Open Access Article
Open Access ArticleChemometric modeling of the lowest observed effect level (LOEL) and no observed effect level (NOEL) for rat toxicity†
Ankur
Kumar
 a,
Probir Kumar
Ojha
a,
Probir Kumar
Ojha
 a and
Kunal
Roy
a and
Kunal
Roy
 *b
*b
aDrug Discovery and Development Laboratory, Department of Pharmaceutical Technology, Jadavpur University, Kolkata 700032, India
bDrug Theoretics and Cheminformatics (DTC) Laboratory, Department of Pharmaceutical Technology, Jadavpur University, Kolkata 700032, India. E-mail: kunal.roy@jadavpuruniversity.in
First published on 6th March 2024
Abstract
Humans and other living species of the ecosystem are constantly exposed to a wide range of chemicals of natural as well as synthetic origin. A multitude of compounds exert profound long-term detrimental health effects. The chronic toxicity profile of chemicals is of utmost importance for long-term risk assessment. Experimental testing of the chronic toxicity of compounds is not always a feasible option considering the magnitude of the number of chemicals, resource intensiveness in terms of time, limited availability of experimental data, and associated cost, which therefore necessitates the use of in silico approaches to overcome the associated limitations. In this work, QSAR (quantitative structure–activity relationship) models were developed employing the regression-based PLS method with strict adherence to OECD guidelines. For this study, chronic and sub-chronic toxicity datasets with LOEL (lowest observed effect levels) and NOEL (no observed effect level) as endpoints were used for model development. The validated models are robust, reliable, and predictable. The statistical results of the models are as follows: R2: 0.6–0.71, QLOO2: 0.51–0.635, and QF12: 0.52–0.658. From the validated models, it was concluded that lipophilicity, electronegativity, the presence of aromatic ethers or aliphatic oxime groups, the presence of complexity in structures, the state of unsaturation in molecules, and the presence of halogen and heavy atoms (phosphate, sulphur, etc.) are responsible for chronic/sub-chronic toxicity. The QSAR models developed in our study can be utilized for the effective gap-filling of toxicity data sets, categorization, and prioritization of chemicals, along with chronic toxicity prediction of new synthetic compounds. Furthermore, we used 2568 approved drugs from the DrugBank and PPDB databases for screening purposes using the validated models, which further corroborated the developed models based on the available toxicity data.
Environmental significanceAt present, chemicals are an essential part of our daily life. These chemicals are either of natural or synthetic origin, and they enter the environment and living beings in different ways. Therefore, there is a chance that these chemicals may have detrimental effects on the environment and human health. The long-term (chronic), sub-chronic, and short-term (acute) toxicities of environmental chemicals are presently of great concern. Due to the scarcity of information on the chronic toxicological effects of most of the compounds, it is difficult to evaluate their potential impact on human health. Therefore, it is necessary to develop an alternative method to identify chemicals that have long-term toxic effects and assess their toxicity. |
1. Introduction
In the present era, synthetic chemicals have vast applications in different fields such as food, healthcare, and the comfort of mankind. Nowadays, chemicals are an essential part of our daily life.1 These chemicals are either of natural or synthetic origin, and they enter the environment and living beings through different ways. Therefore, there is a chance that these chemicals may have detrimental effects on human health.2 At present, their long-term (chronic), sub-chronic, and short-term (acute) toxicities are of great concern. Chronic and sub-chronic toxicity assessment is a challenge for the food and pharmaceutical industries and researchers.1 Several global organizations, viz. the World Health Organization (WHO), the Occupational Safety and Health Administration (OSHA), the Food and Drug Administration (FDA), the United States Environmental Protection Agency (EPA), and the Agency for Toxic Substances and Disease Registry (ATSDR), continuously engage in the determination of the toxicity of compounds. The NOEL (no observed effect level) and LOEL (lowest observed effect level) were used as endpoints for chronic and sub-chronic toxicity assessments. NOEL represents the highest concentration associated with null adverse effects and LOEL represents the lowest concentration associated with an adverse effect (mostly expressed in mg per kg per day). There are around five million synthetic compounds, out of which seventy thousand are meant for daily usage alongside about one million naturally occurring chemicals.2 Due to the scarcity of information on the chronic toxicological effects of most of the compounds, it is difficult to evaluate their potential impact on human health.3 The purpose of chronic toxicological studies is to assess the potential long-term toxicity of chemicals.4,5 Despite the sufficient in vitro and in vivo testing facilities and resources, it is a matter of concern whether these methods are suitable for unbiased testing. Therefore, it is necessary to develop an alternative method to identify chemicals that have long-term toxic effects and assess their toxicity.4To minimize animal testing, test duration, and associated experimental resources, quantitative structure–activity relationship (QSAR) is an alternative in silico technique for the efficient estimation of chronic and sub-chronic toxicity of chemicals.5 QSAR correlates between the response activity/toxicity and numerical description of molecular structures.6
Some attempts were made earlier to develop in silico models for chronic and sub-chronic toxicity prediction in mammals.4,7–12 However, some of the previous studies reported neither different internal and external validation metrics nor mechanistic interpretations. Due to the lack of structural diversity, the generalizability of the models of some previous studies was also restricted. Some of the previous studies reported different endpoints, duration of the study, different numbers and types of compounds, different reference species, and different modeling algorithms.
In the present study, we assessed the chronic (more than 360 days) (pLOEL value: 3.881 to (−1.40); pNOEL value: 4.88 to (−1.88)), and sub-chronic (180 ± 90 days) (pLOEL value: 3.07 to (−1.89), pNOEL value: 3.85 to (−1.32)) toxicity of diverse organic chemicals in rats and mice using the LOEL (lowest observed effect level) and NOEL (no observed effect level) as the endpoints. We have taken maximum experimental (chronic and sub-chronic) toxicity data (NOEL and LOEL value in mg per kg per day) of rats only (around 98%) and very little data for mice. These organic chemicals include a range of diverse chemicals such as pharmaceuticals, industrial waste compounds, food, and agricultural, natural, and compounds meant for daily use. Regression-based partial least squares (PLS) models were developed utilizing only 2D descriptors. Stepwise regression, genetic algorithm, and the best subset selection (BSS) were used as the feature selection methods. We also screened the additional real-world databases, e.g. DrugBank database (https://go.drugbank.com/) and PPDB (Pesticide Properties DataBase), for the estimation of chronic and sub-chronic toxicity using the developed PLS models and checked the quality of the predictions using the Prediction Reliability Indicator (PRI) tool (available from http://teqip.jdvu.ac.in/QSAR_Tools)13 for the developed model. In our present study, the developed QSAR models are accurate, robust, reliable, validated, predictable, wide domain of applicability, and mechanistically interpretable. We introduce here new models for the LOEL and NOEL endpoints based on the collected data. However, we may note that the endpoint values depend on the experimental conditions, inter-laboratory variations, the number of samples, and the exposure details. Furthermore, the values depend on the nature of the species in which the experiment is performed, and the lowest value is usually used in cases when multiple values are available.7–12
2. Materials and methods
2.1. Dataset selection
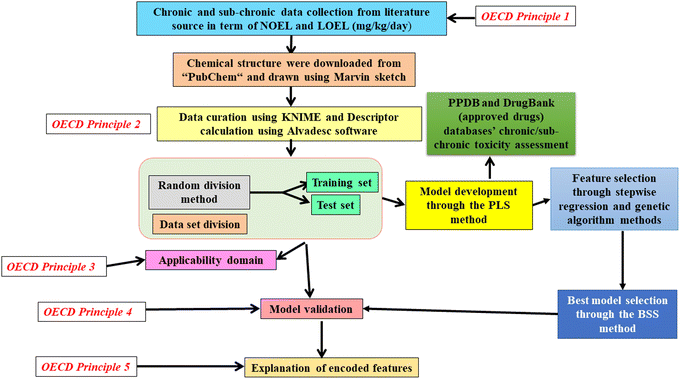
Experimental chronic (≥360 days) and sub-chronic (180 ± 90 days) toxicity data of various organic compounds of rats and mice were collected in terms of NOEL and LOEL. NOEL is the highest tested dose or concentration of a chemical at which no harmful effect is found in exposed test organisms. Doses higher than the NOEL value will result in sub-chronic toxic effects. In contrast, LOEL is the lowest concentration of a contaminant found from an experiment or observation that causes a chronic effect or adverse alteration of morphology, function, capacity, growth, development, or lifespan of a target organism, which is significantly different from the control (mg per kg per day).14 Some compounds were omitted from the dataset due to their high residual values (prediction outlier) and structural outlier behavior to reduce the biases of the data. We collected chronic toxicity data with 176 compounds for the LOEL (mg per kg per day) endpoint and 91 compounds for the NOEL (mg per kg per day) endpoint. We also collected sub-chronic toxicity data with 174 compounds for LOEL (mg per kg per day) and 90 compounds for NOEL (mg per kg per day) and then converted the LOEL and NOEL into their negative logarithmic terms (i.e., pLOEL (−log(LOEL/MW)) and pNOEL (−log(NOEL/MW))). The chemical structures and their SMILES were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), and they were represented in the MarvinSketch (http://www.chemspider.com/) software. The explicit hydrogen atoms were then added to the chemical structures, followed by 2D cleaning. Chemical curation of compounds was carried out using the KNIME workflow (available at https://sites.google.com/site/dtclabdc/) to remove salt and duplicate compounds. Finally, these structures were retrieved in a .sdf file, a recommended format for the alvaDesc15 software for the descriptor calculations.162.2. Calculation of descriptors and data pre-treatment
Descriptors are the numerical representation of chemical structures. They are classified into several sub-categories according to their origin/procedure of calculation. In this investigation, we selected a few classes of descriptors, namely, (i) ring descriptors, (ii) atom-centered fragments, (iii) molecular properties, (iv) functional group count (number of unlike functional groups), (v) connectivity index, (vi) constitutional, (vii) 2D atom pairs, (viii) atom-type E-state indices, and (ix) ETA (extended topo-chemical atom) descriptors. After that, the descriptors with missing values, redundant values, and high inter-correlation (|r| > 0.95) were eliminated. We only used the 2D descriptors for the present study to prevent the complexity associated with 3D descriptors.172.3. Division into training and test sets
In the current study, reliable and robust QSAR models were developed for predicting chronic and sub-chronic chemical toxicities. The datasets were rationally split into training sets and test sets, in a 80/20 ratio for the chronic pLOEL, sub-chronic pNOEL, and sub-chronic pNOEL endpoints, and in 70/30 for the chronic pNOEL endpoint18,19 by a random division method.20 In QSAR modeling, the training set was utilized to develop the model (internal validation) and its predictivity was validated by a test set (external validation). For chronic studies, 140 compounds were taken in the training set and 36 compounds in the test set for the pLOEL endpoint, and 71 compounds in the training set and 20 compounds in the test set for the pNOEL endpoint. For sub-chronic studies, 139 compounds were used as the training set and 35 compounds as the test set for the pLOEL endpoint, and 72 compounds as the training set and 18 compounds as the test set for the pNOEL endpoint.2.4. Selection of descriptors and model development
Feature selection is an important aspect of the QSAR model development through which we can remove the noisy and insignificant input variables from the original variable spaces.21 For our work, feature selection was done using the genetic algorithm (GA) tool (available from http://teqip.jdvu.ac.in/QSAR_Tools/). The GA tool was run multiple times taking different descriptor combinations ranging from 6 to 12 descriptors on the training sets of the four different datasets. The descriptors that occurred maximum in different models were used to generate a more refined pool for further best subset selection. We then obtained a reduced pool of 23 descriptors for the pLOEL endpoint (chronic), 18 descriptors for the pNOEL endpoint (chronic), 22 descriptors for the pLOEL endpoint (sub-chronic), and 25 descriptors for the pNOEL endpoint (sub-chronic) for further development of the QSAR model. The final sets of important features were obtained using the “Best Subset Selection v2.1” tool (available from https://dtclab.webs.com/software-tools). Four PLS models (namely, IM1–IM4 from datasets 1–4 (given in ESI 1†) respectively) were developed using a Java-based tool, ‘Partial Least Squares v1.0’ (available at https://dtclab.webs.com/software-tools).2.5. Model validation
Model validation is a vital aspect of QSAR modelling. The QSAR models were validated based on globally accepted metrics to prove their reliability, predictivity, quality, and fitting ability.17,22,23 Statistical quality and internal validation metrics like the determination coefficient (R2), leave-one-out (LOO) cross-validation (QLOO2), and mean absolute error (MAEtrain) were evaluated for the training set compounds, and external validation metrics such as Rpred2 or QF12, QF22, and MAEtest were estimated for test set compounds.16,24 The R2 and QLOO2 values were utilized to judge the fitting ability of a model, whereas Rpred2 or QF12 and QF22 were utilized to judge the predictivity of the developed models.252.6. Applicability domain (AD) assessment and Y-randomization
The AD is defined as a hypothetical space (biological space, physicochemical, or structural) indicated by the corresponding model descriptors providing reliable prediction,25,26 and it consists of the knowledge of developed training set models.26 In the present study, we used the DModX27 approach (developed using the SIMCA-P software)28 to estimate the AD of the developed PLS models. The Y-scrambling test (Y-randomization)29 was carried out to check the chance correlation of the developed models. The randomized models were developed using 100 permutations. The R2 and Q2 values for the random models (Y-axis) were plotted against the correlation coefficient between the original Y values and the permuted Y values (X-axis). The intercepts should be less than the threshold values (RY2 < 0.3, QY2 < 0.05).2.7. Toxicity assessment of approved drugs from DrugBank and PPDB database
The DrugBank database30 was utilized for screening and ranking the compounds based on their predicted chronic and sub-chronic toxicity toward rats from the developed models. The database includes the following subcategories of the drugs: approved (2568), withdrawn (243), investigational (3660), nutraceuticals (108), illicit (202), and experimental (6221) compounds. Here, we used only approved categories of drugs from the DrugBank database and 1903 pesticides from the PPDB database for toxicity screening with our developed models. The chemical structures of these compounds were downloaded from the DrugBank website (https://go.drugbank.com/) and PubChem, followed by the curation through KNIME data curation platform to remove salts, duplicate compounds, etc., and then the alvaDesc software was employed for the descriptor calculation (https://www.alvascience.com/alvades).15 Finally, we get 1694 compounds from the PPDB database and 2568 approved drugs from the DrugBank database. The prediction reliability indicator (PRI) tool (available at http://teqip.jdvu.ac.in/QSAR_Tools) was utilized to evaluate the toxicity (chronic and sub-chronic) of the approved class of compounds from DrugBank and PPDB databases through validated QSAR models. We screened the approved drugs from the DrugBank database for their chronic and sub-chronic toxicities assessments when these drugs will be exposed to rats via food, water, air, or other ways on a long-term basis. Since any new drug before coming to the market for human/animal use, preclinical toxicity studies are mainly done on rats and mice, these reference species (rats and mice) are considered quite related to humans. Therefore, rats and mice are generally used as model species for chemical toxicity assessment purposes.31 Therefore, if humans/animals are exposed to these drugs or chemicals directly (prescribed for medical purposes for long-term usage (lifetime)) or indirectly (via food, water, air, or other ways) in the long term, they may also show toxicities (chronic and sub-chronic) to the living species. A schematic diagram of the developed QSAR models is presented in Fig. 1.3. Results and discussion
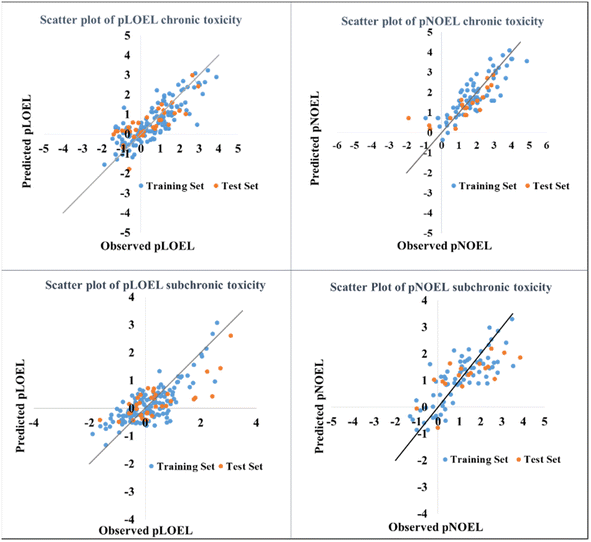
The current work aimed to develop QSAR models which can assess and predict the chronic and sub-chronic toxicities of chemicals towards rats. We have developed four PLS (PLS model reduces the intercorrelation between the descriptors through LVs) models (IM1–IM4) for which the scatter plots, i.e., the experimental versus the predicted value of all four models, are depicted in Fig. 2. The statistical results of the models are as follows: R2: 0.6–0.71, QLOO2: 0.51–0.635 and QF12: 0.52–0.658, which show that the developed models are robust and predictive. The internal and external metrics for all the models pass the threshold values (R2 = 0.6, QLOO2 = 0.6, and QF12 = 0.5), proving the reliability and predictive ability of the models. The developed models are given below (IM1–IM4) and their validation metrics are given in Table 1.| Model | Latent variables | Training set | Test set | ||||
|---|---|---|---|---|---|---|---|
| Model R2 | Model QLOO2 | MAELOO | R pred 2 or Q(F1)2 | Q (F2) 2 | MAEtest | ||
| IM1 | 2 | 0.673 | 0.604 | 0.575 | 0.618 | 0.606 | 0.559 |
| IM2 | 2 | 0.711 | 0.635 | 0.545 | 0.658 | 0.598 | 0.542 |
| IM3 | 4 | 0.607 | 0.547 | 0.464 | 0.562 | 0.537 | 0.546 |
| IM4 | 5 | 0.634 | 0.518 | 0.639 | 0.523 | 0.500 | 0.730 |
IM1: pLOEL chronic toxicity
| pLOEL = −17.04182 + 0.18387 × nHM + 39.64332 × Eta_epsi_3 − 0.13193 × nCb − −0.90898 × nArCOOH − 0.58155 × nOHp − 0.37378 × C − 007 + 0.53198 × B01[N–O] + 0.47263 × B06[C–N] + 0.79515 × B06[C–Cl] + 1.71437 × B01[S–P] − 0.45704 × nArCOOR + 0.88359 × nRSR |
IM2: pNOEL chronic toxicity
| pNOEL = 0.7256 + 0.26063 × nHM + 0.63801 × C − 019 − 0.36212 × O − 058 + 1.5621 × B03[C–P] + 0.86576 × B04[C–N] + 0.60945 × nArOR |
IM3: pLOEL sub-chronic toxicity
| pLOEL = −1.81023 − 0.15185 × nO + 0.55685 × nCsp + 0.18064 × X4v + 2.50839 × nRCNO + 0.10683 × H − 048 − 0.37199 × MaxdssC + 0.54049 × MaxssssC + 2.97984 × B01[C–F] − 0.89772 × B05[O–S] + 0.5201 × SAscore + 0.08844 × C − 026 + 0.18375 × nCconjX |
IM4: pNOEL sub-chronic toxicity
| pNOEL = −2.04357 + 0.98599 × nCrq − 0.25671 × H − 051 − 1.09359 × minssCH2 + 1.84276 × B01[C–C] − 0.71667 × B03[C–C] + 0.76782 × B04[N–N] + 0.81005 × B05[C–O] + 1.10367 × F01[S–P] − 0.32873 × F02[O–O] + 0.36323 × B07[C–C] + 3.75617 × Eta_alpha_A − 0.69496 × nRCONR2 |
3.1. Chronic toxicity assessment
We developed the final QSAR model by the PLS method for chronic toxicity using pLOEL and pNOEL as the endpoints. The developed models are reliable and validated, with good predictivity. The models' statistical validation metrics are given in Table 1. From the VIP plots (Fig. S1 and S2 in ESI 2†), the importance of the descriptors is in the following order: nHM, B01[S–P], B06[C–Cl], Eta_epsi_3, nOHp, nArCOOR, nCb, B06[C–N], nRSR, nArCOOH, C-007, and B01[N–O] in case of final the pLOEL model (IM1) while the importance of the six descriptor model for the pNOEL endpoint is in the following order: nHM, B03[C–P], B04[C–N], O-058, C-019, and nArOR. According to the regression coefficient plot (Fig. S3 and S4 in ESI 2†), descriptors nHM, Eta_epsi_3, B01[N–O], B06[C–N], B06[C–Cl], B01[S–P] and nRSR contribute positively, whereas nCb-, nArCOOH, nOHp, C-007, and nArCOOR contribute negatively to pLOEL, while nHM, B03[C–P], B04[C–N], C-019, and nArOR contribute positively and O-058 contributes negatively to the pNOEL endpoint model. The loading plot shows the effects of descriptors on the toxicity; the descriptor nHM has the maximum impact on the toxicity for the pLOEL and pNOEL endpoint models (Fig. S5 and S6 in ESI 2†). The score plot explains the distribution of dataset compounds in the latent variable space as defined by scores.17,26,27 In our study, compounds 124 (mirex) and 133 (oxadiazon) are situated outside the ellipse of the score plot (outside of AD) for the pLOEL endpoint, and all compounds are inside the ellipse of the score plot (inside the AD) for the pNOEL endpoint model, as shown in Fig. S7 and S8 in ESI 2.†3.2. Sub-chronic toxicity assessment
The sub-chronic toxicities, i.e., pLOEL and pNOEL datasets, were modeled by taking an initial pool of important features through the genetic algorithm, followed by the selection of best features by the best subset selection method. The final model was a 12-descriptor PLS model for both endpoints. The statistical results of the validated, robust, and predictive QSAR models are provided in Table 1. The VIP plots (displayed in Fig. S9 and S10 in ESI 2†) of the models (IM3 and IM4) show the contribution of descriptors in the descending order of magnitude: SAscore, nO, nRCNO, MaxssssC nCconjX, X4v, B01[C–F], B05[O–S], nCsp, H-048-, MaxdssC, and C-026 for the pLOEL endpoint and F01[S–P], Eta_alpha_A, H-051, B07[C–C], minssCH2, F02[O–O], B05[C–O], B01[C–C], B04[N–N], B03[C–C], nCrq, and nRCONR2 for the pNOEL endpoint model. The descriptors exhibiting negative contributions are nO, MaxdssC, and B05[O–S], and the positively contributing descriptors are SAscore, nRCNO, MaxssssC, nCconjX, X4v, B01[C–F], nCsp, H-048, and C-026 for the pLOEL endpoint model, while nCrq, B04[N–N], F01[S–P], B05[C–O], B01[C–C], B07[C–C], and Eta_alpha_A are positively contributing and H-051, minssCH2, B03[C–C], F02[O–O] and nRCONR2 are negatively contributing descriptors for the pNOEL endpoint models, which are demonstrated in regression coefficient plots (shown in Fig. S11 and S12 in ESI 2†). The loading plots of the developed PLS models are illustrated in Fig. S13 and S14 in ESI 2.† The score plots (provided in Fig. S15 and S16 in ESI 2†) of the developed models suggested that compound nos. 60, 61, 85, 108, and 116 of the pLOEL endpoint model and compound nos. 68 (merphos) and 88 (tetraethyldithiopyrophosphate) of the pNOEL endpoint model are outside of the applicability domain.4. Mechanistic interpretation
An attempt has been made to interpret the modeled descriptors for chronic toxicity prediction in a mechanistic approach, catering to principle 5 of OECD guidelines which are given in Tables 2, 3 and Fig. 3–6.| Descriptor with its contribution given in bracket | Description | Type of descriptor | Fragment | Mechanistic interpretation |
|---|---|---|---|---|
| The pLOEL model (IM1) | ||||
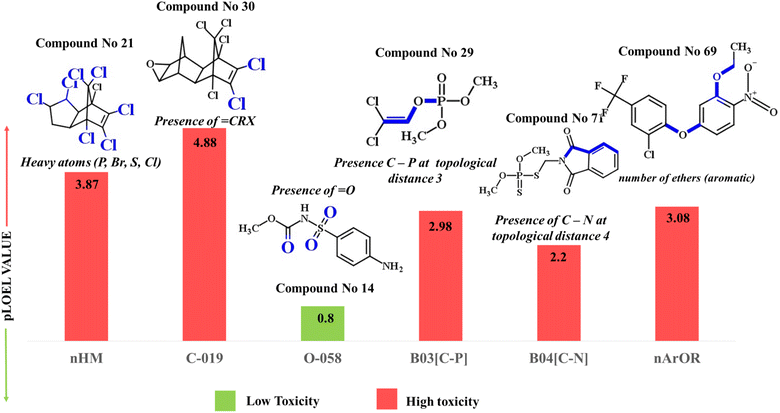
| nHM (+ve) | Number of heavy atoms | Constitutional index | P, Br, S, Cl | Heavy atoms (P, Br, S, Cl) in a chemical structure are associated with chronic chemical toxicity chemicals towards rats, as explained by Kar et al.32 & Singh et al.33 The effect can be observed in compounds 1 (Allura red AC) (given in Fig. 3) and 4 (dibutyl phthalate) |
| Eta_epsi_3 (+ve) | ETA electronegativity measure 3 | ETA descriptor | — | This descriptor is related to the electronegativity of the compound. A compound's toxicity can be attributed to its electronegativity,34,35 as evidenced in compounds 9 (ethylphthalyl ethyl glycolate) (given in Fig. 3) and 10 (FD & C blue no. 2) |
| nCb- (−ve) | Number of substituted benzene C (sp2) | Functional group count descriptor |
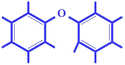
|
The toxicity of substituted benzenes is related to their ability to penetrate the cell through the cell membrane, and the electronic interactions of the chemicals with the active site. In our case, substituted benzenes with hydrophilic groups (prevalent in the present data set) may enhance the tendency of hydrogen bonding of a compound with water which in turn might impart hydrophilicity, reducing compounds' toxicity.36,37 The negative regression coefficient of this (nCb-: the number of substituted groups) descriptor indicates here that it has an inverse correlation with toxicity endpoints as observed in compounds 117 (metalaxyl) and 124 (mirex) (demonstrated in Fig. 3) |
| nArCOOH (−ve) | Number of carboxylic acids (aromatic) | Functional group counts descriptor |
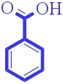
|
The existence of carboxylic acid may be crucial for increasing the compound's hydrophilicity.36,38 Thus, it reduces the toxicity as displayed in compounds 121 (methyl-4-chlorophenoxyacetic acid, 2) (displayed in Fig. 3) and 104 (γ-hexachlorocyclohexane) |
| nOHp (−ve) | Number of primary alcohols | Functional group count |

|
The occurrence of a higher number of hydroxyl groups in compounds increases the solubility of chemicals, thus increasing the excretion rate of these chemicals.39 Phase II metabolism (conjugations) requires primary hydroxyls for chemical detoxification.40 This phenomenon can be explained by compounds 91 (diquat) (provided in Fig. 3) and 97 (ethyl methyl phenyl glycinate), where the presence of more primary alcohols makes the compounds less toxic |
| C-007 (−ve) | CH2X2 | Atom-centered fragment |

|
This descriptor indicates the linkage between the number of methylene groups to electronegative atoms like phosphorus, nitrogen, sulphur, oxygen, and various halogens.41 In our study, an inverse correlation was found between this descriptor and the chronic toxicity of compounds against rats, as evidenced by the least toxic compounds 133 (oxadiazon) (given in Fig. 3) and 140 (phenformin) |
| B06[C–N] (+ve) | Presence/absence of C–N at topological distance 6 | 2D atom pair descriptor |

|
The descriptor is associated with molecular size, and higher values of the same will escalate the compound's lipophilicity.42 The occurrence of nitrogen atoms may also enhance the chronic toxicity of the compounds towards rats by imparting electronegativity (the presence of nitrogen will make the compound more electronegative)16,24,43 as shown in compounds 22 (propyl gallate) (presented in Fig. 3) and 26 (1-naphthyl) ethylene-diamine dihydrochloride, N–) |
| B06[C–Cl] (+ve) | Presence/absence of C–Cl at topological distance 6 | 2D atom pair descriptor |

|
Generally, the presence of a Cl atom (halogen) increases the lipophilicity of chemical compounds. Thus, it can easily cross the cell membranes, resulting in high chronic toxic.44,45 This phenomenon is demonstrated in compounds 43 (aspartame) (demonstrated in Fig. 3) and 20 (methyl salicylate) |
| B01[S–P] (+ve) | Presence/absence of S–P at topological distance 1 | 2D atom pair descriptor |

|
The presence of phosphorus and sulphur atoms may be responsible for the enhancement of chronic toxicity,46,47 as shown in compounds 14 (p-hydroxybenzoic acid methyl ester) (given in Fig. 3), and 19 (methyl methacrylate) |
| nArCOOR (−ve) | Number of esters (aromatic) | Functional group count descriptor |
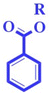
|
The nArCOOR group is polar (hydrogen bonding of oxygen of nArCOOR with water) in nature. Polarity and toxicity are inversely related to each other.42 A functional group with a polar fragment like nArCOOR reduces the toxicity16,24,43 of chemicals in rats as demonstrated in compounds 101 (fluvalinate) (given in Fig. 3), and 105 (hexahydro-1,3,5-trinitro-1,3,5-triazine) |
| nRSR (+ve) | Number of sulfides | Functional group count descriptor |
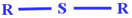
|
The presence of a higher number of sulphurs in molecular structure enhances the toxicity of compounds.48 With the increase in the numerical value of nRSR, the chronic toxicity of a compound is increased, as evidenced in compounds 35 (acitluorin sodium) (illustrated in Fig. 3) and 24 (styrene) |
| B01[N–O] (+ve) | Presence/absence of N–O at topological distance 1 | 2D atom pair descriptor |
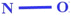
|
The presence of two electronegative atoms in this descriptor may contribute to the chronic toxicity of chemicals in rats, as suggested by Toropov et al.34 in 2008. Notably, this feature amplifies the toxicity of chemicals, as evidenced in compounds 36 (alachlor) (given in Fig. 3) and 17 (lithocholic acid) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| The pNOEL model (IM2) | ||||
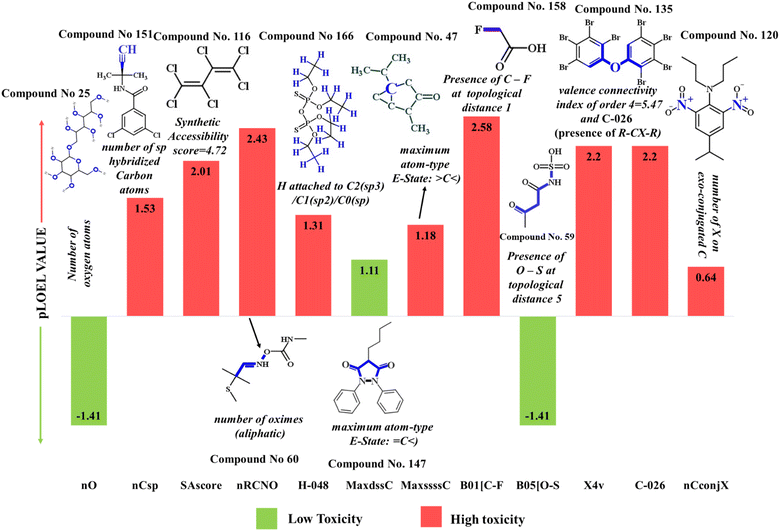
| nHM (+ve) | Number of heavy atoms | Constitutional index | P, Br, S, Cl | The presence of heavy atoms ((P, Br, S, Cl)) in chemical structure is associated with chronic heavy metal toxicity in rats,33 as shown in compounds 21 (chlordane) (given in Fig. 4) and 65 (mirex) |
| C-019 (+ve) | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CRX where R signifies the attachment of any group through a carbon atom, while X represents the occurrence of heteroatoms CRX where R signifies the attachment of any group through a carbon atom, while X represents the occurrence of heteroatoms |
Atom-centered fragment descriptor |
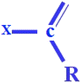
|
The presence of halogens or heteroatoms (generally electronegative) like oxygen, nitrogen, phosphorus, and various halogens may enhance the toxicity of chemicals to rats.33,44 This can be notably demonstrated by compounds 30 (dieldrin) (provided in Fig. 4) and 21 (chlordane) |
| O-058 (−ve) | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (presence of oxygen) O (presence of oxygen) |
Atom-centered fragment descriptor |

|
This descriptor is related to hydrophilicity (high potential to form H-bonding).49 There exists an inverse relationship between hydrophilicity and toxicity.50 Thus, the occurrence of this fragment in the backbone structures does not influence the toxicity, as shown by compound 14 (asulam) (illustrated in Fig. 4) |
| B03[C–P] (+ve) | Presence/absence of C–P at topological distance 3 | 2D atom pair descriptor |

|
The presence of the phosphate group may influence the toxicity of the chemicals.42,51 B03[C–P] is directly correlated with compound toxicity as demonstrated by compound 29 (dichlorvos) (displayed in Fig. 4) |
| B04[C–N] (+ve) | Presence/absence of C–N at topological distance 4 | 2D atom pair descriptor |

|
The presence of highly electronegative atoms like nitrogen may influence the compounds' toxicity, as shown in compounds 71 (phosmet) (displayed in Fig. 4) and 68 (oxamyl)42 |
| nArOR (+ve) | Number of ethers (aromatic) | Functional group count descriptor |
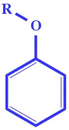
|
Generally, aromatic ethers are toxic in nature.52 Thus, the compound containing such fragments has high pNOEL values (chronic toxicity value), as illustrated in compounds 69 (oxyfluorfen) (displayed in Fig. 4) and 54 (isoxaben) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| The pLOEL model (IM3) | ||||
| SAscore (+ve) | Synthetic accessibility score | Molecular property | — | SAscore signifies the synthetic accessibility score and is linked to the complexity of molecules. The higher value of this descriptor shows that the synthesis of such compounds is complex.42 This, in turn, increases the toxicity of compounds, as shown in compounds 116 (hexachlorobutadiene) (demonstrated in Fig. 5) and 117 (hexachlorocyclopentadiene) |
| nO (−ve) | Number of oxygen atoms | Constitutional descriptor |

|
The presence of oxygen in the structure makes it more hydrophilic by the formation of H-bonding.49 This observation can be demonstrated by compounds with low toxicity, like compounds 25 (isomaltitol) (illustrated in Fig. 5) and 138 (oxytetracycline hydrochloride) |
| nCsp (+ve) | Number of sp hybridized carbon atoms | Constitutional index descriptor |

|
This feature is related to unsaturation in chemical compounds due to the presence of sp hybridized carbon atoms. Unsaturated compounds are more toxic due to their high reactivity53 as demonstrated in compounds 151 (pronamide) (shown in Fig. 5) and 154 (pydrin) |
| X4v (+ve) | Valence connectivity index of order 4 | Connectivity index descriptor | — | This descriptor is related to the molecular size and shape of the compounds.54 The high numerical value of this descriptor makes the compound more toxic, as shown in compounds 135 (octabromodiphenyl ether) (given in Fig. 5) and 165 (tetraethyldithiopyrophosphate) |
| nRCNO (+ve) | Number of oximes (aliphatic) | Functional group count |
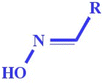
|
The presence of an aliphatic oxime group in the molecular structures might be responsible for the toxicity enhancement.55 This phenomenon is demonstrated by compounds 60 (aldicarb) (shown in Fig. 5) and 61 (aldicarb sulfone) with higher toxicity |
| H-048 (+ve) | H attached to C2(sp3)/C1(sp2)/C0(sp) | Atom-centered fragment descriptor | — | This type of hydrogen atom is very reactive in nature56 and may exhibit toxicity towards rats, as shown by compounds 166 (tetrakis(hydroxymethyl)phosphonium chloride (THPC)) (displayed in Fig. 5) and 167 (tetrakis(hydroxymethyl)phosphonium sulphate (THPS)) |
| MaxdssC (−ve) | Maximum dssC (maximum atom-type E-state: ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C![[double bond splayed right]](https://www.rsc.org/images/entities/char_e00a.gif) ) ) |
Atom-type E-state descriptor |

|
The negative regression coefficient of this descriptor explains its inverse relationship with toxicity as observed in compounds 91 (1,3-dichloro-2-propanol) (given in Fig. 5) and 147 (phenylbutazone) |
| MaxssssC (+ve) | Maximum ssssC (maximum atom-type E-state: ![[double bond splayed left]](https://www.rsc.org/images/entities/char_e009.gif) C C![[double bond splayed right]](https://www.rsc.org/images/entities/char_e00a.gif) ) ) |
Atom-type E-state descriptor |

|
This descriptor is responsible for structure complexity,57 leading to the enhancement of the toxicity as seen in compound 47 (thujone) (given in Fig. 5) |
| B01[C–F] (+ve) | Presence/absence of C–F at topological distance 1 | 2D atom pair descriptor |

|
Fluorine (halogen) atoms in the compound tend to increase the toxicity profile of molecules (due to the high electronegativity of fluorine)58 as shown in compounds 158 (sodium fluoroacetate) (illustrated in Fig. 5) and 112 (fluometuron) |
| B05[O–S] (−ve) | Presence/absence of O–S at topological distance 5 | 2D atom pair descriptor |

|
The presence of oxygen and sulfur increases the hydrophilicity of compounds due to hydrogen bonding,59 resulting in a reduction of toxicity of the chemical compounds. This phenomenon is depicted in compounds 59 (acetoacetamide-N-sulfonic acid) (demonstrated in Fig. 5) and 79 (carmoisine) |
| nCconjX (+ve) | Number of X on exo-conjugated C | Functional group count descriptor |
|
This fragment enhances the electronegativity of molecules due to the presence of a halogen atom (X), thus enhancing toxicity.16,24,43 This can be explained by compounds 120 (isopropalin) (illustrated in Fig. 5) and 14 (dodecyl gallate) |
| C-026 (+ve) | R-CX-R where X represents the existence of an electronegative atom | Atom-centered fragment descriptor |

|
The occurrence of an electronegative atom (P, O, S, N, Se, halogens) makes the compound more electronegative,16,24,43 which in turn enhances the toxicity of compounds as seen in compounds 135 (octabromodiphenyl ether) (given in Fig. 5) and 150 (promethazine hydrochloride) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| The pNOEL model (IM4) | ||||
| nCrq (+ve) | Number of ring quaternary C (sp3) | Functional group count |
|
This group is associated with the lipophilic profile of molecules,16,24,43 enabling easy penetration across the cell membrane, thus causing toxicity. This descriptor contributes positively towards the sub-chronic toxicity against rats, which is explained by compounds 32 (thujone) (provided in Fig. 6) and 29 (isobornyl acetate) and vice versa in compounds 12 (ethylbenzene) and 13 (2-ethylbutyric acid) |
| H-051 (−ve) | Hydrogen atom attached to alpha-C atom | Atom-centered fragment descriptor |
|
This fragment is associated with the polarity of the compounds.24 This descriptor has a negative correlation with the sub-chronic toxicity of compounds, as inferred from the negative value of the regression coefficient. This was evidenced in compounds 2 (acetone) (displayed in Fig. 6) and 35 (acetoacetamide) |
| minssCH2 (−ve) | Minimum ssCH2 (–CH2–) | Atom-type E-state descriptor |

|
The negative regression coefficient associated with minssCH2 (the minimum E-state value of a specific group associated with two single bonds (ss) in a hybrid group (CH2)) indicates a negative correlation with sub-chronic toxicity, as observed in compounds 12 (ethylbenzene) (given in Fig. 6) and 34 (acenaphthene) |
| B01[C–C] (+ve) | Presence/absence of C–C at topological distance 1 |

|
This fragment is correlated with the size (long chain) of molecules. Thus, the presence of these fragments may enhance the lipophilicity of the molecules (easily cross the cell membrane),16,24,43 ultimately increasing toxicity. This observation can be explained by compounds 41 (bentazon) and 68 (merphos) (displayed in Fig. 6) | |
| B07[C–C] (+ve) | Presence/absence of C–C at topological distance 7 | 2D atom pair descriptor |

|
B07[C–C] fragment is directly correlated with the lipophilicity of the molecules (easily crossing the cell membrane),16,24,43 ultimately increasing toxicity. This phenomenon can be shown in compounds 41 (bentazon) and 68 (merphos) (displayed in Fig. 6) |
| B03[C–C] (−ve) | Presence/absence of C–C at topological distance 3 | 2D atom pair descriptor |

|
The presence of such fragments in the molecules reduces the toxicity,49 as shown in compounds 13 (2-ethylbutyric acid) (demonstrated in Fig. 6) and 34 (acenaphthene) |
| B04[N–N] (+ve) | Presence/absence of N–N at topological distance 4 | 2D atom pair descriptor |

|
Electronegative atoms (presence of two nitrogen atoms), if present in the structure, may enhance the toxicity of compounds.60 This phenomenon is described in compounds 77 (m-phenylenediamine) (demonstrated in Fig. 6) and 73 (olaquindox) |
| B05[C–O] (+ve) | Presence/absence of C–O at topological distance 5 | 2D atom pair descriptor |

|
As discussed in the above section (B04 [N–N] section). This phenomenon is described in compounds 74 (paclobutrazol) and 32 (thujone) (shown in Fig. 6) |
| F01[S–P] (+ve) | Frequency of S–P at topological distance 1 | 2D atom pair descriptor |

|
As discussed in the above section (B04 [N–N] section). This phenomenon is described in compounds 88 (tetraethyldithiopyrophosphate) (illustrated in Fig. 6) and 68 (merphos) and vice versa in 48 (cyclodextrin, beta) and 67 (maleic anhydride) |
| F02[O–O] (−ve) | Frequency of O–O at topological distance 2 | 2D atom pair descriptor |

|
The presence of two electron-rich atoms may be responsible for electrostatic repulsion,61 thus can reduce compound toxicity. This feature is inversely related to the toxicity of compounds as explained by the compounds 40 (azorubine) (presented in Fig. 6) and 48 (beta cyclodextrin) |
| Eta_alpha_A (+ve) | ETA average core count | 2D atom pair descriptor | — | The positive regression coefficient of this feature shows that with an increase in the numerical value of this descriptor, the endpoint (pNOEL value) of compounds will also be increased. For example, compound nos. 49 (1,4-dibromobenzene) (shown in Fig. 6) |
| nRCONR2 (−ve) | Number of the tertiary amides (aliphatic) in molecular structure | ETA descriptor |
|
The existence of this group may reduce chemical toxicity (due to hydrophilic interaction since there may be a chance of formation of H-bonding with N, O), as evidenced in compounds 69 (metolachlor) and 79 (propachlor) (shown in Fig. 6) |
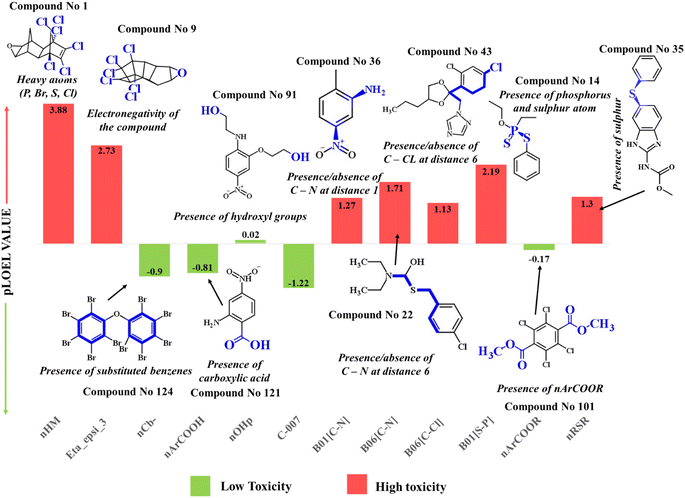 | ||
| Fig. 3 Mechanistic interpretation of the modeled descriptors against chronic toxicity (pLOEL) in rats. | ||
 | ||
| Fig. 4 Mechanistic interpretation of the modeled descriptors against chronic toxicity (pNOEL) in rats. | ||
 | ||
| Fig. 5 Mechanistic interpretation of the modeled descriptors against sub-chronic toxicity (pLOEL) in rats. | ||
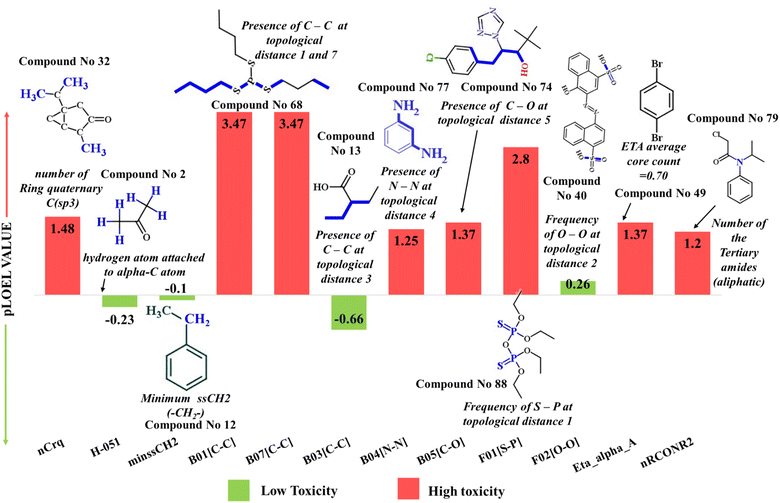 | ||
| Fig. 6 Mechanistic interpretation of the modeled descriptors against sub-chronic toxicity (pNOEL) in rats. | ||
5. Applicability domain assessment
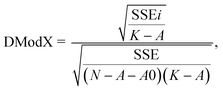
To comply with OECD guideline 3, a developed QSAR model must have a defined chemical domain of application. The applicability domain (AD) of the models was determined by the distance to model in the X space method (DModX).27 The DModX variable represents the unexplained variation (residuals), which corresponds to the X residual standard deviation at some distance from the model X space. A DModX algorithm uses the residuals of Y and X as diagnostic features for ensuring model quality. The SIMCA-P software was used to conduct the applicability domain analysis for DModX, with a 99% confidence level (threshold D-Crit = 0.009999).For the observation i, SSE is the squared sum of the residuals, in a model with A components, K variables, and N observations. A0 is 1 if the model was centered and 0 otherwise. It is claimed that DModX is approximately F-distributed, so it can be used to check if an observation deviates significantly from a normal PLS model.26,27,43
5.1. AD of chronic toxicity models
From the DModX plots (shown in Fig. S17–S20 in ESI 2†), it was found that compounds 2, 24, 35, 38, 67, 77, 91, 101, 104, 121, 133, and 140 from the training set and compound number 168 from the test set for the pLOEL endpoint model and compounds 33 and 80 from the training set and 5, 55, and 74 from the test set for the pNOEL endpoint model are outside the domain of applicability due to their molecular structural dissimilarity.26,27,435.2. AD of sub-chronic toxicity models
For the pLOEL sub-chronic toxicity, compound nos. 45, 60, 61, 88, 112, 117, 116, 151, 158, 166, and 167 of the training set are outliers and compound nos. 125 and 136 in the test set are outside the domain of applicability. For the pNOEL sub-chronic endpoint, compounds nos. 29 (isobornyl acetate), 68 (merphos), 69 (metolachlor), and 79 (propachlor) from the training set were outside the AD due to their molecular structural dissimilarity,26,27,43 as shown in Fig. S21–S24 in ESI 2.†6. The Y-randomization plot
The Y-randomization plot (shown in Fig. S25 and S26 in ESI 2†) shows that the model was not obtained by chance correlation. The randomized model was developed using 100 permutations. It was found that the RY2 and QY2 values of both the chronic models for pLOEL and pNOEL were much lower than the threshold values (RY2 < 0.3 and QY2 < 0.05). Similarly, in the case of sub-chronic toxicity (Fig. S27 and S28 in ESI 2†), the RY2 and QY2 values followed the same trend (i.e., lower than the threshold values).7. Toxicity prediction of the true external dataset
We predicted DrugBank database compounds (approved drugs only) and 1692 compounds from the PPDB database to understand the predictive ability of the models. The predicted response was then segregated into toxic and non-toxic compounds based on the training set mean response (for chronic toxicity, pNOEL ≥ 1.82 and pLOEL ≥ 0.82, and for sub-chronic toxicity, pNOEL ≥ 1.20 and pLOEL ≥ 0.20) for all the datasets.14 We then checked the reliability of prediction and domain of applicability using the Prediction Reliability Indicator tool (https://sites.google.com/site/dtclabpri/). From this prediction, we found that most of the compounds (99.99% for chronic studies and 75–95% for sub-chronic studies) showed good prediction quality and lie within the applicability domain of the model. A list of approved drugs (from the DrugBank database) and 1692 compounds from the PPDB database with predicted toxicity from the developed models is given in ESI 1.† We have provided the top 10 toxic pesticides (chronically) predicted from our developed models in Table S1 in ESI 2.† Their toxicity predictions are also justified by the literature support62 (https://wwwn.cdc.gov/and https://www3.epa.gov/pesticides/). A list of 1692 compounds from the PPDB database with predicted toxicity from the developed models is given in ESI 1.† The top 64 screened drugs belonging to the approved category of the DrugBank database, which are also experimentally known to be highly toxic (chronic and sub-chronic toxicities) in rats exposed to the drugs via food, water, air, or other ways for a long term, are provided in Table 3, and the references are also provided. Since for any new drug before coming to the market for human/animal use, preclinical toxicity studies are mainly done on rats and mice, these reference species (rats and mice) are considered quite related to humans. Therefore, rats and mice are generally used as model organisms for chemical toxicity assessment.31 Therefore, if humans/animals are exposed to these drugs or chemicals directly (prescribed for medical purposes for long-term usage (lifetime)) or indirectly (via food, water, air, or other ways) for the long term, the chemicals may also show toxicities (chronic and sub-chronic) to the living species. These drugs of different classes are reported to be toxic in various literature studies. We have identified a diverse class of toxic chemicals falling under diverse groups: (a) organophosphorus class: Zytron;63 (b) organochlorine class: lindane (EPA (https://www.epa.gov/) has classified lindane as a Class B2/C, possible human carcinogen) and hexachlorophene;64 (c) imidazole class: terconazole, oxiconazole, thiabendazole, tioconazole, maribavir, clotrimazole, tizanidine, econazole, itraconazole, miconazole, sertaconazole, and isoconazole;65 (d) radiopharmaceuticals: propyliodone and technetium Tc-99m sulfur colloid;66 (e) antipsychotic and anti-depressant: zotepine, propericiazine, hydromorphone methotrimeprazine. Morphine,67 and nefazodone;68 (f) beta-blocker: timolol;69 (g) phenanthrene series: nalbuphine,70 polychloro phenoxy phenol class: triclosan,71 disinfectant and antiseptic: chlorhexidine,72 triclocarban, and chloroxine;73 (h) benzodiazepine class: estazolam, fenoldopam, and chlordiazepoxide;74 (i) barbiturates: hydroxyzine;75 (j) antimalarials: proguanil, lumefantrine, and amodiaquine;76 (k) biphenyl tetrazole class: losartan;77 and (l) heteropentacyclic compound: naltrexone78 and other drugs such as lamotrigine79 and carbinoxamine.80| Sl. no. | DrugBank ID | Generic name | Sl. no. | DrugBank ID | Generic name |
|---|---|---|---|---|---|
| 1 | DB11768 | Zytron | 33 | DB00697 | Tizanidine |
| 2 | DB12267 | Brigatinib | 34 | DB00878 | Chlorhexidine |
| 3 | DB00845 | Clofazimine | 35 | DB01243 | Chloroxine |
| 4 | DB00882 | Clomifene | 36 | DB06234 | Maribavir |
| 5 | DB09397 | Technetium Tc-99m sulfur colloid | 37 | DB11327 | Dipyrithione |
| 6 | DB09225 | Zotepine | 38 | DB11632 | Opicapone |
| 7 | DB00251 | Terconazole | 39 | DB01149 | Nefazodone |
| 8 | DB09366 | Propyliodone | 40 | DB01233 | Metoclopramide |
| 9 | DB00239 | Oxiconazole | 41 | DB06237 | Avanafil |
| 10 | DB01007 | Tioconazole | 42 | DB06480 | Prucalopride |
| 11 | DB01110 | Miconazole | 43 | DB06155 | Rimonabant |
| 12 | DB01153 | Sertaconazole | 44 | DB11155 | Triclocarban |
| 13 | DB08943 | Isoconazole | 45 | DB09063 | Ceritinib |
| 14 | DB14201 | 2,2′-Dibenzothiazyl disulfide | 46 | DB11995 | Avatrombopag |
| 15 | DB11691 | Naldemedine | 47 | DB00235 | Milrinone |
| 16 | DB00373 | Timolol | 48 | DB00360 | Sapropterin |
| 17 | DB00539 | Toremifene | 49 | DB04864 | Huperzine A |
| 18 | DB00925 | Phenoxybenzamine | 50 | DB00242 | Cladribine |
| 19 | DB01403 | Methotrimeprazine | 51 | DB00257 | Clotrimazole |
| 20 | DB14881 | Oliceridine | 52 | DB00475 | Chlordiazepoxide |
| 21 | DB01127 | Econazole | 53 | DB00557 | Hydroxyzine |
| 22 | DB06708 | Lumefantrine | 54 | DB00613 | Amodiaquine |
| 23 | DB01167 | Itraconazole | 55 | DB00678 | Losartan |
| 24 | DB00431 | Lindane | 56 | DB00730 | Thiabendazole |
| 25 | DB00756 | Hexachlorophene | 57 | DB00748 | Carbinoxamine |
| 26 | DB00295 | Morphine | 58 | DB00800 | Fenoldopam |
| 27 | DB00844 | Nalbuphine | 59 | DB01131 | Proguanil |
| 28 | DB06230 | Nalmefene | 60 | DB01215 | Estazolam |
| 29 | DB11952 | Duvelisib | 61 | DB01608 | Periciazine |
| 30 | DB08604 | Triclosan | 62 | DB00327 | Hydromorphone |
| 31 | DB00555 | Lamotrigine | 63 | DB00704 | Naltrexone |
| 32 | DB00629 | Guanabenz | 64 | DB06800 | Methylnaltrexone |
8. Comparison to the related studies
Although a strict comparison is not possible due to the different test species, different durations of the study, different compositions of the internal and external sets, different validation metrics reported, and different endpoints as well as the modeling algorithms employed, we attempted to compare the current results to those from earlier reported research work.Mazzatorta et al.7 reported a predictive model of 445 compounds employing multivariate analysis (multiple linear regression or MLR and linear discriminant analysis or LDA) based on two-dimensional physicochemical descriptors. Gadaleta et al.8 reported a study based on the k-NN algorithm for predicting oral sub-chronic toxicity in rats using a training set of 254 chemicals and an external set comprising 179 chemicals. Julian-Ortiz et al.9 reported MLR and LDA models using a diverse set of 234 chemicals (LOAEL values) by using graph-theoretical indices as molecular descriptors. A regression-based model was reported by Mumtaz et al.10 using rat chronic toxicity data and LOAEL as the endpoint. Hisaki et al.11 reported several QSAR models using environmental chemical toxicity data (repeated dose, developmental, and reproductive toxicities) for the NOEL predictions. Toropova et al.4 reported a few regression-based QSAR models for the NOAEL (chronic toxicity) calculation using the Monte Carlo technique. Pradeep et al.12 reported several machine learning-based models (mainly k-nearest neighbors, support vector machine, random forest, and gradient boosting regression) using chronic, sub-chronic, and sub-acute toxicity data. Comparisons with previously reported studies (models) with validation metrics are provided in Table 4.
| Work | Model | Endpoint | Internal validation metrics | External validation metrics | ||||
|---|---|---|---|---|---|---|---|---|
| Model R2 | Model Q(LOO)2 | MAELOO | R pred 2 or Q(F1)2 | Q (F2) 2 | MAEtest | |||
| Present work (regression-based) | IM1 (PLS) | LOEL_CHRONIC (≥360 days) | 0.673 | 0.604 | 0.575 | 0.618 | 0.606 | 0.559 |
| IM2 (PLS) | NOEL_CHRONIC (≥360 days) | 0.711 | 0.635 | 0.545 | 0.658 | 0.598 | 0.542 | |
| IM3 (PLS) | LOEL_SUB-CHRONIC (180 ± 90 days) | 0.607 | 0.547 | 0.464 | 0.562 | 0.537 | 0.546 | |
| IM4 (PLS) | NOEL_SUB-CHRONIC (180 ± 90 days) | 0.634 | 0.518 | 0.639 | 0.523 | 0.500 | 0.730 | |
| Mazzatorta et al.7 | QSAR models | Chronic_LOAELs | 0.54 | — | — | — | — | — |
| Gadaleta et al.8 | k-NN algorithm | Sub-chronic_LOAELs | ≥0.543 | ≥0.632 | — | — | — | — |
| de Julian-Ortiz et al.9 | MLR and LDA | Chronic_LOAELs | 0.524 (whole set) | — | — | — | — | — |
| Mumtaz et al.10 | Regression model | Chronic_(LOAELs) | — | — | — | 0.84 | — | — |
| Hisaki et al.11 | QSAR models | Developmental, and reproductive (NOELs) | — | — | — | — | — | — |
| Toropova et al.4 | Monte Carlo technique_regression models | NOAELs | 0.679–0.718 | 0.672–0.712 | — | 0.610–0.627 | — | — |
| Pradeep et al.12 | Several machine learning algorithms including k nearest neighbors, support vector machines, random forests, and gradient-boosting regression | Chronic, subchronic, reproductive, developmental, subacute (LEL/LOEL/LOAEL and NEL/NOEL/NOAEL) | (−0.19)–0.54 | — | — | (−0.09)–0.57 | — | — |
In this current study, we developed different QSAR models for risk assessment of chronic toxicity (more than 360 days) and sub-chronic (180 ± 90 days) toxicity data using a large available curated dataset of diverse chemicals such as pharmaceuticals, industrial waste compounds, food, agricultural, natural, and compounds meant for daily use in rats and mice using the LOEL and NOEL as the endpoints and strictly following the OECD guidelines. We considered a higher number of compounds than those considered in previously reported models. We used the genetic algorithm as the descriptor thinning method to extract the vital structural features that are important for the endpoints. We have interpreted the models and found the structure–toxicity relationships that are responsible for chronic toxicity and vice versa. The internal and external validation metrics of the predicted PLS models suggest that the models are reliable, predictive, and mechanistically interpretable with a wide domain of applicability representing diverse groups of chemicals compared to the previous works. It can be inferred that lipophilicity, electronegativity, aromatic ethers or aliphatic oxime groups, the complexity of structures, unsaturation in molecules, and the presence of halogen and heavy atoms (phosphate, sulphurs, etc.) are responsible for the chronic or sub-chronic toxicity, whereas the presence of polar and hydroxyl group in molecules (hydrophilic properties) can reduce the chronic and sub-chronic toxicities. Therefore, this information should be useful for the development of safer and greener chemicals that will maintain bio-diversity. The validated models may be employed for screening, and prioritization of chemicals, pharmaceuticals, and other compounds inside the chemical space of the developed models and can be used for screening of chemical databases and data-gap filling.
9. Conclusion
In this work, we assessed the chronic (≥360 days) and sub-chronic (180 ± 90 days) toxicity profiles of chemicals including a wide range of pharmaceuticals, drugs, and daily use chemical products. Several regulatory agencies, researchers, and organizations are deeply concerned about the chronic/sub-chronic toxicity of chemicals. Since there is a large gap in chronic/sub-chronic toxicity data owing to limited experimental data, QSAR modeling can be used as an alternative. The GA-PLS models were validated through globally accepted validation metrics and strictly following the OECD guidelines. From the developed PLS models (valid, accurate, robust, and predictive model), it can be inferred that lipophilicity, electronegativity, aromatic ethers or aliphatic oxime groups, the complexity of structures, unsaturation in molecules, and the presence of halogen and heavy atoms (phosphate, sulphurs, etc.) are responsible for chronic or sub-chronic toxicity, whereas the presence of polar and hydroxyl groups in molecules (hydrophilic properties) can reduce chronic and sub-chronic toxicity. Therefore, this information is beneficial for the development of safer and greener chemicals that will maintain bio-diversity. The validated models may be employed for screening and prioritization of chemicals, pharmaceuticals, and other compounds inside the chemical space (AD) of the developed models. These developed models were utilized for the prediction of chronic and sub-chronic toxicity of the approved category drugs of the DrugBank database and the PPDB database. These validated models enable the assessment of long-term chemical toxicity prior to their synthesis and evaluation. Thus, the developed model will help reduce the time, cost, resources, and frequency of animal testing strictly catering to the “RRR” (reduction, refinement, and replacement) principles.Ethical statement
This is an original article that did not use other information, which requires ethical approval.Consent to participate
All authors participated in this article.Consent for publication
All authors have given consent to the publication of this article.Data availability
Supporting data are available in the ESI† section. Additional data that support the findings of this study are available on request from the corresponding author.Author contributions
The manuscript was written through the contributions of all authors.Conflicts of interest
The authors declare no competing interests.Acknowledgements
AK thanks GPC Regulatory India Private Limited for the financial support in the form of a research assistant (GPC Regulatory India Private Limited sponsored research, Ref No-P-1/RS/171/22, date-07-09.2022).References
- E. Marchiori, J. H. Moore and J. C. Rajapakse, Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics: 5th European Conference, EvoBIO 2007, Valencia, Spain, April, 2007 Search PubMed.
- L. Tilaoui, B. Schilter, L. A. Tran, P. Mazzatorta and M. Grigorov, Integrated Computational Methods for Prediction of the Lowest Observable Adverse Effect Level of Food-Borne Molecules, QSAR Comb. Sci., 2007, 26, 102–108, DOI:10.1002/qsar.200610060.
- N. Basant and S. Gupta, QSAR modeling for predicting mutagenic toxicity of diverse chemicals for regulatory purposes, Environ. Sci. Pollut. Res., 2017, 24, 14430–14444, DOI:10.1007/s11356-017-8903-y.
- A. P. Toropova, A. A. Toropov, J. B. Veselinović and A. M. Veselinović, QSAR as a random event: a case of NOAEL, Environ. Sci. Pollut. Res., 2015, 22, 8264–8271, DOI:10.1007/s11356-014-3977-2.
- R. B. Aher, K. Khan and K. Roy, A Brief Introduction to Quantitative Structure-Activity Relationships as Useful Tools in Predictive Ecotoxicology, in Ecotoxicological QSARs, Methods in Pharmacology and Toxicology, ed. K. Roy, Humana, New York, 2021, DOI:10.1007/978-1-0716-0150-1_2.
- J. C. Dearden, The History and Development of Quantitative Structure-Activity Relationships (QSARs), Oncology: Breakthroughs in Research and Practice, ed. Information Resources Management Association, IGI Global, 2017, pp. 67–117, DOI:10.4018/978-1-5225-0549-5.ch003.
- P. Mazzatorta, M. D. Estevez, M. Coulet and B. Schilter, Modeling oral rat chronic toxicity, J. Chem. Inf. Model., 2008, 48, 1949–1954, DOI:10.1021/ci8001974.
- D. Gadaleta, F. Pizzo, A. Lombardo, A. Carotti, S. E. Escher, O. Nicolotti and E. Benfenati, A k-NN algorithm for predicting oral sub-chronic toxicity in the rat, ALTEX, 2014, 31, 423–432, DOI:10.14573/altex.1405091.
- J. V. De Julian-Ortiz, R. Garcia-Domenech, J. Galvez and L. Pogliani, Predictability and prediction of lowest observed adverse effect levels in a structurally heterogeneous set of chemicals, SAR QSAR Environ. Res., 2005, 16, 263–272, DOI:10.1080/10659360500036927.
- M. M. Mumtaz, L. A. Knauf, D. J. Reisman, W. B. Peirano, C. T. DeRosa, V. K. Gombar, K. Enslein, J. R. Carter, B. W. Blake, K. I. Huque and V. M. S. Ramanujam, Assessment of effect levels of chemicals from quantitative structure-activity relationship (QSAR) models. I. Chronic lowest-observed-adverse-effect level (LOAEL), Toxicol. Lett., 1995, 79, 131–143, DOI:10.1016/0378-4274(95)03365-R.
- T. Hisaki, M. A. née Kaneko, M. Yamaguchi, H. Sasa and H. Kouzuki, Development of QSAR models using artificial neural network analysis for risk assessment of repeated-dose, reproductive, and developmental toxicities of cosmetic ingredients, J. Toxicol. Sci., 2015, 40, 163–180, DOI:10.2131/jts.40.163.
- P. Pradeep, K. P. Friedman and R. Judson, Structure-based QSAR models to predict repeat dose toxicity points of departure, Comput. Toxicol., 2020, 16, 100139, DOI:10.1016/j.comtox.2020.100139.
- K. Roy, P. Ambue and S. Kar, How Precise Are Our Quantitative Structure–Activity Relationship Derived Predictions for New Query Chemicals?, ACS Omega, 2018, 3, 11392–11406, DOI:10.1021/acsomega.8b01647.
- I. C. Munro, R. A. Ford, E. Kennepohl and J. G. Sprenger, Correlation of structural class with no-observed-effect levels: a proposal for establishing a threshold of concern, Food Chem. Toxicol., 1996, 34, 829–867, DOI:10.1016/S0278-6915(96)00049-X.
- A. Mauri, alvaDesc: a tool to calculate and analyze molecular descriptors and fingerprints, in Ecotoxicological QSARs, Humana, New York, NY, 2020, DOI:10.1007/978-1-0716-0150-1_32.
- P. M. Khan, K. Roy and E. Benfenati, Chemometric modeling of Daphnia magna toxicity of agrochemicals, Chemosphere, 2019, 224, 470–479, DOI:10.1016/j.chemosphere.2019.02.147.
- A. Kumar, T. Podder, V. Kumar and P. K. Ojha, Risk assessment of aromatic organic chemicals to T. pyriformis in environmental protection using regression-based QSTR and Read-Across algorithm, Process Saf. Environ. Prot., 2023, 170, 842–854, DOI:10.1016/j.psep.2022.12.067.
- A. Afantitis, G. Melagraki, H. Sarimveis, O. Igglessi-Markopoulou and G. Kollias, A novel QSAR model for predicting the inhibition of CXCR3 receptor by 4-N-aryl-[1,4] diazepane ureas, Eur. J. Med. Chem., 2009, 44, 877–884, DOI:10.1016/j.ejmech.2008.05.028.
- Q. H. Nguyen, H. B. Ly, L. S. Ho, N. Al-Ansari, H. V. Le, V. Q. Tran, I. Prakash and B. T. Pham, Influence of data splitting on performance of machine learning models in prediction of shear strength of soil, Math. Probl. Eng., 2021, 1–15, DOI:10.1155/2021/4832864.
- J. T. Leonard and K. Roy, On selection of training and test sets for the development of predictive QSAR models, QSAR Comb. Sci., 2006, 25, 235–251, DOI:10.1002/qsar.200510161.
- P. M. Khan and K. Roy, Current approaches for choosing feature selection and learning algorithms in quantitative structure–activity relationships (QSAR), Expert Opin. Drug Discovery, 2018, 13, 1075–1089, DOI:10.1080/17460441.2018.1542428.
- R. K. Mukherjee, V. Kumar and K. Roy, Ecotoxicological QSTR and QSTTR modeling for the prediction of acute oral toxicity of pesticides against multiple avian species, Environ. Sci. Technol., 2021, 56, 335–348, DOI:10.1021/acs.est.1c05732.
- A. Kumar, V. Kumar, T. Podder and P. K. Ojha, First report on ecotoxicological QSTR and I-QSTR modeling for the prediction of acute ecotoxicity of diverse organic chemicals against three protozoan species, Chemosphere, 2023, 139066, DOI:10.1016/j.chemosphere.2023.139066.
- K. Khan and K. Roy, Ecotoxicological QSAR modelling of organic chemicals against Pseudokirchneriella subcapitata using consensus predictions approach, SAR QSAR Environ. Res., 2019, 30, 665–681, DOI:10.1080/1062936X.2019.1648315.
- S. Kar and K. Roy, Development and validation of a robust QSAR model for prediction of carcinogenicity of drugs, Indian J. Biochem. Biophys., 2011, 48, 111–112 CAS.
- K. Roy, S. Kar and P. Ambure, On a simple approach for determining applicability domain of QSAR models, Chemom. Intell. Lab. Syst., 2015, 145, 22–29, DOI:10.1016/j.chemolab.2015.04.013.
- S. Wold, M. Sjöström and L. Eriksson, PLS-regression: a basic tool of chemometrics, Chemom. Intell. Lab. Syst., 2001, 58, 109–130, DOI:10.1016/S0169-7439(01)00155-1.
- SIMCA-P UM.16.0, 2021, https://www.umetrics.com/, E-mail: info@umetrics.com Search PubMed.
- C. Rücker, G. Rücker and M. Meringer, Y-randomization and its variants in QSPR/QSAR, J. Chem. Inf. Model., 2007, 47, 2345–2357, DOI:10.1021/ci700157b.
- D. S. Wishart, Y. D. Feunang, A. C. Guo, E. J. Lo, A. Marcu, J. R. Grant, T. Sajed, D. Johnson, C. Li, Z. Sayeeda and N. Assempour, DrugBank 5.0: a major update to the DrugBank database for 2018, Nucleic Acids Res., 2018, 46, D1074–D1082, DOI:10.1093/nar/gkx1037.
- S. A. Saganuwan, Toxicity studies of drugs and chemicals in animals: an overview, Bulg. J. Vet. Med., 2017, 20, 291–318, DOI:10.15547/bjvm.983.
- S. Kar and K. Roy, First report on development of quantitative interspecies structure–carcinogenicity relationship models and exploring discriminatory features for rodent carcinogenicity of diverse organic chemicals using OECD guidelines, Chemosphere, 2012, 87, 339–355, DOI:10.1016/j.chemosphere.2011.12.019.
- W. P. Singh and J. O. Bockris, Toxicity Issues of Organic Corrosion Inhibitors: Applications of QSAR Model, paper presented at the Corrosion 96, Denver, Colorado, 1996 Search PubMed.
- A. A. Toropov, B. F. Rasulev and J. Leszczynski, QSAR modeling of acute toxicity by balance of correlations, Bioorg. Med. Chem., 2008, 16, 5999–6008, DOI:10.1016/j.bmc.2008.04.055.
- E. Amzoiu, P. G. Anoaica and C. Lepadatu, QSAR study of toxicity of aromatic nitroderivatives using the electronegativity of OMO/UMO states as fingerprint descriptors, Rev. Roum. Chim., 2011, 56, 711–716, DOI:10.1016/S0045-6535(00)00214-9.
- S. K. Pandey, P. K. Ojha and K. Roy, Exploring QSAR models for assessment of acute fish toxicity of environmental transformation products of pesticides (ETPPs), Chemosphere, 2020, 252, 126508, DOI:10.1016/j.chemosphere.2020.126508.
- G. H. Lu, X. Yuan and Y. H. Zhao, QSAR study on the toxicity of substituted benzenes to the algae (Scenedesmus obliquus), Chemosphere, 2001, 44, 437–440, DOI:10.1016/S0045-6535(00)00214-9.
- Y. H. Zhao, G. D. Ji, M. T. D. Cronin and J. C. Dearden, QSAR study of the toxicity of benzoic acids to Vibrio fischeri, Daphnia magna and carp, Sci. Total Environ., 1998, 216, 205–215, DOI:10.1016/S0048-9697(98)00157-0.
- D. Gadaleta, K. Vuković, C. Toma, G. J. Lavado, A. L. Karmaus, K. Mansouri, N. C. Kleinstreuer, E. Benfenati and A. Roncaglioni, SAR and QSAR modeling of a large collection of LD50 rat acute oral toxicity data, J. Cheminf., 2019, 11, 1–16, DOI:10.1186/s13321-019-0383-2.
- M. T. D. Cronin, J. C. Dearden, J. C. Duffy, R. Edwards, N. Manga, A. P. Worth and A. D. P. Worgan, The importance of hydrophobicity and electrophilicity descriptors in mechanistically-based QSARs for toxicological endpoints, SAR QSAR Environ. Res., 2002, 13, 167–176, DOI:10.1080/10629360290002316.
- R. Todeschini and V. Consonni, Molecular Descriptors for Chemoinformatics: Volume I:Alphabetical Listing/volume II: Appendices, References, John Wiley & Sons, New Jersey, 2009 Search PubMed.
- A. Kumar, P. K. Ojha and K. Roy, QSAR modeling of chronic rat toxicity of diverse organic chemicals, Comput. Toxicol., 2023, 26, 100270, DOI:10.1016/j.comtox.2023.100270.
- K. Khan, K. Roy and E. Benfenati, Ecotoxicological QSAR modeling of endocrine disruptor chemicals, J. Hazard. Mater., 2019, 369, 707–718, DOI:10.1016/j.jhazmat.2019.02.019.
- R. Mannhold and R. F. Rekker, The hydrophobic fragmental constant approach for calculating log P in octanol/water and aliphatic hydrocarbon/water systems, Perspect. Drug Discovery Des., 2000, 18, 1–18, DOI:10.1023/A:1008782809845.
- G. K. Jillella and K. Roy, QSAR modelling of organic dyes for their acute toxicity in Daphnia magna using 2D-descriptors, SAR QSAR Environ. Res., 2022, 33, 111–139, DOI:10.1080/1062936X.2022.2033318.
- S. Chavan, I. A. Nicholls, B. C. Karlsson, A. M. Rosengren, D. Ballabio, V. Consonni and R. Todeschini, Towards global QSAR model building for acute toxicity: Munro database case study, Int. J. Mol. Sci., 2014, 15, 18162–18174, DOI:10.3390/ijms151018162.
- D. W. Roberts and J. Costello, QSAR and mechanism of action for aquatic toxicity of cationic surfactants, QSAR Comb. Sci., 2003, 22, 220–225, DOI:10.1002/qsar.200390015.
- B. K. Sharma, P. Singh, P. Pilania, M. Shekhawat and Y. S. Prabhakar, QSAR of 2-(4-methylsulphonylphenyl)pyrimidine derivatives as cyclooxygenase-2 inhibitors: simple structural fragments as potential modulators of activity, J. Enzyme Inhib. Med. Chem., 2012, 27, 249–260, DOI:10.3109/14756366.2011.587414.
- K. A. Hossain and K. Roy, Chemometric modeling of aquatic toxicity of contaminants of emerging concern (CECs) in Dugesia japonica and its interspecies correlation with daphnia and fish: QSTR and QSTTR approaches, Ecotoxicol. Environ. Saf., 2018, 166, 92–101, DOI:10.1016/j.ecoenv.2018.09.068.
- D. Mackay, J. A. Arnot, E. P. Petkova, K. B. Wallace, D. J. Call, L. T. Brooke and G. D. Veith, The physicochemical basis of QSARs for baseline toxicity, SAR QSAR Environ. Res., 2009, 20, 393–414, DOI:10.1080/10629360902949153/.
- M. Vervloet, Modifying phosphate toxicity in chronic kidney disease, Toxins, 2019, 11, 522, DOI:10.3390/toxins11090522.
- M. Kimura, H. Miyahara, N. Moritani and Y. Sawaki, Electroreductive dehalogenation of chlorinated aromatic ethers. Unexpected electrogenerated base-catalyzed reactions, J. Org. Chem., 1990, 55, 3897–3902 CrossRef CAS.
- K. Roy and R. N. Das, QSTR with extended topochemical atom (ETA) indices. 16. Development of predictive classification and regression models for toxicity of ionic liquids towards Daphnia magna, J. Hazard. Mater., 2013, 254, 166–178, DOI:10.1016/j.jhazmat.2013.03.023.
- R. Zeng, J. Deng, L. Dang and X. Yu, Correlation between the structure and skin permeability of compounds, Sci. Rep., 2021, 11, 10076, DOI:10.1038/s41598-021-89587-5.
- J. Stenersen, Action of pesticides on earthworms. Part I: the toxicity of cholinesterase-inhibiting insecticides to earthworms as evaluated by laboratory tests, Pestic. Sci., 1979, 10, 66–74, DOI:10.1002/ps.2780100109.
- K. Khan and K. Roy, Ecotoxicological risk assessment of organic compounds against various aquatic and terrestrial species: application of interspecies i-QSTTR and species sensitivity distribution techniques, Green Chem., 2022, 24, 2160–2178, 10.1039/D1GC04320J.
- S. P. Gaudêncio and F. Pereira, Predicting Antifouling Activity and Acetylcholinesterase Inhibition of Marine-Derived Compounds Using a Computer-Aided Drug Design Approach, Mar. Drugs, 2022, 20, 129, DOI:10.3390/md20020129.
- A. Seth and K. Roy, QSAR modeling of algal low level toxicity values of different phenol and aniline derivatives using 2D descriptors, Aquat. Toxicol., 2020, 228, 105627, DOI:10.1016/j.aquatox.2020.105627.
- D. Van der Spoel, P. J. van Maaren, P. Larsson and N. Tîmneanu, Thermodynamics of hydrogen bonding in hydrophilic and hydrophobic media, J. Phys. Chem. B, 2006, 110, 4393–4398, DOI:10.1021/jp0572535.
- R. N. Das, K. Roy and P. L. Popelier, Exploring simple, transparent, interpretable and predictive QSAR models for classification and quantitative prediction of rat toxicity of ionic liquids using OECD recommended guidelines, Chemosphere, 2015, 139, 163–173, DOI:10.1016/j.chemosphere.2015.06.022.
- X. Gironés and R. Carbó-Dorca, Modelling toxicity using molecular quantum similarity measures, QSAR Comb. Sci., 2006, 25, 579–589, DOI:10.1002/qsar.200530128.
- S. Kim, P. A. Thiessen, E. E. Bolton, J. Chen, G. Fu, A. Gindulyte, L. Han, J. He, S. He, B. A. Shoemaker, J. Wang, B. Yu, J. Zhang and S. H. Bryant, PubChem Substance and Compound databases, Nucleic Acids Res., 2016, 44, D1202–D1213, DOI:10.1093/nar/gkv951.
- H. J. De Silva, N. A. Samarawickrema and A. R. Wickremasinghe, Toxicity due to organophosphorus compounds: what about chronic exposure?, Trans. R. Soc. Trop. Med. Hyg., 2006, 100, 803–806, DOI:10.1016/j.trstmh.2006.05.001.
- P. Kaushik and G. Kaushik, An assessment of structure and toxicity correlation in organochlorine pesticides, J. Hazard. Mater., 2007, 143, 102–111, DOI:10.1016/j.jhazmat.2006.08.073.
- R. Ranjith, The chemistry and biological significance of imidazole, benzimidazole, benzoxazole, tetrazole and quinazolinone nucleus, J. Chem. Pharm. Res., 2016, 8, 505–526 Search PubMed.
- D. Marzin, Preclinical evaluation of radiopharmaceutical toxicological prerequisites, Nucl. Med. Biol., 1998, 25, 733–736, DOI:10.1016/S0969-8051(98)00068-7.
- J. M. Davis and D. L. Garver, Neuroleptics: Clinical Use in Psychiatry, in Handbook of Psychopharmacology, ed. L. L. Iversen, S. D. Iversen and S. H. Snyder, Springer, Boston, MA, 1978, DOI:10.1007/978-1-4613-4042-3_4.
- J. A. Dykens, J. D. Jamieson, L. D. Marroquin, S. Nadanaciva, J. J. Xu, M. C. Dunn, A. R. Smith and Y. Will, In vitro assessment of mitochondrial dysfunction and cytotoxicity of nefazodone, trazodone, and buspirone, Toxicol. Sci., 2008, 103, 335–345, DOI:10.1093/toxsci/kfn056.
- F. Barrueto, S. Traub and J. Gragzel, Beta Blocker Poisoning, UnToDate, 2017, https://www.uptodate.com/contents/beta-blocker-poisoning, accessed Oct, 17 Search PubMed.
- V. L. Emery Jr, Chronic Toxicity of Acetone and Phenanthrene on the Marine Polychaete Worm, Nereis (Neanthes) Arenaceodentata, ProQuest Dissertations & Theses Global, 1993, retrieved from https://www.proquest.com/dissertations-theses/chronic-toxicity-acetone-phenanthrene-on-marine/docview/304103748/se-2 Search PubMed.
- E. O. Igbinosa, E. E. Odjadjare, V. N. Chigor, I. H. Igbinosa, A. O. Emoghene, F. O. Ekhaise, N. O. Igiehon and O. G. Idemudia, Toxicological profile of chlorophenols and their class in the environment: the public health perspective, Sci. World J., 2013, 460215, DOI:10.1155/2013/460215.
- F. C. R. Azevedo, I. C. D. Vaz, F. A. R. Barbosa and S. M. S. Magalhães, Toxicological effects of ciprofloxacin and chlorhexidine on growth and chlorophyll a synthesis of freshwater cyanobacteria, Braz. J. Pharm. Sci., 2019, 55 DOI:10.1590/s2175-97902019000217661.
- K. Orito, M. Hashida, K. Hirata, A. Kurokawa, M. Shirai and F. Akahori, Effects of single intratracheal exposure to chlorhexidine gluconate on the rat lung, Drug Chem. Toxicol., 2006, 29, 1–9, DOI:10.1080/01480540500408416.
- D. R. Wesson and S. Camber, Acute and Chronic Toxicity of Benzodiazepines, in The Benzodiazepines: Current Standards for Medical Practice, ed. D. E. Smith and D. R. Wesson, Springer, Dordrecht, 1974, DOI:10.1007/978-94-009-4886-0_17.
- E. C. Klatte, A. L. Brooks and R. K. Rhamy, Toxicity of intra-arterial barbiturates and tranquilizing drugs, Radiology, 1969, 92, 700–704, DOI:10.1148/92.4.700.
- D. H. Wetterholm and F. C. Winter, Histopathology of chloroquine retinal toxicity, Arch. Ophthalmol., 1964, 71, 82–87, DOI:10.1001/archopht.1964.00970010098016.
- F. M. D. Tabak, A. M. D. Mert, R. M. D. Ozaras, M. M. D. Biyikli, R. M. D. Ozturk, G. M. D. Ozbay, H. M. D. Senturk and Y. M. D. Aktuglu, Losartan-Induced Hepatic Injury, J. Clin. Gastroenterol., 2002, 34, 585–586 CrossRef PubMed.
- H. E. Dayton and C. E. Inturrisi, The urinary excretion profiles of naltrexone in man, monkey, rabbit, and rat, Drug Metab. Dispos., 1976, 4, 474–478 CAS.
- L. K. French, N. J. McKeown and R. G. Hendrickson, Complete heart block and death following lamotrigine overdose, Clin. Toxicol., 2011, 49, 330–333, DOI:10.3109/15563650.2011.572555.
- Y. G. Yap and A. J. Camm, Potential Cardiac Toxicity of H1-Antihistamines, Histamine and H1-Antihistamines in Allergic Disease, 2002, pp. 405–436, DOI:10.3109/9780203910375.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3va00265a |
| This journal is © The Royal Society of Chemistry 2024 |