 Open Access Article
Open Access ArticleAutomated fast-flow synthesis of the immune checkpoint receptors PD-1 and PD-L1†
Giulio
Fittolani
a,
Alex J.
Callahan
a,
Andrei
Loas
 a and
Bradley L.
Pentelute
a and
Bradley L.
Pentelute
 *abcd
*abcd
aDepartment of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA. E-mail: blp@mit.edu
bThe Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, 500 Main Street, Cambridge, MA 02142, USA
cCenter for Environmental Health Sciences, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA
dBroad Institute of MIT and Harvard, 415 Main Street, Cambridge, MA 02142, USA
First published on 17th March 2025
Abstract
Programmed cell death protein 1 (PD-1) and programmed cell death ligand 1 (PD-L1) are key targets for cancer therapy. Here, we use automated fast-flow peptide synthesis (AFPS) to rapidly produce these challenging β-sheet-rich proteins in their active forms following oxidative refolding protocols. The methods presented here provide rapid access to synthetic, air-stable mutants of PD-1 and PD-L1 in which L-methionine residues are substituted with L-norleucine, potentially enabling investigation of post-translational modifications and mirror-image analogs for drug discovery.
The programmed cell death protein 1 (PD-1) and programmed cell death ligand 1 (PD-L1) are the essential components of the PD-1/PD-L1 immune regulatory checkpoint pathway.1 PD-1 is located on the surface of T cells and B cells, whereas PD-L1 is expressed in a variety of cells and tissues. When T cell receptors (TCRs) engage with antigen presenting cells through the major histocompatibility complex (MHC), the PD-1/PD-L1 interaction functions as an immune checkpoint inhibiting T cell activation and maintaining self-tolerance.2 Cancer cells overexpressing PD-L1 can hijack this pathway and effectively escape immune surveillance by suppressing the action of T cells.
Immunotherapies directed at inhibiting the PD-1/PD-L1 pathway have significantly advanced cancer treatment modalities over the past decade.3,4 Monoclonal antibodies (mAbs) blocking PD-1, or its ligand PD-L1, disrupt the immunosuppressive PD-1/PD-L1 pathway restoring T cell response against the tumor.4 Despite the success of mAbs as immune checkpoint inhibitors, there is a lack of alternative therapeutic strategies that overcome the typical disadvantages of mAbs such as high cost, no oral bioavailability, immune-related adverse effects, and limited solid-tumor tissue penetration.5,6 Peptide therapeutics have the potential to overcome some of these limitations while preserving target affinity and specificity.7 Mirror-image phage display techniques facilitate the discovery of D-peptides, which offer greater stability and reduced immunogenicity compared to their L-peptide counterparts.8–11 Currently, the only method to produce D-proteins is through chemical synthesis. Developing a rapid and reliable method for synthesizing mirror-image PD-1 and PD-L1 could unlock the discovery of D-peptide immune checkpoint inhibitors. Furthermore, chemical synthesis of glycosylated isoforms of PD-1 could provide a deeper understanding of N-glycosylation, its impact on binding to PD-L1 and its recognition by anti PD-1 mAbs.12,13 The methodologies developed thus far for the chemical synthesis of PD-1 and PD-L1 rely on a combination of solid phase peptide synthesis (SPPS) of peptide fragments and multiple native chemical ligation (NCL) reactions.14,15 In some cases, the use of specific temporary protecting groups designed to inhibit aggregation was necessary to purify intermediates.14 The use of permanent cationic tags (i.e., Arg8 tag) was in some cases required15 to enhance solubility.
In this work, we utilized the automated fast-flow peptide synthesis (AFPS)8,16–19 technology established in our laboratory to rapidly access synthetic PD-1 and PD-L1 ectodomains. The two synthetic proteins were synthesized in a single-shot manner, i.e., a single continuous stepwise synthesis on the solid support, thus bypassing the use of NCL or solubilizing tags. Oxidative refolding protocols were utilized to obtain PD-1 and PD-L1 in their active form as verified by biochemical assays.
The key interaction between human PD-1 and PD-L1 occurs through their immunoglobulin V (IgV)-like domains in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex (Fig. 1A).20 We selected for synthesis residues 33–150 of human PD-1 and residues 18–133 of human PD-L1, both corresponding to the IgV-like domains stabilized by one disulfide bond each. FDA-approved mAbs target diverse locations on the surface of PD-1 and PD-L1 IgV-like domains, making these fragments ideal targets for chemical synthesis.21,22
1 complex (Fig. 1A).20 We selected for synthesis residues 33–150 of human PD-1 and residues 18–133 of human PD-L1, both corresponding to the IgV-like domains stabilized by one disulfide bond each. FDA-approved mAbs target diverse locations on the surface of PD-1 and PD-L1 IgV-like domains, making these fragments ideal targets for chemical synthesis.21,22
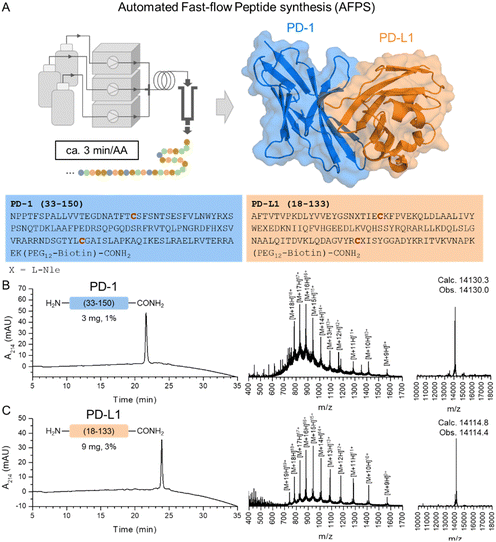 | ||
| Fig. 1 AFPS enables rapid access to synthetic PD-1 and PD-L1. (A) The PD-1/PD-L1 complex (pdb 4ZQK) (left). Amino acid sequence of PD-1 (Uniprot ID Q15116, residues 33–150) and PD-L1 (Uniprot ID Q9NZQ7, residues 18–133) (right). The Cys residues involved in disulfide linkages are highlighted in red. All proteins were synthesized as C-terminal amide and with a biotin-PEG12 handle at the C-terminus. (B) Analytical HPLC trace of purified PD-1 (left). Mass spectrum of purified PD-1 (middle). Deconvoluted MS of purified PD-1 (right). (C) Analytical HPLC trace of purified PD-L1 (left). Mass spectrum of purified PD-L1 (middle). Deconvoluted MS of purified PD-L1 (right). For more details see Section S3 of the ESI.† | ||
The chemical synthesis of PD-1 and PD-L1 is challenging due to solubility issues of the final product or the intermediate peptide fragments required for NCL.14,15 Current methodologies to chemically synthesize PD-1 involve the use of multiple NCL steps, desulfurization, protecting group removal, and require the use of specific solubilizing tags to facilitate HPLC purifications.14 The reported chemical synthesis of PD-L1 relied on a convergent strategy using 4 peptide fragments condensed using NCLs followed by desulfurization and acetamidomethyl (Acm) removal.15 Furthermore, the use of an Arg8 tag was essential to enable sufficient solubility of the final product.
We hypothesized that the AFPS technology available in our laboratory could streamline the access to these proteins in a single-shot format. The high temperature used throughout the synthesis could prevent on-resin aggregation and the single continuous stepwise synthesis process obviates the need for isolation and handling of aggregation-prone peptide fragments. In order to enable surface immobilization using streptavidin-coated beads, we installed a biotin-PEG12 handle at the C-termini of the synthetic proteins. The PEG12 linker enables sufficient spacing between the protein chain and the resin surface and is installed at the C-terminus to prevent any interference with ligand binding at the PD-1/PD-L1 interface (Fig. S2, ESI†).8 We synthesized PD-1 and PD-L1 IgV-like domains by AFPS and purified them using preparative HPLC, obtaining them in 1% and 3% isolated yield, respectively (Fig. 1B and C, see Section S3 of the ESI†). All methionine residues were mutated to L-norleucine (L-Nle) to prevent methionine oxidation during handling and refolding. We also introduced a C93S mutation on PD-1 to aid protein stability as previously reported.20 Aspartimide formation was minimized using formic acid as an additive to the piperidine deprotection solution as previously reported.16
Refolding of PD-1 and PD-L1 was carried out using oxidative refolding protocols.20 Purified synthetic PD-1 was dissolved in a denaturing buffer (typically 6 M Gdn·HCl, 100 mM TRIS·HCl, pH 8.0), diluted in a redox refolding buffer (containing cysteine and cystine), and then dialyzed against a TRIS buffer prior to purification using size exclusion chromatography (SEC) (Fig. 2A, top). In the case of PD-L1, we adopted a slow dialysis refolding protocol allowing reduced aggregate formation when compared to dilution refolding (Fig. S8, ESI†). The optimized protocol for PD-L1 refolding involved dissolving the purified protein in a denaturing buffer and then subjecting it to a two-step dialysis involving a redox buffer (containing GSH and GSSG) (Fig. 2A, bottom). Final SEC purification afforded folded PD-L1. We confirmed the purity of isolated refolded PD-1 and PD-L1 using SDS-PAGE (Fig. 2B) and analytical HPLC, in which a distinct change in retention time was observed upon formation of the disulfide bond (Fig. S4 and S9, ESI†). Upon folding, a distinct change in the mass envelope was observed for both PD-1 and PD-L1 (Fig. 2C) which was accompanied by a loss of 2 Da in the deconvoluted mass spectrum matching with the expected formation of a disulfide bond (see Fig. S7 and S12, ESI†). Importantly, analytical SEC confirmed that both PD-1 and PD-L1 were isolated as monomeric forms (Fig. 2D).
 | ||
| Fig. 2 Oxidative refolding yields active forms of synthetic PD-1 and PD-L1. (A) Refolding protocols used for synthetic PD-1 and PD-L1. (B) SDS-PAGE of synthetic PD-1 and PD-L1. Molecular weight is expressed in kDa. (C) Mass-to-charge spectrum of refolded PD-1 and PD-L1. See Section 4.2 of the ESI.† (D) Analytical SEC of refolded PD-1 and PD-L1 superimposed to a molecular weight standard (Superdex 75 increase 5/150 GL, flow rate 0.25 mL min−1, running buffer: 50 mM HEPES, 150 mM NaCl, pH 7.5). Molecular weight is expressed in kDa. (E) BLI assay of synthetic PD-1 with recombinant PD-L1 (left) and synthetic PD-L1 with Atezolizumab (anti-PD-L1) (right). Synthetic PD-1 and PD-L1 are functionalized with a PEG12-biotin handle for immobilization on the surface of the BLI sensor tip. Concentrations refer to the recombinant protein. Further details are reported in Section S4.3 of the ESI.† | ||
Refolded synthetic PD-1 and PD-L1 were characterized with biophysical binding assays to confirm their bioactive conformation. First, to validate synthetic refolded PD-1 we investigated its binding affinity to recombinant PD-L1 protein. We performed a biolayer interferometry (BLI) assay by immobilizing biotinylated synthetic PD-1 on the surface of a sensor tip coated with streptavidin and then determining the apparent dissociation constant (KD) with recombinant PD-L1 (Fig. 2E, left and Fig. S14, ESI†). In the BLI assay, we observed rapid association and dissociation events, which reached equilibrium within few seconds (Fig. 2E, left). This observation is in line with previous reports on the binding kinetics of the PD-1/PD-L1 complex.23 We calculated a KD value of 3.2 μM from the BLI assay using steady-state analysis which is comparable to previously reported values (KD ∼ 8 μM, determined using a surface plasmon resonance assay).23,24 Second, we analyzed the ability of synthetic refolded PD-L1 to bind recombinant PD-1 protein using a BLI assay. We immobilized the synthetic biotinylated PD-L1 on the sensor tip and assayed it against varying concentrations of recombinant PD-1. As observed in the previous BLI assay, the binding between synthetic PD-L1 and recombinant PD-1 displayed rapid association and dissociation kinetics and we derived a KD of 11.0 μM using steady-state analysis (Fig. S15 and S16, ESI†). The slight difference between the two measured binding affinities could be a result of N-glycosylation of recombinant PD-1 used for the assay. PD-1 N-glycosylation has been reported to have a regulatory function in the PD-1/PD-L1 pathway13,25 and interferes with binding of clinical mAbs.26,27 To further validate the function of synthetic PD-L1 we performed a BLI assay to assess binding of synthetic PD-L1 to Atezolizumab, an anti-PD-L1 mAb (Fig. 2E, right). Dissociation kinetics of Atezolizumab with immobilized synthetic PD-L1 were significantly slower compared to PD-1 binding (Fig. 2E and Fig. S17, ESI†). The dissociation constant for the synthetic PD-L1/Atezolizumab complex was measured to be in the picomolar range using binding curve fit analysis. However, the KD could not be accurately determined due to the antibody slow off-rate posing challenges in the determination of the koff constant.21,26 Crystal structure data indicate that the epitope of Atezolizumab on PD-L1 overlaps with that of its natural binding partner PD-1.21 Thus, Atezolizumab binding to synthetic PD-L1 supports the conclusion that the synthetic protein is folded in an active conformation.
In summary, we have developed a rapid synthetic approach, using AFPS, to access two key immune-checkpoint receptors PD-1 (14.2 kDa) and PD-L1 (14.2 kDa) in their active conformations. The single-shot format of the synthesis bypassed the need for multiple chemical ligation steps and the use of solubilizing tags. Oxidative refolding protocols gave access to PD-1 and PD-L1 in their biologically active form as validated with biochemical binding assays. The protocols developed here can be applied to the production of PD-1 and PD-L1 in their mirror-image form for use in screening campaigns aimed at discovering D-peptide inhibitors such as mirror-image phage display.8–11 Inhibitors based on D-peptides could provide an alternative strategy to block the PD-1/PD-L1 pathway, exhibiting lower production cost compared to mAbs, longer in vivo half-lives and reduced immunogenicity relative to L-peptides.6 Furthermore, rapid access to synthetic immune checkpoint proteins facilitates the preparation of N-glycosylated isoforms for further study.
G. F. acknowledges support from a postdoctoral fellowship from the Ludwig Center at MIT's Koch Institute for Integrative Cancer Research.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
B. L. P. is a co-founder and/or member of the scientific advisory board of several companies focusing on the development of protein and peptide therapeutics. The other authors declare no competing financial interest.Notes and references
- L. Chen and D. B. Flies, Nat. Rev. Immunol., 2013, 13, 227–242 CrossRef PubMed.
- A. H. Sharpe and K. E. Pauken, Nat. Rev. Immunol., 2018, 18, 153–167 CrossRef CAS PubMed.
- S. L. Topalian, C. G. Drake and D. M. Pardoll, Cancer Cell, 2015, 27, 450–461 CrossRef CAS PubMed.
- A. Dömling and T. A. Holak, Angew. Chem., Int. Ed., 2014, 53, 2286–2288 CrossRef PubMed.
- P. Chames, M. Van Regenmortel, E. Weiss and D. Baty, Br. J. Pharmacol., 2009, 157, 220–233 CrossRef CAS PubMed.
- M. Konstantinidou, T. Zarganes-Tzitzikas, K. Magiera-Mularz, T. A. Holak and A. Dömling, Angew. Chem., Int. Ed., 2018, 57, 4840–4848 CrossRef CAS PubMed.
- T. Kremsmayr, A. Aljnabi, J. B. Blanco-Canosa, H. N. T. Tran, N. B. Emidio and M. Muttenthaler, J. Med. Chem., 2022, 65, 6191–6206 CrossRef CAS PubMed.
- A. J. Callahan, S. Gandhesiri, T. L. Travaline, R. M. Reja, L. Lozano Salazar, S. Hanna, Y.-C. Lee, K. Li, O. S. Tokareva, J.-M. Swiecicki, A. Loas, G. L. Verdine, J. H. McGee and B. L. Pentelute, Nat. Commun., 2024, 15, 1813 CrossRef CAS PubMed.
- T. N. M. Schumacher, L. M. Mayr, D. L. Minor, M. A. Milhollen, M. W. Burgess and P. S. Kim, Science, 1996, 271, 1854–1857 CrossRef CAS PubMed.
- Y.-K. Qi, J.-S. Zheng and L. Liu, Chemistry, 2024, 10, 2390–2407 CrossRef CAS.
- K. Harrison, A. S. Mackay, L. Kambanis, J. W. C. Maxwell and R. J. Payne, Nat. Rev. Chem., 2023, 7, 383–404 CrossRef CAS PubMed.
- K. Liu, S. Tan, W. Jin, J. Guan, Q. Wang, H. Sun, J. Qi, J. Yan, Y. Chai, Z. Wang, C. Deng and G. F. Gao, EMBO Rep., 2020, 21, 1–12 Search PubMed.
- L. Sun, C.-W. Li, E. M. Chung, R. Yang, Y.-S. Kim, A. H. Park, Y.-J. Lai, Y. Yang, Y.-H. Wang, J. Liu, Y. Qiu, K.-H. Khoo, J. Yao, J. L. Hsu, J.-H. Cha, L.-C. Chan, J.-M. Hsu, H.-H. Lee, S. S. Yoo and M.-C. Hung, Cancer Res., 2020, 80, 2298–2310 CrossRef CAS PubMed.
- H. Wu, T. Wei, W. L. Ngai, H. Zhou and X. Li, J. Am. Chem. Soc., 2022, 144, 14748–14757 CrossRef CAS PubMed.
- H. N. Chang, B. Y. Liu, Y. K. Qi, Y. Zhou, Y. P. Chen, K. M. Pan, W. W. Li, X. M. Zhou, W. W. Ma, C. Y. Fu, Y. M. Qi, L. Liu and Y. F. Gao, Angew. Chem., Int. Ed., 2015, 54, 11760–11764 CrossRef CAS PubMed.
- N. Hartrampf, A. Saebi, M. Poskus, Z. P. Gates, A. J. Callahan, A. E. Cowfer, S. Hanna, S. Antilla, C. K. Schissel, A. J. Quartararo, X. Ye, A. J. Mijalis, M. D. Simon, A. Loas, S. Liu, C. Jessen, T. E. Nielsen and B. L. Pentelute, Science, 2020, 368, 980–987 CrossRef CAS PubMed.
- M. Jbara, S. Pomplun, C. K. Schissel, S. W. Hawken, A. Boija, I. Klein, J. Rodriguez, S. L. Buchwald and B. L. Pentelute, J. Am. Chem. Soc., 2021, 143, 11788–11798 CrossRef CAS PubMed.
- S. Pomplun, M. Jbara, C. K. Schissel, S. Wilson Hawken, A. Boija, C. Li, I. Klein and B. L. Pentelute, ACS Cent. Sci., 2021, 7, 1408–1418 CrossRef CAS PubMed.
- A. Saebi, J. S. Brown, V. M. Marando, N. Hartrampf, N. M. Chumbler, S. Hanna, M. Poskus, A. Loas, L. L. Kiessling, D. T. Hung and B. L. Pentelute, ACS Chem. Biol., 2023, 18, 518–527 CrossRef CAS PubMed.
- K. M. Zak, R. Kitel, S. Przetocka, P. Golik, K. Guzik, B. Musielak, A. Dömling, G. Dubin and T. A. Holak, Structure, 2015, 23, 2341–2348 CrossRef CAS PubMed.
- S. Tan, K. Liu, Y. Chai, C. W. H. Zhang, S. Gao, G. F. Gao and J. Qi, Protein Cell, 2018, 9, 135–139 CrossRef CAS PubMed.
- H. T. Lee, J. Y. Lee, H. Lim, S. H. Lee, Y. J. Moon, H. J. Pyo, S. E. Ryu, W. Shin and Y.-S. Heo, Sci. Rep., 2017, 7, 5532 CrossRef PubMed.
- X. Cheng, V. Veverka, A. Radhakrishnan, L. C. Waters, F. W. Muskett, S. H. Morgan, J. Huo, C. Yu, E. J. Evans, A. J. Leslie, M. Griffiths, C. Stubberfield, R. Griffin, A. J. Henry, A. Jansson, J. E. Ladbury, S. Ikemizu, M. D. Carr and S. J. Davis, J. Biol. Chem., 2013, 288, 11771–11785 CrossRef CAS PubMed.
- D. Y. Lin, Y. Tanaka, M. Iwasaki, A. G. Gittis, H.-P. Su, B. Mikami, T. Okazaki, T. Honjo, N. Minato and D. N. Garboczi, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3011–3016 CrossRef CAS PubMed.
- C.-W. Li, S.-O. Lim, W. Xia, H.-H. Lee, L.-C. Chan, C.-W. Kuo, K.-H. Khoo, S.-S. Chang, J.-H. Cha, T. Kim, J. L. Hsu, Y. Wu, J.-M. Hsu, H. Yamaguchi, Q. Ding, Y. Wang, J. Yao, C.-C. Lee, H.-J. Wu, A. A. Sahin, J. P. Allison, D. Yu, G. N. Hortobagyi and M.-C. Hung, Nat. Commun., 2016, 7, 12632 CrossRef CAS PubMed.
- J. Benicky, M. Sanda, Z. Brnakova Kennedy, O. C. Grant, R. J. Woods, A. Zwart and R. Goldman, J. Proteome Res., 2021, 20, 485–497 CrossRef CAS PubMed.
- H. H. Lee, Y. N. Wang, W. Xia, C. H. Chen, K. M. Rau, L. Ye, Y. Wei, C. K. Chou, S. C. Wang, M. Yan, C. Y. Tu, T. C. Hsia, S. F. Chiang, K. S. C. Chao, I. I. Wistuba, J. L. Hsu, G. N. Hortobagyi and M. C. Hung, Cancer Cell, 2019, 36, 168–178.e4 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: General methods and materials, additional experimental details on protein synthesis and refolding, analytical characterization. See DOI: https://doi.org/10.1039/d4cc05982d |
| This journal is © The Royal Society of Chemistry 2025 |
