Open Access Article
Open Access ArticlePeptides: potential delivery systems for mRNA
Huiting
Liang
a,
Yun
Xing
b,
Kexin
Wang
c,
Yaping
Zhang
*a,
Feng
Yin
 *ab and
Zigang
Li
*ab and
Zigang
Li
 *ab
*ab
aPingshan Translational Medicine Center, Shenzhen Bay Laboratory, Shenzhen 518118, China. E-mail: yapi_zhang@163.com
bState Key Laboratory of Chemical Oncogenomics, School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen 518055, China. E-mail: yinfeng@pkusz.edu.cn; lizg@pkusz.edu.cn
cDepartment of Chemistry, Southern University of Science and Technology, Shenzhen, 518055, China
First published on 26th February 2025
Abstract
mRNA-based therapies have broad applications in various disease treatments and have been applied in protein replacement therapy, gene editing, and vaccine development. Numerous research studies have been carried out aiming to increase the stability of mRNA, improve its translational efficiency, and reduce its immunogenicity. However, given mRNA's large molecular size and strong electronegativity, the safety and efficient delivery of mRNA into the target cells remains the critical rate-limiting step in current mRNA drug development. Various nanocarriers, such as liposomes, lipid nanoparticles, polyetherimide, and mesoporous silica nanoparticles, have been employed for mRNA delivery in the past few decades. Among them, peptides have demonstrated great potential as promising carrier candidates for mRNA delivery due to their high cell membrane permeability, good biocompatibility, definite chemical structure, and ease of preparation. Here, peptide-based mRNA delivery systems are systematically analyzed, including their construction strategies, mechanisms of action in mRNA delivery, and the application limitations or challenges. It is hoped that this review will guide the design, optimization, and applications of peptide carriers in mRNA-based drug development.
1 Introduction
In the last few decades, ribonucleic acid (RNA)-based therapy has emerged as a promising approach for treating various diseases. Recently, multiple RNA drugs have been developed and applied in several fields, including gene inhibition, gene insertion, gene editing, and protein expression. By the end of 2023, 18 nucleic acid drugs (excluding 3 delisted products) had been approved by the U.S. Food and Drug Administration (FDA), including 9 antisense oligonucleotides (ASOs), 6 small interfering RNAs (siRNAs), 1 aptamer, and 2 messenger ribonucleic acids (mRNAs).1,2 Unlike oligonucleotides (siRNA/ASO/aptamer), mRNAs with longer sequences have unique therapeutic applications due to their ability to produce any peptide/protein in transfected cells transiently. Therefore, mRNA drugs have broad application prospects in protein replacement, gene editing, and vaccine development. mRNA drugs are classified into two main categories: mRNA therapeutic drugs and mRNA vaccines.3 The drugs that utilize the translation function of mRNA molecules to intervene in disease processes are referred to as mRNA therapeutic drugs. Compared with traditional small molecule drugs or recombinant proteins, mRNA drugs have advantages such as relatively simple development and production.4 In comparison to DNA medicines, mRNA drugs have the characteristics of rapid expression and no risk of genomic integration.5 Furthermore, mRNA vaccines encoding antigens can elicit more robust cellular and humoral immune responses than traditional vaccines.6 This results in a higher protection rate. The outbreak of the novel coronavirus SARS-CoV-2 (COVID-19) in 2019 has accelerated the success of mRNA drug development and clinical approval. In 2020, Pfizer developed the first mRNA vaccine based on lipid nanoparticle (LNP) carriers and it was approved for the prevention of SARS-CoV-2 infection.7 This was a major turning point for the development of mRNA therapies. Since then, several nucleic acid drug research and development companies, represented by Moderna and BioNTech, have joined the competition for mRNA drug development, and have continued to expand the potential of mRNA in various scenarios, such as tumor immunotherapies,8 autoimmune diseases,9 and rare genetic diseases.10However, compared to oligonucleotide drugs, mRNA has a larger molecular weight and size, as well as a stronger electronegativity, which makes it more difficult for mRNA to enter cells and exert effects. As a result, the development of safe and efficient mRNA delivery carriers has remained a challenging and crucial step in the progression of mRNA drugs. In the past few decades, researchers have focused on optimizing delivery vectors to protect mRNAs from degradation and promote their cytoplasmic delivery. Commonly, a qualified mRNA delivery carrier should meet the following requirements:11 (i) the carrier needs to be capable of effectively compressing micrometer-sized mRNA into a nanometer-sized complex, which further avoids nuclease-mediated nucleic acid drug degradation and enhances the cellular uptake of mRNA; (ii) the complex can selectively accumulate in the desired tissues and cells, and assist mRNA efficient internalization by the target cells; (iii) the carrier is capable of intracellular escape from the endosomal cytosol, allowing for effective nucleic acid drug delivery to the cytoplasm to exert their effects; (iv) the carrier is safe and stable with low immunogenicity; and (v) the circulation time of mRNA in vivo could be effectively extended by introducing carriers into the system.
To satisfy these requirements, among different types of delivery systems, viral vectors have attracted attention in mRNA delivery due to their high transfection efficiency. At present, numerous viral delivery carriers have been developed, including lentiviral vectors, adenovirus, and adeno-associated virus vectors.12 However, the safe application of virus-based carriers remains a challenge due to the high inherent immunogenicity of viruses and the potential risk of gene insertion. In recent years, a variety of non-viral carriers have been developed as alternatives to viral carriers, which have gradually gained popularity for mRNA delivery, such as lipid-based delivery systems, inorganic nanoparticles, polycationic polymers, extracellular vesicles, hydrogels, peptides, and nucleic acid nanoassemblies.13–17 These novel non-viral carriers not only demonstrate commendable performances in improving mRNA stability and delivery efficiency, but also make important improvements in critical criteria including biocompatibility, toxicity, immunogenicity, and targeting. Mesoporous silica nanoparticles, as a representative of inorganic nanocarriers, can achieve high mRNA loading due to their mesoporous structure and large specific surface area. Their easily modifiable surface also provides the possibility for further improvement in the stability and targeting of mRNA drugs.18 Polyethyleneimine (PEI), as a representative of polycationic polymers, has demonstrated the ability to induce efficient nucleic acid aggregation. Biocompatible molecules such as cyclodextrin and polyethylene glycol (PEG) have been introduced into the carrier to reduce the charge density of PEI, and to improve the safety of PEI carriers while maintaining their nucleic acid delivery performance.19–21 In addition, to improve the biocompatibility of the carrier and reduce its immunogenicity, extracellular vesicles, represented by exosomes, are also developed for mRNA delivery. They have presented good specificity in the targeted delivery of mRNA to the desired tissues and organs.22 Furthermore, nucleic acid nanoassemblies, formed through rolling circle transcription,23 smart-responsive DNA nanogels,24 and mRNA nanoassembly strategies25 have made significant progress in extending mRNA half-life, enhancing protein expression levels, and optimizing intracellular release. As representatives of lipid-based delivery systems, LNPs were the first delivery system approved by the FDA for the novel coronavirus (SARS-CoV-2) mRNA vaccine,7 due to their mRNA protection, improved delivery efficiency, biosafety, and industrialization advantages.
LNP delivery technology is currently the first choice for nucleic acid drug delivery.26 Despite the many advantages of LNP delivery systems, unmodified LNP drug delivery systems have significant limitations, such as potential cytotoxicity, lack of targeting selectivity, short circulation time, poor endosomal escape, and the need for extreme storage conditions (such as freezing). Improved LNP formulations aim to overcome these drawbacks.27–32 Peptides derived from biological sources are expected to circumvent the aforementioned limitations of LNP application and can be excellent mRNA delivery systems. First, peptides, derived from natural products, were widely explored for exhibiting good biocompatibility and biodegradability.33,34 Also, peptides demonstrate remarkable cell-penetrating efficiency, precise targeting specificity, and a versatile array of chemical modification sites,15,35–37 all of which synergistically enhance the targeting efficacy of peptide-based nucleic acid delivery platforms. Furthermore, it was shown that peptide carriers and nucleic acids can be formulated into dry powder preparation via spray drying without compromising their physical integrity and biological activity.38,39 This suggests that peptide-based carriers may address the storage limitations associated with LNPs. In summary, peptide-based carriers hold significant promise for overcoming the limitations of LNPs, particularly in terms of biosafety, targeting specificity, and storage requirements.
A few studies have reviewed the application of peptide carriers in small nucleic acid drugs. However, in this article, we mainly focus on the progress of research on peptide-based carriers in mRNA drug delivery. The construction strategy, mechanism of action, and application limitations of the peptide nano-carrier system are also analyzed in detail. It is hoped that this comprehensive overview will provide a reference for the redesign and optimization of the new generation peptide carriers.
2 Peptides as carriers for mRNA delivery
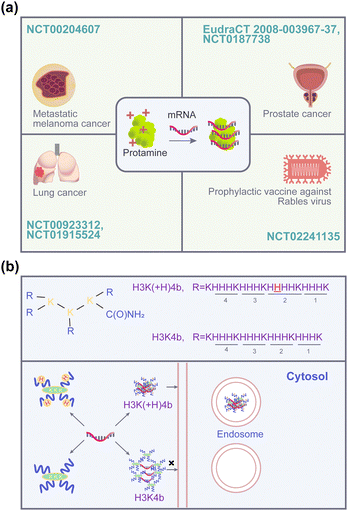
Multifunctional peptides obtained by the arrangement of different amino acids offer the possibility to facilitate efficient cell penetration or deliver nucleic acid drugs to specific organs or organelles.40 Protamine, cell-penetrating peptides (CPPs), and cell membrane protein-targeting peptides have been proven to be effective for mRNA delivery. According to the contribution rate of peptides in the delivery carrier, peptide-based carriers can be divided into two categories: (i) mRNA delivery carriers based on single-component peptides that primarily rely on the peptides for facilitating mRNA delivery; (ii) peptides are combined with other carrier materials for mRNA delivery, primarily to overcome the limitations of the original carriers. Specifically, mRNA delivery dominated by peptides can be subdivided into cationic peptide-driven delivery, amphipathic peptide-driven delivery, mRNA-peptide covalent conjugate delivery, and phase separation-driven delivery. The recent categories of peptides used for efficient mRNA delivery are summarized in Table 1.| Delivery system type | Name of the delivery system | Amino acid sequence | In vivo model | Ref. |
|---|---|---|---|---|
| a O: ornithine. b n.d. = not determined. | ||||
| Cationic peptides | Protamine | MPRRRRSSSRPVRRRRRPRVSRRRRRRGGRRRR | Phase I | 41 |
| H3K(+H)4b | A four branches peptide, KHHHKHHHKHHHHKHHHK in each branch | n.d.b | 42 | |
| Amphipathic cationic peptides | p5RHH | VLTTGLPALISWIRRRHRRHC | Mice | 43 |
| RALA | WEARLARALARALARHLARALARALRACEA | Mice | 44 | |
| LAH4-L1 | KKALLAHALHLLALLALHLAHALKKA | n.d. | 45 | |
| LAH4-L1R | RRALLAHALHLLALLALHLAHALRRA | n.d. | 46 | |
| HELP-4H | GLGTLLTLLHFLLHHLLHFLKRKRQQ | n.d. | 47 | |
| PepFect14 | Stearyl- AGYLLGKLLOOLAAAALOOLLa | Mice | 48 | |
| Covalent binding | — | PFVYLI | n.d. | 49 |
| Phase-separated peptide | HBpep-SR | GHGVY-GHGVY-GHGPY-K-GHGPY-GHGLYW(K was modified by NHS-SS-R, R = Ac/Ph) | n.d. | 50 |
| Lipid/peptide hybrid delivery system | LNP modified by peptide KALA | KALA: WEAKLAKALAKALAKHLAKALAKALKA | Mice | 51 |
| DOTAP liposomes, modified by DP7-C | DP7-C: cholesterol-modified VQWRIRVAVIRK | Mice | 52 | |
| Polymer/peptide hybrid delivery system | PEG12KL4 | PEG12-KLLLLKLLLLKLLLLKLLLLK | Mice | 53 |
| PEG12LAH4-L1 | PEG12-KKALLAHALHLLALLALHLAHALKKA | Mice | 54 | |
| PLA and LAH4-L1 | LAH4-L1: KKALLAHALHLLALLALHLAHALKKA | n.d. | 45 | |
2.1. mRNA delivery carriers based on single-component peptides
However, the presence of high-density positive charges impedes the release of nucleic acids from the complex, which in turn affects the transfection activity of protamine.61 Therefore, to further improve the carrier's transfection efficiency, researchers have focused on the development of synthetic cationic peptide carriers with shorter sequences, which are easier to synthesize with better stability and delivery efficiency. In the early stages, short-chain peptide carriers are effective in the delivery of oligonucleotide drugs such as siRNA.62 Consequently, pioneering researchers attempted to directly utilize the commonly used R8, TAT, and other cationic cell-penetrating peptides for the delivery of mRNA. Kim's group evaluated and compared the delivery effects of short-chain cationic peptides such as R8, TAT, and low molecular weight protamine (LMWP) on mRNA, including encapsulation, cellular uptake, and protein expression.63 Unfortunately, the cell uptake rates of the above peptides in CT26.CL25 cells were poor, with uptake efficiencies of 11.6%, 6.57%, and 24.1%, respectively (Fig. 2a). Furthermore, based on these delivery systems, no reporter genes were detected at the protein level (Fig. 2b). This might be due to the neutralization of the positive charge on the shorter cationic peptide, which in turn inhibits the complex's ability for internalization as well as affecting the ultimate mRNA delivery efficiency.64 Therefore, Mixson's team designed cationic peptides with branching structures to increase the spatial distribution of peptide carriers. It was found that H3K(+H)4b is 10 times more efficient than H3K4b in transfecting luciferase mRNA into MDA-MB-231 cells.42 As shown in Fig. 1b, the difference between the two is that H3K4b has four repeat motifs –HHHK– in each branch, while H3K(+H)4b has a similar repeat pattern but with an extra histidine in the second -HHHK-motif of its branches. The reason for the transfection difference is that H3K(+H)4b forms a more stable complex with mRNA compared to H3K4b. This promotes cellular uptake and endosomal accumulation of complexes. This finding indicates that spatial complexity can achieve efficient mRNA delivery of cationic peptides. However, molecules with complex chemical structures that have such multiple repeats usually face greater difficulty and higher costs in synthesis. A simpler and more efficient delivery system still needs to be developed.
 | ||
| Fig. 2 Representative amphipathic peptides for mRNA delivery. (a) Representative histograms and a quantified graph of FITC-positive CL26.CL25 cells were obtained by flow cytometry, following transfection with CPP/Fluorescein-labelled mCherry mRNA complexes. Reproduced from ref. 63 with permission from MDPI, copyright 2022. (b) Representative histograms and graphs of EGFP-positive in CT26. CL25 by flow cytometry, following transfection with CPP/GFP mRNA complexes. Reproduced from ref. 63 with permission from MDPI, copyright 2022. (c) The sequence and secondary structure of RALA. Arginine residues are shown in blue, and leucine and alanine residues are shown in red. Reproduced from ref. 44 with permission from John Wiley and Sons, copyright 2017. (d) HELP-4H/mRNA transfection into HCT116 cells. For 1 μg mRNA give the indicated amount of HELP-4H peptide. Reproduced from ref. 47 with permission from John Wiley and Sons, copyright 2021. | ||
RALA (WEARLARALARALARHLARALARALRACEA) is a 30-amino acid amphipathic cationic α-helical peptide derived from the pH-sensitive peptide GALA and KALA.69 The α-helix structure enables the sequence's seven positively charged arginine residues to be gathered on one side, interacting with nucleic acids. The other side of the helix shows an arrangement of hydrophobic leucine residues (Fig. 2c). This structural feature endows RALA with robust and efficient cell-penetrating ability and membrane fusion properties. Moreover, RALA displays a higher capacity for selective membrane disruption under acidic conditions, which significantly facilitates the lysosomal escape of mRNA. One study has shown that mRNA encoding the antigen ovalbumin delivered by RALA could induce a significant ovalbumin-specific T-cell response in vivo.44 The efficacy of RALA was superior to that of a standard liposomal delivery system prepared with the cationic lipid DOTAP and the fusion lipid DOPE. Also, RALA could safely and effectively deliver CRISPR cargo to primary stem cells for gene editing, providing a delivery method for the application of regenerative medicine and stem cell treatment of genetic diseases.70
Histidine-rich peptides are often considered to accelerate the endosomal escape process through the proton sponge phenomenon or “flip–flop” effect. To further improve the lysosomal escape capability of the RALA carrier, Liu et al. constructed different vehicles by modulating the ratio of histidine to arginine.71 This strategy was verified in plasmid DNA transfection. Compared with the original peptide, HALA2 (WEARLARALARALARHLARALAHALHACEA), obtained by replacing the two arginines near the C-terminus with histidine, demonstrated enhanced endosome escape ability and transfection efficiency, but not HALA3/4, which contained 4 histidines. Meanwhile, the comparison between RALA and LAH4-L1, another peptide containing four histidines (KKALLAHALHLLALLALHLAHALKKA), revealed superior dendritic cell transfection capabilities of LAH4-L1.45 Moreover, replacing the four lysine residues at the ends of LAH4-L1 with arginine residues (named LAH4-L1R) (RRALLAHALHLLALLALHLAHALRRA) also achieved a similar effect. It was confirmed by D'haese et al. that both LAH4-L1 and LAH4-L1R can deliver antigen-encoding mRNA and activate antigen-specific T cells.46 In addition, both carriers demonstrated self-adjuvant activity for vaccines by inducing the maturation of CD103+ DCs and activating inflammasomes.
HELP-4H is another amphipathic α-helix peptide, with a similar design concept to the RALA delivery carrier. By replacing glutamic acid residues with histidine residues, the HELP peptide was transformed to HELP-4H (GLGTLLTLLHFLLHHLLHFLKRKRQQ). The ability of HELP-4H to effectively increase luciferase expression levels in HCT116 tumor cells was confirmed by Ali et al. (Fig. 2d).47
Meanwhile, different from the above-mentioned amphipathic delivery vehicles composed of complete amino acid sequences, PepFect endows peptide carriers with amphipathic properties by incorporating hydrophobic groups at the terminals of the peptides. For example, PepFect14 (PF14), obtained by replacing the lysine in stearylated TP10 with ornithine, showed stronger resistance to serum proteases. The efficacy of PF14 in delivering diverse nucleic acid therapeutics, including plasmid DNA, siRNA, microRNA (miRNA), and ASO, has been substantiated in multiple studies.72–75 In a mouse model of ovarian cancer, PF14 demonstrated superior mRNA transfection and high protein levels in tumor-associated tissues relative to the commercial Lipofectamine MessengerMAX reagent.48 Furthermore, PF14-mRNA nanoparticles did not cause immune side effects at the injection site in the irritant contact dermatitis (ICD) mouse model.76 Given its favorable biocompatibility, PF14-based mRNA delivery vectors have also been employed in regenerative medicine by introducing them onto porous collagen scaffolds.77
2.2. Hybrid delivery systems involving peptides
Peptides exhibit excellent cell membrane penetration, endosome escape, nuclear targeting, and high target affinity. Thus, peptides can not only be utilized as stand-alone carriers for drug delivery but can also be combined with other carriers to further enhance delivery performance.83,84 For instance, studies have found the incorporation of CPPs into liposomes can significantly improve the mRNA transfection capacity. Specifically, the α-helix cationic peptide KALA was modified on liposomes to construct a novel type of anti-tumor vaccine by Tateshita et al.51 On the one hand, the introduction of peptides can aid in the cellular uptake efficiency of liposomes. On the other hand, peptides can be used as vaccine adjuvants to enhance the efficacy of mRNA tumor immunotherapy. Similarly, a novel cationic peptide DP7 (VQWRIRVAVIRK) with antimicrobial activity and vaccine adjuvant properties was presented by Zhang et al.85 Unfortunately, the introduction of cholesterol modification alone on DP7 did not improve its delivery of mRNA. In 2020, cholesterol-modified DP7 was bound to a liposomal delivery system, successfully achieving personalized mRNA delivery to dendritic cells (Fig. 5a).52 The study also confirmed that the introduction of the peptide played an important role in promoting dendritic cell maturation, improving antigen presentation, and increasing the level of pro-inflammatory factor secretion at the cellular level. It also induced an enhanced antigen-specific lymphocyte response and growth inhibition of LL2 tumors in the animal model.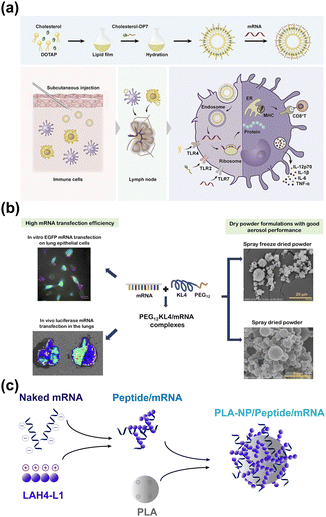 | ||
| Fig. 5 (a) Cholesterol-modified DP7 was incorporated into a liposomal delivery system, enabling mRNA efficient delivery to dendritic cells and stimulating DC maturation. Reproduced from ref. 52 with permission from Elsevier, copyright 2020. (b) The PEG12KL4 peptide enables efficient mRNA expression in the lungs. Reproduced from ref. 53 with permission from Elsevier, copyright 2019. (c) Schematic diagram of the LAH4-L1/mRNA/PLA complex preparation process. | ||
In addition, the combination of peptides with polymers such as polyethylene glycol and polylactic acid has also demonstrated good clinical application potential for the passive targeting delivery of mRNA. This is especially true for local organ delivery, such as to the lungs. Inhalable mRNA drugs have great potential for treating lung diseases and developing vaccines related to respiratory diseases. However, naked mRNA or complexes based on simple lipid carriers have difficulty penetrating the mucus and surfactant barrier of the lungs to reach epithelial cells deep in the lungs, resulting in poor transfection.86,87 In 2017, Qiu et al. demonstrated that the hydrophobic peptide KL4, which is a surfactant protein B (SP-B) mimic with a 21-residue cationic peptide containing repeating KLLLL sequences, could be employed as a siRNA drug carrier for pulmonary delivery.88 Subsequently, the research group modified KL4 with PEG12 to enhance the solubility of the peptide carrier and successfully achieved local expression of luciferase mRNA in the lungs (Fig. 5b).53 Additionally, PEG12 was also introduced by Xu et al. onto the amphipathic LAH4-L1 which had markedly improved the mRNA delivery effect of LAH4-L1 in the lung.54 Also, some researchers have focused on cell lines that are generally challenging for transfection, such as DC cells and stem cells. Poly(lactic acid) (PLA) is a biodegradable, negatively charged polymer that has been approved by the FDA for its good biocompatibility. PLA nanoparticles (PLA-NP) can be effectively taken up by dendritic cells in vivo and in vitro.89,90 However, it is difficult for mRNA to be adsorbed onto the PLA-NP surface due to the same negative charge. To solve this problem, PLA-NP mRNA vaccines were prepared by Coolen et al. via mixing LAH4-L1 with PLA.45 During the preparation process, the mRNA was initially mixed with the amphipathic cationic CPP LAH4-L1 to form a positively charged intermediate complex. Then the intermediate was adsorbed onto the PLA-NPs by electrostatic interaction (Fig. 5c). The PLA-NP/LAH4-L1/mRNA nanocomplexes were unable to transfect into the epithelial cell lines 293T and HeLa but were efficiently taken up by DC2.4 and showed an enhanced transfection effect compared with the LAH4-L1/mRNA complex. The three-component complex was highly taken up by DC cells, which triggered an innate immune response by activating cytoplasmic pattern recognition receptors (PRRs). Besides, it induced an adaptive immune response with a TH1 bias through antigen activation induced by mRNA expression.
Achieving targeted delivery to specific tissues and cells is critical for the clinical translation of mRNA gene therapy. The above-mentioned passive targeting relies on the intrinsic properties of the nanomaterials and the administration routes.91 While active targeting promotes the specific binding of nanoparticles to corresponding cells through modification of the targeting ligands, thereby improving the efficiency and accuracy of delivery.92 Peptides that target cell surface receptors are widely employed to modify lipid- and polymer-based delivery systems, enabling the active targeting delivery of small-molecule drugs and nucleic acids to specific cells and tissues.93,94 The molecular weight of peptides is intermediate between that of small molecule drugs and that of antibodies. Hence, peptides possess both the efficient cell membrane penetration ability of small-molecule drugs and the high target affinity of antibodies. In addition, compared to antibodies, peptides offer abundant chemical modification sites and enhanced stability, facilitating their functionalization across various types of delivery systems. In recent years, researchers have combined targeting peptides with other biomaterials to construct nanocomplexes for mRNA delivery. These efforts have advanced mRNA's applications in gene editing, cancer immunotherapy, and neurological disease treatment. For example, it has been shown that RGD peptide-based lipids can be incorporated into LNPs and used to co-deliver Cas9 mRNA and sgRNA, achieving up to 90% GFP knockdown in HepG2 cells with improved efficiency compared with unmodified LNPs.95 Also, nanomaterials modified with the anti-PD-L1 peptide DPPA, in combination with ultrasound-targeted microbubble disruption, can be used to deliver IL-15 mRNA to tumors, thereby enhancing tumor immunotherapy.96 In addition to tumor targeting, peptides have shown potential in the treatment of other diseases. For instance, studies have shown that PR-targeting peptides can be used to deliver mRNA LNP to retinal neurons, providing a new strategy for the treatment of ocular diseases.97 Moreover, the RVG29-targeted peptide can deliver LNP to brain endothelial and neuronal cells, allowing the mRNA to precisely reach the brain after systemic administration, which is promising for the treatment of neurological diseases.98
Peptides contribute significantly to improving the stability, cellular penetration, delivery efficiency, and targeting capability of lipid- or polymer-based mRNA carriers. However, these composite strategies still face challenges related to manufacturing complexity, altered pharmacokinetics, and in vivo stability.92,99 Nevertheless, these challenges are expected to be positively resolved through further optimization of the peptide and nanoparticle formulations.
3 Internalization mechanism of peptide/mRNA complexes
The mRNA delivered by the carrier needs to go through multiple steps, from adhesion to the cell membrane, cellular uptake via direct membrane penetration or endocytosis, escape from endosomes or lysosomes (if endocytosis occurs), to finally release into the cytoplasm followed by mRNA translation. Each of these steps represents a potential barrier to mRNA delivery. Among these steps, cellular uptake is the first barrier for mRNA to exert its function. A comprehensive understanding of the cellular uptake mechanism of mRNA–peptide nanocomplexes is crucial for the rational design of peptide carriers, improving carrier delivery efficiency, and reducing their toxic side effects. Studies showed that peptides enter the cells mainly through energy-independent direct penetration and endocytosis.100Early studies suggested that most cell-penetrating peptides enter cells through direct penetration.101 The interaction between the positive charge of CPP and the negatively charged cell membrane components leads to membrane instability and peptide folding on the lipid membrane, therefore allowing CPP to directly penetrate the cell. At present, several hypotheses have been put forth regarding the mechanism of direct permeation, including inverted micelle formation, pore formation, the carpet model, and the membrane thinning model (Fig. 6).102 Coarse-grained models were employed by Kawamoto et al. to investigate the impact of the strong attractive forces between CPPs and lipid head groups on the membrane curvature of a lipid bilayer.103 This membrane curvature or indentation could lead to the formation of inverted micelles. It was revealed that the formation of inverted micelles could be a possible mechanism that promotes cell penetration of peptides. The model of transient pore formation is often considered to be the mechanism by which amphipathic peptides are used. For example, the internalization of Pep-1 is carried out by transient pore formation and is dependent on the formation of α-helices.104 In addition, the carpet model is described as the positively charged segments of the peptide, which are oriented parallel to the membrane surface, bind to the acidic phospholipid head groups, and subsequently form a “carpet”-like covering.105 When the concentration of the CPP exceeds a threshold, rotation of the peptide leads to the interactions between the hydrophobic residues of the peptide and the hydrophobic core of the membrane, followed by translocation into the cell. The carpet model is often used to describe the mode of action of antimicrobial peptides that retain a positive charge and a hydrophobic structure, such as dermaseptin natural analogs, cecropins, and the human antimicrobial peptide LL-37.106 An alternative model to the “carpet” model is the “membrane thinning” effect, which was originally proposed for the amphipathic peptide Magainin 2.107 Specifically, disturbances arise due to the interaction of the negatively charged lipids in the outer membrane with the cationic groups of the CPP. This results in a lateral rearrangement of the lipids and a thinning of the membrane. The accumulation of the CPP on the membrane surface leads to a local reduction in surface tension, which promotes the cellular penetration of the peptides.
 | ||
| Fig. 6 Illustration of CPP direct translocation pathways. (a) Inverted micelle formation; (b) pore formation; (c) carpet model; and (d) membrane thinning model. | ||
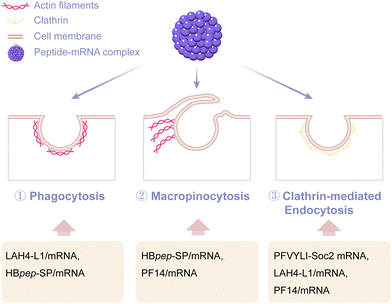
In addition to direct penetration-mediated internalization, more studies have discovered that peptides primarily facilitate the cellular entry of mRNA via endocytosis (Fig. 7). Pathways of endocytosis include common phagocytosis, macropinocytosis, endocytosis dependent on coat proteins such as clathrin or caveolin, and endocytosis independent of clathrin and/or caveolin. For instance, the peptide sequence PFVYLI can form a covalent complex with Soc2 mRNA and assist Soc2 mRNA entry into fibroblasts by clathrin-dependent endocytosis.49 Moreover, the amphipathic peptide delivery system LAH4-L1 was observed to transport mRNA into dendritic cells via phagocytosis and clathrin-dependent endocytosis, therefore inducing a powerful and efficient innate immune response.45 Shebanova et al.108 used scanning electron microscopy and transmission electron microscopy to study HBpep-SP peptide coacervates. Their findings concluded that HeLa and HepG2 cells took up these coacervates through phagocytosis and macropinocytosis. Furthermore, some studies have pointed out that the mechanism of cellular uptake can be closely related to the cell type.76 For example, PF14-mRNA nanoparticles were endocytosed via macropinocytosis into HeLa cells. When HaCaT cells were used as a model, PF14-mRNA nanoparticles were found to internalize via both macropinocytosis and clathrin-mediated endocytosis pathways. Nevertheless, the uptake of PF14-mRNA nanocomplexes by primary keratinocytes was not affected after treatment with different endosomal pathway inhibitors. This indicated that PF14 might internalize into the primary keratinocytes independently of endocytosis. These studies have illustrated that the internalization of mRNA mediated by peptide delivery systems usually cannot be explained by a single pathway, but rather by a simultaneous behavior of multiple pathways. Moreover, other key factors including the sequence composition, chain length of cationic amino acids, hydrophobic properties of the peptide, and the assembly structure of the peptide and mRNA, may also affect the internalization of peptide-based mRNA delivery systems.109–113 However, due to the current limited evaluation methods of cell internalization, there is currently no unified understanding of the mechanism of peptide delivery system internalization. It can be foreseen that with the maturity and application of single-cell visualization analysis technology, AI-assisted simulation, and other technologies, we will have a deeper understanding of the mechanism of peptide internalization in the future.
4 Conclusions and outlook
In summary, this review provides a detailed overview of recent research and application prospects of peptide carriers in mRNA drug delivery. Facing stronger negative charges and larger molecular weight, cationic/amphipathic peptide carriers can effectively compress large mRNA molecules to the nanoscale and protect mRNA drugs from ribonuclease degradation. Furthermore, the high cell membrane permeability and lysosome escape ability of peptide carriers can be utilized to effectively achieve the cytoplasmic delivery of mRNAs and affect the expression of target proteins. In addition to being used alone as a delivery carrier, peptides can also be incorporated with other materials to noticeably improve the cell membrane permeability of traditional delivery carriers while achieving targeted delivery of mRNA to tissues. Moreover, peptide carriers are capable of mRNA drug delivery through non-invasive administration via dry powder inhalation, which has demonstrated a strong ability to penetrate mucus and surfactant barriers. This highlights the potential of peptides in inspiring new research directions for the development of therapeutic mRNA drugs and vaccines for lung-related diseases. Moreover, shorter sequences, well-defined structures, and their commercial availability render peptides highly promising candidates for clinical translation.However, apart from protamine, there have been no other cases of clinical application of peptide-based mRNA carriers. First of all, faced with complex protease systems, peptides still have shortcomings, such as poor stability, easy degradation, and short half-life.114 Optimizing the structure of peptide molecules is expected to circumvent these difficulties. The optimization methods, including replacing the original L-type amino acids with D-type, peptide backbone modifications, and adding protective groups to the N- and C-termini of the molecule, could potentially aid in overcoming these challenges.115 Moreover, various strategies for peptide stapling have been developed to improve the tolerance of peptides to proteases, including ring-closing metathesis between olefinic side chains, lactamization, thiol–ene and thiol–yne, thiol/nitrogen-arylation/alkylation, click-based aide-alkyne cycloaddition, and C–H activation.116,117 In addition to this, the introduction of peptide stapling modification strategies enhances the ability of peptide carriers to penetrate cell membranes. For example, stabilized peptides with methionine residues modified with reducible cross-linkers have been applied to improve the druggability of peptide drugs.118 The peptide stapling strategies are expected to become a promising direction in improving the efficacy of peptide-based mRNA delivery carriers.
Secondly, most of the current peptide-mRNA nanoparticles internalize into cells via endocytosis, which inevitably faces the problem of poor endosome escape. Recently, by incorporating strategies such as introducing histidines with a proton sponge response and forming pH-sensitive structures, mRNA therapeutics that efficiently achieve endosomal disruption have been successfully developed in vitro. Additionally, new transmembrane pathways, such as direct fusion with the cell membrane, are expected to become the design direction for peptide-based mRNA delivery to circumvent the endosome/lysosome metabolic pathway.119
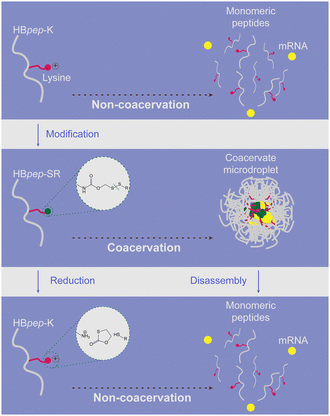
Thirdly, peptide carriers face several potential safety issues.120 Leveraging electrostatic interaction to promote the binding and the subsequent membrane penetration of peptides and nucleic acids will unavoidably lead to cation-induced cytotoxicity. Encapsulation of mRNA and intracellular delivery of the complex based on the principle of phase separation is another promising peptide carrier development strategy to overcome this problem.50 Finally, how to improve the current problem of ultra-low temperature storage and cold chain transportation of mRNA with the help of delivery carriers will also be a key focus for the development of new delivery vehicles. It is believed that the introduction of various modification methods and future AI-assisted rational design will provide new directions for the research and development of peptide-based mRNA delivery vehicles.
Author contributions
Conceptualization: Z. L., F. Y.; investigation: H. L., Y. X., K. W.; writing – original draft preparation: H. L., Y. Z.; writing – review and editing: Z. L., F. Y.; funding acquisition: Z. L., F. Y., Y. Z. All authors have read and agreed to the published version of the manuscript.Data availability
No primary research results, software, or code have been included and no new data were generated or analyzed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (No. 2021YFA0910803), the National Center for Biological Medicine Technology Innovation (NCTIB2022HS01017), the Natural Science Foundation of China grants (22307086), the Natural Science Foundation of Guangdong Province (2022A1515010996), the Shenzhen Science and Technology Program (RCJC20200714114433053, JCYJ20220818095808019), the High-tech Zone Development Special Project of Shenzhen (29853M-KCJ-2023-002-07, GZL202309140842000049), the Shenzhen High-tech Zone Development Special Plan Pingshan District Innovation Platform Construction Project (29853MKCJ202300208), the Shenzhen Medical Research Fund (D2401004, A2403012), the Tian Fu Jin Cheng Laboratory (Advanced Medical Center) Group Racing Project (TFJC2023010008), and the Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions (2019SHIBS0004).References
- Y. Miao, C. Fu, Z. Yu, L. Yu, Y. Tang and M. Wei, Acta Pharm. Sin. B, 2024, 14, 3802–3817 CrossRef CAS PubMed.
- B. Wilson and K. M. Geetha, J. Drug Delivery Sci. Technol., 2022, 74, 103553 CrossRef CAS PubMed.
- Q. Duan, T. Hu, Q. Zhu, X. Jin, F. Chi and X. Chen, Front. Immunol., 2022, 13, 974433 CrossRef CAS PubMed.
- Y. Shi, M. Shi, Y. Wang and J. You, Signal Transduction Targeted Ther., 2024, 9, 1–43 CrossRef.
- C. Wang and F. Yuan, Adv. Drug Delivery Rev., 2024, 210, 115340 CrossRef CAS.
- J. S. Turner, J. A. O’Halloran, E. Kalaidina, W. Kim, A. J. Schmitz, J. Q. Zhou, T. Lei, M. Thapa, R. E. Chen, J. B. Case, F. Amanat, A. M. Rauseo, A. Haile, X. Xie, M. K. Klebert, T. Suessen, W. D. Middleton, P.-Y. Shi, F. Krammer, S. A. Teefey, M. S. Diamond, R. M. Presti and A. H. Ellebedy, Nature, 2021, 596, 109–113 CrossRef CAS.
- F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Pérez Marc, E. D. Moreira, C. Zerbini, R. Bailey, K. A. Swanson, S. Roychoudhury, K. Koury, P. Li, W. V. Kalina, D. Cooper, R. W. Frenck, L. L. Hammitt, Ö. Türeci, H. Nell, A. Schaefer, S. Ünal, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Şahin, K. U. Jansen and W. C. Gruber, N. Engl. J. Med., 2020, 383, 2603–2615 CrossRef CAS PubMed.
- Y. Gu, J. Duan, N. Yang, Y. Yang and X. Zhao, MedComm, 2022, 3, e167 CrossRef CAS.
- C. Krienke, L. Kolb, E. Diken, M. Streuber, S. Kirchhoff, T. Bukur, Ö. Akilli-Öztürk, L. M. Kranz, H. Berger, J. Petschenka, M. Diken, S. Kreiter, N. Yogev, A. Waisman, K. Karikó, Ö. Türeci and U. Sahin, Science, 2021, 371, 145–153 CrossRef CAS PubMed.
- G. Shen, J. Liu, H. Yang, N. Xie and Y. Yang, J. Controlled Release, 2024, 369, 696–721 CrossRef CAS PubMed.
- J. Chen, X. Guan, Y. Hu, H. Tian and X. Chen, Top Curr. Chem., 2017, 375, 32 CrossRef PubMed.
- E. Sasso, A. M. D’Alise, N. Zambrano, E. Scarselli, A. Folgori and A. Nicosia, Semin. Immunol., 2020, 50, 101430 CrossRef CAS PubMed.
- Z. Li, Z. Liu, J. Wu and B. Li, Pharmaceutics, 2022, 14, 2699 CrossRef CAS PubMed.
- M. Mobasher, R. Ansari, A. M. Castejon, J. Barar and Y. Omidi, Biochim. Biophys. Acta, Gen. Subj., 2024, 1868, 130558 CrossRef CAS.
- P. Boisguérin, K. Konate, E. Josse, E. Vivès and S. Deshayes, Biomedicines, 2021, 9, 583 CrossRef PubMed.
- R. D. Kirian, D. Steinman, C. M. Jewell and H. C. Zierden, Theranostics, 2024, 14, 2265 CrossRef CAS PubMed.
- M. Zhao, R. Wang, K. Yang, Y. Jiang, Y. Peng, Y. Li, Z. Zhang, J. Ding and S. Shi, Acta Pharm. Sin. B, 2023, 13, 916–941 CrossRef CAS PubMed.
- W. Zhang, Y. Liu, J. Min Chin and K. K. L. Phua, Eur. J. Pharm. Biopharm., 2021, 163, 179–187 CrossRef CAS PubMed.
- M. Li, M. Zhao, Y. Fu, Y. Li, T. Gong, Z. Zhang and X. Sun, J. Controlled Release, 2016, 228, 9–19 CrossRef CAS.
- M. Li, Y. Li, K. Peng, Y. Wang, T. Gong, Z. Zhang, Q. He and X. Sun, Acta Biomater., 2017, 64, 237–248 CrossRef CAS.
- X. Ke, L. Shelton, Y. Hu, Y. Zhu, E. Chow, H. Tang, J. L. Santos and H.-Q. Mao, ACS Appl. Mater. Interfaces, 2020, 12, 35835–35844 CrossRef CAS PubMed.
- Z. Iqbal, K. Rehman, A. Mahmood, M. Shabbir, Y. Liang, L. Duan and H. Zeng, J. Nanobiotechnol., 2024, 22, 395 CrossRef CAS PubMed.
- H. Kim, Y. Park and J. B. Lee, Sci. Rep., 2015, 5, 12737 CrossRef CAS PubMed.
- X. Fu, T. Chen, Y. Song, C. Feng, H. Chen, Q. Zhang, G. Chen and X. Zhu, Small, 2021, 17, e2101224 CrossRef PubMed.
- N. Yoshinaga, E. Cho, K. Koji, Y. Mochida, M. Naito, K. Osada, K. Kataoka, H. Cabral and S. Uchida, Angew. Chem., Int. Ed., 2019, 58, 11360–11363 CrossRef CAS PubMed.
- I. Urits, D. Swanson, M. C. Swett, A. Patel, K. Berardino, A. Amgalan, A. A. Berger, H. Kassem, A. D. Kaye and O. Viswanath, Neurol. Ther., 2020, 9, 301–315 CrossRef PubMed.
- R. Tenchov, R. Bird, A. E. Curtze and Q. Zhou, ACS Nano, 2021, 15, 16982–17015 CrossRef CAS PubMed.
- S.-H. Bae, S. Yoo, J. Lee, H.-J. Park, S. P. Kwon, H. Jin, S.-I. Park, Y.-S. Lee, Y.-J. Bang, G. Roh, S. Lee, S. B. Youn, I. W. Kim, H. R. Oh, A. K. El-Damasy, G. Keum, H. Kim, H. Youn, J.-H. Nam and E.-K. Bang, Bioact. Mater., 2024, 38, 486–498 CAS.
- X. Wang, S. Liu, Y. Sun, X. Yu, S. M. Lee, Q. Cheng, T. Wei, J. Gong, J. Robinson, D. Zhang, X. Lian, P. Basak and D. J. Siegwart, Nat. Protoc., 2023, 18, 265–291 CrossRef CAS PubMed.
- L. Zheng, S. R. Bandara, Z. Tan and C. Leal, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2301067120 CrossRef CAS PubMed.
- E. Oude Blenke, E. Örnskov, C. Schöneich, G. A. Nilsson, D. B. Volkin, E. Mastrobattista, Ö. Almarsson and D. J. A. Crommelin, J. Pharm. Sci., 2023, 112, 386–403 CrossRef CAS PubMed.
- A.-G. Reinhart, A. Osterwald, P. Ringler, Y. Leiser, M. E. Lauer, R. E. Martin, C. Ullmer, F. Schumacher, C. Korn and M. Keller, Mol. Pharmaceutics, 2023, 20, 6492–6503 CrossRef CAS PubMed.
- S. Al-Halifa, M. Babych, X. Zottig, D. Archambault and S. Bourgault, Pept. Sci., 2019, 111, e24095 CrossRef.
- C. J. C. Edwards-Gayle and I. W. Hamley, Org. Biomol. Chem., 2017, 15, 5867–5876 RSC.
- L. B. De Martini, C. Sulmona, L. Brambilla and D. Rossi, Cells, 2023, 12, 1643 CrossRef CAS PubMed.
- S. H. Nam, J. Park and H. Koo, Arch. Pharm. Res., 2023, 46, 18–34 CrossRef CAS PubMed.
- N. Naowarojna, R. Cheng, J. Lopez, C. Wong, L. Qiao and P. Liu, Synth. Syst. Biotechnol., 2021, 6, 32–49 CrossRef CAS PubMed.
- W. Liang, P. C. L. Kwok, M. Y. T. Chow, P. Tang, A. J. Mason, H.-K. Chan and J. K. W. Lam, Eur. J. Pharm. Biopharm., 2014, 86, 64–73 CrossRef CAS PubMed.
- M. Y. T. Chow and J. K. W. Lam, Curr. Pharm. Des., 2015, 21, 3854–3866 CrossRef CAS PubMed.
- StatPearls, https://www.ncbi.nlm.nih.gov/books/NBK562260/ (accessed November 2024) Search PubMed.
- M. Alberer, U. Gnad-Vogt, H. S. Hong, K. T. Mehr, L. Backert, G. Finak, R. Gottardo, M. A. Bica, A. Garofano, S. D. Koch, M. Fotin-Mleczek, I. Hoerr, R. Clemens and F. von Sonnenburg, The Lancet, 2017, 390, 1511–1520 CrossRef CAS PubMed.
- J. He, S. Xu, Q. Leng and A. J. Mixson, J. Gene Med., 2021, 23, e3295 CrossRef CAS PubMed.
- J. H. Lockhart, J. VanWye, R. Banerjee, S. A. Wickline, H. Pan and H. Totary-Jain, Mol. Ther., 2021, 29, 1744–1757 CrossRef CAS PubMed.
- V. K. Udhayakumar, A. De Beuckelaer, J. McCaffrey, C. M. McCrudden, J. L. Kirschman, D. Vanover, L. Van Hoecke, K. Roose, K. Deswarte, B. G. De Geest, S. Lienenklaus, P. J. Santangelo, J. Grooten, H. O. McCarthy and S. De Koker, Adv. Healthcare Mater., 2017, 6, 1601412 CrossRef PubMed.
- A.-L. Coolen, C. Lacroix, P. Mercier-Gouy, E. Delaune, C. Monge, J.-Y. Exposito and B. Verrier, Biomaterials, 2019, 195, 23–37 CrossRef CAS PubMed.
- S. D’haese, S. Laeremans, S. den Roover, S. D. Allard, G. Vanham and J. L. Aerts, Pharmaceutics, 2022, 14, 1387 CrossRef PubMed.
- S. Ali, C. Dussouillez, B. Padilla, B. Frisch, A. J. Mason and A. Kichler, J. Gene Med., 2022, 24, e3401 CrossRef CAS PubMed.
- D. van den Brand, M. A. J. Gorris, A. H. van Asbeck, E. Palmen, I. Ebisch, H. Dolstra, M. Hällbrink, L. F. A. G. Massuger and R. Brock, Eur. J. Pharm. Biopharm., 2019, 141, 180–190 CrossRef CAS PubMed.
- A. N. Miliotou, I. S. Pappas, G. Spyroulias, E. Vlachaki, A. S. Tsiftsoglou, I. S. Vizirianakis and L. C. Papadopoulou, Mol. Ther. – Nucleic Acids, 2021, 26, 694–710 CrossRef CAS PubMed.
- Y. Sun, S. Y. Lau, Z. W. Lim, S. C. Chang, F. Ghadessy, A. Partridge and A. Miserez, Nat. Chem., 2022, 14, 274–283 CrossRef CAS PubMed.
- N. Tateshita, N. Miura, H. Tanaka, T. Masuda, S. Ohtsuki, K. Tange, Y. Nakai, H. Yoshioka and H. Akita, J. Controlled Release, 2019, 310, 36–46 CrossRef CAS PubMed.
- R. Zhang, L. Tang, Y. Tian, X. Ji, Q. Hu, B. Zhou, Z. Ding, H. Xu and L. Yang, J. Controlled Release, 2020, 328, 210–221 CrossRef CAS PubMed.
- Y. Qiu, R. C. H. Man, Q. Liao, K. L. K. Kung, M. Y. T. Chow and J. K. W. Lam, J. Controlled Release, 2019, 314, 102–115 Search PubMed.
- Y. Xu, Y. Zheng, X. Ding, C. Wang, B. Hua, S. Hong, X. Huang, J. Lin, P. Zhang and W. Chen, Drug Delivery, 2023, 30, 2219870 Search PubMed.
- Y. Gu, J. Chen, Z. Wang, C. Liu, T. Wang, C.-J. Kim, H. Durikova, S. Fernandes, D. N. Johnson, R. De Rose, C. Cortez-Jugo and F. Caruso, Nat. Commun., 2024, 15, 9664 CrossRef CAS PubMed.
- Y.-S. Wang, M. Kumari, G.-H. Chen, M.-H. Hong, J. P.-Y. Yuan, J.-L. Tsai and H.-C. Wu, J. Biomed. Sci., 2023, 30, 84 CrossRef CAS PubMed.
- F. Veider, E. Sanchez Armengol and A. Bernkop-Schnürch, Small, 2024, 20, 2304713 CrossRef CAS PubMed.
- H. Amos, Biochem. Biophys. Res. Commun., 1961, 5, 1–4 CrossRef CAS.
- N. T. Jarzebska, M. Mellett, J. Frei, T. M. Kündig and S. Pascolo, Pharmaceutics, 2021, 13, 877 CrossRef CAS PubMed.
- F. Friedberg, T. Winnick and D. M. Greenberg, J. Biol. Chem., 1947, 169, 763 CrossRef CAS PubMed.
- J. Liu, S. Guo, Z. Li, L. Liu and J. Gu, Colloids Surf., B, 2009, 73, 36–41 CrossRef CAS PubMed.
- L. Falato, M. Gestin and Ü. Langel, Design and Delivery of SiRNA Therapeutic, Humana, New York, 2021 Search PubMed.
- Y. Kim, H. Kim, E. H. Kim, H. Jang, Y. Jang, S.-G. Chi, Y. Yang and S. H. Kim, Pharmaceutics, 2022, 14, 1271 CrossRef CAS PubMed.
- R. H. Mo, J. L. Zaro and W.-C. Shen, Mol. Pharmaceutics, 2012, 9, 299–309 CrossRef CAS PubMed.
- L. Luan, Q. Meng, L. Xu, Z. Meng, H. Yan and K. Liu, J. Mater. Chem. B, 2015, 3, 1068–1078 RSC.
- H. Yan, Y. Hu, A. Akk, M. F. Rai, H. Pan, S. A. Wickline and C. T. N. Pham, Pharmaceutics, 2020, 12, 73 CrossRef CAS PubMed.
- H. Yan, Y. Hu, Y. Lyu, A. Akk, A. C. Hirbe, S. A. Wickline, H. Pan, E. D. O. Roberson and C. T. N. Pham, bioRxiv, 2024, preprint DOI:10.1101/2024.06.17.599454.
- A. U. Kabir, C. Zeng, M. Subramanian, J. Wu, M. Kim, K. Krchma, X. Wang, C. M. Halabi, H. Pan, S. A. Wickline, D. H. Fremont, M. N. Artyomov and K. Choi, Nat. Immunol., 2024, 25, 1546–1554 CrossRef CAS PubMed.
- H. O. McCarthy, J. McCaffrey, C. M. McCrudden, A. Zholobenko, A. A. Ali, J. W. McBride, A. S. Massey, S. Pentlavalli, K.-H. Chen, G. Cole, S. P. Loughran, N. J. Dunne, R. F. Donnelly, V. L. Kett and T. Robson, J. Controlled Release, 2014, 189, 141–149 CrossRef CAS PubMed.
- J. P. Graham, J. G. Castro, L. C. Werba, L. C. Fardone, K. P. Francis, A. Ramamurthi, M. Layden, H. O. McCarthy and T. Gonzalez-Fernandez, bioRxiv, 2024, preprint DOI:10.1101/2024.09.23.614499.
- Y. Liu, H.-H. Wan, D.-M. Tian, X.-J. Xu, C.-L. Bi, X.-Y. Zhan, B.-H. Huang, Y.-S. Xu and L.-P. Yan, Materials, 2021, 14, 4674 CrossRef CAS PubMed.
- K.-L. Veiman, I. Mäger, K. Ezzat, H. Margus, T. Lehto, K. Langel, K. Kurrikoff, P. Arukuusk, J. Suhorutšenko, K. Padari, M. Pooga, T. Lehto and Ü. Langel, Mol. Pharmaceutics, 2013, 10, 199–210 CrossRef CAS PubMed.
- E.-H. Ervin, M. Pook, I. Teino, V. Kasuk, A. Trei, M. Pooga and T. Maimets, Stem Cell Res. Ther., 2019, 10, 43 CrossRef CAS PubMed.
- G. Carreras-Badosa, J. Maslovskaja, K. Periyasamy, E. Urgard, K. Padari, H. Vaher, L. Tserel, M. Gestin, K. Kisand, P. Arukuusk, C. Lou, Ü. Langel, J. Wengel, M. Pooga and A. Rebane, Biomaterials, 2020, 262, 120316 CrossRef CAS PubMed.
- C. Pasquale Cerrato, T. Kivijärvi, R. Tozzi, T. Lehto, M. Gestin and Ü. Langel, J. Mater. Chem. B, 2020, 8, 10825–10836 RSC.
- K. Periyasamy, M. Maloverjan, A. Biswas, A. Remm, M. Pook, A. Rebane and M. Pooga, Front. Pharmacol., 2023, 14, 1219761 CrossRef CAS PubMed.
- R. Oude Egberink, H. M. Zegelaar, N. El Boujnouni, E. M. M. Versteeg, W. F. Daamen and R. Brock, Pharmaceutics, 2022, 14, 1619 CrossRef CAS PubMed.
- S. S. Sommer and J. E. Cohen, J. Mol. Evol., 1980, 15, 37–57 CrossRef CAS PubMed.
- F. Späth, C. Donau, A. M. Bergmann, M. Kränzlein, C. V. Synatschke, B. Rieger and J. Boekhoven, J. Am. Chem. Soc., 2021, 143, 4782–4789 CrossRef PubMed.
- Z. W. Lim, Y. Ping and A. Miserez, Bioconjugate Chem., 2018, 29, 2176–2180 CrossRef CAS PubMed.
- Z. W. Lim, V. B. Varma, R. V. Ramanujan and A. Miserez, Acta Biomater., 2020, 110, 221–230 CrossRef CAS PubMed.
- Y. Sun, X. Xu, L. Chen, W. L. Chew, Y. Ping and A. Miserez, ACS Nano, 2023, 17, 16597–16606 CrossRef CAS PubMed.
- M. Liu, X. Fang, Y. Yang and C. Wang, Front. Bioeng. Biotechnol., 2021, 9, 701504 CrossRef PubMed.
- Y. Wang, L. Zhang, C. Liu, Y. Luo and D. Chen, Pharmaceutics, 2024, 16, 240 CrossRef CAS PubMed.
- R. Zhang, L. Tang, Y. Tian, X. Ji, Q. Hu, B. Zhou, D. Zhenyu, X. Heng and L. Yang, Biomaterials, 2020, 241, 119852 CrossRef CAS PubMed.
- Q. Wang, C. Bu, Q. Dai, J. Chen, R. Zhang, X. Zheng, H. Ren, X. Xin and X. Li, Adv. Sci., 2024, 11, e2309748 CrossRef PubMed.
- X. M. Bustamante-Marin and L. E. Ostrowski, Cold Spring Harbor Perspect. Biol., 2017, 9, a028241 CrossRef PubMed.
- Y. Qiu, M. Y. T. Chow, W. Liang, W. W. Y. Chung, J. C. W. Mak and J. K. W. Lam, Mol. Pharmaceutics, 2017, 14, 4606–4617 CrossRef CAS PubMed.
- C. Primard, N. Rochereau, E. Luciani, C. Genin, T. Delair, S. Paul and B. Verrier, Biomaterials, 2010, 31, 6060–6068 CrossRef CAS PubMed.
- J. Rességuier, E. Delaune, A.-L. Coolen, J.-P. Levraud, P. Boudinot, D. Le Guellec and B. Verrier, Front. Immunol., 2017, 8, 190 Search PubMed.
- R. A. Meyer, S. Y. Neshat, J. J. Green, J. L. Santos and A. D. Tuesca, Mater. Today Adv., 2022, 14, 100240 CrossRef CAS.
- J. Di, P. Huang and X. Chen, Bioconjugate Chem., 2024, 35, 453–456 Search PubMed.
- I. Truebenbach, W. Zhang, Y. Wang, S. Kern, M. Höhn, S. Reinhard, J. Gorges, U. Kazmaier and E. Wagner, Int. J. Pharm., 2019, 569, 118570 CrossRef CAS PubMed.
- B. M. Cooper, J. Iegre, D. H. O. Donovan, M. Ö. Halvarsson and D. R. Spring, Chem. Soc. Rev., 2021, 50, 1480–1494 RSC.
- J. Qin, L. Xue, N. Gong, H. Zhang, S. J. Shepherd, R. M. Haley, K. L. Swingle and M. J. Mitchell, RSC Adv., 2022, 12, 25397–25404 RSC.
- X. Wang, F. Li, J. Zhang, L. Guo, M. Shang, X. Sun, S. Xiao, D. Shi, D. Meng, Y. Zhao, C. Jiang and J. Li, J. Controlled Release, 2024, 367, 45–60 CrossRef CAS PubMed.
- M. Herrera-Barrera, R. C. Ryals, M. Gautam, A. Jozic, M. Landry, T. Korzun, M. Gupta, C. Acosta, J. Stoddard, R. Reynaga, W. Tschetter, N. Jacomino, O. Taratula, C. Sun, A. K. Lauer, M. Neuringer and G. Sahay, Sci. Adv., 2023, 9, eadd4623 CrossRef PubMed.
- E. L. Han, S. Tang, D. Kim, A. M. Murray, K. L. Swingle, A. G. Hamilton, K. Mrksich, M. S. Padilla, R. Palanki, J. J. Li and M. J. Mitchell, Nano Lett., 2025, 25, 800–810 CrossRef CAS PubMed.
- I. Menon, M. Zaroudi, Y. Zhang, E. Aisenbrey and L. Hui, Mater. Today Adv., 2022, 16, 100299 CrossRef CAS.
- I. Ruseska and A. Zimmer, Beilstein J. Nanotechnol., 2020, 11, 101–123 CrossRef CAS PubMed.
- F. Madani, S. Lindberg, U. Langel, S. Futaki and A. Gräslund, J. Biophys., 2011, 2011, 414729 Search PubMed.
- J. Ouyang, Y. Sheng and W. Wang, Cells, 2022, 11, 4016 CrossRef CAS PubMed.
- S. Kawamoto, M. Takasu, T. Miyakawa, R. Morikawa, T. Oda, S. Futaki and H. Nagao, J. Chem. Phys., 2011, 134, 095103 CrossRef PubMed.
- S. Deshayes, M. C. Morris, G. Divita and F. Heitz, Biochim. Biophys. Acta, Biomembr., 2006, 1758, 328–335 CrossRef CAS PubMed.
- Y. Pouny, D. Rapaport, A. Mor, P. Nicolas and Y. Shai, Biochemistry, 1992, 31, 12416–12423 CrossRef CAS PubMed.
- Y. Shai and Z. Oren, Peptides, 2001, 22, 1629–1641 CrossRef CAS PubMed.
- S. Ludtke, K. He and H. Huang, Biochemistry, 1995, 34, 16764–16769 CrossRef CAS PubMed.
- A. Shebanova, Q. M. Perrin, K. Zhu, S. Gudlur, Z. Chen, Y. Sun, C. Huang, Z. W. Lim, E. A. Mondarte, R. Sun, S. Lim, J. Yu, Y. Miao, A. N. Parikh, A. Ludwig and A. Miserez, Adv. Sci., 2024, 11, e2402652 CrossRef PubMed.
- K. D. Saint Jean, K. D. Henderson, C. L. Chrom, L. E. Abiuso, L. M. Renn and G. A. Caputo, Probiotics Antimicrob. Proteins, 2018, 10, 408–419 CrossRef CAS PubMed.
- J. Allen and J.-P. Pellois, Sci. Rep., 2022, 12, 15981 CrossRef CAS PubMed.
- C. Rudolph, C. Plank, J. Lausier, U. Schillinger, R. H. Müller and J. Rosenecker, J. Biol. Chem., 2003, 278, 11411–11418 CrossRef CAS PubMed.
- E. Hinde, K. Thammasiraphop, H. T. T. Duong, J. Yeow, B. Karagoz, C. Boyer, J. J. Gooding and K. Gaus, Nat. Nanotechnol., 2017, 12, 81–89 CrossRef CAS PubMed.
- D. P. Jana, D. K. Samanta, D. M. Ehlers, E. Zellermann, D. S. Bäcker, P. R. H. Stauber, P. D. C. Schmuck and P. S. K. Knauer, ChemBioChem, 2022, 24, e202200519 CrossRef PubMed.
- L. Diao and B. Meibohm, Clin. Pharmacokinet., 2013, 52, 855–868 CrossRef CAS PubMed.
- S. M. Shi and L. Di, in Peptide Therapeutics: Fundamentals of Design, Development, and Delivery, ed. S. D. Jois, Springer, Cham, 1, 2022, vol. 4, 163–182 Search PubMed.
- D. P. Fairlie and A. Dantas de Araujo, Pept. Sci., 2016, 106, 843–852 CrossRef CAS PubMed.
- M. Moiola, M. G. Memeo and P. Quadrelli, Molecules, 2019, 24, 3654 CrossRef CAS PubMed.
- W. Li, D. Wang, X. Shi, J. Li, Y. Ma, Y. Wang, T. Li, J. Zhang, R. Zhao, Z. Yu, F. Yin and Z. Li, Mater. Horiz., 2018, 5, 745–752 RSC.
- D. W. Brown, P. Wee, P. Bhandari, A. Bukhari, L. Grin, H. Vega, M. Hejazi, D. Sosnowski, J. Ablack, E. K. Clancy, D. Pink, J. Kumar, M. P. S. Ares, S. Lamb, R. Quevedo, B. Rawal, F. Elian, N. Rana, L. Morales, N. Govindasamy, B. Todd, A. Delmage, S. Gupta, N. McMullen, D. MacKenzie, P. H. Beatty, H. Garcia, M. Parmar, J. Gyoba, C. McAllister, M. Scholz, R. Duncan, A. Raturi and J. D. Lewis, Cell, 2024, 187, 5357–5375.e24 CrossRef CAS PubMed.
- Y. J. Park, J. F. Liang, K. S. Ko, S. W. Kim and V. C. Yang, J. Gene Med., 2003, 5, 700–711 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |