Expanding the scope of self-assembled supramolecular biosensors: a highly selective and sensitive enzyme-responsive AIE-based fluorescent biosensor for trypsin detection and inhibitor screening†
Jasvir
Kaur
 ab,
Harshad A.
Mirgane
c,
Vrushali S.
Patil
ad,
Geetika M.
Ahlawat
ab,
Harshad A.
Mirgane
c,
Vrushali S.
Patil
ad,
Geetika M.
Ahlawat
 b,
Sheshanath V.
Bhosale
b,
Sheshanath V.
Bhosale
 c and
Prabhat K.
Singh
c and
Prabhat K.
Singh
 *ae
*ae
aRadiation & Photochemistry Division, Bhabha Atomic Research Centre, Mumbai 400 085, India. E-mail: prabhatk@barc.gov.in; prabhatsingh988@gmail.com
bUniversity Institute of Biotechnology, Chandigarh University, Panjab 140 413, India
cDepartment of Chemistry, School of Chemical Sciences, Central University of Karnataka, Kalaburagi 585367, Karnataka, India
dSchool of Nanoscience & Technology, Shivaji University Kolhapur, Vidya Nagar, Kolhapur 416004, Maharashtra, India
eHomi Bhabha National Institute, Anushaktinagar, Mumbai 400085, India
First published on 19th March 2024
Abstract
Trypsin, a pancreatic enzyme associated with diseases like pancreatic cancer and cystic fibrosis, requires effective diagnostic tools. Current detection systems seldom utilize macrocyclic molecules and tetraphenyl ethylene (TPE) derivative-based supramolecular assemblies, known for their biocompatibility and aggregation-induced emission (AIE) properties, for trypsin detection. This study presents an enzyme-responsive, AIE-based fluorescence 'Turn-On' sensing platform for trypsin detection, employing sulfated-β-cyclodextrin (S-βCD), an imidazolium derivative of TPE (TPE-IM), and protamine sulfate (PrS). The anionic S-βCD and cationic TPE-IM formed a strongly fluorescent supramolecular aggregation complex in an aqueous buffer. However, PrS suppresses fluorescence because of its strong binding affinity with S-βCD. The non-fluorescent TPE-IM/S-βCD/PrS supramolecular assembly system exhibits trypsin-responsive properties, as PrS is a known trypsin substrate. Trypsin restores fluorescence in the TPE-IM/S-βCD system through the enzymatic cleavage of PrS, correlating linearly with trypsin catalytic activity in the 0–10 nM concentration range. The limit of detection is 10 pM. This work contributes to the development of self-assembled supramolecular biosensors using charged TPE derivatives and β-cyclodextrin-based host–guest chemistry, offering an innovative fluorescence ‘Turn-On’ trypsin sensing platform. The sensing system is highly stable under various conditions, selective for trypsin, and demonstrates potential for biological analysis and disease diagnosis in human serum. Additionally, it shows promise for the screening of trypsin inhibitors.
1 Introduction
Supramolecular self-assembling systems that are responsive to enzymatic actions have been exploited as a class of materials for the detection of biological analytes, such as enzymes.1–5 Such enzyme-responsive assembly systems are highly specific and efficient, with excellent biocompatibility to external environmental stimuli such as temperature, pH, ionic strength, and light.6,7 Therefore, owing to their various merits, enzyme-responsive self-assembly systems are used for various applications, for example, sensing of biological molecules, drug delivery, and theragnostics.5,8–13 A self-assembly system that is responsive to enzymatic activity typically consists of two primary components: (1) an enzyme-specific substrate/substrate site containing molecule. (2) A molecular assembly system/structure that changes its properties owing to enzymatic action. Therefore, the molecular assembly components that can undergo various types of interactions, such as hydrogen bonding, ionic interactions, π–π stacking, etc., are strongly recommended for fabricating enzyme-responsive supramolecular systems.5,14–18 In this regard, non-covalent interaction-dependent assembly systems have gained considerable attention.17–21 Surface-modified macrocyclic structures, such as cyclodextrins, are particularly noteworthy for their potential use in constructing non-covalent interactions through aggregation/disaggregation assembly systems. This is due to their desirable qualities, such as low cost, high charge density, biocompatibility, and solubility.2,22–24 In this regard, fluorophores named aggregation-induced emission-based luminogens (AIEgens), discovered by Tang and group,25 exhibiting AIE, are suitable co-component molecules to construct non-covalent supra-molecular assembly constructs. The AIEgens themselves show almost negligible emission in the solution at low concentrations; however, a strong emission enhancement known as AIE is reported upon aggregation induced by the oppositely charged co-component. The phenomenon of AIE is based on the fact that intramolecular rotational movements of AIEgens are hindered in the aggregated state26 AIE activable bioprobes have been reported in the literature that emphasizes their high selectivity, sensitivity, and signal-to-noise ratio, which results in their various biosensing applications.27–32Many diseases are associated with abnormal enzyme expression, such as hepatopathy, Alzheimer's disease, pancreatitis, etc.33,34 In this regard, trypsin is also an essential enzyme and disease biomarker.35,36 Trypsin (Enzyme Commission Number 3.4.21.4.) catalytically breaks down the peptide bond on the carboxyl-terminal side of the cationic amino acids, arginine and lysine. Trypsin is a beneficial enzyme with remarkable advantages in many fields, such as proteomics, food biotechnology, biomedicines, immunology, and fundamental research.37–41 The average concentration of trypsin in human serum is 115–350 ng mL−1, however, abnormal expression of trypsin is an indicator of many diseases, for example, pancreatic cancer, cystic fibrosis, biliary cirrhosis, and pancreatitis.42–46 Therefore, developing specific and sensitive strategies for trypsin detection is of great significance. While there are multiple techniques for detecting trypsin, most of these methods rely on traditional strategies such as enzyme-linked immunosorbent assays, gelatine film, electrochemistry, piezoelectric methods, surface plasmon resonance, and colorimetry.36,47–52 Previous methods for detecting trypsin have faced several challenges, including complex designs, high costs, the need for advanced equipment, limited sensitivity, long processing times, and susceptibility to environmental factors. For instance, Surface Plasmon Resonance (SPR) based sensors detect trypsin by measuring changes in refractive index when an analyte binds to a bio-recognition unit immobilized on SPR probe. A drawback of this technique is its reliance on covalent modifications to the substrate, which can lower enzyme activity, impacting the specificity and sensitivity of the detection. Similarly, Fluorescence Resonance Energy Transfer (FRET) based methods are affected by external conditions like metal ions, pH, and temperature. Additionally, selecting suitable donor and acceptor molecules for a FRET system can be difficult. The Enzyme Linked Immunosorbent Assay (ELISA), another traditional method, uses specific antibodies to detect proteins or analytes in a liquid medium. However, ELISA involves both primary and secondary antibodies against the antigen, making it costly and complex. Therefore, there is an evident demand for constructing more easy-to-monitor, direct, cheap, rapid, and sensitive sensing platforms. Interestingly, advanced strategies based on enzyme-responsive macrocyclic molecule-driven supramolecular assembly have seldom been used to develop trypsin detection platforms.53–55 This manuscript presents an innovative study that integrates enzyme-responsive assembly systems with aggregation-induced emission (AIE) technology for the detection of trypsin, a novel approach that is scarcely represented in existing literature. By integrating the specificity of enzyme–substrate interactions with the enhanced sensitivity offered by AIE, our method presents a significant improvement over conventional biosensing techniques. The creation of a supramolecular probe via the self-assembly of a cationic AIEgen with an anionic macrocyclic molecule, alongside its proven selectivity and sensitivity within complex biological matrices, underscores the innovative contributions of this manuscript. The system uses a cationic AIEgen, specifically the imidazolinium derivative of tetraphenylethylene (TPE-IM), along with an anionic macrocyclic molecule known as sulfated-β-cyclodextrin (S-βCD), which contains sulfate groups at the lower and upper rims, and polycationic protamine sulfate (PrS), which is a natural substrate of trypsin.
The supramolecular probe is constructed by the electrostatic interaction-based self-assembly of TPE-IM and S-βCD to form a TPE-IM/S-βCD supramolecular aggregation complex. Subsequently, the TPE-IM/S-βCD supramolecular aggregation dissociates in the presence of the cationic polyelectrolyte protamine (PrS), which is abundant in positive charges and rich in arginine. Protamine is also a natural substrate for the enzyme trypsin. Thus, the TPE-IM/S-βCD/PrS-based supramolecular assembly system is found to be responsive to trypsin because of the specific action of trypsin on protamine (PrS). Therefore, the current paper demonstrates the development of a simple, sensitive, and specific fluorescence ‘Turn-On’ enzyme-responsive supra-molecular assembly/disassembly-based strategy for trypsin detection. In principle, as shown in Scheme 1, a cationic AIEgen, TPE-IM shows a substantial enhancement in fluorescence emission in the presence of an anionic macromolecule, S-βCD, due to electrostatic interactions and subsequent aggregation, leading to AIE of TPE-IM. However, a fluorescence ‘Turn-Off’ signal is observed on adding cationic polyelectrolyte protamine sulfate (PrS), due to predominantly stronger interactions between S-βCD and PrS, which lead to the formation of S-βCD/PrS complex and the release of free TPE-IM in the solution that yields a decrease in emission intensity (Turn-Off). Finally, trypsin-dependent specific digestion of PrS at the carboxyl-terminal of arginine leads to the disintegration of the S-βCD/PrS aggregation complex and allows reformation of the TPE-IM-/S-βCD aggregation complex, which restores fluorescence. Hence, a fluorescence ‘Turn-On’ signal is obtained in response to trypsin. Thus, the TPE-IM/S-βCD/PrS system allows for the quick, sensitive, and specific detection of trypsin. The potential stability of the current sensing system has been monitored in different environments by changing the temperature, pH, ionic strength, etc. The selectivity of enzyme-responsive supramolecular assembly system towards trypsin has been demonstrated in the presence of various other proteins and enzymes. Finally, the potential applicability of the proposed sensing system has been demonstrated in real human serum samples.
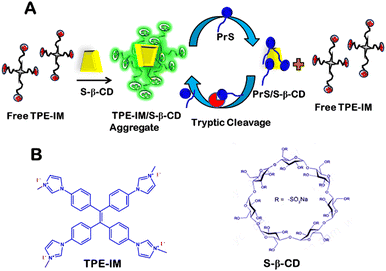 | ||
| Scheme 1 (A) Schematic representation of the working mechanism of the trypsin sensor (B) chemical structure of TPE-IM and S-βCD. | ||
2. Experimental section
2.1 Materials and chemicals
All chemicals, unless specified, were purchased from Sigma Aldrich and used without further purification. The imidazolinium derivative of tetraphenylethylene (TPE-IM) was synthesized following a detailed procedure outlined in the ESI† (Fig. S1) based on previously reported methods.56 The concentration of TPE-IM used in all experiments was accurately determined using its molar absorptivity ∼21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 M−1 cm−1 at 294 nm. Trypsin, sodium salt of sulfated β-cyclodextrin (degree of substitution ∼12 to 14), benzamidine hydrochloride, protamine sulfate, sodium chloride, pepsin, glucose oxidase, horseradish peroxidase, choline oxidase, bovine serum albumin, lysozyme, hemoglobin, alkaline phosphatase, Tris–HCl, and other salts were purchased from Sigma Aldrich. A 10 mM Tris–HCl buffer solution (pH 8.2) was prepared to conduct all experiments at room temperature. The concentration of TPE-IM was maintained at ∼19 μM unless stated otherwise. All measurements were performed in triplicate to ensure reproducibility, with error bars representing the standard deviation.
400 M−1 cm−1 at 294 nm. Trypsin, sodium salt of sulfated β-cyclodextrin (degree of substitution ∼12 to 14), benzamidine hydrochloride, protamine sulfate, sodium chloride, pepsin, glucose oxidase, horseradish peroxidase, choline oxidase, bovine serum albumin, lysozyme, hemoglobin, alkaline phosphatase, Tris–HCl, and other salts were purchased from Sigma Aldrich. A 10 mM Tris–HCl buffer solution (pH 8.2) was prepared to conduct all experiments at room temperature. The concentration of TPE-IM was maintained at ∼19 μM unless stated otherwise. All measurements were performed in triplicate to ensure reproducibility, with error bars representing the standard deviation.
2.2 Spectroscopic measurements
The following equation of the multi-exponential function was used to fit the time-resolved fluorescence decay traces.
 | (1) |
The average fluorescence lifetime (τ) was calculated using the following equation,61
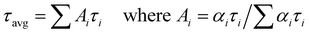 | (2) |
2.3 Enzyme kinetics study
2.4 Morphological characterization
Field-emission scanning electron microscopy (FESEM) was performed using an FEI Nova NanoSEM 450 to analyze the morphology of the TPE-IM/S-βCD aggregates. Samples were prepared by drop-casting the TPE-IM/S-βCD solution onto silicon wafers and allowing them to dry under ambient conditions before imaging.2.5 Rationale for experimental choices
3. Results and discussion
3.1 Fluorescence ‘Turn-On’ assembly of TPE-IM/S-βCD
Fig. 1(A) displays the steady-state fluorescence spectra of TPE-IM in aqueous buffer with varying concentrations of S-βCD present in the solution. As shown in Fig. 1(A), the emission intensity of TPE-IM increases in the presence of various concentrations of the S-βCD solution. A substantial emission enhancement of TPE-IM (∼86 fold) is observed in the presence of nearly 5 μM S-βCD in the solution containing TPE-IM. As shown in the inset of Fig. 1(A), a linear increase in the emission intensity of TPE-IM at 475 nm was observed with increasing concentration of S-βCD. This kind of fluorescence enhancement of AIEgens has previously been reported to be due to aggregation induced by charged derivatives of macrocyclic host molecules, such as anionic derivatives of cyclodextrins.20,62 It has been deduced from previous reports that charged cyclodextrins cause charge neutralization of cationic AIEgens and lead to their aggregation. The free state of tetraphenylethylene (TPE) derivatives has a very weak emission intensity because of the free intramolecular rotations that cause non-radiative de-excitation of their excited states. However, charge neutralization, which causes the aggregation of positively charged TPE molecules on the surface of oppositely charged cyclodextrin derivatives, prevents the fluorescence quenching of free TPE derivatives because of restricted intramolecular rotations in the aggregated state.63 The AIEgen, in the present case, TPE-IM molecules carry positively charged imidazolium groups; thus, there is a natural probability for electrostatic interactions between cationic TPE-IM and anionic S-βCD, due to which charge neutralization occurs for TPE-IM, subsequently leading to the aggregation of TPE-IM molecules in the presence of S-βCD. This leads to the aggregation-induced emission enhancement of TPE-IM, as shown in Fig. 1(A).To further support the results obtained from steady-state emission measurements, excited-state lifetime measurements of TPE-IM have also been performed in the presence of various concentrations of S-βCD, and the results are displayed in Fig. 1(B). It is clear from the results of Fig. 1(B) that the free TPE-IM, in the absence of S-βCD, displays a very fast decay trace that is nearly pulse-limited (i.e., within the resolution of the instrument), whereas the decay traces gradually becomes slower with a gradual increase in the concentration of S-βCD. The decay traces observed in our study were well-described by a 3-exponential function, with time constants (τ) approximately in the range of 0.02 ns, 1–1.2 ns, and 3.8–4.4 ns for population components 1, 2, and 3, respectively. We attribute the faster component (τ1) to free TPE-IM in solution and the intermediate (τ2) and slow (τ3) components to TPE-IM associated with S-βCD. The amplitude of the fast decay component (A1) gradually decreased with increasing concentration of S-βCD, while the sum of the amplitudes of the slow decay components (A2 + A3) increased and was well correlated (Fig. 2, inset). This nice correlation of amplitudes suggests a gradual transformation of the free form of TPE-IM to the aggregated form of TPE-IM upon the addition of S-βCD. Previous reports have shown that TPE molecules, known as molecular rotors, undergo efficient conformational relaxation in the free monomeric form, which causes a rapid decrease in the excited-state population of this molecule.20,64,65 As previously described, the introduction of anionic S-βCD into a solution containing cationic TPE-IM molecules results in the formation of a TPE-IM/S-βCD aggregate complex. In this aggregated state, the unrestricted intramolecular rotational movements of the TPE-IM molecules become limited, leading to a slower excited-state relaxation as S-βCD is gradually added. This observation aligns with the findings of steady-state emission measurements.
To further support the above results and investigate the influence of S-βCD on TPE-IM aggregation, ground-state absorption measurements have also been performed, and the results are displayed in Fig. S2 (ESI†). Fig. S2 (ESI†) displays a broad absorption maximum in the 330–350 nm range. As the concentration of S-βCD increases in the solution containing TPE-IM, the absorbance at 330 nm increases. A saturation behavior is achieved at ∼6 μM of S-βCD. It should be noted that a simultaneous increase in absorbance offset at the longer wavelength is also observed, supporting the proposal of aggregation of cationic TPE-IM molecules induced by anionic S-βCD.65 Thus, in the present case, it can be suggested that S-βCD induces aggregation of TPE-IM molecules. To further support the aggregation of TPE-IM molecules in the presence of S-βCD, we have also collected excitation spectra for both TPE-IM and the TPE-IM/S-βCD complex (Fig. S3, ESI†). These spectra further corroborate our findings, showing a marked increase in emission intensity for the TPE-IM/S-βCD complex compared to TPE-IM alone. These spectral data clearly illustrate the aggregation-induced emission characteristics of our system. In addition, we have also characterized the aggregation using The field emission scanning electron microscopy (FE-SEM) measurements (Fig. S4, ESI†). FE-SEM image depicts a highly dense and homogeneous aggregation of particulates, which is consistent with the formation of an aggregation complex between TPE-IM and S-βCD. The homogeneity of the aggregation pattern across the observed field suggests a consistent interaction between the TPE-IM dye and the sulfated cyclodextrin, leading to the formation of a supramolecular assembly. The morphology of the aggregates, which lack any well-defined shape or size, implies that the aggregation process leads to irregularly shaped, amorphous complexes rather than to crystalline or highly ordered structures.
3.2 Environmental influence on TPE-IM/S-βCD assembly complex
To provide additional evidence for the ionic strength-dependent steady-state emission measurements, excited-state lifetime measurements of the TPE-IM/S-βCD complex were conducted at varying NaCl concentrations, as illustrated in Fig. 2(B). The results in Fig. 2(B) clearly demonstrate that the excited-state decay traces of the TPE-IM/S-βCD complex progressively decrease as the NaCl concentration increases from 0 mM to 25 mM. The inset of Fig. 2(B) presents the amplitude analysis of the decay components, which shows that the a1 (corresponding to τ1, free TPE-IM) gradually increases with increase in the concentration of NaCl in the solution, whereas a2 + a3, corresponding to S-βCD templated aggregates of TPE-IM, decreases with increase in concentration of NaCl in the solution. This clearly indicates the conversion of TPE-IM aggregates to their monomer form upon the addition of NaCl, which can be attributed to the disassembly of the TPE-IM/S-βCD aggregation complex to increase in the ionic strength of the solution, leading to the release of rapidly rotating free TPE-IM molecules in the solution. This kind of salt-induced breakage of electrostatically driven supramolecular aggregate assembly has also been reported earlier.22,68,69 Therefore, the assertion that ionic interactions are vital in the formation of the TPE-IM/S-βCD aggregation complex, as deduced from steady-state emission measurements, can be substantiated by these time-resolved measurements. The dependence on ionic strength highlights the role of electrostatic interactions in the assembly process. However, this also poses a challenge for using these assemblies in biological samples, which usually have high ionic strength. Such a challenge is common in systems that rely on electrostatic forces. A practical way to overcome this problem is by diluting the biological samples to reduce their ionic strength.
Furthermore, the ground-state absorption spectra of the TPE-IM/S-βCD assembly complex also show a similar trend in the reduction in absorbance at 330 nm with increasing temperature, as shown in Fig. S6 (ESI†). The decrease in absorbance can again be assigned to a gradual breakdown of the TPE-IM/S-βCD assembly upon an increase in temperature that causes the release of free TPE-IM in the solution. Consequently, the turbidity of the solution decreases. Hence, the scattering-dependent effect is reduced at high temperatures, as evidenced by the decrease in the offset absorbance of the TPE-IM/S-βCD complex (Fig. S6 (ESI†)). A similar observation was made earlier for a supramolecular assembly upon application of a temperature.70 The temperature-dependent effect on the TPE-IM/S-βCD assembly system was further investigated by performing excited-state lifetime measurements at various temperatures (25–70 °C), and the results are shown in Fig. 3(B). The results shown in Fig. 3(B) indicate that the decay traces progressively became more rapid as the temperature of the solution increased. The analysis of the decay traces suggests that the increase in the temperature of the solution causes an increase in the formation of the free form of TPE-IM, as manifested by the increase in the amplitude of the shorter component (a1). Concomitantly, the amplitude of the slower components (a2 + a3), which represent the aggregate form of TPE-IM, decreased with increasing temperature. These changes can be explained by the fact that an increase in temperature may increase the non-radiative molecular motions of TPE-IM molecules due to the breakage of the non-covalent interaction forces between TPE-IM and S-βCD molecules and the disassociation of the TPE-IM/S-βCD assembly system. A similar effect of temperature on aggregation assembly has also been noted previously.26,70 Thus, from the above temperature-dependent studies, it can be concluded that non-covalent interaction forces are involved in establishing a complex between cationic TPE-IM and anionic S-βCD. The temperature-dependent analysis of the TPE-IM/S-βCD complex reveals a clear correlation between rising temperature and a decrease in fluorescence intensity. This behavior is attributed to the disruption of non-covalent interactions that are central to supramolecular assemblies. As the temperature increases, there is a pronounced disintegration of the TPE-IM/S-βCD complex, evident from both steady-state fluorescence and ground-state absorption spectra. Transient decay measurements further corroborated this observation, with decay traces becoming progressively faster at elevated temperatures. These measurements emphasizes the critical role of non-covalent forces in the stability and behavior of the TPE-IM/S-βCD complex, particularly under varying temperature conditions.
3.3 Fluorescence ‘Turn-Off’ of TPE-IM/S-βCD assembly complex by protamine sulfate (PrS)
With an intention to develop a sensing system for trypsin detection, we investigated the effect of PrS, a poly cationic protein and a natural substrate for trypsin, on the TPE-IM/S-βCD complex. As PrS carries multiple positive charges, it may bind to multiple negative charges bearing S-βCD and cause the disassembly of the TPE-IM/S-βCD complex. To test this hypothesis, the fluorescence response of the TPE-IM/S-βCD complex has been measured at different concentrations of PrS, and the results are presented in Fig. 4(A). Fig. 4(A) shows that, with the addition of different concentrations of PrS, the emission intensity of the TPE-IM/S-βCD aggregation complex decreased and was significantly quenched in the presence of ∼4 μM PrS. It has been reported that PrS interacts electrostatically with anionic molecules and, therefore, can disrupt aggregated assembly systems.71,72Therefore, in the current scenario, it can be assumed that in the presence of PrS, the TPE-IM/S-βCD aggregation complex is disrupted owing to the preferential polycationic and polyanionic complexation between PrS and S-βCD. As a result, the disintegration of the TPE-IM molecules from the TPE-IM/S-βCD assembly is promoted. Hence, an almost negligible fluorescence intensity was observed in the presence of PrS due to the release of free TPE-IM molecules in the solution (Fig. 4(A)). To corroborate the findings of the steady-state emission experiments, excited-state lifetime measurements were conducted, and the results are displayed in Fig. 4(B). As shown in Fig. 4(B), the decay traces of the TPE-IM/S-βCD complex exhibit an increasingly faster decay profile as the concentration of PrS gradually rises. The analysis of the amplitudes of the decay components is presented in the inset of Fig. 4(B). Evidently, the amplitudes of the free form of TPE-IM (a1) increase upon the addition of Protamine Sulfate (PrS), whereas the amplitude for the aggregate form of the dye (a2 + a3) decreases, and the variation of these amplitudes nicely correlates with each other. This clearly suggests a protamine-induced transformation of the aggregate from the TPE-IM/S-βCD association complex to free TPE-IM. As previously hypothesized, the introduction of positively charged PrS molecules competed with TPE-IM molecules to form a complex with S-βCD. This competition causes the disintegration of the TPE-IM/S-βCD assembly complex, resulting in the creation of a PrS/S-βCD complex and the release of free TPE-IM molecules into the solution. Owing to the faster excited-state relaxation associated with free TPE-IM molecules, the gradual disruption of the TPE-IM/S-βCD complex led to a progressively faster decay rate in the observed traces. Thus, the results of the excite-state measurements corroborate those obtained from the steady-state emission measurements. In Section 3.2.3, we have shown that the behaviour of TPE-IM/S-βCD complex is largely unaffected by pH changes within the range of 2–12. However, introducing PrS might change this behavior. To address this, we have conducted experiments examining the fluorescence response of the TPE-IM/S-βCD complex in the presence of PrS under various pH conditions (Fig. S9, ESI†). Our results show that the fluorescence intensity of our system remains stable across different pH levels, demonstrating its reliability even when polyelectrolytes or proteins like PrS are present. This stability is likely due to the nature of protamine, a biomolecule rich in arginine with large cationic charge and an isoelectric point between 12 and 13. Therefore, at pH levels below 12, Protamine retains its positive charge, essential for its interaction with the negatively charged S-βCD, ensuring that the response of the system stays consistent under pH 12.
3.4 Trypsin detection
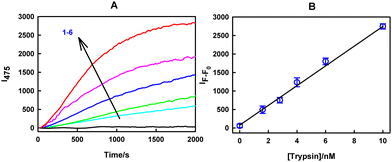
After obtaining a PrS concentration-dependent response from the TPE-IM/S-βCD supramolecular assembly, we intended to obtain a trypsin concentration-dependent response from TPE-IM/S-βCD/PrS system. For this purpose, the TPE-IM/S-βCD/PrS system was subjected to various concentrations of trypsin, and the results are presented in Fig. 5. As shown in Fig. 5(A), recovery of the fluorescence of the TPE-IM/S-βCD/PrS complex is observed in the presence of different trypsin concentrations, which can be attributed to the disassembly of the S-βCD/PrS complex that subsequently triggers the reassembly of the TPE-IM/S-βCD aggregation system. It is well reported that trypsin-specific cleavage of PrS into smaller fragments leads to the loss of its polycationic nature. This, in turn, leads to the reduced affinity of PrS fragments for S-βCD compared to tetra-cationic TPE-IM. Therefore, in this study, it can be inferred that enzymatic cleavage of PrS leads to the re-aggregation of the TPE-IM/S-βCD complex in aqueous buffer, resulting in an increase in fluorescence intensity at 475 nm. Consequently, the TPE-IM/S-βCD/PrS aggregation complex can be considered a promising platform for trypsin detection. To determine the analytical parameters of the TPE-IM/S-βCD/PrS complex assembly system for trypsin detection, changes in the emission intensity at 475 nm were plotted against various trypsin concentrations and linearly fitted. The results are shown in Fig. 5(B). The limit of detection (LOD) for Trypsin using the TPE-IM/S-βCD/PrS complex is 10 pM within a linear detection range of 0–10 nM. This was determined using the linear regression equation I(F − F0) = 273 [trypsin/nM] + 67, which yielded a regression coefficient (R2) of 0.993. The LOD was calculated using the formula 3σ/s, where ‘s’ represents the slope obtained from the fitted line, and ‘σ’ corresponds to the standard deviation of ten blank readings. Consequently, the TPE-IM/S-βCD/PrS complex shows promise as a sensor probe for trypsin detection. Table 1 compares the LOD of the current detection method for trypsin detection with other reported methods. As demonstrated in Table 1, the sensitivity of the TPE-IM/S-βCD/PrS complex system outperforms numerous previously reported methods.| S. no. | Probe/material | LOD | Linearity | Incubation time | Ref. |
|---|---|---|---|---|---|
| 1 | CuNPs/Cyt-C | 42 ng mL−1 | 0.25–1000 μg mL−1 | 30 | 73 |
| 2 | Fe3O4-PDA-HSA/anti-HSA CdTe QDs | 0.25 μg mL−1 | 0.5–30 μg mL−1 | 60 | 74 |
| 3 | Mn: ZnSe d-dots-Arg6 | 40 ng mL−1 | 0.1–12.0 μg mL−1 | 30 | 75 |
| 4 | Arg6-FAM/graphene oxides | 0.1 μg mL−1 | 0–10 μg mL−1 | 15 | 76 |
| 5 | GSH-AuNCs | 0.08 μg mL−1 | 0.2–100 μg mL−1 | 60 | 51 |
| 6 | Poly-Arg polymer nanoparticles/graphene oxides | 0.827 μg mL−1 | 0–25 μg mL−1 | 40 | 77 |
| 7 | SPR | 25.7 μg mL−1 | 0.0–0.30 mg mL−1 | 78 | |
| 8 | BSA-AuNCs/TMB | 0.6 μg mL−1 | 0.9–1 mg mL−1 | 120 | 79 |
| 9 | MPA-AgInZnS | 0.04 μg mL−1 | 0.1–2.0 μg mL−1 | 3.33 | 80 |
| 10 | Alkynylplatinum(II) 2,6-bis(benzimidazol-2′-yl)pyridine | 6.36 ng mL−1 | 0.006 to 0.06 μg mL−1 | 81 | |
| 11 | TPE-IM/S-βCD/PrS | 0.24 ng mL | 0–240 ng mL−1 | None | This method |
3.5 Selectivity
The specificity of a sensing system is of paramount importance, especially when the target analyte co-exists with other similar entities. Therefore, we sought to evaluate the selectivity of the TPE-IM/S-βCD/PrS complex towards trypsin as compared to other enzymes and proteins. To this end, we recorded steady-state emission spectra in the presence of a diverse set of enzymes and proteins, including alkaline phosphatase (ALP), bovine serum albumin (BSA), choline oxidase (ChOx), glucose oxidase (GluOx), hemoglobin (Hb), horseradish peroxidase (HRP), and lysozyme. Our observations, detailed in Fig. 6(A), revealed that only trypsin demonstrated a distinct binding affinity with the TPE-IM/S-βCD/PrS complex. This unique interaction with trypsin facilitated the selective cleavage of PrS. Following this cleavage, there is a consequential re-aggregation of the TPE-IM/S-βCD complex, culminating in a marked enhancement in fluorescence intensity. The molecular basis of this selectivity can be attributed to specificity of tryptic cleavage towards PrS. In conclusion, the TPE-IM/S-βCD/PrS system exhibited remarkable selectivity for trypsin detection in aqueous solutions. Such specificity, especially in the presence of potential interferences, underscores the robustness of the system and its promising application in complex biological environments.3.6 Trypsin inhibition by benzamidine
Next, to determine and demonstrate the potential application of the TPE-IM/S-βCD/PrS aggregation system to detect trypsin inhibitors, a well-known natural inhibitor of trypsin, benzamidine,82 was used as a model inhibitor, and the results are shown in Fig. S10 (ESI†). It is evident from the results that the fluorescence intensity of the TPE-IM/S-βCD/PrS/trypsin complex decreased with time as the concentration of the benzamidine compound increased in the solution (0–160 μM). Next, to measure the inhibitory effect of benzamidine, the percentage inhibition of trypsin activity was evaluated as the recovery of the emission intensity of the TPE-IM/S-βCD/PrS/trypsin complex, and the results are shown in Fig. 6(B). The percentage inhibition was calculated using the formula (F0 − F)/F0, where F0 and F denote the fluorescence intensity of the TPE-IM/S-βCD/PrS/trypsin system at the 1800th second in the absence and presence of benzamidine, respectively. It is visible from the results of Fig. 6(B) that trypsin activity is lost by almost >90% upon the addition of 160 μM of inhibitor. As benzamidine-dependent inhibition of Trypsin has already been well studied,82 the above results demonstrate the potential of the TPE-IM/S-βCD/PrS/trypsin system as a potential sensing platform for detecting trypsin inhibitors.3.7 Trypsin detection in human serum
Trypsin, as highlighted earlier, serves as a biomarker for a myriad of disease.42,43 Recognizing its clinical significance, we endeavored to validate the performance of our sensing system in a complex biological matrix, namely, human serum. For this purpose, we incorporated the TPE-IM/S-βCD/PrS complex into human serum samples (2%) and subsequently introduced varying concentrations of trypsin. The outcomes, as illustrated in Fig. 7, display a discernible fluorescence enhancement at 475 nm, which is amplified in tandem with increasing trypsin concentrations. This fluorescence behavior mirrors our observations from experiments conducted in an aqueous medium, underscoring the robustness of our system. To quantify the sensitivity of the system in this matrix, we analyzed the data and derived a linear relationship described by the equation IF−F0 = 38[trypsin/nM] + 69. This equation yields an impressive linear regression coefficient (R2) of 0.94. Furthermore, we established a detection limit of 78 pM encompassing a linear concentration span of 0–20 nM.Based on these experimental insights, we suggest that the TPE-IM/S-βCD/PrS complex system holds significant promise as an efficient and reliable sensing platform for trypsin detection in real-world biological samples, such as human serum. This capability could potentially update diagnostics, offering a sensitive and rapid method in contrast to existing techniques.
4. Conclusions
In this study, we successfully developed a trypsin-responsive fluorescence turn-on aggregation complex, TPE-IM/S-βCD/PrS. This system leverages the anionic macrocyclic molecule S-βCD and the cationic TPE derivative TPE-IM in conjunction with multi-cationic PrS molecules. Central to this mechanism, a supramolecular assembly (TPE-IM/S-βCD) forms based on electrostatic interactions between polyanionic S-βCD and cationic TPE-IM. Intriguingly, while isolated TPE-IM molecules remain non-fluorescent owing to intramolecular rotational motions, they exhibit strong emission in an S-βCD-induced aggregation state. The presence of PrS, a natural substrate of trypsin, quenched the fluorescence of the TPE-IM/S-βCD complex. This is attributed to the release of free TPE-IM in solution and the consequent formation of an S-βCD/PrS aggregation complex. However, the introduction of trypsin catalyzes the enzymatic cleavage of PrS, prompting the disintegration of the S-βCD/PrS complex and reassembly of the TPE-IM/S-βCD complex. This dynamic system offers a novel approach to trypsin detection. The sensing system showed remarkable linearity across a concentration range of 0–10 nM, with a limit of detection (LOD) as low as 10 pM. In the context of real-world applications, the system effectively detects trypsin in human serum samples, achieving a LOD of 78 pM. Beyond detection, the utility of the platform extends to screening for trypsin inhibitors, as demonstrated with benzamidine, a natural trypsin inhibitor. This platform stands out as a simple, rapid, and sensitive solution, enriching the toolkit available to researchers and clinicians focusing on trypsin detection and related endeavors. The potential of this system in clinical diagnostics, especially in monitoring trypsin-related diseases, is noteworthy and could pave the way for improved patient care. Furthermore, its application in screening trypsin inhibitors provides opportunities for drug discovery, particularly under conditions characterized by trypsin overactivity. We anticipate that this platform will inspire further research into therapeutic agents targeting trypsin or related proteases, marking a significant step in the biochemistry and diagnostics domain.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The authors acknowledge the support from the host institute during the course of this work. Jasvir Kaur thanks the Indian Council of Medical Research, New Delhi, for providing the financial support.References
- L. Peng, G. Zhang, D. Zhang, J. Xiang, R. Zhao, Y. Wang and D. Zhu, A Fluorescence “Turn-On” Ensemble for Acetylcholinesterase Activity Assay and Inhibitor Screening DOI:10.1021/ol9016723, (accessed October 18, 2022).
- Q. Hao, Y. Kang, J.-F. Xu and X. Zhang, Langmuir, 2021, 37, 6062–6068 CrossRef CAS PubMed.
- N. Swaminathan, N. Sharma, Y. Nerthigan and H.-F. Wu, Appl. Surf. Sci., 2021, 554, 149600 CrossRef CAS.
- D. Ding, K. Li, B. Liu and B. Z. Tang, Acc. Chem. Res., 2013, 46, 2441–2453 CrossRef CAS PubMed.
- J. Shi, Y. Li, Q. Li and Z. Li, ACS Appl. Mater. Interfaces, 2018, 10, 12278–12294 CrossRef CAS PubMed.
- Y. Lu, A. A. Aimetti, R. Langer and Z. Gu, Nat. Rev. Mater., 2016, 2, 1–17 Search PubMed.
- A. Tanaka, Y. Fukuoka, Y. Morimoto, T. Honjo, D. Koda, M. Goto and T. Maruyama, J. Am. Chem. Soc., 2015, 137, 770–775 CrossRef CAS PubMed.
- B. Renoux, F. Raes, T. Legigan, E. Péraudeau, B. Eddhif, P. Poinot, I. Tranoy-Opalinski, J. Alsarraf, O. Koniev, S. Kolodych, S. Lerondel, A. L. Pape, J. Clarhaut and S. Papot, Chem. Sci., 2017, 8, 3427–3433 RSC.
- Q. Hu, P. S. Katti and Z. Gu, Nanoscale, 2014, 6, 12273–12286 RSC.
- A. J. Harnoy, I. Rosenbaum, E. Tirosh, Y. Ebenstein, R. Shaharabani, R. Beck and R. J. Amir, J. Am. Chem. Soc., 2014, 136, 7531–7534 CrossRef CAS PubMed.
- J. Mu, J. Lin, P. Huang and X. Chen, Chem. Soc. Rev., 2018, 47, 5554–5573 RSC.
- G. Bharath, R. Madhu, S.-M. Chen, V. Veeramani, A. Balamurugan, D. Mangalaraj, C. Viswanathan and N. Ponpandian, J. Mater. Chem. B, 2015, 3, 1360–1370 RSC.
- Y. Wang, C. Hou, Y. Zhang, F. He, M. Liu and X. Li, J. Mater. Chem. B, 2016, 4, 3695–3702 RSC.
- D.-S. Guo, K. Wang, Y.-X. Wang and Y. Liu, J. Am. Chem. Soc., 2012, 134, 10244–10250 CrossRef CAS PubMed.
- X. Xiao, Z. Xu, W. Wang, S. Sun, Y. Qiao, L. Jiang, Y. Yan and J. Huang, Langmuir, 2021, 37, 8348–8355 CrossRef CAS PubMed.
- D.-S. Guo, T.-X. Zhang, Y.-X. Wang and Y. Liu, Chem. Commun., 2013, 49, 6779–6781 RSC.
- Y. Ding, Y. Kang and X. Zhang, Chem. Commun., 2014, 51, 996–1003 RSC.
- Y.-C. Pan, H. Wang, X. Xu, H.-W. Tian, H. Zhao, X.-Y. Hu, Y. Zhao, Y. Liu, G. Ding, Q. Meng, B. J. Ravoo, T. Zhang and D.-S. Guo, CCS Chem., 2020, 3, 2485–2497 CrossRef.
- J. Kaur and P. K. Singh, Sens. Actuators, B, 2021, 346, 130517 CrossRef CAS.
- J. Kaur, D. N. Nadimetla, S. V. Bhosale and P. K. Singh, J. Phys. Chem. B, 2022, 126, 1147–1155 CrossRef CAS PubMed.
- J. Wang, Y. Zhao, F.-X. Ma, K. Wang, F.-B. Wang and X.-H. Xia, J. Mater. Chem. B, 2013, 1, 1406–1413 RSC.
- N. H. Mudliar and P. K. Singh, Chem. – Eur. J., 2016, 22, 7394–7398 CrossRef CAS PubMed.
- G. Singh and P. K. Singh, Langmuir, 2019, 35, 14628–14638 CrossRef CAS PubMed.
- Y. Chen and Y. Liu, Chem. Soc. Rev., 2010, 39, 495–505 RSC.
- J. Luo, Z. Xie, J. W. Y. Lam, L. Cheng, H. Chen, C. Qiu, H. S. Kwok, X. Zhan, Y. Liu, D. Zhu and B. Z. Tang, Chem. Commun., 2001, 1740–1741 RSC.
- Y. Ren, J. W. Y. Lam, Y. Dong, B. Z. Tang and K. S. Wong, J. Phys. Chem. B, 2005, 109, 1135–1140 CrossRef CAS PubMed.
- T. Zhou, Q. Wang, M. Liu, Z. Liu, Z. Zhu, X. Zhao and W.-H. Zhu, Aggregate, 2021, 2, e22 CrossRef CAS.
- H. Li, H. Kim, J. Han, V.-N. Nguyen, X. Peng and J. Yoon, Aggregate, 2021, 2, e51 CrossRef CAS.
- L. Peng, L. Xiao, Y. Ding, Y. Xiang and A. Tong, J. Mater. Chem. B, 2018, 6, 3922–3926 RSC.
- N. Gu and S. Liu, J. Mater. Chem. B, 2020, 8, 3168–3170 RSC.
- H.-B. Wang, B.-B. Tao, A.-L. Mao, Z.-L. Xiao and Y.-M. Liu, Sens. Actuators, B, 2021, 348, 130729 CrossRef CAS.
- H.-B. Wang, A.-L. Mao, T. Gan and Y.-M. Liu, Analyst, 2020, 145, 7009–7017 RSC.
- S. K. Liew, S. Malagobadan, N. M. Arshad and N. H. Nagoor, Biomolecules, 2020, 10, 138 CrossRef CAS PubMed.
- E. E. Bittar, Arch. Intern. Med., 1962, 109, 601–602 CrossRef CAS PubMed.
- O. L. Tavano, A. Berenguer-Murcia, F. Secundo and R. Fernandez-Lafuente, Compr. Rev. Food Sci. Food Saf., 2018, 17, 412–436 CrossRef PubMed.
- J. Kaur and P. K. Singh, Crit. Rev. Anal. Chem., 2020, 1–19 CAS.
- I. M. E. Lacroix, X.-M. Chen, D. D. Kitts and E. C. Y. Li-Chan, Food Funct., 2017, 8, 701–709 RSC.
- X. Wang, H. Chen, X. Fu, S. Li and J. Wei, LWT--Food Sci. Technol., 2017, 75, 93–99 CrossRef CAS.
- J. Z. Kiser, M. Post, B. Wang and M. Miyagi, J. Proteome Res., 2009, 8, 1810–1817 CrossRef CAS PubMed.
- S. N. Huang, H. Minassian and J. D. More, Lab. Invest., 1976, 35, 383–390 CAS.
- E. Vandermarliere, M. Mueller and L. Martens, Mass Spectrom. Rev., 2013, 32, 453–465 CrossRef CAS PubMed.
- M. Hirota, M. Ohmuraya and H. Baba, J. Gastroenterol., 2006, 41, 832–836 CrossRef CAS PubMed.
- P. Dandona, M. Hodson, J. Bell, L. Ramdial, I. Beldon and J. C. Batten, Thorax, 1981, 36, 60–62 CrossRef CAS PubMed.
- V. Fonseca, O. Epstein, A. Katrak, D. Junglee, D. P. Mikhailidis, N. McIntyre and P. Dandona, J. Clin. Pathol., 1986, 39, 638–640 CrossRef CAS PubMed.
- H. C. Heinrich, E. E. Gabbe and F. Ičagić, Klin. Wochenschr., 1979, 57, 1237–1238 CrossRef CAS PubMed.
- M. Amouzadeh Tabrizi, J. Ferré-Borrull and L. F. Marsal, Sci. Rep., 2020, 10, 2356 CrossRef CAS PubMed.
- A. D. Kersey, T. A. Berkoff and W. W. Morey, Opt. Lett., 1993, 18, 1370–1372 CrossRef CAS PubMed.
- J. A. Braatz, C. Elias, J. G. Finny, H. Tran and M. McCaman, J. Immunol. Methods, 2015, 417, 131–133 CrossRef CAS PubMed.
- M. Dong, H. Qi, S. Ding and M. Li, Microchim. Acta, 2015, 182, 43–49 CrossRef CAS.
- N. A. Karaseva, B. Pluhar, E. A. Beliaeva, T. N. Ermolaeva and B. Mizaikoff, Sens. Actuators, B, 2019, 280, 272–279 CrossRef CAS.
- H. Li, M. Yang, D. Kong, R. Jin, X. Zhao, F. Liu, X. Yan, Y. Lin and G. Lu, Sens. Actuators, B, 2019, 282, 366–372 CrossRef CAS.
- N.-N. Wu, L.-G. Chen, M.-Z. Xiao, R.-Y. Yuan and H.-B. Wang, Microchim. Acta, 2023, 190, 158 CrossRef CAS PubMed.
- P. Li, Y. Liu, X. Wang and B. Tang, Analyst, 2011, 136, 4520–4525 RSC.
- K. Wang, D.-S. Guo, M.-Y. Zhao and Y. Liu, Chemistry, 2016, 22, 1475–1483 CrossRef CAS PubMed.
- X. Dai, Y. Chen and Y. Liu, in Handbook of Macrocyclic Supramolecular Assembly, ed. Y. Liu, Y. Chen and H.-Y. Zhang, Springer, Singapore, 2019, pp. 1–19 Search PubMed.
- C. Kotras, M. Fossépré, M. Roger, V. Gervais, S. Richeter, P. Gerbier, S. Ulrich, M. Surin and S. Clément, Front. Chem, 2019, 7, 493 CrossRef CAS PubMed.
- M. Kumbhakar, P. K. Singh, A. K. Satpati, S. Nath and H. Pal, J. Phys. Chem. B, 2010, 114, 10057–10065 CrossRef CAS PubMed.
- A. M. Pettiwala and P. K. Singh, Spectrochim. Acta, Part A, 2018, 188, 120–126 CrossRef CAS PubMed.
- P. K. Singh, A. K. Satpati, M. Kumbhakar, H. Pal and S. Nath, J. Phys. Chem. B, 2008, 112, 11447–11450 CrossRef CAS PubMed.
- P. K. Singh, S. Nath, A. C. Bhasikuttan, M. Kumbhakar, J. Mohanty, S. K. Sarkar, T. Mukherjee and H. Pal, J. Chem. Phys., 2008, 129, 114504 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Plenum Press, New York, 2006 Search PubMed.
- X. Lou and Y. Yang, Aggregate, 2020, 1, 19–30 CrossRef.
- Y. Cai, L. Du, K. Samedov, X. Gu, F. Qi, H. H. Y. Sung, B. O. Patrick, Z. Yan, X. Jiang, H. Zhang, J. W. Y. Lam, I. D. Williams, D. L. Phillips, A. Qin and B. Z. Tang, Chem. Sci., 2018, 9, 4662–4670 RSC.
- E. Arad, H. Green, R. Jelinek and H. Rapaport, J. Colloid Interface Sci., 2020, 573, 87–95 CrossRef CAS PubMed.
- J. Kaur and P. K. Singh, Sens. Actuators, B, 2021, 130517 CrossRef CAS.
- S. P. Pandey, P. Jha and P. K. Singh, J. Mol. Liq., 2020, 315, 113625 CrossRef CAS.
- J. Kaur and P. K. Singh, Microchem. J., 2022, 183, 108091 CrossRef CAS.
- A. A. Awasthi and P. K. Singh, J. Phys. Chem. B, 2017, 121, 6208–6219 CrossRef CAS PubMed.
- M. M. Cox and D. L. Nelson, Principles of Biochemistry, W H Freeman & Co, 2008 Search PubMed.
- J. Kaur, J. N. Malegaonkar, S. V. Bhosale and P. K. Singh, J. Mol. Liq., 2021, 333, 115980 CrossRef CAS.
- J. Kaur, J. Malegaonkar, S. Bhosale and P. Singh, J. Mol. Liq., 2021, 333, 115980 CrossRef CAS.
- V. R. Singh, J. N. Malegaonkar, S. V. Bhosale and P. K. Singh, Org. Biomol. Chem., 2020, 18, 8414–8423 RSC.
- L.-J. Ou, X.-Y. Li, L.-J. Li, H.-W. Liu, A.-M. Sun and K.-J. Liu, Analyst, 2015, 140, 1871–1875 RSC.
- T. Xia, Q. Ma, T. Hu and X. Su, Talanta, 2017, 170, 286–290 CrossRef CAS PubMed.
- X. Gao, G. Tang, Y. Li and X. Su, Anal. Chim. Acta, 2012, 743, 131–136 CrossRef CAS PubMed.
- X. Gu, G. Yang, G. Zhang, D. Zhang and D. Zhu, ACS Appl. Mater. Interfaces, 2011, 3, 1175–1179 CrossRef CAS PubMed.
- J. Noh, B.-J. Chae, B.-C. Ku and T. S. Lee, J. Polym. Sci., Part A: Polym. Chem., 2014, 52, 1898–1904 CrossRef CAS.
- S. Dutta, K. Saikia and P. Nath, RSC Adv., 2016, 6, 21871–21880 RSC.
- G.-L. Wang, L.-Y. Jin, Y.-M. Dong, X.-M. Wu and Z.-J. Li, Biosens. Bioelectron., 2015, 64, 523–529 CrossRef CAS PubMed.
- Y. Liu, F. Zhang, X. He, P. Ma, Y. Huang, S. Tao, Y. Sun, X. Wang and D. Song, Sens. Actuators, B, 2019, 294, 263–269 CrossRef CAS.
- C. W.-T. Chan, H.-K. Cheng, F. K.-W. Hau, A. K.-W. Chan and V. W.-W. Yam, ACS Appl. Mater. Interfaces, 2019, 11, 31585–31593 CrossRef CAS PubMed.
- R. Mogaki, K. Okuro and T. Aida, Chem. Sci., 2015, 6, 2802–2805 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00264d |
| This journal is © The Royal Society of Chemistry 2024 |