Open Access Article
Open Access ArticleValidating phosphoethanolamine modification as a potential spectral marker of colistin resistance†
Dimple
Saikia
 a,
Cebajel Bhanwarlal
Tanan
a,
Cebajel Bhanwarlal
Tanan
 a,
G.
Dhananjaya
b,
Basavraj S.
Hungund
a,
G.
Dhananjaya
b,
Basavraj S.
Hungund
 c,
Nilkamal
Mahanta
b and
Surya P.
Singh
c,
Nilkamal
Mahanta
b and
Surya P.
Singh
 *a
*a
aDepartment of Biosciences and Bioengineering, Indian Institute of Technology Dharwad, Dharwad 580011, India. E-mail: ssingh@iitdh.ac.in
bDepartment of Chemistry, Indian Institute of Technology Dharwad, Dharwad 580011, India
cDepartment of Biotechnology, KLE Technological University, Hubballi, 580031, India
First published on 18th November 2024
Abstract
The antibiotic colistin is regarded as the final line of defense for treating infections caused by Gram-negative bacteria. The combination of Raman spectroscopy (RS) with diverse machine learning methods has helped unravel the complexity of various microbiology problems. This approach offers a culture-free, rapid, and objective tool for identifying antimicrobial resistance (AMR). In this study, we employed the combinatorial approach of machine learning and RS to identify a novel spectral marker associated with phosphoethanolamine modification in the lipid A moiety of colistin-resistant Gram-negative Escherichia coli. The visible spectral fingerprints of this marker have been validated using partial least squares regression and discriminant analysis. The origin of the spectral feature was confirmed through hyperspectral imaging and K-means clustering of a single bacterial cell. The chemical structure of the modified lipid A moiety was verified by employing gold standard MALDI-TOF mass spectrometry. Our findings support the futuristic applicability of this spectroscopic marker in objectively identifying colistin-sensitive and -resistant strains.
Introduction
Antimicrobial resistance (AMR) associated health crises are increasing silently and require immediate attention. It is estimated that if no proper actions are taken, it could cause 10 million deaths by 2050 and 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sepsis deaths in India alone.1 As per a 2020 report by the Department of Biotechnology (DBT), India, and the World Health Organization, colistin is the last resort antibiotic for tackling Gram-negative resistant infections. Colistin, comprising of cyclic decapeptides linked to a fatty acid chain, is used for treating Gram-negative infections.2 The increased risk of antibiotic resistance towards colistin from critical priority pathogens, such as E. coli, poses a potential risk to public health.3
000 sepsis deaths in India alone.1 As per a 2020 report by the Department of Biotechnology (DBT), India, and the World Health Organization, colistin is the last resort antibiotic for tackling Gram-negative resistant infections. Colistin, comprising of cyclic decapeptides linked to a fatty acid chain, is used for treating Gram-negative infections.2 The increased risk of antibiotic resistance towards colistin from critical priority pathogens, such as E. coli, poses a potential risk to public health.3
Lipopolysaccharides (LPSs), molecules embedded in the outer membrane of E. coli, have three major components: O-polysaccharide (O-antigen), core oligosaccharide, and a lipophilic anchor part “lipid A”. A wide variety of modifications, such as acylation, phosphorylation, and alteration of phosphoethanolamine (PEA), in the lipid A of Gram-negative bacteria have been reported.4 V. Sándor et al. reported different structures of lipid A, which could be present in E. coli. The mobile colistin resistance gene (mcr-1) codes for a transferase enzyme belonging to the family of phosphoethanolamine (PEA) transferases. Resistance to colistin primarily arises from the alteration of the lipid A moiety of LPS. This modification leads to the replacement of phosphate groups of lipid A with 4′-phosphoethanolamine (PEtN) and/or cationic 4-amino-4-deoxy-L-arabinose (L-Ara4N). It leads to a decrease in the net negative charge of the LPS membrane due to blocking of phosphate and carboxyl groups, where colistin binds for its antibacterial action.5–7
The excessive use and misuse of antibiotics have led to increased resistance to nearly all antibiotics, posing a significant threat to last-resort antibiotics, such as carbapenems and colistin/polymyxins.8–10 Conventional methods to identify antimicrobial resistance usually take 2–3 days because of the involvement of long incubation hours and labour-intensive antibiotic susceptibility tests (AST), such as disk diffusion, e-test, or broth microdilution.11 The long waiting time results in more suffering for patients and increases the chances of contagion. This prompts healthcare professionals to use inappropriate antibiotics or sometimes prescribe broad-spectrum antibiotics, which further add to the problem of misuse of antibiotics. E. coli is known to be a highly evolved Gram-negative bacteria that tends to become resistant to multiple classes of antibiotics, including colistin.7 The successful identification of a marker that can help discern sensitive and resistant strains in a non-destructive manner can revolutionize the early screening of colistin resistance, fundamentally altering the current workflow of ASTs.
The utility of Raman spectroscopy (RS) for microbiology applications such as identification, characterization, and tracking of metabolites is widely reported due to attributes such as label-free, fast, culture-free, and objectivity.12,13 Since Raman spectral data consists of a large number of variables, it can be efficiently combined with different machine-learning (ML) modules to help overcome the limitations of traditional decision-making approaches and enable the forecasting of information that would otherwise be beyond the capabilities of traditional approaches. RS coupled with ML can provide fast, effective, and objective analysis of high dimensional datasets and help in identifying complex information and relationships among the data depending upon specific functional groups and compounds.13,14 This exploratory study was conducted by utilizing the exquisite molecular specificity of Raman spectroscopy in combination with different machine learning techniques to identify the alterations in the lipid A fragment of colistin-resistant E. coli as a novel spectral marker. The numerical characteristics from RS profiles were utilized to extract the features from colistin-resistant microorganisms and were complemented with gold-standard mass spectrometry (MS).
Results and discussion
Molecular weight characterization
The extracted lipid from the outer membrane of colistin-resistant and sensitive strains was analysed by MALDI-TOF-MS to confirm the chemical modifications linked to colistin resistance (mcr-1 positive) in comparison to the wild-type sensitive strain. The typical mass-to-charge ratio (m/z) for lipid A from E. coli generally falls within the range of 1700 to 2000 daltons.17Fig. 1A represents the protonated molecular ion [M + H]+ at 1746 Da, corresponding to hexa-acylated monophosphorylated lipid A from the colistin-sensitive strain. The proposed structure suggests the presence of a GlcN disaccharide backbone having six acyl chains of 14 carbons [C4×3-phosphate, C3′–C14:O(3-O-C14:O), C2′–C14:O(3-O-C14:O), C3–C14:O(3-OH), C2–C14:O(3-OH), C1-OH] (3).18,19 Colistin-resistant pathogenic bacteria modify their lipid A to survive antibiotic stress by appending phosphoethanolamine (PEA) to it. Fig. 1B represents the protonated molecular ion [M + H]+ at 1871 Da from the resistant strain containing the modified phosphoethanolamine group at the C1 position of the lipid A structure with a mass difference of 125 Da, which corresponds to the mass of the PEA group. On comparison of both the sensitive and resistant strain lipid A MALDI-TOF-MS data, we saw that the addition of PEA in the resistant strain imparts antibiotic-resistant activity. These results validated the successful generation of mcr-1-positive colistin-resistant strains. | ||
| Fig. 1 Lipid-A structural difference between the sensitive (A) and resistant strains (B) using MALDI-TOF-MS; PEA addition in the resistant strain is indicated by the red rectangle. | ||
Analysis of the average Raman spectral profiles of sensitive and resistant strains
The primary focus of this study was to demonstrate the applicability of Raman spectroscopy coupled with machine learning to identify spectral markers associated with colistin resistance. In the next step, Raman spectra from sensitive and resistant cells were acquired. To avoid any spectral differences caused by cell number and growth phase variations, spectra were acquired by using an equal number of cells collected from the same growth phase.20 Spectra were recorded for both colistin-sensitive and resistant strains, treated with colistin concentrations of 1.2, 3.9, 5, 6.5, and 7.8 μg mL−1. The normalized average spectra from E. coli (sensitive) and resistant isolates are shown in Fig. 2A. The shaded area indicates the standard deviation across four independent experiments. Another strain of colistin-resistant A. baumannii was also tested to establish the generalized applicability of the proposed spectral markers, Fig. 2B. | ||
| Fig. 2 The average Raman spectra of ampicillin-resistant (Amp), sensitive (control) and colistin-treated resistant isolates of (A) E. coli and (B) A. baumannii. | ||
The average spectra from E. coli cells resistant to the antibiotic ampicillin have been used as a control to verify the molecular specificity of Raman spectroscopy. Like colistin, the resistance mechanism for ampicillin also involves the secretion of the enzyme ‘β-lactamase’, which interferes with the binding of the antibiotic to the penicillin-binding proteins (PBPs) involved in the cell wall biosynthesis. The spectral peaks at 757 cm−1 (phosphoethanolamine), 1130 and 1181 cm−1 (C–C skeletal of the acyl backbone in lipids) were observed exclusively in spectra collected from colistin-resistant cells (both E. coli and A. baumannii), Fig. 2. Colistin resistance is often accompanied by lipid A modifications and lengthening of the acyl backbone in highly resistant strains.15,23,24,26 In addition to these exclusive features (associated with colistin resistance), typical Raman signals such as 785 cm−1 (nucleic acids), 1004 cm−1 (phenylalanine), 1094 cm−1 (DNA/RNA), 1245 cm−1 (amide III), 1337 cm−1 (DNA/RNA), 1440 cm−1 (C–H stretch), 1580 cm−1 (DNA/RNA) and 1659 cm−1 (amide I) were also observed.22,23 Most of the signals were found to be superimpositions from similar compounds for each group. Additionally, the spectral features corresponding to glycogen and various amino acids (proline, valine, hydroxyproline) in the below 1000 cm−1 region were found to be strong in resistant cells.15,21 Colistin-resistant A. baumannii resistant isolates showed minor changes in the spectra around 840 cm−1 to 890 cm−1 (acyl chain) and 941 cm−1 (polysaccharide), which could be due to additional glycolipids present in the outer membrane.21 As colistin resistance is known to be associated with the thickening of bacterial capsules via the deposition of polysaccharides, overproduction of the outer membrane vesiclesand additional lipopolysaccharides to change the charge from anionic to zwitterionic.25–28 This is spectrally corroborated by the presence of strong features at 860, 928, and 1103 cm−1 (attributed to lipopolysaccharides). The visible changes in the shape of the amide I band in the resistant strains can be attributed to the structural modifications associated with the induction of resistance, which includes the thickening of the cell wall and changes in the cell shape.29 Our previous study has demonstrated that colistin-resistant E. coli cells tend to acquire a circular shape in contrast to rod-shaped sensitive strains. Also, the induction of resistance leads to a reduction in the membrane fluidity and an increase in surface roughness.20,29,30 The absence of bands associated with cell wall modifications (lipid A) in sensitive and ampicillin control strains supports the exquisite molecular specificity of Raman spectroscopy in identifying chemical modifications associated with colistin resistance.
Feature extraction and regression analysis
Any differences due to independent experiments were evaluated for each strain using principal component analysis (PCA) by combining the experiments into two sets. The PCA score plot obtained from the first two PCs clearly demonstrated the minimal variations across experiments, Fig. S5.† In order to screen specific predictors that can assist in distinguishing resistant and sensitive strains from the pool of spectral features in the 600–1800 cm−1 spectral range, we performed PLS-DA and PLS-regression. Fig. 3A and B show the scatter plot generated from latent variables and the corresponding PLS-DA coefficients, respectively. A clear separation of the sensitive and resistance strains was observed in the scatter plot generated by the score of Factor 1 and Factor 2 of the PLS-DA model. A few important spectral features that could be responsible for the classification are marked in the coefficients shown in Fig. S2†. The visible separation among the spectra appears to be contributed by both factors. A minor misclassification between sensitive and resistant strains treated with low colistin concentration was observed, which could be due to variability across the independent measurements. The spectral features contributing to this classification were further confirmed by the coefficient plot, Fig. 3B. It suggests that the most important variables that were positively correlated to the prediction in the PLS model were the wavenumbers 757 cm−1 (phosphoethanolamine), 980 cm−1 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH bending from lipids), and 1131 cm−1 (lipids). These peaks were found to be strong in the case of high-concentration colistin-treated groups (6.5 and 7.8 μg mL−1). The origin of these bands can be attributed to the earlier-mentioned phenomenon involving the addition of phosphoethanolamine to lipid A or the deposition of extra lipopolysaccharide in the outer membrane to block the interaction between the free negatively charged phosphate groups.31 The successful demonstration of the exclusive contribution of the resistance-associated spectral peaks in the discrimination of sensitive and resistant strains is important with respect to the prospective adaptation of this method in the form of an automated resistance monitoring system. In the next step, to explore the relationship between the resistance-induced spectral features and colistin concentration, a PLS regression model was developed. The primary goal of this model was to predict the antibiotic concentrations using Raman spectra as an input. The model performance was evaluated by different parameters, such as MSE (mean square error), R2 score, RMSE, and MAE (mean absolute error). The MSE measures the mean of the squared differences between the predicted and actual values, quantifying the overall accuracy of the model. The lower the MSE, the better the performance of the predicted model. The RMSE provides the average deviation between the predicted and actual values and helps to interpret the overall model accuracy; MAE represents the average of absolute differences between predicted and actual values rather than squaring them, unlike RMSE. The R2 value explains the proportion of variance in the response values explained by the predicted values, indicating the goodness of individual predictions. R2 value of 1 indicates perfect fit, and 0 indicates poor predictions.33,34 The value of these parameters is shown in Table 1.
CH bending from lipids), and 1131 cm−1 (lipids). These peaks were found to be strong in the case of high-concentration colistin-treated groups (6.5 and 7.8 μg mL−1). The origin of these bands can be attributed to the earlier-mentioned phenomenon involving the addition of phosphoethanolamine to lipid A or the deposition of extra lipopolysaccharide in the outer membrane to block the interaction between the free negatively charged phosphate groups.31 The successful demonstration of the exclusive contribution of the resistance-associated spectral peaks in the discrimination of sensitive and resistant strains is important with respect to the prospective adaptation of this method in the form of an automated resistance monitoring system. In the next step, to explore the relationship between the resistance-induced spectral features and colistin concentration, a PLS regression model was developed. The primary goal of this model was to predict the antibiotic concentrations using Raman spectra as an input. The model performance was evaluated by different parameters, such as MSE (mean square error), R2 score, RMSE, and MAE (mean absolute error). The MSE measures the mean of the squared differences between the predicted and actual values, quantifying the overall accuracy of the model. The lower the MSE, the better the performance of the predicted model. The RMSE provides the average deviation between the predicted and actual values and helps to interpret the overall model accuracy; MAE represents the average of absolute differences between predicted and actual values rather than squaring them, unlike RMSE. The R2 value explains the proportion of variance in the response values explained by the predicted values, indicating the goodness of individual predictions. R2 value of 1 indicates perfect fit, and 0 indicates poor predictions.33,34 The value of these parameters is shown in Table 1.
| PLS-R | MSE | RMSE | MAE | R 2 |
|---|---|---|---|---|
| All concentrations | 1.35 | 1.16 | 0.97 | 0.89 |
| Without 1.2 μg mL−1 | 0.81 | 0.86 | 0.80 | 0.92 |
The regression model was also tested with independent test data, as shown in Fig. 3C. The R2 score of 0.89 was obtained, indicating a strong relationship between specific spectral variations and colistin concentrations. A closer look at the correlation plot suggests that most of the wrong predictions of this model belong to the low-concentration resistance group, specifically, cells treated with the below MIC concentration (1.2 μg mL−1) of colistin.
Removing this group from the regression analysis improves the overall prediction accuracy of the model significantly (R2: 0.92; MSE: 0.81; RMSE 0.86; MAE: 0.80), Table 1. The primary reason for this could be attributed to the fact that cells begin to acquire specific morphological and biochemical characteristics associated with resistance at higher concentrations. This leads to the origin of specific spectral signatures that can help objectively identify colistin-resistant strains. To unravel these spectral signatures, a heat map was generated using pooled normalized spectra obtained from each group of E. coli cells sensitive to colistin, resistant to low colistin-treated concentrations (1.2, 3.9 μg mL−1), and high colistin-treated concentrations (6.5, 7.8 μg mL−1) in the 600–1800 cm−1 range. The heat map was compared to the regression vector to analyse the presence or absence of resistance-specific spectral signatures. As shown in Fig. 3D, peaks such as 757 cm−1, 885 cm−1, 935 cm−1, 981 cm−1, 1095 cm−1, 1124 cm−1, and 1131 cm−1 were prevalent in resistant strains (indicated by bright yellow color). Among these spectral features, the features around 757 cm−1 and 1131 cm−1 were prominent for colistin-resistant groups but were found to be minimal for the sensitive group. These peaks were also present in the regression vector, suggesting their direct involvement in accurately identifying antibiotic concentrations. The findings suggest that the correlation between the intensity of these bands and antibiotic concentrations is not spurious but is due to the spectral peaks related to phosphoethanolamine modification in lipid A for colistin resistant strains.
Localizing the origin of spectral signals
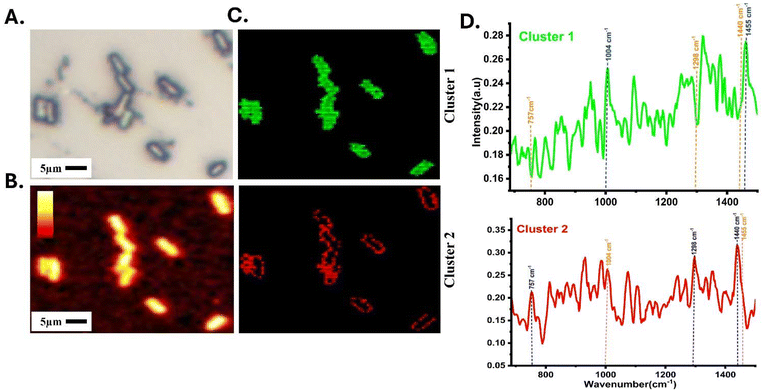
The origin of the spectral signals was validated through hyperspectral imaging of a single colistin-resistant bacterium. The superimposed Raman and optical image is shown in Fig. S6.†Fig. 4A shows the white light optical image of the scanned area with multiple bacteria. The Raman image was constructed after noise removal and putting an intensity filter at the C–H signal (2954 cm−1) for visualization and marking the map boundaries to verify the presence of the hidden biomolecular signals, Fig. 4B. The K-means clustering (K = 3) was performed on the scanned data to mine the biomolecular spectral components. This algorithm performs the calculation for each cluster using a centroid and averages the points within that cluster. This process recalculates the centroids until the centroids stop changing further, resulting in final clusters.38The centroid spectral data extracted from each cluster were evaluated for further understanding of the origin of spectral signatures, Fig. 4D. In cluster 1 (green), it was observed that the spectral wavenumber related to protein 1004 cm−1 and 1455 cm−1 (C–C skeletal stretching of the aromatic ring and CH2/CH3 protein deformation) were found to be strong, suggesting that cluster 1 primarily represents the cytoplasmic region of the cells. However, in cluster 2, it was observed that there was a drastic reduction of peaks at 1004 cm−1 and 1455 cm−1 (shown by an orange dotted line) and any other protein-associated signals. In contrast to cluster 1 (green), cluster 2 (red) had highly intense lipid bands at 1440 cm−1 and 1298 cm−1 (CH2 deformation from lipids and acyl chains) (shown by a blue dotted line), confirming that cluster 2 is a combination of mostly from the outer region of the bacterial cells, which is dominated by lipid bilayer along with some membrane proteins and glycoproteins.35,37
After careful observation of the spectral components of clusters 1 and 2, it was confirmed that they correspond to the cytoplasmic and cell boundary regions of the bacterium. Further, the location for the peak of interest, 757 cm−1, was evaluated. It was found that a highly intensified signal at 757 cm−1 was present in cluster 2 (marked in blue) and a negligible signal in cluster 1 (marked in orange), indicating that phosphoethanolamine is localized in the cell boundary region (cluster 2) of the bacterial cells, Fig. 4D. Cluster 3 (black) has spectral data from the background/substrate, as shown in supplementary (Fig. S3†). Other prominent spectral peaks from cluster 1 (cytoplasm) were at 785, 1094, 1250, and 1320 cm−1 from DNA/RNA molecules. In contrast, in the spectra from cluster 2 (bacterial envelope), major peaks were observed at 1078, 1100, 1130, 1168, and 1298 cm−1, originating from the phospholipids and acyl chains of lipid A.32 We also recorded the Raman spectrum from commercially available purified phosphoethanolamine. As shown in the supplementary (Fig. S4†), a prominent band at 757 cm−1 was observed. These findings further confirmed that the unique spectral features associated with lipid A modifications were primarily localized in the outer membrane of the colistin-resistant bacteria that could be successfully identified using Raman imaging.
Optical spectroscopic approaches, specifically Raman-based methods, have demonstrated significant potential as adjunct tools for different kinds of microbiology applications. As colistin resistance is associated with specific chemical changes in the outer membrane of the bacterium, we aimed to identify specific markers associated with these modifications. The results indicate that Raman spectroscopy can specifically detect spectral changes associated with phosphoethanolamine appendage in lipid A for colistin-resistant strains.
Hyperspectral imaging revealed that the spectral features specific to colistin resistance originated from the outer membrane of the bacterial cells. At higher antibiotic concentrations, the prediction accuracy of the regression models was found to be very good (R2 = 0.92). However, the colistin resistance mechanism also involves the overexpression of polysaccharides and charge-based alteration in lipid A via 4-amino-L-arabinose (L-Ara4N) and galactosamine.36 Theproposed approach using Raman spectroscopy opens the possibility to explore these resistance mechanisms in future studies. The overall findings suggest the prospective adaptation of this combinatorial approach of Raman spectroscopy and machine learning to monitor the specific markers associated with colistin resistance.
Experimental
Microbial strains
The laboratory-grown colistin-resistant strains of E. coli were created by mobile colistin resistance (mcr-1) gene transformation. This was followed by treating the transformed strains with serially higher concentrations of colistin till a stable clone was obtained. The minimum inhibitory concentration (MIC) of 1.9 μg mL−1 determined by the broth microdilution method for the wild-type E. coli Fig. S1†. Raman spectra were collected from concentrations below, intermediate and above the MIC value to monitor the spectral variations associated with colistin resistance. A single colony of colistin-resistant and sensitive strains was inoculated in an LB-broth culture medium with appropriate antibiotic concentrations of 1.2, 3.9, 5, 6.5, and 7.8 μg mL−1.15 The same procedure was followed to generate colistin-resistant A. baumannii strains. An equal number of cells (determined by OD600 value) in the same growth phase were collected for all spectral measurements.Lipopolysaccharide (LPS) extraction and MALDI-TOF-MS analysis of lipid A
E. coli colistin-sensitive and resistant strains were initially plated on the agar medium and incubated for 18 h at 37 °C. A single colony was picked from the sensitive and resistant strains and inoculated in 100 ml of LB-broth overnight with and without antibiotics, respectively. At the OD600 of 0.6, the bacterial cells were pelleted by centrifugation at 5000 rpm for 10 minutes, and the supernatant was discarded. Cell pellets were washed with cold PBS (pH 7.2), and then the weight of the cells was measured. LPS was extracted from E. coli colistin sensitive (wild type) and resistant (mcr-1 positive) strains using an LPS extraction kit (MAK339) purchased from Sigma Aldrich. Briefly, the extraction process consisted of treatment with lysis buffer (100 μl lysis buffer for 10 mg of cells pellet), followed by sonication for complete lysis of the cells at 20 seconds (3×) in continuous pulse at 10 watts followed by 10 min incubation on ice. Then, the samples were centrifuged at 5000 rpm for 10 min at 4 °C. The lysate was collected in a new tube, and 5 μl of proteinase K (1 μl proteinase K for 20 mg of cell mass) was added to each tube and incubated at 60 °C for 1 h. This was followed by centrifugation (5000 rpm, 10 min at 4 °C) and then transferring the supernatant (total LPS) to a new 1.5 mL micro-centrifuge tube. This extracted total LPS was subjected to hydrolysis with 10 μl of 1% acetic acid, followed by heating the samples at 100 °C for 1 hour to obtain lipid A. Finally, lipid A extraction was done using chloroform, methanol, and water in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.25 ratio. The samples were centrifuged, and the chloroform layer (bottom) containing lipid A was collected.16 Lipid A was concentrated using a vacuum concentrator until completely dry. Finally, the lipid A samples were dissolved in chloroform, mixed with the 2,5-dihydroxybenzoic acid (DHB) matrix and were analysed using a RapifleX Matrix assisted laser desorption ionization (MALDI)-time of flight (TOF)-mass spectrometry (MS) instrument (Bruker Daltonics, Germany). The spectra were obtained in the reflectron positive ion mode. The analyses were performed at the mass spectrometry facility, Division of Biological Sciences, IISc, Bangalore, India.
0.25 ratio. The samples were centrifuged, and the chloroform layer (bottom) containing lipid A was collected.16 Lipid A was concentrated using a vacuum concentrator until completely dry. Finally, the lipid A samples were dissolved in chloroform, mixed with the 2,5-dihydroxybenzoic acid (DHB) matrix and were analysed using a RapifleX Matrix assisted laser desorption ionization (MALDI)-time of flight (TOF)-mass spectrometry (MS) instrument (Bruker Daltonics, Germany). The spectra were obtained in the reflectron positive ion mode. The analyses were performed at the mass spectrometry facility, Division of Biological Sciences, IISc, Bangalore, India.
Raman spectra acquisition and pre-processing
To prepare the samples for Raman spectra acquisition, the resistant strains were inoculated in the presence of appropriate colistin concentration, and the sensitive strains were inoculated without antibiotics and incubated at 37 °C until O.D600 reached 0.6–0.8. The cells were pelleted via centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 min, followed by washing with chilled PBS to remove traces of culture media. The experiments were repeated four times under the same experimental conditions. The washed microbial pellets were transferred to a sterilized CaF2 window, and spectra were obtained using the WITec alpha300 Raman microscope. The system is equipped with a 532 nm laser, grating 600 gr mm−1, cooled charged coupled device, and a 100× objective with 0.8 numerical aperture. The spectra were acquired at a power of 10 mW with an exposure time of 5 s and averaged over 3 accumulations. The Raman spectra were imported to MATLAB software for data analysis in the fingerprint region 600–1800 cm−1. The background contributions from the substrate and culture media were removed. Data from each group were pre-processed by removing cosmic spikes, polynomial fitting for baseline correction, and unit normalization.
000 rpm for 3 min, followed by washing with chilled PBS to remove traces of culture media. The experiments were repeated four times under the same experimental conditions. The washed microbial pellets were transferred to a sterilized CaF2 window, and spectra were obtained using the WITec alpha300 Raman microscope. The system is equipped with a 532 nm laser, grating 600 gr mm−1, cooled charged coupled device, and a 100× objective with 0.8 numerical aperture. The spectra were acquired at a power of 10 mW with an exposure time of 5 s and averaged over 3 accumulations. The Raman spectra were imported to MATLAB software for data analysis in the fingerprint region 600–1800 cm−1. The background contributions from the substrate and culture media were removed. Data from each group were pre-processed by removing cosmic spikes, polynomial fitting for baseline correction, and unit normalization.
Data analysis
After pre-processing, the spectra were divided into three groups: sensitive, combining colistin-treated isolates with a low concentration (1.2, 3.9 μg mL−1) and high concentration (6.5, 7.8 μg mL−1). To visualize the spectral differences, the mean spectra were calculated by averaging the variables on the Y-axis, keeping the same X-axis (wavenumber). Then, PLS-DA was utilized to reduce the dimensions of the data by extracting the latent variables to obtain the maximum variations in the spectra.36 The first step of PLS-DA modeling was the construction of PLS components for dimensionality reduction. As a result of this algorithm, the original data were rearranged to a low-dimensional subspace as scores (PLS-DA score) with the highest covariance with class labels. The regression coefficient evaluates the performance of the PLS-DA model. The positive coefficient indicates a strong influence of the corresponding variables on the discrimination between the classes. Then, the relationship between the Raman spectra collected from different groups, sensitive, colistin-treated resistant strains 1.2, 3.9, 6.5, and 7.8 μg mL−1, was established using a partial least squares regression (PLS-R) model. The model performance was validated based on regression coefficients, mean squared error (MSE), root-mean-square error (RMSE), and R2 score for the independent test data. These parameters provide insights into the accuracy, precision, and goodness of fit of regression models. The data analysis was performed using Python (scikit-learn package). Each group had a series of recorded spectra collected on four independent experimental replicates, considering possible variations while keeping the same instrumental and Raman configuration parameters. The entire analysis pipeline employed in this study is shown in Fig. 5.Single-cell hyperspectral imaging for localization of the phosphoethanolamine signal
Microbial cells were spread on a CaF2 window to obtain a single cell, and imaging was performed using the previously mentioned WITec alpha 300 microscope equipped with Zeiss epiplan 100× objective (NA 0.8, WD 1.3 mm). The average laser excitation power was 10 mW with an exposure time of 2 seconds. The total scan area was 25 × 25 μm (width × height), consisting of 100 points per line and 100 lines per image, with a step size of 0.25 μm. Routine pre-processing steps, such as cosmic removal and Savitzky–Golay smoothing, were performed on the Raman images. The image outline was confirmed using a filter on the 2954 cm−1 signal. This was followed by K-means cluster analysis (CA) to partition the data into the nearest centroids-based distance matrix. The Euclidean distance clustering algorithm was used to create the final clusters.Conclusions
In this exploratory study, label-free identification of lipid A modification associated with colistin resistance was demonstrated. Prospective adaptation of the proposed approach can provide an adjunct tool for identifying antibiotic resistance in a short and clinically implementable time.Author contributions
D. S., S. P. S. conceived and designed the project. D. S., T. C. B. performed experiments and data analysis. D. G., N. M. conducted MALDI analysis. B. H. supplied strain. All the authors wrote and corrected the manuscript. S. P. S. acquired the funding and directed the research.Data availability
The data that supports the findings of this study are available from the corresponding author upon reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was carried out under research grant project no (37/1739/23/EMR-II) supported by the Council of Scientific and Industrial Research (CSIR), Government of India and project no. IIRP-2023-1734 from Indian Council of Medical Research (ICMR), Government of India. Dhananjaya G. acknowledges the Prime Minister Research Fellowship (PMRF- 1502705), Government of India. Prof. Nilkamal Mahanta also acknowledges support from CSIR (02/0419/21/EMR II) and ICMR (2021-15271).References
- R. Laxminarayan, A. Duse, C. Wattal, A. K. Zaidi, H. F. Wertheim, N. Sumpradit, E. Vlieghe, G. L. Hara, I. M. Gould, H. Goossens, C. Greko, A. D. So, M. Bigdeli, G. Tomson, W. Woodhouse, E. Ombaka, A. Q. Peralta, F. N. Qamar, F. Mir, S. Kariuki, Z. A. Bhutta, A. Coates, R. Bergstrom, G. D. Wright, E. D. Brown and O. Cars, Antibiotic resistance-the need for global solutions, Lancet Infect. Dis., 2013, 13(12), 1057–1098 CrossRef PubMed.
- M. E. Falagas and S. K. Kasiakou, Colistin: the revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections, Clin. Infect. Dis., 2005, 40(9), 1333–1341 CrossRef CAS PubMed.
- S. S. Sharma, Indian Priority Pathogen List, 2019, Available online:https://cdn.who.int/media/docs/default-source/searo/india/antimicrobial (accessed on 10 September 2022).
- G. Klein, S. Müller-Loennies, B. Lindner, N. Kobylak, H. Brade and S. Raina, Molecular and structural basis of inner core lipopolysaccharide alterations in Escherichia coli: incorporation of glucuronic acid and phosphoethanolamine in the heptose region, J. Biol. Chem., 2013, 288(12), 8111–8127 CrossRef CAS PubMed.
- Y.-Y. Liu, Y. Wang, T. R. Walsh, L.-X. Yi, R. Zhang, J. Spencer, Y. Doi, G. Tian, B. Dong, X. Huang, L.-F. Yu, D. Gu, H. Ren, X. Chen, L. Lv, D. He, H. Zhou, Z. Liang, J.-H. Liu and J. Shen, Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study, Lancet Infect. Dis., 2016, 16(2), 161–168 CrossRef CAS PubMed.
- K. Novović and B. Jovčić, Colistin Resistance in Acinetobacter baumannii: Molecular Mechanisms and Epidemiology, Antibiotics, 2023, 12(3), 516 CrossRef PubMed.
- A. B. Janssen and W. van Schaik, Harder, better, faster, stronger: Colistin resistance mechanisms in Escherichia coli, PLoS Genet., 2021, 17(1), 1–4 Search PubMed.
- B. Li, F. Yin, X. Zhao, Y. Guo, W. Wang, P. Wang, H. Zhu, Y. Yin and X. Wang, Colistin Resistance Gene mcr-1 Mediates Cell Permeability and Resistance to Hydrophobic Antibiotics, Front. Microbiol., 2020, 10, 3015 CrossRef PubMed.
- D. M. P. De Oliveira, B. M. Forde, T. J. Kidd, P. N. A. Harris, M. A. Schembri, S. A. Beatson, D. L. Paterson and M. J. Walker, Antimicrobial Resistance in ESKAPE Pathogens, Clin. Microbiol. Rev., 2020, 33(3), e00181–19 CrossRef CAS PubMed.
- A. N. Schuetz, Antimicrobial Resistance and Susceptibility Testing of Anaerobic Bacteria, Clin. Infect. Dis., 2014, 59(5), 698–705 CrossRef PubMed.
- I. Gajic, J. Kabic, D. Kekic, M. Jovicevic, M. Milenkovic, D. Mitic Culafic, A. Trudic, L. Ranin and N. Opavski, Antimicrobial Susceptibility Testing: A Comprehensive Review of Currently Used Methods, Antibiotics, 2022, 11(4), 427 CrossRef CAS PubMed.
- L. Ashton, K. Lau, C. L. Winder and R. Goodacre, Raman spectroscopy: lighting up the future of microbial identification, Future Microbiol, 2011, 6(9), 991–997 CrossRef CAS PubMed.
- W. Liang, L. Wei, T. Jia-Wei, W. Jun-Jiao, L. Qing-Hua, W. Peng-Bo, W. Meng-Meng, P. Ya-Cheng, G. Bing and Z. Xiao, Applications of Raman Spectroscopy in Bacterial Infections: Principles, Advantages, and Shortcomings, Front. Microbiol., 2021, 12, 683580 CrossRef PubMed.
- S. L. Popa, C. Pop, D. Mo, V. D. Brata, R. Bolchis, Z. Czako, M. M. Saadani, A. Ismaiel, D. I. Dumitrascu, S. Grad, L. David, G. Cismaru and A. M. Padureanu, Deep Learning and Antibiotic Resistance, Antibiotics, 2022, 11(11), 1674 CrossRef CAS PubMed.
- D. Saikia, P. Jadhav, A. R. Hole, C. M. Krishna and S. P. Singh, Unraveling the Secrets of Colistin Resistance with Label-Free Raman Spectroscopy, Biosensors, 2022, 12(9), 749 CrossRef CAS PubMed.
- E. G. Bligh and W. J. Dyer, A rapid method of total lipid extraction and purification, J. Biochem. Physiol., 1959, 37(8), 911–917 CAS.
- C. S. Lee, Y. G. Kim, H. S. Joo and B. G. Kim, Structural analysis of lipid A from Escherichia coli O157:H7:K- using thin-layer chromatography and ion-trap mass spectrometry, J. Mass Spectrom., 2004, 39(5), 514–525 CrossRef CAS PubMed.
- M. Kurogochi and S.-I. Nishimura, Structural Characterization of N-Glycopeptides by Matrix-Dependent Selective Fragmentation of MALDI-TOF/TOF Tandem Mass Spectrometry, Anal. Chem., 2004, 76(20), 6097–6101 CrossRef CAS PubMed.
- V. Sándor, A. Kilár, F. Kilár, B. Kocsis and Á. Dörnyei, Characterization of complex, heterogeneous lipid A samples using HPLC–MS/MS technique II. Structural elucidation of non-phosphorylated lipid A by negative-ion mode tandem mass spectrometry, J. Mass Spectrom., 2016, 51(8), 615–628 CrossRef PubMed.
- D. Saikia, P. Jadhav, A. R. Hole, C. M. Krishna and S. P. Singh, Growth Kinetics Monitoring of Gram-negative Pathogenic Microbes Using Raman Spectroscopy, Appl. Spectrosc., 2022, 76(10), 1263–1271 CrossRef CAS PubMed.
- J. M. Boll, A. T. Tucker, D. R. Klein, A. M. Beltran, J. S. Brodbelt, B. W. Davies and M. S. Trent, Reinforcing Lipid A Acylation on the Cell Surface of Acinetobacter baumannii Promotes Cationic Antimicrobial Peptide Resistance and Desiccation Survival, mBio, 2015, 6(3), e00478–e00415, DOI:10.1128/mBio.00478-15 , PMID: 25991684; PMCID: PMC4442142.
- C. Krafft, L. Neudert, T. Simat and R. Salzer, Near infrared Raman spectra of human brain lipids, Spectrochim. Acta, Part A, 2005, 61(7), 1529–1535 CrossRef PubMed.
- Z. Lin, X. Zhao, J. Huang, W. Liu, Y. Zheng, X. Yang, Y. Zhang, M. L. de la Chapelle and W. Fu, Rapid screening of colistin-resistant Escherichia coli, Acinetobacter baumannii and Pseudomonas aeruginosa by the use of Raman spectroscopy and hierarchical cluster analysis, Analyst, 2019, 144, 2803–2810 RSC.
- A. C. S. Talari, Z. Movasaghi, S. Rehman and I. Rehman, Raman Spectroscopy of Biological Tissues, Appl. Spectrosc. Rev., 2015, 50(1), 46–111 CrossRef CAS.
- F. Gogry, M. T. Siddiqui, I. Sultan, F. M. Husain, A. A. Al-Kheraif, A. Ali and Q. M. R. Haq, Colistin Interaction and Surface Changes Associated with mcr-1 Conferred Plasmid Mediated Resistance in E. coli and A. veronii Strains, Pharmaceutics, 2022, 14, 295 CrossRef CAS PubMed.
- M. Dudek, G. Zajac, E. Szafraniec, E. Wiercigroch, S. Tott, K. Malek, A. Kaczor and M. Baranska, Raman Optical Activity and Raman spectroscopy of carbohydrates in solution, Spectrochim. Acta, Part A, 2019, 206, 597–612 CrossRef CAS PubMed.
- J. Hoque, U. Adhikary, V. Yadav, S. Samaddar, M. Mohan Konai, R. G. Prakash, K. Paramanandham, B. R. Shome, K. Sanyal and J. Haldar, “Chitosan Derivatives Active against Multidrug-Resistant Bacteria and Pathogenic Fungi: In Vivo Evaluation as Topical Antimicrobials, Mol. Pharm., 2016, 13(10), 3578–3589 CrossRef CAS PubMed.
- P. D. A. Rohs and T. G. Bernhardt, Annual Review of Microbiology Growth and Division of the Peptidoglycan Matrix, Annu. Rev. Microbiol., 2021, 75, 315–336 CrossRef CAS PubMed.
- G. Dhanda, Y. Acharya and J. Haldar, Antibiotic Adjuvants: A Versatile Approach to Combat Antibiotic Resistance, ACS Omega, 2023, 8(12), 10757–10783 CrossRef CAS PubMed.
- M. Hartmann, M. Berditsch, J. Hawecker, M. F. Ardakani, D. Gerthsen and A. S. Ulrich, Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy, Antimicrob. Agents Chemother., 2010, 54(8), 3132–3142 CrossRef CAS PubMed.
- R. L. Soon, R. L. Nation, P. G. Hartley, I. Larson and J. Li, Atomic force microscopy investigation of the morphology and topography of colistin-heteroresistant Acinetobacter baumannii strains as a function of growth phase and in response to colistin treatment, Antimicrob. Agents Chemother., 2009, 53(12), 4979–4986 CrossRef CAS PubMed.
- C. M. John, M. Liu, N. J. Phillips, Z. Yang, C. R. Funk, L. I. Zimmerman, J. M. Griffiss, D. C. Stein and G. A. Jarvis, Lack of lipid A pyrophosphorylation and functional lptA reduces inflammation by Neisseria commensals, Infect. Immun., 2012, 80(11), 4014–4026 CrossRef CAS PubMed.
- T. J. Beveridge, Structures of Gram-negative cell walls and their derived membrane vesicles, J. Bacteriol., 1999, 181(16), 4725–4733 CrossRef CAS PubMed.
- D. Chicco, M. J. Warrens and G. Jurman, The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation, PeerJ Comput. Sci., 2021, 7, 1–24 Search PubMed.
- F. S. de Siqueira e Oliveira, H. E. Giana and L. Silveira Jr, Discrimination of selected species of pathogenic bacteria using near-infrared Raman spectroscopy and principal components analysis, Proc. SPIE 8214, Advanced Biomedical and Clinical Diagnostic Systems X, 821409 (22 February 2012).
- H. Abdi, Partial Least Squares (PLS) Regression, Available: https://www.utdallas.edu/.
- S. H. Jun, J. H. Lee, B. R. Kim, S. I. Kim, T. I. Park, J. C. Lee and Y. C. Lee, Acinetobacter baumannii outer membrane vesicles elicit a potent innate immune response via membrane proteins, PLoS One, 2013, 8(8), e71751 CrossRef CAS PubMed.
- R. K. Blashfield and M. S. Aldenderfer, The Methods and Problems of Cluster Analysis, in Handbook of Multivariate Experimental Psychology, ed. J. R. Nesselroade and R. B. Cattell, Perspectives on Individual Differences, Springer, Boston, 1988, 978-1-4613-0893-5 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an01228c |
| This journal is © The Royal Society of Chemistry 2025 |