 Open Access Article
Open Access ArticleRPRD1B's direct interaction with phosphorylated RNA polymerase II regulates polyadenylation of cell cycle genes and drives cancer progression†
Rosamaria Y.
Moreno
,
Svetlana B.
Panina
and
Y. Jessie
Zhang
 *
*
Department of Molecular Biosciences, University of Texas, Austin, Texas, USA. E-mail: jzhang@cm.utexas.edu
First published on 22nd January 2025
Abstract
RNA polymerase II (Pol II) regulates eukaryotic gene expression through dynamic phosphorylation of its C-terminal domain (CTD). Phosphorylation at Ser2 and Thr4 on the CTD is crucial for RNA 3′ end processing and facilitating the recruitment of cleavage and termination factors. However, the transcriptional roles of most CTD-binding proteins remain poorly understood. In this study, we focus on RPRD1B, a transcriptional regulator that interacts with the phosphorylated CTD and has been implicated in various cancers. We investigated its molecular mechanism during transcription and found that RPRD1B modulates alternative polyadenylation of cell growth transcripts by directly interacting with the CTD. RPRD1B is recruited to transcribing Pol II near the 3′ end of the transcript, specifically in response to Ser2 and Thr4 phosphorylation, but only after flanking Ser5 phosphorylation is removed. Transcriptomic analysis of RPRD1B knockdown cells revealed its role in cell proliferation via termination of the key cell growth genes at upstream polyadenylation sites, leading to the production of tumor suppressor transcripts that lack AU-rich elements (AREs) with increased mRNA stability. Overall, our study uncovers previously unrecognized connections between the Pol II CTD and CID, highlighting their influence on 3′ end processing and their contribution to abnormal cell growth in cancer.
Introduction
The C-terminal domain (CTD) of RNA polymerase II is a crucial regulator of transcription in eukaryotes. Comprised of repetitive heptapeptide motifs that undergo extensive post-translational modifications, the CTD conducts an orchestra of transcription regulators by inhibiting or promoting recruitment through phosphorylation on five of the seven residues within this motif.1 Ser5 phosphorylation is associated with the initiation phase of transcription and recruits participants that prime the 5′ cap on nascent RNA while releasing the preinitiation complex at promoter sites.2 On the other hand, Ser2 phosphorylation controls the elongation phase of transcription by recruiting factors involved in mRNA processing, such as splicing, polyadenylation and termination.3,4 Ablation of phosphorylation on either residue is detrimental to cell growth.5,6 Termination is coupled with high levels of Ser2 and Thr4 phosphorylation and modulates 3′ end processing of pre-mRNA through recruitment of the cleavage and polyadenylation complex.7 The precise interactions between the binding motifs of regulatory proteins and the CTD at the 3′ ends of genes has yet to be fully elucidated.Among the several binding modules that connect the varying phosphorylation signatures on the CTD to precise recruitment of transcriptional complexes,8 the C-terminal interaction domain (CID) is the most significant as it has been identified in numerous proteins, conserved through eukaryotic species. The CID has exhibited differing degrees of specificity for phosphorylation patterns within the CTD.8,9 CID domains have been found in several proteins involved in alternative splicing and mRNA processing, such as SCAF4, SCAF8, and CHERP, as well as transcription termination which encompasses proteins like Rtt103, PCF11, and NRD1.10–15 A particularly important CID-containing protein is RPRD1B or CREPT which is a nuclear protein containing an N-terminal CID and a C-terminal coiled-coil domain.16 RPRD1B also binds the 3′ UTR of protein coding RNAs through its CID, suggesting a role in the processing of 3′ ends.17 Significantly, RPRD1B is identified as an oncogene and plays critical functions in cell cycle, cell proliferation, and tumorigenesis.18 RPRD1B promotes gastric cancer proliferation by affecting cyclin B1 expression during mitosis.19 RPRD1B has been shown to enhance melanoma cell proliferation and migration through actin cytoskeleton organization.20 Overexpression of RPRD1B shortens the G1 phase and promotes G1 to S phase transition leading to upregulated cell proliferation by activating Wnt-signaling target genes through interactions with the β-catenin/TCF4 complex.21 In addition, RPRD1B is overexpressed in colorectal cancer cell lines, where it enhances invasion and migration by interacting with p300.22 This interaction stimulates the Wnt/β-catenin signaling pathway by increasing the acetylation of β-catenin and promoting the formation of the β-catenin/TCF4 complex.22 However, the molecular mechanism for RPRD1B's implication in cancers is still poorly understood.
In this study, we identified the structural elements that determine the specificity of RPRD1B binding. We also report the molecular basis underlying cell cycle regulation by RPRD1B through its influence in polyadenylation (polyA) choice at 3′ ends and maintenance of Pol II occupancy on cell cycle genes that are lengthy. Knockdown of RPRD1B promotes proximal (upstream) poly(A) site selection, leading to the removal of AU-rich elements (AREs) from cell cycle inhibitor transcripts, which in turn increases their stability. Thus, while reduced RPRD1B transcriptional activity leads to decreased cell growth, increased RPRD1B expression in colorectal cancer samples correlates with poor patients’ survival and reduced levels of tumor suppressor proteins, as observed in our TCGA (The Cancer Genome Atlas) analysis. Hence, RPRD1B's oncogenic role in colorectal cancer is at least partly explained by regulating the stability of tumor suppressor transcripts through 3′ end processing. Our findings shed light on the engagement of RPRD1B with the CTD and the veiled downstream effects on genes associated with cell proliferation through 3′ end processing and modulating Pol II occupancy, resulting in reduced cell growth when RPRD1B levels are low.
Results
Phosphoryl specificity of the CID domain of RPRD1B
In a recent proteomics analysis investigating the recruitment pattern of Thr4 and Ser2 phosphorylation on the CTD, we identified RPRD1B as highly abundant protein in both phosphorylation mark pulldowns over their respective unmodified control.14,23,24 While RPRD1B has been recognized as an oncogene by promoting cell proliferation in several cancer types,20,22,25–27 how the RPRD1B CID-Pol II axis contributes to the function of RPRD1B in cell cycle regulation is unknown. In order to elucidate this molecular mechanism, we first focused on defining the structural elements that determine the specificity of the CID of RPRD1B towards the phosphoryl species of Pol II CTD.To identify which phosphate marks regulate the recruitment of RPRD1B, we performed fluorescence polarization (FP) to monitor the binding interactions between the purified CID domain from RPRD1B and synthetic CTD peptides in solution (Fig. S1A and B, ESI†). The CID fragment is thermostable and elutes as a monomer (Fig. S1A and C, ESI†). We generated CTD peptides spanning approximately two heptads in length with phosphorylation at either of the five residues modified during transcription since it is well accepted in the field that a diheptide is the function unit of CTD as recruitment motif9 (Fig. 1(A)). Titration of CID binding partner with phosphopeptides with various phosphorylation sites revealed that RPRD1B forms stable interaction with pSer2, pThr4, and pSer7. However, binding was abolished with the presence of phosphorylated Ser5 and Tyr1 (Fig. 1(B)). Specifically, RPRD1B CID exhibited a Kd of 22.5 ± 3 μM towards pSer2, while a four-fold enhancement in binding strength towards pThr4 were found at 7.8 ± 1 μM and a Kd of 32.5 ± 3 μM for the pSer7 CTD peptide (Fig. 1(B)). The dissociation constants are consistent with the results of other laboratories.28,29
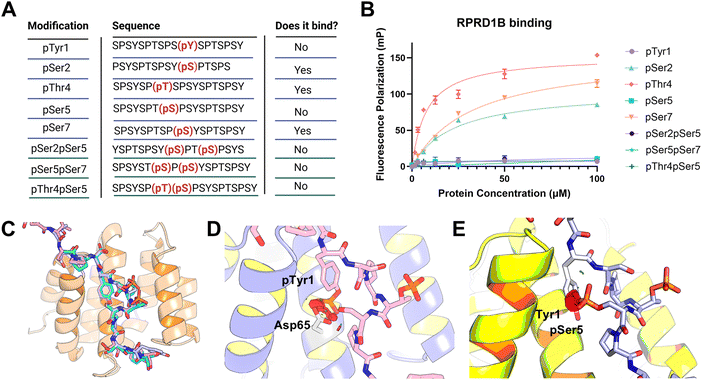 | ||
| Fig. 1 RPRD1B CID recognizes specific phosphorylation states of CTD Peptides. (A) Table of singly or doubly phospho-CTD peptides, spanning two heptads in length, used for fluorescence polarization with the indicated position of phosphorylation in parentheses and highlighted in red. No binding indicates a lack of significant change in polarization across the concentration range tested while yes indicates a significant change in binding. (B) Fluorescence polarization measurements of WT RPRD1B CID with singly phosphorylated and doubly phosphorylated CTD peptides. Experimental isotherms were fitted to a one to one binding model. Binding assays were performed in triplicate. Error bars indicate the standard deviation. (C) Superimposition of RPRD1B CID structures bound to pSer2,pThr4, and unmodified CTD peptide (PDB: 4Q94, 4Q96 and 9B9L). (D) Structural model of RPRD1B CID with pThr4pSer5 CTD peptide. (E) Structural model of RPRD1B with pThr4pTyr1 CTD peptide. For panels D and E, the original structure bound to pThr4 (PDB: 9B9L) was utilized, and phosphorylations at Tyr1 or Ser5 were introduced using the PyTM plugin in PyMol and the resulting model was energy minimized in Maestro. | ||
To understand the specificity of RPRD1B binding, we analyzed the interaction of RPRD1B with the CTD peptides. In all published RPRD1B structures, the backbone position of the CTD is highly preserved (PDB code 9B9L, 4Q94 and 4Q96)) (Fig. 1(C)). We modeled phosphorylated Ser5 using the PyTMs plugin in PyMol to introduce common post translational modifications into protein models30 and chose the likeliest rotamer conformation. The placement of Ser5 is coordinated by a hydrogen bond interaction with the carboxyl group of negatively charged Asp65 (Fig. 1(D)). However, a phosphate group on Ser5 would illicit charge repulsion with Asp65 and steric clashes with the neighboring Tyr1 on the CTD heptad (Fig. 1(D)). Since Ser5 phosphorylation is mostly enriched at the TSS site but prevents RPRD1B from binding to its preferred recognition sites, pSer5 could act as a gatekeeper modification, restricting the recruitment of RPRD1B to certain genomic loci. Similarly, for Tyr1 phosphorylation, as Tyr1 is bound to a hydrophobic pocket fashioned by Val23, Tyr61, Leu62, and Val66, and is in close proximity to Asp65, the addition of a bulky phosphate group would cause steric hindrance at this position as well as same charge repulsion, preventing the CID from binding to pTyr1 (Fig. 1(E)). It is likely phosphorylation of Tyr1 is not accepted by other CID as these hydrophobic interactions are conserved. Overall, the structural analyses of RPRD1B with various CTD peptides phosphorylated at different sites explain its biophysical interaction preferences.
Structural elements conserved in CID for phosphoryl-CTD specificity
CID-containing proteins are the biggest protein family recruited by CTD, exhibiting diverse binding preference for specific phospho-marks on the CTD of Pol II. With the exception of pTyr1, readers of every other phosphorylatable residue have been uncovered across different CID. However, there are astounding similarities in function with many of the CID proteins participate in termination and mRNA processing.10,12,31,32 To better understand the binding preference and function of CID-containing proteins we identified regions of structural variability across them, we aligned the sequences of several yeast and human CID's with published structures (Fig. 2(A)). Our amino acid sequence alignment shows there is significant sequence identity overlap. As shown in red text, there are several conserved hydrophobic patches and hydrophilic residues that participate in backbone and side-chain interactions (Fig. 2(A)).We examined key residues in RPRD1B that interact with the CTD side chain and backbone using the structure of RPRD1B CID bound to a pThr4 CTD peptide24 as a prototype. Conserved across all CIDs, hydrogen bond interactions between Ser19-Ser5 stabilize the heptad residue's position, while the backbone of Ser7 forms additional stabilizing hydrogen bonds with Asn69 (Fig. 2(B)). Additionally, the hydroxyl group of Tyr1 is stabilized by hydrogen bonds with Asn64 and Asp65, while conserved hydrophobic interactions involving Val23, Tyr61, Leu107, and Ile110 anchor Tyr1 and Pro3 in the CID binding pocket (Fig. 2(B)). Other anchoring backbone interactions take place between the carbonyl oxygen of Gln20 and the amide nitrogen of Pro6 and Ser2 (Fig. 2(B)). These interactions result in comparable binding orientation of the CTD backbone across CIDs.
Critical interactions determine phospho-specificity of CID binding, with Arg106 forming key hydrogen bonds that stabilize pSer228 and pThr4 (Fig. 2(C)), a feature conserved across the RPRD family, SCAF4/8,33 and yeast RTT103.13 In contrast, NRD1 lacks a positively charged residue at this position, resulting in weaker binding to pSer2 (Fig. 2(A)). Furthermore, pSer7, positioned at the exit channels of the binding groove, forms a hydrogen bond with the carbonyl oxygen of Asn18 (Fig. 2(D)). In SCAF4/8, Asn18 is substituted with isoleucine, which is unable to participate in hydrogen bonding. Echoing our structural analysis, a recent study showed SCAF4/8 exhibits no binding to pSer7 peptide.33 However, pSer5 binding is exclusive to SCAF4/8 and NRD1 which possess Lys23/Arg23 and Arg28, respectively, that interact with the phosphate group. Our analysis reveals structural features in the RPRD1B CID that serves as a blueprint for identifying the broad recognition mechanisms of the CTD across other CID proteins.
Taking a structure-guided approach, RPRD1B mutants harboring point mutations were designed to test the importance of specific residues in sidechain and phospho–residue interactions (Fig. 2(E)). The mutant CID constructs that were purified exhibited similar thermostability as the wild-type CID (Fig. S2A–C, ESI†). To investigate whether the variants could affect binding towards phosphorylated Ser2 or Thr4, we assessed the binding with fluorescence polarization. As WT CID had a Kd of 7.8 ± 1 μM for pThr4 and 22.5 ± 3 μM for pSer2, mutating Tyr61 to alanine greatly attenuated binding to CTD irrespective of phosphorylation state to 43.4 ± 6 μM for pThr4 and 37.8 ± 6 μM for pSer2 (Fig. 2(E)). While hydrophobic stacking by Tyr61 is conserved in all CID proteins (Fig. 2(A)), the adjacent hydrophobic residues that secure the position of Pro3 and Tyr1 on the heptad contribute incrementally to stronger binding, thereby mitigating the impact of Tyr61 loss on binding affinity. Likewise, the Arg114A mutant retained its ability to associate with the CTD, although the binding was reduced ∼5-fold for pThr4 (41.6 ± 6 μM) and ∼2-fold for pSer2 (37.9 ± 6 μM). The proximity of Arg114 to the phosphate group on pThr4 might contribute favorable electrostatic interactions, but it is too distant from pSer2 (Fig. 2(E)). Removing the negative charge on the Asp65 position, a residue conserved in all CID, by placing an alanine removes critical side chain interactions with Tyr1 and leads to a loss in binding towards both CTD peptides (Fig. 2(E)). Likewise, mutating Arg106 to alanine prevents binding to pSer2/pThr4 peptides as its necessary for phosphate recognition (Fig. 2(E)).
pSer5 prevents binding of RPRD1B to phosphoryl CTD
During active transcription, the CTD is getting hyperphosphorylated and some combination of phosphorylation patterns have been reported. For example, pSer5 and pSer2 within the same heptad is the most abundant double phosphorylation pattern.34,35 Likewise, pSer5/pThr4 is prevalent and pSer5/pSer7 also co-occur and share similar genomic localization, overlapping near the TSS.34 Since Ser5 phosphorylation occurs at the beginning of transcription and it doesn’t bind to RPRD1B, we pondered how the Ser5 phosphorylation affects the ability of RPRD1B to get recruited to Pol II. Specifically, we wonder the binding affinity of the RPRD1B CID towards combinations of pSer5 double phospho-marks. In contrast to strong binding towards singly phosphorylated pSer2, pThr4, or pSer7, the CID of RPRD1B does not exhibit binding towards these marks when pSer5 is present on the same heptad (Fig. 1(B)). Based on structural analyses using RPRD1B structures (Fig. 1(C)), such exclusion of binding is understandable with the steric clashes introduced upon Tyr1 or Ser5 phosphorylation (Fig. 1(D) and (E)). Thus, RPRD1B not only cannot bind to phosphorylated Ser5, but also fails to bind to any phosphoryl CTD forms as long as phosphoryl Ser5 is present in close range. Put in the context of RPRD1B binding to Pol II during transcription, it can only be recruited to Pol II when flanking Ser5 are dephosphorylated, which occurs during transcription elongation when Ser5 phosphorylation level abates. Thus, the recognition profile of RPRD1B suggests a transcriptional role associated with 3′ end events.RPRD1B maintains Pol II occupancy on long cell cycle genes
To understand how RPRD1B recruitment affected eukaryotic transcription, we generated the RPRD1B knockdown of HEK293 cells, using shRNA to knockdown RPRD1B to 19% of its total protein level as shown by western blot analysis (Fig. S3A, ESI†). Specifically, in isolated chromatin fractions we observe reduced RPRD1B levels compared to shControl samples (Fig. S3B, ESI†). We first investigated whether RPRD1B knockdown affected transcription at the promotor initiation or pausing release by conducting ChIP-seq analysis of RNA polymerase II in mutant cells vs. wild-type. Biological replicates of RPB1 ChIP samples exhibited high correlation and reproducibility (Fig. S3C, ESI†). We conducted unsupervised k-means clustering and plotted the intensity profiles of Pol II binding across all protein-coding genes, confirming that Pol II occupancy is predominantly contained within the transcription start site (TSS), with an additional small peak observed at the 3′ end of genes, in both cell types – WT vs. shRPRD1B (Fig. 3(A)). Pol II exhibits significant occupancy near the TSS and shifts from a paused to a processive state during elongation, prompting us to quantify RPB1 distribution following RPRD1B knockdown. We calculated the pausing index, which is the ratio of Pol II signal near the promoter region to the summed signal within the gene body to 3000 bp after the transcription end site (TES).36 Across all expressed genes (ESI,† Table S1), there is no difference in the state of promoter-paused Pol II (Fig. S3D, ESI†). However, when we clustered pausing indices in an unsupervised fashion, there was a significant decrease in pausing index within cluster 2 (n = 3405 genes, p < 2.2 × 10−16) and cluster 3 (n = 1499 genes, p < 2.2 × 10−16) (Fig. 3(B)).To improve rigor of our analysis, we also measured the traveling ratio (TR)37 to quantify RPB1 distribution as a ratio of the read density between the promoter region and the gene body, excluding the region after TES. While there was no observed redistribution of Pol II around the promoter region into the gene body when RPRD1B is knocked down compared to shControl (Fig. S3E, ESI†), clustering revealed that specific genes – belonging to cluster 2 (n = 3146) and cluster 3 (n = 1440) – showed reduced traveling ratio index, whereas the majority of genes (n = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682) retained same values of indices (Fig. 3(C) and ESI,† Table S2). Importantly, gene lists within clusters of pausing and traveling indices overlap by 97.5–99.9%. There is a reduction of Pol II occupancy at the promoter of long genes with a higher number of exons/introns as shown in cluster 2 and 3 (∼5000 genes) when RPRD1B is knocked down (Fig. 3(D)). Our results and analyses were echoed with a recent published dataset of RPRD1B knockout HEK293 cell line.17 When we derived traveling ratio indices of their data, clustering reveals a significant decrease in the traveling ratio in a sole cluster containing longer genes (Fig. S3F, ESI†). While RPRD1B knockdown decreases the traveling ratio in longer genes, it does not affect Pol II processivity (Fig. S3G, ESI†).
682) retained same values of indices (Fig. 3(C) and ESI,† Table S2). Importantly, gene lists within clusters of pausing and traveling indices overlap by 97.5–99.9%. There is a reduction of Pol II occupancy at the promoter of long genes with a higher number of exons/introns as shown in cluster 2 and 3 (∼5000 genes) when RPRD1B is knocked down (Fig. 3(D)). Our results and analyses were echoed with a recent published dataset of RPRD1B knockout HEK293 cell line.17 When we derived traveling ratio indices of their data, clustering reveals a significant decrease in the traveling ratio in a sole cluster containing longer genes (Fig. S3F, ESI†). While RPRD1B knockdown decreases the traveling ratio in longer genes, it does not affect Pol II processivity (Fig. S3G, ESI†).
For downstream analysis, we focused on genes within cluster 3 which contained a significant decrease in pausing and traveling ratio of Pol II. A breakdown of the gene types within cluster 3 indicates that the majority of genes affected are protein-coding genes followed by lncRNAs (1/6 of the subset) (Fig. 3(E)). Gene ontology (GO) analysis highlighted that cluster 3 was enriched in genes that regulate cell cycle transition such as CDK1, CDK7, and CDC7 (Fig. 3(F) and ESI,† Table S2), whereas clusters 1 and 2 are associated with distinct pathways (Fig. S3H, ESI†). Cluster 1 genes were associated with axon guidance and neuron projection guidance while cluster 2 genes are involved in ribosome biogenesis and rRNA processing (Fig. S3H, ESI†). It has been known that RPRD1B is frequently upregulated in endometrial and gastric cancer and promotes cell cycle progression.19,26 The mapping of ChIP-signal across genes within cluster 3 exhibits a decrease in Pol II association on promoters (Fig. 3(G)). In addition, Fig. 3(G) shows examples of reduced Pol II retention at the promoter of cell cycle genes, CDC7 and CCNB2, belonging to cluster 3, when RPRD1B is knocked down. Collectively, analysis of Pol II metrics indicates that RPRD1B maintains Pol II at a poised state on the promoters of longer genes.
RPRD1B knockdown promotes proximal polyA site usage
Our biophysical characterization of RPRD1B physical association with pThr4 and pSer2 phosphorylated forms of RNA Pol II, which are crucial for regulating transcription termination,38,39 suggests a potential role for RPRD1B in influencing the architectural features of 3′ ends. To test that, we first examined the global effects of RPRD1B knockdown in HEK293T on transcription using RNA-seq. Biological replicates were highly consistent, and we investigated the effects of RPRD1B loss on gene expression (Fig. S4A, ESI†). DEG analysis revealed 282 upregulated and 241 downregulated genes (Fold change >1.5 and p.adj. < 0.05) (Fig. 4(A) and ESI,† Table S3). CDK7, which was associated with reduced Pol II pausing (Fig. 3(G)), was also found to be downregulated. Gene expression analysis (qPCR) of selected genes that were up or downregulated was conducted to validate our RNA-Seq analysis (Fig. S4B, ESI†). Similarly, we assessed HA-RPRD1B occupancy at the 3′ end of genes using ChIP-qPCR on a representative target gene, P3H4, which displayed a stronger RPRD1B binding signal compared to the control (Fig. S4C, ESI†).Next we looked at changes in alternative polyadenylation (APA) of shRPRD1B cells compared to shControl cells using LABRAT.40 We identified 411 significant events, of which 111 transcripts favored distal site usage and 300 (73%) transcripts showed proximal site preference (Fig. 4(B) and ESI,† Table S4). Notably, changes in proximal polyadenylation are significantly associated with Pol II pausing, as evidenced by the fact that approximately one-third of the genes (104/300) exhibiting proximal polyadenylation also show a lower pausing index (X-squared = 6.48, p = 0.01). As anticipated, transcripts that showed shorter 3′ UTR usage comprised pathways that regulate cell growth (Fig. 4(C)).
As cell growth was one of the top pathways affected in our polyA analysis and RPRD1B influences Pol II distribution on long cell cycle genes, we evaluated whether RPRD1B knockdown impairs cell growth. RPRD1B deficiency resulted in significant cell growth arrest, consistent with higher proliferation doubling time (Fig. 4(D)). The mean doubling time of control cells was 18 hours, whereas shRPRD1B cells divided on average 1.3-fold slower. These results suggest RPRD1B promotes cell proliferation.
RPRD1B modulates mRNA stability of cell cycle genes
The mRNA expression dynamics can be modulated by the polyA site usage within the 3′-untranslated region of eukaryotic transcripts. Our polyA analysis of 3′ UTR indicates proximal ends were largely on genes involved in cell growth and protein metabolism (Fig. 4(D)). Upon closer inspection of transcripts with proximal polyA sites, roughly 1/3 of transcripts possessed AU-rich elements (ARE) (ESI,† Table S4). ARE elements are bound by RNA binding proteins that recruit deadenylates and exonucleases to regulate the metabolism of target transcripts.41 As an example of gene involved in cell cycle regulation that lost ARE elements upon proximal site switching, STK11 induces cell cycle arrest at G1/S and G2/M checkpoints42 (Fig. 4(E)). As further confirmation of our APA analysis, direct mRNA half-life assessments for STK11 in shRPRD1B HEK293T cells compared to shControl HEK293T were carried out using kinetic studies of mRNA decay. As anticipated, genes with 3′ UTR shortening had altered mRNA turnover and showed longer mRNA half-lives (Fig. 4(F)). For example, STK11 mRNA had a half-life of 0.52 h in shRPRD1B cells that was 2-fold higher than in shControl cells (t1/2 = 0.25 h) (Fig. 4(F)).In addition, we performed gene network analysis (WGCNA) to characterize gene expression shifts in the shRPRD1B vs. Control dataset and found one module, a cluster of densely interconnected genes with correlating co-expression patterns, that were associated with lower levels of RPRD1B (r = −0.96) (Fig. S5A and Table S3, ESI†). Interestingly, within a network of the 30 most highly connected ‘hub’ genes in the module, ELAVL4, a 3′UTR RNA binding protein, was found (Fig. S5B, ESI†). ELAVL4 may preserve the stability of tumor-suppressing genes by binding to their AREs, and its expression is tied with the expression levels of RPRD1B. Likewise, other genes important for cell cycle proliferation in colorectal cancer such as MMP1643 and NUP3744 were also found in the same module (Fig. S5B, ESI†). Taken together, our findings suggest that transitioning to shorter polyadenylation sites stabilizes mRNA encoding cell proliferation repressors by eliminating AU-rich elements (AREs), thereby introducing an additional regulatory mechanism through which RPRD1B influences cell cycle progression.
RPRD1B's oncogenic role in colorectal cancer
As our genomic and transcriptomic analyses have highlighted, RPRD1B via altering Pol II association on promoters of lengthy cell cycle-regulating genes influenced their choice of polyA site in HEK293 cells. Hence, we decided to evaluate the putative oncogenic role of RPRD1B taking advantage of publicly available cancer-related datasets, containing information on both mRNA and protein expression in tumor samples.Analysis of different cancer types from ICGC/TCGA project shows high prevalence (>20%) of RPRD1B alterations in colorectal, esophagogastric, uterine, and lung cancer, followed by other cancer subtypes (Fig. 5(A)). Overall, the most frequently detected genetic alteration was RPRD1B gene amplification, which is consistent with known aberrant overexpression of RPRD1B in colorectal cancer (CRC)22,45 and gastric cancers.46 In CRC subset from the PanCancer Atlas (TCGA, n = 594 samples per patients), overexpression of RPRD1B was the most prevalent alteration and happens in every other patient (50%) with colorectal cancer (Fig. 5(B)). Importantly, overexpression of RPRD1B can serve as a prognostic biomarker for CRC patients.22 Indeed, survival analysis of PanCancer CRC dataset shows that altered RPRD1B tends to be associated with worse disease-free patients’ survival (log-rank p-value = 0.0818) (Fig. 5(C)).
 | ||
| Fig. 5 TCGA database analysis shows frequent RPRD1B alterations in colorectal cancer (CRC). (A) Prevalence of RPRD1B gene alterations in different cancer patients’ cohorts from “Pan-cancer analysis of whole genomes’ project (ICGC/TCGA, n = 2922 samples).47,48 Cancer subtypes with prevalence >10% are shown. (B) Prevalence of RPRD1B gene alterations in colorectal adenocarcinoma samples, n = 594 (TCGA, PanCancer Atlas). (C) Survival analysis of RPRD1B-altered (n = 295) and RPRD1B-unaltered (n = 299) subgroups of colorectal adenocarcinoma samples (TCGA, PanCancer Atlas). Log-rank test p-value = 0.0818. Analysis was done using cBioPortal for cancer genomics tool. (D) Volcano plot showing changes in RPPA/reverse-phase protein array protein expression in RPRD1B-altered vs. RPRD1B-unaltered subgroups (same samples). (E) mRNA (log2 RSEM) and protein (RPPA) expression z-scores of STK11 in RPRD1B-altered vs. RPRD1B-unaltered subgroups (n = 295 and n = 299 samples, respectively). Median (dot) and interquartile range (25–75%) are shown as Tufte's boxplots. Analysis was done using cBioPortal for cancer genomics tool. To compare continuous data, t-test was used. ***p < 0.001, ns – not significant. | ||
Having analyzed effects of RPRD1B on polyA site usage and stability of select mRNA transcripts, we hypothesized that one of the mechanisms linking RPRD1B upregulation with tumor growth may be its ‘destabilizing’ effects on tumor suppressor transcripts via choosing distal 3′-polyA and thereby undergoing potential AU-rich elements-mediated decay (AMD). In this case, mRNA expression of these transcripts will likely stay on the basal level, but there would be changes in protein amount. Indeed, based on our overlap analysis, only a few genes (n = 4) from those preferentially choosing proximal polyA site were simultaneously upregulated on mRNA level. Therefore, we took advantage of RPPA (reverse-phase protein array) data published by TCGA project and compared proteins in CRC samples with altered (= overexpressed, n = 295) vs. unaltered RPRD1B status (n = 299) (Fig. 5(D)). Interestingly, we noticed that several tumor suppressor proteins (CASP7, VHL, CLDN7, PTEN, CDKN1A etc.) that have been already implicated in colorectal cancer progression or its clinical outcome49–53 express at lower levels in RPRD1B-altered samples (Fig. 5(D) and ESI,† Table S5). Moreover, a potent tumor suppressor STK11, which we validated as a transcript being controlled by RPRD1B, was also downregulated in RPRD1B-overexpressed CRC samples on a protein level, but not mRNA level (Fig. 5(D) and (E)). In addition, there was weak, but significant negative correlation (r = −0.23, p = 6.71 × 10−7) between mRNA level of RPRD1B and protein level of STK11 in the same subset of CRC samples (Fig. S6A, ESI†). Inactivation of serine-threonine kinase STK11 (also known as LKB1) is frequently observed in a variety of cancers including CRC.54 Our analysis shows that RPRD1B is overexpressed in colorectal cancer and correlates with reduced stability of tumor suppressor genes such as STK11. While our analysis suggests a potential role for RPRD1B in the development of colorectal cancer through 3′ end processing, it is important to note that this relationship is correlative. Further studies, such as overexpression of RPRD1B in colorectal cancer cell lines, will be necessary to establish a direct mechanistic link between RPRD1B and colorectal cancer progression.
Discussion
As the workhorse for transcribing all mRNA in eukaryotic cells, Pol II must be both highly efficient and precise. This efficiency should at least be partially credited to the coordination of the transcription process by the CTD of RPB1, which undergoes various phosphorylation states to associate with transcriptional regulators. A key player in this regulation is a family of proteins containing the CTD-interaction domain (CID), a reader domain found in all eukaryotes.8 In this study, we focus on RPRD1B, a transcription regulator that contains a CID binding module, to investigate the temporal recruitment to Pol II through CTD phosphorylation combinations, the transcriptional effects of RPRD1B, and to understand its implication in cancer development (Fig. 6). We observed that RPRD1B's CID preferentially binds to Pol II phosphorylated at Ser2 and Thr4, modifications that are highly enriched near the 3′ end of transcripts. Conversely, phosphorylations at Ser5 and Tyr1, which occur early in transcription, prevent binding. Prevalence of Ser5 phosphorylation prohibits the premature recruitment of RPRD1B so that its association with Ser2/Thr4 can only occur when nearby Ser5 is dephosphorylated, which occurs in a later stage of transcription. The preference for later recruitment of RPRD1B via biophysical and structural characterizations led us to explore its role in 3′ end processing (Fig. 6). Transcriptomic analysis revealed that RPRD1B, through its CID, plays a critical role in regulating the transition from proximal pausing to processive elongation and in determining termination site selection for a subset of genes. Reducing RPRD1B levels decreases Pol II pausing at the promoters of longer genes, including those involved in cell cycle regulation. This reduced pausing is associated with upstream polyadenylation and termination, particularly affecting one-third of the genes within the reduced pausing clusters. Since the selection of the polyA site correlates to the half-life of mRNA, the polyA site shift regulated by RPRD1B influences mRNA decay in a subset of genes, primarily those involved in cell cycle regulation (Fig. 6).Our study elucidates the transcriptional function of RPRD1B, providing insights into why it is highly overexpressed in various cancers, particularly colorectal tumors. Transcriptomic analysis of RPRD1B knockdown cells reveals its critical role in promoting cell cycle progression within tumorigenic contexts. Notably, the increased stability of tumor suppressor transcripts following RPRD1B knockdown offers a clue to its pathological role in cancer. Furthermore, previous studies also implicated RPRD1B as a key positive regulator of transcription by facilitating the binding of β-catenin/TCF4 to the promoters of cell cycle genes.21 This is consistent with a recent study showing that RPRD1B localizes to poly(A) sites through its association with the 3′ UTR via its CID.44 This dual localization underscores RPRD1B's role in both the early and late phases of the cell cycle. Overexpression of RPRD1B accelerates the G1 to S phase transition by upregulating cyclins,25 while also expediting the G2/M phase by increasing cyclin B expression.19
Thus, the transcriptional role of RPRD1B suggests it may contribute to cancer development, although this connection remains correlative. Our study provides a layer of mechanistic understanding by illustrating the distinct impact of RPRD1B on Pol II pausing and 3′ end processing, further highlighting its potential as a regulator of gene expression in cancer.
Methods
Cell culture
Human embryonic kidney cells (HEK293 or HEK293T) were purchased from ATCC (Manassas, VA, USA). Cells were routinely cultured in Dulbecco's modified Eagle's media (Sigma-Aldrich, St. Louis, MO, USA, product number #D6429), supplemented with 10% Opti-Gold fetal bovine serum (GenDEPOT, Katy, TX, USA) at 37 °C in humidified atmosphere with 5% CO2. HyClone penicillin and streptomycin mix (Cytiva, Marlborough, MA, USA), was added to the media to reach a final concentration of 1%.shRNA transfection
HEK293T cells were transfected with MISSION shRNA plasmid (Sigma, clone: TRCN0000130891) against RPRD1B using Fugene with a DNA to Fugene ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. Hexadimethrine bromide was added to the cells at a final concentration of 8 μg ml−1. Transduced cells were selected with puromycin at a concentration of 1 μg ml−1 for 7 days. In parallel, the control cells were transfected with MISSION non-mammalian shRNA negative control plasmid (Sigma, Cat: SHC002) using Fugene (Promega, Wadison, WI, USA) at a 1
3. Hexadimethrine bromide was added to the cells at a final concentration of 8 μg ml−1. Transduced cells were selected with puromycin at a concentration of 1 μg ml−1 for 7 days. In parallel, the control cells were transfected with MISSION non-mammalian shRNA negative control plasmid (Sigma, Cat: SHC002) using Fugene (Promega, Wadison, WI, USA) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio with hexadimethrine bromide for the same duration of time and selected using puromycin.
3 ratio with hexadimethrine bromide for the same duration of time and selected using puromycin.
Sequence alignment and constructs
The sequences of CID-containing proteins were obtained from NCBI (RPRD2-Q5VT52, RPRD1A-Q96P16, RPRD1B-Q9NQG5, Scaf4-O95104, and Scaf8-Q9UPN6). The sequences were aligned in Jalview using ClustalO and visualization of the alignment was done with ESPript 3. Residues were highlighted based on high sequence conservation and residues mediating important interactions were denoted based on structural analysis.Cloning
The RPRD1B CID domain (encoding residues 2–133) was ordered as a synthetic gene and cloned into a pET28a (Novogene, Sacramento, CA, USA) derivative vector encoding a 6xHis-tag followed by a GST-tag and a 3C protease site. The full-length RPRD1B cDNA (clone: HG14027-G) encoding residues 1–326 was cloned into a mammalian expression vector containing a CMV promoter and an N-terminal HA tag.Protein expression and purification
For protein expression, BL21 (DE3) cells expressing RPRD1B-CID and mutant variants were grown in one-liter cultures at 37 °C in Luria-Bertani (LB) broth (Thermo Scientific, Waltham, MA, USA) containing 50 μg ml−1 kanamycin. Once the cultures reached an OD 600 value of 0.6–0.8, the protein expression was induced with 0.25 mM isopropyl-β-D-thiogalactopyranoside (IPTG), and the cultures were grown an additional 16 h at 18 °C. The cells were pelleted and resuspended in lysis buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl, 15 mM imidazole, 10% glycerol, 0.1% Triton X-100, and 10 mM 2-mercaptoethanol (BME)) and sonicated at 90 A for 2.5 min of 1 s on/5 s off cycles on ice. The lysate was cleared by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 45 min at 4 °C. The supernatant was loaded over 3 ml of Ni-NTA beads (Qiagen, Germany) equilibrated in lysis buffer, then washed through with wash buffer containing 50 mM Tris–HCl pH 8.0, 500 mM NaCl, 30 mM imidazole, and 10 mM BME. The recombinant protein was eluted with buffer containing 50 mM Tris–HCl pH 8.0, 500 mM NaCl, 300 mM imidazole, and 10 mM BME. Protein fractions were pooled and dialyzed overnight at 4 °C in a 10.0 kDa dialysis membrane (Thermo Scientific) against dialysis buffer (50 mM Tris HCl pH 7.5, 300 mM NaCl, and 10 mM BME). The protein was polished using gel filtration chromatography and loaded onto a Superdex 75 or 200 size exclusion column (GE) in gel filtration buffer (50 mM Tris HCl pH 7.5, 300 mM NaCl, and 10 mM BME). Fractions were collected, analyzed by SDS-PAGE, and fractions containing protein were pooled and flash frozen at −80 °C.
000 rpm for 45 min at 4 °C. The supernatant was loaded over 3 ml of Ni-NTA beads (Qiagen, Germany) equilibrated in lysis buffer, then washed through with wash buffer containing 50 mM Tris–HCl pH 8.0, 500 mM NaCl, 30 mM imidazole, and 10 mM BME. The recombinant protein was eluted with buffer containing 50 mM Tris–HCl pH 8.0, 500 mM NaCl, 300 mM imidazole, and 10 mM BME. Protein fractions were pooled and dialyzed overnight at 4 °C in a 10.0 kDa dialysis membrane (Thermo Scientific) against dialysis buffer (50 mM Tris HCl pH 7.5, 300 mM NaCl, and 10 mM BME). The protein was polished using gel filtration chromatography and loaded onto a Superdex 75 or 200 size exclusion column (GE) in gel filtration buffer (50 mM Tris HCl pH 7.5, 300 mM NaCl, and 10 mM BME). Fractions were collected, analyzed by SDS-PAGE, and fractions containing protein were pooled and flash frozen at −80 °C.
Western blot
Cells were lysed in RIPA lysis buffer (50 mM Tris–Cl pH 8.0, 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM NaCl, NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and 1× protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Protein concentrations were quantified with the Bradford protein assay. Briefly, 50
mM NaCl, NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and 1× protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Protein concentrations were quantified with the Bradford protein assay. Briefly, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μg of protein extracts were loaded and separated by SDS-PAGE gels. Blotting was performed with standard protocols using a PVDF membrane (Bio-Rad, Hercules, CA, USA). Membranes were blocked for 1 h in blocking buffer (5% BSA in PBST) and probed with primary antibodies at 1:1000 dilution at 4 °C overnight. After three washes with PBST, the membranes were incubated with diluted goat anti-rabbit or anti-rat secondary IRDye 680RD antibody at 1:10
μg of protein extracts were loaded and separated by SDS-PAGE gels. Blotting was performed with standard protocols using a PVDF membrane (Bio-Rad, Hercules, CA, USA). Membranes were blocked for 1 h in blocking buffer (5% BSA in PBST) and probed with primary antibodies at 1:1000 dilution at 4 °C overnight. After three washes with PBST, the membranes were incubated with diluted goat anti-rabbit or anti-rat secondary IRDye 680RD antibody at 1:10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (LI-COR, Lincoln, NE, USA) for 1
000 (LI-COR, Lincoln, NE, USA) for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) h at room temperature. After washing, membranes were visualized on LI-COR Odyssey CLx image reader. For western blot analysis, HA antibody (cat: C29F4, 1:1000 dilution for WB and 1:800 for IF), RPRD1B antibody (cat: 74693, 1:1000 dilution for WB), H3 antibody (cat: sc-517576), and beta-tubulin (cat: AB6046) were used.
h at room temperature. After washing, membranes were visualized on LI-COR Odyssey CLx image reader. For western blot analysis, HA antibody (cat: C29F4, 1:1000 dilution for WB and 1:800 for IF), RPRD1B antibody (cat: 74693, 1:1000 dilution for WB), H3 antibody (cat: sc-517576), and beta-tubulin (cat: AB6046) were used.
Subcellular fractionation
To fractionate cellular components, HEK293T cells were seeded to achieve 70–80% confluency in a 10 cm dish, then harvested and processed at 4 °C as described previously.55 Cells were washed with PBS, collected by scraping, and pelleted at 130 × g for 3 minutes. The cell pellet was lysed in cold E1 buffer (50 mM HEPES-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA (pH 8.0), 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1 mM DTT, and 1× protease inhibitor cocktail) and centrifuged at 1100 × g for 2 minutes to obtain the cytoplasmic fraction. The remaining pellet was washed twice with E1 buffer, incubated on ice for 10 minutes, and centrifuged, before being resuspended in cold E2 buffer (10 mM Tris–HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0), and 1× protease inhibitor cocktail) to extract the nuclear fraction. The pellet was washed in E2 buffer, centrifuged, and the supernatant was discarded. For chromatin fraction, the pellet was resuspended in E3 buffer (500 mM Tris–HCl (pH 6.8), 500 mM NaCl, and 1× protease inhibitor cocktail), sonicated for 5 minutes (30 seconds on/off), and centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 minutes. Protein concentrations were measured using a BCA assay and fractions were analyzed by western blot.
000 × g for 10 minutes. Protein concentrations were measured using a BCA assay and fractions were analyzed by western blot.
Cell proliferation
Cells expressing either shControl or shPRD1B were seeded at a density of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in complete media in 24-well plates. Cells were counted every 24 h for a period of four days using Trypan Blue exclusion assay (0.4%) on automated Luna-II automated cell counter (Logos Biosystems, Annandale, VA). Population doubling time (PDT) was estimated with the following formula, PDT = (72 h × ln2)/ln(N4/N1), where N1 and N4 are cell counts in every well on first and fourth days, respectively. Cells were counted for three independent biological replicates at each time interval.
000 cells per well in complete media in 24-well plates. Cells were counted every 24 h for a period of four days using Trypan Blue exclusion assay (0.4%) on automated Luna-II automated cell counter (Logos Biosystems, Annandale, VA). Population doubling time (PDT) was estimated with the following formula, PDT = (72 h × ln2)/ln(N4/N1), where N1 and N4 are cell counts in every well on first and fourth days, respectively. Cells were counted for three independent biological replicates at each time interval.
Fluorescence polarization
CTD peptides with double repeats were labeled with fluorescein isothiocyanate (FITC) or streptavidin conjugated FITC. Protein and peptide concentrations were determined according to their absorbance at 280 nm. Fluorescence polarization values were collected on a Tecan F200 plate reader in buffer (50 mM Tris pH 8.0, 300 mM NaCl and 10 mM BME) at room temperature. Samples were excited with vertically polarized light at 485 nm and at an emission wavelength of 535 nm. RPRD1B-CID and variants were titrated into a reaction mixture containing buffer supplemented with 10 nM of FITC-peptide. Measurements were taken in triplicates and the experimental binding isotherms were analyzed in GraphPad Prism v9 using one to one binding mode to obtain Kd values.Differential scanning fluorometry
Purified recombinant RPRD1B CID domain at a final concentration of 5 μM was incubated with 10X SYPRO Orange (Molecular Probes) in a 96-well low-profile PCR plate (ABgene, Thermo Scientific) and fluorescence was captured in a LightCycler 480 (Roche). Protein melting curves were carried out with a temperature acquisition mode using a total of 10 acquisitions per 1 °C in each cycle from 20 °C to 95 °C. The melting temperature was derived using the Boltzmann equation.RT-qPCR
Total RNA was harvested from HEK293 or HEK293T cells using DirectZol RNA Miniprep kit (Zymo Research, Irvine, CA, USA, product number #R2050). cDNA was generated using AzuraQuant cDNA synthesis kit (Azura Genomics) using manufacturer's instructions. qPCR was done using the AzuraQuant Green Fast qPCR Mix Lo-Rox (Azura Genomics) in a ViiA-7 Real Time PCR system (Applied Biosystems). All qPCR experiments were conducted in biological triplicates, error bars represent mean ± standard error mean. Relative gene expression was assessed using the ΔΔCt method normalized to ACTB expression. To analyze ChIP-qPCR experiments, the fold enrichment was calculated by dividing the ChIP signal (from the experimental IP) by the background signal (from the mock IgG control). Student's t-test was used to compare groups. All primers used in this study can be found in the supplementary section as ESI,† Table S6.mRNA decay
For mRNA stability experiments, 1 × 106 cells in a 6-well plate format of shControl or shRPRD1B HEK293T cells were incubated with 5 μg ml−1 actinomycin D for different time intervals. Cells were collected at several time points (0, 30, 60 and 120 min) and were subject to RNA purification and cDNA synthesis. qPCR was done using the AzuraQuant Green Fast qPCR Mix Lo-Rox (Azura Genomics) in a ViiA-7 Real Time PCR system (Applied Biosystems). The Ct average of each time point was normalized to the Ct average of t = 0 to obtain ΔCt value (ΔCt = (Average Ct of each time point − Average Ct of t = 0). The relative abundance of each time point was calculated as: mRNA abundance = 2(−ΔCt). The relative abundance of mRNA at each time point relative to t = 0 was plotted using GraphPad Prism. The mRNA decay rate was determined by non-linear regression curve fitting (one phase decay). Three biological replicates were used for statistical assessment.RNA isolation, library preparation, and RNA-sequencing
Total RNA was isolated from HEK293T cells (at least ∼106 cells per sample) expressing shControl or shRPRD1B using DirectZol RNA Miniprep kit (Zymo Research). RNA integrity was assessed by Novogene Co. using the RNA Nano 6000 assay kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Libraries were prepared at Novogene Co. according to manufacturer's instructions for the NEBNext Ultra RNA Library kit for Illumina. The resulting libraries tagged with unique dual indices were checked for size and quality using the Agilent Bioanalyzer 2100. Libraries were loaded for sequencing on the NovaSeq 6000 (Illumina, San Diego, CA, USA) instrument (paired-end 2 × 150).Analyses of RNA-seq data
Quality of raw reads was assessed using FastQC read quality reports (https://usegalaxy.org).56 Adapter Illumina sequences were trimmed off by Trimmomatic v.0.38 with default parameters.57 Next, reads were aligned to human reference genome, GRCh38 version, using HISAT2 fast aligner v.2.2.1 with default parameters and library type–unstranded.58 Lastly, mapped fragments were quantified by featureCounts v.2.0.1 in Galaxy.59 Differential expression was analyzed using edgeR v.3.36.0; genes with FDR < 0.05 were considered as differentially expressed.60 Network analysis was performed using ‘WGCNA’ package (v. 1-70.3) in R on rlog-normalized RNA-Seq counts. RNA-seq data was deposited in GEO under the accession number GSE275817.Quantification of differential APA usage was performed using LABRAT.40 For –librarytype, RNAseq was chosen. Calculatepsi was used to calculate the relative usage of these ends, compare across conditions, and Ψ values were calculated for each gene in each sample with an expression level cutoff of 5 TPM. Enrichment analysis of biological processes was performed with ShinyGO v.0.8061 or ‘clusterProfiler’ package in R. AU-rich elements (ARE) search in sequences of select subset of genes preferring proximal polyA sites (ΨshRPRD1B − ΨshControl < 0) was performed using ARED-Plus database.62
Chromatin immunoprecipitation (ChIP) and ChIP-sequencing
For RPB1 samples, HEK293T cells expressing shControl or shRPRD1B cells were crosslinked by 1% formaldehyde for 10 min. Crosslinking was quenched with 0.125 M glycine for 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) min. Cells were successively lysed in lysis buffer LB1 (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1× PI), LB2 (10
min. Cells were successively lysed in lysis buffer LB1 (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1× PI), LB2 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM Tris–HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× PI) and LB3 (10
mM Tris–HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× PI) and LB3 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM Tris–HCl, pH
mM Tris–HCl, pH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 1× PI). Chromatin was sonicated to an average size of ∼200–500 bp using UCD-200 Biorupter (30 s on and 30 s off for 30 min). A total of 5 μg of RPB1 antibody (cat: ab76123) was pre-mixed in a 50 μl volume of Dynabeads protein A (Invitrogen) and was added to each sonicated chromatin sample and incubated overnight at 4 °C. The chromatin-bound beads were washed two times with low salt buffer (0.1% Na deoxycholate, 1% Triton X-100, 1 mM EDTA, 50 mM HEPES pH 7.5, 150 mM NaCl), once with high salt wash buffer (0.1% Na deoxycholate, 1% Triton X-100, 1 mM EDTA, 50 mM HEPES pH 7.5, 500 mM NaCl), once with LiCl wash buffer (250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, 1 mM EDTA, 10 mM Tris–Cl pH 8.0) and twice in TE buffer. The chromatin was reverse crosslinked overnight at 65 °C with shaking at 750 rpm. After DNA extraction using phenol–chloroform, the DNA was resuspended in 10 mM Tris–HCl pH 8.0. The purified DNA was subjected to qPCR to confirm target region enrichment before moving on to deep sequencing library preparation. For sequencing, the extracted DNA was used to construct the ChIP-seq library using the NEBNext Ultra II DNA Library Prep Kit followed by sequencing with an Illumina NovaSeq X Plus system.
8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 1× PI). Chromatin was sonicated to an average size of ∼200–500 bp using UCD-200 Biorupter (30 s on and 30 s off for 30 min). A total of 5 μg of RPB1 antibody (cat: ab76123) was pre-mixed in a 50 μl volume of Dynabeads protein A (Invitrogen) and was added to each sonicated chromatin sample and incubated overnight at 4 °C. The chromatin-bound beads were washed two times with low salt buffer (0.1% Na deoxycholate, 1% Triton X-100, 1 mM EDTA, 50 mM HEPES pH 7.5, 150 mM NaCl), once with high salt wash buffer (0.1% Na deoxycholate, 1% Triton X-100, 1 mM EDTA, 50 mM HEPES pH 7.5, 500 mM NaCl), once with LiCl wash buffer (250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, 1 mM EDTA, 10 mM Tris–Cl pH 8.0) and twice in TE buffer. The chromatin was reverse crosslinked overnight at 65 °C with shaking at 750 rpm. After DNA extraction using phenol–chloroform, the DNA was resuspended in 10 mM Tris–HCl pH 8.0. The purified DNA was subjected to qPCR to confirm target region enrichment before moving on to deep sequencing library preparation. For sequencing, the extracted DNA was used to construct the ChIP-seq library using the NEBNext Ultra II DNA Library Prep Kit followed by sequencing with an Illumina NovaSeq X Plus system.
Analyses of ChIP-Seq data and calculation of Pol II metrics
After initial assessment of read quality, RPB1-Pol II ChIP-Seq data was mapped onto human reference genome, hg38, with Bowtie2 v. 2.5.0 aligner for paired-end reads using default parameters.63 Coverage tracks in.bigwig format were generated from filtered.bam files (mapq > 20) and visualized in IGV v.2.4.16 software. Reproducibility of data was assessed by Pearson correlation analysis cm deepTools.64 BigWig files were generated using the BamCoverage function (–normalize using CPM –smoothLength 150 –binSize 50 –scalefactor 1) from deeptools, and heatmaps were generated on all hg38 protein-coding genes using 50-bp bin matrices obtained with computeMatrix. Gencode.gtf (hg38 version) was used as annotation file.RNA Pol II metrics – Pausing Index (PI),36 Traveling Ratio (TR),37 and Processivity36-were calculated using custom pipeline in R (https://github.com/tailana703/PolII_metrics) and bwtool summary (–with-sum) function (https://github.com/CRG-Barcelona/bwtool). First, we filtered only expressed genes based on RNA-Seq counts with length >2000 bp to focus on transcriptionally active Pol II. Next, we extracted genomic coordinates corresponding to promoter region and gene body (or 5′ and 3′-regions) in.bed format. Then, ChIP-signal of Pol II was summed up over extracted regions and used as input for normalization by region length. Lastly, resulting indices were clustered in an unsupervised fashion using k-means clustering. Indices were compared using paired Wilcoxon test.
PI was defined as follows:
TR was defined as follows:
Processivity Index was defined as follows:
ChIP-seq data was deposited in GEO under the accession number GSE275898.
Analysis of TCGA data
Open TCGA pan-cancer data [PMID:32025007] was accessed using cBioPortal for Cancer genomics (https://cbioportal.org). We queried mutations, CNV alterations, mRNA expression z-scores (RSEM), and protein expression (RPPA) z-scores. Analysis and visualization was performed using cBioPortal and R.Statistical analyses
Statistical analyses were performed using RStudio v4.0.5 and GraphPad Prism v9. Two-tailed, independent sample t-test was used for comparing the two groups (if not stated otherwise). Chi-squared test was used to compare distribution of two categorical variables. p < 0.05 was considered as significant. Correlations were assessed using two-tailed Pearson r coefficients. Protein bands were quantified and compared using ImageJ software.Author contributions
Conceptualization: R. Y. M. & Y. J. Z; investigation: R. Y. M. and S. P.; formal analysis: R. Y. M. & S. P.; funding acquisition: Y. J. Z.; supervision: Y. J. Z; writing – original, review & editing: R. Y. M., S. P., & Y. J. Z.Data availability
The RNA-seq data used in this study is available in GEO under accession code GSE275817. The ChIP-seq data used in this study is available in GEO under accession code GSE275898.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work is supported by grants from the National Institutes of Health (R01GM104896, R01GM125882, R01CA281106 and R35GM148356) and L. Leon Campbell Professorship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.References
- B. Bartkowiak, A. L. MacKellar and A. L. Greenleaf, Genet. Res. Int., 2011, 2011, 623718 Search PubMed.
- C. K. Ho and S. Shuman, Mol. Cell, 1999, 3, 405–411 CrossRef CAS PubMed.
- D. Eick and M. Geyer, Chem. Rev., 2013, 113, 8456–8490 CrossRef CAS PubMed.
- J. L. Corden, Mol. Cell, 2016, 61, 183–184 CrossRef CAS PubMed.
- M. S. Bartolomei, N. F. Halden, C. R. Cullen and J. L. Corden, Mol. Cell. Biol., 1988, 8, 330–339 CAS.
- M. L. West and J. L. Corden, Genetics, 1995, 140, 1223–1233 CrossRef CAS PubMed.
- N. J. Proudfoot, Genes Dev., 2011, 25, 1770–1782 CrossRef CAS PubMed.
- B. M. LeBlanc, R. Y. Moreno, E. E. Escobar, M. K. Venkat Ramani, J. S. Brodbelt and Y. Zhang, RSC Chem. Biol., 2021, 2, 1084–1095 RSC.
- M. K. Venkat Ramani, W. Yang, S. Irani and Y. Zhang, J. Mol. Biol., 2021, 433, 166912 CrossRef CAS PubMed.
- L. H. Gregersen, R. Mitter, A. P. Ugalde, T. Nojima, N. J. Proudfoot, R. Agami, A. Stewart and J. Q. Svejstrup, Cell, 2019, 177, 1797–1813 CrossRef CAS PubMed.
- C. G. Noble, D. Hollingworth, S. R. Martin, V. Ennis-Adeniran, S. J. Smerdon, G. Kelly, I. A. Taylor and A. Ramos, Nat. Struct. Mol. Biol., 2005, 12, 144–151 CrossRef CAS PubMed.
- D. H. Heo, I. Yoo, J. Kong, M. Lidschreiber, A. Mayer, B. Y. Choi, Y. Hahn, P. Cramer, S. Buratowski and M. Kim, J. Biol. Chem., 2013, 288, 36676–36690 CrossRef CAS PubMed.
- O. Jasnovidova, T. Klumpler, K. Kubicek, S. Kalynych, P. Plevka and R. Stefl, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 11133–11138 CrossRef CAS PubMed.
- R. Y. Moreno, K. J. Juetten, S. B. Panina, J. P. Butalewicz, B. M. Floyd, M. K. Venkat Ramani, E. M. Marcotte, J. S. Brodbelt and Y. J. Zhang, iScience, 2023, 26, 107581 CrossRef CAS PubMed.
- A. De Maio, H. K. Yalamanchili, C. J. Adamski, V. A. Gennarino, Z. Liu, J. Qin, S. Y. Jung, R. Richman, H. Orr and H. Y. Zoghbi, Cell Rep., 2018, 25, 726–736 CrossRef CAS PubMed.
- Z. Ni, J. B. Olsen, X. Guo, G. Zhong, E. D. Ruan, E. Marcon, P. Young, H. Guo, J. Li, J. Moffat, A. Emili and J. F. Greenblatt, Transcription, 2011, 2, 237–242 CrossRef PubMed.
- Z. Ni, N. Ahmed, S. Nabeel-Shah, X. Guo, S. Pu, J. Song, E. Marcon, G. L. Burke, A. H. Y. Tong, K. Chan, K. C. H. Ha, B. J. Blencowe, J. Moffat and J. F. Greenblatt, Nucleic Acids Res., 2024, 52, 4483–4501 CrossRef CAS PubMed.
- M. Li, D. Ma and Z. Chang, Oncogene, 2021, 40, 705–716 CrossRef CAS PubMed.
- L. Ding, L. Yang, Y. He, B. Zhu, F. Ren, X. Fan, Y. Wang, M. Li, J. Li, Y. Kuang, S. Liu, W. Zhai, D. Ma, Y. Ju, Q. Liu, B. Jia, J. Sheng and Z. Chang, Cell Death Dis., 2018, 9, 1172 CrossRef PubMed.
- H. Liu, A. L. B. Seynhaeve, R. W. W. Brouwer, I. W. F. J. van, L. Yang, Y. Wang, Z. Chang and T. L. M. Ten Hagen, Cancers, 2019, 12, 33 CrossRef PubMed.
- Y. Zhang, C. Liu, X. Duan, F. Ren, S. Li, Z. Jin, Y. Wang, Y. Feng, Z. Liu and Z. Chang, J. Biol. Chem., 2014, 289, 22589–22599 CrossRef CAS PubMed.
- Y. Zhang, S. Wang, W. Kang, C. Liu, Y. Dong, F. Ren, Y. Wang, J. Zhang, G. Wang, K. F. To, X. Zhang, J. J. Y. Sung, Z. Chang and J. Yu, Oncogene, 2018, 37, 3485–3500 CrossRef CAS PubMed.
- H. A. Hardtke, R. Y. Moreno and Y. J. Zhang, STAR Protoc., 2024, 5, 103277 CrossRef CAS PubMed.
- R. Y. Moreno, S. B. Panina, S. Irani, H. A. Hardtke, R. Stephenson, B. M. Floyd, E. M. Marcotte, Q. Zhang and Y. J. Zhang, Sci. Adv., 2024, 10, eadq0350 CrossRef CAS PubMed.
- D. Lu, Y. Wu, Y. Wang, F. Ren, D. Wang, F. Su, Y. Zhang, X. Yang, G. Jin, X. Hao, D. He, Y. Zhai, D. M. Irwin, J. Hu, J. J. Sung, J. Yu, B. Jia and Z. Chang, Cancer Cell, 2012, 21, 92–104 CrossRef CAS PubMed.
- Y. Wang, H. Qiu, W. Hu, S. Li and J. Yu, Oncol. Rep., 2014, 31, 1389–1395 CrossRef CAS PubMed.
- M. Sun, G. Si, H. S. Sun and F. C. Si, Biochem. Biophys. Res. Commun., 2018, 496, 1183–1190 CrossRef CAS PubMed.
- Z. Ni, C. Xu, X. Guo, G. O. Hunter, O. V. Kuznetsova, W. Tempel, E. Marcon, G. Zhong, H. Guo, W.-H. W. Kuo, J. Li, P. Young, J. B. Olsen, C. Wan, P. Loppnau, M. El Bakkouri, G. A. Senisterra, H. He, H. Huang, S. S. Sidhu, A. Emili, S. Murphy, A. L. Mosley, C. H. Arrowsmith, J. Min and J. F. Greenblatt, Nat. Struct. Mol. Biol., 2014, 21, 686–695 CrossRef CAS PubMed.
- K. Mei, Z. Jin, F. Ren, Y. Wang, Z. Chang and X. Wang, Sci. China: Life Sci., 2014, 57, 97–106 CrossRef CAS PubMed.
- A. Warnecke, T. Sandalova, A. Achour and R. A. Harris, BMC Bioinf., 2014, 15, 370 CrossRef PubMed.
- P. Grzechnik, M. R. Gdula and N. J. Proudfoot, Genes Dev., 2015, 29, 849–861 CrossRef CAS PubMed.
- M. Kim, N. J. Krogan, L. Vasiljeva, O. J. Rando, E. Nedea, J. F. Greenblatt and S. Buratowski, Nature, 2004, 432, 517–522 CrossRef CAS PubMed.
- M. Zhou, F. Ehsan, L. Gan, A. Dong, Y. Li, K. Liu and J. Min, FEBS Lett., 2022, 596, 249–259 CrossRef CAS PubMed.
- R. Schüller, I. Forné, T. Straub, A. Schreieck, Y. Texier, N. Shah, T. M. Decker, P. Cramer, A. Imhof and D. Eick, Mol. Cell, 2016, 61, 305–314 CrossRef PubMed.
- S. Buratowski, Nat. Struct. Biol., 2003, 10, 679–680 CrossRef CAS PubMed.
- Z. Fan, J. R. Devlin, S. J. Hogg, M. A. Doyle, P. F. Harrison, I. Todorovski, L. A. Cluse, D. A. Knight, J. J. Sandow, G. Gregory, A. Fox, T. H. Beilharz, N. Kwiatkowski, N. E. Scott, A. T. Vidakovic, G. P. Kelly, J. Q. Svejstrup, M. Geyer, N. S. Gray, S. J. Vervoort and R. W. Johnstone, Sci. Adv., 2020, 6, eaaz5041 CrossRef CAS PubMed.
- P. C. Boddu, A. K. Gupta, R. Roy, B. De La Peña Avalos, A. Olazabal-Herrero, N. Neuenkirchen, J. T. Zimmer, N. S. Chandhok, D. King, Y. Nannya, S. Ogawa, H. Lin, M. D. Simon, E. Dray, G. M. Kupfer, A. Verma, K. M. Neugebauer and M. M. Pillai, Mol. Cell, 2024, 84, 1475–1495 CrossRef CAS PubMed.
- S. H. Ahn, M. Kim and S. Buratowski, Mol. Cell, 2004, 13, 67–76 CrossRef CAS PubMed.
- B. M. Lunde, S. L. Reichow, M. Kim, H. Suh, T. C. Leeper, F. Yang, H. Mutschler, S. Buratowski, A. Meinhart and G. Varani, Nat. Struct. Mol. Biol., 2010, 17, 1195–1201 CrossRef CAS PubMed.
- R. Goering, K. L. Engel, A. E. Gillen, N. Fong, D. L. Bentley and J. M. Taliaferro, BMC Genomics, 2021, 22, 476 CrossRef CAS PubMed.
- C. Barreau, L. Paillard and H. B. Osborne, Nucleic Acids Res., 2005, 33, 7138–7150 CrossRef CAS PubMed.
- K. D. Scott, S. Nath-Sain, M. D. Agnew and P. A. Marignani, Cancer Res., 2007, 67, 5622–5627 CrossRef CAS PubMed.
- S. Wu, C. Ma, S. Shan, L. Zhou and W. Li, Sci. Rep., 2017, 7, 46531 CrossRef CAS PubMed.
- X. Yang, J. Liu, S. Wang, W. H. A. Al-Ameer, J. Ji, J. Cao, H. M. S. Dhaen, Y. Lin, Y. Zhou and C. Zheng, J. Transl. Med., 2024, 22, 554 CrossRef CAS PubMed.
- Y. S. Kuang, Y. Wang, L. D. Ding, L. Yang, Y. Wang, S. H. Liu, B. T. Zhu, X. N. Wang, H. Y. Liu, J. Li, Z. J. Chang, Y. Y. Wang and B. Q. Jia, World J. Gastroenterol., 2018, 24, 475–483 CrossRef CAS PubMed.
- Y. Jia, Q. Yan, Y. Zheng, L. Li, B. Zhang, Z. Chang, Z. Wang, H. Tang, Y. Qin and X. Y. Guan, J. Exp. Clin. Cancer Res., 2022, 41, 287 CrossRef CAS PubMed.
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium, Nature, 2020, 578, 82–93 CrossRef PubMed.
- K. Tomczak, P. Czerwińska and M. Wiznerowicz, Contemp. Oncol., 2015, 19, A68–77 Search PubMed.
- H. Yang, W. Jin, H. Liu, D. Gan, C. Cui, C. Han and Z. Wang, Front. Genet., 2020, 11, 401 CrossRef PubMed.
- L. Zinnamosca, A. Laudisi, L. Petramala, C. Marinelli, M. Roselli, D. Vitolo, C. Montesani and C. Letizia, Intern. Med., 2013, 52, 1599–1603 CrossRef PubMed.
- C. Xu, Y. H. Ding, K. Wang, M. Hao, H. Li and L. Ding, J. Transl. Med., 2021, 19, 311 CrossRef CAS PubMed.
- I. G. Serebriiskii, V. Pavlov, R. Tricarico, G. Andrianov, E. Nicolas, M. I. Parker, J. Newberg, G. Frampton, J. E. Meyer and E. A. Golemis, Nat. Commun., 2022, 13, 1618 CrossRef CAS PubMed.
- S. Bueno-Fortes, J. K. Muenzner, A. Berral-Gonzalez, C. Hampel, P. Lindner, A. Berninger, K. Huebner, P. Kunze, T. Bäuerle, K. Erlenbach-Wuensch, J. M. Sánchez-Santos, A. Hartmann, J. De Las Rivas and R. Schneider-Stock, Cancers, 2021, 14, 136 CrossRef PubMed.
- S. Wang, K. Ma, C. Zhou, Y. Wang, G. Hu, L. Chen, Z. Li, C. Hu, Q. Xu, H. Zhu, M. Liu and N. Xu, Ther. Adv. Med. Oncol., 2019, 11, 1758835919843736 CrossRef CAS PubMed.
- S. Gillotin, Bio-Protoc., 2018, 8, e3035 CAS.
- E. Afgan, D. Baker, B. Batut, M. van den Beek, D. Bouvier, M. Čech, J. Chilton, D. Clements, N. Coraor, B. A. Grüning, A. Guerler, J. Hillman-Jackson, S. Hiltemann, V. Jalili, H. Rasche, N. Soranzo, J. Goecks, J. Taylor, A. Nekrutenko and D. Blankenberg, Nucleic Acids Res., 2018, 46, W537–W544 CrossRef CAS PubMed.
- A. M. Bolger, M. Lohse and B. Usadel, Bioinformatics, 2014, 30, 2114–2120 CrossRef CAS PubMed.
- D. Kim, B. Langmead and S. L. Salzberg, Nat. Methods, 2015, 12, 357–360 CrossRef CAS PubMed.
- Y. Liao, G. K. Smyth and W. Shi, Bioinformatics, 2014, 30, 923–930 CrossRef CAS PubMed.
- M. D. Robinson, D. J. McCarthy and G. K. Smyth, Bioinformatics, 2010, 26, 139–140 CrossRef CAS PubMed.
- S. X. Ge, D. Jung and R. Yao, Bioinformatics, 2020, 36, 2628–2629 CrossRef CAS PubMed.
- T. Bakheet, E. Hitti and K. S. A. Khabar, Nucleic Acids Res., 2018, 46, D218–d220 CrossRef CAS PubMed.
- B. Langmead and S. L. Salzberg, Nat. Methods, 2012, 9, 357–359 CrossRef CAS PubMed.
- F. Ramírez, D. P. Ryan, B. Grüning, V. Bhardwaj, F. Kilpert, A. S. Richter, S. Heyne, F. Dündar and T. Manke, Nucleic Acids Res., 2016, 44, W160–W165 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00212a |
| This journal is © The Royal Society of Chemistry 2025 |







