Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Feeding secondary fermentations with mammalian and fungal culture waste streams increases productivity and resource efficiency†
Ciara D
Lynch
ab,
Federico
Cerrone
*ab,
Kevin E.
O'Connor
ab and
David J.
O'Connell
 *ab
*ab
aSchool of Biomedical and Biomolecular Sciences, University College Dublin, Belfield, Dublin 4, Ireland. E-mail: federico.cerrone@dcu.ie; david.oconnell@ucd.ie
bBiOrbic, Bioeconomy Research Centre, Belfield, Dublin 4, Ireland
First published on 14th May 2024
Abstract
The evolution of the circular bioeconomy will require the realisation of new value from waste streams generated from all manufacturing processes. Bioprocessing of recombinant proteins and functional foods using animal and microbial fermentations are fast growing industries with increasing volumes of spent culture media waste resulting in significant resource inefficiency. Here Chinese hamster ovary cell spent media (CSM) and Trametes versicolor fungal cell spent media (FSM) were used as model waste streams to feed Escherichia coli in secondary fermentations. Expression of a recombinant fusion protein was used to measure the value of these streams as feedstuff. E. coli cultures fed with either waste stream in bioreactors produced equivalent yields of the protein as those fed with rich microbiological media. Separately CSM was tested as a replacement feed for the fungal cell fermentation producing beta and alpha glucans. This waste-fed fermentation resulted in a 2-fold higher glucan yield than when using standard corn steep liquor feed. Quantitative analysis of the spent media content with mass spectrometry and hplc methods showed significant differences in elemental composition profiles, quantities of individual amino acids and carboxylic acids and levels of carbon sources. These findings underline the versatility of E. coli in utilisation of waste media as a feedstuff and show that the range of applications of CHO cell culture media waste are not limited to feeding bacterial fermentation. Exploration of cellular bioprocesses that can efficiently use cell media waste as fermentation feed will be a valuable step towards increased resource efficiency in bioprocessing industries.
Sustainability spotlightThe expanding range and scale of valuable biomolecular products developed with bioprocessing activities using animal and microbial cells is responsible for the production of hundreds of millions of litres of waste culture media annually. Recycling this waste stream has the potential to decrease the impact of the associated high water and energy consumption contributing to meeting targets for the Paris 2050 Agreement and UN Sustainable Development Goals. This work shows that waste media from distinct bioprocesses can be reused to sustain high-level recombinant protein production in a secondary fermentation and the notable finding that a waste media can add value to a secondary process by increasing its productivity. Efficient valorisation of waste media is an urgent and scientifically valuable objective. |
Introduction
The implementation of a circular bioeconomy is a major goal in the attempt to bring humans back to living within the earth's planetary boundaries, reducing the impact of human activities on climate change. Current research seeks to maximise resource efficiency by maintaining the value and utility of resources for as long as possible e.g., the reuse of waste within a process or use as a starting material for a secondary process.1 This is reflected in a broad array of studies into the conversion of waste to products of value in the last ten years, from generation of valuable biomass to synthesis of bioplastics.2–8 One example of a waste intensive process yet to be assessed for conversion to products of value is the cell culture waste produced by the bioprocessing industry. There are currently 443 individual licensed biopharmaceutical products from vaccines to monoclonal antibodies that are made in cell culture bioprocess using expression hosts ranging from mammalian cells to microbes. Monoclonal antibodies, with over 80% of the total protein-based biopharmaceutical global sales in 2021, are principally made using Chinese hamster ovary cells (CHO) in intensified bioprocesses using chemically defined media at scales from hundreds to tens of thousands of litres.9 Global bioprocessing capacity has rapidly increased to provide millions of litres in production facilities worldwide to support demand for these molecules at tonnes scale. Currently in all of these facilities once the product is harvested the complex media formulation used to grow these cells is immediately sent for treatment as waste for disposal.A newer area of cell culture bioprocessing predicted to undergo rapid growth is that of precision fermentation of synthetic meat and dairy products, designed to replace conventional foods from agricultural production.10–12 Cultured meat using animal stem cells grown in bioreactors for example, has potential benefits proposed to lower greenhouse gas production from animals, reduce land use for animal farming, and reduce energy and water consumption helping to meet those targets for the Paris 2050 Agreement and UN Sustainable Development Goals.13 However, to achieve a scale to meet global demand significant volumes of new bioprocessing waste will be generated that could dwarf those produced by the biopharmaceutical industry in litres per annum.
A number of pilot studies have investigated the principle of recycling spent media waste and show utility when used as a supplement to fresh media in the same bioreactor across a number of cell types and processes.14–17 We have previously shown spent media from CHO cell culture supports growth and recombinant protein production in a secondary fermentation as a standalone feed.18 The nutritional value remaining in spent culture media could potentially be employed to feed multiple, distinct secondary fermentations. CHO cell spent culture media and spent culture media from Trametes versicolor fermentation represent models of biopharmaceutical bioprocess and precision fermentation of functional food ingredient waste streams. Filamentous fungi such as T. versicolor accumulate polysaccharides such as alpha- and beta-glucan in their cell walls.19–21 These molecules have demonstrated immunostimulant activities and are involved in the regulation of the slow absorption of glucose by gut associated lymphoid tissue (GALT) associated microbiota with health benefits against insulin resistance.22–25 Submerged cultivation of filamentous fungi is a recently developed bioprocess strategy to increase the yield of fungal biomass containing bioactive compounds, in comparison to the traditional and unsustainable compost-based methods of fungal production.26 A comparable amount of biomass is obtained in a matter of days in this way rather than weeks in the latter.
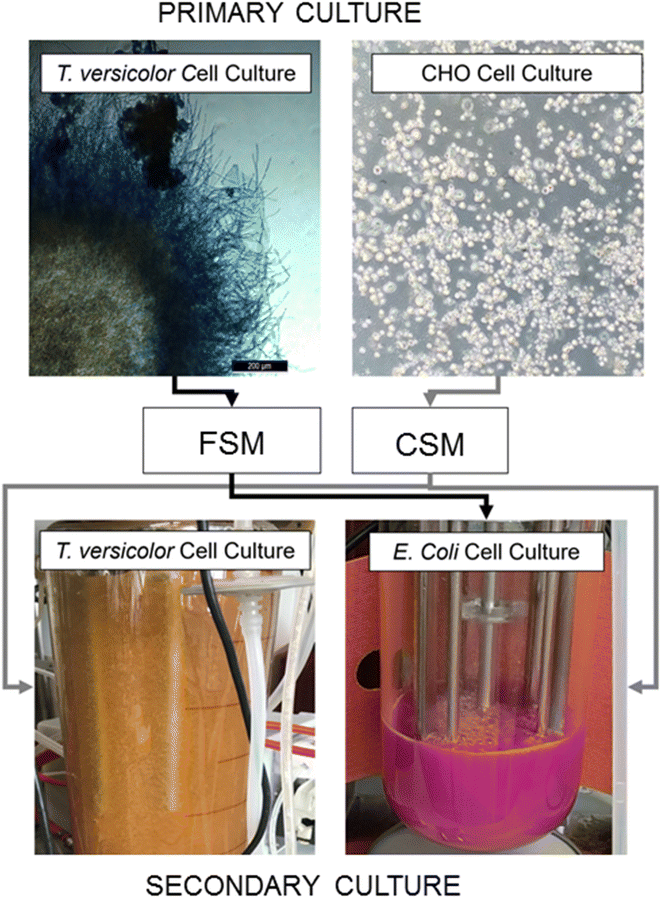
In this study we investigated production of a recombinant protein by E. coli in secondary fermentation in bioreactors fed with rich microbiological media or waste media from the model streams and compared the overall yields of recombinant protein in each. We also compared total glucan production from a T. versicolor secondary fermentation fed with CHO spent media with the yield from the standard fermentation feed. To understand the impact of waste media on each of these bioprocesses we then employed elemental composition analysis with ICP-MS and metabolite composition analysis with LC-MS/MS. Proteomic adaptations made by the E. coli and T. versicolor cells illuminate the versatility of expression hosts in successfully utilising these waste media feeds (Fig. 1).
Results
Spent media from CHO and Trametes versicolor cell culture as nutrients in an E. coli fermentation
CHO spent media (CSM) and fungal spent media (FSM) were compared with respect to the growth rate of the E. coli culture and the recombinant protein expression levels (mCherry-EF2) in shake flask culture. E. coli cultures grown in the standard rich media of LB grew fastest with a specific k of 1.042, while E. coli grown in CSM achieved a growth rate (k) at ∼93% of LB grown cultures. FSM grown cultures had the lowest growth rate (∼88% of LB grown cultures) (Table 1). Recombinant protein yield of the CSM reached 68% of the LB fed cultures, while FSM reached 44% (Table 1).| Measurement | LB | CSM | FSM |
|---|---|---|---|
| Flasks | |||
| Specific growth rate (k) | 1.042 | 0.928 | 0.884 |
| Doubling time (h) | 0.665 | 0.747 | 0.784 |
| R-squared coefficient | 0.998 | 0.995 | 0.996 |
| Protein produced (g L−1 ± SD) | 0.489 ± 0.017 | 0.333 ± 0.002 | 0.216 ± 0.008 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Bioreactors | |||
| Specific growth rate (k) | 0.589 | 0.264 | 0.454 |
| Doubling time (h) | 1.177 | 2.630 | 1.528 |
| R-squared coefficient | 0.991 | 0.951 | 0.950 |
| Cell dry weight (g L−1) | 7.100 | 6.100 | 5.400 |
| Protein produced (g L−1 ± SD) | 1.292 ± 0.093 | 1.353 ± 0.076 | 1.190 ± 0.057 |
Comparison of the cultures in 0.5 L reactors for 20 hours expressions showed the growth of the E. coli in the CSM and FSM conditions remained slower than the control LB rich medium while conversely the recombinant protein yield was greater than or equal to the LB control with 1.35 g L−1 (CSM) and 1.19 g L−1 (FSM) compared to 1.14 g L−1 (LB) (Table 1).
Glycerol consumption was negligible across the conditions, except after induction of expression in the CSM condition, where a further ∼10 g L−1 was consumed by the end of the culture time frame (Fig. 2). Detection of carbon sources by HPLC revealed that, while no glucose was supplemented to any media, the waste media contained glucose (9.08 and 2.92 g L−1 respectively) prior to use in secondary fermentation. LB on the other hand contained no glucose prior to starter culture addition. After addition of the starter culture, the glucose concentrations of the LB, CSM and FSM were 2.76, 9.77 and 5.14 g L−1 respectively. All glucose was fully consumed by the time expression was induced by IPTG addition (Fig. 2).
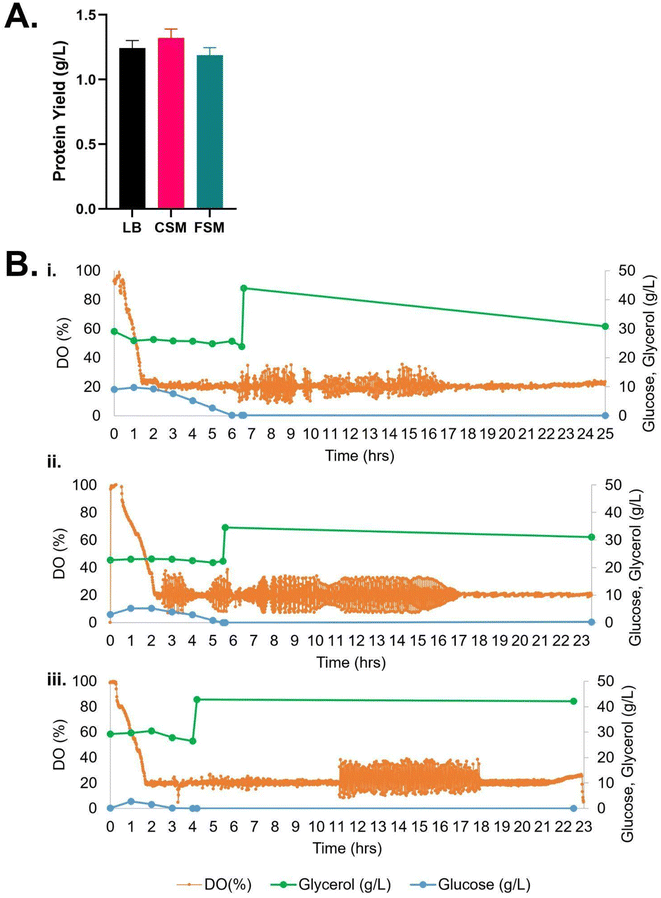 | ||
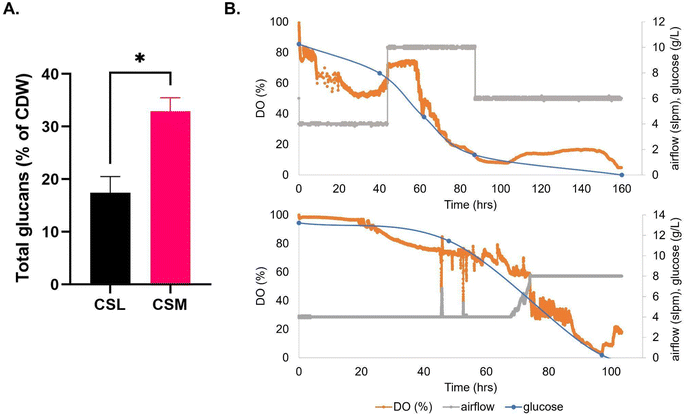
| Fig. 2 (A) Recombinant protein yield of mCherry-EF2 from E. coli cultured in various media in bioreactors, LB = lysogeny broth, CSM = CHO spent media, FSM = fungal spent media. Error bars represent standard deviation, where n = 2. No statistical difference was seen in the conditions. (B) Bioreactor timelapse of dissolved oxygen (DO) and carbon source consumption measured by HPLC (glycerol and glucose), (i) CSM, (ii) FSM, (iii) LB. IPTG induction of protein expression was carried out along with addition of 2% glycerol once cultures reached OD600 nm of 10. For raw data see ESI Table S1.† | ||
Spent media from CHO cell culture as nutrient addition in secondary Trametes versicolor fermentation
The CSM was used to supplement a fungal media composed of 10 g L−1 of glucose and 3.3 g L−1 of KH2PO4. This was added as a 15 ml L−1 sterile solution to shake flask cultures of Trametes versicolor. This spent media-supplemented culture achieved an average CDW of 7.23 ± 0.49 g L−1; this value is 13% higher than cultures grown in the typical corn steep liquor (CSL) control media (Table 2). Glucose was completely consumed after 8 days (the first 15% was consumed within 3 days of growth) and the total protein was reduced by 66%. 20% of the cell dry weight of T. versicolor biomass was composed of glucans after 8 days of growth.| Compound | CSL media | CSM media |
|---|---|---|
| Flasks | ||
| Max total glucans (% as CDW) (w/w) | 18.15 ± 0.57 | 18.75 ± 7.92 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Airlift bioreactor | ||
| Max total glucans (% as CDW) (w/w) | 16.2 ± 3.32 | 32.9 ± 2.55 |
CSM was added as a 15 ml L−1 sterile supplement to the airlift bioreactor medium (Fig. 3) and the maximum average T. versicolor biomass produced was 7.0 ± 0.61 g L−1 after 40 hours of batch growth; this value was also equal to the airlift control using CSL (Fig. 3B). The volumetric productivity (as g L−1 h−1) was higher in the airlift than in the flasks i.e.: 0.17 vs. 0.037, a 4.6-fold improvement. The yields (g of biomass/g of glucose) were similar between the airlift and the flasks (i.e.: 0.7 g g−1). The total glucan production in the bioreactor was 55% higher on average than in the flasks when using CSM as supplement but no difference in levels were seen between flasks and airlift bioreactor when using CSL supplement. Total glucan yield as % cell dry weight in airlift bioreactors were 2-fold higher (32.9 ± 2.55 v 16.2 ± 3.32) in cultures fed with CSM (Table 2).
Spent media composition analysis
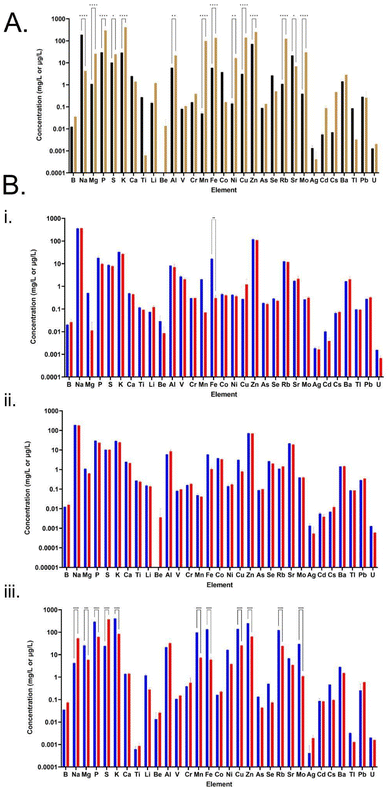
Elemental analysis of the CSM and FSM revealed distinct profiles in each media with 14 of the 31 elements tested demonstrating statistically significant differences in concentration (Fig. 4, panel A). Some elements were also either entirely absent or only present in trace amounts. For example, beryllium was present in the FSM but lacking in the CSM. Every other element tested for was present to some extent in both media, but to varying degrees. Manganese in the FSM (∼98 μg L−1) was at significantly higher levels than in CSM (∼0.05 μg L−1), while sodium was higher in the CSM (∼179 mg L−1) compared to FSM (4 mg L−1). In most of the significantly changed 14 element levels, FSM had higher concentrations than CSM, with the exceptions of sodium and strontium.According to the consumption profile of all media by the E. coli cultures (Fig. 4, panel B), the E. coli cultures grown in both LB and CSM did not change the elemental composition significantly with the only significant change seen in LB where iron was essentially fully consumed. The CSM condition had no significant change, though there was now an increased trace presence of beryllium after the E. coli culture. By contrast, the FSM composition changed significantly, with 11 elements showing significantly altered concentration levels, including two which were increased post-culture; sodium and sulphur. Consumption of potassium was ∼327 mg L−1, and phosphorous was ∼232 mg L−1 compared to LB and CSM fed cultures (K of ∼6 mg L−1, ∼4 mg L−1, and P of ∼8 mg L−1, ∼6 mg L−1). In addition to this increased consumption, the FSM also contained increased sulphur levels post-E. coli culture (difference of ∼350 mg L−1) a higher concentration than was seen in the LB or CSM media (difference of ∼1.5 mg L−1 and ∼0.1 mg L−1 respectively). High levels of zinc were also consumed by the E. coli cultures grown in the FSM, with ∼186 μg L−1 consumed from FSM compared with ∼10.9 μg L−1 and ∼2.1 μg L−1 from LB and CSM respectively. See ESI† for a table of all elemental concentrations before and after E. coli culture (Table S3†).
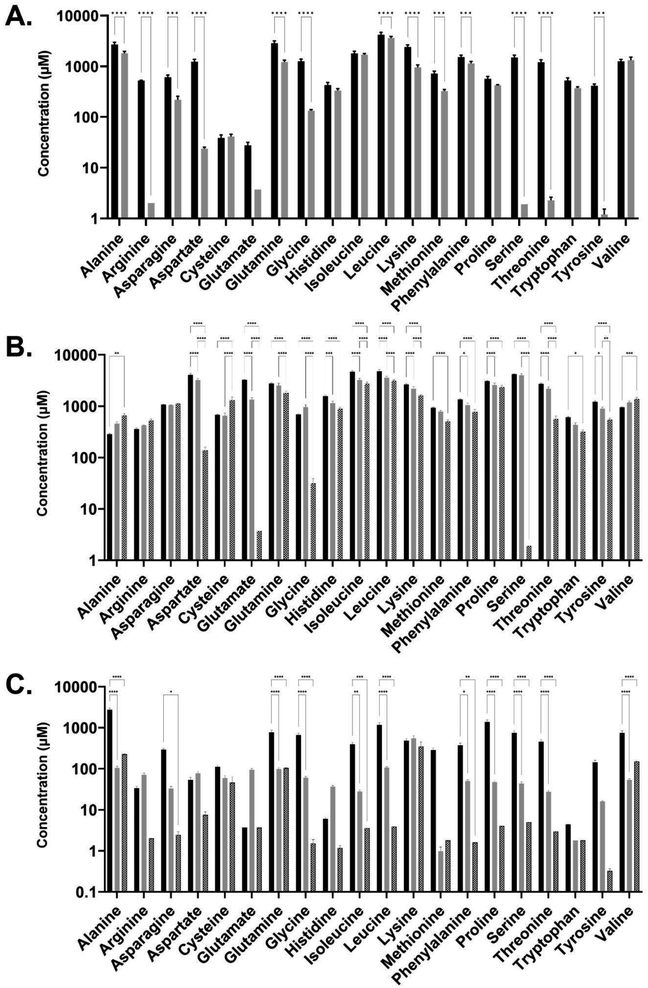
Amino acid composition analysis of the medias revealed that the two spent media conditions were also distinct in the profile of amino acid consumption post E. coli culture compared with pre-culture, and when compared to LB media (Fig. 5). Both the CSM and LB conditions contained higher concentrations of amino acids than the FSM media prior to E. coli growth. FSM had low concentrations of amino acids left after the primary fungal culture. Interestingly, the CSM fed E. coli culture showed increased levels of some amino acids, with higher concentrations seen in amino acids alanine, cysteine, and valine. The FSM fed culture also led to a slight increase in alanine and valine post-secondary culture, but this was not statistically significant (Fig. 5C). In LB fed culture there was significant consumption of amino acids glutamine, serine, lysine, aspartate, threonine, and glycine with over 1 mM of each consumed between the pre and post E. coli measurements. The CSM fed cultures had slight differences with over 1 mM serine, aspartate, threonine, glutamate and glycine consumed by the E. coli, and with 3 mM of serine and aspartate specifically consumed. This was double the amount of serine and almost triple the amount of aspartate consumed by the LB-fed E. coli. Synthesis or release of amino acids was also measured with 0.65 mM of cysteine and approximately 0.2 mM of alanine and valine produced. The FSM-fed E. coli had greatly reduced starting quantities of amino acids and the consumption profile was different with 0.2 mM lysine consumed. Leucine and glutamate were the second and third most consumed with 0.1 and 0.09 mM consumed respectively, and levels at less than 0.07 mM for other amino acids (Fig. 5C).
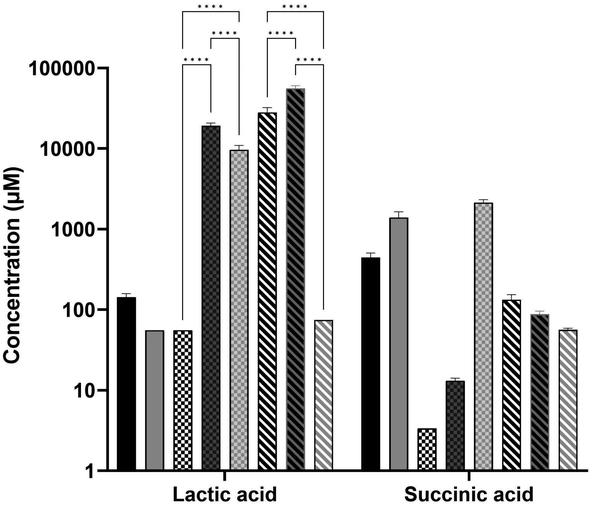
Analysis of carboxylic acids (Fig. 6) showed that lactic acid and succinic acid were both present at varying concentrations in the media, while three other organic acids were not at high enough concentrations to affect the composition (aconitic acid, hippuric acid, and 3-hydroxyglutaric acid). Succinic acid was produced by the E. coli in the CSM and LB-fed cultures though not to a significant extent, while growth in FSM did not show the same production. Lactic acid however was very high in FSM post-primary culture and was likely the reason for the low pH of this media at pH 5.5 before it was adjusted back to 7 prior to secondary culture with E. coli. The CHO cells also produced a large amount of lactic acid, though not to the extent of the fungal culture, and this was not consumed by the E. coli to the same degree as it was in the FSM-fed condition.
Proteomic analysis of Trametes versicolor
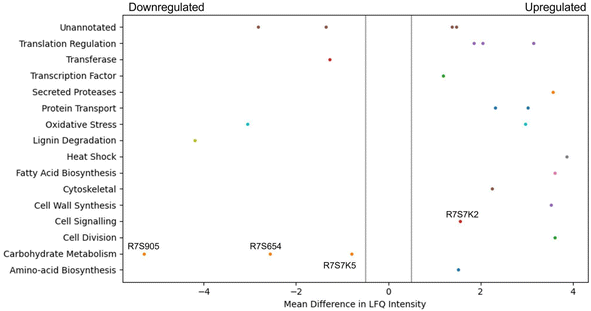
Due to the increased glucan content of the CSM-fed fungal cultures, a proteomic analysis of these cultures was performed and compared to a control (CSL-fed) culture. It was found that 15 proteins were statistically significantly dysregulated after a five-day culture, and 26 proteins after a ten-day culture (Fig. 7). Of particular interest were the annotated proteins of transaldolase (R7S654), phosphoglucomutase (R7S7K5), glycoside hydrolase (R7S9O5), and serine phosphatase (R7S7K2), all of which are indirectly linked to the beta-glucan production pathway. The first three are significantly downregulated in the CSM-supplemented condition, while the last protein, serine phosphatase is upregulated (Fig. 7).Discussion
Cell culture media is a key component of industrial bioprocess that is immediately disposed of as waste after the primary fermentation. It is well described that many nutrients remain within media post-primary culture,27–29 and this is shown in the metabolomic, proteomic and elemental analysis of the two media types used here, that we term CHO spent media (CSM) and fungal spent media (FSM) (Fig. 4–6).CHO spent media (CSM) supports secondary cultures
The CSM used was CHOgro® expression media, a chemically defined media used mainly for monoclonal antibody production, which is both serum and antibiotic free, making it ideal for a secondary culture feed.30–32 Chemically defined media often contain the same base formulae with the addition of certain nutrients necessary for high protein expression such as L-glutamine.33 Many of the elements identified as essential for CHO cell growth are also required by other eukaryotic and prokaryotic cells, such as magnesium, sodium, iron, and zinc.33,34 The elemental analysis of the CSM media showed each of these elements at levels that should support the further growth and protein production in the secondary fermentations. This observation was confirmed with equivalent recombinant protein product yields in the secondary bacterial culture and a 55% increase in glucan yield seen in the CSM-supplemented T. versicolor fermentation (Fig. 3).Fungal spent media (FSM) supports a secondary culture
Trametes versicolor is also termed Coriolus versicolor and is known more colloquially as turkey tail mushroom due to a distinctive half-moon shape found fanning out from tree trunks. T. versicolor is a medically relevant fungus as it produces many useful molecules such as antimicrobials, including some which are secreted from its cells and have been proven to be effective against certain plant pathogens such as against the fungal species F. langsethiae,35 and another protein which was found to have anti-leukemia properties.36 However, no impact of antimicrobial host cell proteins (HCPs) was observed in the secondary E. coli culture with a specific growth rate of 88% of that for the LB condition, and equivalent recombinant protein yields at bioreactor scale as LB fed cultures.CSM is more efficient at feeding shake flask cultures of E. coli than FSM
The primary fermentation feed for Trametes versicolor bioprocess is a 1.5% corn steep liquor-based media.26 Interestingly, the FSM was found to be highly acidic (pH 5.5) after the primary culture of T. versicolor cultures, which had to be adjusted to 7 in order to support the E. coli secondary culture. This low pH was found to be as a result of the high level of lactic acid (∼56 mM) present in the media after primary culture, as shown in the metabolomic analysis of carboxylic acids (Fig. 6). It was also found to have significant levels of glucose (∼3 g L−1) which took over 5 hours to be fully consumed by the bioreactor culture (Fig. 2). Though high glucose presence can inhibit the T7 expression system through catabolite repression, it was fully consumed by the induction point in the bioreactor and therefore did not inhibit the expression.37 The CSM-fed secondary cultures contained even higher levels of glucose post-CHO cell primary culture (∼9 g L−1), and therefore high glucose levels were not the reason for the lower protein yield seen in the FSM shake flasks. Lactic acid on the other hand is well known for its antimicrobial properties, used as protection against E. coli specifically in meat production.38 The high lactic acid levels in the FSM therefore aids in explaining the reduced protein yield obtained in the flask cultures in this media (0.216 g L−1 as opposed to 0.489 g L−1 in LB). There is precedence as it has been shown before that host cell protein (HCP) or leftover metabolite presence can affect the growth of cells in secondary cultures.15,17,39,40 In the shake flask cultures of E. coli fed with either CHO spent media (CSM) or fungal spent media (FSM), there was a noticeable decrease in both growth rate and protein yield compared to the cells grown on nutrient rich LB media (Table 1). This is likely the result of the reduced nutrient value of the media post-primary culture, though high lactic acid concentration had an effect on the FSM-fed secondary cultures at shake flask scale. Interestingly, the bioreactor experiments with the secondary E. coli cultures demonstrated recombinant protein product yields that were equivalent to the rich media control condition (Fig. 2). Therefore the higher dissolved oxygen, stirring, and pH maintenance at 7 is capable of restoring the protein expression performance despite the lower nutrient value and metabolite presence in the spent media feed.The secondary culture in CSM outperformed that grown in FSM in the shake flasks, with a yield of 68% compared to 45% of the recombinant target, mCherry-EF2, compared to the LB condition. It is known that fungal cultures such as T. versicolor tend to excrete metabolites which can be more toxic to bacterial cultures, such as the antimicrobials mentioned before or even just more acidic metabolites in general such as lactic acid.41 The reduced nutrient quantity in terms of available amino acids in the FSM post-primary culture is also a factor to be considered as this would impede protein expression (Fig. 5). Additionally, beta-glucans produced by the T. versicolor primary culture have been shown to exhibit antibacterial activity against E. coli at levels of over 250 mg L−1.42 As T. versicolor is a natural producer of this polysaccharide both within its cell walls and also as a hydrosoluble compound,43 any free beta-glucans that may be present within the FSM during the primary culture would have also affected the secondary E. coli growth, though the slightly reduced growth rate seen in the bioreactor experiments would indicate this was not a significant obstacle for the secondary culture. This finding demonstrates that a waste media containing leftover metabolites can be reused from a primary culture without any detriment to a secondary culture such as recombinant protein producing E. coli cultures.
Primary carbon source choice benefitted protein production
HPLC analysis of the bioreactor experiments revealed that the leftover glucose from both spent media was completely metabolised by the secondary E. coli cultures after 6 hours for the CSM-fed secondary culture and after 5.5 hours for the FSM-fed culture. This aided in the quick initial growth rate in both bioreactors, at approximately 60% of that which was achieved in the LB. This remaining glucose was then completely metabolised by the protein expression induction point, a beneficial consumption point as it is known that the presence of glucose inhibits the expression of recombinant proteins using the Lac operon dependent T7 expression system.44 The cultures then switched to metabolising glycerol which was added to the media at the beginning of the culture, as indicated by the slight decrease in glycerol concentration seen in Fig. 3. Glycerol supplementation in E. coli cultures has been shown to aid in maintenance of high cell density and protein production.45 While glucose is the preferred carbon source for E. coli growth, there are some advantages to using glycerol as the primary carbon source; glycerol has been shown to reduce the accumulation of the growth-inhibiting metabolite acetate,46 is cheaper than glucose as a waste byproduct of the fuel industry itself, and can also reduce the sequestering of subsequently lost recombinant protein product into inclusion bodies.47,48Beta-glucan yield was higher in secondary cultures fed with CSM
CSM supplemented cultures of T. versicolor grown in either shake flask or bioreactor achieved a similar biomass yield to the control corn steep liquor (CSL) supplemented cultures (Table S2†). These cultures generated a 62% higher yield (g of substrate/g of biomass) than previously reported where an olive washing water substrate was used,49 with a similar yield reported by a second study in 2014, and 9% lower than reported by a more recent study in 2016.50,51 However, the volumetric productivity of cells grown in the airlift bioreactor was 4.6-fold higher than the flask grown cultures due to both more efficient aeration and nutrient mass transfer in the airlift compared to the flasks. The volumetric productivity was between 3.36 and 5.5-fold higher than reported by the previous studies.51,52 The volumetric productivity of beta-glucans was 2-fold higher than reported by Rau et al. in 2009.52 Given that there was no inhibition of fungal growth in the flasks nor in the airlift bioreactor, the total protein concentration provided by the CSM was sufficient for the metabolic requirements of the fungal strains. Both media components were added at the same total protein concentration. Therefore, disregarding the qualitative composition of the CSL or CSM, the spectrum of the amino acids is nonetheless sufficient to guarantee the growth of the fungal strain despite one being a waste feed source.Optimal oxygen transfer is often more vital to biomass growth than the source of protein supplement. Even more interesting was the production of bioactives by T. versicolor (namely, glucans); the improved biomass productivity in the airlift bioreactor resulted in a 55% increase in total glucans when CSM was used as a supplement compared with shake flask cultures (Fig. 3). This increase did not occur in the CSL-based media (airlift vs. flasks experiments). This result was believed to be as a result of the specific amino acid composition of the CSM or by some soluble molecules released by the CHO cells inducing a higher glucan production by T. versicolor. It is worth noting that specific amino acids could be a convenient source of glucogenic precursors that could promote the extra production of beta-glucans. One known glucogenic amino acid is serine, which was determined to be at high concentrations in the CSM media (Fig. 5B). The proteomic analysis of the CSM-fed fungal cultures also showed four interesting proteins which are dysregulated in T. versicolor grown on CSM media; transaldolase (R7S654), glycoside hydrolase (R7S9O5), phosphoglucomutase (R7S7K5), and serine phosphatase (R7S7K2), the last of which is upregulated while the former three are downregulated (Fig. 7). These proteins are known to be tied either directly or indirectly to glucan production and this led to the hypothesis that this pathway of serine-originating precursors were actively promoting beta-glucan production.53–55 In particular, the upregulated serine/threonine phosphatase is involved in cell wall maintenance and beta-glucan production in a direct manner via cell wall-mediated stress response, and therefore partially responsible for the increase in glucans seen.56–59 Succinic and lactic acid, two potentially gluconeogenic substrates, were also detected in relevant concentrations in the CSM supernatant prior to secondary culture (Fig. 6). It has been shown that the regulation of the metabolic pathways involving gluconeogenesis in filamentous fungi can utilise these substrates when glucose is diminished in the media;60,61 this aids in explaining why CSM considerably increased the beta-glucan production in the secondary T. versicolor cultures.
FSM was high in zinc and iron elemental composition
The elemental composition of both spent media were very distinct from one another, which is interesting in itself because both managed to successfully support the growth and protein production of subsequent cultures. The elemental composition of E. coli requires that certain elements be scavenged from its environment to ensure cell growth and survival.34 From the BioNumbers database (search BioNumbers – The Database of Useful Biological Numbers (https://www.harvard.edu/) (https://bionumbers.hms.harvard.edu/search.aspx)), the top four elements needed from the environment for E. coli growth are carbon (∼50% of cell biomass), nitrogen (∼15%), phosphorus (∼3%) and sulphur (1%) with various other trace elements making up the remainder such as iron, calcium and zinc.62 The elemental consumptive patterns of the different E. coli cultures is viewable in Fig. 4, panel B. The much higher levels of zinc absorbed by the E. coli cultures grown in the FSM was a particularly interesting finding when taken with the high levels of lactic acid in the media post-primary culture. Zinc, along with iron and magnesium, has been shown to have protective properties for lactic acid-injured E. coli and can aid in cellular healing.63,64 However, high levels of intracellular zinc has also been shown to interfere with iron–sulphur cluster biogenesis, by competing for binding to iron–sulphur cluster sites on certain proteins.65 This offers an explanation for the increased sulphur content of the FSM post-E. coli culture compared to the other two media where the biogenesis of sulphur containing compounds was inhibited and therefore may have increased the secretion of this element into the media.Amino acid consumption patterns show low precursor levels in FSM
In a previous study, it was shown that E. coli grown in CSM-fed cultures underwent a proteomic shift with significant upregulation of amino-acid biosynthetic enzymes.18 This finding is reinforced by our findings here on the amino-acid content of the CSM composition pre and post-secondary culture, with the concentrations of three different amino acids (alanine, cysteine, and valine) increased after the secondary E. coli culture. Interestingly, alanine and valine levels were increased in both spent media conditions but not LB (FSM levels were not statistically significant). In terms of the amino acid consumption, the CSM-fed E. coli consumed very high concentrations of serine and aspartate with over 3 mM consumed. Both of these amino acids are precursor amino acids used to synthesise other amino acids like glycine.66,67 Glutamate, another precursor, was also consumed at a high level in this condition (1.3 mM) and is one of the most essential amino acids as it has many uses outside of protein synthesis including assimilation of nitrogen as ammonium.68 Of the enzymes involved in the amino acid biosynthetic pathway found previously in CSM-fed secondary E. coli cultures, most were involved in the conversion of these precursors into other amino acids, including the most significantly upregulated enzyme, asparagine synthase A, with a difference in LFQ intensity of 2.79.18 These findings suggest that stringent response due to amino acid starvation may influence the CSM-fed E. coli culture in order to upregulate these biosynthetic pathways. These precursor amino acids are present at much lower concentrations in the FSM post-primary culture, having been mostly consumed by the fungal culture. This media may therefore benefit from a targeted supplementation of these precursor amino acids to improve the slightly inhibited growth rate, though the recombinant protein yields obtained were still equivalent to the LB rich media. Overall, these results lead to the conclusion that amino acid biosynthetic pathways are upregulated.The recombinant product of mCherry-EF2 has approximately 10% of its sequence consisting of glutamine, glycine, and lysine. It is therefore unsurprising that the precursors of these amino acids (serine for glycine, glutamate for glutamine, and aspartate for lysine) are all consumed by the CSM-fed secondary cultures at high levels, as serine and aspartate were in the LB-fed condition. The secondary E. coli cultures fed with CSM also demonstrated an increased production of succinic acid according to the metabolomic data, though not statistically significant. A similar increase, though to a lesser extent, was seen in the rich media condition. The FSM-fed secondary cultures did not produce any measurable succinic acid. This was found to be a likely result of the high iron levels (135.65 μg L−1) present in the FSM media prior to secondary culture compared to the CSM (5.92 μg L−1) and LB (16.53 μg L−1). Succinic acid secretion is a known response from iron starvation due to glyoxylate shunt activation in the TCA cycle69 particularly when coupled with glycerol as the primary carbon source.47 FSM could benefit from amino acid precursor supplementation while the CSM may benefit from supplementation of certain essential elements such as iron. These supplementations may increase the growth rate, while the recombinant product yield and the fungal beta-glucan yield are already at optimal levels without further supplementation but could possibly be enhanced with such an approach.
Environmental impact
Reuse of spent media has the potential to have a large impact on the sustainability of many industries, including the biopharmaceutical and the functional food industry. In recent years, a measure of the sustainability of biotechnology processes was introduced called the Process Mass Intensity (PMI).70 This is a measurement of the various resources that go into making a specific product; in the referenced study, it was determined that to make 1 kg of biopharmaceutical product, an average of 551 kg of raw materials is required. This includes both upstream and downstream processing materials such as media, acids, bases, and solvents.How does this relate to new bioprocess development? Life cycle analysis (LCA) performed on current bioprocesses agree that the environmental impact of the culture media is the greatest factor. For example, the life cycle analysis conducted in hollow fibre bioreactors found that culture media ingredients had the highest contribution to environmental impact.71 Other LCAs from various bioprocesses have indicated the same; cell culture media has a huge influence on the environmental impact of a bioprocess and recycling of this waste could only benefit the sustainability and circularity of these processes.72–74
Conclusion
In summary, we have shown that both CSM and FSM are viable waste streams for reuse, either as a sole feed for secondary fermentations or as a protein-rich supplement. This resource has significant potential to be useful for the production of biomolecules from prokaryotic or eukaryotic organisms. In fact, our calculations indicate that this media recycling would save upwards of 800 kg of waste per kg of product. The integration of primary and secondary bioprocesses with carefully selected expression hosts represents an exciting technology capable of driving a pipeline of bioproducts with a sustainable and circular model of bioproduction.Materials and methods
Mammalian cell culture
Chinese Hamster Ovary (CHO) cells were incubated in 20 ml of serum-free CHOgro® Expression media supplemented with 4 mM L-Glutamine in T75 adherent cell line flasks with filter cap, at 37 °C with 5% CO2. Cells were split 1 in 10 once grown to 70–80% confluency every 3–4 days. Spent media was harvested at each split and was clarified of cells and cellular debris by centrifugation at 300×g for 4 minutes prior to storing at 4 °C for up to 14 days.Bacterial cell culture starter and transformation
Competent E. coli BL21 Gold were transformed by heat shock with an mCherry-EF2 fusion protein expression construct with a T7 promoter expression system and ampR, and maintained on an agar plate with 1% glucose and 100 μg ml−1 ampicillin. A 50 ml starter culture of these transformed E. coli was grown by incubating a single, isolated colony in Lysogeny Broth (LB) media, 2% glucose, and 100 μg ml−1 ampicillin. The culture was grown in a shaking incubator at 250 rpm, 37 °C for 16 hours before use in the bioreactor experiments.Flasks-based bacterial expressions
Prior to expression, the harvested spent media, both CHO spent media (CSM) and fungal spent media (FSM), had their pH adjusted to 7. Prior to this step, the pH of the CSM was 7.2, and the pH of the FSM was 5.5. The four hour expressions were carried out in triplicate 50 ml cultures in 250 ml flasks with 2% glycerol, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 μg ml−1 ampicillin and a 1 in 20 dilution of starter culture in the media to be tested (LB, CSM, or FSM) in a shaking incubator at 250 rpm, and 37 °C. Expression of the fusion protein was induced by addition of 1 mM IPTG after the OD600 nm reached 0.6. Cultures were then incubated on a shaking incubator at 250 rpm, 30 °C for 18 hours. Cultures were then spun at 4 °C, 4000 rcf for 20 minutes to harvest the pellets which were weighed and then frozen at −20 °C prior to protein purification.
100 μg ml−1 ampicillin and a 1 in 20 dilution of starter culture in the media to be tested (LB, CSM, or FSM) in a shaking incubator at 250 rpm, and 37 °C. Expression of the fusion protein was induced by addition of 1 mM IPTG after the OD600 nm reached 0.6. Cultures were then incubated on a shaking incubator at 250 rpm, 30 °C for 18 hours. Cultures were then spun at 4 °C, 4000 rcf for 20 minutes to harvest the pellets which were weighed and then frozen at −20 °C prior to protein purification.
Bioreactor-based bacterial expressions
The bioreactor used was a Bionet(R) Baby-F0 stirred tank reactor with a working volume of 1 L. The system was autoclaved prior to use. 500 ml of the media being tested was then added to the bioreactor; if a spent media was used, this was first filter sterilised using 0.22 micron vacuum filters, and 2% glycerol and 100 μg ml−1 ampicillin added. The LB control experiment simply had the addition of the 2% glycerol and 100 μg ml−1 ampicillin. System parameters used were as follows; temperature = 37 °C, pH = 7, DO = 20%, air = 0.5 slpm, and stirring = 500 rpm. A cascade was set up as well for agitation (1500 rpm max) and air (1–10 slpm), when DO drops below 20%, with agitation set up to be activated first. Acid used for pH balancing was 15% (v/v) sulphuric acid, and base used was 1 M sodium hydroxide. Starter culture (50 ml) was added to the bioreactor media (500 ml) once the temperature and pH was balanced, and this was considered as the time point 0. Culture was grown until the OD600 nm was >10, with measurements taken every half an hour. Also taken every half an hour, a 1 ml sample was spun down at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf for 3 minutes, and the pellet kept for freeze drying in order to measure cell dry weight. 400 μl was also taken every half an hour for HPLC analysis of carbon source concentration. Once the correct OD was reached, 1 mM IPTG was added to the culture with an additional 2% glycerol to maintain the cultures carbon source. From this point, an 18 hours expression was carried out with the temperature dropped to 30 °C, and left overnight. At the end of the expression, two 5 ml samples were taken for protein purification and yield measurement, by centrifuging the samples at 4000 rcf for 20 minutes. Supernatant was discarded, and the pellets were frozen at −20 °C until purification.
000 rcf for 3 minutes, and the pellet kept for freeze drying in order to measure cell dry weight. 400 μl was also taken every half an hour for HPLC analysis of carbon source concentration. Once the correct OD was reached, 1 mM IPTG was added to the culture with an additional 2% glycerol to maintain the cultures carbon source. From this point, an 18 hours expression was carried out with the temperature dropped to 30 °C, and left overnight. At the end of the expression, two 5 ml samples were taken for protein purification and yield measurement, by centrifuging the samples at 4000 rcf for 20 minutes. Supernatant was discarded, and the pellets were frozen at −20 °C until purification.
Flasks-based fungal glucans production
T.versicolor strains were grown in 100 ml of a previously autoclaved glucose-based media in 250 ml flasks at 25 °C and 150 RPM provided with a stainless steel spring to facilitate adhesion; these are the starter cultures; these fungal strains were added aseptically into the flasks using 5 mycelia-covered agar plugs, taken from 3 days old mycelia covered malt-agar plates. The glucose based media was composed of: 10 g L−1 glucose, 15 ml L−1 of corn steep liquor (CSL) and 3.3 g L−1 of KH2PO4. After 5 days of growth in submerged conditions the T. versicolor biomass was homogenised for 30 seconds using a T18 Digital Ultraturrax(R) IKA disperser provided with S 18 N 10G dispersing tool, until the grown fungal biomass was dispersed into a homogenous population of approx 0.5 mm individual size pellets; 5 ml of this homogenised biomass was aseptically transferred with a sterile 10 ml pipette into 250 ml flasks (with a 100 ml working volume) of the same media as the starter culture except that the CSL was substituted by filter-sterilised CHO spent media (CSM) supernatant. The addition of CSM was added to the flasks to have the same protein amount as the one provided by the CSL. The strain was grown in triplicate flasks. Total glucans production was evaluated by a specific enzymatic kit (K-YBGL, Megazyme(R), Ireland) assaying the lyophilised fungal biomass post-harvest after the flasks growth.Air-lift bioreactor-based fungal glucans production
A custom manufactured Bionet(R) double-jacketed transparent, 4 L working volume air-lift bioreactor with an internal glass draught tube was used for the growth of T. versicolor strains. The height and the diameter of the glass vessel of the airlift bioreactor were 601 and 236 mm, respectively. The glass draught tube height and diameter were 360 and 65 mm, respectively and the draught tube is suspended inside the vessel by a double stainless-steel ring system at an approximate distance of 40 mm directly above the air sparger. The stainless steel ring air sparger has 8 1 mm holes at 0.8 cm distance from each other. The air sparger allows maximum flow-rate of 18 slpm (max 4.5 vvm). The design of the air-lift bioreactor (especially the height to diameter ratio) allows an internal convective movement of the sparged media with a perfect cyclical mixing approximately every 17 seconds. The media rises from inside the draught-tube due to the air sparging and sinks in the annular shape space (down-comer) externally of the draught tube but internally of the glass vessel. This gradient of media density is maintained throughout the fermentation provided that the sparging does not cease. This movement is the same that allows the pelletised biomass of T. versicolor to constantly recirculate in the airlift bioreactor. Aliquots of pelletised biomass were aseptically sampled at different time intervals from the airlift bioreactor, using a sterile withdrawing 16 mm wide silicon tube and an external wide head peristaltic pump. These biomass aliquots were frozen at −70 °C and then freeze-dried and used for glucans and cell dry weight analyses (CDW). The bioreactor was used in batch mode (i.e.: all the media components are added at the beginning of the fermentation); the media composition was the same as the one in the flasks experiments except that CSL was replaced by filter sterilised CHO cell spent supernatant; the media composition was the following i.e.: 10 g L−1 glucose, 15 ml L−1 of filter sterilised spent CHO cell supernatant and 3.3 g L−1 of KH2PO4. The inocula was taken from a 2 L flask where the T. versicolor pelletised biomass was grown for 72 hours from a homogenised starter culture. The T. versicolor pelletised biomass was homogenised by an Ultraturrax for 30 seconds at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm in aseptic conditions (in a SafefastTM Class II laminar flow cabinet) and added to an empty autoclaved Duran bottle provided with sterile silicone tubing so to be added to the airlift in aseptic conditions.
000 rpm in aseptic conditions (in a SafefastTM Class II laminar flow cabinet) and added to an empty autoclaved Duran bottle provided with sterile silicone tubing so to be added to the airlift in aseptic conditions.
Specific growth rate calculations
Specific growth rate constant (k) and doubling time was calculated by use of GraphPad Prism's exponential (Malthusian) growth equation, version 9.4.1 for Windows, GraphPad Software, San Diego, California USA, http://www.graphpad.com/. Plots were generated from growth curves of OD600nmversus time (h), with triplicate values.Recombinant protein purification and yield calculations
Cell pellets were resuspended using a buffer containing 10 mM Tris and 2 mM CaCl2 pH 8 before lysis by sonication. The lysate was then boiled at 85 °C for one minute and spun at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf for 30 minutes. Supernatant was harvested and referred to as the post-boil (PB) sample. Protein concentration was then measured with a DeNovix DS-11 Spectrophotometer, using the UV-vis application. Measurements were taken at 585 nm for mCherry yield, with an extinction coefficient calculated to be 44
000 rcf for 30 minutes. Supernatant was harvested and referred to as the post-boil (PB) sample. Protein concentration was then measured with a DeNovix DS-11 Spectrophotometer, using the UV-vis application. Measurements were taken at 585 nm for mCherry yield, with an extinction coefficient calculated to be 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 854.2 M cm−1 and a protein molecular weight of 30
854.2 M cm−1 and a protein molecular weight of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 667.4 g mol−1. Total yield was then calculated using Beer–Lambert law.
667.4 g mol−1. Total yield was then calculated using Beer–Lambert law.
High pressure liquid chromatography (HPLC)
The analyses for glucose/glycerol concentrations in the media supernatant were performed using an HPLC unit. This analysis was carried out on a Shimadzu Prominence unit (SIL-20AC HT autosampler, DGU-20A5 degasser, CTO-20A column oven, RID-10A detector) fitted with a Bio-Rad Aminex HPX-87H ion exclusion column. The method used is an isocratic elution with a mobile phase of 0.014 N sulphuric acid. Flow rate is set at 0.55 ml min−1 with a pressure set point of 45 kfg (4.41 MPa). The compounds are detected by a RID (refractive index detector). The RID cell is equilibrated and balanced between each sample injection. Dilutions of the media supernatants were used in Whatman(R) MIni-UniPrepTM syringeless filter vials and 20 μL of the supernatant was injected into the HPLC column from each vial. Standard curves of the aqueous solutions of analytical grade glucose or glycerol were constructed using the same methodology and the dilutions of the supernatant media samples were only considered when they fell into the concentration ranges of the standards.Inductively coupled plasma – mass spectrometry (ICP-MS)
Media samples for ICP-MS were collected by harvesting 1 ml sample volumes in triplicate. Fresh media samples were prepared and harvested immediately, of fresh LB, fresh Corn Steep Liquor (for fungal growth) and fresh CHOgro® expression media (for CHO cell growth) all in triplicate. Fungal spent media (FSM) post fungal culture was harvested after a 72 hours culture of Trametes versicolor in the airlift bioreactor as above. CHO spent media (CSM) samples were harvested from a three day culture of adherent CHO cell culture. LB, CSM, and FSM post E. coli cultures were harvested after the 18 hours expression experiment carried out in shake flask culture. All samples were diluted 1 in 10 with 2% nitric acid in milliQ water, and sent to University of Nottingham for ICP-MS analysis.Author contributions
CL and FC collected experimental data, CL, FC, and DO'C wrote text, and CL, FC, DO'C and KO'C were responsible for editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
CL is funded by the Atoms-2-Products center for doctoral training, which is supported by the Science Foundation Ireland (SFI) and the Engineering and Physical Sciences Research Council (EPSRC) under Grant No. 18/EPSRC-CDT/3582. Our thanks to Katalin Kovacs, Peter License, and the ICP-MS suite at the University of Nottingham for their assistance in the analysis of our media composition by ICP-MS. Our thanks to Lorraine Brennan and Xiaofei Yin at the Core Facilities in the Conway Institute for their assistance in the metabolomic analysis of the media supernatant.References
- P. Stegmann, M. Londo and M. Junginger, Resour., Conserv. Recycl., 2020, 6, 100029 Search PubMed.
- F. Gao, C. Li, Z. H. Yang, G. M. Zeng, L. J. Feng, J. Liu, M. Liu and H. Cai, Ecol. Eng., 2016, 92, 55–61 CrossRef.
- T. Narancic, S. Verstichel, S. R. Chaganti, L. Morales-Gamez, S. T. Kenny, B. De Wilde, R. B. Padamati and K. E. O'Connor, et al. , Environ. Sci. Technol., 2018, 52(18), 10441–10452 CrossRef CAS PubMed.
- T. Narancic, F. Cerrone, N. Beagan and K. E. O’Connor, Polymers, 2020, 12(4), 920 CrossRef CAS PubMed.
- N. Diboune, A. Nancib, N. Nancib, J. Anibal and J. Boudrant, Biotechnol. Appl. Biochem., 2019, 66(5), 744–754 CrossRef CAS PubMed.
- C. Ruiz, S. T. Kenny, T. Narancic, R. Babu and K. O'Connor, J. Biotechnol., 2019, 306, 9–15 CrossRef CAS PubMed.
- O. Khelifi, H. Laksaci, M. A. Merabti, N. D. Zemani, S. Rezzag, M. Ozacar, M. Nacef, M. L. Chelaghmia and A. M. Affoune, Fuel, 2024, 358, 130240 CrossRef CAS.
- M. Edjekouane, F. Lansari, O. Khelifi, I. Boukheteche and H. Laksaci, Alger. J. Renew Energy Sustain., 2020, 2(01), 56–59 Search PubMed.
- G. Walsh and E. Walsh, Nat. Biotechnol., 2022, 40(12), 1722–1760 CrossRef CAS PubMed.
- M. Lübeck and P. S. Lübeck, Microorganisms, 2022, 10(4), 753 CrossRef PubMed.
- F. Boukid, A. Hassoun, A. Zouari, M. C. Tulbek, M. Mefleh, A. Ait-Kaddour and M. Castellari, Foods, 2023, 12(5), 1005 CrossRef CAS PubMed.
- J. H. Dupuis, L. K. Y. Cheung, L. Newman Dr and R. Y. Yada, Compr. Rev. Food Sci. Food Saf., 2022, 22(2), 882–912 CrossRef PubMed.
- M. J. Post, Meat Sci., 2012, 92(3), 297–301 CrossRef PubMed.
- R. Kempken, H. Büntemeyer and J. Lehmann, Cytotechnology, 1991, 7(2), 63–74 CrossRef CAS PubMed.
- T. Y. Hsiao, C. E. Glatz and B. A. Glatz, Biotechnol. Bioeng., 1994, 44(10), 1228–1234 CrossRef CAS PubMed.
- U. Riese, D. Lutkemeyer, R. Heidemann, H. Buntemeyer and J. Lehmann, J. Biotechnol., 1994, 34(3), 247–257 CrossRef CAS PubMed.
- J. Wu, Q. Ruan and H. Y. P. Lam, J. Biotechnol., 1998, 66(2), 109–116 CrossRef CAS.
- C. D. Lynch and D. J. O'Connell, PLoS One, 2022, 17(5), e0266921 CrossRef CAS PubMed.
- M. Lemieszek and W. Rzeski, Contemp. Oncol., 2012, 16(4), 285–289 CAS.
- Y. Maehara, S. Tsujitani, H. Saeki, E. Oki, K. Yoshinga, Y. Emi, M. Morita, S. Kohnoe, Y. Kakeji, T. Yano and H. Baba, Surg. Today, 2012, 42(1), 8–28 CrossRef CAS PubMed.
- M. V. Kyanko, R. S. Canel, V. Ludemann, G. Pose and J. R. Wagner, Appl. Biochem. Microbiol., 2013, 49(1), 41–45 CrossRef CAS.
- G. C.-F. Chan, W. K. Chan and D. M.-Y. Sze, J. Hematol. Oncol., 2009, 2(1), 1–11 CrossRef PubMed.
- V. Vetvicka, M. Novak and R. N. Silva, Biology and Chemistry of Beta Glucan: Beta-Glucan, Structure, Chemistry and Specific Application, 2018 Search PubMed.
- B. Gürünlü, Medical Applications of Beta-Glucan, 2022, pp. 63–72 Search PubMed.
- M. Kozarski, A. Klaus, L. van Griensven, D. Jakovljevic, N. Todorovic, W. Wan-Mohtar and J. Vunduk, Food Sci. Hum. Wellness, 2023, 12(2), 378–396 CrossRef CAS.
- P. A. Gibbs, R. J. Seviour and F. Schmid, Crit. Rev. Biotechnol., 2000, 20(1), 17–48 CrossRef CAS PubMed.
- W. Wang, L. Y. Wang, H. R. Yang, Y. P. Zhang, H. N. Zhang, H. Fan, X. L. Zhao, J. Zhang and W. Jia, Int. J. Med. Mushrooms, 2017, 19(3), 225–232 CrossRef PubMed.
- E. N. O'Neill, J. C. Ansel, G. A. Kwong, M. E. Plastino, J. Nelson, K. Barr and D. E. Block, npj Sci. Food, 2022, 6(1), 46 CrossRef PubMed.
- H. Dodia, A. V. Sunder, Y. Borkar and P. P. Wangikar, Biotechnol. Bioeng., 2023, 120, 10 CrossRef PubMed.
- D. V. Voronina, D. V. Shcheblyakov, I. A. Favorskaya, I. B. Esmagambetov, A. S. Dzharullaeva, A. I. Tukhvatulin, O. V. Zubkova, O. Popova, V. Y. Kan, A. S. Bandelyuk, M. M. Shmarov, D. Y. Logunov, B. S. Naroditskiy and A. L. Gintsburg, Acta Naturae, 2021, 13(4), 33–41 CrossRef CAS PubMed.
- I. A. Favorskaya, D. V. Shcheblyakov, I. B. Esmagambetov, I. V. Dolzhikova, I. A. Alekseeva, A. I. Korobkova, D. V. Voronina, E. I. Ryabova, A. A. Derkaev, A. V. Kovyrshina, A. A. Iliukhina, A. G. Botikov, O. L. Voronina, D. A. Egorova, O. V. Zubkova, N. N. Ryzhova, E. I. Aksenova, M. S. Kunda, D. Y. Logunov, B. S. Naroditsky and A. L. Gintsburg, Front. Immunol., 2022, 13, 822159 CrossRef CAS PubMed.
- C. G. Rappazzo, C. L. Hsieh, S. A. Rush, E. S. Esterman, T. Delgado, J. C. Geoghegan, A. Z. Wec, M. Sakharkar, V. Más, J. S. McLellan and L. M. Walker, Immunity, 2022, 55(9), 1710–1724 CrossRef CAS PubMed.
- F. V. Ritacco, Y. Wu and A. Khetan, Biotechnol. Prog., 2018, 34(6), 1407–1426 CrossRef CAS PubMed.
- N. M. Belliveau, G. Chure, C. L. Hueschen, H. G. Garcia, J. Kondev, D. S. Fisher, J. A. Theriot and R. Phillips, Cell Syst., 2021, 12(9), 924–944 CrossRef CAS PubMed.
- A. Parroni, A. Bellabarba, M. Beccaccioli, M. Scarpari, M. Reverberi and A. Infantino, Int. J. Mol. Sci., 2019, 20(17), 4167 CrossRef PubMed.
- M. R. Ricciardi, R. Licchetta, S. Mirabilii, M. Scarpari, A. Parroni, A. A. Fabbri, P. Cescutti, M. Reverberi, C. Fanelli and A. Tafuri, Oxid. Med. Cell Longev., 2017, 5061639 CAS.
- K. Terpe, Appl. Microbiol. Biotechnol., 2006, 72(2), 211–222 CrossRef CAS PubMed.
- X. Yang, M. Badoni, M. K. Youssef and C. O. Gill, J. Food Prot., 2012, 75(1), 144–149 CrossRef CAS PubMed.
- J. Lowrey, R. E. Armenta and M. S. Brooks, Appl. Microbiol. Biotechnol., 2016, 100(3), 1061–1075 CrossRef CAS PubMed.
- C. Lynch, L. Jordan and D. J. O’ Connell, Cell Culture Engineering and Technology, 2021, 129–146 CAS.
- J. Pereira, A. Cachinho, M. M. R. de Melo, C. M. Silva, P. C. Lemos, A. M. R. B. Xavier and L. S. Serafim, Biomolecules, 2022, 12(9), 1284 CrossRef CAS PubMed.
- A. Chamidah and A. A. Hardoko, Prihanto, AIP Conf. Proc., 2017, 1844(1), 020011 CrossRef.
- J. Vetter, Foods, 2023, 12(5), 1009 CrossRef CAS PubMed.
- G. L. Rosano and E. A. Ceccarelli, Front. Microbiol., 2014, 5, 172 Search PubMed.
- S. D. Ganjave, H. Dodia, A. V. Sunder, S. Madhu and P. P. Wangikar, Biotechnol. Rep., 2022, 33, e00694 CrossRef CAS PubMed.
- K. Martínez-Gómez, N. Flores, H. M. Castañeda, G. Martínez-Batallar, G. Hernández-Chávez, O. T. Ramírez, G. Gosset, S. Encarnación and F. Bolivar, Microb. Cell Factories, 2012, 11, 46 CrossRef PubMed.
- A. Murarka, Y. Dharmadi, S. S. Yazdani and R. Gonzalez, Appl. Environ. Microbiol., 2008, 74(4), 1124–1135 CrossRef CAS PubMed.
- J. Kopp, C. Slouka, S. Ulonska, J. Kager, J. Fricke, O. Spadiut and C. Herwig, Bioengineering, 2017, 5(1), 1 CrossRef PubMed.
- F. Cerrone, P. Barghini, C. Pesciaroli and M. Fenice, Chemosphere, 2011, 84(2), 254–259 CrossRef CAS PubMed.
- F. Wang, M. Zhang, W. Peng, Y. He, H. Lin, J. Chen, H. Hong, A. Wang and H. Yu, Bioresour. Technol., 2014, 166, 602–605 CrossRef CAS PubMed.
- K. F. Wang, J. H. Hu, C. Guo and C. Z. Liu, Bioprocess Biosyst. Eng., 2016, 39(7), 1041–1049 CrossRef CAS PubMed.
- U. Rau, A. Kuenz, V. Wray, M. Nimtz, J. Wrenger and H. Cicek, Appl. Microbiol. Biotechnol., 2009, 81(5), 827–837 CrossRef CAS PubMed.
- C. R. Santos, P. A. C. R. Costa, P. S. Vieira, S. E. T. Gonzalez, T. L. R. Correa, E. A. Lima, F. Mandelli, R. A. S. Pirolla, M. N. Domingues, L. Cabral, M. P. Martins, R. L. Cordeiro, A. T. Junior, B. P. Souza, É. T. Prates, F. C. Gozzo, G. F. Persinoti, M. S. Skaf and M. T. Murakami, Nat. Chem. Biol., 2020, 16(8), 920–929 CrossRef CAS PubMed.
- A. Tripathi, E. Liverani, A. Y. Tsygankov and S. Puri, J. Biol. Chem., 2020, 295(29), 10032–10044 CrossRef CAS PubMed.
- Y. Duan, H. Han, J. Qi, J. M. Gao, Z. Xu, P. Wang, J. Zhang and C. Liu, BMC Genom., 2022, 23(1), 314 CrossRef CAS PubMed.
- B. Jiang, A. F. Ram, J. Sheraton, F. M. Klis and H. Bussey, Mol. Gen. Genet., 1995, 248(3), 260–269 CrossRef CAS PubMed.
- B. B. Fuchs and E. Mylonakis, Eukaryotic Cell, 2009, 8(11), 1616–1625 CrossRef CAS PubMed.
- J. Ariño, D. Velázquez and A. Casamayor, Microb. Cell Factories, 2019, 6(5), 217–256 CrossRef PubMed.
- M. J. Hynes, E. Szewczyk, S. L. Murray, Y. Suzuki, M. A. Davis and H. M. Sealy-Lewis, Genetics, 2007, 176(1), 139–150 CrossRef CAS PubMed.
- S. Y. Chew and L. T. L. Than, Encyclopedia of Mycology, 2021, pp. 220–229 Search PubMed.
- R. Milo, P. Jorgensen, U. Moran, G. Weber and M. SpringeR, Nucleic Acids Res., 2010, 38, D750 CrossRef CAS PubMed.
- H. Shi, R. Zhang, L. Lan, Z. Chen and J. Kan, J. Appl. Microbiol., 2019, 127(6), 1741–1750 CrossRef CAS PubMed.
- H. C. Zhang, R. Zhang and H. Shi, Lett. Appl. Microbiol., 2022, 75(1), 161–170 CrossRef CAS PubMed.
- J. Li, C. Luo, D. Zhang, X. Cai, L. Jiang and G. Zhang, Appl. Environ. Microbiol., 2019, 85(14), e00511 CAS.
- L. I. Pizer and M. L. Potochny, J. Bacteriol., 1964, 88(3), 611–619 CrossRef CAS PubMed.
- L. I. Pizer, J. Bacteriol., 1965, 89(4), 1145–1150 CrossRef CAS PubMed.
- R. E. Viola, Acc. Chem. Res., 2001, 34(5), 339–349 CrossRef CAS PubMed.
- M. C. Walker and W. A. van der Donk, J. Ind. Microbiol. Biotechnol., 2016, 43(2–3), 419–430 CrossRef CAS PubMed.
- S. C. Andrews, A. K. Robinson and F. Rodríguez-Quiñones, FEMS Microbiol. Rev., 2003, 27(2–3), 215–237 CrossRef CAS PubMed.
- K. Budzinski, M. Blewis, P. Dahlin, D. D'Aquila, J. Esparza, J. Gavin, S. V. Ho, C. Hutchens, D. Kahn, S. G. Koenig, R. Kottmeier, J. Millard, M. Snyder, B. Stanard and L. Sun, New Biotechnol., 2019, 49, 37–42 CrossRef CAS PubMed.
- H. L. Tuomisto, S. J. Allan and M. J. Ellis, Sci. Total Environ., 2022, 851(Pt 1), 158051 CrossRef CAS PubMed.
- E. Amasawa, H. Kuroda, K. Okamura, S. Badr and H. Sugiyama, ACS Sustain. Chem. Eng., 2021, 9(42), 14012–14021 CrossRef CAS.
- M. Pietrzykowski, W. Flanagan, V. Pizzi, A. Brown, A. Sinclair and M. Monge, J. Cleaner Prod., 2013, 41, 150–162 CrossRef CAS.
- A. L. Cataldo, B. Sissolak, K. Metzger, K. Budzinski, O. Shirokizawa, M. Luchner, A. Jungbauer and P. Satzer, Chem. Eng. Sci.: X, 2020, 8, 100083 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3su00483j |
| This journal is © The Royal Society of Chemistry 2024 |