Open Access Article
Open Access ArticleVisible light-mediated photocatalytic coupling between tetrazoles and carboxylic acids for biomolecule labelling†
Takuro
Matsuoka
a,
Ryosuke
Takasaki
a,
Hiroki
Akiba
 ab,
Kosuke
Ogata
ab,
Kosuke
Ogata
 a,
Akira
Hattori
a,
Akira
Hattori
 a,
Norihito
Arichi
a,
Norihito
Arichi
 a,
Hideaki
Kakeya
a,
Hideaki
Kakeya
 a,
Sho
Yamasaki
cd,
Yasushi
Ishihama
a,
Sho
Yamasaki
cd,
Yasushi
Ishihama
 ab,
Hiroaki
Ohno
ab,
Hiroaki
Ohno
 *ab and
Shinsuke
Inuki
*ab and
Shinsuke
Inuki
 *ae
*ae
aGraduate School of Pharmaceutical Sciences, Kyoto University, Sakyo-ku, Kyoto, Kyoto 606-8501, Japan
bCenter for Drug Design Research, National Institutes of Biomedical Innovation, Health and Nutrition, Ibaraki, Osaka 567-0085, Japan
cResearch Institute for Microbial Diseases, Osaka University, Suita, Osaka 565-0871, Japan
dImmunology Frontier Research Center (IFReC), Osaka University, Suita, Osaka 565-0871, Japan
eGraduate School of Biomedical Sciences, Tokushima University, Tokushima, Tokushima 770-8505, Japan
First published on 24th March 2025
Abstract
Photocatalytic biomolecular labelling is gaining attention as a foundational technique for analyzing biological phenomena. However, photocatalytic reactions compatible with physiological conditions remain limited. Here, we present a photocatalytic reaction of diaryltetrazoles to generate nitrile imines, which readily couple with carboxylic acids in aqueous environments. This reaction is applied for photocatalyst-dependent labelling of proteins and cells.
Detecting and tracking biomolecular interactions are essential for a deep understanding of biological phenomena,1 leading to the development of numerous biological and chemical techniques.2 Proximity labelling is a robust method that enables profiling physiological interactions with high spatial and temporal precision.3 This technique facilitates the selective labelling of biomolecules near a target of interest within the cellular milieu. Enzyme-catalysed labelling platforms, such as BioID4 and APEX5 have recently been established. However, these require genetic engineering or specific surface glycans to introduce cell-tagging enzymes. Photocatalytic biomolecular labelling has also garnered attention for proximity labelling owing to its simplicity and versatility.6 Photocatalysts can be placed at a desired position on the cell; then, light irradiation triggers the production of reactive intermediates that covalently modify proximal molecules with a label, e.g., biotin tag or fluorescent dye, thereby enabling monitoring and enrichment of the target molecules or cells. In this method, the lifetime and reactivity of the reactive intermediate are crucial for controlling labelling radius or efficiency. Several photocatalytic reactions that enable labelling under physiological conditions have been developed using singlet oxygen,6a carbenes6b or phenoxy radicals6c as reactive intermediates. However, there are limitations in terms of labelling efficiency and the functional groups in biomolecules that can be labelled. Thus, further exploration of reaction types is required to facilitate the analysis of various targets in cells and tissues. To develop a photocatalytic reaction enabling protein and cell labelling under physiological condition, we focused on biomolecular labelling using photolysis of diaryltetrazole.
Diaryltetrazoles can be excited to triplet states under UV irradiation and then converted to nitrile imines via N2 elimination (Fig. 1).7 Nitrile imines are highly reactive species that mainly react with alkenes to afford pyrazoline cycloadducts or with carboxylic acids to form hydrazide via 1,5-acyl shift.8 Because carboxylic acid moieties are abundant in biomolecules, this type of reaction has been applicable to various photoaffinity-based labelling methods, e.g., drug target identification and proteome-wide profiling.9 However, despite the utility of nitrile imines, photocatalytic production of nitrile imine has not been explored. We speculated that tetrazoles could be excited via energy transfer from an excited photocatalyst,10 rather than UV irradiation, and could serve as an efficient photocatalytic labelling agent for biomolecules (Fig. 1 and Fig. S1, ESI†). Herein, we have developed a visible light-mediated photocatalytic coupling reaction between tetrazoles and carboxylic acids and applied it for protein and cell labelling.
First, we sought tetrazole derivatives that could be converted to nitrile imines in the presence of a photocatalyst under visible-light irradiation. Various 2,5-disubstituted tetrazoles were investigated in the reaction with 3-phenylpropionic acid and Ir[dF(CF3)ppy]2(dtbpy)PF6 in H2O/MeCN (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (Table 1). Tetrazole 1a, which is known to photodegrade under UV irradiation,8c did not give the desired hydrazide. Methoxy benzene-containing tetrazole 1b afforded the desired carboxylic acid adduct in 37% yield.8c Notably, the desired adduct was not detected without photocatalyst. In general, energy transfer occurs when the triplet excitation energy of the acceptor is sufficiently lower than that of the photocatalyst, i.e., the energy donor.10 Blasco et al. showed that extending the conjugation system of the substituent at the 2-position of the tetrazole lowers the triplet excitation energy.11 Thus, we synthesized tetrazoles 1c–g with extended conjugation systems (Schemes S1–S3, ESI†), and investigated their reactivity. Tetrazoles 1c and 1d with 1-naphthyl and 5-isoquinolinyl groups at the 2-positions gave the desired carboxylic acid adducts 3c and 3d in 70% and 73% yields, respectively. Density functional theory calculations indicated that the triplet excitation energies of 1c and 1d were nearly 10 kcal mol−1 lower than those of 1a and 1b (Table 1). Only trace amounts of hydrazide were detected when 1c and 1d were irradiated with LEDs in the absence of a photocatalyst. An 8-quinolinyl group at the 2-position (1e) or a carbamoyl group at the 5-position (1f) had a negative effect on the photocatalytic reaction. Introducing 1-naphthyl group at the 5-position (1g) resulted in low conversion to the desired adduct, likely due to the poor solubility of 1g. These results indicated that tetrazoles with a 1-naphthyl or 5-isoquinolinyl group at the 2-position and a phenyl group at the 5-position were optimal for photocatalyst-dependent nitrile imine formation. Additionally, a comparison of photocatalysts revealed that Ir[dF(CF3)ppy]2(dtbpy)PF6 (ET = 60.1 kcal mol−1) was well suited for tetrazole excitation (ET of 1d = 51.95 kcal mol−1) (Table S1, ESI†). From a mechanistic perspective, the UV-Vis spectra showed that 1b and 1d absorbed only below 340 nm, indicating that the direct photolysis of tetrazoles is unlikely (Fig. S2, ESI†). Stern–Volmer experiments demonstrated that the tetrazole quenched the luminescence of the photocatalyst (Fig. S3, ESI†). The reactions proceeded efficiently when using tetrazoles with sufficiently low triplet excitation energies compared to the Ir catalyst (Table 1 and Fig. S2, ESI†). These results suggested that the generation of nitrile imine takes place via energy transfer to tetrazoles.
1) (Table 1). Tetrazole 1a, which is known to photodegrade under UV irradiation,8c did not give the desired hydrazide. Methoxy benzene-containing tetrazole 1b afforded the desired carboxylic acid adduct in 37% yield.8c Notably, the desired adduct was not detected without photocatalyst. In general, energy transfer occurs when the triplet excitation energy of the acceptor is sufficiently lower than that of the photocatalyst, i.e., the energy donor.10 Blasco et al. showed that extending the conjugation system of the substituent at the 2-position of the tetrazole lowers the triplet excitation energy.11 Thus, we synthesized tetrazoles 1c–g with extended conjugation systems (Schemes S1–S3, ESI†), and investigated their reactivity. Tetrazoles 1c and 1d with 1-naphthyl and 5-isoquinolinyl groups at the 2-positions gave the desired carboxylic acid adducts 3c and 3d in 70% and 73% yields, respectively. Density functional theory calculations indicated that the triplet excitation energies of 1c and 1d were nearly 10 kcal mol−1 lower than those of 1a and 1b (Table 1). Only trace amounts of hydrazide were detected when 1c and 1d were irradiated with LEDs in the absence of a photocatalyst. An 8-quinolinyl group at the 2-position (1e) or a carbamoyl group at the 5-position (1f) had a negative effect on the photocatalytic reaction. Introducing 1-naphthyl group at the 5-position (1g) resulted in low conversion to the desired adduct, likely due to the poor solubility of 1g. These results indicated that tetrazoles with a 1-naphthyl or 5-isoquinolinyl group at the 2-position and a phenyl group at the 5-position were optimal for photocatalyst-dependent nitrile imine formation. Additionally, a comparison of photocatalysts revealed that Ir[dF(CF3)ppy]2(dtbpy)PF6 (ET = 60.1 kcal mol−1) was well suited for tetrazole excitation (ET of 1d = 51.95 kcal mol−1) (Table S1, ESI†). From a mechanistic perspective, the UV-Vis spectra showed that 1b and 1d absorbed only below 340 nm, indicating that the direct photolysis of tetrazoles is unlikely (Fig. S2, ESI†). Stern–Volmer experiments demonstrated that the tetrazole quenched the luminescence of the photocatalyst (Fig. S3, ESI†). The reactions proceeded efficiently when using tetrazoles with sufficiently low triplet excitation energies compared to the Ir catalyst (Table 1 and Fig. S2, ESI†). These results suggested that the generation of nitrile imine takes place via energy transfer to tetrazoles.
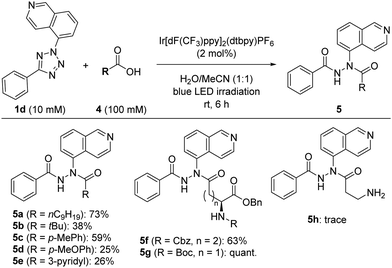
Using the optimized conditions for photocatalytic modification in aqueous solvents, we examined the scope of carboxylic acid substrates that react with photocatalytically-generated nitrile imines (Fig. 2) to assess suitability for protein or cell labelling. The reaction with a medium-length saturated fatty acid gave hydrazide 5a in 73% yield. The sterically-bulky pivalic acid led to a low yield of the carboxylic acid adduct 5b. Coupling with aromatic p-toluic acid gave the desired compound 5c in moderate yield, whereas the yields were much lower when 4-methoxy benzoic acid (5d) or nicotinic acid (5e) was used. Next, we investigated the photoreaction with proteinogenic amino acid derivatives to potentially expand the application scope to protein labelling. The glutamic acid and aspartic acid derivatives with unprotected side-chain carboxyl groups gave the desired compounds 5f and 5g, respectively, in moderate to good yields. In contrast, when β-alanine was used as a substrate, only a small amount of the desired adduct (5h) was detected. These results suggest structural limitations for carboxylic acids in this coupling reaction. Nevertheless, photocatalytically-generated nitrile imines can readily react with the side chains of proteinogenic acidic amino acids, such as glutamic acid and aspartic acid, thus highlighting the potential of this reaction for protein labelling applications.
To evaluate the applicability of this reaction to protein labelling, we tested whether tetrazoles could efficiently undergo photolysis and react with carboxylic acids even under diluted conditions. The reaction of 100 μM tetrazole and 10 mM carboxylic acid was nearly complete within 30 min, yielding the desired adduct 3d (Fig. S4, ESI†). Still, the reaction did not proceed in the absence of photocatalyst (Fig. S4, ESI†). Moreover, this reaction tolerated many proteinogenic amino acids (except tryptophan or tyrosine; Table S2, ESI†).12
We then attempted to protein modification. Chymotrypsinogen A (25 kDa; Fig. 3a and Fig. S5, ESI†), which contains 14 acidic amino acids, underwent photocatalytic labelling with biotin-linked tetrazole 6 (Scheme S4, ESI†), and the labelling efficiency was assessed using western blot analysis (Fig. 3b and Fig. S6, S7, ESI†). The reaction proceeded efficiently with a photocatalyst but was limited in its absence (Fig. 3b and Fig. S8, ESI†). Further MS analysis of the tryptic digests showed that the peptide containing Asp153 was modified with the oxidized biotin tag 6 (Fig. 3c). Next, we attempted to label proteins with varying molecular weights and numbers of acidic amino acids. Upon LED irradiation in the presence of a photocatalyst and tetrazole 6, various proteins [GST, aldolase, BSA (Fig. 4 and Fig. S9 and 10, ESI†), ovalbumin, catalase (Fig. S9, ESI†)] were biotin-labelled. Total GST including biotinylated proteins decreased from 5 to 15 min, and the cause is under investigation. MS analysis of labeled GST, aldolase, and BSA detected peptides containing Asp or Glu residues modified with the biotin tag 6 (Fig. S11 and S12, ESI†). Since Asp and Glu residues are widely present on various protein surfaces, this method enables the labelling of a broad range of proteins.
 | ||
| Fig. 4 Photocatalytic labelling of GST (0.28 mg mL−1), aldolase (1.0 mg mL−1) and BSA (1.0 mg mL−1). | ||
Next, we investigated the feasibility of this reaction for cell labelling. Cells treated with antibody-photocatalyst conjugates were irradiated with blue LED in the presence of biotin-linked tetrazole 6, and the amount of biotin present on the cell surface was quantified by flow cytometry (Fig. S13, ESI†). Anti-CD30 antibody-photocatalyst conjugate (anti-CD30/PC+) and anti-TNFR2 antibody-photocatalyst conjugate (anti-TNFR2/PC+) were prepared (Fig. S14, ESI†) and used to label CD30-expressing Ramos-Blue (RB.CD30) cells and TNFR2-expressing RB (RB.TNFR2) cells. The RB.CD30 cells treated with anti-CD30/PC+ exhibited a substantial enhancement in biotin labelling relative to anti-TNFR2/PC+ treatment (Fig. 5a and Fig. S15a, ESI†). Similarly, RB.TNFR2 cells treated with anti-TNFR2/PC+ resulted in a significant increase in biotin labelling compared with anti-CD30 antibody treatment (Fig. 5b and Fig. S15b, ESI†). In contrast, labelling reactions using antibodies conjugated with PEG linkers instead of photocatalysts (anti-CD30/PC− and anti-TNFR2/PC−) led to limited cell labelling (Fig. 5a,b and Fig. S15, ESI†). These observations imply that cell labelling occurs in an antigen- and photocatalyst-dependent manner. Western blot analysis of CD30-targeted cell labelling on RB.CD30 cells was also consistent with antigen- and photocatalyst-dependent protein biotinylation (Fig. S16, ESI†).13
Finally, we examined the cell labelling reaction using a cell line that overexpresses MHC class-I related protein 1 (MR1), which is an antigen-presenting molecule. Cell surface expression levels of MR1 are altered in a ligand-dependent manner, e.g., Ac-6-FP. In the absence of ligands, MR1 exists mainly in unliganded forms stabilized by chaperones in the ER. With few exceptions, MR1 usually changes its conformation upon ligand capture and translocates to the cell surface, where the MR1–ligand complexes accumulate.14 MR1-expressing HeLa (HeLa.MR1) cells treated with Ac-6-FP as the MR1 ligand were incubated with anti-MR1 antibody and the corresponding secondary antibody (anti-IgG)-photocatalyst conjugate and then irradiated with blue LED in the presence of biotin-linked tetrazole 6. As shown in Fig. 5c and Fig. S17 (ESI†), both photocatalyst and Ac-6-FP addition increased the quantity of biotin tags on the cell surface. This labelling reaction proceeded in a light irradiation time-dependent manner only in the presence of photocatalyst (Fig. S18, ESI†). In contrast, the photoreaction did not occur under isotype control as a primary antibody (Fig. S19, ESI†). Notably, similar outcomes were observed with HEK293 cells expressing MR1 (Fig. S20, ESI†). Overall, these results indicated that the photocatalytic reaction of tetrazole would be applicable for surface labelling of various cells expressing different antigens.
In this study, we found that tetrazoles with 1-naphthyl or 5-isoquinolinyl groups at the 2-position can be photocatalytically converted to nitrile imines, which can then react with carboxylic acids to form hydrazides in aqueous solution. This reaction was successfully applied to label various proteins and cells in an antigen- and photocatalyst-dependent manner. We are currently exploring the application of this method for proximity labelling.
This work was supported by JSPS KAKENHI (JP22H05183, JP24K02147 and JP24H01769), Research Support Project for Life Science and Drug Discovery [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED (22ama121042), JST GteX Program (JPMJGX23B3) and JST FOREST Program (JPMJFR2303). We thank Ms Y. Hiranuma (Kyoto Univ.) for her assistance with the LC/MS/MS analysis.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- B. Belardi, S. Son, J. H. Felce, M. L. Dustin and D. A. Fletcher, Nat. Rev. Mol. Cell Biol., 2020, 21, 750–764 CAS
.
- T. J. Bechtel, T. Reyes-Robles, O. Fadeyi and R. C. Oslund, Nat. Chem. Biol., 2021, 17, 641–652 CAS
.
- W. Qin, K. F. Cho, P. E. Cavanagh and A. Y. Ting, Nat. Methods, 2021, 18, 133–143 CAS
.
- K. J. Roux, D. I. Kim, M. Raida and B. Burke, J. Cell Biol., 2012, 196, 801–810 CAS
.
- H.-W. Rhee, P. Zou, N. D. Udeshi, J. D. Martell, V. K. Mootha, S. A. Carr and A. Y. Ting, Science, 2013, 339, 1328–1331 CAS
.
-
(a) T.-L. To, K. F. Medzihradszky, A. L. Burlingame, W. F. DeGrado, H. Jo and X. Shu, Bioorg. Med. Chem. Lett., 2016, 26, 3359–3363 CAS
; (b) J. B. Geri, J. V. Oakley, T. Reyes-Robles, T. Wang, S. J. McCarver, C. H. White, F. P. Rodriguez-Rivera, D. L. Parker Jr., E. C. Hett, O. O. Fadeyi, R. C. Oslund and D. W. C. MacMillan, Science, 2020, 367, 1091–1097 CAS
; (c) R. C. Oslund, T. Reyes-Robles, C. H. White, J. H. Tomlinson, K. A. Crotty, E. P. Bowman, D. Chang, V. M. Peterson, L. Li, S. Frutos, M. Vila-Perello, D. Vlerick, K. Cromie, D. H. Perlman, S. Ingale, S. D. O. Hara, L. R. Roberts, G. Piizzi, E. C. Het, D. J. Hazuda and O. O. Fadeyi, Nat. Chem. Biol., 2022, 18, 850–858 CAS
; (d) S. D. Knutson, B. F. Buksh, S. W. Huth, D. C. Morgan and D. W. C. MacMillan, Cell Chem. Biol., 2024, 31, 1145–1161 CAS
.
-
(a) J. P. Menzel, B. B. Noble, A. Lauer, M. L. Coote, J. P. Blinco and C. Barner-Kowollic, J. Am. Chem. Soc., 2017, 139, 15812–15820 CAS
; (b) K. Livingstone, G. Little and C. Jamieson, Synthesis, 2021, 2395–2407 CAS
; (c) C. Guerra, L. Ayarde-Henríquez, Y. A. Rodríguez-Neúñz, A. Ensuncho and E. Chamorro, Chem. Phys. Chem., 2023, 24, e202200867 CAS
.
-
(a) H. Meier and H. Heimgartner, Helv. Chim. Acta, 1985, 68, 1283–1300 CAS
; (b) Y. Wang, C. I. Vera and Q. Lin, Org. Lett., 2007, 9, 4155–4158 CrossRef CAS PubMed
; (c) S. Zhao, J. Dai, M. Hu, C. Liu, R. Meng, X. Liu, C. Wang and T. Luo, Chem. Commun., 2016, 52, 4702–4705 RSC
.
-
(a) A. Herner, J. Marjanovic, T. M. Lewandowski, V. Marin, M. Patterson, L. Miesbauer, D. Ready, J. Williams, A. Vasudevan and Q. Lin, J. Am. Chem. Soc., 2016, 138, 14609–14615 CrossRef CAS PubMed
; (b) Z. Li, L. Qian, L. Li, J. C. Bernhammer, H. V. Huynh, J.-S. Lee and S. Q. Yao, Angew. Chem., Int. Ed., 2016, 55, 2002–2006 CrossRef CAS PubMed
; (c) K. Cheng, J.-S. Lee, P. Hao, S. Q. Yao, K. Ding and Z. Li, Angew. Chem., Int. Ed., 2017, 56, 15044–15048 CAS
; (d) Y. Tian, P. Jacinto, Y. Zheng, Z. Yu, J. Qu, W. R. Liu and Q. Lin, J. Am. Chem. Soc., 2017, 139, 6078–6081 CrossRef CAS PubMed
; (e) K. Bach, B. L. H. Beerkens, P. R. A. Zanon and S. M. Hacker, ACS Cent. Sci., 2020, 6, 546–554 CAS
.
-
(a) S. Dutta, J. E. Erchinger, F. Strieth-Kalthoff, R. Kleinmans and F. Glorius, Chem. Soc. Rev., 2024, 53, 1068–1089 CAS
; (b) F. Strieth-Kalthoff and F. Glorius, Chem, 2020, 6, 1888–1903 CAS
.
- E. Blasco, Y. Sugawara, P. Lederhose, J. P. Blinco, A.-M. Kelterer and C. Barner-Kowollik, ChemPhotoChem, 2017, 1, 159–163 CAS
.
- Trp or Tyr may function as quenchers by reacting with the photocatalyst, thereby inhibiting the reaction. In contrast, in protein labelling experiments (Fig. 3 and 4 and Fig. S12, ESI†), Asp and Glu carboxy groups were successfully labelled even in proteins containing multiple Trp and Tyr residues.
- A clear difference in biotin labelling was observed at 30 minutes with and without PC, while at 60 minutes, a substantial amount of non-specific modification was detected even without PC, suggesting that the reaction time is an important factor in photocatalyst-dependent labelling.
- H. E. G. McWilliam, J. Y. W. Mak, W. Awad, M. Zorkau, S. Cruz-Gomez, H. J. Lim, Y. Yan, S. Wormald, L. F. Dagley, S. B. G. Eckle, A. J. Corbett, H. Liu, S. Li, S. J. J. Reddiex, J. D. Mintern, L. Liu, J. McCluskey, J. Rossjohn, D. P. Fairlie and J. Villadangos, Prod. Natl. Acad. Sci. U. S. A., 2020, 117, 24974–24985 CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc04452e |
| This journal is © The Royal Society of Chemistry 2025 |