Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The role of spatial arrangement of aromatic rings on the binding of N,N′-diheteroaryl guanidine ligands to the G2C4/G2C4 motif DNA†
Eitaro
Murakami
 a,
Tomonori
Shibata
a,
Tomonori
Shibata
 *a,
Megumi
Tomemori
b,
Gota
Kawai
*a,
Megumi
Tomemori
b,
Gota
Kawai
 b and
Kazuhiko
Nakatani
b and
Kazuhiko
Nakatani
 *a
*a
aDepartment of Regulatory Bioorganic Chemistry, SANKEN (the Institute of Science and Industrial Research), Osaka University, 8-1, Mihogaoka, Ibaraki, Osaka, 567-0047, Japan. E-mail: nakatani@sanken.osaka-u.ac.jp
bDepartment of Life Science, Faculty of Advanced Engineering, Chiba Institute of Technology, 2-17-1, Tsudanuma, Narashino, Chiba, 275-0016, Japan
First published on 13th January 2025
Abstract
Non-canonical DNA structures formed by aberrantly expanded repeat DNA are implicated in promoting repeat instability and the onset of repeat expansion diseases. Small molecules that target these disease-causing repeat DNAs hold promise as therapeutic agents for such diseases. Specifically, 1,3-di(quinolin-2-yl)guanidine (DQG) has been identified to bind to the disease-causing GGCCCC (G2C4) repeat DNA associated with amyotrophic lateral sclerosis and frontotemporal dementia (ALS/FTD). In this study, we investigate the structure-binding relationships between DQG analogs and double-stranded DNA (dsDNA) containing a G2C4/G2C4 unit. Our findings, derived from UV melting temperature, circular dichroism spectra, and surface plasmon resonance (SPR) analyses of DQG analogs, highlight the crucial role of the spatial arrangements of aromatic rings in binding to the G2C4/G2C4 unit. Among the tested DQG analogs, N,N′-di(quinazolin-2-yl)guanidine (DQzG) stands out for its ability to form seven planar conformers. These conformers enable ADD–DAA hydrogen bonding with cytosine and multiple spatial arrangements of aromatic rings, including those resembling DQG. Our binding analyses revealed that DQzG exhibits the highest affinity binding for the G2C4/G2C4 unit. NMR analysis of the DQzG-bound G2C4/G2C4-dsDNA further suggested that DQzG binds to the G2C4/G2C4 unit via hydrogen bonding. Moreover, SPR analysis demonstrated that DQzG binds more strongly to G2C4 repeat DNA compared to DQG. These results position DQzG as a promising lead compound for targeting the G2C4 repeat, offering potential therapeutic avenues for the treatment of ALS/FTD and other repeat expansion diseases.
Introduction
Aberrant expansion of repeat DNA in the human genome is associated with neurodegeneration and repeat expansion diseases.1,2 A prominent cause of amyotrophic lateral sclerosis and frontotemporal dementia (ALS/FTD) is the expansion of the GGGGCC (G4C2) repeat DNA sequence located within the first intron of the chromosome 9 open reading frame 72 (C9orf72) gene. This expansion is observed in over 40% of familial and 8% of sporadic ALS/FTD cases.3,4 In ALS/FTD patients, the number of G4C2 repeats can surpass 250, while unaffected individuals typically have fewer than 20 repeats.5,6 The pathogenesis involves RNA gain-of-function due to the formation of abnormal RNA aggregates, known as RNA foci, leading to the sequestration of RNA-binding proteins. Additionally, repeat-associated non-AUG translation (RAN translation) produces abnormal peptide repeat protein aggregates.7–9 The presence of RNA foci leads to loss of function of RNA-binding proteins, causing impaired splicing and nuclear-cytoplasmic transport. Another reported pathological mechanism is the repression of C9ORF72-coding mRNA expression in affected individuals, termed haploinsufficiency.10 Reduced levels of the C9ORF72 protein exacerbate the toxicity mediated by the repeat expansions, contributing to ALS/FTD.11 Despite these insights, the molecular mechanisms underlying the root cause of repeat diseases, specifically the aberrant expansion, remain to be fully elucidated.We have developed mismatch-binding ligands (MBLs) capable of recognizing mismatched base pairs in both DNAs and RNAs through complementary hydrogen bonding with nucleobases.12–15 Among these ligands, naphthyridine-azaquinolone (NA) was identified to selectively bind to the CAG/CAG motif, a key component of the hairpin structures formed by CAG repeats. NMR structural analysis revealed that NA forms hydrogen bonds with nucleobases and engages in stacking interactions with adjacent nucleobases. In preclinical models, NA induced the contraction of expanded CAG repeats in Huntington's disease mice and improved motor phenotypes in dentatorubral–pallidoluysian atrophy (DRPLA) mice.12,16,17 These findings suggest that small molecules capable of binding to disease-causing repeat DNA have therapeutic potential by contracting aberrantly expanded repeat sequences.
Previous studies have explored small molecules that target the hairpin and G quadruplex formed by the G4C2 repeat.18–21 Our study focused on the antisense repeat GGCCCC (G2C4), which is predicted to form hairpins and i-motif structures.22 Based on a design concept utilizing hydrogen bond complementarity and stacking with adjacent nucleobases, we identified 1,3-di(quinolin-2-yl)guanidine (DQG; Fig. 1a) as a promising core unit for binding to the G2C4 repeat.23DQG significantly increased the melting temperature (Tm) of the model duplex 5′-GCAT GGCCCC TACG-3′/3′-CGTA CCCCGG ATGC-5′ containing the G2C4/G2C4 unit. Given that the guanidyl group of DQG is protonated at neutral pH, an intramolecular hydrogen bond forms between the N1 nitrogen of quinoline and the guanidium N–H, resulting in several conformers with donor–donor (DD) hydrogen bonding groups. This pattern partially complements the hydrogen bonding surface of cytosine with donor–acceptor–acceptor (DAA) groups. The lowest energy conformation of DQG (DQG conf-1) binds to cytosine with two hydrogen bonds without steric repulsion (Fig. 1b). This hydrogen-bonding model suggests that by changing the substitution position of the guanidyl group on quinoline, the aromatic ring arrangement required for stacking interactions with adjacent base pairs can be adjusted. Substitution of quinoline with quinazoline may yield hydrogen-bonded surfaces with an acceptor–donor–donor (ADD) pattern. As the association constant of the ADD/DAA hydrogen-bonded complex is reported to be approximately 50-fold higher than that of the AA/DD hydrogen-bonded complex,24 designing a DQG analog with an ADD hydrogen bonding pattern may enhance its binding property. Here, we present the synthesis of DQG analogs and evaluate their binding properties to the G2C4/G2C4 unit in dsDNA. Our analyses encompassed UV melting, circular dichroism (CD) spectra, surface plasmon resonance (SPR) assays, and nuclear magnetic resonance (NMR) spectra measurements. Notably, among the tested DQG analogs, N,N′-di(quinazolin-2-yl)guanidine (DQzG) demonstrated the highest binding affinity. Conformational analysis of DQG analogs, along with spatial alignment of conformers, revealed a similar spatial orientation of aromatic rings for both DQG and DQzG upon hydrogen bonding to cytosine. This similarity underscores the critical role of stacking interactions, complementing hydrogen bonding, in the high-affinity binding of DQzG to the G2C4/G2C4 unit.
Results
Molecular design of DQG analogs
To develop DQG analogs that adopt different orientation of two heteroaromatic rings in planar conformation, we designed N,N′-di(isoquinolin-3-yl)guanidine (D3iQG), N,N′-di(isoquinolin-1-yl)guanidine (D1iQG), and N-(isoquinolin-3-yl)-N′-(quinolin-2-yl)guanidine (Q3iQG) as structural isomers of DQG (Fig. 1c). These DQG analogs containing isoquinoline can adopt planar conformations with intramolecular hydrogen bonding between N2 position of isoquinoline and guanidyl group in a manner similar with DQG. N,N′-di(naphthalen-2-yl)guanidine (DNpG) and N,N′-di(quinolin-2-yl)urea (DQU) were synthesized as control compounds. Furthermore, we designed DQzG, which consisted of two quinazoline rings linked by a guanidine. The ADD hydrogen bonding surface in planar conformations of DQzG can be complementary to DAA hydrogen boding surface of cytosine. The synthesis of these molecules is described in the Experimental section. Briefly, DQG analogs were synthesized in one or two steps by a Buchwald–Hartwig reaction of guanidine nitrate with the corresponding haloarene.25DQU was synthesized from triphosgene and 2-aminoquinoline.26 In these reactions, multiple byproducts, including monomers and trimers, were detected by mass spectrometry (MS), along with the presence of unreacted haloarenes, which contributed to the low yield. Further investigation into the synthetic method is required to improve the reaction efficiency.Conformational analysis of DQG analogs
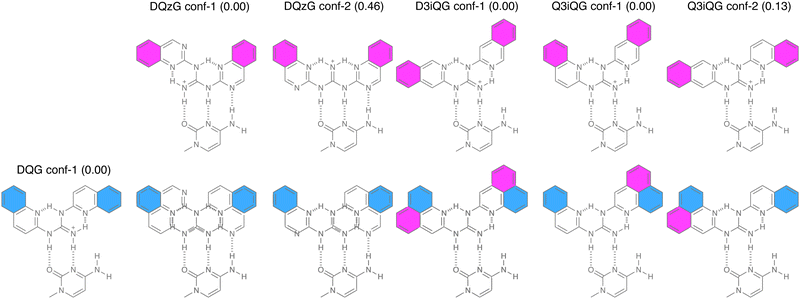
To gain insights into the formation of planar conformations in DQG and DQG analogs, conformational analysis was performed using a force field, OPLS4 in an aqueous environment. The conformations of DQG analogs within 5 kcal mol−1 of the most stable conformer were obtained from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 conformations generated by molecular mechanics (MM) calculations. Furthermore, quantum mechanical calculations were performed for all conformers obtained by MM calculation using DFT ωb97xd/6-31G+(d,p) in Gaussian 16 (Table 1 and Table S1, ESI†). The numbers of conformers within 3.5 kcal mol−1 of the lowest energy conformer obtained for DQG, D3iQG, D1iQG, Q3iQG, and DQzG were 2, 2, 2, 3, and 7, respectively, and all these conformers are planar structures. The conformations are shown in Fig. S1 (ESI†). While DQG has two planar conformations, the conf-2 of DQG could not form stable hydrogen bonds to cytosine due to the steric repulsion between the amino N–H and aromatic C–H (Fig. S1a, ESI†), indicating that the conf-1 of DQG would be likely responsible for the binding to C in the G2C4/G2C4 unit to form two hydrogen bonds (cf.Fig. 1b). Likewise, D3iQG conf-2, D1iQG conf-1, D1iQG conf-2, and Q3iQG conf-3 were ineligible for the hydrogen bonding with cytosine. In contrast, all seven conformers of DQzG were eligible to form three hydrogen bonds with cytosine. DQzG conf-6 and conf-7 are more than 2 kcal mol−1 above the lowest energy conf-1, and it is reasonable to assume that these conformers have little contribution for the complexation with cytosine. Thus, further discussion on the spatial orientation of heteroaromatic rings in the bound complexes would be focused on the ten conformers of DQG conf-1, D3iQG conf-1, D1iQG conf-1, Q3iQG conf-1 and 2, and DQzG conf-1, 2, 3, 4, and 5. To discuss the special orientation of heteroaromatic rings, the ring directly attached to the guanidyl group is identified as ring A, and the other is referred as ring B (cf.Fig. 1a).
000 conformations generated by molecular mechanics (MM) calculations. Furthermore, quantum mechanical calculations were performed for all conformers obtained by MM calculation using DFT ωb97xd/6-31G+(d,p) in Gaussian 16 (Table 1 and Table S1, ESI†). The numbers of conformers within 3.5 kcal mol−1 of the lowest energy conformer obtained for DQG, D3iQG, D1iQG, Q3iQG, and DQzG were 2, 2, 2, 3, and 7, respectively, and all these conformers are planar structures. The conformations are shown in Fig. S1 (ESI†). While DQG has two planar conformations, the conf-2 of DQG could not form stable hydrogen bonds to cytosine due to the steric repulsion between the amino N–H and aromatic C–H (Fig. S1a, ESI†), indicating that the conf-1 of DQG would be likely responsible for the binding to C in the G2C4/G2C4 unit to form two hydrogen bonds (cf.Fig. 1b). Likewise, D3iQG conf-2, D1iQG conf-1, D1iQG conf-2, and Q3iQG conf-3 were ineligible for the hydrogen bonding with cytosine. In contrast, all seven conformers of DQzG were eligible to form three hydrogen bonds with cytosine. DQzG conf-6 and conf-7 are more than 2 kcal mol−1 above the lowest energy conf-1, and it is reasonable to assume that these conformers have little contribution for the complexation with cytosine. Thus, further discussion on the spatial orientation of heteroaromatic rings in the bound complexes would be focused on the ten conformers of DQG conf-1, D3iQG conf-1, D1iQG conf-1, Q3iQG conf-1 and 2, and DQzG conf-1, 2, 3, 4, and 5. To discuss the special orientation of heteroaromatic rings, the ring directly attached to the guanidyl group is identified as ring A, and the other is referred as ring B (cf.Fig. 1a).
| DQG | D3iQG | D1iQG | Q3iQG | DQzG | |
|---|---|---|---|---|---|
| a Relative potential energy (kcal mol−1) of the conformers of DQG, D3iQG, D1iQG, Q3iQG, and DQzG calculated at the ωB97XD/6-31G+(d,p) level. All conformers listed here have the planar conformation. The entire list of conformational energies can be found in ESI. b These conformers are ineligible for stable hydrogen bonds formation with cytosine due to steric repulsion. | |||||
| conf-1 | 0.00 | 0.00 | 0.00b | 0.00 | 0.00 |
| 2 | 2.24b | 2.17b | 3.41b | 0.13 | 0.46 |
| 3 | 2.28b | 1.03 | |||
| 4 | 1.18 | ||||
| 5 | 1.42 | ||||
| 6 | 2.22 | ||||
| 7 | 2.41 | ||||
UV melting temperature analysis
To examine the effect of the DQG analogs on the thermal stability of a 14-mer double-stranded DNA 5′-d(GCAT GGCCCC TACG)-3′/3′-d(CGTA CCCCGG ATGC)-5′ containing the G2C4/G2C4 motif (G2C4/G2C4-dsDNA), we measured the Tm of the G2C4/G2C4-dsDNA without and with DQG analogs (Fig. S2, ESI†). The differences in the melting temperature (ΔTm) in the presence and absence of DQG analogs are shown in Table 2. As reported previously, the Tm of the G2C4/G2C4-dsDNA in the absence and presence of DQG were 29.8 and 49.4 °C (ΔTm = 19.5 °C), respectively.23 To examine the influence of quinoline and guanidyl group moieties on the interaction between DQG and G2C4/G2C4-dsDNA, we performed Tm measurements using DNpG and DQU. The control compounds, DNpG and DQU, did not show any significant increase in the Tm of G2C4/G2C4-dsDNA, suggesting that quinoline and guanidyl group are indispensable for DQG binding to G2C4/G2C4-dsDNA. The ΔTm of D3iQG, D1iQG, and Q3iQG were 11.4, 13.4, and 18.7 °C, respectively, showing a decreased Tm compared with DQG. In contrast, DQzG showed ΔTm of 20.2 °C, indicating that the substitution of quinoline on DQG by quinazoline slightly increased the thermal stability of the ligand-bound complex.| Ligand | T m (+)b | ΔTmc |
|---|---|---|
| a Thermal melting curves were measured for dsDNA [5′-d(GCATGGCCCCTACG)-3′/3′-d(CGTACCCCGGATGC)-5′] (5 μM) in sodium phosphate buffer (10 mM, pH 7) containing sodium chloride (100 mM) and 5% DMSO. b T m values of DNA duplexes in the presence of DQG or DQG analogs (20 μM). The melting temperature is calculated by using the median method. All measurements were made three times or more and standard deviations are shown in parentheses. c ΔTm (°C) is calculated as the difference of Tm (°C) in the absence Tm (−) (29.8 °C (0.5)) and presence Tm (+) of ligands. | ||
| DQG | 49.4 (0.3) | 19.5 |
| D3iQG | 41.3 (1.7) | 11.4 |
| D1iQG | 43.2 (0.7) | 13.4 |
| Q3iQG | 48.5 (0.4) | 18.7 |
| DQzG | 50.0 (0.7) | 20.2 |
| QG | 33.2 (0.8) | 3.4 |
| DNpG | 31.1 (0.4) | 1.3 |
| DQU | 30.3 (1.0) | 0.4 |
CD spectral change on ligand binding
To obtain insights into structural information of complexes bound by DQG analogs, we carried out CD titration experiments with DQG analogs against G2C4/G2C4-dsDNA (Fig. 2). The CD spectrum of G2C4/G2C4-dsDNA showed negative and positive bands around 250 and 275 nm, respectively. As previously reported, the addition of DQG resulted in the appearance of a positive band around 260 nm and characteristic induced CD bands around 300–345 nm (Fig. 2a).23 In the case of D3iQG and Q3iQG, the CD spectra showed two distinctive positive bands around 250 and 275 nm in the DNA region (Fig. 2b and d). The induced CD bands of D3iQG and Q3iQG were observed around 305 and 300–360 nm, respectively. Broad induced CD bands around 320–360 nm and a positive CD band around 255 nm with shoulder were observed in the presence of DQzG (Fig. 2e). In contrast, the CD spectrum in the presence of D1iQG showed a ligand-derived induced CD band in the 330–360 nm region and the disappearance of the negative CD band around 250 nm, but the positive CD band in DNA region around 275 nm did not show any significant changes (Fig. 2c). These data suggested that the binding of DQG analogs except for D1iQG induced large conformational change of G2C4/G2C4-dsDNA. The mode of D1iQG binding to the G2C4/G2C4 unit would be different from those of other DQG analogs.Binding analysis by SPR
SPR measurements were performed to assess the binding affinity of DQG analogs for the G2C4/G2C4 unit using sensor chip with immobilized 32-mer hairpin-DNA containing the G2C4/G2C4 unit (Fig. 3). All ligands were sequentially injected at increasing concentrations, resulting in concentration-dependent increases in response unit (RU). The sensorgrams showed a similar shape of SPR response curves among all ligands except for DQzG. The injection of DQG, D3iQG, and Q3iQG at a concentration of 4 μM resulted in the RU of 30–35 (Fig. 3a, b, and d). On the other hand, sequential injection of D1iQG reached a plateau of RU at 15 when the concentration was 2 μM (Fig. 3c). The difference in RU at the saturation point was likely due to a different mode of binding in D1iQG. When DQG and Q3iQG were injected, the RU was almost the same under the condition of same concentrations, suggesting that the binding affinity of Q3iQG was comparable with that of DQG. In contrast, the injection of D3iQG exhibited lower RU than DQG under the condition of same concentrations, implying that the binding affinity of D3iQG was lower than that of DQG. The sensorgram obtained for DQzG showed slow dissociation kinetics, which is different from those of the other DQG analogs (Fig. 3e). In the case of DQzG, the significant increase in RU was observed under the condition of one order of magnitude lower concentrations than that of DQG. We also confirmed that saturation point for RU after injection of DQzG was approximately 30 at a concentration of 0.4 μM. These results suggested that DQzG showed the highest binding affinity among DQG analogs we tested. We confirmed that the injection of all DQG analogs for fully matched DNA duplex-immobilized sensor chip did not show any significant increases in RU, indicating the preferential binding of DQG analogs to the G2C4/G2C4 unit (Fig. S3, ESI†). The apparent dissociation constants (KDapp) of ligands binding to the hairpin DNA containing the G2C4/G2C4 unit were determined using BIAevaluation software integrated into the SPR equipment. The KDapp values were calculated based on the assumption of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model and are as follows: DQG, 1.0 μM; D3iQG, 10 μM; D1iQG, 3.4 μM; Q3iQG, 1.4 μM; and DQzG, 63 nM. These values are fully consistent with the trends of Tm measurements. It is important to emphasize, however, that this assumption was made solely for the purpose of comparing the affinities of the five molecules. The possibility of other binding stoichiometries cannot be excluded, as discussed later.
1 binding model and are as follows: DQG, 1.0 μM; D3iQG, 10 μM; D1iQG, 3.4 μM; Q3iQG, 1.4 μM; and DQzG, 63 nM. These values are fully consistent with the trends of Tm measurements. It is important to emphasize, however, that this assumption was made solely for the purpose of comparing the affinities of the five molecules. The possibility of other binding stoichiometries cannot be excluded, as discussed later.
NMR analysis of DQzG-bound G2C4/G2C4-dsDNA
To obtain further insights into the interaction between DQzG and G2C4/G2C4-dsDNA, we performed NMR titration experiments with molar ratios from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 (DNA
0 (DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DQzG) to 1
DQzG) to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 in 20 mM phosphate buffer (pH 6.5) containing 50 mM NaCl and 10% D2O and monitored the changes in signals of exchangeable protons from 10 to 14 ppm (Fig. 4). In the absence of DQzG, the signals of imino protons in A–T and G–C base pairs appeared at 13–14 ppm and 12–13 ppm, respectively. As the concentration of DQzG was increased, the appearance of signals around 10–12 ppm was observed with concomitant changes in the signal around 12–14 ppm. These new signals around 10–12 ppm were likely to correspond to guanidine protons of DQzG. These results suggested that DQzG bound to G2C4/G2C4-dsDNA resulting in the formation of hydrogen bonded complex.
4 in 20 mM phosphate buffer (pH 6.5) containing 50 mM NaCl and 10% D2O and monitored the changes in signals of exchangeable protons from 10 to 14 ppm (Fig. 4). In the absence of DQzG, the signals of imino protons in A–T and G–C base pairs appeared at 13–14 ppm and 12–13 ppm, respectively. As the concentration of DQzG was increased, the appearance of signals around 10–12 ppm was observed with concomitant changes in the signal around 12–14 ppm. These new signals around 10–12 ppm were likely to correspond to guanidine protons of DQzG. These results suggested that DQzG bound to G2C4/G2C4-dsDNA resulting in the formation of hydrogen bonded complex.
SPR analysis of the binding to G2C4 repeat
Having confirmed the superior properties of DQzG in the binding to the G2C4/G2C4 unit in dsDNA compared with the parent molecule DQG, we investigated the DQzG binding to the d(G2C4)9 repeat by SPR. DQzG showed a significant response at 0.2 μM with a characteristic slow binding and dissociation profile (Fig. 5b). The SPR profile is similar to that obtained for the G2C4/G2C4 unit in dsDNA (cf.Fig. 3e). DQG showed SPR response only at 1 μM (Fig. 5a). Q3iQG showed a similar response to DQG (Fig. S4, ESI†).Discussion
Hydrogen bonding and π-stacking play crucial roles in the interactions between small molecules and nucleic acids.27,28 Using molecular design based on complementary hydrogen bonding with mismatched nucleotide bases and π-stacking with adjacent bases, we have developed mismatch-binding ligands (MBLs).12–15,29–33 In our ongoing studies focused on nucleic acid-targeted ligands, we previously reported on DQG, which binds to the G2C4/G2C4-dsDNA. DQG adopts planar conformations due to a π-conjugated system comprising two quinoline rings and the guanidyl group, facilitated by intramolecular hydrogen bonding. Conformational analysis revealed that the DQG conf-1 conformation is the only one suitable for complex formation with cytosine in the G2C4/G2C4 unit of dsDNA. To investigate the relationship between hydrogen bonding capability and stacking interactions with adjacent bases, we designed and synthesized six DQG analogs.A critical factor influencing the binding of DQG analogs is the planarity of the ligand molecule. Conformational analyses revealed that, except for DNpG, the lowest energy state for six analogs is a planar conformer (Fig. S1, ESI†). For instance, DQG exhibits two planar conformers, conf-1 and conf-2 (Fig. S1a, ESI†). The non-planar conformer, conf-3, is 9.37 kcal mol−1 higher in energy than conf-1. DNpG does not have a planar conformer due to steric repulsion between the guanidyl N–H and C–H in the naphthyl ring. While this non-planar conformer offers a hydrogen-bonding group with a DD arrangement (Fig. S1g, ESI†), UV-melting analyses show a minimal ΔTm (1.3 °C) for DNpG (Table 2). In contrast, DQG exhibits a ΔTm of 19.5 °C under the same conditions, indicating that a planar conformation is essential for binding to the G2C4/G2C4 unit. Among the five analogs with planar conformers, DQU fails to bind to G2C4/G2C4-dsDNA. Its lowest-energy conformer is planar but lacks hydrogen-bonding donors with a DD arrangement (Fig. S1f, ESI†). Although the next stable conformer has the required hydrogen-bonding arrangement, it is 4.33 kcal mol−1 higher in energy than conf-1, suggesting limited binding potential. Additionally, DQU lacks a positive charge essential for nucleic acid binding. The monomeric ligand QG has a planar conformation and hydrogen-bonding donors with a DD arrangement but only weakly stabilizes G2C4/G2C4-dsDNA (ΔTm: 3.4 °C). Based on these findings for DNpG, DQU, and QG, the requirements for DQG analogs to bind effectively to G2C4/G2C4-dsDNA include planar conformations with two heterocycles and a DD arrangement of hydrogen-bonding donors. Five molecules, namely DQG, D3iQG, D1iQG, Q3iQG, and DQzG, meet these criteria.
Among the five molecules tested, D3iQG and D1iQG exhibited a significant decrease in ΔTm compared to the other three molecules. D1iQG displayed marked differences from the other molecules in both CD spectral changes and SPR analyses. In CD measurements, D1iQG induced fewer structural changes in G2C4/G2C4-dsDNA upon binding (Fig. 2c). The SPR response with D1iQG was lower than that of other DQG analogs at a ligand concentration of 4 μM (Fig. 3c). Additionally, the UV melting profile of D1iQG showed a steeper rise in absorbance upon the melting of the ligand-bound DNA duplex compared to the other DQG analogs (Fig. S2c, ESI†). These observations suggest that the binding mode of D1iQG differs from that of the other four molecules. The likely determinant of D1iQG's binding mode is the orientation of its two isoquinoline rings relative to the hydrogen-bonding donors. The lowest-energy conformer of D1iQG has DD hydrogen bonding groups but cannot form stable hydrogen bonds with cytosine due to interference by the isoquinoline C–H at C8 (Fig. S1c, ESI†). The second lowest-energy conformer experiences even stronger steric repulsion. These steric hindrances likely contribute to the distinct binding mode of D1iQG compared to the other DQG analogs.
The low ΔTm observed for D3iQG provides valuable insights into the favorable arrangement of aromatic rings in the ligand-bound complex. Both DQG and D3iQG possess two planar conformers with similar energy differences. However, the conf-2 conformer of both molecules cannot bind to cytosine due to steric repulsion between the cytosine C4–NH2 group and the C3–H of quinoline in DQG conf-2, and between the cytosine C4–H and the C4–H of isoquinoline in D3iQG conf-2 (Fig. S1, ESI†). Consequently, DQG and D3iQG bind to cytosine using their conf-1 conformers. When both molecules bind to cytosine, the spatial arrangement of aromatic ring A is consistent between them, but not for ring B. This discrepancy suggests that the spatial arrangements of rings A and B may contribute differently to the complex stability of DQG analogs.
The spatial arrangements of ring A and B of each conformer of DQG analogs were compared by superimposing the expected hydrogen-bonded pair with cytosine. Conformers of DQzG (conf-1 and 2), D3iQG (conf-1), and Q3iQG (conf-1 and 2) were superimposed onto DQG conf-1 with cytosine as the reference. The three minor conformers (conf-3, 4, and 5) of DQzG exhibited properties similar to conf-1 and 2 (Fig. S5, ESI†). In Fig. 6, ring A of the six conformers was colored magenta, while DQG's ring A was colored cyan. In the superimposed figures, the overlapped ring A would be shown only in cyan, otherwise both cyan and magenta colored ring As were visible. Consistent with previous discussions, two ring As in D3iQG conf-1 are overlapped with those of DQG conf-1, therefore only cyan-colored ring A was identified. Interestingly, both conf-1 and conf-2 of Q3iQG showed full overlap of ring A with DQG conf-1. In contrast, ring As of DQzG conf-1 and conf-3 did not overlap with those of DQG at all. The conf-2 of DQzG showed overlap of one of two ring As.
Fig. 7 illustrates the overlap of ring Bs. Contrary to the results for ring A superimposition, DQzG exhibited a notable similarity in spatial arrangement to DQG for ring B. Both conf-1 and conf-2 of DQzG showed complete overlap of both ring Bs, while conf-3 overlapped with one of its two ring Bs. In contrast, D3iQG showed no overlap for ring B. Q3iQG's conf-1 and conf-2 displayed overlap with one of their two ring Bs. The alignment of ring A and B in DQG analogs with those in DQG is summarized in Table 3.
| DQG | DQzG | D3iQG | Q3iQG | |||
|---|---|---|---|---|---|---|
| conf-1 | conf-1 | conf-2 | conf-1 | conf-1 | conf-2 | |
| a The number of ring A and B overlapped with that of DQG conf-1. b Hydrogen bonding pattern between DQG analogs and cytosine. | ||||||
| Ring Aa | — | 0 | 1 | 2 | 2 | 2 |
| Ring Ba | — | 2 | 2 | 0 | 1 | 1 |
| H-bondingb | DD/AA | ADD/DAA | ADD/DAA | DD/AA | DD/AA | DD/AA |
| ΔTm (°C) | 19.5 | 20.2 | 11.4 | 18.7 | ||
Table 3 highlights the relationships between the spatial orientation of rings A and B, the hydrogen-bonding pattern with cytosine, and the stability of the ligand-bound complex. Both D3iQG and DQG share the orientation of ring A relative to cytosine and the DD/AA hydrogen-bonding pattern, but differ in ring B arrangement. The significant ΔTm difference (19.5 °C vs. 11.4 °C) suggests that the spatial arrangement of ring B likely accounts for this variation. Q3iQG, which overlaps one of its two ring Bs with DQG and shares the ring A orientation and hydrogen-bonding pattern with D3iQG, exhibits only a slight decrease in stability compared to DQG. This underscores the importance of ring B arrangement in the Q3iQG-bound complex. DQzG's conformers feature ADD hydrogen-bonding groups and are susceptible to hydrogen bonding with cytosine. The lowest-energy conf-1 of DQzG shares the ring B spatial arrangement with DQG, while conf-2 also shares ring A arrangement. Conf-3 of DQzG shared the arrangement of one of two ring Bs similarly to conf-1 and conf-2 (Table S2, ESI†). A slight increase in stability can be attributed to the ADD–DAA hydrogen-bonding with cytosine. Thus, DQzG has an advantage over DQG due to its ADD hydrogen-bonding groups while maintaining the spatial arrangement of heteroaromatic rings. Conf-4 and conf-5 also share similar features with conf-3. Further discussion on these minor conformers of DQzG is provided in ESI.†
In summary, the pronounced and consistent SPR responses of DQzG to G2C4 repeat DNA, when compared to those of DQG, strongly indicate the potential of DQzG as a superior lead compound for targeting G2C4 repeat DNA. The SPR response of DQzG towards G2C4 repeat DNA closely parallels that observed for the G2C4/G2C4 unit in double-stranded DNA. This suggests that double-stranded DNA containing the G2C4/G2C4 unit is a suitable model for drug discovery studies focused on G2C4 repeat.
In the SPR data analysis of DQzG binding to the hairpin DNA containing G2C4/G2C4, the binding stoichiometry could be estimated using the following equation:
| Rmax = MWA × RL/MWL × n, |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DQzG ratio of 1
DQzG ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (Fig. 2), and NMR titration experiments similarly indicated a DNA
4 (Fig. 2), and NMR titration experiments similarly indicated a DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DQzG ratio of 1
DQzG ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 as peaks corresponding to the initial DNA duplex disappeared (Fig. 4). These discrepancies likely arise from differences in DQzG concentrations used in each measurement: ∼4 μM for SPR, 50 μM for CD titration, and 1200 μM for NMR titration. This suggested that at lower concentrations (below 4 μM), the binding stoichiometry is 2, while at higher concentrations it increases to 4. The G2C4/G2C4 unit contains four mismatched cytosines, which could potentially bind up to four DQzG molecules via hydrogen bonding. At lower concentrations, two of these cytosines are likely bound by DQzG to form the initial complex. At higher concentrations, the binding transitions to a complex involving all four DQzG molecules. The interaction between two DQzG molecules in the initial complex and their potential stacking interactions with neighbouring base pairs are key questions. However, structural determination of the complex will be necessary for further discussion and validation of these hypotheses.
4 as peaks corresponding to the initial DNA duplex disappeared (Fig. 4). These discrepancies likely arise from differences in DQzG concentrations used in each measurement: ∼4 μM for SPR, 50 μM for CD titration, and 1200 μM for NMR titration. This suggested that at lower concentrations (below 4 μM), the binding stoichiometry is 2, while at higher concentrations it increases to 4. The G2C4/G2C4 unit contains four mismatched cytosines, which could potentially bind up to four DQzG molecules via hydrogen bonding. At lower concentrations, two of these cytosines are likely bound by DQzG to form the initial complex. At higher concentrations, the binding transitions to a complex involving all four DQzG molecules. The interaction between two DQzG molecules in the initial complex and their potential stacking interactions with neighbouring base pairs are key questions. However, structural determination of the complex will be necessary for further discussion and validation of these hypotheses.
Conclusions
In conclusion, our comparative analysis of binding characteristics among DQG analogs underscores the importance of the spatial arrangement of aromatic rings. The interplay of electrostatic attraction, hydrogen bonding, and stacking interactions with neighboring bases demonstrates a compensatory relationship, with DQzG exhibiting an optimal combination of these interactions, resulting in enhanced binding to the G2C4 repeat compared with the parent ligand, DQG. This study offers valuable insights into the molecular design of ligands targeting GGCCCC repeats, leveraging guanidine and heteroaromatic rings. Such insights pave the way for developing next-generation ligands with potential applications in the study and treatment of ALS/FTD and other repeat diseases. The elucidation of the DQzG-bound structure to G2C4 repeat DNA is pivotal for deeper understanding of the critical factors influencing molecule binding to the repeat. Ongoing research in our laboratories is focused on further exploring these aspects, promising further advancements in this field.Experimental section
Synthesis of ligands
Reagents and solvents were purchased from standard suppliers and used without further purification. Reactions were monitored with TLC plates precoated with Merch Silica Gel 60 F254. Spots were visualized with UV light or ninhydrin. Wako gel C-200 was used for silica gel flash chromatography. High performance liquid chromatography (HPLC) was performed by a Gilson 811C Dynamic Mixer system with a UV detector set at 254 nm using a Cosmosil 5C18-MS-II column (150 × 20 mm) with a dual solvent system of 0.1% AcOH/H2O (solvent A) and CH3CN (solvent B). 1H NMR and 13C NMR spectra were measured with ECS400 (JEOL), ECA600 (JEOL), and Avance III 700 (BRUKER). The chemical shifts are expressed in parts per million (ppm) relative to a residual solvent as an internal standard. Chemical shifts were reported in ppm and coupling constants (J values) in hertz. 1H NMR chemical shifts were referenced to the residual solvent peak at 3.31 ppm in CD3OD and at 2.50 ppm in DMSO-d6. 13C NMR chemical shifts were referenced to the center solvent peak at 49.00 ppm for CD3OD and at 39.52 ppm for DMSO-d6. High-resolution mass spectra (HRMS) were recorded on a Thermo LTQ Orbitrap XL mass spectrometer in electron spray ionization mode.Conformational searches
Conformational searches were performed for the initially selected structures of DQG, D3iQG, D1iQG, Q3iQG, DQzG, DQU, and DNpG by using Maestro version 13.0.135 (Schrödinger lnc.) under the following conditions: low-frequency-mode conformational search with probability of TORS/MOLS steps of 0.5 and OPLS4 force field in solvent water. Among 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 conformations generated, the lowest conformer within 5 kcal mol−1 after energy minimization were saved. The saved conformers of DQG, D3iQG, D1iQG, Q3iQG, DQzG, DQU, and DNpG were found 21, 19, 32, 36, 11, 9, and 32 conformers, respectively. Structure optimization of DQG, D3iQG, D1iQG, Q3iQG, DQzG, DQU, and DNpG was performed by Gaussian 16 using parameter of ωB97XD/6-31G+(d,p). All conformers saved in conformational searches of DQG, D3iQG, D1iQG, Q3iQG, DQzG, DQU, and DNpG were used as the initial structures.
000 conformations generated, the lowest conformer within 5 kcal mol−1 after energy minimization were saved. The saved conformers of DQG, D3iQG, D1iQG, Q3iQG, DQzG, DQU, and DNpG were found 21, 19, 32, 36, 11, 9, and 32 conformers, respectively. Structure optimization of DQG, D3iQG, D1iQG, Q3iQG, DQzG, DQU, and DNpG was performed by Gaussian 16 using parameter of ωB97XD/6-31G+(d,p). All conformers saved in conformational searches of DQG, D3iQG, D1iQG, Q3iQG, DQzG, DQU, and DNpG were used as the initial structures.
UV-melting analysis
All samples were mixed with 5 μM solutions of DNA, 10 mM sodium phosphate buffer (pH 7.0), 100 mM NaCl, 20 μM ligand, and 5.0% DMSO. The melting temperature was determined by measuring the absorbance of samples at λ = 260 nm from 2 to 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C (1 °C min−1). Thermal denaturation profiles were monitored on a UV-2700 spectrophotometer (Shimadzu) equipped with a 10 mm path-length cell and a TMSPC-8 temperature controller. Tm was calculated by using the median method.
°C (1 °C min−1). Thermal denaturation profiles were monitored on a UV-2700 spectrophotometer (Shimadzu) equipped with a 10 mm path-length cell and a TMSPC-8 temperature controller. Tm was calculated by using the median method.
Circular dichroism (CD) measurements
CD spectra were recorded with a J-725 CD spectrometer (JASCO) using a 10 mm path length cell. CD spectra of DNA (5 μM DNA duplex) in the absence and presence of ligand (DQG, D3iQG, D1iQG, Q3iQG (10, 20, 30, 40, 50 μM), and DQzG (5, 10, 15, 20, 25 μM)) were measured in sodium phosphate buffer (10 mM, pH 7.0), NaCl (100 mM), and 5.0% DMSO.Surface plasmon resonance (SPR) analysis
5′-Biotinylated DNAs (G2C4/G2C4, (G2C4)9, and fullmatch) were immobilized on the streptavidine-coated surface of Series S Sensor chip SA (GE Healthcare, Life Science). All immobilization reactions were performed in HBS-EP + buffer (0.01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M HEPES, pH 7.4, 0.15
M HEPES, pH 7.4, 0.15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M NaCl, 3 mM ethylenediaminetetraacetic acid (EDTA), 0.05% (v/v) surfactant P20) by using a Biacore T200 instrument (GE Healthcare, Life Science) at 25
M NaCl, 3 mM ethylenediaminetetraacetic acid (EDTA), 0.05% (v/v) surfactant P20) by using a Biacore T200 instrument (GE Healthcare, Life Science) at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C. The amounts of immobilized G2C4/G2C4, (G2C4)9, and fullmatch on the chip surface were 515, 521, and 490 response units (RU), respectively. Sensorgrams were obtained in the ligand concentrations of 0.25, 0.50, 1.0, 2.0, 4.0 μM and 0.025, 0.050, 0.1, 0.2, 0.4 μM in single-cycle mode (flow rate 60 μM min−1, contact time 30 s, and dissociation time 120 s). All sensorgrams were corrected by reference subtraction of blank flow cell response and buffer injection response. The data collected were analyzed with the Biacore T200 evaluation software (version 2.0), and kinetic parameters were derived through the application of both the kinetic analysis and curve fitting techniques, assuming a 1
°C. The amounts of immobilized G2C4/G2C4, (G2C4)9, and fullmatch on the chip surface were 515, 521, and 490 response units (RU), respectively. Sensorgrams were obtained in the ligand concentrations of 0.25, 0.50, 1.0, 2.0, 4.0 μM and 0.025, 0.050, 0.1, 0.2, 0.4 μM in single-cycle mode (flow rate 60 μM min−1, contact time 30 s, and dissociation time 120 s). All sensorgrams were corrected by reference subtraction of blank flow cell response and buffer injection response. The data collected were analyzed with the Biacore T200 evaluation software (version 2.0), and kinetic parameters were derived through the application of both the kinetic analysis and curve fitting techniques, assuming a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding interaction.
1 binding interaction.
Sample preparation and NMR measurement
The chemically synthesized DNA oligomers 5′-GCATGGCCCCTACG-3′/3′-CGTACCCCGGATGC-5′ were purchased from commercial suppliers (FASMAC). DQzG at molar ratios of 0, 1, 2, 3, and 4 was dissolved in 20 mM sodium phosphate buffer (pH 6.5), 100 mM NaCl, 300 μM DNA duplex, and 10% D2O. The solution was heated at 80 °C for 1 h and cooled to 4 °C overnight. Using these solutions, 1H-NMR were measured on BRUKER Avance III 700.Data availability
Raw data are available from the corresponding author upon request.Conflicts of interest
Conflict of interest statement. None declared.Acknowledgements
This work was supported by Grant-in-Aid for Scientific research (A) [19H00924], [22H00351] to K. N., for Scientific Research (B) [20H02880] and for Challenging Exploratory Research [21K19049] to T. S. We thank for Ms Yu Kawakami for experimental assistance.Notes and references
- C. E. Pearson, K. N. Edamura and J. D. Cleary, Nat. Rev. Genet., 2005, 6, 729–742 CrossRef CAS PubMed.
- I. Malik, C. P. Kelley, E. T. Wang and P. K. Todd, Nat. Rev. Mol. Cell Biol., 2021, 22, 589–607 CrossRef CAS PubMed.
- M. DeJesus-Hernandez, I. R. Mackenzie, B. F. Boeve, A. L. Boxer, M. Baker, N. J. Rutherford, A. M. Nicholson, N. A. Finch, H. Flynn, J. Adamson, N. Kouri, A. Wojtas, P. Sengdy, G.-Y. R. Hsiung, A. Karydas, W. W. Seeley, K. A. Josephs, G. Coppola, D. H. Geschwind, Z. K. Wszolek, H. Feldman, D. S. Knopman, R. C. Petersen, B. L. Miller, D. W. Dickson, K. B. Boylan, N. R. Graff-Radford and R. Rademakers, Neuron, 2011, 72, 245–256 CrossRef CAS PubMed.
- A. E. Renton, E. Majounie, A. Waite, J. Simón-Sánchez, S. Rollinson, J. R. Gibbs, J. C. Schymick, H. Laaksovirta, J. C. van Swieten, L. Myllykangas, H. Kalimo, A. Paetau, Y. Abramzon, A. M. Remes, A. Kaganovich, S. W. Scholz, J. Duckworth, J. Ding, D. W. Harmer, D. G. Hernandez, J. O. Johnson, K. Mok, M. Ryten, D. Trabzuni, R. J. Guerreiro, R. W. Orrell, J. Neal, A. Murray, J. Pearson, I. E. Jansen, D. Sondervan, H. Seelaar, D. Blake, K. Young, N. Halliwell, J. B. Callister, G. Toulson, A. Richardson, A. Gerhard, J. Snowden, D. Mann, D. Neary, M. A. Nalls, T. Peuralinna, L. Jansson, V.-M. Isoviita, A.-L. Kaivorinne, M. Hölttä-Vuori, E. Ikonen, R. Sulkava, M. Benatar, J. Wuu, A. Chiò, G. Restagno, G. Borghero, M. Sabatelli, D. Heckerman, E. Rogaeva, L. Zinman, J. D. Rothstein, M. Sendtner, C. Drepper, E. E. Eichler, C. Alkan, Z. Abdullaev, S. D. Pack, A. Dutra, E. Pak, J. Hardy, A. Singleton, N. M. Williams, P. Heutink, S. Pickering-Brown, H. R. Morris, P. J. Tienari and B. J. Traynor, Neuron, 2011, 72, 257–268 CrossRef CAS PubMed.
- M. Van Blitterswijk, M. DeJesus-Hernandez, E. Niemantsverdriet, M. E. Murray, M. G. Heckman, N. N. Diehl, P. H. Brown, M. C. Baker, N. A. Finch, P. O. Bauer, G. Serrano, T. G. Beach, K. A. Josephs, D. S. Knopman, R. C. Petersen, B. F. Boeve, N. R. Graff-Radford, K. B. Boylan, L. Petrucelli, D. W. Dickson and R. Rademakers, Lancet Neurol., 2013, 12, 978–988 CrossRef CAS PubMed.
- B. Swinnen, W. Robberecht and L. Van Den Bosch, EMBO J., 2020, 39, e101112 CrossRef CAS PubMed.
- K. Mori, S.-M. Weng, T. Arzberger, S. May, K. Rentzsch, E. Kremmer, B. Schmid, H. A. Kretzschmar, M. Cruts, C. Van Broeckhoven, C. Haass and D. Edbauer, Science, 2013, 339, 1335–1338 CrossRef CAS PubMed.
- Y.-B. Lee, H.-J. Chen, J. N. Peres, J. Gomez-Deza, J. Attig, M. Štalekar, C. Troakes, A. L. Nishimura, E. L. Scotter, C. Vance, Y. Adachi, V. Sardone, J. W. Miller, B. N. Smith, J.-M. Gallo, J. Ule, F. Hirth, B. Rogelj, C. Houart and C. E. Shaw, Cell Rep., 2013, 5, 1178–1186 CrossRef CAS PubMed.
- J. Cooper-Knock, M. J. Walsh, A. Higginbottom, J. Robin Highley, M. J. Dickman, D. Edbauer, P. G. Ince, S. B. Wharton, S. A. Wilson, J. Kirby, G. M. Hautbergue and P. J. Shaw, Brain, 2014, 137, 2040–2051 CrossRef PubMed.
- Y. Shi, S. Lin, K. A. Staats, Y. Li, W.-H. Chang, S.-T. Hung, E. Hendricks, G. R. Linares, Y. Wang, E. Y. Son, X. Wen, K. Kisler, B. Wilkinson, L. Menendez, T. Sugawara, P. Woolwine, M. Huang, M. J. Cowan, B. Ge, N. Koutsodendris, K. P. Sandor, J. Komberg, V. R. Vangoor, K. Senthilkumar, V. Hennes, C. Seah, A. R. Nelson, T.-Y. Cheng, S.-J. J. Lee, P. R. August, J. A. Chen, N. Wisniewski, V. Hanson-Smith, T. G. Belgard, A. Zhang, M. Coba, C. Grunseich, M. E. Ward, L. H. Van Den Berg, R. J. Pasterkamp, D. Trotti, B. V. Zlokovic and J. K. Ichida, Nat. Med., 2018, 24, 313–325 CrossRef CAS PubMed.
- Q. Zhu, J. Jiang, T. F. Gendron, M. McAlonis-Downes, L. Jiang, A. Taylor, S. Diaz Garcia, S. Ghosh Dastidar, M. J. Rodriguez, P. King, Y. Zhang, A. R. La Spada, H. Xu, L. Petrucelli, J. Ravits, S. Da Cruz, C. Lagier-Tourenne and D. W. Cleveland, Nat. Neurosci., 2020, 23, 615–624 CrossRef CAS PubMed.
- K. Nakatani, S. Hagihara, Y. Goto, A. Kobori, M. Hagihara, G. Hayashi, M. Kyo, M. Nomura, M. Mishima and C. Kojima, Nat. Chem. Biol., 2005, 1, 39–43 CrossRef CAS PubMed.
- K. Nakatani, S. Sando, H. Kumasawa, J. Kikuchi and I. Saito, J. Am. Chem. Soc., 2001, 123, 12650–12657 CrossRef CAS PubMed.
- T. Yamada, K. Furuita, S. Sakurabayashi, M. Nomura, C. Kojima and K. Nakatani, Nucleic Acids Res., 2022, 50, 9621–9631 CrossRef CAS PubMed.
- K. Nakatani, Proc. Jpn. Acad., Ser. B, 2022, 98, 30–48 CrossRef CAS PubMed.
- M. Nakamori, G. B. Panigrahi, S. Lanni, T. Gall-Duncan, H. Hayakawa, H. Tanaka, J. Luo, T. Otabe, J. Li, A. Sakata, M.-C. Caron, N. Joshi, T. Prasolava, K. Chiang, J.-Y. Masson, M. S. Wold, X. Wang, M. Y. W. T. Lee, J. Huddleston, K. M. Munson, S. Davidson, M. Layeghifard, L.-M. Edward, R. Gallon, M. Santibanez-Koref, A. Murata, M. P. Takahashi, E. E. Eichler, A. Shlien, K. Nakatani, H. Mochizuki and C. E. Pearson, Nat. Genet., 2020, 52, 146–159 CrossRef CAS PubMed.
- Y. Hasuike, H. Tanaka, T. Gall-Duncan, M. Mehkary, K. Nakatani, C. E. Pearson, S. Tsuji, H. Mochizuki and M. Nakamori, Neurobiol. Dis., 2022, 163, 105604 CrossRef CAS PubMed.
- Y. Lu, C. Dohno and K. Nakatani, Chem. Commun., 2020, 56, 754–757 RSC.
- C.-R. Jhan, R. Satange, S.-C. Wang, J.-Y. Zeng, Y.-C. Horng, P. Jin, S. Neidle and M.-H. Hou, Nucleic Acids Res., 2021, 49, 9526–9538 CrossRef CAS PubMed.
- R. Simone, R. Balendra, T. G. Moens, E. Preza, K. M. Wilson, A. Heslegrave, N. S. Woodling, T. Niccoli, J. Gilbert-Jaramillo, S. Abdelkarim, E. L. Clayton, M. Clarke, M. Konrad, A. J. Nicoll, J. S. Mitchell, A. Calvo, A. Chio, H. Houlden, J. M. Polke, M. A. Ismail, C. E. Stephens, T. Vo, A. A. Farahat, W. D. Wilson, D. W. Boykin, H. Zetterberg, L. Partridge, S. Wray, G. Parkinson, S. Neidle, R. Patani, P. Fratta and A. M. Isaacs, EMBO Mol. Med., 2018, 10, 22–31 CrossRef CAS PubMed.
- A. Cheng, C. Liu, W. Ye, D. Huang, W. She, X. Liu, C. P. Fung, N. Xu, M. C. Suen, W. Ye, H. H. Y. Sung, I. D. Williams, G. Zhu and P.-Y. Qian, J. Med. Chem., 2022, 65, 12825–12837 CrossRef CAS PubMed.
- A. Kovanda, M. Zalar, P. Šket, J. Plavec and B. Rogelj, Sci. Rep., 2015, 5, 17944 CrossRef CAS PubMed.
- T. Shibata, E. Murakami and K. Nakatani, Bioorg. Med. Chem. Lett., 2018, 28, 2364–2368 CrossRef CAS PubMed.
- S. C. Zimmerman and T. J. Murray, Philos. Trans. R. Soc., A, 1993, 345, 49–56 CrossRef CAS.
- B. P. Fors, K. Dooleweerdt, Q. Zeng and S. L. Buchwald, Tetrahedron, 2009, 65, 6576–6583 CrossRef CAS PubMed.
- P. S. Corbin, S. C. Zimmerman, P. A. Thiessen, N. A. Hawryluk and T. J. Murray, J. Am. Chem. Soc., 2001, 123, 10475–10488 CrossRef CAS PubMed.
- J. Rebek, B. Askew, P. Ballester, C. Buhr, S. Jones, D. Nemeth and K. Williams, J. Am. Chem. Soc., 1987, 109, 5033–5035 CrossRef CAS.
- A. D. Hamilton and N. Pant, Chem. Commun., 1988, 765–766 RSC.
- J. Li, A. Sakata, H. He, L.-P. Bai, A. Murata, C. Dohno and K. Nakatani, Chem. – Asian J., 2016, 11, 1971–1981 CrossRef CAS PubMed.
- A. Murata, T. Otabe, J. Zhang and K. Nakatani, ACS Chem. Biol., 2016, 11, 2790–2796 CrossRef CAS PubMed.
- T. Shibata and K. Nakatani, Chem. Commun., 2018, 54, 7074–7077 RSC.
- S. Hagihara, H. Kusama, Y. Goto, G. Hayashi, A. Kobori, I. Saito and K. Nakatani, Nucleic Acids Res., 2004, 32, 278–286 CrossRef CAS PubMed.
- T. Shibata, K. Nagano, M. Ueyama, K. Ninomiya, T. Hirose, Y. Nagai, K. Ishikawa, G. Kawai and K. Nakatani, Nat. Commun., 2021, 12, 236 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cp03213f |
| This journal is © the Owner Societies 2025 |