Open Access Article
Open Access ArticleReaction-free mitochondrial membrane potential independent luminogens with aggregation-induced emission characteristics for live neuron imaging†
Hojeong
Park
 a,
Guangle
Niu
a,
Guangle
Niu
 b,
Alex Y. H.
Wong
b,
Alex Y. H.
Wong
 c,
Eric Y.
Yu
a,
Ryan T. K.
Kwok
ad and
Ben Zhong
Tang
c,
Eric Y.
Yu
a,
Ryan T. K.
Kwok
ad and
Ben Zhong
Tang
 *ade
*ade
aDepartment of Chemistry, Institute for Advanced Study, State Key Laboratory of Neuroscience and Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, China
bSchool of Chemistry and Chemical Engineering, Beijing Institute of Technology, Beijing 100081, China
cMing Wai Lau Centre for Reparative Medicine, Karolinska Institutet, Hong Kong, China
dHKUST-Shenzhen Research Institute, No. 9 Yuexing 1st RD, South Area, Hi-tech Park, Nanshan, Shenzhen 518057, China
eSchool of Science and Engineering, Shenzhen Institute of Aggregate Science and Technology, The Chinese University of Hong Kong, Shenzhen, Guangdong 518172, China
First published on 30th January 2025
Abstract
We developed novel photostable mitochondria targeting probes based on aggregation-induced emission (AIE) luminogens with a cyanostilbene core. The introduction of an alkyl chain onto the pyridinium moiety enhanced their interaction with the mitochondrial membrane. This design effectively prevents probe leakage following mitochondrial membrane depolarization while significantly reducing cytotoxicity.
Mitochondria, evolved by endosymbiosis, play a vital role as a double membrane organelle involved in ATP synthesis by oxidative phosphorylation, intracellular calcium homeostasis, generation of free radical species, and apoptotic cell death.1,2 Dysfunction of the mitochondria has devastating effects on the integrity of cells, which is implicated in aging, metabolic and neurodegenerative diseases, and cancer in higher organisms.3 Tools aiding in the study of the mitochondria are needed to gain a deeper understanding of its pivotal role as a key cellular organelle.
Fluorescence microscopy has been a robust tool for visualising cellular organelles with high sensitivity and temporal/spatial resolution. One way to visualize mitochondria in a live cell is by expressing fluorescent proteins with mitochondrial targeting sequences, such as the cytochrome c oxidase subunit VIII.4–6 Nevertheless, the disadvantage of genetic manipulation is that it requires tedious, long, and expensive biological experiments. In addition, the over-expression of fluorescent protein may interfere with normal biological functions in the mitochondria due to proteomic stress.7 Small molecule fluorescent dyes have become useful tools for labeling mitochondria in both live and dead cells. The staining procedure with chemical dyes is relatively simple and convenient, and unlike fluorescent proteins, they do not require maturation time.8–11 Furthermore, fluorescent dyes show advantageous photophysical properties in terms of higher photostability and brightness while providing tuneable emissions by structural modifications.12 Mitochondrial functions help neurons establish membrane excitability and play a pivotal role in neurotransmission and plasticity.13 Neurons require dyes that are less cytotoxic, especially when compared with cancer cells.
Several cationic and lipophilic dyes have been developed to selectively stain the mitochondria.14 These fluorescent probes can be divided into electrostatic attraction based cationic probes and reaction based MitoTracker probes.15 Tetramethylrhodamine falls under the class of electrostatic attraction based cationic probes.16 The major problem with this class of dye is that its mitochondria targetability is highly dependent on mitochondrial membrane potential (Δψm).17 Δψm fluctuates across the life cycle of an organism, within cells, and mitochondria.18–20 As the Δψm depolarises, the probes leak out from the mitochondria. Therefore, reaction based MitoTracker probes are an attractive alternative which were developed to prevent dyes from leaking out, even if there is a change in Δψm. Cyanine based MitoTracker series are the most frequently used dyes for mitochondrial staining. MitoTracker Deep Red (MTDR) is one of the most frequently utilized probe for visualising mitochondria.17 Unfortunately, MTDR suffers from poor photostability and high concentrations lead to unspecific signals from the cytoplasm. The high cytotoxicity observed in MTDR staining comes from its reaction with cysteine in the mitochondria to form a covalent bond, disrupting important protein functions and morphological abnormalities in addition to cell death.15,21
The general strategy to synthesize plasma membrane probes that can stain the membrane in fixed cells is to increase the hydrophobic interaction between a long alkyl chain and the lipid bilayer of the membrane.22 Inspired by this design strategy, attaching a long alkyl chain to a cationic probe can lead to a long retention time in mitochondria, even if there is Δψm depolarisation. Over the years, several works have been published on generating reaction free mitochondria targeting dyes by incorporating a long alkyl chain onto the pyridinium moiety. A mitochondria targeting probe Mito-MOI with a similar chemical structure to that of ECPI-12 (but with a long alkyl chain attached to the carbazole moiety) was reported to be dependent on Δψm (Fig. 1).23,24 This indicates that the location of the alkyl chain may play a critical role in designing mitochondria targeting dyes that are not influenced by Δψm.
 | ||
| Fig. 1 The chemical structures of MT-PVIM, ECPI-12, IVPI-12, Mito-MOI, CS-Py+, CS-Py-PF6, CS-Py-ClO4, and CS-Py-I. | ||
Although MT-PVIM, ECPI-12, and IVPI-12 demonstrated the concept that a long lipophilic aliphatic chain helps to increase the retention time in mitochondria, it was only investigated in aggregation-caused quenching (ACQ) probes, and not in AIE probes.23–25 Compared to ACQ probes (Fig. 1), AIE probes are non- or weakly emissive in solution but emit strongly in the solid state and as an aggregate. Molecules exhibiting AIE properties have been used extensively in various fields such as biological and chemical probes, and optoelectronic devices.26,27 A highly photostable mitochondria targeting AIE luminogen, CS-Py+, based on cyanostilbene was developed in 2019 for in vitro and ex vivo bioimaging (Fig. 1).28 However, a limitation of this dye is that even though it has a long alkyl chain attached to the cationic probe, its ability to retain in the mitochondria after Δψm depolarization is rather lacking (Fig. S17, ESI†).
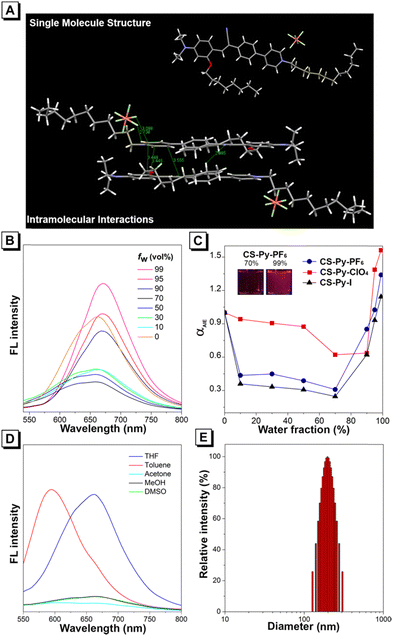
We developed CS-Py-PF6, CS-Py-ClO4, and CS-Py-I; improved versions of CS-Py+ with different counterions by incorporating the alky chain onto the pyridinium moiety to enhance the interaction with the mitochondrial membrane. Detailed synthetic procedures are reported in the ESI.† All the final products were characterized by 1H NMR, 13C NMR, 19F NMR, HRMS, and HPLC to confirm their structures (Scheme S1 and Fig. S1–S10, S21, ESI†). The structure of CS-Py-PF6 was further confirmed by X-ray crystallography (Fig. 2A). Single crystal of CS-Py-PF6 was obtained by slow evaporation in a mixture of CH2Cl2 and MeOH (CH2Cl2/MeOH = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) at room temperature. The molecules are arranged in a head-to-tail arrangement, resulting in strong intramolecular interactions between the donor and acceptor of CS-Py-PF6. Multiple intramolecular interactions were observed, such as C–H⋯N, C–H⋯F, P–F⋯π, and C–H⋯π. The crystal structure of CS-Py-ClO4 and CS-Py-I could not be obtained even after multiple rounds of concentration and solvent system optimization.
1, v/v) at room temperature. The molecules are arranged in a head-to-tail arrangement, resulting in strong intramolecular interactions between the donor and acceptor of CS-Py-PF6. Multiple intramolecular interactions were observed, such as C–H⋯N, C–H⋯F, P–F⋯π, and C–H⋯π. The crystal structure of CS-Py-ClO4 and CS-Py-I could not be obtained even after multiple rounds of concentration and solvent system optimization.
Photophysical properties of CS-Py-PF6, CS-Py-ClO4, and CS-Py-I were investigated, and data is summarized in Table S1 (ESI†). The maximum absorbance of CS-Py-PF6, CS-Py-ClO4, and CS-Py-I are 483, 478, and 468 nm (Fig. S11, ESI†). We evaluated the AIE property of these three compounds by adding different ratios of THF and water to form aggregates (Fig. 2B–D and Fig. S12, ESI†). All three compounds show relatively high fluorescence in pure THF compared to the 70% water fraction. As the water fraction of the solvent system increases to 70%, the fluorescence intensity decreases (Fig. 2C). Because these are intramolecular charge transfer molecules, an increase in the polarity of the solvent system leads to a decrease in fluorescence intensity. However, as the water fraction further increases, the AIE effect dominates. As all three molecules show the highest intensity in 99% water fraction, we confirmed the formation of nanoparticles by measuring the size by dynamic light scattering. The sizes of nanoparticles for CS-Py-PF6, CS-Py-ClO4, and CS-Py-I were 196, 228, and 119 nm, respectively (Fig. 2E and Fig. S13, ESI†).
We investigated the cytotoxicity of CS-Py-PF6, CS-Py-ClO4, CS-Py-I, and MTDR by standard MTT assay under a range of concentrations from 0 to 2.5 μM. The cell viabilities of HeLa cells treated with CS-Py-PF6, CS-Py-ClO4, and CS-Py-I only reduced by approximately 20%, whereas the cell viability of cells treated with MTDR decreased to 51.7% (Fig. 3A). We observed a similar pattern of decrease in cell viability in primary rat hippocampal neurons, indicating that MTDR is more cytotoxic compared to CS-Py-PF6, CS-Py-ClO4, and CS-Py-I (Fig. 4A). Photostability comparisons (Fig. 3B) of these three compounds and MTDR were conducted. As shown, CS-Py-PF6, CS-Py-ClO4, and CS-Py-I exhibit only a 20% decrease in fluorescence intensity after continuous irradiation of light for 16 min. For MTDR, we observed a decrease in fluorescence intensity to 33.8% after continuous light irradiation, making these three newly synthesized probes ideal for live fluorescence imaging. Fluorescence images of HeLa cells indicate that CS-Py-PF6, CS-Py-ClO4, and CS-Py-I can all target mitochondria, which has been confirmed by colocalisation with MTDR (Fig. 3C, D and Fig. S14, S15, ESI†).
Subsequently, we also examined whether CS-Py-PF6, CS-Py-ClO4, and CS-Py-I could stain mitochondria even after mitochondrial membrane depolarization. First, we verified whether our method of decreasing mitochondrial membrane potential was feasible. A ratiometric probe JC-1 was used to monitor changes in mitochondrial membrane potentials. As shown in Fig. S16 (ESI†), cells without CCCP treatment were observed with red, fluorescent J-aggregates in the mitochondria, indicating high Δψm. However, in cells treated with CCCP, the red aggregates were no longer observed in the mitochondria which is a clear sign of mitochondrial depolarization. We performed colocalisation experiment with MTDR in HeLa cells with CCCP. CS-Py-PF6, CS-Py-ClO4, and CS-Py-I show good overlaps with MTDR even with CCCP addition, indicating that they can tolerate the change in mitochondrial membrane potential; unlike their derivative, CS-Py+ (Fig. 3D and Fig. S17, ESI†).
Although the previously developed TPE-based mitochondrial probes, TPE-TPP and TPE-Py effectively targeted mitochondria in cancer cell lines, they failed to do so in primary neurons due to high cytotoxicity (Fig. S18 and S19, ESI†).29–31 This limitation may be attributed to the greater sensitivity of neurons compared to cancer cells.32 In a similar way to Fig. 3C, we performed a colocalisation experiment with MTDR and all three dyes showed good fluorescence intensity overlaps (Fig. 4B). As CS-Py-ClO4, CS-Py-PF6, and CS-Py-I are highly biocompatible, we were keen in demonstrating their use in the long-term live cell imaging, particularly in cell types susceptible to cytotoxicity. We stained live neurons (7 days in vitro) with CS-Py-PF6 as there were no distinct differences in terms of brightness in cells between the three AIE luminogens (Fig. 4C). After 24 h, we observed that neurons stained with MTDR went through a severe morphological change. On the other hand, neurons stained with CS-Py-PF6 did not show any apparent morphological changes. We envision Py-ClO4, CS-Py-PF6, and CS-Py-I as potential replacements for MTDR for live cell imaging.
In conclusion, we developed the highly photostable Δψm-independent and reaction-free mitochondrial probes, CS-Py-PF6, CS-Py-ClO4, and CS-Py-I. Our dyes also show higher cell viability than the leading commercial mitochondria probe MTDR, making them more suitable for live cell imaging and studies in neuroscience. The probes we explicitly designed are derived from a previously reported mitochondria targeting AIE luminogen based on the cyanostilbene core, CS-Py+. This demonstrates for the first time the importance of alkyl chain location for mitochondria membrane interaction by turning Δψm-dependent, CS-Py+ into Δψm-independent probes. We expect that this design strategy can be utilized to benefit both AIE and ACQ probes that are currently being made into mitochondria-targeting dyes.
Hojeong Park: conceptualization, methodology, data curation, visualization, investigation, writing – original draft preparation Guangle Niu: conceptualization, methodology, data curation, supervision, writing – reviewing and editing Alex. Y. H. Wong: data curation, visualization, investigation, writing – original draft preparation Eric Y. Yu data curation, visualization Ryan T. K. Kwok supervision, funding acquisition, writing – reviewing and editing Ben Zhong Tang conceptualization, supervision, funding acquisition, writing – reviewing and editing.
This work was supported by the Research Grant Council of Hong Kong (16303221 and C6014-20W), the Innovation and Technology Commission (ITC-CNERC14SC01 and ITCPD/17-9), and Shenzhen Key Laboratory of Functional Aggregate Materials (ZDSYS20211021111400001).
Data availability
The data that support the findings of this study are openly available in Zenodo with https://doi.org/10.5281/zenodo.14089005.Conflicts of interest
There are no conflicts to declare.Notes and references
- M. W. Gray, G. Burger and B. F. Lang, Genome Biol., 2001, 2, reviews1018.1 CrossRef.
- H. M. McBride, M. Neuspiel and S. Wasiak, Curr. Biol., 2006, 16, R551–R560 CrossRef CAS PubMed.
- D. C. Wallace, Annu. Rev. Genet., 2005, 39, 359–407 CrossRef CAS PubMed.
- R. Rizzuto, P. Pinton, W. Carrington, F. S. Fay, K. E. Fogarty, L. M. Lifshitz, R. A. Tuft and T. Pozzan, Science, 1998, 280, 1763–1766 CrossRef CAS.
- S. Jakobs, Biochim. Biophys. Acta, Mol. Cell Res., 2006, 1763, 561–575 CrossRef CAS.
- M. J. Kim, K. H. Kang, C. H. Kim and S. Y. Choi, Biotechniques, 2008, 45, 331–334 CrossRef CAS PubMed.
- M. S. Gonçalves, Chem. Rev., 2009, 109, 190–212 CrossRef PubMed.
- R. Y. Tsien, Nat. Rev. Mol. Cell Biol., 2003, 16–21 Search PubMed.
- J. Ordóñez-Hernández, D. Ceballos-Ávila, F. H. Real, L. B. Tovar-Y-Romo and A. Jiménez-Sánchez, Chem. Commun., 2024, 60, 5062–5065 RSC.
- C. Hernández-Juárez, G. Morales-Villafaña, F. López-Casillas and A. Jiménez-Sánchez, ACS Sens., 2023, 8, 3076–3085 CrossRef PubMed.
- J. Chen, C. Wang, W. Liu, Q. Qiao, H. Qi, W. Zhou, N. Xu, J. Li, H. Piao, D. Tan, X. Liu and Z. Xu, Angew. Chem., Int. Ed., 2021, 60, 25104–25113 CrossRef CAS PubMed.
- L. D. Lavis and R. T. Raines, ACS Chem. Biol., 2008, 3, 142–155 CrossRef CAS PubMed.
- O. Kann and R. Kovács, Am. J. Physiol., 2007, 292, C641–C657 CrossRef CAS PubMed.
- G. Battogtokh, Y. S. Choi, D. S. Kang, S. J. Park, M. S. Shim, K. M. Huh, Y. Y. Cho, J. Y. Lee, H. S. Lee and H. C. Kang, Acta Pharm. Sin. B, 2018, 8, 862–880 CrossRef PubMed.
- K. Neikirk, A. G. Marshall, B. Kula, N. Smith, S. LeBlanc and A. Hinton, Eur. J. Cell Biol., 2023, 102, 151371 CrossRef CAS PubMed.
- C. Lee, D. C. Wallace and P. J. Burke, ACS Nano, 2024, 18, 1345–1356 CrossRef CAS PubMed.
- S. W. Perry, J. P. Norman, J. Barbieri, E. B. Brown and H. A. Gelbard, Biotechniques, 2011, 50, 98–115 CrossRef CAS PubMed.
- R. A. Gottlieb, H. Piplani, J. Sin, S. Sawaged, S. M. Hamid, D. J. Taylor and J. de Freitas Germano, Cell. Mol. Life Sci., 2021, 78, 3791–3801 CrossRef CAS PubMed.
- U. AL-Zubaidi, J. Liu, O. Cinar, R. L. Robker, D. Adhikari and J. Carroll, Mol. Hum. Reprod., 2019, 25, 695–705 CrossRef CAS PubMed.
- D. M. Wolf, M. Segawa, A. K. Kondadi, R. Anand, S. T. Bailey, A. S. Reichert, A. M. van der Bliek, D. B. Shackelford, M. Liesa and O. S. Shirihai, EMBO J., 2019, 38, e101056 CrossRef CAS PubMed.
- C. Sargiacomo, S. Stonehouse, Z. Moftakhar, F. Sotgia and M. P. Lisanti, Front Oncol, 2021, 11, 678343 CrossRef CAS PubMed.
- M. Collot, P. Ashokkumar, H. Anton, E. Boutant, O. Faklaris, T. Galli, Y. Mely, L. Danglot and A. S. Klymchenko, Cell Chem Biol, 2019, 26, 600–614 CrossRef CAS PubMed.
- R. Zhang, Y. Sun, M. Tian, G. Zhang, R. Feng, X. Li, L. Guo, X. Yu, J. Z. Sun and X. He, Anal. Chem., 2017, 89, 6575–6582 CrossRef CAS PubMed.
- R. Zhang, G. Niu, X. Li, L. Guo, H. Zhang, R. Yang, Y. Chen, X. Yu and B. Z. Tang, Chem. Sci., 2019, 10, 1994–2000 RSC.
- Y. Liu, F. Meng, Y. Tang, X. Yu and W. Lin, New J. Chem., 2016, 40, 3726–3731 RSC.
- G. Niu, R. Zhang, X. Shi, H. Park, S. Xie, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, TrAC, Trends Anal. Chem., 2020, 123, 115769 CrossRef CAS.
- X. He, H. Xie, L. Hu, P. Liu, C. Xu, W. He, W. Du, S. Zhang, H. Xing, X. Liu, H. Park, T. S. Cheung, M.-H. Li, R. T. K. Kwok, J. W. Y. Lam, J. Lu and B. Z. Tang, Aggregate, 2023, 4, e239 CrossRef CAS.
- G. Niu, R. Zhang, Y. Gu, J. Wang, C. Ma, R. T. K. Kwok, J. W. Y. Lam, H. H. Y. Sung, I. D. Williams, K. S. Wong, X. Yu and B. Z. Tang, Biomaterials, 2019, 208, 72–82 CrossRef CAS PubMed.
- C. W. T. Leung, Y. Hong, S. Chen, E. Zhao, J. W. Y. Lam and B. Z. Tang, J. Am. Chem. Soc., 2013, 135, 62–65 CrossRef CAS PubMed.
- J. Qian, Z. Zhu, C. W. T. Leung, W. Xi, L. Su, G. Chen, A. Qin, B. Z. Tang and S. He, Biomed. Opt. Express, 2015, 6, 1477–1486 CrossRef CAS PubMed.
- N. Zhao, M. Li, Y. Yan, J. W. Y. Lam, Y. L. Zhang, Y. S. Zhao, K. S. Wong and B. Z. Tang, J. Mater. Chem. C, 2013, 1, 4640–4646 RSC.
- H. Park, G. Niu, C. Wu, C. Park, H. Liu, H. Park, R. T. K. Kwok, J. Zhang, B. He and B. Z. Tang, Chem. Sci., 2022, 13, 2965–2970 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, NMR and HRMS spectra, photophysical data, and imaging data. CCDC 2402029. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4cc06022a |
| This journal is © The Royal Society of Chemistry 2025 |