Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Regulation of bacterial phosphorelay systems
Daniel M.
Foulkes
,
Daniel M.
Cooper
 ,
Catherine
Westland
,
Catherine
Westland
 and
Dominic P.
Byrne
and
Dominic P.
Byrne
 *
*
Department of Biochemistry, Cell and Systems Biology, Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK. E-mail: bs0u4193@liverpool.ac.uk
First published on 19th June 2025
Abstract
In terms of biomass, bacteria are the most successful organisms on earth. This is partly attributed to their tremendous adaptive capabilities, which allows them to sense and rapidly organise responses to changing environmental stimuli. Using complex signalling mechanisms, bacteria can relay cellular information to fine-tune their metabolism, maintain homeostasis, and trigger virulence processes during infection. Across all life, protein phosphorylation represents the most abundant signalling mechanism, which is controlled by a versatile class of enzymes called protein kinases and their cognate phosphatases. For many years, histidine kinase (HK)-containing two-component systems (TCSs) were considered the canonical instruments of bacterial sensing. However, advances in metagenomics has since proven that bacterial phosphorelay is in fact orchestrated by a functionally diverse array of integrated protein kinase types, including Ser, Thr, Tyr and Arg-targeting enzymes. In this review, we provide an up-to-date appraisal of bacterial kinase signalling, with an emphasis on how these sensing pathways are regulated to modulate kinase output. Finally, we explore how selective kinase inhibitors may be exploited to control infections and combat the looming health emergency of multidrug resistant bacteria.
Life has evolved an abundance of protein-based molecular machines to detect and coordinate responses to environmental stimuli. These biological processes are collectively referred to as signal transduction events and are controlled, in part, by a series of functionally divergent enzymes that post-translationally introduce covalent chemical modifications onto target proteins in a tightly regulated manner. One such class of enzymes are the protein kinases, which catalyse the reversible addition of a phosphate group from ATP to conserved amino acid residues within a protein substrate, in a process called phosphorylation. Since the discovery of protein kinase activity in 1954,1 and the first protein kinase (phosphorylase kinase) a few years later,2–4 it is now accepted that protein phosphorylation controls virtually every aspect of life. Indeed, protein phosphorylation is the most extensively studied post-translational modification in bacteria, as evidenced by the wealth of literature written on the topic.5–13 Bacterial kinases can be classified into 5 groups: His kinases (HKs), non-Hanks type Tyr kinases, Arg kinases, Hanks-type Ser/Thr kinases (the so-called ‘eukaryotic-like Ser/Thr kinases’ [eSTKs]), and atypical Ser/Thr kinases.14 HK-containing two-component systems (TCS) constitute the dominant signal transduction circuitry across all prokaryote lineages and function as conduits between extracellular sensing and intracellular responses. Individual bacterial species can encode multiple TCSs, which enables sensing of a diverse and ever-expanding repertoire of known environmental signals.15–17 The ubiquitous TCSs are thus considered master regulators of prokaryotic metabolism,18–20 adaptation to environmental changes,21–23 virulence,18,24–27 antimicrobial resistance,28–33 motility and chemotaxis.34–37 Although TCSs are not components of mammalian cellular signalling networks, similar systems have been identified in some eukaryotic fungi, and higher plants.38 These eukaryotic TCSs are structurally more divergent, often supplemented with additional regulatory modules, and can tune the outputs of downstream signalling proteins, such as other kinases.38–41
In addition to HKs, phosphorylation of protein Ser/Thr and Tyr residues (O-phosphorylation), by enzymes that share striking sequence or structural similarities with canonical eukaryotic Hanks-type kinases (Fig. 1), has also emerged as a pervasive phosphosignalling mechanism in bacteria and archaea.42,43 Since the discovery of the first eSTK, Pkn1 in Myxococcus xanthus,44 it is now established that the genomes of bacteria can contain multiple eSTKs, with the prevalence of these kinases approximately correlating with the complexity of the bacteria's replicative niche.45 For example, Sorangium cellulosum encodes 317 eSTKs, which may imply a multi-directional signalling network reminiscent of eukaryotic systems.46 In fact, >80% of the Mycobacterium tuberculosis proteome (which encodes 11 eSTKs, including PknG (Fig. 1B)), and 63% of the theoretical proteome of Clostridium difficile is predicted to be O-phosphorylated.7,12 Several bacteria also encode atypical Ser/Thr protein kinases which diverge from the traditional Hanks-type kinase fold. For example, the HPr kinase/phosphorylase (HprK/P) of Bacillus subtilis is unrelated to Hanks-type kinases (instead binding nucleotides via a Walker A motif located in the P-loop) and regulates catabolite repression by phosphorylating (and dephosphorylating) the central HPr protein of the phosphoenolpyruvate: sugar phosphotransferase system.47–49 YihE is another example of an atypical Ser/Thr kinase of E. coli that does not resemble canonical eSTKs in terms of structure or sequence homology, but plays critical role in regulating stress-related programmed cell death in bacteria.50,51 Interestingly, CotH, an atypical protein kinase of B. subtilis and B. cereus (Fig. 1C) shares sequence similarity with the human Golgi casein kinase, Fam20C.52 Although CotH is not known to serve sensing or signalling functions per se, it is required to maintain the integrity of the spore-coat structure.52 CotH retains some features of canonical Ser/Thr protein kinases (including conserved active site residues corresponding to D166, N171, and D184 of the prototypical protein kinase, PKA) but diverges from the typical kinase fold in several crucial ways (Fig. 1C and D); it lacks a putative metal binding DFG motif and utilises an unusual mode of ATP binding involving two aromatic residues, Tyr142 and Trp245, that ‘sandwich’ the adenine moiety of ATP.52 In contrast to eSTKs, bacterial tyrosine kinases (BY tyrosine kinases) display limited homology with their mammalian equivalents, and are characterised by Walker A and B ATP/GTP binding motifs.53 Finally, while “classical” sensing and signalling roles have yet to be identified, arginine phosphorylation is known to regulate several cellular processes, particularly in Gram-positive bacteria, such as B. subtillus and Staphylococcus aureus, where it functions as a protein degradation signal.54,55 For example, phosphorylation of proteins by the arginine kinase, McsB of B. subtilis, targets them for recycling by the ClpCP protease system.54 In contrast to the other phosphotransfer systems, N-phosphorylation of arginine is a relatively understudied PTM, owing to the intrinsic lability of the phosphoamidate bond impeding conventional proteomics investigations.56 As a result, our broader understanding of arginine phosphorylation and its roles within bacterial signaling networks remains limited. Collectively, the varied phosphorelay systems encoded by bacterial species provide a finely tuned and tightly regulated series of signal cascades that govern multiple aspects of bacterial life. This review provides an up-to-date account of the mechanisms that regulate bacterial phosphotransfer events.
 | ||
| Fig. 1 Structural similarities between eukaryotic and prokaryotic Ser/Thr kinases. Crystal structures of (A) human PKA in complex with ATP (PDB: 4WB5), (B) M. tuberculosis PknG in complex with ADP (4Y0X) and (C) Bacillus atypical protein kinase, CotH, in complex with AMP. Insets show ATP/AMP coordinating amino acid residues (white sticks). α-Helices are shaded blue, β-strands are shaded red, loops are shaded grey, Mg2+ ions are represented as turquoise spheres and ATP/AMP is represented by green sticks. (D) Sequence alignment of PKA with PknG and CotH (using MUSCLE). Canonical amino acid residues (or functional equivalents) involved in ATP binding or phosphate transfer are colour coordinated. Note that the atypical kinase, CotH, aligns poorly due to its highly diverged sequence. | ||
Regulation of TCS phosphorelay signals
A prototypical bacterial histidine kinase (HK) is a multidomain His to Asp phosphorelay enzyme (Fig. 2A), consisting of a variable N-terminal “sensing region”, transmembrane (TM) domain, and a conserved catalytic transmitter domain (containing catalytic ATPase [CA domain] and homodimeric phosphoacceptor/phosphotransferase [DHp] subdomains57). HKs also commonly contain one or more intracellular signalling domains; including HAMP (found in histidine kinases, adenylyl cyclases, methyl binding proteins, and phosphatases), PAS (Per-ARNT-Sim), STAC (SLC and TCST-Associated Component) or GAF (found in cGMP-specific phosphodiesterase, adenylyl cyclases, and FhlA) domains,57,58 that augment HK regulation by serving as sensing conductors of extracellular stimuli. Upon stimulation, the signal is transduced to the TCS catalytic core, resulting in ATP binding by the CA domain and autophosphorylation of the conserved DHp acceptor His residue. The phosphate moiety is then relayed to the receiver (REC) domain of a response regulator (RR) module, which catalyses its own aspartic acid phosphorylation.38 Following phosphorylation, most RRs undergo a conformational switch (typically leading to homodimerisation) which facilitates binding to regulatory genomic motifs and activation or repression of gene expression.59–62 Fascinatingly, there is also growing evidence to suggest that some RRs also possess phosphorylation-independent regulatory functions.63–65 For example, the phosphorylated form of the Salmonella typhimurium RR, RcsB, can homodimerize or form a heterodimer with the ancillary regulator, RcsA, to modulate gene transcription.66 However, recent structural investigations have revealed that an active RcsB dimer can be stabilized just by binding to DNA even in the absence of phosphorylation67 and the unphosphorylated form of RcsB can also interact with other transcription factors to regulate gene expression68–71 adding an extra layer of regulatory complexity to the traditional TCS model. Some HKs are also ‘hybrid kinases’ that are supplemented with carboxy-terminal REC and/or histidine phosphotransfer (HPt) domains, which are frequently involved in multi-step phosphorelay systems with contiguous signalling elements, such as DHp and RR proteins.72 Although TCS-HKs are canonically recognized to adopt a dimeric architecture, monomeric species have also been reported, including EL346 of Erythrobacter litoralis.73 In the inactive EL346 state, the typical DHp dimer interface of the kinase domain is blocked by direct interaction with a blue-light sensitive light–oxygen–voltage (LOV) domain, which competes with the CA domain for access to the phosphorylatable histidine. | ||
| Fig. 2 Overview of bacterial kinase signal transduction mechanisms. (A) Asymmetrical TCS kinase state: classical two component signalling systems (TCSs) respond to extracellular (through periplasmic receptor) or cytosolic (through HAMP/PAS/GAF) sensor domains, leading to dimerization. In a multistep phosphorelay, the catalytic CA domains phosphorylate, in trans, a conserved His in the DHp domain. The phosphate is then shuttled (dashed arrows) to a conserved Asp residue in cognate response regulators (RRs). RRs dimerize and bind to a specific DNA binding domain (DBD) to elicit transcription of target genes. Symmetrical TCS phosphatase state: in the absence of stimuli, the CA domain undergoes a conformation switch to a phosphatase-active state, dephosphorylating RRs in cis, releasing phosphate into the cytosol. (B) Simplified schematic of a B. subtilis eSTK, PrkC, responding to extracytosolic stimuli via PASTA sensor domains. During stationary phase, binding of muropeptides to PASTA repeats induces dimerization of PrkC TM and eSTK domains, leading to in trans autophosphorylation and subsequent phosphorylation of a range of target cytosolic proteins. Substrates include, but are not limited to, GpsB (a cell division protein74), YkwC (an oxidoreductase75) and CpgA (a GTPase76). PrkC also regulates the WalRK TCS by directly phosphorylating the WalR RR, influencing expression of genes relating to cell wall biosynthesis.77 | ||
Extra/intracellular or periplasmic signals are typically sensed by N-terminal located sensory PAS domains and propagated along the HK through a series of transient structural rearrangements and inter-domain interactions that regulate catalytic activity and phosphotransfer.58,78 Although the amino acid composition between different PAS domains of HKs is extremely variable (which drives substrate specificity), they adopt a highly conserved multi-β-sheet core structure with adjoining α-helices that forms a ligand-accepting cleft region.57,78,79 Sensing can also be achieved by PAS-like domains, all α-helix type structures, and Venus flytrap (VFT) domains.57 Through these mechanisms, TCSs can perceive and respond to a range of chemical and physical stimuli. In the absence of an activation signal, many HKs function as putative phosphatases of RRs, allowing a molecular reset of TCS signalling.80–83 Shifts in the equilibrium of kinase and phosphatase-active states are therefore necessary to balance the net physiological outcomes of TCS signalling.84,85 The molecular mechanisms of HK signal sensing, as it pertains to the structural dynamics of these mutually exclusive kinase and phosphatase competent states, have been extensively reviewed elsewhere.8,58,86 In brief, the activity of HKs is contingent on signal-dependent domain reorganisations, whereby the conserved kinase core adopts an asymmetrical homodimer configuration, containing both active and inactive catalytic subunits.8 The CA domain of the inactive conformation can bind to ATP, whilst the DHp domain remains accessible for phosphotransfer to the REC domain of an RR. In the active conformation, both the DHp and CA domains form a tight complex to catalyse in-cis or in-trans transfer of the γ-ATP phosphate onto the eponymous His site of the DHp domain, which is inaccessible to the RR.8,86,87 Bacteria also encode a range of non-TCS kinase signalling relay systems. A related group of non-TCS HKs are associated with the core signalling complexes of chemoreceptors in motile bacteria and archaea,88 which incorporate soluble cytoplasmic HKs (such as CheA in E. coli) as part of a supramolecular sensing array. Of note, several other HKs that are soluble enzymes, lacking transmembrane segments, have been identified.34,89,90
Regulation of Ser/Thr protein kinases
Membrane-associated (and cytoplasmic) Hanks-type eSTKs serve regulatory functions in numerous cell processes and have also been implicated in the virulence of pathogens.10,91–96 eSTK-based signalling systems typically rely on the opposing activities of a protein kinase and an associated phosphatase, that (in contrast to HKs) can concurrently (de)phosphorylate Ser/Thr residues of a broad range of protein classes.10 However, mechanistic details of how branching eSTK signalling cascades are organised are relatively scarce. In addition to a surprisingly well-conserved Hanks-type fold in the kinase catalytic core,45,94 many eSTKs are buttressed with additional modular regulatory domains that serve as sensor regions, mediate protein–protein interactions and are sites of ligand binding. These include penicillin-binding and Ser/Thr kinase-associated repeats (PASTA) and forkhead-associated (FHA) domains that recognize phosphothreonines.95,97,98 In particular, extracellular sensory PASTA domains of transmembrane eSTKs (the “PASTA kinase” family), which can interact with ligands (usually peptidoglycan or peptidoglycan precursors) and induce activated kinase domain homodimers, are reasonably well characterised.45,95,99 PrkC, a membrane-associated eSTK of B. subtilis, contains three PASTA domains and plays crucial roles in spore germination, vegetative growth, modulation of cell wall synthesis and antibiotic resistance.5,100 PrkC activity is differentially regulated by bacterial growth phases. During the stationary phase, binding of peptidoglycan fragments to the extracellular PASTA domain results in PrkC dimerisation and activation of the cytosolic kinase domain (Fig. 2B),101 whilst during exponential growth, activation is attained by binding to the cell division proteins GpsB, DivIVA, and EzrA.102 Peptidoglycan binding by PASTA repeats of S. aureus STK similarly induces dimerization and activation of the kinase,103 which goes on to phosphorylate and inhibit several cell wall synthesis enzymes, such as FemX.104 PASTA domains also enable ligand-induced modulation of kinase activity and proper localisation of PknB from M. tuberculosis105,106 with dimerisation of the PknB kinase domain N-lobe resulting in allosteric activation of catalytic function.107A common feature of eukaryotic protein kinases and several eSTKs is a requirement for phosphorylation of conserved sites within their putative activation loop to switch on catalysis. For example, the PASTA-kinase of Enterococcus faecalis, IreK, autophosphorylates at three positions on the activation loop (Thr163,![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 166,
166,![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) and
and![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 168) and an additional site on the C-lobe in response to cephalosporin treatment, resulting in enhanced activity.108 Autophosphorylation of activation loop amino acid residues appears to be a conserved regulatory mechanism for many eSTKs, including PrkA of Listeria monocytogenes,109 PrkC from B. subtilis,110 YegI of E. coli111 and PknB from M. tuberculosis.112 The molecular basis of mycobacterial cytoplasmic PknG activation is also predicted to involve N-terminal domain autophosphorylation, which creates a recruitment site for the substrate GarA, and displaces a substrate-occlusive N-terminal rubredoxin domain.113 Phosphorylation of juxtamembrane regions of PknG is also predicted to initiate interaction with the cell wall recruitment protein, FhaA.114 Other unique modes of eSTK regulation have also been discovered, including for YabT of B. subtilis, which is essential for a robust DNA-damage response, and harbours a DNA-binding motif that activates the kinase upon ds- or ss-DNA binding.115
168) and an additional site on the C-lobe in response to cephalosporin treatment, resulting in enhanced activity.108 Autophosphorylation of activation loop amino acid residues appears to be a conserved regulatory mechanism for many eSTKs, including PrkA of Listeria monocytogenes,109 PrkC from B. subtilis,110 YegI of E. coli111 and PknB from M. tuberculosis.112 The molecular basis of mycobacterial cytoplasmic PknG activation is also predicted to involve N-terminal domain autophosphorylation, which creates a recruitment site for the substrate GarA, and displaces a substrate-occlusive N-terminal rubredoxin domain.113 Phosphorylation of juxtamembrane regions of PknG is also predicted to initiate interaction with the cell wall recruitment protein, FhaA.114 Other unique modes of eSTK regulation have also been discovered, including for YabT of B. subtilis, which is essential for a robust DNA-damage response, and harbours a DNA-binding motif that activates the kinase upon ds- or ss-DNA binding.115
Antagonistic regulation of kinase activity by phosphatases
A responsive signalling network requires rapid and reversible addition of phosphate moieties to target proteins to selectively ‘switch on’ and ‘switch off’ signalling cascades. Across life, this involves the concerted opposing actions of protein kinases and protein phosphatases. For this purpose, dual-activity HKs can adopt phosphatase-competent states to ‘turn-over’ their signalling outputs. Moreover, RR dephosphorylation can be further accelerated by the actions of dedicated phosphatases, including CheZ of E. coli, Rap and Spo0E of B. subtills, and CheX of Borrelia burgdorferi.58 Bacteria have also evolved additional mechanisms to degrade phosphate-dependent signals in response to environmental cues. For example, bacterial phytochrome photoreceptors usually transmit photosensory input through a HK TCS system,82 including Agp1 bacteriophytochrome from Agrobacterium fabrum which exhibits light-sensitive HK-autophosphorylation activity.116 In contrast, no kinase activity has been detected for a structurally homologous bacteriophytochrome, DrBphP, from Deinococcus radiodurans which functions exclusively as a light-dependent phosphatase for the RR DrRR.117 In fact, bespoke bacterial phosphatases are prevalent in bacterial genomes, particularly in the context of eSTK- Ser/Thr phosphatase (STP) pairs10,14,93,118 and even arginine phosphorylation is counteracted by a cognate phosphatase, YwlE.119M. tuberculosis expresses 11 eSTKs (PknA-L, two of which, PknG and PknK, are soluble proteins) which collectively phosphorylate a wide range of protein substrates and play a dominant role in bacterial signalling, virulence and viability.7,120–122 Full activity of the membrane-anchored eSTK, PknB, depends on phosphorylation of two activation loop residues (Thr171 and Thr173), which can be reversed by dephosphorylation by the cognate STP, PstP.112 PstP is the only M. tuberculosis phosphatase discovered to-date and is predicted to dephosphorylate all members of the PknA-L family and their substrates.45 Interestingly, phosphorylation of PstP by PknA/B also regulates its catalytic activity, suggesting a regulatory feedback loop.123,124The conserved S. aureus eSTK-phosphatase pair (STK-STP) is co-transcribed on a regulatory operon, with STP dephosphorylating STK.125,126 Notably, STK-STP has been implicated in controlling expression of secreted virulence factors, including hemolysins α, β and γ, which are drivers of S. aureus infection127 and the kinase/phosphatase pair also have antithetical activities on cell wall biosynthesis.104Streptococcus agalactiae phosphatase, Stp1, is also required for appropriate regulation of Stk1 eSTK function.128 eSTK-STP pairs of Streptococci, including Stk1-Stp1 in S. agalactiae, Stk-Stp in S. pyogenes, StkP-PhpP in S. pneumoniae, and PknB-PppL in S. mutans are similarly co-transcribed, to ensure integrative reversible control of Ser/Thr phosphate-signalling.118 An antagonistic relationship has also been observed between an eSTK of Pseudomonas aeruginosa, PpkA (which is essential for assembly of a type-VI secretion system [H-T6SS] and secretion of hemolysin-coregulated protein 1 [Hcp1]) and PppA, a Ser/Thr phosphatase.129 The lifecycle of Bacillus anthracis, the aetiological agent of human anthrax, comprises vegetative and sporulating phases. Spore germination is partly controlled by the co-expressed kinase-phosphatase pair PrkC-PrpC,101 with PrkC kinase activity inhibited by PrpC-dependent dephosphorylation. Curiously, PrpC is a dual specificity phosphatase capable of removing a phosphate moiety from Ser, Thr and Tyr residues of PrkD and PrkG kinases.130 Likewise, although a putative tyrosine kinase of P. aeruginosa has yet to be discovered, TpbA is also believed to be a novel dual specificity Try-phosphatase that negatively regulates biofilm formation in P. aeruginosa.131
Regulation of kinase function through protein binding and subcellular distribution
Interactions with accessory proteins can determine the catalytic outputs of bacterial kinases, either through direct modulation of catalysis or organisation of their spatial distribution. For example, the McsABB.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) subtilis arginine kinase holoenzyme complex is comprised of the McsB kinase domain and an allosteric activator subunit, McsA.132 As previously discussed, interaction of the eSTK PrkCB.
subtilis arginine kinase holoenzyme complex is comprised of the McsB kinase domain and an allosteric activator subunit, McsA.132 As previously discussed, interaction of the eSTK PrkCB.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) subtilis with GpsB, DivIVA, and EzrA is responsible for appropriate localisation and activation of the kinase. Interestingly, PrkC is variably regulated by GpsB, which can function as both an activator and suppressor of PrkC, with the phosphorylated form of GpsB (itself a substrate of PrkC) providing a negative feedback loop to PrkC activity.102 EmbR is a transcriptional regulator, and a substrate protein of the mycobacterial eSTK, PknH. Curiously, a structural homolog, EmbR2 (which is a substrate of PknE and PknF) physically interacts with and inhibits PknH.133 Accessory proteins, GlnX and GlnH, are also required for PknG activation (a cytoplasmic eSTK of mycobacteria that lacks extracellular or transmembrane sensing regions) in response to extracellular amino acid concentration.134 HipA adopts an atypical protein kinase fold,135–137 and has been identified in the genomes of E. coli, and several paralogs are also found in other Gram-negative bacteria.138 HipA functions as a dormancy-driving toxin by inhibiting protein synthesis and is allosterically suppressed under normal growth conditions via direct complexation with the HipB antitoxin.136,139 Remarkably, HipA activity is also negatively regulated by intermolecular autophosphorylation of Ser150, which stabilises a P-loop ‘out-state’ and disrupts the ATP-binding pocket.140 HipT of Legionella pneumophila, which has homology to the N-terminal region of HipA, similarly adopts an inactivate conformation when in complex with its cognate antitoxin, HipS, which leads to the steric occlusion of ATP from the kinase core.138
subtilis with GpsB, DivIVA, and EzrA is responsible for appropriate localisation and activation of the kinase. Interestingly, PrkC is variably regulated by GpsB, which can function as both an activator and suppressor of PrkC, with the phosphorylated form of GpsB (itself a substrate of PrkC) providing a negative feedback loop to PrkC activity.102 EmbR is a transcriptional regulator, and a substrate protein of the mycobacterial eSTK, PknH. Curiously, a structural homolog, EmbR2 (which is a substrate of PknE and PknF) physically interacts with and inhibits PknH.133 Accessory proteins, GlnX and GlnH, are also required for PknG activation (a cytoplasmic eSTK of mycobacteria that lacks extracellular or transmembrane sensing regions) in response to extracellular amino acid concentration.134 HipA adopts an atypical protein kinase fold,135–137 and has been identified in the genomes of E. coli, and several paralogs are also found in other Gram-negative bacteria.138 HipA functions as a dormancy-driving toxin by inhibiting protein synthesis and is allosterically suppressed under normal growth conditions via direct complexation with the HipB antitoxin.136,139 Remarkably, HipA activity is also negatively regulated by intermolecular autophosphorylation of Ser150, which stabilises a P-loop ‘out-state’ and disrupts the ATP-binding pocket.140 HipT of Legionella pneumophila, which has homology to the N-terminal region of HipA, similarly adopts an inactivate conformation when in complex with its cognate antitoxin, HipS, which leads to the steric occlusion of ATP from the kinase core.138
HKs can similarly be regulated by accessory proteins. For example, two B. subtilis HKs, KinA and KinB, are essential for initiation of sporulation, and are respectively inhibited by Sda and KipI, which are predicted to repress autokinase activity by sterically blockading interaction between the CA and DHp subdomains.141,142 In contrast, binding of the accessory protein, PtsN to the DHp domain of the KdpD HK of E. coli stimulates trans-autophosphorylation between protomers of a KdpD dimer.143 Interaction with protein subunits can also shift the equilibrium of TCS kinase and phosphatase states. For example, the core SaeRS TCS of S. aureus is composed of a sensor kinase (SaeS), RR (SaeR), and two auxiliary proteins, SaeP and SaeQ, which regulate genes associated with biofilm formation.144 SaeQ and SaeP form a complex with SaeS and induce a phosphatase-competent state in the HK, effectively silencing signalling by the TCS. Finally, HK sensing of environmental stimuli (and subsequent activation) can also be mediated through accessory proteins. For example, the BceS HK of B. subtilis forms an activating intermolecular sensory complex with BceAB.145 In this system, the BceS sensor kinase is indirectly activated by the peptide-based antibiotic, bacitracin, through a flux-sensing mechanism which detects conformational cycling of the BceAB transporter.146,147 The PhoQP system can also be indirectly regulated by the acid sensing EvgSA system of E. coli via an intermediary membrane protein, SafA. SafA expression is induced following activation of EvgSA, and directly interacts with the PhoQ periplasmic sensor domain to stimulate autophosphorylation.148–150 The Rcs envelope stress response phosphorelay system (comprising RcsC HK, RcsD phosphotransfer protein, and RcsB RR) is conserved in enterobacteria (reviewed in ref. 66, 67 and 151) and is regulated by the outer-membrane sensor lipoprotein, RcsF.152 Under normal growth conditions, another inner membrane protein, IgaA, represses the RcsCDB phosphorelay cascade through an inhibitory interaction with RcsD. However, detection of envelope stress by RcsF induces complex formation with AgaA, relieving repression and resulting in autophosphorylation of RcsC.153,154
Subcellular localisation can also be critical for kinase function. Cell pole organising protein PopZ of Caulobacter crescentus self-assembles into a polymeric superstructure that recruits several signalling protein clients (such as the cell fate HKs CckA and DivJ) into a regulatory hub that coordinates cellular polarity.155,156 DivJ expression at the cell poles is critical for propagating chromosomal replication and stalk appendage biogenesis,157 and this is controlled by PopZ-dependent recruitment factor, SpmX.156 A second C. crescentus scaffold protein, PodJ, assembles at the opposite cell pole to PopZ and sequesters several additional proteins, including the HK PleC via its sensory PAS domain.158 CckA and PleC of C. crescentus are bifunctional HKs that oscillate between kinase and phosphatase activity.159 CckA exhibits density-dependent activity coordinated through its tandem PAS domains;159 functioning as an indirect kinase of the CtrA transcription factor when accumulated at high density at a newly formed cell pole,160 but transitioning to a phosphatase of CtrA in the presence of high levels of the secondary messenger c-di-GMP (cdG).160–162 Recruitment to the PopZ microdomain, which defines CckA surface density, is partly regulated through complexation with a catalytically inactive pseudohistidine kinase, DivL.163–165 Curiously, DivL is capable of transforming information relating to subcellular localisation into conformational switches that can toggle CckA between its kinase or phosphatase modes, suggesting a highly dynamic regulatory profile for this HK complex.166–168 PleC is also mechanistically density-regulated through PAS domains and is recruited to polar PodJ biomolecular condensates, which simultaneously stimulates PleC phosphatase function and suppresses kinase activity.158 Spatial organisation may also be an evolutionary strategy to limit cross-talk between TCSs, and has been observed in Rhodobacter sphaeroides and for the CheZYA chemotaxis system of E. coli, as reviewed by Sourjik and Armitage, 2010.169
Cross-talk between bacterial phosphorelay systems
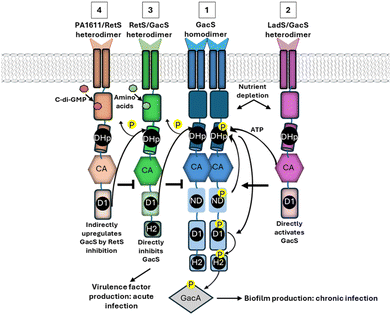
Cognate HK-RR pairs are usually highly monogamous and are generally co-expressed under a single promotor.170,171 As such, encoded TCS paralogs typically serve distinct cellular functions through linear signal transmission processes.61,171 Retention of signal fidelity is primarily dictated by specificity-determining amino acids that flank the conserved phosphotransfer/acceptor sites in the DHp and REC domains.172 However, convergence between non-cognate TCS pairs has also been observed in some bacteria.173–176 For example, in E. coli, NarQ can target its cognate RR, NarP, and also cross-phosphorylate NarL of the NarX-NarL TCS axis,177 and CreC can interact with PhoB, the cognate RR of PhoR.178 Far from representing an evolutionary redundancy, crosstalk between TCS may enhance the complexity of responses to individual input events, and promote bacterial growth, metabolism and survival.173,176 Relaxed specificity is potentially less detrimental in unorthodox ‘hybrid’ TCS, where spatial tethering of transmitter and receiver domains effectively increases their local concentrations to one another.179–181 For example, the GacSA hybrid TCS of P. aeruginosa is part of a multi-kinase communication network with LadS, RetS and PA1611 hybrid HKs182,183 (Fig. 3). Following signal-mediated activation, GacS autophosphorylates at His294 (H1), and the phosphate moiety is then shuttled to Asp717 (D1) and His863 (H2).18 Heterodimersiation of RetS and GacS downregulates GacSA signal transduction by inhibiting GacS autophosphorylation, siphoning phosphate groups from the DHp site, and through direct dephosphorylation of the GacS D1 site by RetS.184,185 Moreover, RetS-dependent inhibition can subsequently be relieved through formation of a competitive RetS-PA1611 heterodimer.186 In contrast, LadS forms a multi-component phosphorelay system with GacS, with the transmitter and REC domains of LadS facilitating trans-phosphorylation of the GacS H2 domain.182 Through these mechanisms, RetS and LadS can reciprocally regulate virulence factor expression controlled by the GacSA regulon and influence initiation of chronic P. aeruginosa infection.182 Cross-communication has also been detected between HssSR and HitSR of B. anthracis,187 NtrBC and NtrYX of Rhodobacter capsulatus,188 RscS and SypF of Vibrio fisheri189 HpArsRS and HpNikR of Helicobacter pylori190 and GraSR and ArlSR of S. aureus.191 Interestingly, RR heterodimers have also been described that can enable differential coactivation of target genes, as reviewed by Agrawal et al., 2016.172HKs and the other major phosphorelay system, eSTKs, may also intertwine to potentiate the output of adaptive signalling responses (Fig. 2B). This is perhaps not surprising given recent studies revealing that eSTKs exert a broad regulatory influence; phosphorylating 80% of the M. tuberculosis proteome and modulating expression of ∼30% of its genes.7 Such an expansive regulatory footprint may facilitate overlapping regulons between eSTKs and TCS. Indeed, 170 O-phosphorylation sites were identified on TCSs of M. tuberculosis, suggesting extensive intersection of these two phosphosignalling pathways.192 Notably, phosphorylation of the M. tuberculosis TCS-HK, NarS, at Thr380 by an eSTK, PknL, was sufficient to stimulate its autokinase activity and downstream signalling.192 The AlgRZ TCS, which is comprised of AlgR, and it's putative HK AlgZ, is an important regulator of P. aeruginosa gene expression and pathogenesis.193 Strikingly, Ser143 of the RR, AlgR, is also phosphorylated by the eSTK Stk1, which adjusts its physiological functions.194 Cross talk between STK of S. aureus and TCSs involved in cell wall metabolism has also been described, through direct phosphorylation of several Thr residues in VraTSR,195 WalRK,196 and GraSR.197 The StkP-PhdP eSTK-STP pair of S. pneumoniae can also reversibly regulate the RRs, RitR,198 RR06,199 and PknBM.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) tuberculosis can downregulate DevSR and SenX3/RegX3 signalling by phosphorylating their RRs.200,201 Fascinatingly, another P. aeruginosa, RR PA3346, contains an N-terminal receiver domain and an active STP domain that dephosphorylates Ser56 of another RR, PA3347. The Ser/Thr phosphatase activity of PA3346 is in turn potently stimulated following phosphorylation of the RES domain by a HK, PA2824.202 Direct modulation of HK kinase activity by eSTKs has also been described. For example, the CroS HK of the opportunistic pathogen, Enterococcus faecalis, is phosphorylated at a Thr by a transmembrane PASTA-eSTK, IreK, thereby enhancing HK activity. Of note, this site (Thr346) is situated in the ATP lid of the CroS CA domain, a crucial structure for appropriate nucleotide positioning and phosphate exchange during autophosphorylation.203 An eSTK of B. subtilis, YbdM, was also reported to directly phosphorylate the sensing domain of an atypical cytoplasmic HK, DegS, and enhance its kinase activity in vitro.204 Fascinatingly, the S. aureus tyrosine protein kinase CapAB (which positively regulates capsule biosynthesis) can be negatively regulated by PknB-dependent phosphorylation205 suggesting communication between eSTK and tyrosine kinase phosphotransfer networks.
tuberculosis can downregulate DevSR and SenX3/RegX3 signalling by phosphorylating their RRs.200,201 Fascinatingly, another P. aeruginosa, RR PA3346, contains an N-terminal receiver domain and an active STP domain that dephosphorylates Ser56 of another RR, PA3347. The Ser/Thr phosphatase activity of PA3346 is in turn potently stimulated following phosphorylation of the RES domain by a HK, PA2824.202 Direct modulation of HK kinase activity by eSTKs has also been described. For example, the CroS HK of the opportunistic pathogen, Enterococcus faecalis, is phosphorylated at a Thr by a transmembrane PASTA-eSTK, IreK, thereby enhancing HK activity. Of note, this site (Thr346) is situated in the ATP lid of the CroS CA domain, a crucial structure for appropriate nucleotide positioning and phosphate exchange during autophosphorylation.203 An eSTK of B. subtilis, YbdM, was also reported to directly phosphorylate the sensing domain of an atypical cytoplasmic HK, DegS, and enhance its kinase activity in vitro.204 Fascinatingly, the S. aureus tyrosine protein kinase CapAB (which positively regulates capsule biosynthesis) can be negatively regulated by PknB-dependent phosphorylation205 suggesting communication between eSTK and tyrosine kinase phosphotransfer networks.
Regulation of kinases by cofactors, secondary messengers, and quorum sensing
Much like their eukaryotic counterparts, bacterial kinases are also regulated by cofactors that can induce conformational changes and interdomain interactions or augment catalytic function. Coordination of haem or flavin adenine-dinucleotide (FAD) cofactors by HK PAS domains enables sensing of cellular redox state. For example, the kinase activity of FixL (found in nitrogen-fixing rhizobia), is potently inhibited by O2 binding to a PAS-B located haem group.206 Redox sensing cysteines have also been discovered in the CA domain of the S. aureus HK, SrrB, which form a unique intramolecular disulfide bond that tunes autokinase function.207 Furthermore, oxidation or S-nitrosylation of a Salmonella enterica RR, SsrB, at Cys203 lowers DNA-binding affinity and thus signalling by the SsrA/SsrB TCS,208 which may represent a defensive adaptation to promote Salmonella fitness when exposed to nitric oxides (NOs) produced by the host immune response. Similarly, Cys67, located in the REC domain of the redox-sensitive WalR RR, can be S-nitrosylated to modulate WalKR signalling and promote S. aureus NO-mediated vancomycin resistance.209 The cyanobacterium Synechocystis sp. PCC 6803, encodes six eSTKs (SpkA-D, G), one pseudokinase (SpkE), and five atypical kinases (SpkH-L). Of note, SpkB was shown to be inactivated by oxidation of an N-terminal Cys motif, as an adaptation to oxidative stress tolerance.210 As previously discussed, the secondary messenger c-di-GMP binds within the interface between the PAS-B site and CA domain of the HK, CckA, to stimulate phosphatase activity.159,211 In contrast, the kinase activities of ShKa, a cytosolic hybrid HK of C. crescentus, and RavS, a HK of Xanthomonas campestris, are increased following binding to c-di-GMP.212,213 The activity of a DNA-damage responsive eSTK of the extremophilic bacteria Deinococcus radiodurans, RqkA, is also enhanced by binding to pyrroloquinoline-quinone (PQQ).214S. aureus biofilm formation is partially regulated by intercellular communication and density sensing, commonly known as quorum sensing.215 In this manner, bacteria can detect changes in population numbers based on the local concentration of specific autoinducer signals. Direct binding of an autoinducer peptide (AIP) to the AgrC HK stimulates phosphorylation of the RR AgrA, resulting in positive regulation of toxin genes, autoinduction of AIP biosynthesis, and reduced expression of several surface adhesins, as reviewed by Jenul & Horswill, 2019.216 The Lux system in Vibrio species is another well-studied TCS that is regulated by quorum sensing.217 Low autoinducer concentrations at low cell density induces LuxQ activity and results in phosphorylation of the LuxO RR. In contrast, high autoinducer concentrations (detected by the periplasmic receptor LuxP) activate HK phosphatase activities and reverts the TCS to a pre-activated state.218
Regulation of secreted effector kinases
In order to establish an infection and subvert host defences, pathogenic bacteria can also deploy an arsenal of signal pathway-hijacking virulence effectors, including eSTKs, into the cytoplasm of target cells.219 PknBM.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) tuberculosis (and the phosphatase SapM) was the first discovered effector eSTK, which functions to promote intracellular survival of the bacteria in macrophages by blocking phagosome maturation.220,221 Since then, secreted bacterial effector kinases have been identified in the genomes of numerous pathogens, including Yersinia, Salmonella, Legionella and Shigella219 but little is understood about how they are regulated. To avoid premature activation of cytotoxicity, the activities of virulence factors and exotoxins are extremely tightly controlled, only ‘switched on’ following secretion or exposure to specific host-derived activation signals. Similar host-dependent activation mechanisms have also been observed for a handful of effector eSTKs. Legionella pneumophila (L.p.) is an opportunistic intracellular pathogen of human alveolar macrophages and the causative agent of Legionnaire's disease. L.p. translocates over 300 survival-promoting effector proteins, including at least 6 experimentally validated eSTKs (LegK1-4, LegK7, and Lem28/Lpg2603) into invaded host cells via the Dot/Icm type IV secretion system (T4SS).222–231 Fascinatingly, LegK7, which manipulates the host Hippo pathway by functioning as a molecular mimic of Mammalian Ste20-like kinases 1/2 (MST1/2), is allosterically activated by N-terminal binding to a host scaffold protein, MOB kinase activator 1A (MOB1A).227,228 Similarly, Lem28 is a remote member of the protein kinase superfamily that is allosterically activated by the eukaryotic-specific ligand inositol hexaphosphate (IP6).230 Members of the pathogenic Yersinia genus secrete an eSTK domain containing protein, YpkA, which directly phosphorylates the heterotrimeric Gαq protein and vasodilator-stimulated phosphoprotein (VASP) to disrupt macrophage cytoskeletal dynamics and phagocytosis.232,233 YpkA is delivered into host cells in an inactive form via a type III secretion system, where it is allosterically activated following actin binding to the actin binding domain (ABD) and undergoes extensive autophosphorylation.234,235 The Shigella type-III secretion system effector OspG is an atypical serine/threonine protein kinase that lacks several canonical structural features, including a regulatory activation loop.236 Despite the extensive degradation of the catalytic core, OspG exhibits intrinsic kinase activity, but only upon binding to components of the host ubiquitin system.236 In contrast, homologous atypical kinases of E. coli, NleH1, and NleH2, display ubiquitin-independent autophosphorylation which promotes interaction with target proteins.236
tuberculosis (and the phosphatase SapM) was the first discovered effector eSTK, which functions to promote intracellular survival of the bacteria in macrophages by blocking phagosome maturation.220,221 Since then, secreted bacterial effector kinases have been identified in the genomes of numerous pathogens, including Yersinia, Salmonella, Legionella and Shigella219 but little is understood about how they are regulated. To avoid premature activation of cytotoxicity, the activities of virulence factors and exotoxins are extremely tightly controlled, only ‘switched on’ following secretion or exposure to specific host-derived activation signals. Similar host-dependent activation mechanisms have also been observed for a handful of effector eSTKs. Legionella pneumophila (L.p.) is an opportunistic intracellular pathogen of human alveolar macrophages and the causative agent of Legionnaire's disease. L.p. translocates over 300 survival-promoting effector proteins, including at least 6 experimentally validated eSTKs (LegK1-4, LegK7, and Lem28/Lpg2603) into invaded host cells via the Dot/Icm type IV secretion system (T4SS).222–231 Fascinatingly, LegK7, which manipulates the host Hippo pathway by functioning as a molecular mimic of Mammalian Ste20-like kinases 1/2 (MST1/2), is allosterically activated by N-terminal binding to a host scaffold protein, MOB kinase activator 1A (MOB1A).227,228 Similarly, Lem28 is a remote member of the protein kinase superfamily that is allosterically activated by the eukaryotic-specific ligand inositol hexaphosphate (IP6).230 Members of the pathogenic Yersinia genus secrete an eSTK domain containing protein, YpkA, which directly phosphorylates the heterotrimeric Gαq protein and vasodilator-stimulated phosphoprotein (VASP) to disrupt macrophage cytoskeletal dynamics and phagocytosis.232,233 YpkA is delivered into host cells in an inactive form via a type III secretion system, where it is allosterically activated following actin binding to the actin binding domain (ABD) and undergoes extensive autophosphorylation.234,235 The Shigella type-III secretion system effector OspG is an atypical serine/threonine protein kinase that lacks several canonical structural features, including a regulatory activation loop.236 Despite the extensive degradation of the catalytic core, OspG exhibits intrinsic kinase activity, but only upon binding to components of the host ubiquitin system.236 In contrast, homologous atypical kinases of E. coli, NleH1, and NleH2, display ubiquitin-independent autophosphorylation which promotes interaction with target proteins.236
TCSs as potential therapeutic targets for the treatment of microbial infections
Multi-drug resistant (MDR) bacterial infections rank globally amongst the leading causes of mortality, and are projected to cause 40 million deaths by 2050.237 Most clinically approved antibiotics are derivatives of small molecules that disrupt a narrow range of essential microbial biosynthetic pathways. Compounds exhibiting alternative modes of action are scarce,238 which has exacerbated the spread of MDR bacteria. Accordingly, the many crucial roles that TCSs and kinases serve in bacterial growth, viability and pathogenic processes (including antibiotic resistance mechanisms and biofilm formation) make them attractive alternative therapeutic targets to develop novel antimicrobials. Moreover, the CA and DHp domains of TCS (which are crucially absent in mammalian signalling pathways) are highly conserved across the bacterial kingdom,84,239 potentially enabling development of compounds with broad-spectrum activity.240,241 In this final section we discuss some recent advances in kinase-targeting antimicrobial drug strategies.Several studies have investigated and confirmed TCS inhibition by small-molecule compounds. For example, WalK is an essential HK conserved among Gram-positive bacteria, including S. aureus and B. subtilis.242 Walkmycins A, B, and C were demonstrated to exhibit strong antibacterial activity against B. subtilis (a model organism for studying pathogenic B. anthracis and B. cereus) and walkmycin B was revealed to specifically inhibit WalK autophosphorylation.243 Interestingly, walkmycin C was also an effective inhibitor of three other Streptococcus mutans HKs: VicK, CiaH, and LiaS in vitro,244 and isatin derivatives also exhibit potent antimicrobial activity against S. aureus, which was attributed to direct binding to and inhibition of WalK.245 Other inhibitors of HK autophosphorylation have now been identified for several different bacterium, targeting VicK and AgrC of S. pneumoniae,246 DosS and DosT of M. tuberculosis,247 and PhoQ of S. typhimurium,248 suggesting that TCSs are viable antibacterial therapeutic targets.
Most identified TCS-targeting inhibitors, including walkmycin C,244 are functional ATP-competitive molecules that ‘blockade’ ATP binding and thus HK catalysis (HKs such as HK853Thermotoga![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) maritima, CheAE.
maritima, CheAE.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) coli and VicKS.
coli and VicKS.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) pneumoniae can all be inactivated by ATP obstruction249). The ATP-binding pocket of the HK CA domain adopts a distinctive α/β sandwich, consisting of four conserved regions (the N-box, the G1-box, the G2-box, and the G3-box) and a highly variable ATP-lid.250,251 This structural architecture is known as the Bergerat fold, and is also observed in the ATP-binding domains of the diverse GHKL protein superfamily, which also includes DNA gyrases, HSP90, and the MutL mismatch repair enzyme.250 Guarnieri et al. (2008) demonstrated that the human HSP90 inhibitor, radicicol, binds to the ATP-binding pocket of the Salmonella HK, PhoQ (albeit weakly).252 Since then, optimised derivatives of ATP-competitive HSP90 inhibitors have been explored as potential HK active agents.251,253 For example, a fragment of an established HSP90, 3,4-diphenylpyrazole (DPP)-based inhibitor was shown to bind to the CA domain of CheA from Thermotoga maritima, and several DDP analogues are effective inhibitors of EnvZE.
pneumoniae can all be inactivated by ATP obstruction249). The ATP-binding pocket of the HK CA domain adopts a distinctive α/β sandwich, consisting of four conserved regions (the N-box, the G1-box, the G2-box, and the G3-box) and a highly variable ATP-lid.250,251 This structural architecture is known as the Bergerat fold, and is also observed in the ATP-binding domains of the diverse GHKL protein superfamily, which also includes DNA gyrases, HSP90, and the MutL mismatch repair enzyme.250 Guarnieri et al. (2008) demonstrated that the human HSP90 inhibitor, radicicol, binds to the ATP-binding pocket of the Salmonella HK, PhoQ (albeit weakly).252 Since then, optimised derivatives of ATP-competitive HSP90 inhibitors have been explored as potential HK active agents.251,253 For example, a fragment of an established HSP90, 3,4-diphenylpyrazole (DPP)-based inhibitor was shown to bind to the CA domain of CheA from Thermotoga maritima, and several DDP analogues are effective inhibitors of EnvZE.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) coli, PhoQSalmonella
coli, PhoQSalmonella![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) typhimurium, and CckAC.
typhimurium, and CckAC.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) crescentus.251,253 Repurposing human protein kinase inhibitors (for which there are ∼100 FDA approved clinical compounds) is another desirable strategy to identify novel therapeutic agents to target bacterial kinases. Carabajal et al. (2020) screened 686 compounds from the Published Kinase Inhibitor Set (PKIS [by GlaxoSmithKline]), and identified quinazoline-based ATP-competitive inhibitors of S. typhimurium PhoQ that repressed autokinase kinase activity.248 However, despite these promising results, potential toxicity associated with ‘off-target’ inhibition of human kinase signalling pathways may necessitate a more refined, structure-guided design approach to develop selective inhibitors which only target bacterial HKs before such agents can be considered therapeutically viable.
crescentus.251,253 Repurposing human protein kinase inhibitors (for which there are ∼100 FDA approved clinical compounds) is another desirable strategy to identify novel therapeutic agents to target bacterial kinases. Carabajal et al. (2020) screened 686 compounds from the Published Kinase Inhibitor Set (PKIS [by GlaxoSmithKline]), and identified quinazoline-based ATP-competitive inhibitors of S. typhimurium PhoQ that repressed autokinase kinase activity.248 However, despite these promising results, potential toxicity associated with ‘off-target’ inhibition of human kinase signalling pathways may necessitate a more refined, structure-guided design approach to develop selective inhibitors which only target bacterial HKs before such agents can be considered therapeutically viable.
Interestingly, several non-ATP competitive bacterial kinase inhibitors have also been discovered. The HK PhoQ (of the conserved PhoP/PhoQ TCS) plays pivotal roles in Salmonella, Shigella, and Pseudomonas pathogenesis, regulating virulence factor production and bacterial survival within host cells.254 Moreover, PhoP/PhoQ contributes to ‘last-resort’ polymyxin antibiotic resistance by indirectly activating the PmrA/PmrB TCS.255,256 Fascinatingly, PhoQ (Salmonella typhimurium) HK activity can be repressed by allosteric targeting its second transmembrane region using a hydrazone derivative compound, N′-(thiophen-2-ylmethylene)benzohydrazide (designated A16B1).257–259 Watanabe et al., (2012) also identified signermycin B as a potent allosteric inhibitor of WalK that disrupted autokinase activity by sterically blocking the HK dimerization domain.260 Moreover waldiomycin (and a methyl ester derivative) can effectively inhibit WalK from both S. aureus and B. subtilis (and PhoR and ResE from B. subtilis) by binding to the phosphoacceptor region and blocking autophosphorylation.261,262
Conclusions
TCSs and bacterial kinases represent a promising frontier for the development of novel therapeutics for microbial infections, offering a versatile platform for addressing both antimicrobial resistance and bacterial virulence. Phosphate-based PTMs, regulated by kinases and phosphatases, are essential for bacterial homeostasis, viability and virulence and clinical compounds with the ability to inhibit conserved domains across multiple protein kinases could provide broad-spectrum antimicrobial activity. Conceptually, TCS inhibitors also show promise as therapeutic adjuvants, potentially synergizing with conventional antibiotics to overcome resistance mechanisms.263 For example, compounds like PhoQ-targeting A16B1 can enhance the bactericidal activity of polymyxins.259 However, despite significant interest in this area, no bacterial kinase-targeting small molecule have advanced to clinical approval. To progress this area of research, a combination of structure-guided drug design and innovative screening methodologies will be essential to develop optimised and selective kinase-targeting compounds with robust antimicrobial activity.Author contributions
Writing – original draft: DPB, DMF, DMC & CW; writing – review and editing: DPB, DMF, DMC & CW; visualisation: DMF, DMC & DPB.Conflicts of interest
There are no conflicts of interest.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.References
- G. Burnett and E. P. Kennedy, The enzymatic phosphorylation of proteins, J. Biol. Chem., 1954, 211(2), 969–980 CrossRef CAS.
- E. H. Fischer and E. G. Krebs, Conversion of phosphorylase b to phosphorylase a in muscle extracts, J. Biol. Chem., 1955, 216(1), 121–132 CrossRef CAS.
- E. G. Krebs and E. H. Fischer, The phosphorylase b to a converting enzyme of rabbit skeletal muscle, Biochem. Biophys. Acta, 1956, 20, 150–157 CAS.
- E. H. Fischer, D. J. Graves, E. R. S. Crittenden and E. G. Krebs, Structure of the site phosphorylated in the phosphorylase b to a reaction, J. Biol. Chem., 1959, 234(7), 1698–1704 CrossRef CAS.
- S. N. Nagarajan, C. Lenoir and C. Grangeasse, Recent advances in bacterial signaling by serine/threonine protein kinases, Trends Microbiol., 2022, 30(6), 553–566 CrossRef CAS.
- S. Lim, A Review of the Bacterial Phosphoproteomes of Beneficial Microbes, Microorganisms, 2023, 11(4), 931 CrossRef CAS PubMed.
- A. Frando, V. Boradia, M. Gritsenko, C. Beltejar, L. Day and D. R. Sherman, et al., The Mycobacterium tuberculosis protein O-phosphorylation landscape, Nat. Microbiol., 2023, 8(3), 548–561 CrossRef CAS PubMed.
- M. Kansari, F. Idiris, H. Szurmant, T. Kubař and A. Schug, Mechanism of activation and autophosphorylation of a histidine kinase, Commun. Chem., 2024, 7(1), 196 CrossRef CAS PubMed.
- A. Frando and C. Grundner, More than two components: complexities in bacterial phosphosignaling, mSystems, 2024, 9(5), e00289-24 CrossRef PubMed.
- J. Dworkin, Ser/Thr phosphorylation as a regulatory mechanism in bacteria, Curr. Opin. Microbiol., 2015, 24, 47–52 CrossRef CAS PubMed.
- P. Yagüe, N. Gonzalez-Quiñonez, G. Fernánez-García, S. Alonso-Fernández and A. Manteca, Goals and Challenges in Bacterial Phosphoproteomics, Int. J. Mol. Sci., 2019, 20(22), 5678 CrossRef PubMed.
- T. Garcia-Garcia, T. Douché, Q. Giai Gianetto, S. Poncet, N. El Omrani and W. K. Smits, et al., In-Depth Characterization of the Clostridioides difficile Phosphoproteome to Identify Ser/Thr Kinase Substrates, Mol. Cell. Proteomics, 2022, 21(11), 100428 CrossRef CAS.
- S. Lim, A Review of the Bacterial Phosphoproteomes of Beneficial Microbes, Microorganisms, 2023, 11(4), 931 CrossRef CAS.
- I. Mijakovic, C. Grangeasse and K. Turgay, Exploring the diversity of protein modifications: special bacterial phosphorylation systems, FEMS Microbiol. Rev., 2016, 40(3), 398–417 CrossRef CAS.
- T. Parish, Two-component regulatory systems of mycobacteria, Mol. Genet. Mycobact., 2014, 209–223 Search PubMed.
- A. Rajput, Y. Seif, K. S. Choudhary, C. Dalldorf, S. Poudel and J. M. Monk, et al., Pangenome analytics reveal two-component systems as conserved targets in ESKAPEE pathogens, mSystems, 2021, 6(1), e00981-20 CrossRef PubMed.
- A. Rodrigue, Y. Quentin, A. Lazdunski, V. Méjean and M. Foglino, Cell signalling by oligosaccharides. Two-component systems in Pseudomonas aeruginosa: why so many?, Trends Microbiol., 2000, 8(11), 498–504 CrossRef CAS.
- H. Song, Y. Li and Y. Wang, Two-component system GacS/GacA, a global response regulator of bacterial physiological behaviors, Eng. Microbiol., 2023, 3(1), 100051 CrossRef CAS PubMed.
- A. N. Brown, M. T. Anderson, M. A. Bachman and H. L. Mobley, The ArcAB two-component system: function in metabolism, redox control, and infection, Microbiol. Mol. Biol. Rev., 2022, 86(2), e00110–e00121 CrossRef PubMed.
- P. Bisicchia, D. Noone, E. Lioliou, A. Howell, S. Quigley and T. Jensen, et al., The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis, Mol. Microbiol., 2007, 65(1), 180–200 CrossRef CAS PubMed.
- A. F. Alvarez and D. Georgellis, Environmental adaptation and diversification of bacterial two-component systems, Curr. Opin. Microbiol., 2023, 76, 102399 CrossRef CAS PubMed.
- Y. Qiu, D. Xu, X. Xia, K. Zhang, R. M. Aadil and Z. Batool, et al., Five major two components systems of Staphylococcus aureus for adaptation in diverse hostile environment, Microb. Pathog., 2021, 159, 105119 CrossRef CAS.
- D. Burbulys, K. A. Trach and J. A. Hoch, Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay, Cell, 1991, 64(3), 545–552 CrossRef CAS PubMed.
- Y. M. Ibrahim, A. R. Kerr, J. McCluskey and T. J. Mitchell, Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae, J. Bacteriol., 2004, 186(16), 5258–5266 CrossRef CAS.
- V. Malhotra, D. Sharma, V. Ramanathan, H. Shakila, D. K. Saini and S. Chakravorty, et al., Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis, FEMS Microbiol. Lett., 2004, 231(2), 237–245 CrossRef CAS.
- N. Sengupta, K. Paul and R. Chowdhury, The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae, Infect. Immun., 2003, 71(10), 5583–5589 CrossRef CAS.
- C. Shaw, M. Hess and B. C. Weimer, Two-component systems regulate bacterial virulence in response to the host gastrointestinal environment and metabolic cues, Virulence, 2022, 13(1), 1666–1680 CrossRef CAS.
- T. Mascher, S. L. Zimmer, T.-A. Smith and J. D. Helmann, Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis, Antimicrob. Agents Chemother., 2004, 48(8), 2888–2896 CrossRef CAS PubMed.
- A. Hiron, M. Falord, J. Valle, M. Débarbouillé and T. Msadek, Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters, Mol. Microbiol., 2011, 81(3), 602–622 CrossRef CAS PubMed.
- A. Kato, H. D. Chen, T. Latifi and E. A. Groisman, Reciprocal control between a bacterium's regulatory system and the modification status of its lipopolysaccharide, Mol. Cell, 2012, 47(6), 897–908 CrossRef CAS PubMed.
- H. Hussain, P. Branny and E. Allan, A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans, J. Bacteriol., 2006, 188(4), 1628–1632 CrossRef CAS PubMed.
- A. R. Tierney and P. N. Rather, Roles of two-component regulatory systems in antibiotic resistance, Future Microbiol., 2019, 14(6), 533–552 CrossRef PubMed.
- M. Hu, X. Huang, X. Xu, Z. Zhang, S. He and J. Zhu, et al., Characterization of the role of two-component systems in antibiotic resistance formation in Salmonella enterica serovar Enteritidis, mSphere, 2022, 7(6), e00383-22 CrossRef PubMed.
- T. Mascher, J. D. Helmann and G. Unden, Stimulus perception in bacterial signal-transducing histidine kinases, Microbiol. Mol. Biol. Rev., 2006, 70(4), 910–938 CrossRef CAS.
- V. Sperandio, A. G. Torres and J. B. Kaper, Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli, Mol. Microbiol., 2002, 43(3), 809–821 CrossRef CAS PubMed.
- J. F. Hess, R. B. Bourret and M. I. Simon, Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis, Nature, 1988, 336(6195), 139–143 CrossRef CAS PubMed.
- B. M. Prüβ, Involvement of two-component signaling on bacterial motility and biofilm development, J. Bacteriol., 2017, 199(18), e00259-17 Search PubMed.
- S. Kabbara, A. Hérivaux, T. Dugé de Bernonville, V. Courdavault, M. Clastre and A. Gastebois, et al., Diversity and Evolution of Sensor Histidine Kinases in Eukaryotes, Genome Biol. Evol., 2019, 11(1), 86–108 CrossRef CAS PubMed.
- A. Hérivaux, Y.-S. So, A. Gastebois, J.-P. Latgé, J.-P. Bouchara and Y.-S. Bahn, et al., Major sensing proteins in pathogenic fungi: the hybrid histidine kinase family, PLoS Pathog., 2016, 12(7), e1005683 CrossRef PubMed.
- K. L. Clark, P. B. Larsen, X. Wang and C. Chang, Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors, Proc. Natl. Acad. Sci. U. S. A., 1998, 95(9), 5401–5406 CrossRef CAS PubMed.
- K. Jung, L. Fried, S. Behr and R. Heermann, Histidine kinases and response regulators in networks, Curr. Opin. Microbiol., 2012, 15(2), 118–124 CrossRef CAS PubMed.
- I. A. Stancik, M. S. Šestak, B. Ji, M. Axelson-Fisk, D. Franjevic and C. Jers, et al., Serine/Threonine Protein Kinases from Bacteria, Archaea and Eukarya Share a Common Evolutionary Origin Deeply Rooted in the Tree of Life, J. Mol. Biol., 2018, 430(1), 27–32 CrossRef CAS PubMed.
- P. J. Kennelly, Protein Ser/Thr/Tyr phosphorylation in the Archaea, J. Biol. Chem., 2014, 289(14), 9480–9487 CrossRef CAS PubMed.
- J. Muñoz-Dorado, S. Inouye and M. Inouye, A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a Gram-negative bacterium, Cell, 1991, 67(5), 995–1006 CrossRef PubMed.
- M. Janczarek, J.-M. Vinardell, P. Lipa and M. Karaś, Hanks-Type Serine/Threonine Protein Kinases and Phosphatases in Bacteria: Roles in Signaling and Adaptation to Various Environments, Int. J. Mol. Sci., 2018, 19(10), 2872 CrossRef PubMed.
- S. Schneiker, O. Perlova, O. Kaiser, K. Gerth, A. Alici and M. O. Altmeyer, et al., Complete genome sequence of the myxobacterium Sorangium cellulosum, Nat. Biotechnol., 2007, 25(11), 1281–1289 CrossRef CAS PubMed.
- S. Fieulaine, S. Morera, S. Poncet, V. Monedero, V. Gueguen-Chaignon and A. Galinier, et al., X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain, EMBO J., 2001, 20(15), 3917–3927 CrossRef CAS PubMed.
- A. Galinier, M. Kravanja, R. Engelmann, W. Hengstenberg, M.-C. Kilhoffer and J. Deutscher, et al., New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression, Proc. Natl. Acad. Sci. U. S. A., 1998, 95(4), 1823–1828 CrossRef CAS PubMed.
- S. Fieulaine, S. Morera, S. Poncet, I. Mijakovic, A. Galinier and J. Janin, et al., X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(21), 13437–13441 CrossRef CAS PubMed.
- J. Zheng, C. He, V. K. Singh, N. L. Martin and Z. Jia, Crystal structure of a novel prokaryotic Ser/Thr kinase and its implication in the Cpx stress response pathway, Mol. Microbiol., 2007, 63(5), 1360–1371 CrossRef CAS PubMed.
- A. Dorsey-Oresto, T. Lu, M. Mosel, X. Wang, T. Salz and K. Drlica, et al., YihE kinase is a central regulator of programmed cell death in bacteria, Cell Rep., 2013, 3(2), 528–537 CrossRef CAS PubMed.
- K. B. Nguyen, A. Sreelatha, E. S. Durrant, J. Lopez-Garrido, A. Muszewska and M. Dudkiewicz, et al., Phosphorylation of spore coat proteins by a family of atypical protein kinases, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(25), E3482–E3491 CrossRef CAS PubMed.
- C. Grangeasse, S. Nessler and I. Mijakovic, Bacterial tyrosine kinases: evolution, biological function and structural insights, Philos. Trans. R. Soc., B, 2012, 367(1602), 2640–2655 CrossRef CAS PubMed.
- A. K. Elsholz, K. Turgay, S. Michalik, B. Hessling, K. Gronau and D. Oertel, et al., Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(19), 7451–7456 CrossRef CAS PubMed.
- S. Junker, S. Maaβ, A. Otto, S. Michalik, F. Morgenroth and U. Gerth, et al., Spectral Library Based Analysis of Arginine Phosphorylations in Staphylococcus aureus, Mol. Cell. Proteomics, 2018, 17(2), 335–348 CrossRef CAS PubMed.
- B. Huang, Z. Zhao, Y. Zhao and S. Huang, Protein arginine phosphorylation in organisms, Int. J. Biol. Macromol., 2021, 171, 414–422 CrossRef CAS PubMed.
- E. Ishii and Y. Eguchi, Diversity in Sensing and Signaling of Bacterial Sensor Histidine Kinases, Biomolecules, 2021, 11(10), 1524 CrossRef CAS PubMed.
- C. P. Zschiedrich, V. Keidel and H. Szurmant, Molecular Mechanisms of Two-Component Signal Transduction, J. Mol. Biol., 2016, 428(19), 3752–3775 CrossRef CAS PubMed.
- L. Rajeev, M. E. Garber and A. Mukhopadhyay, Tools to map target genes of bacterial two-component system response regulators, Environ. Microbiol. Rep., 2020, 12(3), 267–276 CrossRef PubMed.
- R. Gao, S. Bouillet and A. M. Stock, Structural basis of response regulator function, Annu. Rev. Microbiol., 2019, 73(1), 175–197 CrossRef CAS PubMed.
- A. M. Stock, V. L. Robinson and P. N. Goudreau, Two-component signal transduction, Annu. Rev. Biochem., 2000, 69, 183–215 CrossRef CAS PubMed.
- A. Hernández-Eligio, Á. Andrade, L. Soto, E. Morett and K. Juárez, The unphosphorylated form of the PilR two-component system regulates pilA gene expression in Geobacter sulfurreducens, Environ. Sci. Pollut. Res., 2017, 24, 25693–25701 CrossRef PubMed.
- E. Hong, H. M. Lee, H. Ko, D.-U. Kim, B.-Y. Jeon and J. Jung, et al., Structure of an atypical orphan response regulator protein supports a new phosphorylation-independent regulatory mechanism, J. Biol. Chem., 2007, 282(28), 20667–20675 CrossRef CAS PubMed.
- Y. Wen, Z. Ouyang, Y. Yu, X. Zhou, Y. Pei and B. Devreese, et al., Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region, Nucleic Acids Res., 2017, 45(16), 9773–9787 CrossRef CAS PubMed.
- M. M. Ali, A. Provoost, K. Mijnendonckx, R. Van Houdt and D. Charlier, DNA-binding and transcription activation by unphosphorylated response regulator AgrR from Cupriavidus metallidurans involved in silver resistance, Front. Microbiol., 2020, 11, 1635 CrossRef PubMed.
- E. Wall, N. Majdalani and S. Gottesman, The complex Rcs regulatory cascade, Annu. Rev. Microbiol., 2018, 72(1), 111–139 CrossRef CAS PubMed.
- J. Huesa, J. Giner-Lamia, M. G. Pucciarelli, F. Paredes-Martínez, F. G.-D. Portillo and A. Marina, et al., Structure-based analyses of Salmonella RcsB variants unravel new features of the Rcs regulon, Nucleic Acids Res., 2021, 49(4), 2357–2374 CrossRef CAS PubMed.
- M.-P. Castanie-Cornet, K. Cam, B. Bastiat, A. Cros, P. Bordes and C. Gutierrez, Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE, Nucleic Acids Res., 2010, 38(11), 3546–3554 CrossRef CAS PubMed.
- G. R. Venkatesh, F. C. Kembou Koungni, A. Paukner, T. Stratmann, B. Blissenbach and K. Schnetz, BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS, J. Bacteriol., 2010, 192(24), 6456–6464 CrossRef CAS PubMed.
- T. A. Lehti, J. Heikkinen, T. K. Korhonen and B. Westerlund-Wikström, The response regulator RcsB activates expression of Mat fimbriae in meningitic Escherichia coli, J. Bacteriol., 2012, 194(13), 3475–3485 CrossRef CAS PubMed.
- C. Kühne, H. M. Singer, E. Grabisch, L. Codutti, T. Carlomagno and A. Scrima, et al., RflM mediates target specificity of the RcsCDB phosphorelay system for transcriptional repression of flagellar synthesis in Salmonella enterica, Mol. Microbiol., 2016, 101(5), 841–855 CrossRef PubMed.
- M. Brüderlin, R. Böhm, F. Fadel, S. Hiller, T. Schirmer and B. N. Dubey, Structural features discriminating hybrid histidine kinase Rec domains from response regulator homologs, Nat. Commun., 2023, 14(1), 1002 CrossRef PubMed.
- I. Dikiy, U. R. Edupuganti, R. R. Abzalimov, P. P. Borbat, M. Srivastava and J. H. Freed, et al., Insights into histidine kinase activation mechanisms from the monomeric blue light sensor EL346, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(11), 4963–EL372 CrossRef CAS PubMed.
- F. Pompeo, E. Foulquier, B. Serrano, C. Grangeasse and A. Galinier, Phosphorylation of the cell division protein GpsB regulates PrkC kinase activity through a negative feedback loop in Bacillus subtilis, Mol. Microbiol., 2015, 97(1), 139–150 CrossRef CAS PubMed.
- V. Ravikumar, L. Shi, K. Krug, A. Derouiche, C. Jers and C. Cousin, et al., Quantitative phosphoproteome analysis of Bacillus subtilis reveals novel substrates of the kinase PrkC and phosphatase PrpC, Mol. Cell. Proteomics, 2014, 13(8), 1965–1978 CrossRef CAS PubMed.
- C. Absalon, M. Obuchowski, E. Madec, D. Delattre, I. B. Holland and S. J. Seror, CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis, Microbiology, 2009, 155(3), 932–943 CrossRef CAS PubMed.
- E. A. Libby, L. A. Goss and J. Dworkin, The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in Bacillus subtilis, PLoS Genet., 2015, 11(6), e1005275 CrossRef PubMed.
- E. C. Stuffle, M. S. Johnson and K. J. Watts, PAS domains in bacterial signal transduction, Curr. Opin. Microbiol., 2021, 61, 8–15 CrossRef CAS PubMed.
- A. Möglich, R. A. Ayers and K. Moffat, Structure and Signaling Mechanism of Per-ARNT-Sim Domains, Structure, 2009, 17(10), 1282–1294 CrossRef PubMed.
- L. J. Kenney, Structure/function relationships in OmpR and other winged-helix transcription factors, Curr. Opin. Microbiol., 2002, 5(2), 135–141 CrossRef CAS PubMed.
- M. Y. Galperin, Structural classification of bacterial response regulators: diversity of output domains and domain combinations, J. Bacteriol., 2006, 188(12), 4169–4182 CrossRef CAS PubMed.
- A. Möglich, Signal transduction in photoreceptor histidine kinases, Protein Sci., 2019, 28(11), 1923–1946 CrossRef PubMed.
- T. N. Huynh and V. Stewart, Negative control in two-component signal transduction by transmitter phosphatase activity, Mol. Microbiol., 2011, 82(2), 275–286 CrossRef CAS PubMed.
- R. Gao and A. M. Stock, Biological insights from structures of two-component proteins, Annu. Rev. Microbiol., 2009, 63(1), 133–154 CrossRef CAS PubMed.
- F. Trajtenberg, J. A. Imelio, M. R. Machado, N. Larrieux, M. A. Marti and G. Obal, et al., Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action, eLife, 2016, 5, e21422 CrossRef PubMed.
- F. Jacob-Dubuisson, A. Mechaly, J.-M. Betton and R. Antoine, Structural insights into the signalling mechanisms of two-component systems, Nat. Rev. Microbiol., 2018, 16(10), 585–593 CrossRef CAS PubMed.
- O. Ashenberg, A. E. Keating and M. T. Laub, Helix Bundle Loops Determine Whether Histidine Kinases Autophosphorylate in cis or in trans, J. Mol. Biol., 2013, 425(7), 1198–1209 CrossRef CAS PubMed.
- J. S. Parkinson, G. L. Hazelbauer and J. J. Falke, Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update, Trends Microbiol., 2015, 23(5), 257–266 CrossRef CAS PubMed.
- G. Rivera-Cancel, W. H. Ko, D. R. Tomchick, F. Correa and K. H. Gardner, Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(50), 17839–17844 CrossRef CAS PubMed.
- G. S. Wright, A. Saeki, T. Hikima, Y. Nishizono, T. Hisano and M. Kamaya, et al., Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system, Sci. Signaling, 2018, 11(525), eaaq0825 CrossRef PubMed.
- K. Burnside and L. Rajagopal, Regulation of prokaryotic gene expression by eukaryotic-like enzymes, Curr. Opin. Microbiol., 2012, 15(2), 125–131 CrossRef CAS PubMed.
- D. P. Wright and A. T. Ulijasz, Regulation of transcription by eukaryotic-like serine-threonine kinases and phosphatases in Gram-positive bacterial pathogens, Virulence, 2014, 5(8), 863–885 CrossRef PubMed.
- M. Janczarek, J. M. Vinardell, P. Lipa and M. Karaś, Hanks-Type Serine/Threonine Protein Kinases and Phosphatases in Bacteria: Roles in Signaling and Adaptation to Various Environments, Int. J. Mol. Sci., 2018, 19(10), 2872 CrossRef PubMed.
- S. Manuse, A. Fleurie, L. Zucchini, C. Lesterlin and C. Grangeasse, Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis, FEMS Microbiol. Rev., 2016, 40(1), 41–56 CrossRef CAS PubMed.
- D. A. Pensinger, A. J. Schaenzer and J.-D. Sauer, Do Shoot the Messenger: PASTA Kinases as Virulence Determinants and Antibiotic Targets, Trends Microbiol., 2018, 26(1), 56–69 CrossRef CAS PubMed.
- J. Bonne Køhler, C. Jers, M. Senissar, L. Shi, A. Derouiche and I. Mijakovic, Importance of protein Ser/Thr/Tyr phosphorylation for bacterial pathogenesis, FEBS Lett., 2020, 594(15), 2339–2369 CrossRef PubMed.
- M. Pallen, R. Chaudhuri and A. Khan, Bacterial FHA domains: neglected players in the phospho-threonine signalling game?, Trends Microbiol., 2002, 10(12), 556–563 CrossRef CAS PubMed.
- A. W. Almawi, L. A. Matthews and A. Guarné, FHA domains: Phosphopeptide binding and beyond, Prog. Biophys. Mol. Biol., 2017, 127, 105–110 CrossRef CAS PubMed.
- M. S. Bluma, K. M. Schultz, C. J. Kristich and C. S. Klug, Conformational changes in the activation loop of a bacterial PASTA kinase, Protein Sci., 2023, 32(7), e4697 CrossRef CAS PubMed.
- F. Pompeo, E. Foulquier and A. Galinier, Impact of serine/threonine protein kinases on the regulation of sporulation in Bacillus subtilis, Front. Microbiol., 2016, 7, 568 Search PubMed.
- I. M. Shah, M.-H. Laaberki, D. L. Popham and J. Dworkin, A Eukaryotic-like Ser/Thr Kinase Signals Bacteria to Exit Dormancy in Response to Peptidoglycan Fragments, Cell, 2008, 135(3), 486–496 CrossRef CAS PubMed.
- F. Pompeo, E. Foulquier, B. Serrano, C. Grangeasse and A. Galinier, Phosphorylation of the cell division protein GpsB regulates PrkC kinase activity through a negative feedback loop in B acillus subtilis, Mol. Microbiol., 2015, 97(1), 139–150 CrossRef CAS PubMed.
- P. Barthe, G. V. Mukamolova, C. Roumestand and M. Cohen-Gonsaud, The structure of PknB extracellular PASTA domain from Mycobacterium tuberculosis suggests a ligand-dependent kinase activation, Structure, 2010, 18(5), 606–615 CrossRef CAS PubMed.
- M. Jarick, U. Bertsche, M. Stahl, D. Schultz, K. Methling and M. Lalk, et al., The serine/threonine kinase Stk and the phosphatase Stp regulate cell wall synthesis in Staphylococcus aureus, Sci. Rep., 2018, 8(1), 13693 CrossRef PubMed.
- P. Kaur, M. Rausch, B. Malakar, U. Watson, N. P. Damle and Y. Chawla, et al., LipidII interaction with specific residues of Mycobacterium tuberculosis PknB extracytoplasmic domain governs its optimal activation, Nat. Commun., 2019, 10(1), 1231 CrossRef PubMed.
- M. Mir, J. Asong, X. Li, J. Cardot, G. J. Boons and R. N. Husson, The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization, PLoS Pathog., 2011, 7(7), e1002182 CrossRef CAS PubMed.
- T. N. Lombana, N. Echols, M. C. Good, N. D. Thomsen, H.-L. Ng and A. E. Greenstein, et al., Allosteric Activation Mechanism of the Mycobacterium tuberculosis Receptor Ser/Thr Protein Kinase, PknB, Structure, 2010, 18(12), 1667–1677 CrossRef CAS PubMed.
- B. D. Labbe, C. L. Hall, S. L. Kellogg, Y. Chen, O. Koehn and A. M. Pickrum, et al., Reciprocal regulation of PASTA kinase signaling by differential modification, J. Bacteriol., 2019, 201(10), e00016-19 CrossRef PubMed.
- A. Lima, R. Durán, G. E. Schujman, M. J. Marchissio, M. M. Portela and G. Obal, et al., Serine/threonine protein kinase PrkA of the human pathogen Listeria monocytogenes: Biochemical characterization and identification of interacting partners through proteomic approaches, J. Proteomics, 2011, 74(9), 1720–1734 CrossRef CAS PubMed.
- E. Madec, A. Stensballe, S. Kjellström, L. Cladière, M. Obuchowski and O. N. Jensen, et al., Mass Spectrometry and Site-directed Mutagenesis Identify Several Autophosphorylated Residues Required for the Activity of PrkC, a Ser/Thr Kinase from Bacillus subtilis, J. Mol. Biol., 2003, 330(3), 459–472 CrossRef CAS PubMed.
- K. Rajagopalan and J. Dworkin, Escherichia coli YegI is a novel Ser/Thr kinase lacking conserved motifs that localizes to the inner membrane, FEBS Lett., 2020, 594(21), 3530–3541 CrossRef CAS PubMed.
- B. Boitel, M. Ortiz-Lombardía, R. Durán, F. Pompeo, S. T. Cole and C. Cerveñansky, et al., PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis, Mol. Microbiol., 2003, 49(6), 1493–1508 CrossRef CAS PubMed.
- M. N. Lisa, M. Gil, G. André-Leroux, N. Barilone, R. Durán and R. M. Biondi, et al., Molecular Basis of the Activity and the Regulation of the Eukaryotic-like S/T Protein Kinase PknG from Mycobacterium tuberculosis, Structure, 2015, 23(6), 1039–1048 CrossRef CAS PubMed.
- C. Roumestand, J. Leiba, N. Galophe, E. Margeat, A. Padilla and Y. Bessin, et al., Structural insight into the Mycobacterium tuberculosis Rv0020c protein and its interaction with the PknB kinase, Structure, 2011, 19(10), 1525–1534 CrossRef CAS PubMed.
- V. Bidnenko, L. Shi, A. Kobir, M. Ventroux, N. Pigeonneau and C. Henry, et al., B acillus subtilis serine/threonine protein kinase YabT is involved in spore development via phosphorylation of a bacterial recombinase, Mol. Microbiol., 2013, 88(5), 921–935 CrossRef CAS PubMed.
- B. Karniol and R. D. Vierstra, The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(5), 2807–2812 CrossRef CAS PubMed.
- E. Multamäki, R. Nanekar, D. Morozov, T. Lievonen, D. Golonka and W. Y. Wahlgren, et al., Comparative analysis of two paradigm bacteriophytochromes reveals opposite functionalities in two-component signaling, Nat. Commun., 2021, 12(1), 4394 CrossRef PubMed.
- A. Sajid, G. Arora, A. Singhal, V. C. Kalia and Y. Singh, Protein Phosphatases of Pathogenic Bacteria: Role in Physiology and Virulence, Annu. Rev. Microbiol., 2015, 69, 527–547 CrossRef CAS PubMed.
- J. Zhang, P. Yang, Q. Zeng, Y. Zhang, Y. Zhao and L. Wang, et al., Arginine kinase McsB and ClpC complex impairs the transition to biofilm formation in Bacillus subtilis, Microbiol. Res., 2024, 127979 Search PubMed.
- P. Fernandez, B. Saint-Joanis, N. Barilone, M. Jackson, B. Gicquel and S. T. Cole, et al., The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth, J. Bacteriol., 2006, 188(22), 7778–7784 CrossRef CAS PubMed.
- J. Chao, D. Wong, X. Zheng, V. Poirier, H. Bach and Z. Hmama, et al., Protein kinase and phosphatase signaling in Mycobacterium tuberculosis physiology and pathogenesis, Biochim. Biophys. Acta, 2010, 1804(3), 620–627 CrossRef CAS PubMed.
- Y. Av-Gay and M. Everett, The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis, Trends Microbiol., 2000, 8(5), 238–244 CrossRef CAS PubMed.
- A. Sajid, G. Arora, M. Gupta, S. Upadhyay, V. K. Nandicoori and Y. Singh, Phosphorylation of Mycobacterium tuberculosis Ser/Thr phosphatase by PknA and PknB, PLoS One, 2011, 6(3), e17871 CrossRef CAS PubMed.
- F. Shamma, E. H. Rego and C. C. Boutte, Mycobacterial serine/threonine phosphatase PstP is phosphoregulated and localized to mediate control of cell wall metabolism, Mol. Microbiol., 2022, 118(1–2), 47–60 CrossRef CAS PubMed.
- M. Beltramini Amanda, D. Mukhopadhyay Chitrangada and V. Pancholi, Modulation of Cell Wall Structure and Antimicrobial Susceptibility by a Staphylococcus aureus Eukaryote-Like Serine/Threonine Kinase and Phosphatase, Infect. Immun., 2009, 77(4), 1406–1416 CrossRef CAS PubMed.
- M. Débarbouillé, S. Dramsi, O. Dussurget, M.-A. Nahori, E. Vaganay and G. Jouvion, et al., Characterization of a Serine/Threonine Kinase Involved in Virulence of Staphylococcus aureus, J. Bacteriol., 2009, 191(13), 4070–4081 CrossRef PubMed.
- K. Burnside, A. Lembo, M. de Los Reyes, A. Iliuk, N. T. Binhtran and J. E. Connelly, et al., Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase, PLoS One, 2010, 5(6), e11071 CrossRef PubMed.
- K. Burnside, A. Lembo, M. I. Harrell, M. Gurney, L. Xue and N. T. BinhTran, et al., Serine/threonine phosphatase Stp1 mediates post-transcriptional regulation of hemolysin, autolysis, and virulence of group B Streptococcus, J. Biol. Chem., 2011, 286(51), 44197–44210 CrossRef CAS PubMed.
- J. D. Mougous, C. A. Gifford, T. L. Ramsdell and J. J. Mekalanos, Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa, Nat. Cell Biol., 2007, 9(7), 797–803 CrossRef CAS PubMed.
- G. Arora, A. Sajid, M. D. Arulanandh, A. Singhal, A. R. Mattoo and A. P. Pomerantsev, et al., Unveiling the novel dual specificity protein kinases in Bacillus anthracis: identification of the first prokaryotic dual specificity tyrosine phosphorylation-regulated kinase (DYRK)-like kinase, J. Biol. Chem., 2012, 287(32), 26749–26763 CrossRef CAS PubMed.
- K. Xu, S. Li, W. Yang, K. Li, Y. Bai and Y. Xu, et al., Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1, PLoS One, 2015, 10(4), e0124330 CrossRef PubMed.
- M. Arifuzzaman, E. Kwon and D. Y. Kim, Structural insights into the regulation of protein-arginine kinase McsB by McsA, Proc. Natl. Acad. Sci. U. S. A., 2024, 121(17), e2320312121 CrossRef CAS PubMed.
- V. Molle, C. Reynolds Robert, J. Alderwick Luke, S. Besra Gurdyal, J. Cozzone Alain and K. Fütterer, et al., EmbR2, a structural homologue of EmbR, inhibits the Mycobacterium tuberculosis kinase/substrate pair PknH/EmbR, Biochem. J., 2008, 410(2), 309–317 CrossRef CAS PubMed.
- N. Bhattacharyya, I. N. Nkumama, Z. Newland-Smith, L.-Y. Lin, W. Yin and R. E. Cullen, et al., An aspartate-specific solute-binding protein regulates protein kinase G activity to control glutamate metabolism in mycobacteria, mBio, 2018, 9(4) DOI:10.1128/mbio.00931-18.
- M. A. Schumacher, K. M. Piro, W. Xu, S. Hansen, K. Lewis and R. G. Brennan, Molecular Mechanisms of HipA-Mediated Multidrug Tolerance and Its Neutralization by HipB, Science, 2009, 323(5912), 396–401 CrossRef CAS PubMed.
- I. Kaspy, E. Rotem, N. Weiss, I. Ronin, N. Q. Balaban and G. Glaser, HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase, Nat. Commun., 2013, 4, 3001 CrossRef PubMed.
- E. Germain, D. Castro-Roa, N. Zenkin and K. Gerdes, Molecular mechanism of bacterial persistence by HipA, Mol. Cell, 2013, 52(2), 248–254 CrossRef CAS PubMed.
- X. Zhen, Y. Wu, J. Ge, J. Fu, L. Ye and N. Lin, et al., Molecular mechanism of toxin neutralization in the HipBST toxin-antitoxin system of Legionella pneumophila, Nat. Commun., 2022, 13(1), 4333 CrossRef CAS PubMed.
- M. A. Schumacher, P. Balani, J. Min, N. B. Chinnam, S. Hansen and M. Vulić, et al., HipBA-promoter structures reveal the basis of heritable multidrug tolerance, Nature, 2015, 524(7563), 59–64 CrossRef CAS PubMed.
- A. Schumacher Maria, J. Min, M. Link Todd, Z. Guan, W. Xu and Y.-H. Ahn, et al., Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance, Cell Rep., 2012, 2(3), 518–525 CrossRef CAS PubMed.
- M. J. Bick, V. Lamour, K. R. Rajashankar, Y. Gordiyenko, C. V. Robinson and S. A. Darst, How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the inhibitor Sda, J. Mol. Biol., 2009, 386(1), 163–177 CrossRef CAS PubMed.
- A. E. Whitten, D. A. Jacques, B. Hammouda, T. Hanley, G. F. King and J. M. Guss, et al., The Structure of the KinA-Sda Complex Suggests an Allosteric Mechanism of Histidine Kinase Inhibition, J. Mol. Biol., 2007, 368(2), 407–420 CrossRef CAS PubMed.
- M. Mörk-Mörkenstein, R. Heermann, Y. Göpel, K. Jung and B. Görke, Non-canonical activation of histidine kinase KdpD by phosphotransferase protein PtsN through interaction with the transmitter domain, Mol. Microbiol., 2017, 106(1), 54–73 CrossRef PubMed.
- Q. Peng, X. Tang, W. Dong, N. Sun and W. Yuan, A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism, Antibiotics, 2023, 12(1), 12 CrossRef CAS PubMed.
- G. Fritz, S. Dintner, S. Treichel Nicole, J. Radeck, U. Gerland and T. Mascher, et al., A New Way of Sensing: Need-Based Activation of Antibiotic Resistance by a Flux-Sensing Mechanism, mBio, 2015, 6(4), e00975 CrossRef CAS PubMed.
- N. L. George, A. L. Schilmiller and B. J. Orlando, Conformational snapshots of the bacitracin sensing and resistance transporter BceAB, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(14), e2123268119 CrossRef CAS PubMed.
- N. L. George and B. J. Orlando, Architecture of a complete Bce-type antimicrobial peptide resistance module, Nat. Commun., 2023, 14(1), 3896 CrossRef CAS PubMed.
- K. Yoshitani, E. Ishii, K. Taniguchi, H. Sugimoto, Y. Shiro and Y. Akiyama, et al., Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control, Biosci., Biotechnol., Biochem., 2019, 83(4), 684–694 CrossRef CAS PubMed.
- Y. Eguchi, E. Ishii, M. Yamane and R. Utsumi, The connector SafA interacts with the multi-sensing domain of PhoQ in Escherichia coli, Mol. Microbiol., 2012, 85(2), 299–313 CrossRef CAS PubMed.
- Y. Eguchi, J. Itou, M. Yamane, R. Demizu, F. Yamato and A. Okada, et al., B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(47), 18712–18717 CrossRef CAS PubMed.
- A. Petchiappan, N. Majdalani, E. Wall and S. Gottesman, RcsF-independent mechanisms of signaling within the Rcs phosphorelay, PLoS Genet., 2024, 20(12), e1011408 CrossRef CAS PubMed.
- M.-P. Castanié-Cornet, K. Cam and A. Jacq, RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli, J. Bacteriol., 2006, 188(12), 4264–4270 CrossRef PubMed.
- N. A. Hussein, S.-H. Cho, G. Laloux, R. Siam and J.-F. Collet, Distinct domains of Escherichia coli IgaA connect envelope stress sensing and down-regulation of the Rcs phosphorelay across subcellular compartments, PLoS Genet., 2018, 14(5), e1007398 CrossRef PubMed.
- S. R. Lach, S. Kumar, S. Kim, W. Im and A. Konovalova, Conformational rearrangements in the sensory RcsF/OMP complex mediate signal transduction across the bacterial cell envelope, PLoS Genet., 2023, 19(1), e1010601 CrossRef CAS PubMed.
- J. A. Holmes, S. E. Follett, H. Wang, C. P. Meadows, K. Varga and G. R. Bowman, Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(44), 12490–12495 CrossRef CAS PubMed.
- A. M. Perez, T. H. Mann, K. Lasker, D. G. Ahrens, M. R. Eckart and L. Shapiro, A Localized Complex of Two Protein Oligomers Controls the Orientation of Cell Polarity, mBio, 2017, 8(1), e02238-16 CrossRef PubMed.
- R. T. Wheeler and L. Shapiro, Differential Localization of Two Histidine Kinases Controlling Bacterial Cell Differentiation, Mol. Cell, 1999, 4(5), 683–694 CrossRef CAS PubMed.
- C. Zhang, W. Zhao, S. W. Duvall, K. A. Kowallis and W. S. Childers, Regulation of the activity of the bacterial histidine kinase PleC by the scaffolding protein PodJ, J. Biol. Chem., 2022, 298(4), 101683 CrossRef CAS PubMed.
- T. H. Mann, W. Seth Childers, J. A. Blair, M. R. Eckart and L. Shapiro, A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues, Nat. Commun., 2016, 7(1), 11454 CrossRef CAS PubMed.
- E. G. Biondi, S. J. Reisinger, J. M. Skerker, M. Arif, B. S. Perchuk and K. R. Ryan, et al., Regulation of the bacterial cell cycle by an integrated genetic circuit, Nature, 2006, 444(7121), 899–904 CrossRef CAS PubMed.
- C. Lori, S. Ozaki, S. Steiner, R. Böhm, S. Abel and B. N. Dubey, et al., Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication, Nature, 2015, 523(7559), 236–239 CrossRef CAS PubMed.
- M. Christen, H. D. Kulasekara, B. Christen, B. R. Kulasekara, L. R. Hoffman and S. I. Miller, Asymmetrical Distribution of the Second Messenger c-di-GMP upon Bacterial Cell Division, Science, 2010, 328(5983), 1295–1297 CrossRef CAS PubMed.
- A. A. Iniesta, N. J. Hillson and L. Shapiro, Cell pole–specific activation of a critical bacterial cell cycle kinase, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(15), 7012–7017 CrossRef CAS PubMed.
- J. Wu, N. Ohta, J.-L. Zhao and A. Newton, A novel bacterial tyrosine kinase essential for cell division and differentiation, Proc. Natl. Acad. Sci. U. S. A., 1999, 96(23), 13068–13073 CrossRef CAS PubMed.
- S. J. Reisinger, S. Huntwork, P. H. Viollier and K. R. Ryan, DivL Performs Critical Cell Cycle Functions in Caulobacter crescentus Independent of Kinase Activity, J. Bacteriol., 2007, 189(22), 8308–8320 CrossRef CAS PubMed.
- G. Tsokos Christos, S. Perchuk Barrett and T. Laub Michael, A Dynamic Complex of Signaling Proteins Uses Polar Localization to Regulate Cell-Fate Asymmetry in Caulobacter crescentus, Dev. Cell, 2011, 20(3), 329–341 CrossRef CAS PubMed.
- W. S. Childers, Q. Xu, T. H. Mann, I. I. Mathews, J. A. Blair and A. M. Deacon, et al., Cell fate regulation governed by a repurposed bacterial histidine kinase, PLoS Biol., 2014, 12(10), e1001979 CrossRef PubMed.
- T. H. Mann and L. Shapiro, Integration of cell cycle signals by multi-PAS domain kinases, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(30), E7166-e73 CrossRef PubMed.
- V. Sourjik and J. P. Armitage, Spatial organization in bacterial chemotaxis, EMBO J., 2010, 29(16), 2724–2733 CrossRef CAS PubMed.
- R. H. Williams and D. E. Whitworth, The genetic organisation of prokaryotic two-component system signalling pathways, BMC Genomics, 2010, 11, 1–12 CrossRef PubMed.
- M. T. Laub and M. Goulian, Specificity in two-component signal transduction pathways, Annu. Rev. Genet., 2007, 41(1), 121–145 CrossRef CAS PubMed.
- R. Agrawal, B. K. Sahoo and D. K. Saini, Cross-Talk and Specificity in Two-Component Signal Transduction Pathways, Future Microbiol., 2016, 11(5), 685–697 CrossRef CAS PubMed.
- B. Vemparala, A. Valiya Parambathu, K. Saini Deepak and M. Dixit Narendra, An Evolutionary Paradigm Favoring Cross Talk between Bacterial Two-Component Signaling Systems, mSystems, 2022, 7(6), e00298-22 CrossRef PubMed.
- K. Yamamoto, K. Hirao, T. Oshima, H. Aiba, R. Utsumi and A. Ishihama, Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli, J. Biol. Chem., 2005, 280(2), 1448–1456 CrossRef CAS PubMed.
- R. Agrawal, A. Pandey, M. P. Rajankar, N. M. Dixit and D. K. Saini, The two-component signalling networks of Mycobacterium tuberculosis display extensive cross-talk in vitro, Biochem. J., 2015, 469(1), 121–134 CrossRef CAS PubMed.
- L. Ali and H. Abdel Aziz May, Crosstalk involving two-component systems in Staphylococcus aureus signaling networks, J. Bacteriol., 2024, 206(4), e00418–e00423 CrossRef PubMed.
- C. E. Noriega, H. Y. Lin, L. L. Chen, S. B. Williams and V. Stewart, Asymmetric cross-regulation between the nitrate-responsive NarX-NarL and NarQ-NarP two-component regulatory systems from Escherichia coli K-12, Mol. Microbiol., 2010, 75(2), 394–412 CrossRef CAS PubMed.
- J. H. Baek, Y. J. Kang and S. Y. Lee, Transcript and protein level analyses of the interactions among PhoB, PhoR, PhoU and CreC in response to phosphate starvation in Escherichia coli, FEMS Microbiol. Lett., 2007, 277(2), 254–259 CrossRef CAS PubMed.
- E. J. Capra, B. S. Perchuk, O. Ashenberg, C. A. Seid, H. R. Snow and J. M. Skerker, et al., Spatial tethering of kinases to their substrates relaxes evolutionary constraints on specificity, Mol. Microbiol., 2012, 86(6), 1393–1403 CrossRef CAS PubMed.
- G. E. Townsend, V. Raghavan, I. Zwir and E. A. Groisman, Intramolecular arrangement of sensor and regulator overcomes relaxed specificity in hybrid two-component systems, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(2), E161–E169 CrossRef CAS PubMed.
- S. Wegener-Feldbrügge and L. Søgaard-Andersen, The atypical hybrid histidine protein kinase RodK in Myxococcus xanthus: spatial proximity supersedes kinetic preference in phosphotransfer reactions, J. Bacteriol., 2009, 191(6), 1765–1776 CrossRef PubMed.
- G. Chambonnier, L. Roux, D. Redelberger, F. Fadel, A. Filloux and M. Sivaneson, et al., The Hybrid Histidine Kinase LadS Forms a Multicomponent Signal Transduction System with the GacS/GacA Two-Component System in Pseudomonas aeruginosa, PLoS Genet., 2016, 12(5), e1006032 CrossRef PubMed.
- V. I. Francis, E. M. Waters, S. E. Finton-James, A. Gori, A. Kadioglu and A. R. Brown, et al., Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa, Nat. Commun., 2018, 9(1), 2219 CrossRef PubMed.
- V. I. Francis, E. M. Waters, S. E. Finton-James, A. Gori, A. Kadioglu and A. R. Brown, et al., Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa, Nat. Commun., 2018, 9(1), 2219 CrossRef PubMed.
- J. M. Mancl, W. K. Ray, R. F. Helm and F. D. Schubot, Helix Cracking Regulates the Critical Interaction between RetS and GacS in Pseudomonas aeruginosa, Structure, 2019, 27(5), 785–793.e5 CrossRef CAS PubMed.
- A. Y. Bhagirath, S. P. Pydi, Y. Li, C. Lin, W. Kong and P. Chelikani, et al., Characterization of the Direct Interaction between Hybrid Sensor Kinases PA1611 and RetS That Controls Biofilm Formation and the Type III Secretion System in Pseudomonas aeruginosa, ACS Infect. Dis., 2017, 3(2), 162–175 CrossRef CAS PubMed.
- L. A. Mike, J. E. Choby, P. R. Brinkman, L. Q. Olive, B. F. Dutter and S. J. Ivan, et al., Two-component system cross-regulation integrates Bacillus anthracis response to heme and cell envelope stress, PLoS Pathog., 2014, 10(3), e1004044 CrossRef PubMed.
- T. Drepper, J. Wiethaus, D. Giaourakis, S. Gross, B. Schubert and M. Vogt, et al., Cross-talk towards the response regulator NtrC controlling nitrogen metabolism in Rhodobacter capsulatus, FEMS Microbiol. Lett., 2006, 258(2), 250–256 CrossRef CAS PubMed.
- A. N. Norsworthy and K. L. Visick, Signaling between two interacting sensor kinases promotes biofilms and colonization by a bacterial symbiont, Mol. Microbiol., 2015, 96(2), 233–248 CrossRef CAS PubMed.
- B. M. Carpenter, A. L. West, H. Gancz, S. L. Servetas, O. Q. Pich and J. J. Gilbreath, et al., Crosstalk between the HpArsRS two-component system and HpNikR is necessary for maximal activation of urease transcription, Front. Microbiol., 2015, 6, 558 Search PubMed.
- M. Villanueva, B. García, J. Valle, B. Rapún, I. Ruiz de Los Mozos and C. Solano, et al., Sensory deprivation in Staphylococcus aureus, Nat. Commun., 2018, 9(1), 523 CrossRef PubMed.
- A. Frando, V. Boradia and C. Grundner, Regulatory Intersection of Two-component System and Ser/Thr Protein Kinase Signaling in Mycobacterium tuberculosis, J. Mol. Biol., 2024, 436(2), 168379 CrossRef CAS PubMed.
- S. E. Lizewski, D. S. Lundberg and M. J. Schurr, The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis, Infect. Immun., 2002, 70(11), 6083–6093 CrossRef CAS PubMed.
- R. Li, X. Zhu, P. Zhang, X. Wu, Q. Jin and J. Pan, Ser/Thr protein kinase Stk1 phosphorylates the key transcriptional regulator AlgR to modulate virulence and resistance in Pseudomonas aeruginosa, Virulence, 2024, 15(1), 2367649 CrossRef PubMed.
- M. J. Canova, G. Baronian, S. Brelle, M. Cohen-Gonsaud, M. Bischoff and V. Molle, A novel mode of regulation of the Staphylococcus aureus Vancomycin-resistance-associated response regulator VraR mediated by Stk1 protein phosphorylation, Biochem. Biophys. Res. Commun., 2014, 447(1), 165–171 CrossRef CAS PubMed.
- P. Hardt, I. Engels, M. Rausch, M. Gajdiss, H. Ulm and P. Sass, et al., The cell wall precursor lipid II acts as a molecular signal for the Ser/Thr kinase PknB of Staphylococcus aureus, Int. J. Med. Microbiol., 2017, 307(1), 1–10 CrossRef CAS PubMed.
- M. Fridman, G. D. Williams, U. Muzamal, H. Hunter, K. W. Siu and D. Golemi-Kotra, Two unique phosphorylation-driven signaling pathways crosstalk in Staphylococcus aureus to modulate the cell-wall charge: Stk1/Stp1 meets GraSR, Biochemistry, 2013, 52(45), 7975–7986 CrossRef CAS PubMed.
- A. T. Ulijasz, S. P. Falk and B. Weisblum, Phosphorylation of the RitR DNA-binding domain by a Ser-Thr phosphokinase: implications for global gene regulation in the streptococci, Mol. Microbiol., 2009, 71(2), 382–390 CrossRef CAS PubMed.
- S. Agarwal, S. Agarwal, P. Pancholi and V. Pancholi, Strain-Specific Regulatory Role of Eukaryote-Like Serine/Threonine Phosphatase in Pneumococcal Adherence, Infect. Immun., 2012, 80(4), 1361–1372 CrossRef CAS PubMed.
- H.-J. Bae, H.-N. Lee, M.-N. Baek, E.-J. Park, C.-Y. Eom and I.-J. Ko, et al., Inhibition of the DevSR two-component system by overexpression of Mycobacterium tuberculosis PknB in Mycobacterium smegmatis, Mol. Cells, 2017, 40(9), 632–642 CrossRef CAS PubMed.
- E.-J. Park, Y.-M. Kwon, J.-W. Lee, H.-Y. Kang and J.-I. Oh, Dual control of RegX3 transcriptional activity by SenX3 and PknB, J. Biol. Chem., 2019, 294(28), 11023–11034 CrossRef CAS PubMed.
- J. L. Hsu, H. C. Chen, H. L. Peng and H. Y. Chang, Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1, J. Biol. Chem., 2008, 283(15), 9933–9944 CrossRef CAS PubMed.
- S. L. Kellogg and C. J. Kristich, Convergence of PASTA kinase and two-component signaling in response to cell wall stress in Enterococcus faecalis, J. Bacteriol., 2018, 200(12), e00086-18 CrossRef PubMed.
- C. Jers, A. Kobir, E. O. Søndergaard, P. R. Jensen and I. Mijakovic, Bacillus subtilis two-component system sensory kinase DegS is regulated by serine phosphorylation in its input domain, PLoS One, 2011, 6(2), e14653 CrossRef CAS PubMed.
- M. Rausch, J. P. Deisinger, H. Ulm, A. Müller, W. Li and P. Hardt, et al., Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus, Nat. Commun., 2019, 10(1), 1404 CrossRef PubMed.
- J. Key and K. Moffat, Crystal Structures of Deoxy and CO-Bound bjFixLH Reveal Details of Ligand Recognition and Signaling, Biochemistry, 2005, 44(12), 4627–4635 CrossRef CAS PubMed.
- N. Tiwari, M. López-Redondo, L. Miguel-Romero, K. Kulhankova, M. P. Cahill and P. M. Tran, et al., The SrrAB two-component system regulates Staphylococcus aureus pathogenicity through redox sensitive cysteines, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(20), 10989–10999 CrossRef CAS PubMed.
- M. Husain, J. Jones-Carson, M. Song, B. D. McCollister, T. J. Bourret and A. Vázquez-Torres, Redox sensor SsrB Cys203 enhances Salmonella fitness against nitric oxide generated in the host immune response to oral infection, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(32), 14396–14401 CrossRef CAS PubMed.
- X. Shu, Y. Shi, Y. Huang, D. Yu and B. Sun, Transcription tuned by S-nitrosylation underlies a mechanism for Staphylococcus aureus to circumvent vancomycin killing, Nat. Commun., 2023, 14(1), 2318 CrossRef CAS PubMed.
- A. Mata-Cabana, M. García-Domínguez, F. J. Florencio and M. Lindahl, Thiol-based redox modulation of a cyanobacterial eukaryotic-type serine/threonine kinase required for oxidative stress tolerance, Antioxid. Redox Signaling, 2012, 17(4), 521–533 CrossRef CAS PubMed.
- B. N. Dubey, C. Lori, S. Ozaki, G. Fucile, I. Plaza-Menacho and U. Jenal, et al., Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking, Sci. Adv., 2016, 2(9), e1600823 CrossRef PubMed.
- B. N. Dubey, E. Agustoni, R. Böhm, A. Kaczmarczyk, F. Mangia and C. von Arx, et al., Hybrid histidine kinase activation by cyclic di-GMP–mediated domain liberation, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(2), 1000–1008 CrossRef CAS PubMed.
- S. T. Cheng, F. F. Wang and W. Qian, Cyclic-di-GMP binds to histidine kinase RavS to control RavS-RavR phosphotransfer and regulates the bacterial lifestyle transition between virulence and swimming, PLoS Pathog., 2019, 15(8), e1007952 CrossRef CAS PubMed.
- Y. S. Rajpurohit and H. S. Misra, Characterization of a DNA damage-inducible membrane protein kinase from Deinococcus radiodurans and its role in bacterial radioresistance and DNA strand break repair, Mol. Microbiol., 2010, 77(6), 1470–1482 CrossRef CAS PubMed.
- K. Vadakkan, K. Sathishkumar, S. Kuttiyachan Urumbil, S. Ponnenkunnathu Govindankutty, A. Kumar Ngangbam and B. Devi Nongmaithem, A review of chemical signaling mechanisms underlying quorum sensing and its inhibition in Staphylococcus aureus, Bioorg. Chem., 2024, 148, 107465 CrossRef CAS PubMed.
- C. Jenul and A. R. Horswill, Regulation of Staphylococcus aureus virulence, Microbiol. Spectrum, 2019, 7(2) DOI:10.1128/microbiolspec.gpp3-0031-2018.
- M. Liu, X. Zhu, C. Zhang and Z. Zhao, LuxQ-LuxU-LuxO pathway regulates biofilm formation by Vibrio parahaemolyticus, Microbiol. Res., 2021, 250, 126791 CrossRef CAS PubMed.
- Q. Fan, H. Wang, C. Mao, J. Li, X. Zhang and D. Grenier, et al., Structure and Signal Regulation Mechanism of Interspecies and Interkingdom Quorum Sensing System Receptors, J. Agric. Food Chem., 2022, 70(2), 429–445 CrossRef CAS PubMed.
- B. M. St Louis, S. M. Quagliato and P. C. Lee, Bacterial effector kinases and strategies to identify their target host substrates, Front. Microbiol., 2023, 14, 1113021 CrossRef PubMed.
- A. Walburger, A. Koul, G. Ferrari, L. Nguyen, C. Prescianotto-Baschong and K. Huygen, et al., Protein kinase G from pathogenic mycobacteria promotes survival within macrophages, Science, 2004, 304(5678), 1800–1804 CrossRef CAS PubMed.
- K. E. Zulauf, J. T. Sullivan and M. Braunstein, The SecA2 pathway of Mycobacterium tuberculosis exports effectors that work in concert to arrest phagosome and autophagosome maturation, PLoS Pathog., 2018, 14(4), e1007011 CrossRef PubMed.
- D. C. Lockwood, H. Amin, T. R. D. Costa and G. N. Schroeder, The Legionella pneumophila Dot/Icm type IV secretion system and its effectors, Microbiology, 2022, 168(5) DOI:10.1099/mic.0.001187.
- J. Ge, Y. Wang, X. Li, Q. Lu, H. Yu and H. Liu, et al., Phosphorylation of caspases by a bacterial kinase inhibits host programmed cell death, Nat. Commun., 2024, 15(1), 8464 CrossRef CAS PubMed.
- E. Hervet, X. Charpentier, A. Vianney, J.-C. Lazzaroni, C. Gilbert and D. Atlan, et al., Protein Kinase LegK2 Is a Type IV Secretion System Effector Involved in Endoplasmic Reticulum Recruitment and Intracellular Replication of Legionella pneumophila, Infect. Immun., 2011, 79(5), 1936–1950 CrossRef CAS PubMed.
- C. Michard, D. Sperandio, N. Baïlo, J. Pizarro-Cerdá, L. LeClaire and E. Chadeau-Argaud, et al., The Legionella kinase LegK2 targets the ARP2/3 complex to inhibit actin nucleation on phagosomes and allow bacterial evasion of the late endocytic pathway, mBio, 2015, 6(3), e00354-15 CrossRef PubMed.
- A. Flayhan, C. Bergé, N. Baïlo, P. Doublet, R. Bayliss and L. Terradot, The structure of Legionella pneumophila LegK4 type four secretion system (T4SS) effector reveals a novel dimeric eukaryotic-like kinase, Sci. Rep., 2015, 5(1), 14602 CrossRef CAS PubMed.
- P.-C. Lee and M. P. Machner, The Legionella effector kinase LegK7 hijacks the host Hippo pathway to promote infection, Cell Host Microbe, 2018, 24(3), 429–438.e6 CrossRef CAS PubMed.
- S. C. Park, S. Y. Cho, T. H. Kim, K. Y. Ko, W. S. Song and S. G. Kang, et al., Activation of the Legionella pneumophila LegK7 effector kinase by the host MOB1 protein, J. Mol. Biol., 2021, 433(3), 166746 CrossRef CAS PubMed.
- J. Ge, H. Xu, T. Li, Y. Zhou, Z. Zhang and S. Li, et al., A Legionella type IV effector activates the NF-κB pathway by phosphorylating the IκB family of inhibitors, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(33), 13725–13730 CrossRef CAS PubMed.
- A. Sreelatha, C. Nolan, B. C. Park, K. Pawłowski, D. R. Tomchick and V. S. Tagliabracci, A Legionella effector kinase is activated by host inositol hexakisphosphate, J. Biol. Chem., 2020, 295(18), 6214–6224 CrossRef CAS PubMed.
- D. T. Patel, P. J. Stogios, L. Jaroszewski, M. L. Urbanus, M. Sedova and C. Semper, et al., Global atlas of predicted functional domains in Legionella pneumophila Dot/Icm translocated effectors, Mol. Syst. Biol., 2024, 1–31 Search PubMed.
- S. J. Juris, A. E. Rudolph, D. Huddler, K. Orth and J. E. Dixon, A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption, Proc. Natl. Acad. Sci. U. S. A., 2000, 97(17), 9431–9436 CrossRef CAS PubMed.
- L. Navarro, A. Koller, R. Nordfelth, H. Wolf-Watz, S. Taylor and J. E. Dixon, Identification of a molecular target for the Yersinia protein kinase A, Mol. Cell, 2007, 26(4), 465–477 CrossRef CAS PubMed.
- W. L. Lee, J. M. Grimes and R. C. Robinson, Yersinia effector YopO uses actin as bait to phosphorylate proteins that regulate actin polymerization, Nat. Struct. Mol. Biol., 2015, 22(3), 248–255 CrossRef CAS PubMed.
- K. Pha, M. E. Wright, T. M. Barr, R. A. Eigenheer and L. Navarro, Regulation of Yersinia protein kinase A (YpkA) kinase activity by multisite autophosphorylation and identification of an N-terminal substrate-binding domain in YpkA, J. Biol. Chem., 2014, 289(38), 26167–26177 CrossRef CAS PubMed.
- Y. Zhou, N. Dong, L. Hu and F. Shao, The Shigella type three secretion system effector OspG directly and specifically binds to host ubiquitin for activation, PLoS One, 2013, 8(2), e57558 CrossRef CAS PubMed.
- S. Kariuki, Global burden of antimicrobial resistance and forecasts to 2050, Lancet, 2024, 404(10459), 1172–1173 CrossRef CAS PubMed.
- M. K. Gugger and P. J. Hergenrother, A new type of antibiotic targets a drug-resistant bacterium, Nature, 2024, 625(7995), 451–452 CrossRef CAS PubMed.
- T. Krell, J. Lacal, A. Busch, H. Silva-Jiménez, M.-E. Guazzaroni and J. L. Ramos, Bacterial sensor kinases: diversity in the recognition of environmental signals, Annu. Rev. Microbiol., 2010, 64(1), 539–559 CrossRef CAS PubMed.
- A. E. Bem, N. Velikova, M. T. Pellicer, P. V. Baarlen, A. Marina and J. M. Wells, Bacterial Histidine Kinases as Novel Antibacterial Drug Targets, ACS Chem. Biol., 2015, 10(1), 213–224 CrossRef CAS PubMed.
- D. Beier and R. Gross, Regulation of bacterial virulence by two-component systems, Curr. Opin. Microbiol., 2006, 9(2), 143–152 CrossRef CAS PubMed.
- H. Takada and H. Yoshikawa, Essentiality and function of WalK/WalR two-component system: the past, present, and future of research, Biosci., Biotechnol., Biochem., 2018, 82(5), 741–751 CrossRef CAS PubMed.
- A. Okada, M. Igarashi, T. Okajima, N. Kinoshita, M. Umekita and R. Sawa, et al., Walkmycin B targets WalK (YycG), a histidine kinase essential for bacterial cell growth, J. Antibiot., 2010, 63(2), 89–94 CrossRef CAS PubMed.
- Y. Eguchi, N. Kubo, H. Matsunaga, M. Igarashi and R. Utsumi, Development of an antivirulence drug against Streptococcus mutans: repression of biofilm formation, acid tolerance, and competence by a histidine kinase inhibitor, walkmycin C, Antimicrob. Agents Chemother., 2011, 55(4), 1475–1484 CrossRef CAS PubMed.
- A. A. Radwan, F. K. Aanazi, M. Al-Agamy and G. M. Mahrous, Design, synthesis and molecular modeling study of substituted indoline-2-ones and spiro [indole-heterocycles] with potential activity against Gram-positive bacteria, Acta Pharm., 2022, 72(1), 79–95 CrossRef CAS PubMed.
- S. Zhang, J. Wang, W. Xu, Y. Liu, W. Wang and K. Wu, et al., Antibacterial effects of Traditional Chinese Medicine monomers against Streptococcus pneumoniae via inhibiting pneumococcal histidine kinase (VicK), Front. Microbiol., 2015, 6, 479 Search PubMed.
- H. Zheng, C. J. Colvin, B. K. Johnson, P. D. Kirchhoff, M. Wilson and K. Jorgensen-Muga, et al., Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence, Nat. Chem. Biol., 2017, 13(2), 218–225 CrossRef CAS PubMed.
- M. A. Carabajal, C. R. M. Asquith, T. Laitinen, G. J. Tizzard, L. Yim and A. Rial, et al., Quinazoline-Based Antivirulence Compounds Selectively Target Salmonella PhoP/PhoQ Signal Transduction System, Antimicrob. Agents Chemother., 2019, 64(1) DOI:10.1128/aac.01744-19.
- K. E. Wilke, S. Francis and E. E. Carlson, Inactivation of Multiple Bacterial Histidine Kinases by Targeting the ATP-Binding Domain, ACS Chem. Biol., 2015, 10(1), 328–335 CrossRef CAS PubMed.
- R. Dutta and M. Inouye, GHKL, an emergent ATPase/kinase superfamily, Trends Biochem. Sci., 2000, 25(1), 24–28 CrossRef CAS PubMed.
- B. Fernandez-Ciruelos, M. Albanese, A. Adhav, V. Solomin, A. Ritchie-Martinez and F. Taverne, et al., Repurposing Hsp90 inhibitors as antimicrobials targeting two-component systems identifies compounds leading to loss of bacterial membrane integrity, Microbiol. Spectrum, 2024, 12(8), e00146-24 CrossRef PubMed.
- M. T. Guarnieri, L. Zhang, J. Shen and R. Zhao, The Hsp90 Inhibitor Radicicol Interacts with the ATP-Binding Pocket of Bacterial Sensor Kinase PhoQ, J. Mol. Biol., 2008, 379(1), 82–93 CrossRef CAS PubMed.
- C. D. Vo, H. L. Shebert, S. Zikovich, R. A. Dryer, T. P. Huang and L. J. Moran, et al., Repurposing Hsp90 inhibitors as antibiotics targeting histidine kinases, Bioorg. Med. Chem. Lett., 2017, 27(23), 5235–5244 CrossRef CAS PubMed.
- E. A. Groisman, A. Duprey and J. Choi, How the PhoP/PhoQ System Controls Virulence and Mg2+ Homeostasis: Lessons in Signal Transduction, Pathogenesis, Physiology, and Evolution, Microbiol. Mol. Biol. Rev., 2021, 85(3), e0017620 CrossRef PubMed.
- A. O. Olaitan, S. Morand and J. M. Rolain, Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria, Front. Microbiol., 2014, 5, 643 Search PubMed.
- A. P. Zavascki, L. Z. Goldani, J. Li and R. L. Nation, Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review, J. Antimicrob. Chemother., 2007, 60(6), 1206–1215 CrossRef CAS PubMed.
- S. Rollas and S. G. Küçükgüzel, Biological activities of hydrazone derivatives, Molecules, 2007, 12(8), 1910–1939 CrossRef CAS PubMed.
- I. Cabezudo, C. A. Lobertti, E. G. Véscovi and R. L. E. Furlan, Effect-Directed Synthesis of PhoP/PhoQ Inhibitors to Regulate Salmonella Virulence, J. Agric. Food Chem., 2022, 70(22), 6755–6763 CrossRef CAS PubMed.
- C. A. Lobertti, I. Cabezudo, F. O. Gizzi, V. Blancato, C. Magni and R. L. E. Furlán, et al., An allosteric inhibitor of the PhoQ histidine kinase with therapeutic potential against Salmonella infection, J. Antimicrob. Chemother., 2024, 79(8), 1820–1830 CrossRef CAS PubMed.
- T. Watanabe, M. Igarashi, T. Okajima, E. Ishii, H. Kino and M. Hatano, et al., Isolation and Characterization of Signermycin B, an Antibiotic That Targets the Dimerization Domain of Histidine Kinase WalK, Antimicrob. Agents Chemother., 2012, 56(7), 3657–3663 CrossRef CAS PubMed.
- M. Igarashi, T. Watanabe, T. Hashida, M. Umekita, M. Hatano and Y. Yanagida, et al., Waldiomycin, a novel WalK-histidine kinase inhibitor from Streptomyces sp. MK844-mF10, J. Antibiot., 2013, 66(8), 459–464 CrossRef CAS PubMed.
- A. Kato, S. Ueda, T. Oshima, Y. Inukai, T. Okajima and M. Igarashi, et al., Characterization of H-box region mutants of WalK inert to the action of waldiomycin in Bacillus subtilis, J. Gen. Appl. Microbiol., 2017, 63(4), 212–221 CrossRef CAS PubMed.
- A. R. Tierney and P. N. Rather, Roles of two-component regulatory systems in antibiotic resistance, Future Microbiol., 2019, 14(6), 533–552 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2025 |