Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Engineering bacterial biomanufacturing: characterization and manipulation of Sphingomonas sp. LM7 extracellular polymers†
Ellen W.
van Wijngaarden
 a,
Alexandra G.
Goetsch
b,
Ilana L.
Brito
c,
David M.
Hershey
b and
Meredith N.
Silberstein
a,
Alexandra G.
Goetsch
b,
Ilana L.
Brito
c,
David M.
Hershey
b and
Meredith N.
Silberstein
 *a
*a
aSibley School of Mechanical and Aerospace Engineering, Cornell University, Ithaca, NY, USA. E-mail: meredith.silberstein@cornell.edu
bDepartment of Bacteriology, University of Wisconsin-Madison, Madison, WI, USA
cMeinig School of Biomedical Engineering, Cornell University, Ithaca, NY, USA
First published on 26th July 2024
Abstract
Biologically produced materials are an attractive alternative to traditional materials such as metals and plastics and offer improved functionalities such as better biodegradability and biocompatibility. Polysaccharides are an example of biologically produced materials that can have a range of chemical and physical properties including high stiffness to weight ratios and thermal stability. Polysaccharides synthesized by bacteria can come with many advantages such as being non-toxic and are mechanically robust relative to proteins and lipids, which are also secreted by bacteria to generate a biofilm. Biomanufacturing offers benefits compared to traditional manufacturing including low resource investment and equipment requirements, providing an alternative to sourcing fossil fuel byproducts, and relatively low temperatures needed for production. However, many biologically produced materials require complex and lengthy purification processes before use. This paper (1) identifies the material properties of a novel polysaccharide, dubbed promonan, isolated from the extracellular polymeric substances of Sphingomonas sp. LM7; (2) demonstrates that these properties can be manipulated to suit specific applications; and (3) presents two alternative methods of processing to shorten purification time by more than 50% while maintaining comparable material properties.
1 Introduction
Researchers are increasingly looking towards biomanufacturing to find alternative methods of material production that eliminate using fossil fuel byproducts and have low resource investment, mild growth temperatures, and comparatively less equipment required than traditional materials. Biomanufacturing can help achieve these goals by harnessing existing cellular machinery to produce desired materials. Progress in the field has been facilitated by recent advances in synthetic biology and genetic modification techniques such as modern sequencing technologies1 and molecular genetic tools.2 However, in order to successfully replace traditional materials, biomanufactured materials must demonstrate satisfactory mechanical and chemical properties, as well as be easy to produce in terms of time and required resources.Materials synthesized by plants, fungi and bacteria come with a broad range of physical and chemical properties, making them ideal for a wide array of applications.3 Bacteria are the most genetically tractable of the three options, with a wide variety of applicable synthetic biology tools.4 Bacterial cells naturally produce an array of biopolymers including polysaccharides, polyphosphates, extracellular DNA, and proteins that make up an extracellular matrix, known as a biofilm.5 Secreted polysaccharides aid cells with adhesion, aggregation, and structure.6 Well known producers of bacterial polysaccharides include Xanthamonas campestris,7Pseudomonas aeruginosa8 and Sphingomonas sp.9 Polysaccharides can be isolated and purified from a biofilm using enzymes and filtration methods.7,9
Bacterial polysaccharides have gained attention for their innovative uses. Common applications where bacterial polysaccharides are used include food processing,10 healthcare,5,11 and the oil and gas industry.12 Xanthan and dextran are two examples of bacterial polymers that are used in medicine, and as additives in cosmetics, food, and packaging.13 The unique rheological properties of bacterial polysaccharides enable them to be mixed homogeneously with other compounds to act as a thickening agent, achieving high viscosities at low concentrations. Furthermore, high biodegradability and non-toxicity make these materials ideal for use in consumable products.14 Xanthan gum, for example, is used as a thickening agent in chocolate, a water binding agent in meat, and an additive to shampoo for suspending insolubles.7 Another family of bacterial polysaccharides, called sphingans, enhances oil recovery from high temperature and high salinity reservoirs when used in a technique known as polymer flooding.15 Finally, high resistance to degradation from salt and/or heat (often stable up to 300 °C) make bacterial polysaccharides versatile and ideal for broad use.14,16
Many bacteria produce polysaccharides that can be extracted for use. The ideal bacteria for polysaccharide manufacturing produce a medium amount of material under reasonable temperatures, between 20 °C and 40 °C, enabling growth in a standard lab incubator.17 It is important to note that sometimes too much material or too high of a viscosity can make extracting the polymers challenging within a bioreactor setup.18 Bacterial polymers are usually first precipitated with an alcohol such as methanol, ethanol, or isopropanol.19,20 Other methods of separating bacterial polymers include metal coordination, quaternary ammonium salt precipitation, and column chromatography. However, these methods require difficult to source reagents, accurate pH control and are time-consuming.21 Following isolation of the material, impurities are typically removed using a combination of enzymes, liquid–liquid extractions, using solvents such as phenol and chloroform, and dialysis.21 The type of material and quantity can be altered genetically to facilitate biomanufacturing if the organism is genetically tractable. In addition to genetically adjusting overall yield, the material properties can be fine-tuned genetically.22 However, additional work is needed to understand how these extraction techniques and genetic modifications alter material properties.
Polysaccharides produced by the Sphingomonas genus, known as sphingans, can provide a broad range of properties including molecular weight (MW), surface charge,14,15,23 and thermal stability, making these bacteria ideal for material production. The genus Sphingomonas was first established in 1990 and has since been put to use in many applications. These bacteria are known for producing high MW polysaccharides such as welan gum, gellan gum, and diutan gum.14 Sphingans serve as good thickening agents, similar to other biopolysaccharides, and can also be used for chelating, emulsifying, and stabilizing.14 Unique rheological properties and stability at high temperatures and salinity have made Sphingans popular for use in oil recovery.15 One example, welan gum, exhibits a storage modulus of approximately 2 Pa when hydrated in water at a weight to volume (w/v) ratio of only 0.175%.24 Another example, gellan gum, is commonly used as a food thickener and has a storage modulus that is strongly frequency dependent, ranging from 0.01 to 10 Pa at a concentration of 1% w/v in water.25 Sphingans have also been recently applied in bioremediation due to their hydrophobicity, minimal nutrient requirements for production, and ability to degrade various pollutants. Furthermore, many Sphingomonas strains are genetically tractable, holding potential for tailor-made sphingans.26
This study investigates the properties of a newly isolated material, named promonan, from a novel strain, Sphingomonas sp. LM7, to identify possible uses and potential for biomanufacturing. Specifically, we address the following research questions: (a) How do the material properties of promonan compare with similarly structured bacterial polysaccharides? (b) Can these properties be manipulated to suit specific applications? (c) Can the purification process be shortened to improve manufacturability?
2 Materials and methods
2.1 Bacterial culturing
A 10 mL peptone yeast extract media (PYE: 0.2% w/v peptone w/v yeast extract) with 3% sucrose starter culture was innoculated with LM7 cells from a frozen glycerol stock and grown at 30 °C overnight on a shaker. 10 mL of starter culture was added to 1 L of PYE and 3% w/v sucrose solution. The culture was grown until approximately the end of day 4 at 18 °C with vigorous shaking. All cultures were grown until an optical density at 600 nm of 0.5 was reached.2.2 Enzymatic purification process
The full enzymatic purification process spans 8 days, following 2 days of culturing time, as outlined in Fig. 1. Following growth, the cultures were then centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 90 minutes to remove cells. The supernatant was transferred to new containers in 150 mL aliquots. 250 mL ethanol was added to each tube and incubated overnight at 4 °C to perform cold ethanol precipitation. On day 5, the tubes were centrifuged at 16 k × g for 1 hour to result in the polysaccharide adhering to the bottom of the tube. The supernantant was decanted and the remaining pellet was left to dry for 30 minutes with the tube inverted. Each pellet was then resuspended in 7.5 mL deionized H2O. The solution was then transferred to a 50 mL falcon tube so that each falcon tube has a total volume of 15 mL.
000 × g for 90 minutes to remove cells. The supernatant was transferred to new containers in 150 mL aliquots. 250 mL ethanol was added to each tube and incubated overnight at 4 °C to perform cold ethanol precipitation. On day 5, the tubes were centrifuged at 16 k × g for 1 hour to result in the polysaccharide adhering to the bottom of the tube. The supernantant was decanted and the remaining pellet was left to dry for 30 minutes with the tube inverted. Each pellet was then resuspended in 7.5 mL deionized H2O. The solution was then transferred to a 50 mL falcon tube so that each falcon tube has a total volume of 15 mL.
To perform nuclease digestion, 1 M Tris buffer and 1 M MgCl2 were added to the tubes to yield a final concentration of 10 mM and 2 mM respectively. Benzonuclease (Turbonuclease) (Accelagen Inc., San Diego, CA) was added at a concentration of 1 μL mL−1 to the tubes. The tubes were inverted and incubated at 37 °C overnight.
A peptide digest was performed by adding proteinase K at a concentration of 10 mg mL−1. Tris-HCl was added to reach a final concentration of 25 mM. The tubes were inverted and incubated at 37 °C overnight.
Phenol-chloroform extraction was performed by dividing volumes in half, 18 mL phenol and 2 mL chloroform were added to each 20 mL volume. The tubes were gently mixed and left to sit for 30 minutes to allow layers to separate. The upper aqueous layer was removed and tubes were combined in a new 50 mL falcon tube. The solution was then added to pre-wetted dialysis tubing with a molecular weight cutoff (MWCO) of 100 kDa.
Dialysis was conducted in a 4 L large beaker in deionized H2O. The water was changed daily for 3 days. Following dialysis, the solutions were pipetted in equal volumes not exceeding 4.5 mL into clean 11 mL glass tubes. The tubes were frozen at −20 °C for 2 hours before being transferred to the −80 °C freezer overnight. The caps were replaced with parafilm with a hole punched out or a piece of tissue for lyophilization. Samples were left in the lyophilizer (FreeZone 2.5 L Benchtop Freeze dryer, Labconco, Kansas City, MO) for 24 hours or until they were fully dry.
2.3 Dialysis-based alternative biomanufacturing process
The dialysis-based biomanufacturing process spanned 3 days, less than half the time required for the production process using enzymatic purification. After initial culture growth, the cells were removed via centrifugation as described in the enzymatic purification process. The solutions were then directly dialyzed for 24 hours, with 4 water changes approximately 6 hours apart (MWCO = 3.5, 50 or 100 kDa) and lyophilized for 24 hours.2.4 Ultracentrifugation
Ultracentrifugation was used as an alternative physical purification method to investigate the effect of removing outer membrane vesicles, which usually contain proteins. Samples were centrifuged at 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rotations per minute using an S-28 rotor at 4 °C in an Optima XL-100 K Ultracentrifuge (Beckman Coulter, Brea, CA). The supernatant was decanted, frozen and lyophilized to isolate the material.
000 rotations per minute using an S-28 rotor at 4 °C in an Optima XL-100 K Ultracentrifuge (Beckman Coulter, Brea, CA). The supernatant was decanted, frozen and lyophilized to isolate the material.
2.5 Protein knockout biomanufacturing process
The protein knockout biomanufacturing process consisted of culturing the strain ΔfliC, removing cells as per the previously described enzymatic purification processes and then lyophilizing the material.Wild-type LM7 or the ΔprmJ mutant were transformed with pDH1329 by electroporation, and primary integrants were identified by selection on PYE plates supplemented with 25 μg mL−1 kanamycin. Colonies were restreaked on PYE plates containing 25 μg mL−1 kanamycin and used to inoculate a liquid culture in PYE that was grown overnight under non-selective conditions. The outgrowth was then spread on PYE plates supplemented with 3% (w/v) sucrose to select for plasmid excision. Individual colonies were replica plated to confirm sensitivity to kanamycin and KanS colonies were screened using PCR to identify fliC deletion mutants.
Dried spent medium extracts were resuspended in 250 μL 2X SDS-LOAD buffer, heated at 70 °C for 15 minutes and centrifuged to remove insoluble material. Samples were loaded on polyacrylamide (10% w/v) gels, separated using the Tris-glycine buffer system and stained with Coomassie Brilliant Blue R. Individual bands were excised with clean razor blades and transferred to fresh tubes for protein identification.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 folllwed by CID-type MS/MS with 2.0 AMU isolation and 10 ms activation time with 35% normalized collision energy fragmentation of 30 most intense peptides detected in the MS1 scan from 350 to 1800 m/z. Redundancy was limited by dynamic exclusion. Monoisotopic precursor selection and charge state screening were enabled and both +1 and undefined charge states were rejected.
000 folllwed by CID-type MS/MS with 2.0 AMU isolation and 10 ms activation time with 35% normalized collision energy fragmentation of 30 most intense peptides detected in the MS1 scan from 350 to 1800 m/z. Redundancy was limited by dynamic exclusion. Monoisotopic precursor selection and charge state screening were enabled and both +1 and undefined charge states were rejected.
2.6 Lectin binding
A dot blot was conducted to check that the polysaccharide was still present after dialysis. 5 μL of 50% w/v sample was pipetted onto a nitrocellulose membrane and allowed to dry for 15 minutes. The membrane was blocked with 25 mL of 5% bovine serum albumin (BSA) solution in TBST (100 mL 10× TBS buffer, 900 mL deionized water, 1 mL Tween-20). The membrane was then stained for 1 hour in 12 mL of 1 μg mL−1 fluorescein conjugated Griffonia simplicifolia-II lectin solution with 1.25% BSA in TBST. Following the incubation, the membrane was rinsed twice with TBST solution for 10 minutes before imaging at a wavelength of 302 nm using an Azure 300 imager (Azure Biosystems, Dublin, CA).2.7 Zeta potential and molecular weight measurements
Zeta potential and MW were measured using a Malvern Nano Zs Zetasizer (Malvern Pananalytical, Malvern, U.K.). Solutions were prepared in 2 mL tubes and given three hours to equilibrate before being transferred to polystyrene or zeta potential folded capillary cuvettes for measurement of either MW or zeta potential. Zeta potential was measured at a concentration of 0.1% with deionized water as the solvent. The zetasizer calculates the zeta potential by finding the electrophoretic mobility and then applying the Henry equation.28 The MW was measured using five consecutive concentrations, 0%, 0.025%, 0.05%, 0.075%, and 0.1%. Static light scattering was used to obtain a time-averaged intensity of scattered light. The MW was computed using the Rayleigh equation describing the relationship between the intensity of light scattered by a sample, its size, and MW.292.8 Fourier-transform infrared spectroscopy
Fourier-transform infrared spectroscopy (FTIR) measurements were performed on a Bruker Vertex FT-IR spectrometer (Bruker Corp., Billerica, MA). A spectral range of 600 cm−1 to 3500 cm−1 was selected. Approximately 5 mg of sample was loaded into the sample compartment to sufficiently cover the attenuated total reflectance (ATR) crystal. The sample chamber was vented with nitrogen before taking all reference and sample measurements.2.9 Bicinchoninic acid (BCA) assay
A BCA protein assay was conducted to quantify protein content in the samples. A colorimetric kit was used measure the reduction of Cu2+ to Cu1+ by protein (Thermo Fisher Scientific). The color change was determined by measuring the optical density at a wavelength of 562 nm using a SpectraMax platereader (Danaher Life Sciences, Washington, DC).2.10 Tensile testing
Films were cast from 1% w/v promonan dissolved in deionized water and dried in ambient conditions. The dry sample mass was 1.7 ± 0.5 mg. The dry films had a density of 0.7 ± 0.1 g cm−3. Uniaxial tensile tests were conducted on a Zwick-Roell (Zwick-Roell, Germany) Z010 system with a 20 N capacity load cell. All tests were carried out using a strain rate of 0.05 s−1 with a pre-load of 0.1 N. Films were cast on a leveled surface and given 24 hours to dry at room temperature and humidity before being punched into dog bone shapes following ASTM D638 Type 5.Digital image correlation (DIC) analysis was conducted by speckling tensile samples with white spray paint and back lighting the sample for imaging. Imaging was carried out using VIC-Snap-8 software and analyzed using VIC-2D Digital Image Correlation (Correlated Solutions, Irmo, SC).
2.11 Rheometry
Experiments were conducted using a TA Instruments DHR3 rheometer (Texas Instruments, Dallas, TA). A strain sweep at a frequency of 1 rad s−1 was used to determine the linear viscoelastic region and select a strain of 5% to be used for frequency sweeps. Both a strain sweep and a frequency sweep were conducted for each sample. A 40 mm parallel plate fixture was used with the gap set to 300 μM, requiring a sample volume of approximately 400 μL. The fixtures were thoroughly cleaned before use and between samples with deionized water and isopropyl alchohol and dried with compressed air. The sample was pipetted onto the bottom plate and the upper fixture lowered to the set gap value. A visual check confirmed that the amount of sample adequately filled the gap. Sweeps of the strain rate were used to produce the viscosity and stress graphs. Graphs presented are the average of three independent trials.2.12 Differential scanning calorimetry (DSC)
Thermal analysis was conducted using a TA Instruments DSC Auto 2500 differential scanning calorimeter. The sample was first weighed before being placed in a pan and sealed with a press. The temperature range of 90 °C to 400 °C was selected with a ramp rate of 10 °C min−1.2.13 Thermogravimentric analysis (TGA)
Thermogravimetric analysis was completed on a TA Instruments 5500 thermogravimetric analyzer (Texas Instruments, Dallas, TA). The sample was initially weighed and loaded using a platinum pan. The ramp rate was set to 10 °C min−1 and the gas supply was set to nitrogen.2.14 Statistical analysis
ANOVA tests were conducted to determine the probability of significant differences between groups. Two-tailed student t-tests were used to identify which groups were significant with a p-value of 0.01 to determine significance.3 Results and discussion
3.1 Pure promonan material properties
Comparison between the chemical composition of promonan and other sphingans can provide insight into material properties including stiffness and thermal stability.30 Promonan is composed of a linear backbone of 3- and 4-linked glucose, and galactose and glucuronic acid residues with functional groups following the pattern seen for many sphingans and biopolysaccharides.26 The structure of promonan is similar to that of gellan gum, however, one of the main features of sphingans is the presence of rhamnose which is notably absent in promonan. Most Sphingomonas strains produce polysaccharides that have a generally conserved backbone consisting of a tetrasaccharide with rhamnose, glucuronic acid and two glucose molecules. Rhamnose is typically connected with an alpha linkage which may contribute to a less planar structure, compared to a beta glycosidic linkage, for these polymer chains and less dense packing between chains overall.14 Sphingans typically differ in their side chains and functional groups. The functional groups for promonan indicate high hydrogen bonding, a characteristic of carbohydrates. The carboxylic acid group gives the glucuronic acid a negative charge at pH 4.5 or higher and enables ionic cross-linking in the presence of positively charged bivalent ions.31 Despite the apparent similarity in chemical structures, analysis of the biosynthetic genes indicates that promonan has distinct biosynthesis pathways from other sphingans, such as gellan gum, and that the polymers represent distinct polysaccharide families.Sphingans with similar chemical structure to promonan are used in healthcare applications due to their stiffness and biocompatibility. Gellan gum, with a Young's modulus of 111 MPa, has been used as a stiffening component in antimicrobial films for wound healing.19,32 We determined the suitability of promonan for similar applications through casting the material into a film and conducting uniaxial tensile testing as shown in Fig. 2a and ESI† Video S1. For our material, promonan, the Young's modulus is 282.06 ± 35.74 MPa while the fracture strength is 4.49 ± 1.28 MPa, with large variation due to sample defects initiating tearing, as shown in ESI† Fig. S1. The high Young's modulus for pure promonan films indicates that the material may be useful for providing mechanical structure in composite materials, such as in antimicrobial films for use in healthcare.
Sphingans have also been selected for many applications because of their thermal stability due to their ability to covalently cross-link, high MW, and stable glycosidic bonds.33–35 Both TGA and DSC were conducted to establish the thermal properties of this novel material and to enable comparison to fellow sphingans. The results of TGA show that weight loss occurs in three stages. There is a 12% decrease in weight in the first stage due to loss of absorbed and structural water as the temperature increases from 23.7 °C to 93.9 °C (Fig. 2b).36 The main mass loss of 56% of the initial weight due to polysaccharide decomposition occurs between 248.1 °C and 380.9 °C. The third mass loss of 9.1% at approximately 480 °C is likely due to decomposition of high molecular weight fragments. The sample weight gradually levels off with approximately 5% of the initial value remaining as ash. Therefore, the oxidation onset temperature is 248.1 °C and the maximum oxidation temperature is 380.9 °C. These results suggest that our material has good thermal stability, slightly greater than gellan gum. The higher Young's modulus and improved thermal stability may be due to the presence of exclusively beta glycosidic linkages in the proposed promonan structure which allows for a more planar structure than sphingans with alpha linkages present.37,38 DSC tests further support thermal stability and show consistency with the TGA data. Fig. 2c shows an endothermic event at 104.4 °C possibly due to water evaporation, which would correspond to the TGA results. The DSC tests also identify decomposition around 277.5 °C with crystallization as occurring between 104.4 °C and 277.5 °C. These results correspond well with the second mass loss event in TGA with the exothermic peak of 277.5 °C as seen in the Fig. 2c. These temperatures follow the patterns for other thermally stable biologically produced polysaccharides for both TGA and DSC.39
3.2 Material properties of hydrated promonan
Bacterial polysaccharides are widely used in a hydrated state. Structural characteristics including high MW and a negative surface charge provide emulsion stability for many applications.40,41 The MW and zeta potential of promonan were analyzed to gain insight into the properties of the material in a hydrated state. The MW of promonan, 579 ± 120 kDa is similar to gellan gum, which has an MW of approximately 500 kDa.40 High MW typically correlates with chemical resistance as it takes more main chain scission events before the material strength is affected.42,43 Lastly, the high MW indicates a higher viscosity at room temperature for water based solutions at a given w/v ratio, as longer chains resist flow due to higher entanglement. The zeta potential is −31.8 ± 2.7 mV, similar to other sphingans such as gellan gum, which has a zeta potential of −29.1 ± 3.0 mV.41 A negative zeta potential value is expected due to the carboxylic acid groups. The zeta potential indicates the potential stability of the colloidal system. Typically, a value below −30 mV or above +30 mV is considered stable, whereas materials with zeta potentials within this range tend to aggregate within a solution. A zeta potential value below −30 mV for promonan therefore, indicates that promonan particles are likely to repel each other rather than aggregating and flocculating. This is ideal for many applications where aggregation would not be desirable and demonstrates that the material dispersion is stable in a hydrated state, expanding the possible uses of promonan.Bacterial polysaccharides are often used as thickening agents where large changes in viscosity due to small increases in the w/v ratio are of interest. The experiments show that promonan acts as a gel at w/v ratios over 1%. Fig. 3a shows the effect of varying water content on the dynamic moduli of promonan measured using rheology. A 1% w/v promonan gel can be conveniently pipetted and handled for testing while a 3% w/v gel is too viscous to be pipetted. In contrast, the 0.5% ratio does not gelate and maintains solution like behavior throughout the frequency sweep. A small increase from 0.5% to 3% w/v ratio changes the storage modulus by several orders of magnitude.
The changes in viscosity as water content is altered can be compared to study the changes in the material behavior. These properties are of particular interest for polysaccharides used as stabilizers, binders and preservatives in the food industry.10,13,14 A flow sweep was conducted by varying the shear rate, as shown in Fig. 3b for the 0.5%, 1% and 3% w/v fractions. The figure indicates shear thinning viscosity for all three concentrations. The negative surface charge causes molecules to repel each other, inhibiting flow and increasing the viscosity at low shear rates. Promonan would make an effective thickening agent and could be used in food-processing, bioprinting, and cosmetic applications.
3.3 Tailoring rheological properties with ions
In many applications it is desirable to be able to tune properties such as material stiffness. One prominent example is bioprinting, which requires a shear thinning material and a stiffness of approximately 0.5 kPa.44–46 Previous studies have explored bioprinting of various polysaccharides, like alginate, cross-linked with ions, such as calcium, for tissue culturing scaffolds due to their biocompatibility and rheological properties.44,47 We demonstrate in this study that the dynamic moduli of promonan increases with the addition of ions by adding a salt to hydrated samples to be comparable to gels used for bioprinting.47 Bivalent calcium ions cross-link carboxylic acid sites resulting in an increase to both the storage and loss moduli by approximately one order of magnitude (Fig. 4a). Furthermore, we show that ethylenediamine tetraacetic acid (EDTA) can be used to chelate ions and decrease the dynamic moduli, a desirable functionality for tuning a bioink to be a substrate for a particular cell type. As expected, the monovalent sodium ions, which should not enable ionic cross-linking, do not cause a statistically significant increase to the dynamic moduli (Fig. 4b).3.4 The impact of the purification process on material properties
Biomanufactured materials typically undergo many purification steps. However, it is unclear how the purification process alters the material properties and whether the process could be shortened while still yielding a usable material. We investigated methods of shortening the purification process to accelerate production, increase the material yield and facilitate potential future manufacturing of the material. The first method follows the bacteria culturing step with dialysis to remove undesired low MW proteins and culture media components and eliminate the need for enzymatic purification steps (Fig. 5a). The material yield, shown in Fig. 5b, drops significantly when salts and sucrose from the growth media are removed via dialysis with an MWCO of 3.5 kDa. Dialysis significantly decreased the material yield through removing culture media components. Although the value of 100 mg L−1 for the final purified material yield is higher than for the 100 kDa MWCO dialyzed sample with a yield of 77 mg L−1, the values were not significantly different. A dialysis MWCO of 50 kDa or higher greatly reduces the protein content by over 15%, as shown in Fig. 5c. A graph without the unpurified sample is included in Fig. S4a (ESI†). A dot blot was also performed to demonstrate that the polysaccharide is still present following dialysis for each MWCO (Fig. S4b, ESI†). Note that the sample dialyzed at 100 kDa was too viscous to permeate into the nitrocellulose membrane.The changes in composition were further analyzed via FTIR for each of the dialyzed, unpurified and purified samples (Fig. 5d). Regions for lipids, proteins and polysaccharides are labeled based on literature on unpurified biofilms.48 Clear peaks are seen for the protein, lipid and carbohydrate regions. The spectra show a decrease in the polysaccharide to protein ratio between the unpurified to the purified samples with the dialyzed samples between these values, indicating removal of proteins and other substances. The typical spectral pattern for polysaccharides49 is apparent in all cases. The polysaccharide peak is co-located with signatures from sucrose and nucleic acids. The unpurified sample closely matches peaks of the known spectra for sucrose, which is not surprising given the high amount of sucrose used in the growth media for Sphingomonas sp. LM7.50 The purified, 50 kDa dialyzed, and 100 kDa dialyzed spectra all appear to have higher protein content than the unpurified samples, but this is an artifact of the choice to normalize to the peak at 1050 cm−1, since all these procedures remove sugar associated with the growth media. The peaks in Fig. 5d indicate that the functional groups found include both primary and secondary alcohols (separate peaks) at 3350 cm−1 and 2918 cm−1, amine salts and strong O–H, C–H, C–O stretches in the spectra (1031 cm−1). The peak at approximately 3200 cm−1, likely from water absorption, remains small indicating that there is likely little effect on O–H bond stretching.
Rheology was used to measure how compositional changes altered stiffness. Fig. 5e shows that the rheological stiffness of the unpurified material is significantly lower than the purified material, but does remain gel-like with the storage modulus above the loss modulus throughout the frequency sweep. The presence of proteins, sugars, deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) with lower MW values than promonan likely results in a lower storage and loss modulus for the unpurified sample. However, the components of the material may also interact to increase material stiffness. Sucrose, for example, has been shown to stabilize proteins and increase the viscosity of protein solutions through the concept of preferential exclusion and hydration.51 The sugars are excluded from the volume around the protein leading to preferential hydration of the protein shell region. This results in a more compact and stable protein folding state and increases the viscosity. Protein in the unpurified sample accounts for of 21.5% of the solid material. In contrast, the protein concentration in the purified material is estimated to be 1%. The protein concentrations for the dialyzed samples with MWCOs of 3.5 kDa, 50 kDa and 100 kDa are 20%, 3% and 3% respectively. Fig. 5e shows a further decrease in moduli and transition to fluid-like behavior when the large amount of sucrose is taken out with dialysis at 3.5 kDa MWCO. The dynamic moduli are found to increase relative to the 3.5 kDa MWCO material when a higher MWCO for dialysis is used. Fig. 5e shows a transition back to gel-like behavior when 50 kDa is used. The values for the storage and loss moduli are largely recovered, relative to the enzymatically purified material, when using the 100 kDa MWCO. These results indicate that mechanical properties close to the enzymatically purified material could be obtained using a shorter, dialysis-based purification process, especially if the ability to further increase dynamic moduli with divalent ions or decreasing water content is taken into account.
3.5 Purification via ultracentrifugation
The presence of proteins, which have a lower MW than promonan, may be contributing to the decrease in dynamic moduli of the dialyzed material compared to the enzymatically-purified material. To understand which proteins might alter the material stiffness, we analyzed proteins present in the spent medium of LM7 cultures by performing mass spectrometry-based identification of bands excised from an SDS-PAGE gel (Fig. 6a). Many of the proteins identified in the analysis were predicted to be located in the Gram-negative outer membrane, suggesting that they were released from the cell in outer membrane vessicles (OMVs). The most abundant proteins identified were three different TonB-dependent receptor proteins, an OmpA family protein, Flagellin, outer membrane (OM) lipoprotein pal and a 17 kDa surface antigen. We therefore explored ultracentrifugation as a means of removing OMVs to lower the amount of protein in the sample. Although ultracentrifugation did not significantly alter the material yield compared to the unpurified material, it was found to successfully decrease the protein content Fig. 6b and c. The protein content for the unpurified sample was 2150 μg mL−1 compared with 1750 μg mL−1 for the ultracentrifuged sample. Coupling ultracentrifugation with dialysis significantly decreased the overall yield and protein content compared with only ultracentrifugation. The ultracentrifuged and 100 kDa dialyzed process produced a yield of 0.16 g L−1 and a protein content of 600 μg mL−1. However, the reduction in yield and protein content is primarily due to dialysis, rather than ultracentrifugation, with values similar to the 100 kDa dialyzed sample (yield of 0.27 g L−1 and 325 μg mL−1). Combined ultracentrifugation and 100 kDa dialysis did result in a decrease in protein abundance relative to the amount of polysaccharide of interest as seen in Fig. 6d. The protein to polysaccharide ratio for the ultracentrifuged 100 kDa dialyzed sample was 0.66, slightly lower than that of the 100 kDa dialyzed sample at 0.70.The changes in material composition due to ultracentrifugation alter the rheological properties as seen in Fig. 6e. Results of the frequency sweep show a slight decrease in the moduli between the unpurified material and the ultracentrifuged material which could be due to the high shear stresses applied during centrifugation (Fig. 6e).52 High shear stresses can decrease the MW and viscosity of polymer solutions which may decrease the dynamic moduli. The shear stresses may also facilitate subsequent dialysis by shearing polymer chains. Lectin binding demonstrates that the polysaccharide is not removed by ultracentrifugation (Fig. S5a, ESI†). Fig. 6e shows that when ultracentrifugation is combined with dialysis, the material moduli improve significantly while yield significantly decreases, likely due to the removal of protein, lipids and DNA as shown in Fig. 6c, d and Fig. S7 (ESI†).
3.6 Protein knockout purification pathway
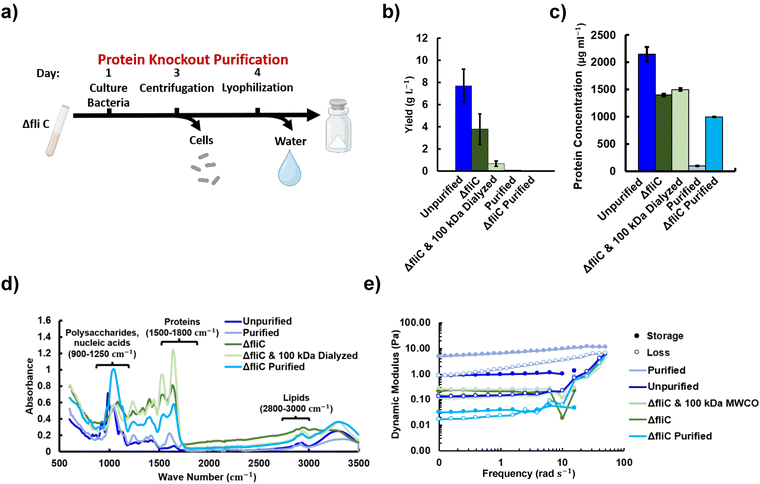
One advantage to producing materials in genetically tractable organisms is the potential to make mutations that simplify the purification process. Biosynthetic tools, such as sequencing and cloning strategies like Gibson assembly, have the potential to target and alter protein production to streamline purification processes by eliminating the need for filtration, enzymatic breakdown, and extraction of undesired protein content. However, due to the recent development of these technologies, they remain underutilized in the field of materials science. We explored the potential of this strategy by deleting the gene for a protein that is secreted in high abundance by LM7 based on the identified secreted proteins shown in Fig. 6a. We chose to delete the gene for ΔfliC, as this was the only protein identified in the supernatant that was not predicted to be contained in OMVs. A ΔfliC strain was developed that still produced promonan, but not fliC, the protein that polymerizes to form filaments for bacterial flagella. Analysis of the overall material yield and protein content indicate a decrease in the total material and protein abundance (Fig. 7b and c). The purified fliC sample yield, 0.02 μg L−1 was approximately one fifth that of the wild type purified material. Interestingly, in this case, dialysis did not further decrease the protein content but is shown to decrease yield through the removal of solute from the growth media. Further study of which proteins are produced by the strain, as shown in Fig. S6a (ESI†), and the resulting properties may lend insight into what additional protein knockouts or modifications could be used to alter the material composition. FTIR, shown in Fig. 7d, indicates that the protein to polysaccharide ratio is much greater for both the ΔfliC and the ΔfliC dialyzed samples than the enzymatically purified material. The lipid to polysaccharide ratio for both the ΔfliC and the ΔfliC dialyzed samples is also higher than both the purified and unpurified samples however, the intensities of the lipid peaks remain low relative to the protein and polysaccharide peaks.The changes in composition between the wild type and ΔfliC produced material offer insight into the different rheological behaviors observed. Fig. 7e shows that the dynamic moduli of the protein knockout material are decreased compared to the unpurified material, possibly due to the increased protein to polysaccharide ratio and lipid to polysaccharide ratio or the production of the signalling molecule cyclic dimeric guanosine monophosphate (c-di-GMP). Mutating genes that affect the flagellum have been shown to influence mechanosensing pathways and activate the production of c-di-GMP,53,54 which can alter enzymes related to biofilm production.54 The purified ΔfliC material also had decreased dynamic moduli compared to the purified material despite having similar protein to polysaccharide ratios. This may point to the role of signalling molecules in mechanics however, the ΔfliC mutant may also be secreting a less soluble form of promonan as has been seen with other mutants of the LM7 strain.26 Dialyzing the spent media from the ΔfliC strain removes the viscosity enhancing effect of sucrose but also eliminates low molecular weight species, such as charged ions, resulting in little overall change in stiffness. Charged ions are thought to alter protein–protein interactions and ultimately decrease solution viscosity.51 The 100 kDa MWCO dialyzed wild type strain had dynamic moduli that were approximately two orders of magnitude stiffer than the 100 kDa dialyzed ΔfliC strain. These results demonstrate that genetic modification can be applied to alter the material properties and reduce the required purification steps. However, additional research is needed to identify genetic modifications capable of maintaining or increasing the rheological stiffness.
4 Conclusions
In this paper we have successfully characterized a novel bacterial polysaccharide, providing an overview of thermal, rheological and mechanical properties. It is interesting that promonan and other sphingans, such as gellan gum, display similar material parameters. This suggests that polysaccharides with divergent biosynthesis pathways can produce polymers with similar properties. The potential applications of the material for films and bioinks was demonstrated via tensile testing of films and rheological tests of hydrogels. Salt was added to hydrated promonan samples to investigate the effectiveness of ions in crosslinking the gel to tailor the stiffness. This study demonstrated that the lengthy purification process can be shortened by over 50% while still yielding gel state polymers at 1% w/v ratio. Alternative purification processes using dialysis and genetic modification were compared to improve the purification time and user-friendliness. Purification via dialysis with a MWCO of 100 kDa provided the closest rheological properties to those of the enzymatically purified material. This paper provides an overview of material properties, identifies advantages such as tailorable properties for applications and highlights the material's manufacturability through various processing methods.Our work identified material properties that might be suitable for an array of applications in healthcare, biomanufacturing and food processing. Future research is required to demonstrate these applications as well as investigate further material modifications, such as functionalization for bioremediation or as an antimicrobial film for use in environmental or healthcare applications.
Author contributions
Conceptualization, I. L. B., D. M. H., M. N. S.; data curation, E. W. v. W., A. G. G.; formal analysis, E. W. v. W.; writing – original draft preparation, E. W. v. W.; writing – review and editing, I. L. B., D. M. H., M. N. S.; visualization, E. W. v. W.; supervision, I. L. B., M. N. S.; project administration, I. L. B., D. M. H., M. N. S.; funding acquisition, I. L. B., D. M. H., M. N. S. All authors have read and agreed to the published version of the manuscript.Data availability
Data for this article are available at BOX at [https://cornell.box.com/s/147k53yra25vjgs8grpudspvnkkwpmy7].Conflicts of interest
The authors have no conflict of interest to declare.Acknowledgements
This work was supported through the NSERC PGS-D scholarship (E. W. v. W.), the Beckman Young Investigator award (D. M. H.) and NIH R35GM150652 (D. M. H). This work was supported in part by and performed in part at the Engineered Living Materials Institute at Cornell University. This work was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (Grant NNCI69 2025233). The authors acknowledge the use of facilities and instrumentation supported by NSF through the Cornell University Materials Research Science and Engineering Center DMR-1719875.References
- F. Meng and T. Ellis, Nat. Commun., 2020, 11, 5174 CrossRef CAS PubMed
.
- W. Wang, G. Zheng and Y. Lu, Front. Bioeng. Biotechnol., 2021, 9, 692797 CrossRef PubMed
.
- Z. Zhang, M. Parker, K. Liao, J. Cao, A. Waghmare, J. Breda, C. Matsumura, S. Eley, E. Roumeli, S. Patel and V. Iyer, Biodegradable Interactive Materials, arXiv, preprint, 2024, https://arxiv.org/abs/2404.03130 Search PubMed.
- P. Q. Nguyen, N.-M. D. Courchesne, A. Duraj-Thatte, P. Praveschotinunt and N. S. Joshi, Adv. Mater., 2018, 30, 1704847 CrossRef PubMed
.
- B. H. A. Rehm, Nat. Rev. Microbiol., 2010, 8, 578–592 CrossRef CAS PubMed
.
- D. H. Limoli, C. J. Jones and D. J. Wozniak, Microbiol. Spectrum, 2015, 3(3), 223–247 Search PubMed
.
- A. Palaniraj and V. Jayaraman, J. Food Eng., 2011, 106, 1–12 CrossRef CAS
.
- S. Singh, Y. Almuhanna, M. Y. Alshahrani, D. W. Lowman, P. J. Rice, C. Gell, Z. Ma, B. Graves, D. Jackson, K. Lee, R. Juarez, J. Koranteng, S. Muntaka, D. A. Mitchell, A. C. da Silva, F. Hussain, G. Yilmaz, F. Mastrotto, Y. Irie, P. Williams, D. L. Williams, M. Cámara and L. Martinez-Pomares, npj Biofilms Microbiomes, 2021, 7, 1–14 CrossRef PubMed
.
- V. Kaur, M. B. Bera, P. S. Panesar, H. Kumar and J. F. Kennedy, Int. J. Biol. Macromol., 2014, 65, 454–461 CrossRef CAS PubMed
.
- J. C. McGuffey, D. Leon, E. Z. Dhanji, D. M. Mishler and J. E. Barrick, J. Microbiol. Biol. Educ., 2018, 19(2), 74 Search PubMed
.
- F. Wahid, X.-J. Zhao, X.-Q. Zhao, X.-F. Ma, N. Xue, X.-Z. Liu, F.-P. Wang, S.-R. Jia and C. Zhong, ACS Appl. Mater. Interfaces, 2021, 13, 32716–32728 CrossRef CAS PubMed
.
- V. Brar and G. Kaur, Int. J. Biomater., 2018, 2018, 4856287 Search PubMed
.
- M. F. Moradali and B. H. A. Rehm, Nat. Rev. Microbiol., 2020, 18, 195–210 CrossRef CAS PubMed
.
- H. Huang, J. Lin, W. Wang and S. Li, Polymers, 2022, 14, 1920 CrossRef CAS PubMed
.
- Y. Li, L. Xu, H. Gong, B. Ding, M. Dong and Y. Li, Energy Fuels, 2017, 31, 3960–3969 CrossRef CAS
.
-
R. B. Seymour and C. E. Carraher, in Structure-Property Relationships in Polymers, ed. R. B. Seymour and C. E. Carraher, Springer US, 1984, pp. 83–93 Search PubMed
.
- G. R. Stouten, C. Hogendoorn, S. Douwenga, E. S. Kilias, G. Muyzer and R. Kleerebezem, ISME J., 2019, 13, 3112 CrossRef CAS PubMed
.
- R. J. Seviour, B. McNeil, M. L. Fazenda and L. M. Harvey, Crit. Rev. Biotechnol., 2011, 31, 170–185 CrossRef CAS PubMed
.
- F. Freitas, V. D. Alves and M. A. M. Reis, Trends Biotechnol., 2011, 29, 388–398 CrossRef CAS PubMed
.
- H. Li, X. Jiao, Y. Sun, S. Sun, Z. Feng, W. Zhou and H. Zhu, Sci. Rep., 2016, 6, 37899 CrossRef CAS PubMed
.
- L. Shi, Int. J. Biol. Macromol., 2016, 92, 37–48 CrossRef CAS PubMed
.
- R. R. Singhania, A. K. Patel, M.-L. Tsai, C.-W. Chen and C. Di Dong, Bioengineered, 2021, 12, 6793–6807 CrossRef CAS PubMed
.
- L. K. Jennings, K. M. Storek, H. E. Ledvina, C. Coulon, L. S. Marmont, I. Sadovskaya, P. R. Secor, B. S. Tseng, M. Scian, A. Filloux, D. J. Wozniak, P. L. Howell and M. R. Parsek, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11353–11358 CrossRef CAS PubMed
.
- L. Xu, G. Xu, T. Liu, Y. Chen and H. Gong, Carbohydr. Polym., 2013, 92, 516–522 CrossRef CAS PubMed
.
- E. Miyoshi, T. Takaya and K. Nishinari, Food Hydrocolloids, 1994, 8, 505–527 CrossRef CAS
.
- A. G. Goetsch, D. Ufearo, G. Keiser, C. Heiss, P. Azadi and D. M. Hershey, A novel exopolysaccharide pathway from a freshwater Sphingomonas isolate, bioRxiv, preprint, 2023, https://www.biorxiv.org/content/10.1101/2023.11.03.565537v1 Search PubMed.
- A. I. Nesvizhskii, A. Keller, E. Kolker and R. Aebersold, Anal. Chem., 2003, 75, 4646–4658 CrossRef CAS PubMed
.
- M. Kaszuba, J. Corbett, F. M. Watson and A. Jones, Philos. Trans. R. Soc., A, 2010, 368, 4439–4451 CrossRef CAS PubMed
.
-
B. Kruppa, G. Strube and C. Gerlach, in Optical Measurements: Techniques and Applications, ed. F. Mayinger and O. Feldmann, Springer, 2001, pp. 99–116 Search PubMed
.
- T. P. West, Polysaccharides, 2021, 2, 234–244 CrossRef CAS
.
- I. Nakashima, A. Kishida, Y. Takaoka, S. Morisada, K. Ohto, H. Kawakita, W. Iwasaki, R. R. Sathuluri and M. Miyazaki, J. Appl. Glycosci., 2016, 63, 69–75 CrossRef CAS PubMed
.
- N. A. N. Mohd Azam and K. A. M. Amin, IOP Conf. Ser.: Mater. Sci. Eng., 2017, 209, 012027 Search PubMed
.
- Z. Xu, Z. Li, S. Jiang and K. M. Bratlie, ACS Omega, 2018, 3, 6998–7007 CrossRef CAS PubMed
.
- X. Lu, eFood, 2023, 4, e82 CrossRef
.
- J. Zhang, H. Huo, L. Zhang, Y. Yang, H. Li, Y. Ren and Z. Zhang, Polymers, 2022, 14, 2053 CrossRef CAS PubMed
.
- C. W. Vendruscolo, C. Ferrero, E. A. G. Pineda, J. L. M. Silveira, R. A. Freitas, M. R. Jiménez-Castellanos and T. M. B. Bresolin, Carbohydr. Polym., 2009, 76, 86–93 CrossRef CAS
.
- S. Sukumar, S. Arockiasamy and M. Chemmattu Moothona, J. App. Biol. Biotech., 2021, 9, 58–67 CAS
.
- E. I. Nep and B. R. Conway, J. Excipients Food Chem., 2010, 1, 30–40 CAS
.
- C.-P. Lin and S.-Y. Tsai, Molecules, 2019, 24, 2856 CrossRef CAS PubMed
.
- Z. Feketshane, S. Alven and B. A. Aderibigbe, Polymers, 2022, 14, 4098 CrossRef CAS PubMed
.
- L. G. Duarte, W. M. Alencar, R. Iacuzio, N. C. Silva and C. S. Picone, Curr. Res. Food. Sci., 2022, 5, 642–652 CrossRef CAS PubMed
.
- G. Vaganov, M. Simonova, M. Romasheva, A. Didenko, E. Popova, E. Ivankova, A. Kamalov, V. Elokhovskiy, V. Vaganov, A. Filippov and V. Yudin, Polymers, 2023, 15, 2922 CrossRef CAS PubMed
.
- A. Levy, F. Wang, A. Lang, O. Galant and C. E. Diesendruck, Angew. Chem., Int. Ed., 2017, 56, 6431–6434 CrossRef CAS PubMed
.
- A. GhavamiNejad, N. Ashammakhi, X. Y. Wu and A. Khademhosseini, Small, 2020, 16, e2002931 CrossRef PubMed
.
- R. Sharma, R. Kirsch, K. P. Valente, M. R. Perez and S. M. Willerth, Processes, 2021, 9, 1205 CrossRef CAS
.
- R. K. Bay, A. M. Hancock, A. S. Dill-Macky, H. N. Luu and S. S. Datta, JoVE, 2024, e66166 CAS
.
- I. Gorroñogoitia, U. Urtaza, A. Zubiarrain-Laserna, A. Alonso-Varona and A. M. Zaldua, Polymers, 2022, 14, 354 CrossRef PubMed
.
- P. Di Martino, AIMS Microbiol., 2018, 4, 274–288 Search PubMed
.
- E. Gómez-Ordóñez and P. Rupérez, Food Hydrocolloids, 2011, 25, 1514–1520 CrossRef
.
- N. V. Patil and A. N. Netravali, Cellulose, 2020, 27, 5407–5420 CrossRef CAS
.
- F. He, C. E. Woods, J. R. Litowski, L. A. Roschen, H. S. Gadgil, V. I. Razinkov and B. A. Kerwin, Pharm. Res., 2011, 28, 1552–1560 CrossRef CAS
.
- A. Ram and A. Kadim, J. Appl. Polym. Sci., 1970, 14, 2145–2156 CrossRef CAS
.
- V. Zorraquino, B. García, C. Latasa, M. Echeverz, A. Toledo-Arana, J. Valle, I. Lasa and C. Solano, J. Bacteriol., 2013, 195, 417–428 CrossRef CAS PubMed
.
- D. C. Wu, D. Zamorano-Sánchez, F. A. Pagliai, J. H. Park, K. A. Floyd, C. K. Lee, G. Kitts, C. B. Rose, E. M. Bilotta, G. C. L. Wong and F. H. Yildiz, PLoS Genet., 2020, 16, e1008703 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sm00712c |
| This journal is © The Royal Society of Chemistry 2024 |