, 123060 CrossRef PubMed.
- S. Chen, T. Liu, D. Xu, Y. Huo and Y. Yang, 2019 Chinese Control Conference (CCC), IEEE, Guangzhou, China, 2019, 7933–7938 Search PubMed.
- H. Salami, M. A. McDonald, A. S. Bommarius, R. W. Rousseau and M. A. Grover, Org. Process Res. Dev., 2021, 25, 1670–1679 CrossRef CAS.
- S. Zong, G. Zhou, M. Li and X. Wang, Particuology, 2023, 74, 173–183 CrossRef CAS.
- A. Jaeggi, A. K. Rajagopalan, M. Morari and M. Mazzotti, Ind. Eng. Chem. Res., 2021, 60, 473–483 CrossRef CAS.
- Z. Zhou, X. Li and R. N. Zare, ACS Cent. Sci., 2017, 3, 1337–1344 CrossRef CAS PubMed.
- W. C. Wong, E. Chee, J. Li and X. Wang, Mathematics, 2018, 6, 242 CrossRef.
- B. Benyahia, P. D. Anandan and C. Rielly, Computer Aided Chemical Engineering, Elsevier, 2021, vol. 50, pp. 1371–1376 Search PubMed.
- X. Xie and R. Schenkendorf, Processes, 2019, 7, 509 CrossRef.
- Q. Sun and Z. Ge, IEEE Trans. Ind. Inform., 2021, 17, 5853–5866 Search PubMed.
- G. W. Stockdale and A. Cheng, Qual. Technol. Quant. Manag., 2009, 6, 391–408 CrossRef.
- F. L. Gonzalez, J. E. Tabora, E. C. Huang, S. R. Wisniewski, R. Carrasquillo-Flores, T. Razler and B. Mack, Org. Process Res. Dev., 2019, 23, 211–219 CrossRef CAS.
- Y. Korteby, K. Kristó, T. Sovány and G. Regdon, Powder Technol., 2018, 331, 286–295 CrossRef CAS.
- C. Humer, R. Nicholls, H. Heberle, M. Heckmann, M. Pühringer, T. Wolf, M. Lübbesmeyer, J. Heinrich, J. Hillenbrand, G. Volpin and M. Streit, J. Cheminformatics, 2024, 16, 51 CrossRef.
- H. Heberle, L. Zhao, S. Schmidt, T. Wolf and J. Heinrich, J. Cheminformatics, 2023, 15, 2 CrossRef.
- C. Humer, H. Heberle, F. Montanari, T. Wolf, F. Huber, R. Henderson, J. Heinrich and M. Streit, J. Cheminformatics, 2022, 14, 21 CrossRef PubMed.
- S. Moosavi, M. Farajzadeh-Zanjani, R. Razavi-Far, V. Palade and M. Saif, Electronics, 2024, 13, 3497 CrossRef.
- D. E. Fitzpatrick, C. Battilocchio and S. V. Ley, Org. Process Res. Dev., 2016, 20, 386–394 CrossRef CAS.
- D. Reker, E. A. Hoyt, G. J. L. Bernardes and T. Rodrigues, Cell Rep. Phys. Sci., 2020, 1, 100247 CrossRef.
- Z. Crandall, K. Basemann, L. Qi and T. L. Windus, React. Chem. Eng., 2022, 7, 416–428 RSC.
- B. J. Shields, J. Stevens, J. Li, M. Parasram, F. Damani, J. I. M. Alvarado, J. M. Janey, R. P. Adams and A. G. Doyle, Nature, 2021, 590, 89–96 CrossRef CAS PubMed.
- Active Learning UK - 2024, https://www.active-learning.uk/2024, (accessed April 16, 2025).
- J. P. Dürholt, T. S. Asche, J. Kleinekorte, G. Mancino-Ball, B. Schiller, S. Sung, J. Keupp, A. Osburg, T. Boyne, R. Misener, R. Eldred, W. S. Costa, C. Kappatou, R. M. Lee, D. Linzner, D. Walz, N. Wulkow and B. Shafei, arXiv, 2024, preprint, arxiv.2408.05040 DOI:10.48550/arxiv.2408.05040.
- C. J. Van Der Westhuizen, J. Du Toit, N. Neyt, D. Riley and J.-L. Panayides, Digit. Discov., 2022, 1, 596–604 RSC.
- M. Sim, M. G. Vakili, F. Strieth-Kalthoff, H. Hao, R. J. Hickman, S. Miret, S. Pablo-García and A. Aspuru-Guzik, Matter, 2024, 7, 2959–2977 CrossRef CAS.
- R. A. Bourne, K. K. (Mimi) Hii and B. J. Reizman, React. Chem. Eng., 2019, 4, 1504–1505 RSC.
- S. Sadeghi, R. B. Canty, N. Mukhin, J. Xu, F. Delgado-Licona and M. Abolhasani, ACS Sustainable Chem. Eng., 2024, 12, 12695–12707 CrossRef CAS.
- S. Callaghan, Patterns, 2021, 2, 100221 CrossRef CAS PubMed.
- D. Fysikopoulos, B. Benyahia, A. Borsos, Z. K. Nagy and C. D. Rielly, Comput. Chem. Eng., 2019, 122, 275–292 CrossRef CAS.
- C. Waldron, A. Pankajakshan, M. Quaglio, E. Cao, F. Galvanin and A. Gavriilidis, React. Chem. Eng., 2019, 4, 1623–1636 RSC.
- B. J. Reizman and K. F. Jensen, Org. Process Res. Dev., 2012, 16, 1770–1782 CrossRef CAS.
- X. Yuan and B. Benyahia, Computer Aided Chemical Engineering, Elsevier, 2023, vol. 52, pp. 255–260 Search PubMed.
- X. Yuan and B. Benyahia, Computer Aided Chemical Engineering, Elsevier, 2024, vol. 53, pp. 391–396 Search PubMed.
- F. Galvanin, E. Cao, N. Al-Rifai, A. Gavriilidis and V. Dua, Comput. Chem. Eng., 2016, 95, 202–215 CrossRef CAS.
- K. Matsunami, J. Meyer, M. Rowland, N. Dawson, T. De Beer and D. Van Hauwermeiren, Int. J. Pharm., 2023, 646, 123481 CrossRef CAS PubMed.
- F. Cenci, A. Pankajakshan, P. Facco and F. Galvanin, Comput. Chem. Eng., 2023, 177, 108353 CrossRef CAS.
- I. Bouchkira and B. Benyahia, Computer Aided Chemical Engineering, Elsevier, 2023, vol. 52, pp. 349–354 Search PubMed.
- L. T. Biegler and I. E. Grossmann, Comput. Chem. Eng., 2004, 28, 1169–1192 CrossRef CAS.
- F. Trespalacios and I. E. Grossmann, Chem. Ing. Tech., 2014, 86, 991–1012 CrossRef CAS.
- E. Sheikholeslamzadeh, C.-C. Chen and S. Rohani, Ind. Eng. Chem. Res., 2012, 51, 13792–13802 CrossRef CAS.
- C. Zhang and J. Huang, Pol. J. Environ. Stud., 2015, 24, 391–395 CrossRef CAS.
- P.-F. Chavez, P. Lebrun, P.-Y. Sacré, C. De Bleye, L. Netchacovitch, S. Cuypers, J. Mantanus, H. Motte, M. Schubert, B. Evrard, P. Hubert and E. Ziemons, Int. J. Pharm., 2015, 486, 13–20 CrossRef CAS.
- J. P. McMullen and B. M. Wyvratt, React. Chem. Eng., 2023, 8, 137–151 RSC.
- Y. Wang, R. D. Snee, W. Meng and F. J. Muzzio, Powder Technol., 2016, 294, 22–29 CrossRef CAS.
- Z. Wang and M. Ierapetritou, Comput. Chem. Eng., 2018, 118, 210–223 CrossRef CAS.
- P. Mueller, A. Vriza, A. D. Clayton, O. S. May, N. Govan, S. Notman, S. V. Ley, T. W. Chamberlain and R. A. Bourne, React. Chem. Eng., 2023, 8, 538–542 RSC.
- C. Hu, C. J. Testa, S. C. Born, W. Wu, K. Shvedova, R. Sayin, B. S. Halkude, F. Casati, A. Ramnath, P. Hermant, B. Takizawa, T. F. O’Connor, X. Yang, S. Ramanujam and S. Mascia, Green Chem., 2020, 22, 4350–4356 RSC.
- C. Ding, H. Ardeshna, C. Gillespie and M. Ierapetritou, Biotechnol. Bioeng., 2022, 119, 3567–3583 CrossRef CAS PubMed.
- D. Ott, D. Kralisch, I. Denčić, V. Hessel, Y. Laribi, P. D. Perrichon, C. Berguerand, L. Kiwi-Minsker and P. Loeb, ChemSusChem, 2014, 7, 3521–3533 CrossRef CAS.
- Y. Chen, L. Kotamarthy, A. Dan, C. Sampat, P. Bhalode, R. Singh, B. J. Glasser, R. Ramachandran and M. Ierapetritou, Int. J. Pharm., 2023, 631, 122487 CrossRef CAS PubMed.
- A. A. Mirani, G. Velasco-Hernandez, A. Awasthi and J. Walsh, Sensors, 2022, 22, 5836 CrossRef.
- S. Latif, M. Driss, W. Boulila, Z. E. Huma, S. S. Jamal, Z. Idrees and J. Ahmad, Sensors, 2021, 21, 7518 CrossRef.
- A. Echtermeyer, Y. Amar, J. Zakrzewski and A. Lapkin, Beilstein J. Org. Chem., 2017, 13, 150–163 CrossRef CAS.
- B. J. Reizman and K. F. Jensen, Chem. Commun., 2015, 51, 13290–13293 RSC.
- K. Y. Nandiwale, R. P. Pritchard, C. T. Armstrong, S. M. Guinness and K. P. Girard, React. Chem. Eng., 2024, 9, 2460–2468 RSC.
- J. A. Nelder and R. Mead, Comput. J., 1965, 7, 308–313 CrossRef.
- L. M. Rios and N. V. Sahinidis, J. Glob. Optim., 2013, 56, 1247–1293 CrossRef.
- W. Huyer and A. Neumaier, ACM Trans. Math. Softw., 2008, 35, 1–25 CrossRef.
- M. I. Jeraal, N. Holmes, G. R. Akien and R. A. Bourne, Tetrahedron, 2018, 74, 3158–3164 CrossRef CAS.
- A. D. Clayton, L. A. Power, W. R. Reynolds, C. Ainsworth, D. R. J. Hose, M. F. Jones, T. W. Chamberlain, A. J. Blacker and R. A. Bourne, J. Flow Chem., 2020, 10, 199–206 CrossRef.
- A.-C. Bédard, A. Adamo, K. C. Aroh, M. G. Russell, A. A. Bedermann, J. Torosian, B. Yue, K. F. Jensen and T. F. Jamison, Science, 2018, 361, 1220–1225 CrossRef.
- C. Avila, C. Cassani, T. Kogej, J. Mazuela, S. Sarda, A. D. Clayton, M. Kossenjans, C. P. Green and R. A. Bourne, Chem. Sci., 2022, 13, 12087–12099 RSC.
- A. D. Clayton, A. M. Schweidtmann, G. Clemens, J. A. Manson, C. J. Taylor, C. G. Niño, T. W. Chamberlain, N. Kapur, A. J. Blacker, A. A. Lapkin and R. A. Bourne, Chem. Eng. J., 2020, 384, 123340 CrossRef CAS.
- S. Sano, T. Kadowaki, K. Tsuda and S. Kimura, J. Pharm. Innov., 2020, 15, 333–343 CrossRef.
- J. H. Dunlap, J. G. Ethier, A. A. Putnam-Neeb, S. Iyer, S.-X. L. Luo, H. Feng, J. A. Garrido Torres, A. G. Doyle, T. M. Swager, R. A. Vaia, P. Mirau, C. A. Crouse and L. A. Baldwin, Chem. Sci., 2023, 14, 8061–8069 RSC.
- R. Liu, Z. Wang, W. Yang, J. Cao and S. Tao, Digit. Discov., 2024, 3, 1958–1966 RSC.
- R. Liang, X. Duan, J. Zhang and Z. Yuan, React. Chem. Eng., 2022, 7, 590–598 RSC.
- F. Häse, L. M. Roch and A. Aspuru-Guzik, Chem. Sci., 2018, 9, 7642–7655 RSC.
- F. Häse, L. M. Roch, C. Kreisbeck and A. Aspuru-Guzik, ACS Cent. Sci., 2018, 4, 1134–1145 CrossRef.
- F. Häse, M. Aldeghi, R. J. Hickman, L. M. Roch and A. Aspuru-Guzik, Appl. Phys. Rev, 2021, 8, 031406 Search PubMed.
- A. Yewale, Y. Yang, N. Nazemifard, C. D. Papageorgiou, C. D. Rielly and B. Benyahia, Computer Aided Chemical Engineering, Elsevier, 2024, vol. 53, pp. 3031–3036 Search PubMed.
- D. Cortés-Borda, K. V. Kutonova, C. Jamet, M. E. Trusova, F. Zammattio, C. Truchet, M. Rodriguez-Zubiri and F.-X. Felpin, Org. Process Res. Dev., 2016, 20, 1979–1987 CrossRef.
- B. E. Walker, J. H. Bannock, A. M. Nightingale and J. C. deMello, React. Chem. Eng., 2017, 2, 785–798 RSC.
- H.-W. Hsieh, C. W. Coley, L. M. Baumgartner, K. F. Jensen and R. I. Robinson, Org. Process Res. Dev., 2018, 22, 542–550 CrossRef CAS.
- B. J. Reizman, Y.-M. Wang, S. L. Buchwald and K. F. Jensen, React. Chem. Eng., 2016, 1, 658–666 RSC.
- A. M. Schweidtmann, A. D. Clayton, N. Holmes, E. Bradford, R. A. Bourne and A. A. Lapkin, Chem. Eng. J., 2018, 352, 277–282 CrossRef CAS.
- O. J. Kershaw, A. D. Clayton, J. A. Manson, A. Barthelme, J. Pavey, P. Peach, J. Mustakis, R. M. Howard, T. W. Chamberlain, N. J. Warren and R. A. Bourne, Chem. Eng. J., 2023, 451, 138443 CrossRef CAS.
- K. C. Felton, J. G. Rittig and A. A. Lapkin, Chem. Methods, 2021, 1, 116–122 CrossRef CAS.
- F. Häse, M. Aldeghi, R. J. Hickman, L. M. Roch, M. Christensen, E. Liles, J. E. Hein and A. Aspuru-Guzik, Mach. Learn. Sci. Technol., 2021, 2, 035021 CrossRef.
- P. Müller, A. D. Clayton, J. Manson, S. Riley, O. S. May, N. Govan, S. Notman, S. V. Ley, T. W. Chamberlain and R. A. Bourne, React. Chem. Eng., 2022, 7, 987–993 RSC.
- J. Orehek, D. Teslić and B. Likozar, Org. Process Res. Dev., 2021, 25, 16–42 CrossRef CAS.
- Y. Zhang and N. Abatzoglou, Chem. Eng. Sci., 2022, 255, 117678 CrossRef CAS.
- A. Chindrus, D. Copot and C.-F. Caruntu, Processes, 2023, 11, 1258 CrossRef.
- Y. Ma, S. Wu, E. G. J. Macaringue, T. Zhang, J. Gong and J. Wang, Org. Process Res. Dev., 2020, 24, 1785–1801 CrossRef CAS.
- J. Palmer, C. J. O’Malley, M. J. Wade, E. B. Martin, T. Page and G. A. Montague, J. Pharm. Innov., 2020, 15, 26–40 CrossRef.
- A. Burggraeve, T. Monteyne, C. Vervaet, J. P. Remon and T. D. Beer, Eur. J. Pharm. Biopharm., 2013, 83, 2–15 Search PubMed.
- P. Srisuma, G. Barbastathis and R. D. Braatz, Comput. Chem. Eng., 2023, 177, 108318 CrossRef CAS.
- B. Benyahia, R. Lakerveld and P. I. Barton, Ind. Eng. Chem. Res., 2012, 51, 15393–15412 CrossRef CAS.
- R. Lakerveld, B. Benyahia, R. D. Braatz and P. I. Barton, AIChE J., 2013, 59, 3671–3685 CrossRef CAS.
- D. M. Roberge, B. Zimmermann, F. Rainone, M. Gottsponer, M. Eyholzer and N. Kockmann, Org. Process Res. Dev., 2008, 12, 905–910 CrossRef CAS.
- A. Mesbah, J. A. Paulson, R. Lakerveld and R. D. Braatz, in 2015 American Control Conference (ACC), IEEE, Chicago, IL, USA, 2015, 4301–4307 Search PubMed.
- D. Copot, in 2022 IEEE International Conference on Automation, Quality and Testing, Robotics (AQTR), IEEE, Cluj-Napoca, Romania, 2022, 1–6 Search PubMed.
- M. Kirchengast, S. Celikovic, J. Rehrl, S. Sacher, J. Kruisz, J. Khinast and M. Horn, Int. J. Pharm., 2019, 567, 118457 CrossRef CAS.
- R. Singh, Computer Aided Chemical Engineering, Elsevier, 2018, vol. 41, pp. 317–351 Search PubMed.
- N. T. Haas, M. Ierapetritou and R. Singh, J. Pharm. Innov., 2017, 12, 110–123 CrossRef.
- A. Bhaskar, F. N. Barros and R. Singh, Int. J. Pharm., 2017, 534, 159–178 CrossRef CAS PubMed.
- A. Mesbah, J. A. Paulson, R. Lakerveld and R. D. Braatz, Org. Process Res. Dev., 2017, 21, 844–854 CrossRef CAS.
- S. Sacher, S. Celikovic, J. Rehrl, J. Poms, M. Kirchengast, J. Kruisz, M. Sipek, S. Salar-Behzadi, H. Berger, G. Stark, M. Horn and J. G. Khinast, Eur. J. Pharm. Sci., 2020, 142, 105097 CrossRef CAS PubMed.
- R. Singh, A. Sahay, K. M. Karry, F. Muzzio, M. Ierapetritou and R. Ramachandran, Int. J. Pharm., 2014, 473, 38–54 CrossRef CAS.
- S. Celikovic, M. Kirchengast, J. Rehrl, J. Kruisz, S. Sacher, J. Khinast and M. Horn, Chem. Eng. Res. Des., 2020, 154, 101–114 CrossRef CAS.
- J. Rehrl, J. Kruisz, S. Sacher, J. Khinast and M. Horn, Int. J. Pharm., 2016, 510, 100–115 CrossRef CAS PubMed.
- M. Patrascu and P. I. Barton, Chem. Eng. Process. Process Intensif., 2018, 125, 298–310 CrossRef CAS.
- M. Patrascu and P. I. Barton, Chem. Eng. Process. Process Intensif., 2018, 125, 124–132 CrossRef CAS.
- J. A. Paulson, S. Streif, R. Findeisen, R. D. Braatz and A. Mesbah, Computer Aided Chemical Engineering, Elsevier, 2018, vol. 41, pp. 353–378 Search PubMed.
- S. Zomer, J. Zhang, S. Talwar, S. Chattoraj and C. Hewitt, Int. J. Pharm., 2018, 547, 506–519 CrossRef CAS.
- Y.-S. Huang, S. Medina-González, B. Straiton, J. Keller, Q. Marashdeh, M. Gonzalez, Z. Nagy and G. V. Reklaitis, J. Pharm. Sci., 2022, 111, 69–81 CrossRef CAS.
- J. Liu and B. Benyahia, Comput. Chem. Eng., 2022, 159, 107671 CrossRef CAS.
- J. Liu and B. Benyahia, Ind. Eng. Chem. Res., 2024, 63, 7300–7314 CAS.
- A. C. Dimian, C. S. Bildea and A. A. Kiss, in Computer Aided Chemical Engineering, ed A. C. Dimian, C. S. Bildea and A. A. Kiss, Elsevier, 2014, vol. 35, pp. 449–488 Search PubMed.
- E. Santandrea, C. Waldraff, G. Gerber, M. Moreau and P. Beney, Org. Process Res. Dev., 2021, 25, 1190–1205 CrossRef CAS.
- J. Chen, B. Sarma, J. M. B. Evans and A. S. Myerson, Cryst. Growth Des., 2011, 11, 887–895 CrossRef CAS.
- K. Plumb, Chem. Eng. Res. Des., 2005, 83, 730–738 CrossRef CAS.
- I. Kekessie, K. Wegner, I. Martinez, M. E. Kopach, T. D. White, J. K. Tom, M. N. Kenworthy, F. Gallou, J. Lopez, S. G. Koenig, P. R. Payne, S. Eissler, B. Arumugam, C. Li, S. Mukherjee, A. Isidro-Llobet, O. Ludemann-Hombourger, P. Richardson, J. Kittelmann, D. Sejer Pedersen and L. J. van den Bos, J. Org. Chem., 2024, 89, 4261–4282 CrossRef CAS PubMed.
- J. Coffman, M. Brower, L. Connell-Crowley, S. Deldari, S. S. Farid, B. Horowski, U. Patil, D. Pollard, M. Qadan, S. Rose, E. Schaefer and J. Shultz, Biotechnol. Bioeng., 2021, 118, 1735–1749 CrossRef.
- R. P. V. Faria and A. E. Rodrigues, J. Chromatogr. A, 2015, 1421, 82–102 CrossRef CAS.
- S. Aldington and J. Bonnerjea, J. Chromatogr. B, 2007, 848, 64–78 CrossRef CAS PubMed.
- D. A. L. Otte, K. Basu, L. Jellett, M. Whittington, G. Spencer, M. Burris, E. B. Corcoran, K. Stone, J. Nappi, R. A. Arvary, D. Donoghue, H. Ren, K. M. Maloney and J. R. Naber, Org. Process Res. Dev., 2020, 24, 2478–2490 CrossRef CAS.
- F. Meng, S.-R. Chae, A. Drews, M. Kraume, H.-S. Shin and F. Yang, Water Res., 2009, 43, 1489–1512 CrossRef CAS.
- M. Razali, C. Didaskalou, J. F. Kim, M. Babaei, E. Drioli, Y. M. Lee and G. Szekely, ACS Appl. Mater. Interfaces, 2017, 9, 11279–11289 CrossRef CAS.
- A. Sahu, A. B. Vir, L. N. S. Molleti, S. Ramji and S. Pushpavanam, Chem. Eng. Process. Process Intensif., 2016, 104, 190–200 CrossRef CAS.
- R. A. Milescu, M. L. Segatto, A. Stahl, C. R. McElroy, T. J. Farmer, J. H. Clark and V. G. Zuin, ACS Sustainable Chem. Eng., 2020, 8, 18245–18257 CrossRef CAS.
- F. M. Antony, D. Pal and K. Wasewar, Phys. Sci. Rev., 2021, 6, 20180064 Search PubMed.
- H. Lorenz and A. Seidel-Morgenstern, Angew. Chem., Int. Ed., 2014, 53, 1218–1250 CrossRef CAS.
- N. Weeranoppanant and A. Adamo, ACS Med. Chem. Lett., 2020, 11, 9–15 CrossRef CAS.
- Z. Lei, H. T. Ang and J. Wu, Org. Process Res. Dev., 2024, 28, 1355–1368 CrossRef CAS.
- J. García-Lacuna and M. Baumann, Beilstein J. Org. Chem., 2022, 18, 1720–1740 CrossRef.
- T. Wang, H. Lu, J. Wang, Y. Xiao, Y. Zhou, Y. Bao and H. Hao, J. Ind. Eng. Chem., 2017, 54, 14–29 CrossRef CAS.
- G. Capellades, C. Neurohr, N. Briggs, K. Rapp, G. Hammersmith, D. Brancazio, B. Derksen and A. S. Myerson, Org. Process Res. Dev., 2021, 25, 1534–1546 CrossRef CAS.
- A. Eren, F. Civati, W. Ma, J. C. Gamekkanda and A. S. Myerson, J. Cryst. Growth, 2023, 601, 126958 CrossRef CAS.
- S. D. Schaber, D. I. Gerogiorgis, R. Ramachandran, J. M. B. Evans, P. I. Barton and B. L. Trout, Ind. Eng. Chem. Res., 2011, 50, 10083–10092 CrossRef CAS.
- A. Cashmore, R. Miller, H. Jolliffe, C. J. Brown, M. Lee, M. D. Haw and J. Sefcik, Cryst. Growth Des., 2023, 23, 4779–4790 CrossRef CAS.
- L. R. Agnew, T. McGlone, H. P. Wheatcroft, A. Robertson, A. R. Parsons and C. C. Wilson, Cryst. Growth Des., 2017, 17, 2418–2427 CrossRef CAS.
- G. Hou, G. Power, M. Barrett, B. Glennon, G. Morris and Y. Zhao, Cryst. Growth Des., 2014, 14, 1782–1793 CrossRef CAS.
- R. Achermann, A. Košir, B. Bodák, L. Bosetti and M. Mazzotti, Cryst. Growth Des., 2023, 23, 2485–2503 CrossRef CAS.
- Y. Gao, T. Zhang, Y. Ma, F. Xue, Z. Gao, B. Hou and J. Gong, Crystals, 2021, 11, 221 CrossRef CAS.
- K. Galan, M. J. Eicke, M. P. Elsner, H. Lorenz and A. Seidel-Morgenstern, Cryst. Growth Des., 2015, 15, 1808–1818 CrossRef CAS.
- C. De Luca, Y. Krauke, S. Stephan, G. Greco, G. Compagnin, A. Buratti, A. Cavazzini, M. Catani and S. Felletti, Green Anal. Chem., 2023, 6, 100066 CrossRef.
- S. Peper, M. Johannsen and G. Brunner, J. Chromatogr. A, 2007, 1176, 246–253 CrossRef CAS.
- J. Andersson and B. Mattiasson, J. Chromatogr. A, 2006, 1107, 88–95 CrossRef CAS.
- J. W. Lee, A. Kienle and A. Seidel-Morgenstern, Chem. Eng. Sci., 2020, 225, 115810 CrossRef CAS.
- R. S. Arafah, A. E. Ribeiro, A. E. Rodrigues and L. S. Pais, Chirality, 2019, 31, 62–71 CrossRef CAS PubMed.
- W.-S. Lee and C.-H. Lee, Sep. Purif. Technol., 2022, 288, 120597 CrossRef CAS.
- J. F. Kim, G. Székely, I. B. Valtcheva and A. G. Livingston, Green Chem., 2013, 16, 133–145 RSC.
- D. L. Gin and R. D. Noble, Science, 2011, 332, 674–676 CrossRef CAS PubMed.
- B. Van der Bruggen, A. Verliefde, L. Braeken, E. R. Cornelissen, K. Moons, J. Q. Verberk, H. J. van Dijk and G. Amy, J. Chem. Technol. Biotechnol., 2006, 81, 1166–1176 CrossRef CAS.
- S. Shin, O. Shardt, P. B. Warren and H. A. Stone, Nat. Commun., 2017, 8, 15181 CrossRef PubMed.
- B. Van der Bruggen, M. Mänttäri and M. Nyström, Sep. Purif. Technol., 2008, 63, 251–263 CrossRef CAS.
- L. Peeva, J. da, S. Burgal, I. Valtcheva and A. G. Livingston, Chem. Eng. Sci., 2014, 116, 183–194 CrossRef CAS.
- S. Divakar, M. Padaki and R. G. Balakrishna, ACS Omega, 2022, 7, 44495–44506 CrossRef CAS.
- I. Abdiaj, S. Cañellas, A. Dieguez, M. L. Linares, B. Pijper, A. Fontana, R. Rodriguez, A. Trabanco, E. Palao and J. Alcázar, J. Med. Chem., 2023, 66, 716–732 CrossRef CAS PubMed.
- Dipl. Ing. F. S. Beetz, Inkjet Printing in Industry, ed. W. Zapka, Wiley, 1st edn, 2022, pp. 1427–1460 Search PubMed.
- S. Guo, T.-M. Choi and S.-H. Chung, Eur. J. Oper. Res., 2022, 299, 883–897 CrossRef.
- E. J. Hurst, J. Hosp., 2016, 16, 255–267 Search PubMed.
- A. Jandyal, I. Chaturvedi, I. Wazir, A. Raina and M. I. U. Haq, Comput, 2022, 3, 33–42 CAS.
- C. W. J. Lim, K. Q. Le, Q. Lu and C. H. Wong, IEEE Potentials, 2016, 35, 18–22 Search PubMed.
- G. Prashar, H. Vasudev and D. Bhuddhi, J. Interact. Manuf., 2023, 17, 2221–2235 CrossRef.
- B. A. Praveena, N. Lokesh, A. Buradi, N. Santhosh, B. L. Praveena and R. Vignesh, Today Proc, 2022, 52, 1309–1313 Search PubMed.
- G. Bayram and B. G. Çetiner, J. Adv. Eng. Pure Sci., 2023, 35, 54–65 Search PubMed.
- M. Despeisse, M. Baumers, P. Brown, F. Charnley, S. J. Ford, A. Garmulewicz, S. Knowles, T. H. W. Minshall, L. Mortara, F. P. Reed-Tsochas and J. Rowley, Soc Change, 2017, 115, 75–84 Search PubMed.
- I. Gibson, D. W. Rosen and B. Stucker, Additive manufacturing technologies: rapid prototyping to direct digital manufacturing, Springer, New York, 2010 Search PubMed.
- D. T. Pham and R. S. Gault, J. Mach. Tools Manuf., 1998, 38, 1257–1287 CrossRef.
- M. B. Balletti and J. F. Guerra, Herit, 2017, 26, 172–182 Search PubMed.
- 3D Printing of Pharmaceuticals, ed. A. W. Basit and S. Gaisford, Springer, Cham, 2018, vol. 31 Search PubMed.
- B. Berman, Bus. Horiz., 2012, 55, 155–162 CrossRef.
- A. Rindfleisch, Letter, 2020, 31, 13–17 Search PubMed.
- M. Jayakrishna, M. Vijay and B. Khan, J. Eng., 2023, 2023, 7465737 Search PubMed.
- I. Karakurt and L. Lin, Opin Chem Eng, 2020, 28, 134–143 CrossRef.
- M. Livesu, S. Ellero, J. Martínez, S. Lefebvre and M. Attene, Graph Forum, 2017, 36, 537–564 CrossRef.
- A. Melocchi, M. Uboldi, M. Cerea, A. Foppoli, A. Maroni, S. Moutaharrik, L. Palugan, L. Zema and A. Gazzaniga, Drug Delivery Rev., 2021, 173, 216–237 CrossRef CAS.
- L. Zema, A. Melocchi, A. Maroni and A. Gazzaniga, Pharm. Sci., 2017, 106, 1697–1705 CrossRef CAS.
- A. Perrot and S. Amziane, in 3D Printing of Concrete, ed. A. Perrot, Wiley, 1st edn, 2019, pp. 1–40 Search PubMed.
- I. Poudel, N. Mita and R. J. Babu, Drug Delivery, 2024, 21, 1595–1614 CAS.
- M. Ahmed, S. Tomlin, C. Tuleu and S. Garfield, Pharmaceutics, 2024, 16, 1212 CrossRef.
- A. Melocchi, M. Uboldi, M. Cerea, A. Foppoli, A. Maroni, S. Moutaharrik, L. Palugan, L. Zema and A. Gazzaniga, Pharm. Sci., 2020, 109, 2943–2957 CrossRef CAS PubMed.
- M. Preis and H. Öblom, AAPS PharmSciTech, 2017, 18, 303–308 CrossRef.
- J. Wang, Y. Zhang, N. H. Aghda, A. R. Pillai, R. Thakkar, A. Nokhodchi and M. Maniruzzaman, Drug Delivery Rev., 2021, 174, 294–316 CrossRef CAS.
- C. Parra-Cabrera, C. Achille, S. Kuhn and R. Ameloot, Chem. Soc. Rev., 2018, 47, 209–230 RSC.
- A. Bubliauskas, D. J. Blair, H. Powell-Davies, P. J. Kitson, M. D. Burke and L. Cronin, Angew. Chem., Int. Ed., 2022, 61, e202116108 CrossRef CAS PubMed.
- D. Albani, G. Vilé, M. A. Beltran Toro, R. Kaufmann, S. Mitchell and J. Pérez-Ramírez, React. Chem. Eng., 2016, 1, 454–462 RSC.
- B. Gutmann, M. Köckinger, G. Glotz, T. Ciaglia, E. Slama, M. Zadravec, S. Pfanner, M. C. Maier, H. Gruber-Wölfler and C. Oliver Kappe, React. Chem. Eng., 2017, 2, 919–927 RSC.
- M. Ciriani, B. Ritzen, T. Schmitges, R. Reintjens, P. Hermsen and R. Van Summeren, Org. Process Res. Dev., 2025, 29, 640–649 CrossRef CAS.
- T. Furrer, M. Levis, B. Berger, M. Kandziora and A. Zogg, Org. Process Res. Dev., 2023, 27, 1365–1376 CrossRef CAS.
- C. H. Hornung, X. Nguyen, A. Carafa, J. Gardiner, A. Urban, D. Fraser, M. D. Horne, D. R. Gunasegaram and J. Tsanaktsidis, Org. Process Res. Dev., 2017, 21, 1311–1319 CrossRef CAS.
- CSIRO develops low-cost titanium wire for additive manufacturing, https://www.csiro.au/en/news/All/News/2021/September/CSIRO-develops-low-cost-titanium-wire-for-additive-manufacturing, (accessed July 5, 2025).
- G. Vilé, D. Ng, Z. Xie, I. Martinez-Botella, J. Tsanaktsidis and C. H. Hornung, ChemCatChem, 2022, 14, e202101941 CrossRef.
- J. Luo, V. Ruta, I. S. Kwon, J. Albertazzi, N. Allasia, O. Nevskyi, V. Busini, D. Moscatelli and G. Vilé, Adv. Funct. Mater., 2024, 34, 2404794 CrossRef CAS.
- S. J. T. Awad, S. Gaisford and A. W. Basit, J. Pharm., 2018, 548, 586–596 Search PubMed.
- H. Bagga, A. Aime, G. Chaudhari and U. Vyas, J. Drug Delivery Technol., 2024, 14, 1791–1800 CrossRef.
- S. K. Debnath, M. Debnath, R. Srivastava and A. Omri, Drug Delivery, 2021, 18, 1659–1672 Search PubMed.
- N. A. Elkasabgy, A. A. Mahmoud and A. Maged, J. Pharm., 2020, 588, 119732 CAS.
- A. Goyanes, A. B. M. Buanz, A. W. Basit and S. Gaisford, J. Pharm., 2014, 476, 88–92 CAS.
- A. Goyanes, A. B. M. Buanz, G. B. Hatton, S. Gaisford and A. W. Basit, J. Pharm. Biopharm., 2015, 89, 157–162 CrossRef CAS.
- A. Gazzaniga, A. Foppoli, M. Cerea, L. Palugan, M. Cirilli, S. Moutaharrik, A. Melocchi and A. Maroni, J. Pharm. X, 2023, 5, 100171 CAS.
- U. Garg, N. Jain, S. Kaul, V. K. Rai, M. Pandey, U. Nagaich and K. Dua, Drug Delivery Sci. Technol., 2022, 76, 103798 CrossRef.
- H. Liu, A. Nail, D. Meng, L. Zhu, X. Guo, C. Li and H.-J. Li, J. Pharm., 2025, 668, 124995 CAS.
- A. Melocchi, F. Briatico-Vangosa, M. Uboldi, F. Parietti, M. Turchi, D. von Zeppelin, A. Maroni, L. Zema, A. Gazzaniga and A. Zidan, J. Pharm., 2021, 592, 119901 CAS.
- G. Mora-Castaño, J. Domínguez-Robles, A. Himawan, M. Millán-Jiménez and I. Caraballo, J. Pharm., 2024, 663, 124543 Search PubMed.
- C. Muehlenfeld, P. Duffy, F. Yang, D. Z. Pérez, F. El-Saleh and T. Durig, Pharmaceutics, 2024, 16, 317 CrossRef CAS PubMed.
- S. Narala, A. A. A. Youssef, S. R. Munnangi, N. Narala, P. Lakkala, S. K. Vemula and M. Repka, Drug Delivery, 2024, 21, 1543–1557 CAS.
- S. K. Patel, M. Khoder, M. Peak and M. A. Alhnan, Drug Delivery Rev., 2021, 174, 369–386 CrossRef CAS.
- I.-R. Turac, A. Porfire, S. Iurian, A. G. Crisan, T. Casian, R. Iovanov and I. Tomută, Pharmaceutics, 2024, 16, 790 CrossRef CAS.
- M. Uboldi, A. Melocchi, S. Moutaharrik, L. Palugan, M. Cerea, A. Foppoli, A. Maroni, A. Gazzaniga and L. Zema, Release, 2022, 348, 537–552 CrossRef CAS PubMed.
- L. Alzoubi, A. A. A. Aljabali and M. M. Tambuwala, AAPS PharmSciTech, 2023, 24, 228 CrossRef CAS PubMed.
- K. K. Jain and O. Curr, J. Ther., 2002, 4, 548–558 CAS.
- M. Deng, S. Wu and M. Ning, J. Pharm., 2025, 669, 125089 CAS.
- J. Norman, R. D. Madurawe, C. M. V. Moore, M. A. Khan and A. Khairuzzaman, Adv. Drug Delivery Rev., 2017, 108, 39–50 CrossRef CAS.
- F. F. Awad, A. Goyanes, S. Gaisford and A. W. Basit, J. Pharm., 2020, 586, 119594 Search PubMed.
- J. Goole and K. Amighi, J. Pharm., 2016, 499, 376–394 Search PubMed.
- H. Ragelle, S. Rahimian, E. A. Guzzi, P. D. Westenskow, M. W. Tibbitt, G. Schwach and R. Langer, Drug Delivery Rev., 2021, 178, 113990 CrossRef CAS PubMed.
- M. C. Simon, K. Laios, I. Nikolakakis and T. G. Papaioannou, Medicine, 2024, 14, 1080 Search PubMed.
- M. Tiboni, M. Tiboni, A. Pierro, M. D. Papa, S. Sparaventi, M. Cespi and L. Casettari, J. Pharm., 2021, 599, 120464 CAS.
- M. Tiboni, G. Curzi, A. Aluigi and L. Casettari, Drug Delivery Sci. Technol., 2021, 65, 102661 CrossRef CAS.
- H.-G. Yi, H. Lee and D.-W. Cho, Bioengineering, 2017, 4, 10 CrossRef PubMed.
- E. Fuenmayor, C. O’Donnell, N. Gately, P. Doran, D. M. Devine, J. G. Lyons, C. McConville and I. Major, J. Pharm., 2019, 569, 118611 CAS.
- M. Enke, K. Kühne, S. Seyferth, D. Fischer and A. Schneeberger, J. Sci. Tech. Res., 2021, 41, 32889–32895 Search PubMed.
- S. J. Trenfield, H. X. Tan, A. Awad, A. Buanz, S. Gaisford, A. W. Basit and A. Goyanes, J. Pharm., 2019, 567, 118443 CAS.
- K. Yang, L. Wang, S. Vijayavenkataraman, Y. Yuan, E. C. K. Tan and L. Kang, Drug Delivery Rev., 2024, 214, 115456 CrossRef CAS.
- Y. Yang, D. Tilman, Z. Jin, P. Smith, C. B. Barrett, Y.-G. Zhu, J. Burney, P. D’Odorico, P. Fantke, J. Fargione, J. C. Finlay, M. C. Rulli, L. Sloat, K. Jan van Groenigen, P. C. West, L. Ziska, A. M. Michalak, The Clim-Ag Team and D. B. Lobell, Science, 2024, 385, eadn3747 CrossRef CAS PubMed.
- P. E. Antezana, S. Municoy, G. Ostapchuk, P. N. Catalano, J. G. Hardy, P. A. Evelson, G. Orive and M. F. Desimone, Pharmaceutics, 2023, 15, 2743 CrossRef CAS.
- P. Pingale, S. Dawre, V. Dhapte-Pawar, N. Dhas and A. Rajput, Transl. Res., 2023, 13, 164–188 Search PubMed.
- D. B. Mahmoud and M. Schulz-Siegmund, Adv. Healthc. Mater., 2023, 12, 2202631 CrossRef CAS.
- B. Muñiz Castro, M. Elbadawi, J. J. Ong, T. Pollard, Z. Song, S. Gaisford, G. Pérez, A. W. Basit, P. Cabalar and A. Goyanes, J. Controlled Release, 2021, 337, 530–545 CrossRef.
- X. Rodríguez-Maciñeiras, C. Bendicho-Lavilla, C. Rial, K. Garba-Mohammed, A. Worsley, E. Díaz-Torres, C. Orive-Martínez, Á. Orive-Mayor, A. W. Basit, C. Alvarez-Lorenzo and A. Goyanes, Int. J. Pharm., 2025, 671, 125251 CrossRef.
- CurifyLabs Home Page, https://curifylabs.com, (accessed July 23, 2025).
- Triastek raises $20.4 million to 3D print pharmaceuticals, https://www.voxelmatters.com/triastek-raises-20-4-million-to-3d-print-pharmaceuticals/, (accessed March 28, 2025).
- T. G. West and T. J. Bradbury, 3D and 4D printing in biomedical applications: process engineering and additive manufacturing, 2019 Search PubMed.
- Aprecia Home Page, https://aprecia.com/, (accessed March 4, 2025).
- K. Sen, T. G. West and B. Chaudhuri, Additive Manufacturing in Pharmaceuticals, Springer, Singapore, 2023 Search PubMed.
- V. Ianno, S. Vurpillot, S. Prillieux and P. Espeau, Pharmaceutics, 2024, 16, 441 CrossRef CAS.
- T. Tracy, L. Wu, X. Liu, S. Cheng and X. Li, J. Pharm., 2023, 631, 122480 CrossRef CAS.
- S. Wang, X. Chen, X. Han, X. Hong, X. Li, H. Zhang, M. Li, Z. Wang and A. Zheng, Pharmaceutics, 2023, 15, 416 CrossRef CAS.
- A. Schneeberger, S. Seyferth and D. Fischer, Arch. Dent., 2021, 3, 19–21 Search PubMed.
- D. Moldenhauer, D. C. Y. Nguyen, L. Jescheck, F. Hack, D. Fischer and A. Schneeberger, J. Pharm., 2021, 592, 120096 CrossRef CAS.
- N. Schwarz, M. Enke, F. V. Gruschwitz, D. Winkler, S. Franzmann, L. Jescheck, F. Hanf and A. Schneeberger, Pharmaceutics, 2024, 16, 368 CrossRef CAS PubMed.
- H. Eswaran, R. D. Ponnuswamy and R. P. Kannapan, 3D Print Med., 2023, 12, 100125 CrossRef.
- R.-A. Varvara, K. Szabo and D. C. Vodnar, Nutrients, 2021, 13, 3617 CrossRef CAS.
- H. Shi, M. Zhang, A. S. Mujumdar and C. Li, Rev Food Sci Food Saf, 2024, 23, e70005 CrossRef.
- N. Beer, S. Kaae, N. Genina, S. K. Sporrong, T. L. Alves, J. Hoebert, M. L. D. Bruin and I. Hegger, Innov. Regul. Sci., 2023, 57, 26–36 CrossRef.
- J. T. Y. Cheng, E. C. K. Tan and L. Kang, Biofabrication, 2025, 17, 012002 CrossRef CAS PubMed.
- K. Huanbutta, K. Burapapadh, P. Sriamornsak and T. Sangnim, Pharmaceutics, 2023, 15, 1877 CrossRef CAS PubMed.
- L. R.-P. Bendicho-Lavilla, P. Januskaite, C. Rial, C. Alvarez-Lorenzo, A. W. Basit and J. D. D. A. Goyanes, Technology, 2024, 92, 105337 Search PubMed.
- S. J. Trenfield, P. Januskaite, A. Goyanes, D. Wilsdon, M. Rowland, S. Gaisford and A. W. Basit, Pharmaceutics, 2022, 14, 589 CrossRef CAS PubMed.
- S. J. Trenfield, X. Xu, A. Goyanes, M. Rowland, D. Wilsdon, S. Gaisford and A. W. Basit, J. Pharm. X, 2023, 5, 100148 CAS.
- M. Elbadawi, B. M. Castro, F. K. H. Gavins, J. J. Ong, S. Gaisford, G. Pérez, A. W. Basit, P. Cabalar and A. Goyanes, J. Pharm., 2020, 590, 119837 CrossRef CAS.
- A. Goyanes, C. M. Madla, A. Umerji, G. D. Piñeiro, J. M. G. Montero, M. J. L. Diaz, M. G. Barcia, F. Taherali, P. Sánchez-Pintos, M.-L. Couce, S. Gaisford and A. W. Basit, J. Pharm., 2019, 567, 118497 CrossRef CAS PubMed.
- L. Rodríguez-Pombo, C. Gallego-Fernández, A. K. Jørgensen, C. J. Parramon-Teixidó, C. Cañete-Ramirez, M. J. Cabañas-Poy, A. W. Basit, C. Alvarez-Lorenzo and A. Goyanes, Drug Delivery, 2024, 21, 1665–1681 Search PubMed.
- L. Rodríguez-Pombo, M. J. de Castro-López, P. Sánchez-Pintos, J. M. Giraldez-Montero, P. Januskaite, G. Duran-Piñeiro, M. D. Bóveda, C. Alvarez-Lorenzo, A. W. Basit, A. Goyanes and M. L. Couce, J. Pharm., 2024, 657, 124140 Search PubMed.
- Curifylab and Natural Machines to launch affordable, pharma-compliant drug 3D printing technology in 2022, https://3dprintingindustry.com/news/curifylabs-and-natural-machines-to-launch-affordable-pharma-compliant-drug-3d-printing-technology-in-2022-205130/, (accessed March 3, 2025).
- H. Öblom, E. Sjöholm, M. Rautamo and N. Sandler, Pharmaceutics, 2019, 11, 334 CrossRef.
- M. Rautamo, K. Kvarnström, M. Sivén, M. Airaksinen, P. Lahdenne and N. Sandler, Pharmaceutics, 2020, 12, 229 CrossRef PubMed.
- E. Sjöholm, R. Mathiyalagan, X. Wang and N. Sandler, Pharmaceutics, 2022, 14, 1339 CrossRef.
- N. S. Topelius, F. Shokraneh, M. Bahman, J. Lahtinen, N. Hassinen, S. Airaksinen, S. Verma, L. Hrizanovska, J. Lass, U. Paaver, J. Tähnas, C. Kern, F. Lagarce, D. Fenske, J. Malik, H. Scherliess, S. P. Cruz, M. Paulsson, J. Dekker, K. Kammonen, M. Rautamo, H. Lück, A. Pierrot, S. Stareprawo, M. Tubic-Grozdanis, S. Zibolka, U. Lösch, M. Jeske, U. Griesser, K. Hummer, A. Thalmeier, A. Harjans, A. Kruse, R. Heimke-Brinck, K. Khoukh and F. Bruno, Pharmaceutics, 2024, 16, 678 CrossRef.
- M. Rautamo, H. M. Tolonen, N. Asinger, H. Ruutiainen, S. Kuitunen, S. S. K. Sporrong, M. Sivén and M. Paulsson, J. Hosp. Pharm., 2024, 31, A43 Search PubMed.
- M. Palo, K. Kogermann, N. Genina, D. Fors, J. Peltonen, J. Heinämäki and N. Sandler, Drug Delivery Sci. Technol., 2016, 34, 60–70 CrossRef CAS.
- H. Vakili, H. Wickström, D. Desai, M. Preis and N. Sandler, J. Pharm., 2017, 524, 414–423 CAS.
- Cellink announces third-year extension of collaboration with a global biopharmaceutical company for drug discovery, https://www.cellink.com/cellink-announces-third-year-extension-of-collaboration-with-a-global-biopharmaceutical-company-for-drug-discovery/, (accessed March 3, 2025).
- Pandemic Calls 3D Printers Into Action to Make Everything from Parts to Prescriptions Vitae Industries Helps Automate Compounding Pharmacies, https://rxinsider.com/market-buzz/?p=5950-pandemic-calls-3d-printers-into-action-to-make-everything-from-parts-to-prescriptions-vitae-industries-helps-automate-compounding-pharmacies, (accessed March 4, 2025).
- 3DStartup: Craft Health Is Personalizing Medicines With 3D Printing, https://www.3dnatives.com/en/startup3d-craft-health-personalizing-medicines-3d-printing-051220234/, (accessed March 4, 2025).
- DiHeSys takes steps towards application, https://www.gesundheitsindustrie-bw.de/en/article/news/dihesys-takes-steps-towards-application, (accessed March 4, 2025).
- M. Pistone, G. F. Racaniello, I. Arduino, V. Laquintana, A. Lopalco, A. Cutrignelli, R. Rizzi, M. Franco, A. Lopedota and N. Denora, Research, 2022, 12, 1895–1910 CAS.
- M. Pistone, G. F. Racaniello, R. Rizzi, R. M. Iacobazzi, I. Arduino, A. Lopalco, A. A. Lopedota and N. Denora, J. Pharm., 2023, 632, 122592 CAS.
- G. F. Racaniello, M. Pistone, C. Meazzini, A. Lopedota, I. Arduino, R. Rizzi, A. Lopalco, U. M. Musazzi, F. Cilurzo and N. Denora, J. Pharm., 2023, 643, 123214 CAS.
- N. Beer, I. Hegger, S. Kaae, M. L. D. Bruin, N. Genina, T. L. Alves, J. Hoebert and S. K. Sporrong, Res. Clin. Soc Pharm., 2021, 4, 100073 Search PubMed.
- K. Englezos, L. Wang, E. C. K. Tan and L. Kang, J. Pharm., 2023, 635, 122785 CAS.
- R. Parhi and J. D. Deliv, Technology, 2021, 64, 102571 CAS.
- J. M. Stevens, J. M. Ganley, M. J. Goldfogel, A. Furman and S. R. Wisniewski, Org. Process Res. Dev., 2025, 29, 189–199 CrossRef CAS.
- C. P. Haas, M. Lübbesmeyer, E. H. Jin, M. A. McDonald, B. A. Koscher, N. Guimond, L. Di Rocco, H. Kayser, S. Leweke, S. Niedenführ, R. Nicholls, E. Greeves, D. M. Barber, J. Hillenbrand, G. Volpin and K. F. Jensen, ACS Cent. Sci., 2023, 9, 307–317 CrossRef CAS.
- M. P. Maloney, C. W. Coley, S. Genheden, N. Carson, P. Helquist, P.-O. Norrby and O. Wiest, J. Org. Chem., 2023, 88, 5239–5241 CrossRef CAS.
- S. M. Kearnes, M. R. Maser, M. Wleklinski, A. Kast, A. G. Doyle, S. D. Dreher, J. M. Hawkins, K. F. Jensen and C. W. Coley, J. Am. Chem. Soc., 2021, 143, 18820–18826 CrossRef CAS.
- W. Heyndrickx, L. Mervin, T. Morawietz, N. Sturm, L. Friedrich, A. Zalewski, A. Pentina, L. Humbeck, M. Oldenhof, R. Niwayama, P. Schmidtke, N. Fechner, J. Simm, A. Arany, N. Drizard, R. Jabal, A. Afanasyeva, R. Loeb, S. Verma, S. Harnqvist, M. Holmes, B. Pejo, M. Telenczuk, N. Holway, A. Dieckmann, N. Rieke, F. Zumsande, D.-A. Clevert, M. Krug, C. Luscombe, D. Green, P. Ertl, P. Antal, D. Marcus, N. Do Huu, H. Fuji, S. Pickett, G. Acs, E. Boniface, B. Beck, Y. Sun, A. Gohier, F. Rippmann, O. Engkvist, A. H. Göller, Y. Moreau, M. N. Galtier, A. Schuffenhauer and H. Ceulemans, J. Chem. Inf. Model., 2024, 64, 2331–2344 CrossRef CAS.
- A. M. Bran, S. Cox, O. Schilter, C. Baldassari, A. D. White and P. Schwaller, Nat. Mach. Intell., 2024, 6, 525–535 CrossRef.
- M. C. Ramos, C. J. Collison and A. D. White, Chem. Sci., 2025, 16, 2514–2572 RSC.
- M. Schilling-Wilhelmi, M. Ríos-García, S. Shabih, M. V. Gil, S. Miret, C. T. Koch, J. A. Márquez and K. M. Jablonka, Chem. Soc. Rev., 2025, 54, 1125–1150 RSC.
- J. Krueger, A. P. Dieskau, J. Hassfeld, J. Gries, O. Block, H. Weinmann, D. Kaufmann, S. Hildbrand, V. Kraft, R. Moeckel, J. R. Dehli, U. Scholz and C. F. Nising, Angew. Chem., Int. Ed., 2025, e202420719 CAS.
- S. Caille, S. Cui, M. M. Faul, S. M. Mennen, J. S. Tedrow and S. D. Walker, J. Org. Chem., 2019, 84, 4583–4603 CrossRef CAS.
- J. C. Hethcox, J. Kim, H. C. Johnson, Y. Ji, M. Chow, J. A. Newman, D. A. DiRocco and J. P. McMullen, Org. Process Res. Dev., 2024, 28, 413–421 CrossRef CAS.
- J. Kim, V. Zhang, K. Abe, Y. Qin, D. A. DiRocco, J. P. McMullen, A. Sun, R. Gangam, M. Chow, A. Pitts-McCoy and A. S. Malkani, Org. Process Res. Dev., 2024, 28, 422–431 CrossRef CAS.
- S. D. McCann, S. H. Dubina, B. Kosjek, E. Alwedi, T. Behre, S. A. Burgess, G. Dal Poggetto, D. A. DiRocco, C. Hartmanshenn, J. P. McMullen, N. Padivitage and A. C. Sun, Org. Process Res. Dev., 2024, 28, 441–450 CrossRef CAS.
- D. N. Le, T. J. Wright, E. Alwedi, C. Hartmanshenn, H. Li, J. P. McMullen and D. A. DiRocco, Org. Process Res. Dev., 2024, 28, 451–459 CrossRef CAS.
- Y. Qin, K. A. Mattern, V. Zhang, K. Abe, J. Kim, M. Zheng, R. Gangam, A. Kalinin, J. N. Kolev, S. Axnanda, Z. E. X. Dance, U. Ayesa, Y. Ji, S. T. Grosser, E. Appiah-Amponsah and J. P. McMullen, Org. Process Res. Dev., 2024, 28, 432–440 CrossRef CAS.
- F. Peng, L. Tan, L. Chen, S. M. Dalby, D. A. DiRocco, J. Duan, M. Feng, G. Gong, H. Guo, J. C. Hethcox, L. Jin, H. C. Johnson, J. Kim, D. Le, Y. Lin, W. Liu, J. Shen, Y. Wan, C. Xiao, B. Xiang, Q. Xiang, J. Xu, L. Yan, W. Yang, H. Ye, Y. Yu and J. Zhang, Org. Process Res. Dev., 2022, 26, 508–515 CrossRef CAS.
- C. Bottecchia, F. Lévesque, J. P. McMullen, Y. Ji, M. Reibarkh, F. Peng, L. Tan, G. Spencer, J. Nappi, D. Lehnherr, K. Narsimhan, M. K. Wismer, L. Chen, Y. Lin and S. M. Dalby, Org. Process Res. Dev., 2022, 26, 516–524 CrossRef CAS.
- Z. Chen, N. Salehi Marzijarani, S. Quirie, G. F. Pirrone, S. M. Dalby, T. Wang, J. Kim, F. Peng and A. J. Fine, Org. Process Res. Dev., 2022, 26, 525–532 CrossRef CAS.
- T. Wang, E. M. Phillips, S. M. Dalby, E. Sirota, S. Axnanda, C. S. Shultz, P. Patel, J. H. Waldman, E. Alwedi, X. Wang, K. Zawatzky, M. Chow, N. Padivitage, M. Weisel, M. Whittington, J. Duan and T. Lu, Org. Process Res. Dev., 2022, 26, 543–550 CrossRef CAS.
- M. Pirnot, K. Stone, T. J. Wright, D. J. Lamberto, J. Schoell, Y. Lam, K. Zawatzky, X. Wang, S. M. Dalby, A. J. Fine and J. P. McMullen, Org. Process Res. Dev., 2022, 26, 551–559 CrossRef CAS.
- D. A. DiRocco, Y.-L. Zhong, D. N. Le, S. D. McCann, J. C. Hethcox, J. Kim, J. N. Kolev, B. Kosjek, S. M. Dalby, J. P. McMullen, R. Gangam and W. J. Morris, Org. Process Res. Dev., 2024, 28, 404–412 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |





