Open Access Article
Open Access ArticleProbing the nucleobase-specific binding interaction of hydroxychloroquine sulfate with RNA and subsequent sequestration by a water-soluble molecular basket†
Rahul
Yadav
,
Subhasis
Das
,
Madhumita
Mukherjee
and
Saptarshi
Mukherjee
 *
*
Department of Chemistry, Indian Institute of Science Education and Research Bhopal, Bhopal Bypass Road, Bhauri, Bhopal, 462 066, Madhya Pradesh, India. E-mail: saptarshi@iiserb.ac.in
First published on 10th March 2025
Abstract
A thorough understanding of the binding interactions of small molecules with genetic materials of the cell (DNA/RNA) has a persistent importance in pharmaceutical industries for the development of new drugs for treating various life-threatening ailments. Hydroxychloroquine sulfate (HCQS), an antimalarial drug, was potentially used for clinical trials with the hope of treating patients suffering from SARS-CoV-2 during the COVID-19 pandemic. Herein, we have extensively delineated the binding interactions of HCQS with RNA under physiological conditions using multi-spectroscopic and calorimetric approaches. Our results demonstrated that HCQS binds to RNA through the groove-binding mode in uridine- and cytidine-rich regions. The mode of binding was meticulously characterized by fluorescence quenching studies and circular dichroism spectroscopy, well complemented by other experiments. Our results obtained from isothermal titration calorimetry reveal the phenomenon of the release of bound water molecules when HCQS binds at the groove position of RNA, the process being entropically driven. Furthermore, we have employed the concept of host–guest chemistry for the sequestration of RNA-bound HCQS using a water-soluble, non-toxic, 4-sulfocalix[4]arene as a basket-type macrocyclic host. This investigation may be conducive to the development of safe RNA-based therapeutics like RNA-based vaccines that comprise small molecule–RNA interactions.
Introduction
The exploration of the associated interactions between small molecules and biomolecules like proteins, DNA, RNA, lipid membranes, etc. is an indispensable domain of contemporary research in biochemistry and biotechnology. Awareness of mechanistic insights into these interactions is of utmost importance for the strategic design of new drug molecules to combat fatal diseases, like SARS-CoV-2, malaria, etc.1–4 Chloroquine (CLQ) and hydroxychloroquine (HCQ) drugs, which are the derivatives of the 4-aminoquinoline family, have potential antimalarial properties and hence are used for preventing and curing these diseases.5,6 Both these drugs are administered in the body in the form of chloroquine phosphate (CLQP) and hydroxychloroquine sulfate (HCQS) salts and get adsorbed in the upper intestinal tract.6 HCQ is more polar and less lipophilic than CLQ; however, both are water-soluble and permeable to the cell membrane.6HCQ is used more frequently than CLQ because of its reduced cytotoxicity.7 Additionally, it has been proven to be an anti-inflammatory, non-steroidal medicine for the treatment of other severe diseases like rheumatoid arthritis, systemic lupus erythematosus, dermatomyositis, etc.8,9 HCQ came to the focus again in 2020 when it was anticipated to treat coronavirus disease 2019 (COVID-19). After the approval of the FDA, the drug was globally repurposed as an emergency antiviral drug for the treatment of patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, the drug was not completely successful in treating COVID-19.1,10–13 The mechanistic activities of CLQ and HCQ depend on their binding interactions with nucleic acids like DNA and RNA. Significant scientific attempts have been devoted to the comprehensive understanding of the underlying mechanistic aspects of such interactions between these drugs and nucleic acids. For instance, Irvin et al. proposed that the binding of CLQ is facilitated by an intercalative mode of binding with DNA.14 On the other hand, Cohen et al. and Kwakye-Berko et al. independently contradicted that the binding is electrostatic in nature.15,16 Further investigations of CLQ and HCQS binding interaction with nucleic acids have been done using experimental and computational approaches.17 Compared to CLQ, the binding mechanism of HCQS was studied extensively during the COVID-19 pandemic period. Rocha and co-workers reported that HCQS strongly interacts with double-stranded DNA (dsDNA), and the mode of binding was proved to be (i) groove binding at lower drug concentrations and (ii) intercalative binding at higher drug concentrations utilizing single-molecule force spectroscopy and bulk gel-electrophoresis approaches.18 Singh and co-workers reported the specific binding efficacy of HCQS with AT-rich and GC-rich dsDNA using multi-spectroscopic techniques and molecular dynamics (MD) simulations.19 They concluded that HCQS binds to AT-rich DNA at the minor groove while binding to GC-rich DNA happens at the major groove. Using MD simulations, they proposed the binding mechanism of HCQS with DNA involving hydrogen bond formation of different nucleobases with the N atom of the quinoline ring of HCQS, side chain N atoms of HCQS, and the O atom of the hydroxy group of HCQS.19 The same group also reported the binding of HCQS with different topological structures of G-quadruplex DNA (Gq-DNA) and revealed that HCQS preferentially binds at hybrid (3+1) parallel conformation over antiparallel conformation of Gq-DNA.20 Although substantial progress has been made in elucidating the binding interaction between the small molecules and DNA, the mechanistic investigations of the binding interaction between small molecules and RNA were not extensively carried out heretofore. RNA is valuable in many fields, from molecular and cellular biology to medicine, catalysis, and nanotechnology.21 Biswal and co-workers elucidated the binding interactions of RNA with choline-based ionic liquids and demonstrated that ionic liquid molecules bind at minor grooves of RNA. Their results were consolidated by dye-displacement assay and many other spectroscopic and computational approaches.21 Our group has reported the nucleobase-specific interaction of RNA with an antimalarial drug cryptolepine hydrate (CRYP), which binds to RNA through the intercalative mode. It has been established that hydrophobic interactions between the probe molecule and RNA nucleobases were the governing factors modulating the said interaction.22
In the present investigation, we have thoroughly examined the interaction of HCQS with torula yeast RNA (please refer to the ESI† for more details, Table S1) at physiological pH, a topic which is not much explored. The mode of binding of the drug with RNA has been explored by fluorescence quenching methods using an external quenching agent. Additionally, we have elucidated the nucleobase-specific binding of HCQS by titrating the four monomeric RNA nucleobases (A, U, G, and C) independently as a function of their increasing concentrations. All the fluorescence quenching results were analyzed using the Benesi–Hildebrand binding model.22 The associated thermodynamics of binding interactions was studied using isothermal titration calorimetry (ITC). The binding constants obtained from our ITC results were in excellent agreement with the steady-state spectroscopic results. Furthermore, to circumvent the cytotoxic and adverse effects of the excess use of HCQS, the sequestration of the drug from the binding site is essential for its secured medicinal usage.23 Physiologically, extraction of excess bound drugs from biomolecular/biomimicking assemblies is momentous to curtail their cytotoxic effects.23 Successful efforts for the sequestration of DNA-bound drugs have been reported by applying various strategies like micelle formation, mixed micelle formation, liposome encapsulation, etc.23 Our group has also reported deintercalation of RNA-bound CRYP using a biocompatible water-soluble host molecule, cucurbit[7]uril hydrate, implementing the concept of host–guest chemistry.22 Herein, we have also attempted the application of host–guest chemistry for the dissociation of the RNA–HCQS complex by a water-soluble molecular basket type molecule, 4-sulfocalix[4]arene (SCX4). Importantly, we could completely sequester the RNA-bound HCQS through host–guest complexation. The binding constants of host–guest interactions of free HCQS and RNA-bound HCQS with SCX4 were quantitatively analyzed by fluorescence spectroscopy and ITC experiments. Additionally, Circular Dichroism (CD) spectra of RNA in the presence of SCX4 suggested that the sequestrating agent does not hamper the secondary structure of RNA, thereby affirming the biocompatible nature of SCX4.
Results and discussion
Photophysical properties of hydroxychloroquine sulfate
The absorption spectrum of hydroxychloroquine sulfate (HCQS, Fig. 1(A)) exhibits two absorption peaks at 330 nm and 342 nm, which are typical features of π → π* and n → π* transitions, respectively, occurring in the quinoline ring present in the molecule (Fig. 1(B)).24,25 The molar extinction coefficients of HCQS at 330 nm and 342 nm were estimated to be 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 960 M−1 cm−1 and 17
960 M−1 cm−1 and 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 720 M−1 cm−1, respectively, determined spectrophotometrically (Fig. S1A and B, ESI†). The photophysical features of this molecule are highly sensitive to the pH of the medium. At physiological pH ∼ 7.4 upon excitation at 330 nm, HCQS displays a characteristic fluorescence peak at 388 nm (Fig. 1(B)).
720 M−1 cm−1, respectively, determined spectrophotometrically (Fig. S1A and B, ESI†). The photophysical features of this molecule are highly sensitive to the pH of the medium. At physiological pH ∼ 7.4 upon excitation at 330 nm, HCQS displays a characteristic fluorescence peak at 388 nm (Fig. 1(B)).
The excitation spectrum of HCQS also almost overlaps with the absorption spectrum, affirming the existence of a single excited state in the system (Fig. 1(B)). Additionally, the non-variant nature of the excitation-dependent emission spectra of HCQS indicated that HCQS exhibits a single excitation wavelength (Fig. S1C, ESI†). Upon variations in the pH of the buffer, the photophysical properties altered considerably (Fig. S1D and E, ESI†). At physiological pH (∼7.4), the HCQS molecule exists in diprotonated form with sulfate as a counterion (Fig. 1(A)).19,26 Upon increasing the pH above 9, the two absorption peaks (observed at lower pH values) merge into a single broad absorption peak as shown in Fig. S1D (ESI†). Additionally, with the increased pH, due to deprotonation, a huge augmentation in fluorescence intensity was also observed (Fig. S1E, ESI†). On the other hand, at lower pH values, due to protonation, the molecule becomes poorly fluorescent because of increased non-radiative rate constant.24 A detailed study of the photophysical properties of hydroxychloroquine has been previously reported by Nord et. al. and our observations are in line with the literature.24,25
Steady-state and time-resolved spectroscopic studies of interaction of HCQS with RNA
UV-visible spectroscopy is one of the most pervasive tools to study the interaction of small molecules with DNA/RNA, their complex formation, and their mode of binding. Generally, modulations in the magnitude of absorbance and shift in peak positions are correlated with the strength of complex formation and mode of binding, respectively. Usually, when the binding of small molecules with DNA/RNA is facilitated through the intercalative mode, it results in a significant bathochromic shift in absorption spectra due to a change in the microenvironment of the probe and stabilization of the helix by insertion of the probe molecule into the strands. On the other hand, groove binding or electrostatic mode involves the binding of small molecules outside the strands of DNA/RNA, resulting in a non-significant peak shift in the absorption maxima along with a hypochromic effect.22,27As mentioned earlier, HCQS exhibits two characteristic absorption peaks at 330 nm and 342 nm. Upon incremental addition of RNA to HCQS, the absorbance of both these peaks decreased (RNA does not display any interfering absorbance in this region beyond 330 nm), which suggests the possible interaction of HCQS with the nucleic acid.15 Additionally, no discernible shift in the absorption spectra was observed, which suggests that the probable binding mode of HCQS with RNA is groove binding or electrostatic in nature (Fig. 1(C)). Most probably, upon binding with RNA, π orbitals of RNA nucleobases bind to π* orbitals of the drug molecule, which subsequently suppresses the transition probability of the HOMO to the LUMO in the quinoline ring, resulting in the hypochromic effect.28,29 The binding of HCQS to RNA was quantified using the Benesi–Hildebrand model and the binding constant was estimated to be 1.56 ± 0.29 × 103 M−1 (Fig. 1(D)). Additionally, we estimated the fraction of bound HCQS with RNA and free HCQS present in the system using two different quantitative approaches. It was estimated that when the initial concentration of HCQS was kept at 20 μM, then upon the addition of 500 μM RNA, approximately 44% of the drug binds to RNA, and the rest remained free in the buffer (please see ESI† for details, Fig. S1F).
The interaction of HCQS with RNA was further explained by fluorescence spectroscopy. Since RNA is poorly fluorescent, the fluorescence intensity of HCQS was monitored instead. As mentioned earlier, HCQS displays an emission peak centered around 388 nm. The fluorescence intensity of HCQS dramatically decreased without any discernible shift in the emission maximum as a function of increasing concentration of RNA, when excited at 330 nm (Fig. 1(E)). The extent of quenching affirms the complex formation between the nucleic acid and the drug. Additionally, a strong interaction between the drug molecules and a particular nucleobase(s) present in the nucleic acid may be responsible for quenching the fluorescence intensity of the drug.19 The non-variant nature of the emission maxima indicates that the probe (HCQS) experiences the same microenvironment in the presence of the added RNA. Such an observation substantiates the possibility of groove binding of HCQS to RNA.
If the binding involved an intercalative mode, a blue shift was expected as observed in the case of the binding of another drug, CRYP, to RNA.22 The binding constant for the HCQS–RNA complexation was further estimated using the Benesi–Hildebrand equation and it was estimated to be 1.61 ± 0.17 × 103 M−1 (Fig. 1(F)). The binding constants obtained from steady-state absorption and fluorescence approaches were almost of the same magnitude suggesting moderately weak binding affinity of HCQS with RNA. A schematic representation of the binding interaction of HCQS with RNA is depicted in Scheme 1. We also performed similar steady-state spectroscopic investigations to elucidate the binding affinity of HCQS towards calf thymus DNA (ct-DNA) and one duplex RNA, namely (polyadenylic acid–polyuridylic acid sodium salt {Poly(A)·Poly(U)}) (Fig. S2A–D and S3A–D, ESI†). A comparative plot of binding constants obtained from absorption and fluorescence spectroscopic techniques is summarized in Fig. S4 (ESI†). From these studies, it can be interpreted that the binding constant for HCQS with ct-DNA and duplex RNA was slightly higher in magnitude than what we observed for the torula yeast RNA and these results are in good agreement with a previous literature report.19
 | ||
| Scheme 1 Schematic representation of binding interaction of HCQS with RNA and its sequestration by SCX4. | ||
Moreover, we also performed time-resolved fluorescence studies to have a better understanding of the microenvironment of the probe in the excited state. We have recorded the fluorescence lifetime of HCQS with the increasing concentration of RNA (please refer to Fig. S5, ESI†), and the corresponding decay parameters are listed in Table S2 (ESI†). HCQS has an average lifetime of 1.80 ns in 20 mM PB. However, upon the gradual addition of RNA, the average lifetime of HCQS remained unchanged. This suggested that the microenvironment of the drug remains intact subsequent to the interaction with RNA. HCQS forms a complex with RNA, and it binds at the exterior surface of RNA, complying with the groove binding mechanism. The variation in the average lifetime of probes with the addition of DNA/RNA indicates the intercalation mode of binding.22,30 These observations further substantiate the possibility of groove binding of HCQS with RNA, as correlated by our earlier observations.
Understanding the mode of binding by circular dichroism (CD) spectroscopy
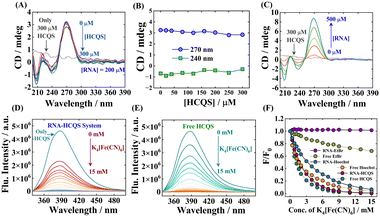
Circular dichroism (CD) spectroscopy is a convenient technique to obtain information about the secondary structure of biomacromolecules and biopolymers.31,32 The CD spectra of the native conformation of RNA display a positive peak at 270 nm and a negative peak at 240 nm.33 The characteristic positive and negative peaks originated due to the RNA's base-stacking and helical nature, respectively.34 These bands are exceedingly sensitive to the intervention of small molecules in the secondary structure of RNA.Typically, in the intercalative mode of binding, the drug enters inside the duplex of RNA and perturbs the base-stacking. Thus, a substantial change in CD spectra of native conformation of RNA is expected due to elongation and stabilization of strands. In contrast, only groove or electrostatic binding of small molecules to the RNA duplex will lead to insignificant changes in the CD spectra.34,35 As depicted in Fig. 2(A) and (B), the intensity of the positive peak of the CD spectra does not alter significantly, and no considerable shift was observed in the lower concentration regime of HCQS, which substantiates that the addition of HCQS does not perturb the base-stacking of RNA. These results suggest that the binding of HCQS to RNA appears to primarily involve the groove binding mode. Similarly, the change in the negative peak was also not very prominent, suggesting that the helical structure of RNA also remains intact upon the addition of HCQS (Fig. 2(A) and (B)). Additionally, we recorded induced circular dichroism (ICD) spectra of HCQS as a function of increasing concentration of RNA. Since no additional peak was generated at the absorbance region of HCQS (330 to 342 nm), we therefore rationally discard the possibility of the intercalative mode of binding (Fig. 2(C)). These results are in line with previous reports of groove binding of esculetin with DNA.35
Understanding the mode of binding by fluorescence quenching studies
Fluorescence quenching studies using extrinsic quenchers like potassium iodide (KI), potassium ferrocyanide (K4[Fe(CN)6]), etc. are some of the most reliable methods to distinguish the mode of binding of small molecules with nucleic acids. Highly negatively charged quencher molecules are expected to be repelled from the negatively charged polyanionic phosphate backbone of DNA/RNA. In the case of binding proceeding through the intercalation mode, the binder molecule inserts inside the strands of nucleic acid duplexes and becomes shielded from being quenched by an anionic quencher. However, the binder molecules when present at the exterior of nucleic acids get readily quenched by the anionic quencher through the dynamic/collisional quenching mechanism.36–38 For our case, we have chosen potassium ferrocyanide (K4[Fe(CN)6]) as an external quenching agent. Fig. 2(D) and (E) respectively present the quenching of the drug when bound to RNA and when free induced by the quencher. Additionally, we have also performed similar quenching experiments with ethidium bromide (EtBr, a well-established intercalator dye, Fig. S6A–C, ESI†) and Hoechst 33258 (Hoechst, a well-established groove binder dye, Fig. S6D–F, ESI†).39 A comparative result of all the quenching studies is provided in Fig. 2(F). Free EtBr exhibits a small quenching in the presence of K4[Fe(CN)6], but when it intercalates within the RNA strands, no further quenching was observed due to the non-accessibility of quencher ions. In contrast, free Hoechst and RNA–Hoechst complexes exhibit relatively high fluorescence quenching as compared to free EtBr and RNA–EtBr complexes. As shown in Fig. 2(F), the fluorescence quenching profiles of free HCQS and the RNA–HCQS complex are similar to those of free Hoechst and RNA–Hoechst complexes, which suggests that HCQS binds to RNA in a similar manner to Hoechst. This study therefore establishes that the binding mode of HCQS with RNA follows the groove binding/electrostatic mechanism, ruling out the possibility of intercalative mode of binding. The rationale behind this mode of binding may be attributed to the electrostatic forces of attraction involved between the negatively charged polyphosphate backbone of RNA and the divalent positively charged hydroxychloroquine moiety. Apart from this, π–π stacking between RNA nucleobases and the aromatic quinoline moiety of HCQS may also govern the said process.Salt-dependent fluorescence spectral studies
At physiological pH (∼7.4), since the HCQ moiety possesses a positive charge and the polyphosphate backbone of RNA has a negative charge, the effect of ionic strength has been employed to further decipher the influence of electrostatic interaction between RNA and HCQS.28,33,35,40 To elucidate the impact of salt content on the drug–RNA interaction, we have chosen NaCl as a strong electrolyte. The relative changes in the fluorescence intensity of HCQS in the absence and presence of RNA with varying concentrations of NaCl have been displayed in Fig. S7A–C (ESI†).In the absence of RNA, the fluorescence intensity of free HCQS was markedly quenched by NaCl. The fluorescence quenching of quinoline-like molecules in the presence of halides occurs due to the charge transfer.41 Interestingly, we can see that, in the presence of RNA, the fluorescence intensity of HCQS quenches to a minimal extent as compared to the free form, with the variation of NaCl (Fig. S7C, ESI†). This observation may be attributed to the fact that when NaCl is present in the system having RNA–HCQS, the electrostatic screening of the polyanionic phosphate backbone of RNA is operational owing to which those electrostatic interactions get shielded/attenuated in the presence of Na+ ions. In the case of the intercalative mode of binding, the drug molecules reside in the interior strands of the nucleic acid, but in the case of groove binding, the bound drug is much more exposed to the bulk. Thus, small molecules bound at the groove position can be easily released from the exterior of RNA in the presence of NaCl, or these molecules will not be able to efficiently approach the exterior of RNA, and hence, the overall binding of the drug will be reduced or almost nullified (Fig. S7D, ESI†). Here, a substantial weakening of RNA–HCQS binding strength with an increased ionic strength of the media highlights the involvement of electrostatic interactions. These observations are consistent with the phenomenon of groove binding, categorically ruling out the possibility of intercalation.35 Additionally, similar observations were further fortified by ITC experiments (discussed later).
Nucleobase-specific interactions of HCQS
The results obtained so far revealed that HCQS binds to RNA involving the groove binding mechanism. To further substantiate this, we explored the interactions of HCQS with the nucleobases comprising the RNA, namely, adenosine-5′-monophosphate (AMP), guanosine-5′-monophosphate (GMP), cytidine-5′-monophosphate (CMP) and uridine-5′-monophosphate (UMP). We have independently titrated all four nucleobases with HCQS and recorded their absorbance and fluorescence spectra upon excitation at 330 nm. Although the absorption of HCQS did not alter much by the addition of the nucleobases (Fig. S8A–D, ESI†), significant modulations in the fluorescence signatures were obtained. Fig. 3(A), (B) and Fig. S9A, B (ESI†) display the fluorescence spectra of HCQS with the addition of different monophosphate nucleobases. Fig. 3(C) displays a comparative change in the relative fluorescence intensity (F/F0) for all four nucleobases. A substantial quenching in the fluorescence intensity was observed for all four nucleobases, which followed the order of quenching: UMP > CMP > AMP > GMP. Purines and pyrimidine nucleobases are known to quench the fluorescence intensity of organic molecules through the involvement of stacking interactions and electron transfer processes.42–44 These results suggest that HCQS strongly binds at UMP- and CMP-specific regions of the RNA duplex. We have also estimated the binding constants from quenching results (please refer to the ESI† for details), which were found to be 2.1 ± 0.24 × 103 M−1 for UMP, and 1.55 ± 0.32 × 103 M−1 for CMP (Fig. S9C and D, ESI†). We would like to state that these results were in excellent agreement with our ITC experimental results.Molecular docking studies of interaction of HCQS with RNA
Molecular docking studies help in predicting the plausible binding site(s) of a drug to protein or DNA/RNA based on computational approaches. Abbas and co-workers reported the binding interactions of CLQ and HCQ with SARS-CoV-2 spike (S) protein by molecular docking studies.45 The S protein of SARS-CoV-2 acts as a key to bind with the angiotensin-converting enzyme 2 (ACE2) receptor of host cells, thereby facilitating the entry of the virus inside the cellular environments.46 From molecular docking results, they have proposed that the CLQ and HCQ molecules attack at the S-protein-ACE2 binding site, which SARS-CoV-2 uses to enter the host cells and display inhibitory action.45 We have also explored the binding locale of HCQ with the torula yeast RNA by molecular docking studies. The secondary structure of RNA has a cloverleaf-type structure and the tertiary structure has an L-shaped geometry. The 6TNA PDB entry was used for molecular docking studies.47,48 As presented in Fig. 3(D), molecular docking results suggested that the HCQS molecule binds in between the D-loop and acceptor stem at the groove position of RNA. The nucleobases surrounding the HCQS (with a distance of 5 Å) were found to be uridine-5′-monophosphate, cytidine-5′-monophosphate, adenosine-5′-monophosphate, 2-methylguanosine-5′-monophosphate and 5-methylcytidine-5′ monophosphate. This suggests that HCQS binds at the U-C-rich region of the RNA, which is also corroborated by our specific fluorescence quenching experiments using the nucleobases. Thus, using an independent theoretical approach, we could attest to our experimental observations that the binding of HCQS happens via the groove binding mechanism.Elucidation of thermodynamics of binding interactions
In this section, the associated thermodynamics corresponding to the HCQS–RNA interactions have been extensively explored using the isothermal titration calorimetric (ITC) technique. The ITC heat burst spikes for the interaction of HCQS with RNA over a varying range of temperatures from 293 K to 313 K have been displayed in Fig. 4(A) and S10A–D (ESI†) and the corresponding thermodynamic parameters are listed in Table S3 (ESI†). At a particular temperature say, 298 K, it can be interpreted that the binding of HCQS with RNA is an exothermic process (ΔH < 0). Along with this, the interaction seemed to be entropically governed and characterized by positive entropy changes (TΔS > 0). The overall negative Gibbs free energy change (ΔG < 0) authenticates the thermodynamic spontaneity of the said interaction. The current observation is in close agreement with the hypothesis that intercalative binding is an enthalpically driven process, while groove-binding is primarily entropically driven.39,49Hence, the thermodynamic parameters obtained from our studies also correlate to the fact that the binding of HCQS with RNA is facilitated by the groove binding mode. A strong positive entropic contribution is linked with the phenomenon of the “release of bound water molecules” after the binding of the drug at the exterior of the RNA. The obtained binding constant from ITC at 298 K was found to be 1.529 ± 0.07 × 103 M−1, which attests to our steady-state experiments. To have a better understanding of the issue related to the specificity/selectivity, we have done a reverse titration control experiment using ITC (Fig. S11, ESI†), wherein we titrated 100 μM RNA (in the cell) with 1000 μM HCQS (in the syringe). The binding constant and other thermodynamic parameters obtained by this reverse titration approach are in excellent agreement with what was obtained earlier (Table S4, ESI†). A saturation takes place at a lower molar ratio, with a Kd value of 324 μM. The fractional saturation was estimated to be 0.76 (please refer to Fig. S11 and the ESI† section for more details), which indicates a good specificity, albeit weak binding as found from the magnitude of the binding constant (obtained from steady-state spectroscopic and ITC data). Additionally, such a weak binding obtained in our case is in good agreement with the literature, wherein groove binding has been proposed to be operational when characterized by such a magnitude of the binding constant.19 Furthermore, in the presence of 100 mM NaCl, RNA–HCQS titration did not yield any significant binding, which indicated the intervention of electrostatics involved in the said interaction (Fig. S12, ESI†). It is also pertinent to mention here that with the increase in temperature, the binding constant marginally decreases (Table S3, ESI†), indicating the instability of the complex at elevated temperatures. With the rise in temperature, the enthalpy changes (ΔH) increased toward a more negative magnitude, whereas a reverse trend was obtained for the entropy changes (TΔS). As seen from Fig. 4(B) and Table S3 (ESI†), an overall exothermic binding was observed at all temperatures. Fig. 5 shows the alterations in ΔH values against temperature for the said interaction. According to the standard thermodynamic relationship,  , a negative slope of the ΔH versus T plot provides the value of heat capacity change at constant pressure, which was estimated to be, ΔCp = −0.21 ± 0.02 kJ mol−1 K−1.22 The magnitude of the ΔCp value provides information about the energetics of the associated interaction. A typical negative value of ΔCp describes the decisive role of the hydrophobic effect and release of ordered water molecules that is coupled with the transfer of the hydrophobic moiety on RNA resulting in the drug–RNA complex formation.33 We also studied the interactions of all four nucleobases (UMP, CMP, GMP, and AMP) with HCQS at 298 K using ITC (Fig. S13A–D, ESI†), and the relevant thermodynamic parameters are tabulated in Table S3 (ESI†). A negative change in enthalpy (ΔH) indicates that the interaction of UMP and CMP with HCQS is exothermic in nature and negative Gibbs free energy changes (ΔG) attest to the feasibility of the process. Additionally, positive entropic contribution (TΔS) ascertains the involvement of stacking interactions between pyrimidine bases and the hydroxychloroquine moiety. However, the binding of GMP and AMP with HCQS was not found to be conspicuous by ITC experiments. This observation further reveals that HCQS binds at the U–C-rich region of the RNA. Thus, all our experimental techniques based on spectroscopic and calorimetric approaches and our molecular docking results not only complement each other but also unequivocally substantiate that HCQS binds to the RNA involving a groove binding mechanism.
, a negative slope of the ΔH versus T plot provides the value of heat capacity change at constant pressure, which was estimated to be, ΔCp = −0.21 ± 0.02 kJ mol−1 K−1.22 The magnitude of the ΔCp value provides information about the energetics of the associated interaction. A typical negative value of ΔCp describes the decisive role of the hydrophobic effect and release of ordered water molecules that is coupled with the transfer of the hydrophobic moiety on RNA resulting in the drug–RNA complex formation.33 We also studied the interactions of all four nucleobases (UMP, CMP, GMP, and AMP) with HCQS at 298 K using ITC (Fig. S13A–D, ESI†), and the relevant thermodynamic parameters are tabulated in Table S3 (ESI†). A negative change in enthalpy (ΔH) indicates that the interaction of UMP and CMP with HCQS is exothermic in nature and negative Gibbs free energy changes (ΔG) attest to the feasibility of the process. Additionally, positive entropic contribution (TΔS) ascertains the involvement of stacking interactions between pyrimidine bases and the hydroxychloroquine moiety. However, the binding of GMP and AMP with HCQS was not found to be conspicuous by ITC experiments. This observation further reveals that HCQS binds at the U–C-rich region of the RNA. Thus, all our experimental techniques based on spectroscopic and calorimetric approaches and our molecular docking results not only complement each other but also unequivocally substantiate that HCQS binds to the RNA involving a groove binding mechanism.
Sequestration of HCQS from RNA binding sites
In the earlier sections of the manuscript, we have successfully discussed the binding interaction of HCQS with the torula yeast RNA. Keeping in mind the perspective of drug overdose and the cytotoxic effects of HCQS, we have also attempted the implementation of the concept of host–guest chemistry to achieve the sequestration of bound HCQS from RNA binding sites. For this attempt, we have judiciously chosen a water-soluble molecular basket, 4-sulfocalix[4]arene (SCX4) (Fig. 6(A)), and the dissociation process was quantitatively analyzed by fluorescence spectroscopy and ITC studies.50 The family of p-sulfocalix[n]arenes are some of the special classes of host molecules that are known to be more polar and water soluble compared to native calix[n]arenes that have profound capability to bind to the guest in their cavities in aqueous medium. The governing forces for the formation of inclusion complexes of p-sulfocalix[n]arenes with guest molecules are hydrophobic and π-stacking interactions that are much more prominent in aqueous media than organic media. The presence of sulfonate groups on the upper rim of these molecular receptors facilitates the incoming of the guest molecule to its π-electron rich cavity, which renders these host molecules to be highly selective toward organic cations (Fig. 6(A)). As compared to other conventional water-soluble macrocycles, like cyclodextrins and cucurbiturils, p-sulfocalix[n]arenes have a different cavity structure and rigid framework.51 These macrocycles are biocompatible and completely non-toxic, have a variety of biological applications, such as drug solubilization and targeted drug delivery, and exhibit antiviral and antibacterial activities.52Recently, Li et al. reported that a complex of hydroxychloroquine (HCQ) with sulfonated azocalix[4]arene (HCQ@SAC4A) can be used for treating rheumatoid arthritis in a combined fashion via a drug-in-drug approach.53 Using fluorescence spectroscopy, they have reported a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest complex formulation between sulfonated azocalix[4]arene and HCQ, with a binding constant on the order of 107 M−1.53 We added SCX4 to the free HCQS and RNA–HCQS complex solution and the modulations in the fluorescence properties are displayed in Fig. 6(B) and (C). In the case of free HCQS in buffer, fluorescence intensity got quenched upon successive addition of SCX4, which signifies the formation of a host–guest complex involving SCX4 as a host and HCQS as a guest molecule. However, since no shift in the emission peak was observed, the fluorescence intensity recorded at 388 nm was utilized to obtain the binding parameters. It must be mentioned here that only SCX4 does not exhibit any fluorescence intensity upon excitation at 330 nm (Fig. 6(B) and (C)). The data analyzed from fluorescence titrations were fitted to a 1
1 host–guest complex formulation between sulfonated azocalix[4]arene and HCQ, with a binding constant on the order of 107 M−1.53 We added SCX4 to the free HCQS and RNA–HCQS complex solution and the modulations in the fluorescence properties are displayed in Fig. 6(B) and (C). In the case of free HCQS in buffer, fluorescence intensity got quenched upon successive addition of SCX4, which signifies the formation of a host–guest complex involving SCX4 as a host and HCQS as a guest molecule. However, since no shift in the emission peak was observed, the fluorescence intensity recorded at 388 nm was utilized to obtain the binding parameters. It must be mentioned here that only SCX4 does not exhibit any fluorescence intensity upon excitation at 330 nm (Fig. 6(B) and (C)). The data analyzed from fluorescence titrations were fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding non-linear curve fitting model and the estimated binding constant was found to be 8.4 ± 0.92 × 105 M−1 (Fig. 6(B), (D) and Table S5, ESI†)54 Similarly, when we added SCX4 in RNA–HCQS solution, further quenching of the fluorescence intensity was observed without any shift. This indicates the disruption of the RNA–HCQS complex and the subsequent formation of the HCQS–SCX4 complex. Similarly, the binding constant for the HCQS–SCX4 complex in the presence of RNA was found to be 6.9 ± 1.5 × 105 M−1 (Fig. 6(C), (D) and Table S5, ESI†). Additionally, time-dependent emission spectra of free HCQS, RNA-bound HCQS, and RNA-bound HCQS in the presence of SCX4 were also monitored. We noted that the fluorescence intensities do not change significantly with time (Fig. S14A–C, ESI†). It is worth mentioning here that the association constant for RNA–HCQS binding was on the order of 103 M−1 (discussed in earlier sections), and the association constant for HCQS–SCX4 formulation was on order of 105 M−1. Considerably, a higher binding constant for the latter verifies the disruption of the RNA–HCQS complex, and the released HCQS concertedly gets encapsulated in the SCX4 cavity. The driving forces for the complexation of HCQS with SCX4 are effective, most probably because of the (i) electrostatic interactions between the dicationic hydroxychloroquine ion and the sulfonate rim of SCX4 and (ii) π-stacking between the aromatic quinoline moiety and the hydrophobic π-electron rich cavity of SCX4. The molecular docking results also clearly show the encapsulation of HCQS into the SCX4 cavity (Fig. 6(E)). Additionally, the distance monitored between the centroid of the quinoline ring of HCQS and four phenyl rings of SCX4 was found to be in the range of 4 to 5.5 Å, which satisfies the hypothesis of slanted π–π interactions among the aromatic guest moiety and phenyl rings of the host.55 A schematic representation of the binding of HCQS with RNA and its subsequent sequestration is depicted in Scheme 1.
1 binding non-linear curve fitting model and the estimated binding constant was found to be 8.4 ± 0.92 × 105 M−1 (Fig. 6(B), (D) and Table S5, ESI†)54 Similarly, when we added SCX4 in RNA–HCQS solution, further quenching of the fluorescence intensity was observed without any shift. This indicates the disruption of the RNA–HCQS complex and the subsequent formation of the HCQS–SCX4 complex. Similarly, the binding constant for the HCQS–SCX4 complex in the presence of RNA was found to be 6.9 ± 1.5 × 105 M−1 (Fig. 6(C), (D) and Table S5, ESI†). Additionally, time-dependent emission spectra of free HCQS, RNA-bound HCQS, and RNA-bound HCQS in the presence of SCX4 were also monitored. We noted that the fluorescence intensities do not change significantly with time (Fig. S14A–C, ESI†). It is worth mentioning here that the association constant for RNA–HCQS binding was on the order of 103 M−1 (discussed in earlier sections), and the association constant for HCQS–SCX4 formulation was on order of 105 M−1. Considerably, a higher binding constant for the latter verifies the disruption of the RNA–HCQS complex, and the released HCQS concertedly gets encapsulated in the SCX4 cavity. The driving forces for the complexation of HCQS with SCX4 are effective, most probably because of the (i) electrostatic interactions between the dicationic hydroxychloroquine ion and the sulfonate rim of SCX4 and (ii) π-stacking between the aromatic quinoline moiety and the hydrophobic π-electron rich cavity of SCX4. The molecular docking results also clearly show the encapsulation of HCQS into the SCX4 cavity (Fig. 6(E)). Additionally, the distance monitored between the centroid of the quinoline ring of HCQS and four phenyl rings of SCX4 was found to be in the range of 4 to 5.5 Å, which satisfies the hypothesis of slanted π–π interactions among the aromatic guest moiety and phenyl rings of the host.55 A schematic representation of the binding of HCQS with RNA and its subsequent sequestration is depicted in Scheme 1.
Further to substantiate the fluorescence spectroscopy results, we have performed ITC experiments for host–guest binding. Fig. 7(A) and (B) display the binding profiles obtained from ITC for the complexation of SCX4 with RNA-bound HCQS and free HCQS in an aqueous/buffer medium. Fig. 7(A) displays the binding profile of SCX4 with the RNA–HCQS complex and the binding constant was found to be 0.94 ± 0.05 × 105 M−1. Similarly, Fig. 7(B) presents the binding profile of SCX4 with free HCQS. The sigmoidal nature of the binding curve shows the strong binding potentiality of SCX4 with HCQS with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry and a binding constant of 3.26 ± 0.17 × 105 M−1 (Table S5, ESI†). The magnitude of binding constants acquired from ITC was in excellent agreement with that obtained from fluorescence spectroscopy.
1 stoichiometry and a binding constant of 3.26 ± 0.17 × 105 M−1 (Table S5, ESI†). The magnitude of binding constants acquired from ITC was in excellent agreement with that obtained from fluorescence spectroscopy.
 | ||
| Fig. 7 ITC heat burst spikes (upper panel, 298 K) and thermodynamic enthalpograms (lower panel) for the addition of SCX4 to (A) the RNA–HCQS system and (B) the free HCQS system. | ||
The strong binding was characterized by a large enthalpy change (thereby enabling the process to be enthalpically controlled). Furthermore, the negative signs of ΔH and TΔS (Table S5, ESI†) indicate the involvement of π-stacking and electrostatic interactions. The negative sign of ΔH and positive ΔS for RNA-bound HCQS with SCX4 substantiate the disruption of the RNA–HCQS complex, resulting in the release of bound water molecules contributing to a positive value of ΔS. Thus, this further validates the concept and proposal of the disruption of the RNA–HCQS complex in the presence of the sequestrating agent, SCX4. An interplay of the various interactions involved modulates the overall energetics of the process. As a control experiment, to ascertain the effect of the sequestrating agent (SCX4) on the native structure of RNA, we have recorded the CD spectra of RNA in the presence of a maximum concentration of SCX4 (70 μM) (Fig. S15A, ESI†). We have found that the addition of SCX4 does not alter the secondary structure of RNA. A similar investigation was also performed by the ITC experiment of SCX4 and RNA (in the absence of HCQS), which also confirmed the non-interacting nature of both SCX4 and RNA (Fig. S15B, ESI†). These findings make SCX4 a promising candidate for drug sequestration and certify its biocompatible nature.
Conclusions
In this article, we have explored the nucleobase-specific binding of an antimalarial drug, hydroxychloroquine sulfate (HCQS), with torula yeast RNA. The associated interactions were monitored through the significant variations in steady-state spectroscopic properties. A decrease in the absorbance and quenching in fluorescence spectra of HCQS upon the addition of RNA without any discernible peak shift suggested the groove-binding propensity of HCQS with RNA. Fluorescence quenching using the ferrocyanide ion (as the external quencher) indicated the groove-binding nature of HCQS with RNA. Furthermore, the non-variant nature of circular dichroism spectra of RNA upon the addition of HCQS revealed the groove-binding mode. The absence of any induced peak in induced CD spectra of RNA–HCQS ruled out the possibility of intercalative mode of binding. Next, we explored the specific binding efficacy of HCQS at uridine and cytidine-rich regions of RNA, with the results being confirmed collectively by fluorescence spectroscopy, molecular docking, and ITC experiments. A detailed study of the thermodynamics of the associated interaction was also carried out using the ITC experiments at variable temperatures. Entropically driven binding of HCQS with RNA showed the “release of water molecules” from the exterior of RNA upon binding with HCQS. This further supported the hypothesis of groove-binding, as obtained from other experimental approaches. The undertaken work also illustrates the applications of host–guest chemistry for the sequestration of RNA-bound HCQS using a water-soluble, biocompatible 4-sulfocalix[4]arene scaffold. This study may be beneficial for the basic understanding of drug–RNA interactions and for developing safe RNA-based therapeutics.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
R. Y. thanks CSIR, Govt. of India for fellowship. S. D. thanks DST INSPIRE, Govt. of India, for fellowship. S. M. thanks SERB-ANRF, Govt. of India (project no.: SERB-CRG/2023/000473) and the Ministry of Education, Govt. of India (project no.: MoE-STARS/STARS-2/2023-0109) for financial assistance. We thank Central Instrumentation Facility (CIF) IISER Bhopal for the ITC instrument.References
- G. Li, R. Hilgenfeld, R. Whitley and E. De Clercq, Nat. Rev. Drug Discovery, 2023, 22, 449–475 CrossRef CAS PubMed.
- P. C. Robinson, D. F. L. Liew, H. L. Tanner, J. R. Grainger, R. A. Dwek, R. B. Reisler, L. Steinman, M. Feldmann, L.-P. Ho, T. Hussell, P. Moss, D. Richards and N. Zitzmann, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2119893119 CrossRef PubMed.
- C. Gil, T. Ginex, I. Maestro, V. Nozal, L. Barrado-Gil, M. Á. Cuesta-Geijo, J. Urquiza, D. Ramírez, C. Alonso, N. E. Campillo and A. Martinez, J. Med. Chem., 2020, 63, 12359–12386 CrossRef CAS PubMed.
- J. L. Siqueira-Neto, K. J. Wicht, K. Chibale, J. N. Burrows, D. A. Fidock and E. A. Winzeler, Nat. Rev. Drug Discovery, 2023, 22, 807–826 CrossRef CAS PubMed.
- S. Yalduz, Ö. Eyilcim, N. Kılıç, S. Gülyüz, O. Denizoğlu, Y. Zorlu and E. Ertürk, Org. Process Res. Dev., 2024, 28, 137–151 CrossRef CAS.
- I. Faraone, F. Labanca, M. Ponticelli, N. De Tommasi and L. Milella, Molecules, 2020, 25, 5318–5362 CrossRef CAS PubMed.
- J. Liu, R. Cao, M. Xu, X. Wang, H. Zhang, H. Hu, Y. Li, Z. Hu, W. Zhong and M. Wang, Cell Discovery, 2020, 6, 16–19 CAS.
- C. Xu, L. Zhu, T. Chan, X. Lu, W. Shen, M. C. Madigan, M. C. Gillies and F. Zhou, J. Pharm. Sci., 2016, 105, 884–890 CAS.
- K. D. Rainsford, A. L. Parke, M. Clifford-Rashotte and W. F. Kean, Inflammopharmacology, 2015, 23, 231–269 CAS.
- M. Wang, R. Cao, L. Zhang, X. Yang, J. Liu, M. Xu, Z. Shi, Z. Hu, W. Zhong and G. Xiao, Cell Res., 2020, 30, 269–271 CAS.
- J. Fantini, C. Di Scala, H. Chahinian and N. Yahi, Int. J. Antimicrob. Agents, 2020, 55, 105960 CAS.
- X. Li, Y. Wang, P. Agostinis, A. Rabson, G. Melino, E. Carafoli, Y. Shi and E. Sun, Cell Death Dis., 2020, 11, 512 CAS.
- P. Maisonnasse, J. Guedj, V. Contreras, S. Behillil, C. Solas, R. Marlin, T. Naninck, A. Pizzorno, J. Lemaitre, A. Gonçalves, N. Kahlaoui, O. Terrier, R. H. T. Fang, V. Enouf, N. Dereuddre-Bosquet, A. Brisebarre, F. Touret, C. Chapon, B. Hoen, B. Lina, M. R. Calatrava, S. van der Werf, X. de Lamballerie and R. Le Grand, Nature, 2020, 585, 584–587 CAS.
- J. L. Irvin, E. M. Irvin and F. S. Parker, Science, 1949, 110, 426–428 CAS.
- S. N. Cohen and K. L. Yielding, J. Biol. Chem., 1965, 240, 3123–3131 Search PubMed.
- F. Kwakye-Berko and S. R. Meshnick, Mol. Biochem. Parasitol., 1989, 35, 51–55 CAS.
- A. Bisoi, S. Sarkar, T. Lai and P. C. Singh, J. Phys. Chem. B, 2023, 127, 3341–3351 CAS.
- R. F. Bazoni, T. A. Moura and M. S. Rocha, J. Phys. Chem. Lett., 2020, 11, 9528–9534 Search PubMed.
- S. Sarkar, S. Roy and P. C. Singh, J. Phys. Chem. B, 2021, 125, 6889–6896 CrossRef CAS PubMed.
- S. Sarkar, A. Bisoi and P. C. Singh, J. Phys. Chem. B, 2022, 126, 5241–5249 CrossRef CAS PubMed.
- K. D. Tulsiyan, S. Jena, M. González-Viegas, R. K. Kar and H. S. Biswal, ACS Cent. Sci., 2021, 7, 1688–1697 CrossRef CAS PubMed.
- A. Nandy, S. Shekhar, B. K. Paul and S. Mukherjee, Langmuir, 2021, 37, 11176–11187 CrossRef CAS PubMed.
- R. Yadav, B. K. Paul and S. Mukherjee, Colloids Interfaces, 2023, 7, 51 CrossRef CAS.
- K. Nord, J. Karlsen and H. W. Tosnnesen, Photochem. Photobiol., 1994, 60, 427–431 CrossRef CAS.
- G. Viola, A. Salvador, L. Cecconet, G. Basso, D. Vedaldi, F. Dall’Acqua, G. G. Aloisi, M. Amelia, A. Barbafina, L. Latterini and F. Elisei, Photochem. Photobiol., 2007, 83, 1415–1427 Search PubMed.
- E. Mamontov, Y. Cheng, L. L. Daemen, J. K. Keum, A. I. Kolesnikov, D. Pajerowski, A. Podlesnyak, A. J. Ramirez-Cuesta, M. R. Ryder and M. B. Stone, ACS Omega, 2020, 5, 21231–21240 CrossRef CAS PubMed.
- M. Rahban, A. Divsalar, A. A. Saboury and A. Golestani, J. Phys. Chem. C, 2010, 114, 5798–5803 CrossRef CAS.
- S. Luikham, A. Mavani, D. Sinha and J. Bhattacharyya, J. Phys. Chem. B, 2023, 127, 4966–4978 Search PubMed.
- C. C. Ju, A. G. Zhang, C. L. Yuan, X. L. Zhao and K. Z. Wang, J. Inorg. Biochem., 2011, 105, 435–443 Search PubMed.
- S. Kumar, C. Ghosh, P. Roy and M. S. Nair, J. Mol. Struct., 2025, 1323, 140765 CrossRef CAS.
- R. Yadav, A. Nandy, A. Bisoi and S. Mukherjee, Langmuir, 2024, 40, 6172–6186 CrossRef CAS PubMed.
- A. Nandy and S. Mukherjee, J. Phys. Chem. Lett., 2022, 13, 6701–6710 CrossRef CAS PubMed.
- B. K. Paul, N. Ghosh and S. Mukherjee, J. Colloid Interface Sci., 2019, 538, 587–596 CrossRef CAS PubMed.
- M. Z. Akhter and M. R. Rajeswari, J. Photochem. Photobiol., B, 2014, 141, 36–46 CrossRef CAS PubMed.
- T. Sarwar, M. A. Husain, S. U. Rehman, H. M. Ishqi and M. Tabish, Mol. BioSyst., 2015, 11, 522–531 RSC.
- N. Kundu, A. Roy, D. Banik and N. Sarkar, J. Phys. Chem. B, 2016, 120, 1106–1120 CrossRef CAS PubMed.
- F. Arjmand, G. C. Sharma, F. Sayeed, M. Muddassir and S. Tabassum, J. Photochem. Photobiol., B, 2011, 105, 167–174 CrossRef CAS PubMed.
- X. Yin, S. Wei, C. Zhai, B. Wang, H. Zhang, C. Wang, X. Song, G. Sun and C. Jiang, Food Chem., 2024, 432, 137207 Search PubMed.
- A. Das, G. S. Kumar and S. Dutta, J. Mol. Recognit., 2019, 32, e2786 Search PubMed.
- B. K. Paul, N. Ghosh and S. Mukherjee, J. Colloid Interface Sci., 2016, 470, 211–220 CrossRef CAS PubMed.
- S. Jayaraman and A. S. Verkman, Biophys. Chem., 2000, 85, 49–57 CAS.
- B. Y. Michel, D. Dziuba, R. Benhida, A. P. Demchenko and A. Burger, Front. Chem., 2020, 8, 112 CAS.
- S. Doose, H. Neuweiler and M. Sauer, ChemPhysChem, 2009, 10, 1389–1398 Search PubMed.
- C. A. M. Seidel, A. Schulz and M. H. M. Sauer, J. Phys. Chem., 1996, 100, 5541–5553 Search PubMed.
- M. Amin and G. Abbas, J. Biomol. Struct. Dyn., 2020, 1, 1–11 Search PubMed.
- G. Negi, A. Sharma, M. Chaudhary, D. Gupta, K. H. Harshan and N. Parveen, ACS Infect. Dis., 2023, 9, 1346–1361 Search PubMed.
- T. Suzuki, Nat. Rev. Mol. Cell Biol., 2021, 22, 375–392 Search PubMed.
- S. E. Butcher and A. M. Pyle, Acc. Chem. Res., 2011, 44, 1302–1311 CAS.
- J. B. Chaires, Arch. Biochem. Biophys., 2006, 453, 26–31 Search PubMed.
- A. S. Pires, K. Droguett Muñoz, V. de Freitas, N. Basílio and L. Cruz, J. Agric. Food Chem., 2024, 72, 3719–3729 Search PubMed.
- D. S. Guo and Y. Liu, Acc. Chem. Res., 2014, 47, 1925–1934 CrossRef CAS PubMed.
- F. Perret, A. N. Lazar and A. W. Coleman, Chem. Commun., 2006, 2425–2438 Search PubMed.
- S. Li, R. Ma, X. Y. Hu, H. B. Li, W. C. Geng, X. Kong, C. Zhang and D. S. Guo, Adv. Mater., 2022, 34, e2203765 CrossRef PubMed.
- Y. Xu, S. Gsänger, M. B. Minameyer, I. Imaz, D. Maspoch, O. Shyshov, F. Schwer, X. Ribas, T. Drewello, B. Meyer and M. Vondelius, J. Am. Chem. Soc., 2019, 141, 18500–18507 CrossRef CAS PubMed.
- A. Husain and C. L. Oliver, CrystEngComm, 2014, 16, 3749–3757 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cp04687k |
| This journal is © the Owner Societies 2025 |