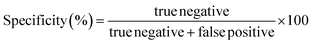Open Access Article
Open Access ArticleRapid quantification and classification of five vinca alkaloids used in cancer therapy by flow injection analysis with UV detection and chemometrics†
Eric
Caudron
 ab,
Cécile
Boughanem
a,
Marion
Berge
ab,
Cécile
Boughanem
a,
Marion
Berge
 ab,
Jehanne
Saidi
a,
Antoine
Dowek
ab,
Jehanne
Saidi
a,
Antoine
Dowek
 *ab and
Laetitia Minh Mai
Lê
ab
*ab and
Laetitia Minh Mai
Lê
ab
aPharmacy Department, Georges Pompidou European Hospital, AP-HP, Paris, France
bLip(Sys)2, Faculty of Pharmacy, Paris-Saclay University, Orsay, France. E-mail: antoine.dowek@universite-paris-saclay.fr
First published on 25th March 2025
Abstract
Vinca alkaloids represent a major class of antineoplastic agents used against cancer. They are prepared in centralized production units by pharmacy technicians. Control of the preparation is indispensable to secure their preparation and avoid any errors, which can have serious consequences for the patient because antineoplastic agents are very toxic. Analytical quality control was proven to be the most efficient control to ensure the right drug at the right dose to the patient. The study focused on vinca alkaloids: vinblastine, vincristine, vindesine, vinflunine, and vinorelbine, in the form of commercially diluted solutions in 0.9% NaCl at therapeutic concentrations. The aim of this study was to develop an analytical methodology for quality control capable of discriminating and quantifying these molecules. The primary objective was to assess the capability of Flow Injection Analysis with UV detection (FIA-UV), combined with chemometrics, for rapid classification and quantification of these alkaloids. A rapid High-Performance Liquid Chromatography with UV-visible detection (HPLC-UV) method was also developed and established as a reference standard. HPLC-UV discrimination was based on retention time, whereas FIA-UV relied on spectral analysis. Therefore, to improve discrimination in FIA-UV, Partial Least Squares Discriminant Analysis (PLS-DA) was incorporated. FIA-UV achieved 100% sensitivity and specificity in discriminating the five alkaloids, demonstrating non-inferiority to HPLC-UV. This method offers a streamlined workflow, reduces iatrogenic risk, and is well suited for antineoplastic agent preparation environments.
1. Introduction
Cancer chemotherapy is a treatment consisting of the administration of drugs for systemic use which have an action on tumor cells by killing them or preventing them from proliferating.Antineoplastics are considered high-risk drugs. They may have a narrow therapeutic range associated with high toxicity. Vinca-alkaloid drugs (vincristine, vinblastine, vinorelbine, vindesine, and vinflunine) hold a crucial place in the therapeutic arsenal against cancer.1 They are the earliest developed microtubule-targeting agents and approved for clinical use, and have been used as agents for many years in the treatment of such cancers as hematological, lymphatic neoplasms and solid tumors of the breast, lung, colorectal or urothelial cancers.2 Short and long-term side effects constitute a substantial drawback of vinca-alkaloids with high hematologic toxicities or peripheral neuropathy for example. Regular surveillance strategies are necessary and modification of the administered dose, use of alternative treatment or cessation of chemotherapy in case of severe, life-threatening toxicities seem to be the most effective strategies to control the adverse effects. Ensuring the quality of the preparations and limiting the risk of medication errors are of paramount importance in the hospital setting, where the accuracy of treatments is decisive. Safety measures have been taken at each stage of the chemotherapy circuit from prescription to administration. For many years, injectable chemotherapy treatments have been prepared in centralized production units under pharmaceutical responsibility.3–5 This centralized production in dedicated pharmaceutical units allowed safety in terms of healthcare exposure, management of good preparation practices, cost reduction and patient safety to guarantee the right drug at the right dose and at the right patient. Analytical quality control (AQC) of injectable preparations, performed prior to patient administration, ensures safety by verifying correct drug and dosage, and detecting preparation errors. This involves two primary objectives: precise quantification and accurate molecular discrimination. Because AQC is performed between the end of the preparation and the administration to the patient, rapid analytical techniques are essential to maintain workflow efficiency and avoid delaying chemotherapy. As a result, ultra-fast analytical methods for quality control are developed to cope with the high production flows. Among them, spectroscopic techniques such as UV-visible, infrared, or Raman have been applied to the quality control of cytotoxic drugs6–13 and were rapid approaches providing spectral information related to the structure of molecules from which discriminative analysis should be based. However, discriminating between molecules with very similar structures using UV-visible spectroscopy can be challenging.14,15 To address this challenge, chemometrics can be a valuable approach. Chemometrics involves the application of mathematical and statistical tools to chemical analysis. A prominent example is partial least squares (PLS) regression, a widely employed chemometric method for analyzing spectral data. Therefore, separation techniques can be associated to discriminate molecules by their retention time.16 The choice of a method must be carefully considered, taking into account its quality, reliability, and security.17,18 These approaches require a rigorous validation of the discrimination before its routine application to avoid the discrimination error corresponding to two situations. The first one corresponds to the non-identification of the wrong drug in the preparation with serious potential consequences for the patient. The second corresponds to a lack of identification of the drug in the preparation, leading to the destruction of the preparation with a significant financial impact regarding the high cost of the anticancer drugs.
In this context, the five commonly prescribed and approved for clinical use vinca-alkaloids were studied: vincristine, vinblastine, vinorelbine, vindesine and vinflunine. Their discrimination by the UV-visible spectrum is challenging due to the close structure of these molecules. Although several HPLC methods have been developed for the analysis for plant or plasma samples,19–21 they do not simultaneously analyze all five vinca alkaloids used in cancer therapy. More recently, a high-performance thin-layer chromatographic method was proposed for the quantification of vinca alkaloids;22 however, it requires a minimum analysis time of 30 minutes. These methods predominately focused on quantification, while neglecting the classification of the compounds. Therefore, two distinct analytical approaches using UV-visible spectroscopy were employed. First, a High-Pressure Liquid Chromatography technique with UV detection (HPLC-UV) was established as a reference method, achieving discrimination of the five vinca alkaloids through retention time analysis. Second, a Flow Injection Analysis method with UV detection (FIA-UV) was developed, utilizing spectral discrimination. FIA enables direct UV spectral measurement of molecules injected into a flow stream without prior separation, using the same materials as HPLC but without a column. These two techniques facilitate Analytical Quality Control (AQC) within a closed system, allowing pharmaceutical technicians to perform direct sampling from sealed vials within an isolator at the end of preparation, minimizing the risk of contamination to the user.
The objective of this study was to develop and assess two analytical methodologies, FIA-UV and HPLC-UV, for the simultaneous discrimination and quantification of the five vinca alkaloids. We then aimed to establish the non-inferiority of FIA-UV relative to HPLC-UV for the analytical quality control of chemotherapy preparations within a hospital setting.
2. Experimental
2.1. Chemicals
All the solvents are of analytical grade. Di-sodium hydrogen phosphate dihydrate (Na2HPO4, 2H2O) and sodium dihydrogen phosphate dihydrate (NaH2PO4, 2H2O) were obtained from Prolabo VWR (Fontenay-sous-Bois, France). Acetonitrile (ACN) was from Sigma-Aldrich (Saint-Louis, USA). Ultrapure water was obtained from a MiliQ® purification station (Millipore, USA). All solutions were filtered through a 0.45 μm membrane (Durapore® 0.45 μm HV, Merck, Darmstadt, Germany) before use.The following pharmaceutical products of the vinca-alkaloids are ready-to-use preparations and their chemical structures are shown in Fig. 1: vincristine sulfate at 2 mg mL−1 (VCST from Teva, France), vinflunine ditartrate at 25 mg mL−1 (VINF: Javlor®, Pierre Fabre, France) and vinorelbine ditartrate at 10 mg mL−1 (VINO: Navelbine®, Pierre Fabre, France). Vinblastine sulfate at 10 mg mL−1 (VBST: Velbe®, EG Labo, France) and vindesine sulfate at 1 mg mL−1 (VDSN: Eldisine®, EG Labo, France) were reconstituted in 0.9% sodium chloride aqueous solution (NaCl 0.9%) (FreeFlex®, Fresenius Kabi, France).
The excipients of pharmaceutical products were water for injection (VCST, VINF, VINO), sulfuric acid (VBST), sodium hydroxide (VCST) and mannitol (VCST, VBST, VDSN).
2.2. Instrumentation and analytical conditions
The analysis by FIA and by HPLC was carried out on two Ultimate 3000 LC® HPLC systems named “HPLC 1” and “HPLC 2” (Thermo-Fisher Scientific, USA) respectively. The detection was carried out using a UV-visible diode array detector (DAD) equipped with a detection cell with an optical path of 0.4 mm for HPLC 1 and 10.0 mm for HPLC 2. Spectral acquisitions were performed from 200 to 400 nm. HPLC 1 and HPLC 2 were used for HPLC or FIA analysis depending on the concentrations of therapeutic range to avoid sample dilution before analysis. Analysis and data collection were performed using Chromeleon® 7.2 software (Thermo-Fisher Scientific, USA).2.3. Sample preparation
Several commonly used vinca-alkaloids drugs have been prepared by diluting the stock solution (ready-to-use or extemporaneously reconstituted solution) with 0.9% chloride sodium at different concentrations corresponding to the therapeutic range: 0.030–0.120 mg mL−1 for VBST, 0.010–0.046 mg mL−1 for VCST, 0.015–0.070 mg mL−1 for VDSN, 2.0–6.0 mg mL−1 for VINF and 0.25–0.70 mg mL−1 for VINO (see ESI, Table S1†). Dilutions were made with a Microlab 600 series dilutor (Hamilton, Switzerland). All the samples were gathered in glass vials (Interchim, France) and stored at +4 °C before analysis.Three sets of data were acquired: i.e. the calibration set (n = 75, 15 for each molecule) corresponding to the calibration range to develop the quantification and discrimination models with 5 concentration levels, each range being performed every day for 3 consecutive days, (ii) an internal validation set (n = 135, 27 for each molecule) corresponding to the quality controls to validate and optimize the model. It includes 3 levels of concentration repeated 3 times, each sample being analysed each day for 3 consecutive days and (iii) an external validation set (n = 150, 30 for each molecule) to validate the predictive capacity of the quantification and discrimination models. This dataset includes true-life samples within the therapeutic range.
72 samples per vinca-alkaloid giving a total of 360 samples were analyzed by the two methods to assess the performance of FIA and HPLC methods for discriminative and quantitative analyses.
2.4. Data analysis
By FIA, the discriminant analysis was carried out using the open-source R Studio® software: R core team (2023) (R: a language and environment for statistical computing, Vienna, Austria), version 2023.06.0+421. The qualitative analysis was performed using a multivariate analysis approach. Various normalizations of spectra were considered: normalization by the mean spectrum, by the total area and by the absorbance at 269 nm. Partial Least Squares Discriminant Analysis (PLS-DA)23 was used to reduce the spectral data and thus differentiate the samples of the five vinca-alkaloids. Discriminant analysis, including PLS-DA, was conducted using the Caret24 and mdatools25 packages.
PLS-DA, a linear, supervised classification algorithm, enhances the differentiation of sample groups by maximizing the covariance between independent UV spectra (X) and dependent vinca alkaloid classes (Y). A linear subspace of the explanatory variables is identified, enabling the prediction of Y based on a reduced set of latent variables (LVs).26 To prevent the risk of overfitting, the optimal number of LVs is determined using the training set to achieve maximum prediction accuracy. A key advantage of PLS-DA lies in its ability to effectively handle highly collinear and noisy data, which are common characteristics of spectral measurements.27 To determine the optimal number of latent variables, leave-one-out cross-validation was performed on the internal validation set. The number of LVs yielding the highest accuracy, defined as the percentage of correctly classified samples, was selected.
After the selection of the spectral range and the pretreatment by the chemometric approach developed on R Studio®, the method was transposed to the spectral correlation algorithm of the Chromeleon® software. The degree of similarity (match) between two spectra was determined using numerical methods and subtle difference can be quantified allowing correct identification of the five vinca-alkaloids based on spectral matching. Several algorithms are suitable to quantify spectral similarity.28 Among them, the least square matching method is widely available on HPLC software. This method yields the sum of the squared absorbance deviations at each wavelength to determine an average square deviation between the reference and unknown spectra. This step-by-step wavelength analysis offers precision to measure the similarity between two spectra. For each vinca-alkaloid, 15 spectra of the calibration set were recorded in the library spectra as reference spectra. For each unknown sample, the UV spectrum is compared to the set of 75 spectra recorded in the spectral library on a scale from 0 to 1000. A match score varying from 0 (no match) to 1000 (perfect match) is calculated according to the similarity between the unknown spectrum and the library spectra.
The closer the score obtained is to 1000, the greater the probability of the sample matching the molecule identified by the software. The performance of the model was evaluated using a confusion matrix constructed from the first likelihood score of each sample in the external validation set.
The performance parameters corresponding to the sensitivity, specificity and accuracy were evaluated through a matrix of confusion carried out on the external validation set. Sensitivity is the ability of a method to correctly classify a positive as a positive sample. Specificity measures the ability of a method to correctly classify a negative sample as negative. Then, accuracy assesses the overall performance of the method. They are calculated as follows:
In addition, on this external validation set and for each sample, the relative error was calculated between the concentration measured by analytical methods and the theoretical concentration. The relative error on the concentrations predicted to release a chemotherapy preparation was routinely set at ±15%. This interval was established based on the variability inherent in the diluent infusion bag filling (5% dextrose or 0.9% sodium chloride), the precision of the syringe, and the analytical error associated with the method.
3. Results and discussion
A total of 72 samples per vinca-alkaloid corresponding to 360 samples were analyzed by HPLC and FIA methods.3.1. Discriminant analysis
Two analytical strategies have been studied to allow the discrimination of vinca-alkaloids by a separative technique based on retention times and by a non-separative technique based on a multivariate analysis of spectral data by chemometrics.Based on the calibration set data, specific ranges of retention times were defined for each drug to guarantee their discrimination. As shown in Fig. 2, the order of the five vinca-alkaloids elution was VDSN (0.913 ± 0.001 min), VCST (1.076 ± 0.002 min), VINO (1.251 ± 0.034 min), VBST (1.570 ± 0.004 min) and VINF (1.602 ± 0.007 min). Despite close retention times between VBST and VINF, their retention times were significatively different at 5% (p < 0.001).
 | ||
| Fig. 3 Mean normalized UV spectra at 269 nm from 220 to 400 nm after analysis by FIA (A) and HPLC (B) for the five vinca-alkaloids. | ||
To develop discriminative analysis, the spectral data were pretreated to overcome variations linked to concentration. The spectral region between 200 and 220 nm exhibiting a non-specific absorption range with random spectral variations was excluded. The spectra were then normalized by the spectral band at 269 nm to limit the intra-group variations linked to the concentration (Fig. 3).
The discriminant analysis was performed by a method supervised using PLS-DA to separate the five vinca-alkaloids. The best predictive model was obtained after normalization by the wavelength at 269 nm from 220 to 400 nm. Five latent variables (LVs) were selected to develop the model and represent 99.98% of the total variability of the original variables (98.0% for the first LV, 1.6% for the second) (Fig. 4). Despite achieving 99% explained variance with the initial two latent variables (LVs), five LVs were necessary for accurate classification. This finding is consistent with the hypothesis that five LVs are needed to differentiate five distinct molecules. All unknown samples from the external validation set were correctly assigned with 100% accuracy, sensitivity and specificity.
 | ||
| Fig. 4 Score plot of the PLS-DA model for samples of the five vinca alkaloids after FIA analysis (normalization at 269 nm, spectra from 220 nm to 400 nm). | ||
The discriminant analysis by the least square matching method was then based on the results of PLS-DA, and qualitative models of discrimination were developed using Chromeleon® software for each vinca alkaloid.
In Table 1, very high match scores were obtained for each of the five vinca-alkaloid samples analyzed under their own analytical conditions with values >999.0. From this experiment, we concluded the excellent relevance of the highest match score. As a first step, a threshold limit is determined for the processing of the least square matching method. Indeed, as a matching is operated from the spectral library, a limit to specify whether the sample is accepted or rejected is obvious. The value of the threshold was set at 998.0 for the five vinca-alkaloids. As a second step, the qualitative parameters of the method were assessed through specificity and sensitivity with an external validation set of vinca-alkaloids (n = 150). The set was analyzed in parallel by the chromatographic method (discrimination by the retention time) and no discordant result was obtained between the two methods and excellent (100%) specificity and sensitivity were achieved under these conditions leading to excellent accuracy (100%).
3.2. Quantitative analysis
A convenient calibration process was carried out for the five vinca-alkaloids with the FIA method and HPLC method (ESI, Fig. S1†). These distinct analytical conditions were validated in terms of linearity, accuracy, and precision according to ICH requirements (Q2R1). Based on the FIA results, the linearity of each of the five analytical conditions was very satisfactory (R2 > 0.997) under the FIA conditions. Satisfactory RSDs of both the repeatability and the precision were reached with values <0.8% and 2.3% respectively (ESI, Table S2†).From these results, we showed that FIA and HPLC conditions provided accurate quantification of the five vinca-alkaloids.
3.3. Comparison of FIA and HPLC
To demonstrate the transposability of the FIA and HPLC methods and the applicability of the FIA spectral method coupled with chemometric analysis, samples of the external validation set were analyzed with FIA and HPLC procedures. Using the Bland–Altman plot (Fig. 5), the five vinca alkaloid drugs satisfied the predefined limits for SE as well as TE and the methods were interchangeable.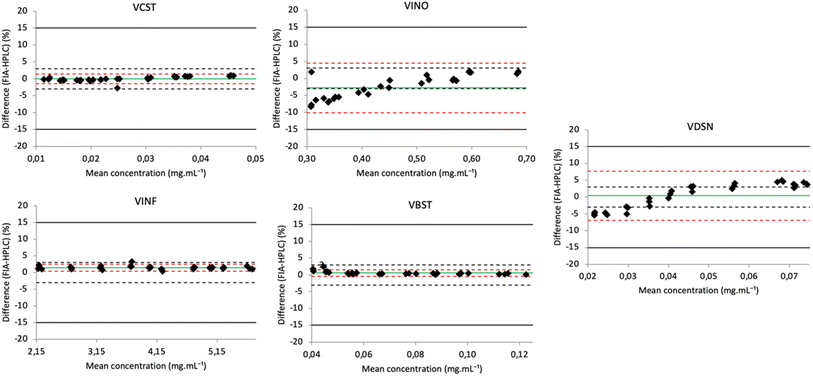 | ||
| Fig. 5 Bland–Altman plot showing the relative difference between FIA/UV and HPLC/UV (UV) against the mean predicted concentration for the external validation set. The green dotted line is the bias (d), red dashed lines are the limits of agreement (±1.96SD) and the dark dashed and dotted lines are respectively the predefined limits of systematic error (±SE) and total error (±TE). The advantages and drawbacks of the methods and the comments are summarized in Table 2. | ||
| FIA | HPLC | ||
|---|---|---|---|
| Optimization | Chemometric optimization by PLS-DA or by the least square matching method | Chromatographic optimization (resolution and retention time) | |
| Short time and easy optimization | Long time and tedious optimization | ||
| Easy to apply in current practice | Relatively easy to apply in current practice | ||
| In current practice | Time of analysis: 30 s | Time of analysis: 2 min | |
| HPLC system | HPLC system | ||
| No consumable | Chromatographic column | ||
| Solvent: ultrapure water | Solvent: buffer and organic | ||
| Low cost | Medium cost | ||
| Environmental footprint | Low | Medium (organic solvent) | |
| Robustness | Risk of low spectral shift | Alteration of chromatographic support (peak broadening, retention time variations…) | |
| Risk of variation of the UV lamp intensity | Risk of variation of the UV lamp intensity | ||
| Global cost | Equipment | Expensive | Expensive |
| Consumable | Cheap | Relatively expensive | |
4. Conclusions
This research achieved the development of two analytical techniques for the classification and quantification of five vinca alkaloids, critical in cancer treatment. HPLC-UV was established as the reference method for comparison with FIA-UV. FIA-UV exhibited excellent accuracy in both classification and quantification, demonstrating performance comparable to HPLC-UV. The FIA method provides several benefits over HPLC, such as faster processing, lower costs, reduced environmental burden, and ease of routine use, making it highly suitable for integration into anticancer drug preparation workflows.Data availability
Data, i.e. raw spectra and R code are available at: https://doi.org/10.5281/zenodo.14924579 (version 2).Author contributions
Conceptualization E. Caudron and L. M. M. Lê; data curation: E. Caudron, A. Dowek, M. Berge, C. Boughanem and L. M. M. Lê; formal analysis: C. Boughanem; investigation: E. Caudron, C. Boughanem and L. M. M. Lê; methodology: E. Caudron, C. Boughanem and L. M. M. Lê; project administration: E. Caudron and L. M. M. Lê; resources: E. Caudron, J. Saidi and L. M. M. Lê; software: C. Boughanem, A. Dowek and M. Berge; supervision: E. Caudron and L. M. M. Lê; validation: E. Caudron, C. Boughanem and A. Dowek; visualization: E. Caudron, C. Boughanem, L. M. M. Lê and A. Dowek; roles/writing – original draft: C. Boughanem, E. Caudron; and writing – review & editing: C. Boughanem, E. Caudron, A. Dowek, M. Berge and L. M. M. Lê.Conflicts of interest
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.Notes and references
- A. Banyal, S. Tiwari, A. Sharma, I. Chanana, S. K. S. Patel, S. Kulshrestha and P. Kumar, 3 Biotech, 2023, 13, 211 Search PubMed.
- E. Martino, G. Casamassima, S. Castiglione, E. Cellupica, S. Pantalone, F. Papagni, M. Rui, A. M. Siciliano and S. Collina, Bioorg. Med. Chem. Lett., 2018, 28, 2816–2826 Search PubMed.
- J. Cazin and P. Gosselin, Pharm. World Sci., 1999, 21, 177–183 Search PubMed.
- F. Drapeau, G. Claustre, S. Gaimard and C. Rossard, Bull. Cancer, 2023, 110, 301–307 CrossRef PubMed.
- American Society of Health System Pharmacists, Am. J. Health Syst. Pharm., 2000, 57(12), 1150–1169 CrossRef.
- F. Dziopa, G. Galy, S. Bauler, B. Vincent, S. Crochon, M. L. Tall, F. Pirot and C. Pivot, J. Oncol. Pharm. Pract., 2013, 19, 121–129 Search PubMed.
- A. A. Makki, S. Elderderi, V. Massot, R. Respaud, H. J. Byrne, C. Tauber, D. Bertrand, E. Mohammed, I. Chourpa and F. Bonnier, Talanta, 2021, 228, 122137 CrossRef CAS.
- A. A. Makki, V. Massot, H. J. Byrne, R. Respaud, D. Bertrand, E. Mohammed, I. Chourpa and F. Bonnier, Vib. Spectrosc., 2021, 113, 103200 CrossRef CAS.
- L. Lê, M. Berge, A. Tfayli, P. Prognon and E. Caudron, BioMed Res. Int., 2018, 2018, 1–7 CrossRef.
- L. M. M. Lê, M. Berge, A. Tfayli, J. Zhou, P. Prognon, A. Baillet-Guffroy and E. Caudron, Eur. J. Pharm. Sci., 2018, 111, 158–166 Search PubMed.
- Y. Lee, J. Kim, H. Jeong and H. Chung, Appl. Spectrosc. Rev., 2023, 58, 509–524 Search PubMed.
- F. Nardella, M. Beck, P. Collart-Dutilleul, G. Becker, C. Boulanger, L. Perello, A. Gairard-Dory, B. Gourieux and G. Ubeaud-Séquier, Int. J. Pharm., 2016, 499, 343–350 CrossRef CAS PubMed.
- I. Bennani, M. A. Chentoufi, A. Cheikh, M. E. Karbane and M. Bouatia, J. Oncol. Pharm. Pract., 2021, 27, 99–107 Search PubMed.
- A. Dowek, M. Annereau, L. Denis, T. Fleury, E. Daguet, H. Aboudagga, A. Rieutord, E. Caudron and L. Lê, Microchem. J., 2024, 205, 111285 CrossRef CAS.
- L. M. H. Reinders, M. D. Klassen, C. vom Eyser, T. Teutenberg, M. Jaeger, T. C. Schmidt and J. Tuerk, Anal. Bioanal. Chem., 2021, 413, 2587–2596 CrossRef CAS.
- A. Delmas, J. B. Gordien, J. M. Bernadou, M. Roudaut, A. Gresser, L. Malki, M. C. Saux and D. Breilh, J. Pharm. Biomed. Anal., 2009, 49, 1213–1220 CrossRef CAS PubMed.
- M. Savelli, M. Roche, C. Curti, C. Bornet, P. Rathelot, M. Montana and P. Vanelle, Pharmazie, 2018, 73, 251–259 CAS.
- C. Bazin, B. Cassard, E. Caudron, P. Prognon and L. Havard, Int. J. Pharm., 2015, 494, 329–336 Search PubMed.
- O. van Tellingen, J. H. Beijnen and W. J. Nooyen, J. Pharm. Biomed. Anal., 1991, 9, 1077–1082 Search PubMed.
- M. De Smet, S. J. P. Van Belle, G. A. Storme and D. L. Massart, J. Chromatogr. B Biomed. Sci. Appl., 1985, 345, 309–321 CrossRef CAS PubMed.
- Z. Liu, H.-L. Wu, Y. Li, H.-W. Gu, X.-L. Yin, L.-X. Xie and R.-Q. Yu, J. Chromatogr. B, 2016, 1026, 114–123 Search PubMed.
- A. Paci, L. Mercier and P. Bourget, J. Pharm. Biomed. Anal., 2003, 30, 1603–1610 Search PubMed.
- M. Barker and W. Rayens, J. Chemom., 2003, 17, 166–173 CrossRef CAS.
- M. Kuhn, J. Stat. Softw., 2008, 28, 1–26 Search PubMed.
- S. Kucheryavskiy, Chemom. Intell. Lab. Syst., 2020, 198, 103937 CrossRef CAS.
- S. Wold, M. Sjöström and L. Eriksson, Chemom. Intell. Lab. Syst., 2001, 58, 109–130 CrossRef CAS.
- P. S. Gromski, H. Muhamadali, D. I. Ellis, Y. Xu, E. Correa, M. L. Turner and R. Goodacre, Anal. Chim. Acta, 2015, 879, 10–23 Search PubMed.
- P. R. Griffiths and L. Shao, Appl. Spectrosc., 2009, 63, 916–919 Search PubMed.
- P. Borman and D. Elder, in ICH Quality Guidelines, John Wiley & Sons, Ltd, 2017, pp. 127–166 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ay00325c |
| This journal is © The Royal Society of Chemistry 2025 |