Open Access Article
Open Access ArticleChemoenzymatic synthesis of sialylated and fucosylated mucin analogs reveals glycan-dependent effects on protein conformation and degradation†
Amanda M.
Wood
 ,
Casia L.
Wardzala
and
Jessica R.
Kramer
,
Casia L.
Wardzala
and
Jessica R.
Kramer
 *
*
Department of Biomedical Engineering, University of Utah, Salt Lake City, UT 84112, USA. E-mail: jessica.kramer@utah.edu
First published on 14th July 2025
Abstract
Mucin proteins are essential for life but are challenging to study due to their complex glycosylation patterns. Synthetic mimics have become vital tools for understanding and modulating the roles of mucins in human health and disease. These materials also have diverse biomedical applications as lubricants and anti-infectives, in vaccine formulations, and more. We developed a chemoenzymatic approach to prepare polypeptide-based synthetic mucins displaying a variety of glycans with native linkages and orientations. By combining the polymerization of glycosylated amino acid N-carboxyanhydrides with enzymatic sialylation and fucosylation, we produced a tunable panel of synthetic mucins. These polymers were recognized by natural glycan-binding and glycan-degrading enzymes, providing insights into the structural preferences of these proteins. Glycan- and linkage-dependent effects on proteolysis were observed. Further, investigation of the influence of glycans on peptide backbone secondary structure revealed that both sialylation and linkage at Ser vs. Thr have profound effects on hierarchical conformation. Overall, our methodology offers versatile tools for exploring the diverse glycobiology of mucins.
Introduction
Mucins are a family of glycoproteins that coat every wet surface in the human body and are foundational to mucus, tears, saliva, and the epithelial and endothelial glycocalyces.1,2 More than twenty human mucins have been identified with differing polypeptide sequences, but all characteristically feature a densely O-glycosylated domain rich in Pro, Thr, and Ser (PTS domains).3–5 Mucin glycosylation initiates with α-N-acetylgalactosamine (αGalNAc) attached to Ser or Thr. Additional sugars can be appended to the 2-, 3-, 4-, and/or 6-hydroxyls, with glycan chains frequently terminating in fucose (Fuc) or sialic acid (Sia) (Fig. 1).6–8 Due to their terminal location and multivalent display, Sia and Fuc are primed for diverse binding interactions. For example, these sugar groups play essential roles in regulation of extravasation, immune function, cancer, pathogen defense and interaction with symbiotic microbes.9–11 Yet, probing molecular details of these interactions has been challenging due to mucins’ inherent heterogeneity. Here, we present tunable and well-defined synthetic mucin analogs that display Sia and Fuc in their native chemical forms.Biosynthesis of glycan-mature mucins takes place in the Golgi via glycosyltransferase enzymes.12 Eight core mucin di- and tri-saccharide structures have been identified, featuring αGalNAc linked with various regio- and stereo-chemistries to N-acetylglucosamine (GlcNAc), GalNAc, and/or galactose (Gal). These structures can be further diversified and commonly terminate with Sia and Fuc moieties, again linked with unique regio- and stereo- chemistries. The result is an impressive and densely packed array of chemical complexity.
These discrete mucin glycan structures, each possessing differing molecular shapes and chemical properties, confer unique biology as they vary with species, tissue, and disease. For example, distinct human tissue distributions of Sia α2,3- and α2,6-linked to Gal or GalNAc appear to contribute differently to viral infection, cancer progression, and receptor binding on both adaptive and innate immune cells.13–20 Similarly, Fuc α1,2-, α1,3-, and α1,4-linked to Gal or GlcNAc play differing roles in human blood group antigens, cancer, chronic respiratory disorders, and coronavirus infection.21–24
Due to the complexity of glycan chemistry and biology, glycan-dependent effects are typically correlative rather causative. Detailed molecular studies have been roadblocked by the intricacies of protein glycosylation biosynthesis which integrates diet, environment, and genetics into hundreds or even thousands of enzymes.25–27 Advances in genetic engineering have made strides in overcoming the challenges in expressing the large, highly repetitive protein sequences characteristic of mucins.28,29 Recent work tackled glycoengineering of mucins in a model human cell line and remarkable control over simple glycan patterning was reported.30 Despite these advances, precise control over glycan density and pattern, particularly with respect to regio- and stereo-specific sialylation and fucosylation, remains problematic. Characterization of glycan patterns in biologically-derived mucins is a challenge in and of itself, and scalability of niche recombinant materials has limitations. Synthetic mimics are poised to address these challenges and serve as tools to probe mucin biology. Further, mucin mimics have biomedical applications as lubricants, vaccine components, or as anti-infectives.31,32
A variety of mucin surrogates have been explored including short peptides, polysaccharides, and synthetic polymers.31,32 Polymeric mimics are particularly promising tools for further development since they can capture the glycan multivalency long-known to play key roles in diverse glycan binding events.33–35 However, these prior surrogates are generally poor substitutes due to their inability to achieve the molecular weights, multivalency, conformations, chemical linkages, or specific glycans found in nature.
To address the need for improved mucin surrogates, we have been developing peptide polymers that feature natural chemical linkages and adopt native protein secondary structures.36–42 These tunable synthetic mucins (synMUCs) are derived from α-amino acid N-carboxyanhydride (NCA) polymerization, which is a rapid and scalable (mg–kg) technique used in research, commodities, and pharmaceuticals.43–47 With application of modern polymerization initiators, polypeptides from ca. 10–1000 residues and with low dispersity (Đ < 1.2) can be reproducibly prepared. Considering the importance of terminal Sia and Fuc glycans in mammalian biology, we sought to develop synMUCs that display these glycans with their various native linkage orientations and with tunable density.
Efforts towards multivalent Sia48–52 and Fuc53–55 display have mainly focused on non-native backbones such as methacrylate, norbornene, methylvinylketone, lactic-co-glycolic acid. Fuc- and Sia-modified short peptides have also been described.24,56–58 As previously described, such surrogates have inherent limitations based on use of non-natural chemical groups or low molecular weight, thus affecting biological interactions from glycan orientation to lack of multivalency. For example, solid phase synthesis (SPPS)-derived glycopeptides typically have ca. 20 residues or less and only 1–3 glycosylation sites.59–64 Natural mucins are hundreds, or even thousands, of residues long and can display glycans at 50% or more of these sites.3–5 Similarly, natural mucin proteins take on ordered secondary structures that flexible synthetic polymers cannot adopt, surely affecting glycan display. We could find only two examples of Sia display from long-chain polypeptide backbones. One was limited by non-native p-nitrophenyl linkages to glutamic acid and the other could not achieve α2,3 linkages.65,66 We could find no reported long-chain polypeptide systems for Fuc.
To achieve our goal of Sia- and Fuc-presenting synMUCs, we utilized NCA chemistry to prepare polypeptides bearing simple glycans which were substrates for regio- and stereo-selective sialylation and fucosylation via an efficient one-pot multi enzyme (OPME) strategy. After optimization of this chemoenzymatic strategy, we validated synMUC binding to native targets, probed glycan-dependent effects on peptide conformation, and investigated their proteolytic susceptibility to mucinase enzymes.
Characterization of glycan-dependent effects on conformation, binding, and degradation are highly relevant to human health and disease. For example, glycosylation changes associated with cancer, chronic respiratory diseases, or infections,67–70 could affect protein structure and have undiscovered functional outcomes in these pathologies. Similarly, mucinases, identified in diverse mucus-resident pathogenic and commensal microbes,71–77 impact mucin-microbe relationships and offer therapeutic opportunities. There has been a recent explosion of interest in mucinases for application as therapeutics and analytical tools.78–82 Here, we report glycan-dependent effects on glycopeptide conformation and mucinase degradation which may shed light on the structure and function of natural mucin glycoproteins and inform on the broader application of mucinases in research and therapeutics.
Results and discussion
Chemical synthesis of synMUCs with simple glycosylation
For preparation of the synMUC backbones, we chose NCA polymerization since the resulting materials are composed entirely of amino acids and sugars with their natural linkages, and native protein secondary structures can be adopted.36–38 The NCA method also allows precise tuning of molecular weight (MW) and peptide composition, including glycan density and glycan identity. For this study, we selected Ser/Thr conjugates of αGalNAc, βGal, and β1,3Gal-αGalNAc (core 1) (Fig. 2A). These glycans are abundant in mucins, both as standalone moieties and components of core structures, and are substrates for enzymatic sialylation and fucosylation. Further, these glycans are relevant to human disease. For example, in carcinoma tissues the Thomsen-nouveau (Tn) antigen, αGalNAc, the Thomsen–Friedenreich (Tf) antigen, β1,3Gal-αGalNAc, and sialyl-Tn appear to play roles in tumorigenesis, immune evasion, and more.83–85Established methodology was utilized to prepare the Ser and Thr glyco-conjugates and convert them to NCAs (Fig. 2A).36–42 To initiate NCA polymerization, we employed nickel or cobalt based organometallic complexes known to offer fast kinetics, side reaction suppression, controlled and living polymerization, and to yield high molecular weight (MW) polymers of low dispersity.45,86 The nickel complexes also allow for installation of clickable groups at the initiation site,37,41 which we used later for attachment to multiwell plates for binding assays. With both initiators, the peptide termini are amine groups primed for conjugation to biotin, fluorophores, or other molecules of interest.
We hypothesized that glycan density and proximity to charged groups or sterically restricted residues could affect activity of mucin-modifying glycosyltransferases and mucin-degrading glycoproteases. To address these questions, we prepared synMUC variants with differing backbone structures. βGal-, αGalNAc-, and β1,3Gal-αGalNAcSer/Thr NCAs were singularly polymerized to yield homopolypeptides with 100% glycosylation. We also mixed the glyco-NCAs in various ratios with “spacer” amino acid NCAs to prepare polypeptides with varying densities of glycosylation, differing ionic charges, and different steric complexity (Table 1).
| SynMUC | Abbrv.[c] | M n [d] | DP[e] | Đ [f] |
|---|---|---|---|---|
| (GalβSer)n[a] | 100% βGalS | 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070 070 |
108 | 1.34 |
| (GalβThr)n[a] | 100% βGalT | 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
101 | 1.18 |
| (GalβSer0.5-s-Glu0.25-s-Ala0.25)n[a] | 50% βGalSEA | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760 760 |
120 | 1.04 |
| (GalβThr0.5-s-Glu0.25-s-Ala0.25)n[b] | 50% βGalTEA | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 590 590 |
112 | 1.17 |
| (GalβThr0.5-s-K0.25-s-Ala0.25)n[a] | 50% βGalTKA | 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 940 940 |
110 | 1.33 |
| (GalβThr0.25-s-Glu0.38-s-Ala0.38)n[b] | 25% βGalTEA | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
112 | 1.03 |
| (GalβThr0.25-s-Glu0.25-s-Ala0.25-s-Pro0.25)n[b]* | 25% βGalTEAP | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 880 880 |
106 | 1.12 |
| (GalNAcαSer)n[a] | 100% αGalNAcS | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 690 690 |
103 | 1.30 |
| (GalNAcαThr)n[a] | 100% αGalNAcT | 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 550 550 |
101 | 1.30 |
| (GalNAcαSer0.5-s-Glu0.25-s-Ala0.25)n[a] | 50% αGalNAcSEA | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650 650 |
98 | 1.18 |
| (GalNAcαSer0.5-s-K0.25-s-Ala0.25)n[a] | 50% αGalNAcSKA | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070 070 |
92 | 1.32 |
| (GalNAcαSer0.25-s-Glu0.38-s-Ala0.38)n[a] | 25% αGalNAcSEA | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
115 | 1.24 |
| (GalNAcαSer0.25-s-Glu0.25-s-Ala0.25-s-Pro0.25)n[b]* | 25% αGalNAcSEAP | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970 970 |
104 | 1.06 |
| (Galβ-GalNAcαThr0.25-s-Glu0.38-s-Ala0.38)n[b]* | 25% βGal-αGalNAcTEA | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 290 290 |
99 | 1.53 |
For spacer residues, we chose Ala, Glu, Lys, and Pro, which naturally occur in mucin PTS domains. Ala NCA was selected due to its small, neutral, and non-reactive side-chain, Glu to achieve anionic charge, Lys to achieve cationic charge, and Pro for its highly restrictive φ and ψ angles and abundance in native mucin glycodomains. For consistent comparison of glycopolypeptides with varied compositions, we prepared all structures with chain lengths of ca. 100 residues.
Polymerizations were monitored by attenuated total reflectance Fourier-transformed infrared spectroscopy (ATR-FTIR). After complete conversion of all NCA monomer to polypeptide chains, reaction products were directly characterized by size exclusion chromatography coupled with multi-angle light scattering and refractive index detectors (SEC/MALS/RI). As expected from previously established methodology,36–42 polymerizations were quantitative in yield, gave low dispersity materials, and amino acid compositions matched relative NCA monomer feed ratios as confirmed by 1H NMR36–42 (see Fig. S1–S4, ESI†). We note that these are statistical copolymers with sequential distribution of monomeric units dictated by monomer reaction rates. We previously measured kinetics of a variety of NCA monomers in this polymerization system,37,87 including mono- and di-saccharide conjugates, and that the rates are remarkably similar. Therefore, we expect residues to be incorporated with near-random frequencies.
While such materials cannot be expected to recapitulate properties requiring a specific peptide sequence (i.e. peptide-dependent antibody binding), these materials can harness many important features of natural mucins. They form rigid, extended structures with mucin conformations and persistence lengths relevant to native mucins and they present glycans in their native orientations and densities.37 For many functions of mucins, the peptide backbone is actually buried in under the dense glycosylation. Additionally, heterogeneity is inherent in mucin-based structures like mucus and the glycocalyx since natural mucins are subject to splice variation and differing expression of the many mucin genes.2,88 While we recognize lack of sequence specificity is a limitation for certain applications, overall the synMUCs are very promising and unique mimetics since SPPS-derived materials cannot currently capture length-dependent mucin properties of high glycan density or large persistence lengths.59–64
Enzymatic modification of synMUCs with Sia and Fuc glycans
Extensive literature has associated α2,3- and α2,6-linked Sia (N-acetylneuraminic acid, Neu5Ac) groups with cancer, immunity, and human and zoonotic infections.13–15,20,84,89 Therefore, we selected these structures to append via native linkages to αGalNAc (Fig. 2B). Similarly, overexpression of Fuc-bearing glycans has been correlated to asthma, cystic fibrosis, aggressive pancreatic and lung cancers, and altered microbial interactions.11,90–99 Hence, we targeted installation of native α1,2-, α1,3-, and α1,4-Fuc linkages to βGal (Fig. 2B). Considering the complexity of selectively installing Sia and Fuc regio- and stereo- selectively via chemical synthesis, we investigated enzymatic glycosylation of our NCA-derived mono- and di- saccharide bearing synMUCs.Identified in both mammals and bacteria, sialyltransferases (STs) and fucosyltransferases (FTs) utilize cytidine 5′-monophosphate-Sia (CMP-Sia) or guanosine 5′-diphosphate-Fuc (GDP-Fuc), respectively, to sialylate or fucosylate glycan substrates. We first investigated the in vitro reaction of our synMUCs with commercially available sialyltransferases (Photobacterium damsela α-2,6-sialyltransferase and Pasteurella multocida α-2,3-sialyltransferase from Sigma) and CMP-Sia (Nacalai USA). However, we found most commercial enzymes were of poor quality and could not be dissolved in water. Further, we noted literature reports that the activated sugar has poor shelf stability due to hydrolysis.100,101 To address such issues, optimize yields, and improve economic viability of enzymatic glycosylations, Chen and coworkers pioneered one-pot multi-enzyme (OPME) glycosylation reactions that generate the activated CMP-Sia or GDP-Fuc donors in situ from economical glycan precursors.102–110 In some cases, they report remarkable gram-scale production. Inspired by their work, we sought to expand their OPME methodology from free glycans to polypeptide-bound glycan substrates.
In the OPME sialylations, N-acetyl mannosamine (ManNAc) is first converted to Sia by sialic acid aldolase (PmNanA), followed by conversion to CMP-Sia by CMP-sialic acid synthetase (NmCSS) in the presence of cytidine 5′-triphosphate (CTP). In fucosylation reactions, GDP-Fuc is formed from L-fucose, adenosine 5′-triphosphate (ATP), and guanosine 5′-triphosphate (GTP) using bifunctional enzyme (BfFKP) that has both L-fucokinase and GDP-Fuc pyrophosphorylase activities. Inorganic pyrophosphatase (PmPpA) is also present to drive the reaction forward. Necessary in each case is a relevant FT or ST enzyme. For STs, we selected proteins from Pasteurella multocida and Photobacterium damsela since expression has been previously reported in E. coli and their α2,3- and α2,6-linkage specificities have been established.105,108 Similarly, we selected FTs originating from Thermosynechococcus elongatus, Helicobacter hepaticus, and Helicobacter pylori for their ability to install α1,2-, α1,3-, or α1,4-Fuc, respectively.102,111 We recombinantly expressed the set of nine enzymes on the mg scale using modified literature protocols (Table S4, ESI†). All enzymes were 6x-His-tagged to allow for purification via Ni2+ or Co2+ affinity columns (Fig. S10, ESI†).
With purified enzymes in hand, OPME sialylation and fucosylation of chemically synthesized synMUCs was examined. Considering the large number of possible combinations, we did not attempt enzymatic glycosylation of each of the 14 synMUC compositions with each of the 5 enzyme systems. Instead, we chose representative samples with varied amino acid identity, initiating glycan identity, and glycan density. Detailed optimized conditions for the OPME reactions are reported in the ESI.† In brief, we used 1.5 equiv. of monosaccharide donor and 1.3–1.5 equiv. of NTP per mole of acceptor glycan. For fucosylations, we used 0.067 mg mL−1 PmPpA and 0.1 mg mL−1 BfFKP with 0.1 mg mL−1 Te2FT, HhFT1, or HpFT4. For sialylations, we used 0.1 mg mL−1 NmCSS and 0.2 mg mL−1 PmNanA with 0.2 mg mL−1 PmST1 or Pd2,6ST. In all cases, reactions were in Tris buffer with MgCl2. For all synMUC glycosylation reactions, 6xHis tagged enzymes were removed from the reaction mixture via incubation with Ni-NTA magnetic beads. SynMUCs were purified by dialysis against ultrapure water and lyophilized to yield white powders.
Key to characterizing the synMUCs was an accurate method of glycan analysis. Therefore, we undertook multiple approaches to tackle this non-trivial task. Analysis by NMR and ATR-FTIR confirmed the presence of the correct functional groups and we could see appropriate increases in the glycan integrals (Fig. S1–S7, ESI†). However, the overlap of glycan and amino acid spectral signatures and broad peaks due to the polymeric nature of the materials convoluted accurate calculation of the extent of fucosylation and sialylation from peak integrations. We also explored commercially available glycan analysis kits for Sia and Fuc (Sigma, Megazyme). The kits were useful to detect relative changes between reactions with differing compositions. However, the results were qualitative as Sia assays consistently reported higher than theoretical yields and the Fuc assay kit suffered from specificity issues where control samples containing only Gal yielded results positive for Fuc.
To more accurately confirm the predicted structures and to quantify the amounts of added carbohydrates, we turned to gas chromatography-mass spectrometry (GC-MS), high-performance anion exchange chromatography (HPAEC), X-ray photoelectron spectrometry (XPS), and SEC/MALS/RI. These techniques are attractive since only μg quantities are required.
For GC-MS, amino and neutral sugars were analyzed after treatment of synMUCs with acidic methanolysis and conversion to methyl glycosides followed by trimethylsilyl (TMS) derivatization. Since Sia degradation can occur during acidic methanolysis, detection of this glycan was attempted using HPAEC. To avoid interference of amino acids on HPAEC, synMUCs were treated with an α-2-3,6,8,9-neuraminidase A to release Sia residues from the polypeptide prior to analysis. MS results confirmed that Sia and Fuc groups had indeed been enzymatically appended to our polymers (Table S7, ESI†). However, we could not accurately determine Sia and Fuc content using this method. Glycan ratios were inconsistent for our core 1 synMUC samples where 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratios of Gal
1 ratios of Gal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GalNAc were expected and glycan yields were consistently lower than observed by XPS, SEC, NMR and the commercial assays. We suspect this is due to variability in efficiency of methanolysis, silylation, or sialidase cleavage.
GalNAc were expected and glycan yields were consistently lower than observed by XPS, SEC, NMR and the commercial assays. We suspect this is due to variability in efficiency of methanolysis, silylation, or sialidase cleavage.
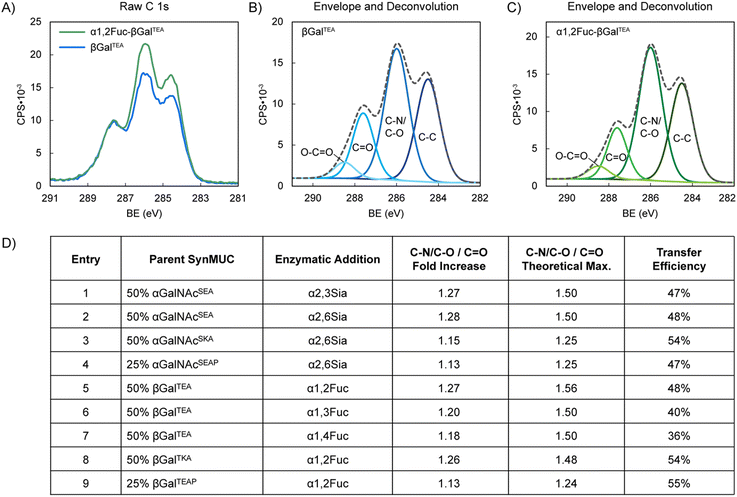
XPS was an attractive alternative since the technique can use intact glycopolypeptides with no chemical reactions needed prior to analysis, and very small sample mass is required. Intact synMUCs were analyzed for elemental composition and high-resolution scans were deconvoluted into characteristic peaks for various bond species. The results of these analyses and representative spectra are shown in Fig. 3. Additional data can be found in the ESI.† Appending of Fuc or Sia residues generates changes in the number, and consequently ratios, of C–C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–O, and C–N bonds. Therefore, we compared the spectra of the synMUCs before and after the enzymatic reactions and calculated the fold-change in these bonds. C 1s curves were deconvoluted into their constituent C–C, C–N/C–O, and C
O, C–O, and C–N bonds. Therefore, we compared the spectra of the synMUCs before and after the enzymatic reactions and calculated the fold-change in these bonds. C 1s curves were deconvoluted into their constituent C–C, C–N/C–O, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peaks and plotted under their calculated C 1s envelope curve. O–C
O peaks and plotted under their calculated C 1s envelope curve. O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peaks were added when apparent. To correct for adventitious carbon contamination, C
O peaks were added when apparent. To correct for adventitious carbon contamination, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peaks were normalized between samples when appropriate. Glycosylation efficiency was defined as the observed number of Sia or Fuc groups as compared to the total number of potential Gal or GalNAc acceptors and was calculated via the observed fold change in C–N/C–O/C
O peaks were normalized between samples when appropriate. Glycosylation efficiency was defined as the observed number of Sia or Fuc groups as compared to the total number of potential Gal or GalNAc acceptors and was calculated via the observed fold change in C–N/C–O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vs. the theoretical maximum.
O vs. the theoretical maximum.
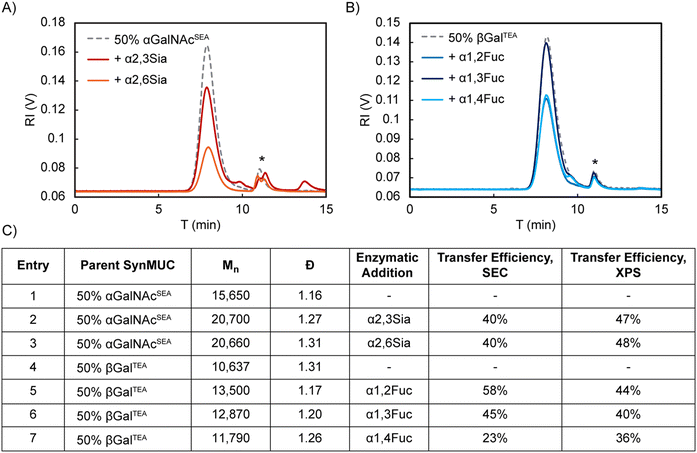
To further confirm the accuracy of the XPS data, we selected a subset of samples to analyze by aqueous SEC/MALS/RI before and after the OPME reactions. For this technique, we expected to see a molecular weight shift due to the extra mass of the Fuc or Sia residues. Number average molecular weight (Mn) and dispersity were calculated from the MALS and RI data using Astra 7 software. Fig. 4 reports the results of these analyses with representative chromatograms included. Due to similar interactions of the glycopolypeptides with the column stationary phase, major shifts in elution time are not expected and elution time is not a factor in the calculations. We were pleased to see unimodal glycopolypeptide distributions after the OPME reactions, indicating chains remain intact and only one species is present. Gratifyingly, side by side comparison of the SEC and XPS data revealed the two methodologies are in relatively close agreement.
For the identical 50% glycosylated anionic αGalNAcSEA acceptor polypeptide, efficiency of Sia transfer was essentially identical for both the α2,3 and α2,6 linkages generated by PmST1 and Pd2,6ST (47% and 48%, respectively, by XPS and both 40% by SEC, Fig. 3D, entries 1, 2 and Fig. 4C, entries 2, 3). To probe the effect of neighboring residue charge or steric effect, we used Pd2,6ST to sialylate acceptor polypeptides where anionic Glu was fully replaced with cationic Lys or where 25% of the residues were replaced by Pro (Fig. 3B, entries 2, 3). Glycan transfer efficiency was unaffected by the addition of Pro (47%, Fig. 3D, entry 3), which is rational considering that mucins are naturally rich in Pro. Sialylation efficiency improved slightly for the Lys containing peptide (54%, Fig. 3D, entry 4). We speculate this could be due to direct or allosteric stabilization of the enzyme active site, or simply favorable electrostatic interaction since the ST enzyme has an isoelectric point of 4.88 (Sigma-Aldrich112). Our results align nicely with those of the aforementioned report of α2,6-sialylation a 50%-lactose-Ser/Ala copolypeptide by Bertozzi and coworkers65 and that of Totani et al. who sialylated p-nitrophenylester linked glycans on polyglutamic acid backbones.66 They used a two-enzyme system of Pd2,6ST and NmCSS in the presence of Sia and reported ca. 50% efficiency as determined by SEC and NMR. They also reported unsuccessful attempts at α2,3-sialylation with a variety of STs, including PmST1. In our hands, using the OPME system with in-house expressed PmST1, we achieved excellent α2,3 sialylation yields for our synMUCs.
Fucosylation of an identical 50% βGalTEA acceptor glycopolypeptide by the enzymes Te2FT, HhFT1, and HpFT4, which yield α1,2-, α1,3-, and α1,4-linkages, respectively, were relatively consistent (36–44% by XPS, Fig. 3D, entries 5–7; 23–58% by SEC, Fig. 4C, entries 5–7). To again probe steric or charge effects, we used Te2FT to fucosylate similar Lys or Pro containing backbones (Fig. 3D, entries 8, 9). Proline appeared to increase the favorability of the acceptor peptide since efficiency of fucosylation 25% βGalTEAP increased slightly as compared to 50% βGalTEA (55% vs. 48%), though we acknowledge this could be due to the lower glycan density also reducing steric hinderance. Similar to the sialylation case, lysine modified 50% βGalTKA was a slightly more favorable substrate for Te2FT than the Glu containing polymer with 54% vs. 48% of the sites fucosylated (Fig. 3D, entries 3, 8).
The combination of NCA polymerization with OPME methodology is robust for generation of a variety of glycopolypeptide compositions. We are pleased to present first-in-kind materials with regio- and stereo-specific multivalent display of sialyl-Tn and fucose from native polypeptide backbones. In native mucins, reported Sia and Fuc content, as quantified by NMR or gas or liquid chromatography coupled to MS, is variable depending upon the MUC type and source tissue.113–116 Representative values for Sia and Fuc content, reported as % of total monosaccharide composition, ranged from 6–24% for Sia and 5–30% for Fuc. However, as seen in our chemically-defined materials, these data can be affected by the differing abilities of glycan structures to derivatize, enzymatically cleave, or ionize. In any case, we expect that the Sia and Fuc levels in our synMUCs are biologically relevant. Overall, we conclude the OPME reactions open doors to functional and tunable mucin mimics with application in probing the diverse biology of Fuc and Sia.
Evaluation of functional glycan binding
Considering our proposed application of synMUCs as mucin mimics, we sought to evaluate functional binding of the presented glycans. For proof-of-concept experiments, we selected commercially available biotinylated lectin proteins wheat germ agglutinin (WGA) and Ulex Europaeus agglutinin I (UEA-I) since these have previously been applied to mucin studies.117,118 UEA-I has specificity for fucosylated glycans,119–121 while WGA is reported to have moderate specificity for Sia.120–123 For binding assays, we utilized a dot blot format where synMUCs or control reagents were spotted on nitrocellulose in triplicate at varied concentrations. Membranes were washed, blocked with bovine serum albumin (BSA), and then incubated with biotinylated lectin. After washing, dot blots were incubated with streptavidin-IR680 fluorophore, washed, and imaged at 700 nm. Blot images were analyzed using ImageJ and fluorescence intensity was quantified. A representative dot bot is shown in Fig. 5A and additional data can be found in the ESI.†For the WGA dot blot, we examined binding to the 50% glycosylated αGalNAcSEA parent synMUC as compared to the α2,3Sia-αGalNAcSEA and α2,6Sia-αGalNAcSEA enzymatic products (Fig. 5B). For controls, we chose commercially-available fetuin and asialofetuin and all samples were compared to membrane background signal. Signal from the controls was as expected, where binding was substantially higher for fetuin than asialofetuin. Signal above background for asialofetuin was expected, as WGA also has reported affinity for GlcNAc residues. We found that WGA had excellent binding to both the parent synMUC and the sialylated synMUCs. No preference for α2,3Sia vs. α2,6Sia was observed and the lectin actually had a slightly stronger binding preference for the parent synMUC displaying only αGalNAc. This is not entirely unexpected considering literature reports of promiscuous binding to GlcNAc and GalNAc.119–121
Lectin dot blots with UEA-I utilized 50% glycosylated βGalTEA as the parent synMUC, which was compared to the α1,2-, α1,3-, and α1,4-fucosylated products (Fig. 5C). For controls, we spotted BSA, which has no expected fucosylation; human milk, which is rich in diverse fucosylated glycans; lactose (Lac); and 2-fucosylLac (2′FucLac). For the controls, UEA-1 bound strongly to human milk as compared to BSA. Surprisingly, the lectin did not have any preference for 2′FucLac over Lac. We found that UEA-1 strongly preferred our α1,3Fuc-αGalTEA polypeptide over the α1,2- and α1,4-Fuc regioisomers, or the parent βGalTEA polymer.119–121 We found this result interesting because UEA-I is reported to prefer α1,2Fuc.121,124 However, we note prior lectin-glycan binding assays may not have directly compared these regioisomers or may have used free glycans not attached to a peptide where conformational effects could play a role.
Over the course of examining glycan-specific binding, we also attempted enzyme-linked lectin and immunosorbent assays (ELLA, ELISA) where azide-terminal glycopolypeptides were conjugated to alkyne-modified 96-well plates. However, we found higher background for the ELLAs and ELISAs than for the dot blot assays (Fig. S13 and S14, ESI†). We also examined a commercial antibody with listed specificity to Sia, and lectins reported have linkage-specific preferences for Sia (Maakia amurensis I for α2,3Sia and Sambucus nigra I for α2,6Sia) but these exhibited poor specificity and high background.
Overall, we found that our sialylated and fucosylated synMUCs are indeed substrates for binding by natural glycan-targeting proteins. These data confirm the utility of our synMUCs as surrogates for native mucins, and as such, we expect these polymers to find broad utility in probing the diverse and fascinating biology of mucins. Further, considering the widespread use of glycan-binding lectins and antibodies in glycobiology research, we expect the synMUCs to be useful as standards or controls since the glycosylation can be controlled with exquisite precision.
Conformational analysis of synMUC glycoforms
We utilized circular dichroism spectroscopy (CD) to examine the glycan-dependent secondary structures of our synMUC panel. This technique relies on the absorption of circularly polarized light by peptide bonds. The wavelengths at which the absorptions occur, and their intensities, reveal characteristics about the orientation of those bonds and the conformation of the peptide. Distinct CD signatures have been shown for the η → π* and π → π* amide bond transitions of disordered, β-sheet, α-helical, or polyproline II (PPII) helical conformations.125,126 SynMUC CD spectra were obtained in Milli-Q water and data were converted to molar ellipticity (Fig. 5D–F). Homopolymers were utilized to hone in on the effects of glycosylation at Ser/Thr without convoluting effects of other amino acids. As structural controls, we prepared polySer and polyThr from AcOSer and AcOThr using the same NCA methodology. However, the polypeptides precipitated out of water due to formation of insoluble beta sheets.38,127,128Data in Fig. 5D–F indicate that peptide conformation is strongly dependent upon glycan identity and amino acid linkage type. We observed a range of structures including α-helix, disordered, and extended or PPII. We note that the CD spectra of PPII helices (left-handed, 3 residues per turn), extended rod-like structures typical of polyelectrolytes, and disordered “random coil” conformations are quite similar. In fact, for many decades all of these structures were mistakenly assigned as random coils.129,130 Even now, despite their ubiquity in nature, PPII reference data are still absent from common deconvolution algorithms.131–133 However, careful comparison of the spectra of collagen, denatured collagen, intrinsically disordered proteins, and polyelectrolytes have shed light on the relationship between secondary structure and spectral absorbance wavelengths and intensities.129,130,134–137 Of particular focus is the η → π* region between ca. 205–220 nm. Both polyanionic polyGlu and intact collagen have positive maxima, with the relative absorbance intensity increasing with degree of helicity or charge-repulsion-induced chain extension. By contrast, intrinsically disordered proteins have no positive maxima; absorbance is negative. The GalNAc amide contributes a positive absorbance between 190–205 nm in the π → π* region.38,40
Fig. 5D shows an overlay of spectra obtained for various chemically-synthesized glycosylated polySer vs. polyThr backbones. Based on the η → π* transition intensity and blueshift, we observed that chains of galactosylated-Thr are more extended, rigid, and PPII-like than those of galactosylated-Ser, likely due to steric contributions from the Thr methyls. Similarly, GalNAc results in more rigid, extended structures than Gal regardless of linkage to Ser vs. Thr. This is likely due to hydrogen bonding between glycan amides and the peptide backbone.138 Extension from GalNAc to the disaccharide core 1 did not have a strong effect (Fig. S11, ESI†).38,127,128
Sialylation had profound effects on glycopolypeptide secondary structure (Fig. 5E). After addition of either α2,3Sia or α2,6Sia, to αGalNAcS homopolymers, we observed distinct shifts in the spectra. We noted loss of the η → π* positive maximum at 220.8 nm and appearance of two new minima at ca. 212 and 223 nm, which is the classic signature of a canonical α-helix. Our data align with a prior report by Thornton and coworkers, who used CD and Raman techniques to characterize native MUC5B and observed helical and extended/disordered structures.139 To check if the drastic conformational shift is unique to our synthetic materials, we also collected CD spectra of bovine mucin before and after treatment with sialidase. Again, we observed an increased helical character with increased sialylation (Fig. S12, ESI†). Therefore, we conclude that sialylation induces mucin α-helicity. Uncovering the molecular drivers of this shift warrants detailed analyses in a future study. However, we hypothesize that the Sia carboxylic acid group could form a glycan–glycan hydrogen bond with a NAc group, and/or the steric bulk of the hydrated glycerol unit on Sia could disrupt the αGalNAc-to-peptide hydrogen bond which would free the peptide units for energetically favorable internal hydrogen bonds in the helix.
Fucosylation had less dramatic impacts on synMUC structure. All of the fucosylated βGalT glycopolypeptides maintained the rigid, extended, PPII-like conformations observed for the monosaccharide-bearing precursors (Fig. 5C). We observed slight shifts in the wavelength of the η → π* positive maxima from 220.8 nm for βGalT to 218.2, 216.1, and 215.6 nm for α1,2-, α1,3-, α1,4-Fuc modified βGalT, respectively. Following the same trend of increasing energy of absorbance wavelength, the magnitude of the absorption also increased with conjugation site. Together, these data suggest that peptide chain extension and rigidity is highest when Fuc is conjugated to C4 of the Gal ring, that fucosylation at C3, and even more so C2, results in slightly more flexible chains. Effects were greater for the Ser-based glycopolypeptide. For α1,2Fuc-βGalS, we noted loss of the η → π* maxima and shift to a spectrum analogous to that of truly disordered and flexible structures. Presumably, the relative hydrophobicity of the Fuc moiety and its proximity to the peptide backbone based on Gal linkage orientation, all affect the resulting structure. Again, the Thr methyl group appears to play a dominant role since Ser conjugates were more affected.
Collectively, the results of the structural analysis have implications for understanding evolution of mucin structure, interactions between mucins and their partners, as well as biophysical functions of the glycocalyx and mucus. Variations in glycoprotein secondary structures would result in differing presentations of glycans via their orientation in single or multivalent displays. Indeed, there are multiple reports of differences in the ability of antibodies and lectins to bind αGalNAc (Tn antigens) presented on Ser vs. Thr peptides.83,140,141 Considering that mucins are candidates for cancer vaccines and anti-infectives, understanding such effects is crucial. Further, the mechanics of the glycocalyx and mucus are intertwined with their biological roles and are affected by the rigidity and persistence length of component glycoproteins.142,143 Our data indicate that chain conformation and rigidity in mucin-type structures is glycan- and amino acid-dependent and is the likely result of an intricate balance of steric and hydrophobic effects, charge repulsion, and hydrogen bonding.
Mucinase degradation
Proteolytic degradation of mucins occurs naturally in mucosal epithelial tissues that are colonized by microbes. Secreted mucinase enzymes play a variety of roles for commensal and pathogenic microbes alike, including in nutrition and infection. To shed light on the role of glycosylation in mucin degradation, we subjected our panel of synMUCs to two mucin-specific proteases of microbial origin. We chose secreted protease of C1 esterase (StcE), a zinc metalloprotease produced by E. coli, and Serratia marcescens Enhancin (SmE). StcE proteolysis occurs at Ser/Thr*-X-Ser/Thr sites, where cleavage occurs before the second Ser/Thr, X is any amino acid, and the (*) position must be glycosylated.144 SmE is reported to cleave N-terminally to a glycosylated Ser or Thr residue and between S/T–S/T sites.78,79 For both species, these mucinases contribute to cleavage of the protective mucus layers of the gut and, presumably, opportunistic infection.145,146 Further, these enzymes are under investigation for application as new research tools and therapeutics.We examined the susceptibility of various synMUCs to StcE and SmE degradation (Fig. 6A). SynMUCs were incubated with the enzymes at an enzyme-to-substrate ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 for 48 hours at 37 °C, followed by heat denaturation of the mucinases. Control samples were exposed to the same conditions but without enzyme. Samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with either silver stain or a glycoprotein-specific fluorescent stain. Representative gel data is shown in Fig. 6B and C and complete gel data is in the ESI.† Gels were analyzed with ImageJ147 to quantify remaining intact polypeptide as compared to no-enzyme controls within the same gel.
10 for 48 hours at 37 °C, followed by heat denaturation of the mucinases. Control samples were exposed to the same conditions but without enzyme. Samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with either silver stain or a glycoprotein-specific fluorescent stain. Representative gel data is shown in Fig. 6B and C and complete gel data is in the ESI.† Gels were analyzed with ImageJ147 to quantify remaining intact polypeptide as compared to no-enzyme controls within the same gel.
Both StcE and SmE preferred to cleave GalNAc-peptides as compared to Gal-peptides (Fig. 6D entries 1–9). We speculate the NAc group could have a stabilizing interaction within the active sites, or peptide secondary structure could be more favorable. We also observed that proteolysis was more efficient for Ser vs. Thr conjugates of GalNAc (Fig. 6D, entries 3 vs. 4), and that SmE was more affected than StcE. We presume this is due to the higher steric constraints of the Thr methyl. Spacing of GalNAcylated residues to 50% did not affect proteolytic efficiency for either StcE or SmE, while spacing of Gal residues slightly increased proteolysis. Further decrease of glycan density to 25% reduced chain cleavage for both proteases over the time period examined, aligning with the fact that glycans are required for recognition by these enzymes (Fig. 6D, entries 3, 6, 7). Extension of the GalNAc residue to the disaccharide core 1 glycan did not appear to affect enzyme efficiency and the polypeptide was partially degraded during the time period examined (Fig. 6D, entry 9). Sialylation of GalNAc at C6 did not affect proteolysis by either StcE or SmE. Sialylation at C3 reduced proteolysis by 9% and 18% for SmE and StcE, respectively (Fig. 6D, entries 6 vs. 13 and 14) presumably due to altered access to the enzyme active site. By contrast, fucosylation of Gal at C2, C3, or C4 essentially eliminated the ability of both mucinases to cleave the peptide backbone.
Collectively, the mucinase data confirm our synMUCs can be recognized and degraded by natural enzymes and shed light on the proteolytic preferences of these two important enzymes. Prior mass spectrometry analyses eluded to glycan-dependent proteolysis by StcE and SmE; however, it was unclear if this was an artifact of detection methodology.30,78,79 SmE could accommodate a variety of O-linked glycans adjacent to the cleavage site, but might prefer smaller structures such as GalNAc or core 1 (Gal-GalNAc). StcE efficiently cleaved mucins bearing GalNAc or core 1, but sialylated-GalNAc mucins were reported as essentially resistant to proteolysis.30 By contrast, our data show that synMUCs bearing either α2,3 or α2,6-sialyated GalNAcs can be efficiently degraded. These data highlight the utility of chemically-defined mucin surrogates and their unique position to deconvolute structure–function relationships.
Conclusions
Mucins are the primary protein component of mucus, saliva, tears, and the epithelial and endothelial glycocalyces. The heterogenous and dynamic nature of mucin glycosylation has been an obstacle to probing the biology of these tissues. We tackled this challenge by developing a chemoenzymatic route to prepare mucin-mimicking polypeptides. We used NCA polymerization to form glycopolypeptides bearing mono- and di-saccharides and then regio- and stereo-selectively diversified the glycans by enzymatic sialylation and fucosylation. After validation that these synthetic mucins are recognized by native glycan-binding proteins, we probed glycan-dependent effects on peptide backbone conformation and proteolytic susceptibility to mucinase enzymes. We found that glycosylation at Thr results in greater chain extension, and presumably rigidification, as compared to Ser residues. Similarly, GalNAc resulted in greater chain rigidification than Gal when linked at Ser or Thr. Fucosylation had only minor effects on secondary structure, while sialylation drove a conformation shift to alpha helices. We observed increased mucinase-catalyzed proteolysis for Ser- vs. Thr-based glycopolypeptides and for GalNAcylated vs. galactosylated and fucosylated structures. In summation, we present mucin mimics composed entirely of natural chemical groups and linkages, and that present multivalent displays of key bioactive fucose and sialic acid glycans. We expect these materials to serve as valuable tools for probing the diverse biology of mucins and in mucin-based therapeutics.Author contributions
Conceptualization: J. R. K., A. M. W., C. L. W., data curation: J. R. K., A. M. W., C. L. W., formal analysis: J. R. K., A. M. W., C. L. W., funding acquisition: J. R. K., investigation: J. R. K., A. M. W., C. L. W., methodology: J. R. K., A. M. W., C. L. W., project administration: J. R. K., resources: J. R. K., A. M. W., C. L. W., software: J. R. K., A. M. W., C. L. W., supervision: J. R. K., validation: J. R. K., A. M. W., C. L. W., visualization: J. R. K., A. M. W., C. L. W., writing – original draft: J. R. K., writing – review & editing: J. R. K.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the ESI.† Full experimental details, additional data, and characterization of compounds can be found in the ESI.†Acknowledgements
This work was funded by NIH NIGMS 1R35GM147262-01 to J. R. K. This work made use of Nanofab EMSAL shared facilities of the Micron Technology Foundation Inc. Microscopy Suite sponsored by the John and Marcia Price College of Engineering, Health Sciences Center, and Office of the Vice President for Research for XPS experiments. We thank the lab of Prof. Michael Yu for the use of their CD spectrophotometer. We thank the lab of Prof. Russell Stewart for use of their aqueous SEC/MALS/RI system and Monika Sima and Dr Rachel Detwiler for time and expertise on the related experiments. We thank the lab of Prof. Stacy Malaker for the gift of the mucinase enzymes, StcE and SmE. Finally, we thank Parastoo Azadi, Li Tan, and Christian Heiss at the Complex Carbohydrate Research Center for time and expertise with analysis by mass spectrometry.References
- G. Petrou and T. Crouzier, Mucins as multifunctional building blocks of biomaterials, Biomater. Sci., 2018, 6(9), 2282–2297, 10.1039/c8bm00471d.
- C. E. Wagner, K. M. Wheeler and K. Ribbeck, Mucins and Their Role in Shaping the Functions of Mucus Barriers, Annu. Rev. Cell Dev. Biol., 2018, 34, 189–215, DOI:10.1146/annurev-cellbio-100617-062818.
- S. E. Baldus, K. Engelmann and F.-G. G. Hanisch, MUC1 and the MUCs: A family of human mucins with impact in cancer biology, Crit. Rev. Clin. Lab. Sci., 2004, 41(2), 189–231, DOI:10.1080/10408360490452040.
- R. Bansil and B. S. Turner, Mucin structure, aggregation, physiological functions and biomedical applications, Curr. Opin. Colloid Interface Sci., 2006, 11, 164–170, DOI:10.1016/j.cocis.2005.11.001.
- J. A. Voynow and B. K. R. Mengr, Mucins, mucus, and sputum, Chest, 2009, 135, 505–512, DOI:10.1378/chest.08-0412.
- S. J. Gendler, MUC1, The renaissance molecule, J. Mammary Gland Biol. Neoplasia, 2001, 6(3), 339–353, DOI:10.1023/a:1011379725811.
- L. Arike and G. C. Hansson, The Densely O-Glycosylated MUC2 Mucin Protects the Intestine and Provides Food for the Commensal Bacteria, J. Mol. Biol., 2016, 428(16), 3221–3229, DOI:10.1016/j.jmb.2016.02.010.
- C. R. Bertozzi and D. Rabuka, Essentials of Glycobiology, Cold Spring Harbor (NY), 2nd edn, 2009, pp. 2015–2017 Search PubMed.
- S. Pinzón Martín, P. H. Seeberger and D. Varón Silva, Mucins and Pathogenic Mucin-Like Molecules Are Immunomodulators During Infection and Targets for Diagnostics and Vaccines, Front. Chem., 2019, 7, 710, DOI:10.3389/fchem.2019.00710.
- A. Varki, Sialic acids in human health and disease, Trends Mol. Med., 2008, 14(8), 351–360, DOI:10.1016/j.molmed.2008.06.002.
- M. Schneider, E. Al-Shareffi and R. S. Haltiwanger, Biological functions of fucose in mammals, Glycobiology, 2017, 27(7), 601–618, DOI:10.1093/glycob/cwx034.
- H. C. Hang and C. R. Bertozzi, The chemistry and biology of mucin-type O-linked glycosylation, Bioorg. Med. Chem., 2005, 13(17), 5021–5034, DOI:10.1016/j.bmc.2005.04.085.
- C. Krempl, M. L. Ballesteros, G. Zimmer, L. Enjuanes, H. D. Klenk and G. Herrler, Characterization of the sialic acid binding activity of transmissible gastroenteritis coronavirus by analysis of haemagglutination-deficient mutants, J. Gen. Virol., 2000, 81(Pt 2), 489–496, DOI:10.1099/0022-1317-81-2-489.
- K. N. Barnard, B. R. Wasik, J. R. Laclair, D. W. Buchholz, W. S. Weichert, B. K. Alford-Lawrence, H. C. Aguilar and C. R. Parrish, Expression of 9-o-and 7,9-o-acetyl modified sialic acid in cells and their effects on influenza viruses, mBio, 2019, 10(6), 1–17, DOI:10.1128/mBio.02490-19.
- K. N. Barnard, B. K. Alford-Lawrence, D. W. Buchholz, B. R. Wasik, J. R. LaClair, H. Yu, R. Honce, S. Ruhl, P. Pajic, E. K. Daugherity, X. Chen, S. L. Schultz-Cherry, H. C. Aguilar, A. Varki and C. R. Parrish, Modified Sialic Acids on Mucus and Erythrocytes Inhibit Influenza A Virus Hemagglutinin and Neuraminidase Functions, J. Virol., 2020, 94(9), e01567–19, DOI:10.1128/JVI.01567-19.
- M. A. Langereis, M. J. G. Bakkers, L. Deng, V. Padler-Karavani, S. J. Vervoort, R. J. G. Hulswit, A. L. W. van Vliet, G. J. Gerwig, S. A. H. de Poot, W. Boot, A. M. van Ederen, B. A. Heesters, C. M. van der Loos, F. J. M. van Kuppeveld, H. Yu, E. G. Huizinga, X. Chen, A. Varki, J. P. Kamerling and R. J. de Groot, Complexity and Diversity of the Mammalian Sialome Revealed by Nidovirus Virolectins, Cell Rep., 2015, 11(12), 1966–1978, DOI:10.1016/j.celrep.2015.05.044.
- H. S. Y. Leung, O. T. W. Li, R. W. Y. Chan, M. C. W. Chan, J. M. Nicholls and L. L. M. Poon, Entry of Influenza A Virus with a 2,6-Linked Sialic Acid Binding Preference Requires Host Fibronectin, J. Virol., 2012, 86(19), 10704–10713, DOI:10.1128/JVI.01166-12.
- J. E. Stencel-Baerenwald, K. Reiss, D. M. Reiter, T. Stehle and T. S. Dermody, The sweet spot: Defining virus-sialic acid interactions, Nat. Rev. Microbiol., 2014, 12(11), 739–749, DOI:10.1038/nrmicro3346.
- C. Wardzala, A. Wood, D. Belnap, J. Kramer, C. L. Wardzala, A. M. Wood, D. M. Belnap and J. R. Kramer, Mucins Inhibit Coronavirus Infection in a Glycan-Dependent Manner, ACS Cent. Sci., 2022, 8, 351–360, DOI:10.1021/acscentsci.1c01369.
- P. R. Crocker, J. C. Paulson and A. Varki, Siglecs and their roles in the immune system, Nat. Rev. Immunol., 2007, 7(4), 255–266, DOI:10.1038/nri2056.
- C. Zhao, Y. Wu, H. Yu, I. M. Shah, Y. Li, J. Zeng, B. Liu, D. A. Mills and X. Chen, The one-pot multienzyme (OPME) synthesis of human blood group H antigens and a human milk oligosaccharide (HMOS) with highly active Thermosynechococcus elongatus α1-2-fucosyltransferase, Chem. Commun., 2016, 52(20), 3899–3902, 10.1039/c5cc10646j.
- H. H. Lu, S. Y. Lin, R. R. Weng, Y. H. Juan, Y. W. Chen, H. H. Hou, Z. C. Hung, G. A. Oswita, Y. J. Huang, S. Y. Guu, K. H. Khoo, J. Y. Shih, C. J. Yu and H. C. Tsai, Fucosyltransferase 4 shapes oncogenic glycoproteome to drive metastasis of lung adenocarcinoma, EBioMedicine, 2020, 57, 102846, DOI:10.1016/j.ebiom.2020.102846.
- A. Kousathanas, E. Pairo-Castineira, K. Rawlik, A. Stuckey and C. A. Odhams, et al., Whole-genome sequencing reveals host factors underlying critical COVID-19, Nature, 2022, 607(7917), 97–103, DOI:10.1038/s41586-022-04576-6.
- S. Behren, J. Yu, C. Pett, M. Schorlemer, V. Heine, T. Fischöder, L. Elling and U. Westerlind, Fucose Binding Motifs on Mucin Core Glycopeptides Impact Bacterial Lectin Recognition, Angew. Chem., Int. Ed., 2023, 62(32), e202302437, DOI:10.1002/anie.202302437.
- C. L. Hattrup and S. J. Gendler, Structure and Function of the Cell Surface (Tethered) Mucins, Annu. Rev. Physiol., 2008, 70, 431–457, DOI:10.1146/annurev.physiol.70.113006.100659.
- A. Guzman-Aranguez, A. M. Woodward, J. Pintor and P. Argüeso, Targeted disruption of core 1 β1,3-galactosyltransferase (C1galt1) induces apical endocytic trafficking in human corneal keratinocytes, PLoS One, 2012, 7(5), e36628, DOI:10.1371/journal.pone.0036628.
- F.-G. G. Hanisch and S. Müller, MUC1: The polymorphic appearance of a human mucin, Glycobiology, 2000, 10(5), 439–449, DOI:10.1093/glycob/10.5.439.
- H. Pan, M. J. Colville, N. T. Supekar, P. Azadi and M. J. Paszek, Sequence-Specific Mucins for Glycocalyx Engineering, ACS Synth. Biol., 2019, 8(10), 2315–2326, DOI:10.1021/acssynbio.9b00127.
- C. R. Shurer, M. J. Colville, V. K. Gupta, S. E. Head, F. Kai, J. N. Lakins and M. J. Paszek, Genetically Encoded Toolbox for Glycocalyx Engineering: Tunable Control of Cell Adhesion, Survival, and Cancer Cell Behaviors, ACS Biomater. Sci. Eng., 2017, 4(2), 388–399, DOI:10.1021/acsbiomaterials.7b00037.
- R. Nason, C. Büll, A. Konstantinidi, L. Sun, Z. Ye, A. Halim, W. Du, D. M. Sørensen, F. Durbesson, S. Furukawa, U. Mandel, H. J. Joshi, L. A. Dworkin, L. Hansen, L. David, T. M. Iverson, B. A. Bensing, P. M. Sullam, A. Varki, E. de Vries, C. A. M. de Haan, R. Vincentelli, B. Henrissat, S. Y. Vakhrushev, H. Clausen and Y. Narimatsu, Display of the human mucinome with defined O-glycans by gene engineered cells, Nat. Commun., 2021, 12(1), 4070, DOI:10.1038/s41467-021-24366-4.
- V. R. Kohout, C. L. Wardzala and J. R. Kramer, Synthesis and biomedical applications of mucin mimic materials, Adv. Drug Delivery Rev., 2022, 191, 114540, DOI:10.1016/j.addr.2022.114540.
- R. E. Detwiler and J. R. Kramer, Preparation and applications of artificial mucins in biomedicine, Curr. Opin. Solid State Mater. Sci., 2022, 26(6), 101031, DOI:10.1016/j.cossms.2022.101031.
- C. R. Bertozzi and M. D. Bednarski, Antibody Targeting to Bacterial Cells Using Receptor-Specific Ligands, J. Am. Chem. Soc., 1992, 114(6), 2242–2245, DOI:10.1021/ja00032a046.
- K. G. Leslie, S. S. Berry, G. J. Miller and C. S. Mahon, Sugar-Coated: Can Multivalent Glycoconjugates Improve upon Nature’s Design?, J. Am. Chem. Soc., 2024, 146(40), 27215–27974, DOI:10.1021/jacs.4c08818.
- C. Müller, G. Despras and T. K. Lindhorst, Organizing Multivalency in Carbohydrate Recognition, Chem. Soc. Rev., 2016, 45(11), 3275–3302, 10.1039/C6CS00165C.
- R. E. Detwiler, A. E. Schlirf and J. R. Kramer, Rethinking Transition Metal Catalyzed N-Carboxyanhydride Polymerization: Polymerization of Pro and AcOPro N-Carboxyanhydrides, J. Am. Chem. Soc., 2021, 143(30), 11482–11489, DOI:10.1021/jacs.1c03338.
- J. R. Kramer, B. Onoa, C. Bustamante and C. R. Bertozzi, Chemically tunable mucin chimeras assembled on living cells, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(41), 12574–12579, DOI:10.1073/pnas.1516127112.
- A. C. Deleray and J. R. Kramer, Biomimetic Glycosylated Polythreonines by N-Carboxyanhydride Polymerization, Biomacromolecules, 2022, 23(3), 1453–1461, DOI:10.1021/acs.biomac.2c00020.
- A. C. Deleray, S. S. Saini, A. C. Wallberg and J. R. Kramer, Synthetic Antifreeze Glycoproteins with Potent Ice-Binding Activity, Chem. Mater., 2024, 36(7), 3424–3434, DOI:10.1021/acs.chemmater.4c00266.
- V. R. Kohout, C. L. Wardzala and J. R. Kramer, Mirror Image Mucins and Thio Mucins with Tunable Biodegradation, J. Am. Chem. Soc., 2023, 145(30), 16573–16583, DOI:10.1021/jacs.3c03659.
- Z. S. Clauss, C. L. Wardzala, A. E. Schlirf, N. S. Wright, S. S. Saini, B. Onoa, C. Bustamante and J. R. Kramer, Tunable, biodegradable grafting-from glycopolypeptide bottlebrush polymers, Nat. Commun., 2021, 12, 6472, DOI:10.1038/s41467-021-26808-5.
- Z. S. Clauss and J. R. Kramer, Polypeptoids and Peptoid-Peptide Hybrids by Transition Metal Catalysis, ACS Appl. Mater. Interfaces, 2022, 14(20), 22781–22789, DOI:10.1021/acsami.1c19692.
- H. R. Kricheldorf, Polypeptides and 100 years of chemistry of α-amino acid N-carboxyanhydrides, Angew. Chem., Int. Ed., 2006, 45(35), 5752–5784, DOI:10.1002/anie.200600693.
- T. J. Deming, Synthetic polypeptides for biomedical applications, Prog. Polym. Sci., 2007, 32(8), 858–875, DOI:10.1016/j.progpolymsci.2007.05.010.
- T. J. Deming, Facile synthesis of block copolypeptides of defined architecture, Nature, 1997, 390(6658), 386–389, DOI:10.1038/37084.
- T. J. McPartlon and J. R. Kramer, An Ultra-Potent, Ultra-Economical, Antifreeze Polypeptide, ChemRxiv, 2025, preprint DOI:10.26434/chemrxiv-2025-gvb4b.
- T. J. Deming, Synthesis of Side-Chain Modified Polypeptides, Chem. Rev., 2016, 116(3), 786–808, DOI:10.1021/acs.chemrev.5b00292.
- J. E. Hudak, H. H. Yu and C. R. Bertozzi, Protein glycoengineering enabled by the versatile synthesis of aminooxy glycans and the genetically encoded aldehyde tag, J. Am. Chem. Soc., 2011, 133(40), 16127–16135, DOI:10.1021/ja206023e.
- H. Zhang, J. Zhao, X. Gu and Y. Wen, Targeted treatment of CD22-positive non-Hodgkin’s lymphoma with sialic acid–modified chitosan-PLGA hybrid nanoparticles, J. Nanopart. Res., 2019, 21(7), 154, DOI:10.1007/s11051-019-4595-1.
- Y. Cao, W. Song and X. Chen, Multivalent sialic acid materials for biomedical applications, Biomater. Sci., 2022, 11(8), 2620–2638, 10.1039/D2BM01595A.
- A. H. Courtney, E. B. Puffer, J. K. Pontrello, Z. Q. Yang and L. L. Kiessling, Sialylated multivalent antigens engage CD22 in trans and inhibit B cell activation, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(8), 2500–2505, DOI:10.1073/pnas.0807207106.
- J. E. Hudak, S. M. Canham and C. R. Bertozzi, Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion, Nat. Chem. Biol., 2014, 10(1), 69–75, DOI:10.1038/nchembio.1388.
- P. Zhang, C. Li, X. Ma, J. Ye, D. Wang, H. Cao, G. Yu, W. Wang, X. Lv and C. Cai, Glycopolymer with Sulfated Fucose and 6′-Sialyllactose as a Dual-Targeted Inhibitor on Resistant Influenza A Virus Strains, ACS Macro Lett., 2024, 13(7), 874–881, DOI:10.1021/acsmacrolett.4c00221.
- J. Cervin, A. Boucher, G. Youn, P. Björklund, V. Wallenius, L. Mottram, N. S. Sampson and U. Yrlid, Fucose-Galactose Polymers Inhibit Cholera Toxin Binding to Fucosylated Structures and Galactose-Dependent Intoxication of Human Enteroids, ACS Infect. Dis., 2020, 6(5), 1192, DOI:10.1021/acsinfecdis.0c00009.
- H. Yu, J. V. Rowley, D. C. Green and P. D. Thornton, Fucose-modified thermoresponsive poly(2-hydroxypropyl methacrylate) nanoparticles for controlled doxorubicin release from an injectable depot, Mater. Adv., 2020, 1(5), 1293–1300, 10.1039/D0MA00280A.
- A. L. Sørensen, C. A. Reis, M. A. Tarp, U. Mandel, K. Ramachandran, V. Sankaranarayanan, T. Schwientek, R. Graham, J. Taylor-Papadimitriou, M. A. Hollingsworth, J. Burchell and H. Clausen, Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance, Glycobiology, 2006, 16(2), 96–107, DOI:10.1093/glycob/cwj044.
- M. Fumoto, H. Hinou, T. Ohta, T. Ito, K. Yamada, A. Takimoto, H. Kondo, H. Shimizu, T. Inazu, Y. Nakahara and S. I. Nishimura, Combinatorial synthesis of MUC1 glycopeptides: Polymer blotting facilitates chemical and enzymatic synthesis of highly complicated mucin glycopeptides, J. Am. Chem. Soc., 2005, 127(33), 11804–11818, DOI:10.1021/ja052521y.
- T. Matsushita, R. Sadamoto, N. Ohyabu, H. Nakata, M. Fumoto, N. Fujitani, Y. Takegawa, T. Sakamoto, M. Kurogochi, H. Hinou, H. Shimizu, T. Ito, K. Naruchi, H. Togame, H. Takemoto, H. Kondo and S. I. Nishimura, Functional neoglycopeptides: synthesis and characterization of a new class of MUC1 glycoprotein models having core 2-based O-glycan and complex-type N-glycan chains, Biochemistry, 2009, 48(46), 11117–11133, DOI:10.1021/bi901557a.
- C. S. Kwan, A. R. Cerullo and A. B. Braunschweig, Design and Synthesis of Mucin-Inspired Glycopolymers, ChemPlusChem, 2020, 85(12), 2704–2721, DOI:10.1002/cplu.202000637.
- L. A. Marcaurelle and C. R. Bertozzi, Recent advances in the chemical synthesis of mucin-like glycoproteins, Glycobiology, 2002, 12(6), 69R–77R, DOI:10.1093/glycob/12.6.69R.
- O. Seitz and C. H. Wong, Chemoenzymatic solution- and solid-phase synthesis of O-glycopeptides of the mucin domain of MAdCAM-1. A general route to O-LacNAc, O-sialyl-LacNAc, and O-Sialyl-Lewis-X peptides, J. Am. Chem. Soc., 1997, 119(38), 8766–8776, DOI:10.1021/ja971383c.
- C. Bello, E. Pranzini, E. Piemontese, M. Schrems, M. L. Taddei, L. Giovannelli, M. Schubert, C. F. W. Becker, P. Rovero and A. M. Papini, Chemoenzymatic Synthesis of Glycopeptides to Explore the Role of Mucin 1 Glycosylation in Cell Adhesion, ChemBioChem, 2023, 24(12), e202200741, DOI:10.1002/cbic.202200741.
- I. Nagashima and H. Shimizu, Preparation of Mucin Glycopeptides by Organic Synthesis, Methods Mol. Biol., 2024, 2763, 187–199, DOI:10.1007/978-1-0716-3670-1_16.
- S. Peters, T. Bielfeldt, M. Meldal, K. Bock and H. Paulsen, Solid phase peptide synthesis of mucin glycopeptides, Tetrahedron Lett., 1992, 33(43), 6445–6448, DOI:10.1016/S0040-4039(00)79011-3.
- C. S. Delaveris, E. R. Webster, S. M. Banik, S. G. Boxer and C. R. Bertozzi, Membrane-tethered mucin-like polypeptides sterically inhibit binding and slow fusion kinetics of influenza A virus, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(23), 12643–12650, DOI:10.1073/pnas.1921962117.
- K. Totani, T. Kubota, T. Kuroda, T. Murata, K. I. P. J. Hidari, T. Suzuki, Y. Suzuki, K. Kobayashi, H. Ashida, K. Yamamoto and T. Usui, Chemoenzymatic synthesis and application of glycopolymers containing multivalent sialyloligosaccharides with a poly(L-glutamic acid) backbone for inhibition of infection by influenza viruses, Glycobiology, 2003, 13(5), 315–326, DOI:10.1093/glycob/cwg032.
- Q. Zhao and C. L. Maynard, Mucus, commensals, and the immune system, Gut Microbes, 2022, 14(1), 2041342, DOI:10.1080/19490976.2022.2041342.
- N. Aguilera Montilla, M. Pérez Blas, M. López Santalla and J. M. Martín Villa, Mucosal immune system: A brief review, Inmunologia, 2004, 23(2), 207–216 Search PubMed.
- R. A. Dwek, Glycobiology: ‘Towards understanding the function of sugars’, Biochem. Soc. Trans., 1995, 23(1), 1–25, DOI:10.1042/bst0230001.
- A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar and P. H. Seeberger, Essentials of Glycobiology, 3rd Edition, Cold Spring Harbor (NY), 2017 Search PubMed.
- L. E. Tailford, E. H. Crost, D. Kavanaugh and N. Juge, Mucin glycan foraging in the human gut microbiome, Front. Genet., 2015, 6, 1 CAS.
- J. M. Flynn, D. Niccum, J. M. Dunitz and R. C. Hunter, Evidence and Role for Bacterial Mucin Degradation in Cystic Fibrosis Airway Disease, PLoS Pathog., 2016, 12(8), e1005846, DOI:10.1371/journal.ppat.1005846.
- M. Derrien, E. E. Vaughan, C. M. Plugge and W. M. de Vos, Akkermansia municiphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium, Int. J. Syst. Evol. Microbiol., 2004, 54(Pt 5), 1469–1476, DOI:10.1099/ijs.0.02873-0.
- H. Yuan, J. Zhou, N. Li, X. Wu, S. Huang and S. Park, Isolation and identification of mucin-degrading bacteria originated from human faeces and their potential probiotic efficacy according to host–microbiome enterotype, J. Appl. Microbiol., 2022, 133(2), 362–374, DOI:10.1111/jam.15560.
- K. S. Kim, E. Tiffany, J. Y. Lee, A. Oh, H. S. Jin, J. S. Kim, J. S. Lee, M. H. Nam, S. J. Hong, S. Park, H. Koh, B. S. Kim, Y. K. Lee and D. W. Lee, Genome-wide multi-omics analysis reveals the nutrient-dependent metabolic features of mucin-degrading gut bacteria, Gut Microbes, 2023, 15(1), 2221811, DOI:10.1080/19490976.2023.2221811.
- J. S. Glover, T. D. Ticer and M. A. Engevik, Characterizing the mucin-degrading capacity of the human gut microbiota, Sci. Rep., 2022, 12(1), 8456, DOI:10.1038/s41598-022-11819-z.
- M. A. Engevik, A. C. Engevik, K. A. Engevik, J. M. Auchtung, A. L. Chang-Graham, W. Ruan, R. A. Luna, J. M. Hyser, J. K. Spinler and J. Versalovic, Mucin-Degrading Microbes Release Monosaccharides That Chemoattract Clostridioides difficile and Facilitate Colonization of the Human Intestinal Mucus Layer, ACS Infect. Dis., 2021, 7(5), 1126–1142, DOI:10.1021/acsinfecdis.0c00634.
- A. Helms, V. Chang, S. A. Malaker and J. S. Brodbelt, Unraveling O-Glycan Diversity of Mucins: Insights from SmE Mucinase and Ultraviolet Photodissociation Mass Spectrometry, Anal. Chem., 2024, 96(49), 19230–19237, DOI:10.1021/acs.analchem.4c02011.
- J. Chongsaritsinsuk, A. D. Steigmeyer, K. E. Mahoney, M. A. Rosenfeld, T. M. Lucas, C. M. Smith, A. Li, D. Ince, F. L. Kearns, A. S. Battison, M. A. Hollenhorst, D. Judy Shon, K. H. Tiemeyer, V. Attah, C. Kwon, C. R. Bertozzi, M. J. Ferracane, M. A. Lemmon, R. E. Amaro and S. A. Malaker, Glycoproteomic landscape and structural dynamics of TIM family immune checkpoints enabled by mucinase SmE, Nat. Commun., 2023, 14(1), 6169, DOI:10.1038/s41467-023-41756-y.
- K. Pedram, D. J. Shon, G. S. Tender, N. R. Mantuano, J. J. Northey, K. J. Metcalf, S. P. Wisnovsky, N. M. Riley, G. C. Forcina, S. A. Malaker, A. Kuo, B. M. George, C. L. Miller, K. M. Casey, J. G. Vilches-Moure, M. J. Ferracane, V. M. Weaver, H. Läubli and C. R. Bertozzi, Design of a mucin-selective protease for targeted degradation of cancer-associated mucins, Nat. Biotechnol., 2024, 42(4), 597–607, DOI:10.1038/s41587-023-01840-6.
- S. Park, M. J. Colville, J. H. Paek, C. R. Shurer, A. Singh, E. J. Secor, C. J. Sailer, L. T. Huang, J. C. H. Kuo, M. C. Goudge, J. Su, M. Kim, M. P. DeLisa, S. Neelamegham, J. Lammerding, W. R. Zipfel, C. Fischbach, H. L. Reesink and M. J. Paszek, Immunoengineering can overcome the glycocalyx armour of cancer cells, Nat. Mater., 2024, 23(3), 429–438, DOI:10.1038/s41563-024-01808-0.
- C. Liu, A. C. Madl, D. Cirera-Salinas, W. Kress, F. Straube, D. Myung and G. G. Fuller, Mucin-Like Glycoproteins Modulate Interfacial Properties of a Mimetic Ocular Epithelial Surface, Advanced Science, 2021, 8(16), 2100841, DOI:10.1002/advs.202100841.
- H. Coelho, T. Matsushita, G. Artigas, H. Hinou, F. J. Cañada, R. Lo-Man, C. Leclerc, E. J. Cabrita, J. Jiménez-Barbero, S. I. Nishimura, F. Garcia-Martín and F. Marcelo, The Quest for Anticancer Vaccines: Deciphering the Fine-Epitope Specificity of Cancer-Related Monoclonal Antibodies by Combining Microarray Screening and Saturation Transfer Difference NMR, J. Am. Chem. Soc., 2015, 137(39), 12438–12441, DOI:10.1021/jacs.5b06787.
- S. Julien, P. A. Videira and P. Delannoy, Sialyl-Tn in cancer: (How) did we miss the target?, Biomolecules, 2012, 2(4), 435–466, DOI:10.3390/biom2040435.
- S. Sell, Cancer-associated carbohydrates identified by monoclonal antibodies, Hum. Pathol., 1990, 21(10), 1003–1019, DOI:10.1016/0046-8177(90)90250-9.
- T. J. Deming, Living polymerization of α-amino acid-N-carboxyanhydrides, J. Polym. Sci., Part A: Polym. Chem., 2000, 38(17), 3011–3018 CrossRef CAS.
- Z. S. Clauss, C. L. Wardzala, A. E. Schlirf, N. S. Wright, S. S. Saini, B. Onoa, C. Bustamante and J. R. Kramer, Tunable, biodegradable grafting-from glycopolypeptide bottlebrush polymers, Nat. Commun., 2021, 12, 6472, DOI:10.1038/s41467-021-26808-5.
- D. J. Thornton, From Mucins to Mucus: Toward a More Coherent Understanding of This Essential Barrier, Proc. Am. Thorac. Soc., 2004, 1(1), 54–61, DOI:10.1513/pats.2306016.
- E. Gianchecchi, A. Arena and A. Fierabracci, Sialic acid-siglec axis in human immune regulation, involvement in autoimmunity and cancer and potential therapeutic treatments, Int. J. Mol. Sci., 2021, 22(11), 5774, DOI:10.3390/ijms22115774.
- Z. Gao, Z. Wu, Y. Han, X. Zhang, P. Hao, M. Xu, S. Huang, S. Li, J. Xia, J. Jiang and S. Yang, Aberrant Fucosylation of Saliva Glycoprotein Defining Lung Adenocarcinomas Malignancy, ACS Omega, 2022, 7(21), 17894–17906, DOI:10.1021/acsomega.2c01193.
- A. Martínez-Antón, C. DeBolós, M. Garrido, J. Roca-Ferrer, C. Barranco, I. Alobid, A. Xaubet, C. Picado and J. Mullol, Mucin genes have different expression patterns in healthy and diseased upper airway mucosa, Clin. Exp. Allergy, 2006, 36(4), 448–457, DOI:10.1111/j.1365-2222.2006.02451.x.
- C. Liu, S. Deng, K. Jin, Y. Gong, H. Cheng, Z. Fan, Y. Qian, Q. Huang, Q. Ni, G. Luo and X. Yu, Lewis antigen-negative pancreatic cancer: An aggressive subgroup, Int. J. Oncol., 2020, 56(4), 900–908, DOI:10.3892/ijo.2020.4989.
- V. Pieri, A. L. Gallotti, D. Drago, M. Cominelli, I. Pagano, V. Conti, S. Valtorta, A. Coliva, S. Lago, D. Michelatti, L. Massimino, F. Ungaro, L. Perani, A. Spinelli, A. Castellano, A. Falini, A. Zippo, P. L. Poliani, R. M. Moresco, A. Andolfo and R. Galli, Aberrant L-Fucose Accumulation and Increased Core Fucosylation Are Metabolic Liabilities in Mesenchymal Glioblastoma, Cancer Res., 2023, 83(2), 195–218, DOI:10.1158/0008-5472.CAN-22-0677.
- C. A. Werlang, W. G. Chen, K. Aoki, K. M. Wheeler, C. Tymm, C. J. Mileti, A. C. Burgos, K. Kim, M. Tiemeyer and K. Ribbeck, Mucin O-glycans suppress quorum-sensing pathways and genetic transformation in Streptococcus mutans, Nat. Microbiol., 2021, 6(5), 574–583, DOI:10.1038/s41564-021-00876-1.
- N. L. Kavanaugh, A. Q. Zhang, C. J. Nobile, A. D. Johnson and K. Ribbeck, Mucins suppress virulence traits of Candida albicans, mBio, 2014, 5(6), 1–8, DOI:10.1128/mBio.01911-14.
- K. M. Wheeler, G. Cárcamo-Oyarce, B. S. Turner, S. Dellos-Nolan, J. Y. Co, S. Lehoux, R. D. Cummings, D. J. Wozniak and K. Ribbeck, Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection, Nat. Microbiol., 2019, 4(12), 2146–2154, DOI:10.1038/s41564-019-0581-8.
- J. Takagi, K. Aoki, B. S. Turner, S. Lamont, S. Lehoux, N. Kavanaugh, M. Gulati, A. Valle Arevalo, T. J. Lawrence, C. Y. Kim, B. Bakshi, M. Ishihara, C. J. Nobile, R. D. Cummings, D. J. Wozniak, M. Tiemeyer, R. Hevey and K. Ribbeck, Mucin O-glycans are natural inhibitors of Candida albicans pathogenicity, Nat. Chem. Biol., 2022, 18(7), 762–773, DOI:10.1038/s41589-022-01035-1.
- B. X. Wang, J. Takagi, A. McShane, J. H. Park, K. Aoki, C. Griffin, J. Teschler, G. Kitts, G. Minzer, M. Tiemeyer, R. Hevey, F. Yildiz and K. Ribbeck, Host-derived O-glycans inhibit toxigenic conversion by a virulence-encoding phage in Vibrio cholerae, EMBO J., 2022, 42, e111562, DOI:10.15252/embj.2022111562.
- M. C. Glick, V. A. Kothari, A. Liu, L. I. Stoykova and T. F. Scanlin, Activity of fucosyltransferases and altered glycosylation in cystic fibrosis airway epithelial cells, Biochimie, 2001, 83(8), 743–747, DOI:10.1016/s0300-9084(01)01323-2.
- D. G. Comb, D. R. Watson and S. Roseman, The sialic acids. IX. Isolation of cytidine 5′-monophospho-N-acetylneuraminic acid from Escherichia coli K-235, J. Biol. Chem., 1966, 241(23), 5637–5642 CrossRef CAS PubMed.
- P. A. Gilormini, C. Lion, M. Noel, M. A. Krzewinski-Recchi, A. Harduin-Lepers, Y. Guérardel and C. Biot, Improved workflow for the efficient preparation of ready to use CMP-activated sialic acids, Glycobiology, 2016, 26(11), 1151–1156, DOI:10.1093/glycob/cww084.
- C. Zhao, Y. Wu, H. Yu, I. M. Shah, Y. Li, J. Zeng, B. Liu, D. A. Mills and X. Chen, The one-pot multienzyme (OPME) synthesis of human blood group H antigens and a human milk oligosaccharide (HMOS) with highly active Thermosynechococcus elongatus α1-2-fucosyltransferase, Chem. Commun., 2016, 52(20), 3899–3902, 10.1039/c5cc10646j.
- H. Yu, J. Zeng, Y. Li, V. Thon, B. Shi and X. Chen, Effective one-pot multienzyme (OPME) synthesis of monotreme milk oligosaccharides and other sialosides containing 4-: O -acetyl sialic acid, Org. Biomol. Chem., 2016, 14(36), 8586–8597, 10.1039/C6OB01706A.
- H. Yu, H. Yu, R. Karpel and X. Chen, Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: Comparison of substrate flexibility of three microbial CMP-sialic acid synthetases, Bioorg. Med. Chem., 2004, 12(24), 6427–6435, DOI:10.1016/j.bmc.2004.09.030.
- H. Yu, S. Huang, H. Chokhawala, M. Sun, H. Zheng and X. Chen, Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural α-2,6-linked sialosides: A P. damsela α-2,6- sialyltransferase with extremely flexible donor-substrate specificity, Angew. Chem., Int. Ed., 2006, 45(24), 3938–3944, DOI:10.1002/anie.200600572.
- X. Yang, H. Yu, X. Yang, A. Singh Kooner, Y. Yuan, B. Luu and X. Chen, One-pot Multienzyme (OPME) Chemoenzymatic Synthesis of Brain Ganglioside Glycans with Human ST3GAL II Expressed in E. coli, ChemCatChem, 2022, 14(2), e202101498, DOI:10.1002/cctc.202101498.
- L. Ding, H. Yu, K. Lau, Y. Li, S. Muthana, J. Wang and X. Chen, Efficient chemoenzymatic synthesis of sialyl Tn-antigens and derivatives, Chem. Commun., 2011, 47(30), 8691–8693, 10.1039/c1cc12732b.
- H. H. Yu, H. Chokhawala, R. Karpel, H. H. Yu, B. Wu, J. Zhang, Y. Zhang, Q. Jia and X. Chen, A multifunctional Pasteurella multocida sialyltransferase: A powerful tool for the synthesis of sialoside libraries, J. Am. Chem. Soc., 2005, 127(50), 17618–17619, DOI:10.1021/ja0561690.
- L. Zhang, K. Lau, J. Cheng, H. Yu, Y. Li, G. Sugiarto, S. Huang, L. Ding, V. Thon, P. G. Wang and X. Chen, Helicobacter hepaticus Hh0072 gene encodes a novel α1-3-fucosyltransferase belonging to CAZy GT11 family, Glycobiology, 2010, 20(9), 1077–1088, DOI:10.1093/glycob/cwq068.
- H. Yu, Y. Li, Z. Wu, L. Li, J. Zeng, C. Zhao, Y. Wu, N. Tasnima, J. Wang, H. Liu, M. R. Gadi, W. Guan, P. G. Wang and X. Chen, H. pylori α1-3/4-fucosyltransferase (Hp3/4FT)-catalyzed one-pot multienzyme (OPME) synthesis of Lewis antigens and human milk fucosides, Chem. Commun., 2017, 53(80), 11012–11015, 10.1039/C7CC05403C.
- L. Ding, H. Yu, K. Lau, Y. Li, S. Muthana, J. Wang and X. Chen, Efficient chemoenzymatic synthesis of sialyl Tn-antigens and derivatives, Chem. Commun., 2011, 47(30), 8691–8693, 10.1039/C1CC12732B.
- Sigma Aldrich, α(2 → 6) Sialyltransferase from Photobacterium damsela, https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/277/753/s2076dat.pdf.
- Sigma Aldrich, Mucin from bovine submaxillary glands, https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/230/079/m3895pis.pdf.
- C. Robbe, C. Capon, E. Maes, M. Rousset, A. Zweibaum, J. P. Zanetta and J. C. Michalski, Evidence of regio-specific glycosylation in human intestinal mucins: Presence of an acidic gradient along the intestinal tract, J. Biol. Chem., 2003, 278(47), 46337–46348, DOI:10.1074/jbc.M302529200.
- A. Wesley, J. Forstner, R. Qureshi, M. Mantle and G. Forstner, Human intestinal mucin in cystic fibrosis, Pediatr. Res., 1983, 17(1), 65–69, DOI:10.1203/00006450-198301000-00013.
- C. Jin, D. T. Kenny, E. C. Skoog, M. Padra, B. Adamczyk, V. Vitizeva, A. Thorell, V. Venkatakrishnan, S. K. Lindén and N. G. Karlsson, Structural diversity of human gastric mucin glycans, Mol. Cell. Proteomics, 2017, 16(5), 743–758, DOI:10.1074/mcp.M117.067983.
- E. S. Harris, H. J. McIntire, M. Mazur, H. Schulz-Hildebrandt, H. M. Leung, G. J. Tearney, S. Krick, S. M. Rowe and J. W. Barnes, Reduced sialylation of airway mucin impairs mucus transport by altering the biophysical properties of mucin, Sci. Rep., 2024, 14(1), 1–16, DOI:10.1038/s41598-024-66510-2.
- E. E. L. Nyström, B. Martinez-Abad, L. Arike, G. M. H. Birchenough, E. B. Nonnecke, P. A. Castillo, F. Svensson, C. L. Bevins, G. C. Hansson and M. E. V. Johansson, An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function, Science, 2021, 372(6539), eabb1590, DOI:10.1126/science.abb1590.
- M. E. A. Pereira, E. C. Kisailus, F. Gruezo and E. A. Kabat, Immunochemical studies on the combining site of the blood group H-Specific lectin 1 from Ulex europeus seeds, Arch. Biochem. Biophys., 1978, 185(1), 108–115, DOI:10.1016/0003-9861(78)90149-2.
- J. C. Manimala, T. A. Roach, Z. Li and J. C. Gildersleeve, High-throughput carbohydrate microarray analysis of 24 lectins, Angew. Chem., Int. Ed., 2006, 45(22), 3607–3610, DOI:10.1002/anie.200600591.
- D. Bojar, L. Meche, G. Meng, W. Eng, D. F. Smith, R. D. Cummings and L. K. Mahal, A Useful Guide to Lectin Binding: Machine-Learning Directed Annotation of 57 Unique Lectin Specificities, ACS Chem. Biol., 2022, 17, 2993–3012, DOI:10.1021/acschembio.1c00689.
- A. K. Allen, A. Neuberger and N. Sharon, The purification, composition and specificity of wheat-germ agglutinin, Biochem. J., 1973, 131(1), 155, DOI:10.1042/bj1310155.
- P. Schick and W. J. Filmyer, Sialic acid in mature megakaryocytes: detection by wheat germ agglutinin, Blood, 1985, 65(5), 120–126 CrossRef.
- M. Sato, S. Yonezawa, H. Uehara, Y. Arita, E. Sato and T. Muramatsu, Differential distribution of receptors for two fucose-binding lectins in embryos and adult tissues of the mouse, Differentiation, 1986, 30(3), 211–219, DOI:10.1111/j.1432-0436.1986.tb00783.x.
- I. H. van Stokkum, H. J. Spoelder, M. Bloemendal, R. van Grondelle and F. C. Groen, Estimation of protein secondary structure and error analysis from circular dichroism spectra, Anal. Biochem., 1990, 191(1), 110–118, DOI:10.1016/0003-2697(90)90396-q.
- S. W. Provencher and J. Glöckner, Estimation of Globular Protein Secondary Structure from Circular Dichroism, Biochemistry, 1981, 20(1), 33–37, DOI:10.1021/bi00504a006.
- M. Frankel, M. Breuer and S. Cordova, Synthesis of poly-O-acetylserine and of polyserine, Experientia, 1952, 8(8), 299–300, DOI:10.1007/BF02153298.
- R. Owada, S. Mitsui and K. Nakamura, Exogenous polyserine and polyleucine are toxic to recipient cells, Sci. Rep., 2022, 12(1), 1685, DOI:10.1038/s41598-022-05720-y.
- R. W. Woody, Optical Rotatory Properties of Biopolymers, J. Polym. Sci., Macromol. Rev., 1977, 12(1), 181–320, DOI:10.1002/pol.1977.230120104.
- Z. Shi, R. W. Woody and N. R. Kallenbach, Is polyproline II a major backbone conformation in unfolded proteins?, Adv. Protein Chem., 2002, 62, 163–240, DOI:10.1016/s0065-3233(02)62008-x.
- N. Sreerama and R. W. Woody, Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set, Anal. Biochem., 2000, 287(2), 252–260, DOI:10.1006/abio.2000.4880.
- A. J. Miles, S. G. Ramalli and B. A. Wallace, DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data, Protein Sci., 2022, 31(1), 37–46, DOI:10.1002/pro.4153.
- L. Whitmore and B. A. Wallace, Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases, Biopolymers, 2008, 89(5), 392–400, DOI:10.1002/bip.20853.
- A. J. Miles, E. D. Drew and B. A. Wallace, DichroIDP: a method for analyses of intrinsically disordered proteins using circular dichroism spectroscopy, Commun. Biol., 2023, 6(1), 823, DOI:10.1038/s42003-023-05178-2.
- L. B. Chemes, L. G. Alonso, M. G. Noval and G. De Prat-Gay, Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains, Methods Mol. Biol., 2012, 895, 387–404, DOI:10.1007/978-1-61779-927-3_22.
- J. L. S. Lopes, A. J. Miles, L. Whitmore and B. A. Wallace, Distinct circular dichroism spectroscopic signatures of polyproline II and unordered secondary structures: Applications in secondary structure analyses, Protein Sci., 2014, 23(12), 1765–1772, DOI:10.1002/pro.2558.
- R. E. Detwiler, T. J. McPartlon, C. S. Coffey and J. R. Kramer, Clickable Polyprolines from Azido-proline N -Carboxyanhydride, ACS Polym. Au, 2023, 3, 383–393, DOI:10.1021/acspolymersau.3c00011.
- D. M. Coltart, A. K. Royyuru, L. J. Williams, P. W. Glunz, D. Sames, S. D. Kuduk, J. B. Schwarz, X. T. Chen, S. J. Danishefsky and D. H. Live, Principles of mucin architecture: Structural studies on synthetic glycopeptides bearing clustered mono-, di-, tri-, and hexasaccharide glycodomains, J. Am. Chem. Soc., 2002, 124(33), 9833–9844, DOI:10.1021/ja020208f.
- H. S. Davies, P. Singh, T. Deckert-Gaudig, V. Deckert, K. Rousseau, C. E. Ridley, S. E. Dowd, A. J. Doig, P. D. A. Pudney, D. J. Thornton and E. W. Blanch, Secondary structure and glycosylation of mucus glycoproteins by Raman spectroscopies, Anal. Chem., 2016, 88(23), 11609–11615, DOI:10.1021/acs.analchem.6b03095.
- N. Martínez-Sáez, J. Castro-Lõpez, J. Valero-González, D. Madariaga, I. Compañõn, V. J. Somovilla, M. Salvadõ, J. L. Asensio, J. Jiménez-Barbero, A. Avenoza, J. H. Busto, G. J. L. Bernardes, J. M. Peregrina, R. Hurtado-Guerrero and F. Corzana, Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an anti-MUC1 Antibody, Angew. Chem., Int. Ed., 2015, 54(34), 9830–9834, DOI:10.1002/anie.201502813.
- D. Madariaga, N. Martínez-Sáez, V. J. Somovilla, L. García-García, M. Á. Berbis, J. Valero-Gónzalez, S. Martín-Santamaría, R. Hurtado-Guerrero, J. L. Asensio, J. Jiménez-Barbero, A. Avenoza, J. H. Busto, F. Corzana and J. M. Peregrina, Serine versus threonine glycosylation with α-o-GalNac: Unexpected selectivity in their molecular recognition with lectins, Chem. – Eur. J., 2014, 20(39), 12616–12627, DOI:10.1002/chem.201403700.
- M. J. Paszek, C. C. DuFort, O. Rossier, R. Bainer, J. K. Mouw, K. Godula, J. E. Hudak, J. N. Lakins, A. C. Wijekoon and L. Cassereau, et al., The cancer glycocalyx mechanically primes integrin-mediated growth and survival, Nature, 2014, 511(7509), 319–325, DOI:10.1038/nature13535.
- J. C. H. Kuo, J. G. Gandhi, R. N. Zia and M. J. Paszek, Physical biology of the cancer cell glycocalyx, Nat. Phys., 2018, 14(7), 658–669, DOI:10.1038/s41567-018-0186-9.
- S. A. Malaker, K. Pedram, M. J. Ferracane, B. A. Bensing, V. Krishnan, C. Pett, J. Yu, E. C. Woods, J. R. Kramer, U. Westerlind, O. Dorigo and C. R. Bertozzi, The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(15), 7278–7287, DOI:10.1073/pnas.1813020116.
- R. L. Szabady and R. A. Welch, StcE Peptidase and the StcE-Like Metalloendopeptidases, Handbook of Proteolytic Enzymes, 2013, vol. 1, pp. 1272–1280 Search PubMed.
- W. W. Lathem, T. E. Grys, S. E. Witowski, A. G. Torres, J. B. Kaper, P. I. Tarr and R. A. Welch, StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor, Mol. Microbiol., 2002, 45(2), 277–288, DOI:10.1046/j.1365-2958.2002.02997.x.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, NIH Image to ImageJ: 25 years of image analysis, Nat. Methods, 2012, 9(7), 671–675, DOI:10.1038/nmeth.2089.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5cb00111k |
| This journal is © The Royal Society of Chemistry 2025 |