Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Resource limitation and population fluctuation drive spatiotemporal order in microbial communities†
Rohit
Khandoori
 a,
Kaustav
Mondal
a,
Kaustav
Mondal
 b and
Pushpita
Ghosh
b and
Pushpita
Ghosh
 *ab
*ab
aSchool of Chemistry, Indian Institute of Science Education and Research, Thiruvananthapuram, Kerala 695551, India. E-mail: pushpita@iisertvm.ac.in
bCenter for High-Performance Computing, Indian Institute of Science Education and Research, Thiruvananthapuram, Kerala 695551, India
First published on 15th April 2024
Abstract
Microbial communities display complex spatiotemporal behaviors leading to spatially-structured and ordered organization driven by species interactions and environmental factors. Resource availability plays a pivotal role in shaping the dynamics of bacterial colonies. In this study, we delve into the intricate interplay between resource limitation and the emergent properties of a growing colony of two visually distinct bacterial strains having similar growth and mechanical properties. Employing an agent-based modeling and computer simulations, we analyze the resource-driven effect on segregation and sectoring, cell length regulation and nematic ordering within a growing colony. We introduce a dimensionless parameter referred to as the active layer thickness, derived from nutrient diffusion equations, indicating effective population participation due to local resource availability. Our results reveal that lower values of active layer thickness arising from decreased resource abundance lead to rougher colony fronts, fostering heightened population fluctuations within the colony and faster spatial genetic diversity loss. Our temporal analyses unveil the dynamics of mean cell length and fluctuations, showcasing how initial disturbances evolve as colonies are exposed to nutrients and subsequently settle. Furthermore, examining microscopic details, we find that lower resource levels yield diverse cell lengths and enhanced nematic ordering, driven by the increased prevalence of longer rod-shaped cells. Our investigation sheds light on the multifaceted relationship between resource constraints and bacterial colony dynamics, revealing insights into their spatiotemporal organization.
1 Introduction
The formation of microbial colonies represents a fundamental process with far-reaching implications across various domains, from biofilm development to tumor growth and ecological phenomena in nature.1–5 Previous investigations into microbial colonies have explored the intricate factors that influence colony morphology and genetic diversity. These factors encompass a wide range of variables, including nutrient concentration, diffusion rates, cell motility, growth rates, cell death dynamics, and various physicochemical parameters. Numerous studies have elucidated how these factors collectively shape colony characteristics.6–13 Recent research has also highlighted the significance of stochasticity within these systems manifesting as demographic noise, variations in bacterial division conditions, and randomness in cell alignment post-division.10,14–16 In parallel, computational models, such as cellular automata, agent-based models, and continuum models, have emerged as invaluable tools for unraveling the intricate mechanisms underpinning microbial colony formation.10,11,17–20 These models have illuminated the dynamics of microbial colonies, explaining emergent features observed in these systems.21–23Research on bacterial colonies, especially those formed by rod-shaped bacteria like E. coli and P. aeruginosa, has extensively delved into topics such as competition, cooperation, selection, and mutation.12,13,24–32 Numerous studies have previously modeled bacteria as spherocylinders, detailing their growth and expansion dynamics. However, our emphasis lies in the particular context of modeling bacterial colonies comprising two neutral bacterial strains that differ in color only. We note a limited body of literature addressing this specific scenario, as the majority of studies predominantly focus on bacteria as spherical entities12,13 overlooking critical microscopic and stochastic dynamics specific to rod-shaped bacteria. Our motivation stems from the recognition of the necessity for a more nuanced approach, particularly when investigating the dynamics inherent to rod-shaped bacteria, which are functionally identical but may vary genetically. Furthermore, investigations into bacterial colony morphology, genetic diversity, and system stochasticity have often been conducted in isolation, neglecting their interconnected nature.
Our present study focuses on a comprehensive investigation of spatial morphology, genetic diversity, and cell size variability, as well as nematic ordering within rod-shaped bacterial colonies. We employ an agent-based model that represents two bacterial cells as rod-shaped entities differing genetically or only in demographic stochasticity and otherwise identical. Here, the phrase “identical” corresponds to the two bacteria having similar growth and mechanical properties. This is a key point in our study, and it necessitates the use of an agent-based model with similar morphology and characteristics for both the visually distinct bacterial cells. This intentional design choice is pivotal to our study, as it allows us to investigate how spatial patterns emerge during growth and expansion when these functionally identical bacteria are subjected to resource limitations. The decision to model these bacteria with similar morphology and characteristics in the agent-based model is crucial to isolate the impact of inherent population fluctuation on spatial sectoring. Our focus is on exploring how these two seemingly identical bacteria labeled in two colors, with inherent population fluctuations, exhibit spatial sectoring when resources are limited during growth and expansion. This model enables us to integrate all these essential features within a single cohesive framework. Notably, our agent-based approach accounts for microscopic details and inherent stochasticity in single-cell dynamics. Each bacterium within the colony is treated as an individual entity, allowing us to introduce random fluctuations associated with individual cell properties, including stochasticity during cell division conditions modeled through probability distributions.
In particular, we investigate how the availability of resources influences the spatial and temporal dynamics of bacterial colonies. By analyzing the intricate relationship between resource constraints and colony organization, we uncover essential principles governing microbial communities. A novel dimensionless parameter, “the active layer thickness,” is introduced to quantify the effects of resource availability, shedding light on colony morphology and expansion dynamics. Notably, lower values of the active layer thickness emerge as a critical factor, leading to rougher colony fronts, heightened population fluctuations, and a rapid loss of genetic diversity. In essence, our study aims to comprehensively address spatial morphology, genetic diversity, cell size variability, and nematic ordering in rod-shaped bacterial colonies by incorporating the impact of resource availability and the active layer thickness. Our current study promises to advance our understanding of complex biological systems, shedding light on the intricate interplay of factors that shape microbial communities and their evolution.
2 Model and methods
In our investigation of colony growth on a hard agar surface, we employ an agent-based model adopted from previous studies7,10 of rod-shaped nonmotile bacterial cells in a two-dimensional box. Each cell is represented by a spherocylindrical particle with a fixed diameter do and of variable length l, where l represents the particle's cylindrical length. Our system is a mixture of two types of bacterial cells differing only in color. In the model, we distinguish them as blue and red colored cells.The position of each bacterial cell is denoted by a two-dimensional spatial coordinate, represented as ![[r with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/i_char_0072_20d1.gif) = (x,y). Additionally, the axial orientation of the bacteria is indicated by the unit vector having components (ûx, ûy). We established the spatial grid by discretizing the simulation box with dimensions Lx = 200 μm and Ly = 400 μm, incorporating 100 bins along the x-axis and 200 bins along the y-axis. This configuration resulted in a grid size of 2 × 2 μm2 across the entire simulation box. At the start of the simulation, the nutrient concentration at each grid point on the agar surface is set to a uniform value denoted as C0.
= (x,y). Additionally, the axial orientation of the bacteria is indicated by the unit vector having components (ûx, ûy). We established the spatial grid by discretizing the simulation box with dimensions Lx = 200 μm and Ly = 400 μm, incorporating 100 bins along the x-axis and 200 bins along the y-axis. This configuration resulted in a grid size of 2 × 2 μm2 across the entire simulation box. At the start of the simulation, the nutrient concentration at each grid point on the agar surface is set to a uniform value denoted as C0.
Our model takes into account the dependence of a cell's growth on its size and the availability of diffusing nutrients from the local concentration. This interaction is captured by the following equation, which describes the dynamics of nutrient consumption by bacteria:
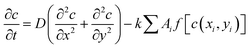 | (1) |
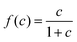 , with a half-saturation constant of 1. Concentrations are measured in units of the half-saturation constant. The empirical Monod equation, introduced by Monod et al. in 1949,33 remains the most commonly utilized rate expression for characterizing the growth of microorganisms, despite the availability of various precise growth rate equations in the literature.34 This hyperbolic function describes the specific growth rate, which is solely dependent on one limiting substrate concentration. Both experimental and computational studies in previous literature have consistently employed this rate equation to model similar systems and have obtained coherent results.7,12,13,35 Bacterial growth along its major axis is described by:
, with a half-saturation constant of 1. Concentrations are measured in units of the half-saturation constant. The empirical Monod equation, introduced by Monod et al. in 1949,33 remains the most commonly utilized rate expression for characterizing the growth of microorganisms, despite the availability of various precise growth rate equations in the literature.34 This hyperbolic function describes the specific growth rate, which is solely dependent on one limiting substrate concentration. Both experimental and computational studies in previous literature have consistently employed this rate equation to model similar systems and have obtained coherent results.7,12,13,35 Bacterial growth along its major axis is described by: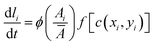 | (2) |
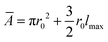 represents the average area, with lmax being the maximum length of the cell. For bacteria, the rate of length growth typically falls between a constant and a linear function of the cell length.7,36 Cell division occurs when the cell length reaches a critical length of lc = 3 μm. Division is modeled with a Gaussian distribution having a standard deviation of 0.0055, and both types of cells divide at a rate of kdiv into two independent daughter cells.37 Furthermore, to account for environmental factors like slight cell bending and elastic pressures between cells, each daughter cell's orientation deviates slightly from that of the mother cell after division. This random orientation change prevents cells from forming long, filamentous structures as they expand. After division, the length of the daughter cells is chosen such that the combined length of the two daughter cells is equal to the length of the mother cell.
represents the average area, with lmax being the maximum length of the cell. For bacteria, the rate of length growth typically falls between a constant and a linear function of the cell length.7,36 Cell division occurs when the cell length reaches a critical length of lc = 3 μm. Division is modeled with a Gaussian distribution having a standard deviation of 0.0055, and both types of cells divide at a rate of kdiv into two independent daughter cells.37 Furthermore, to account for environmental factors like slight cell bending and elastic pressures between cells, each daughter cell's orientation deviates slightly from that of the mother cell after division. This random orientation change prevents cells from forming long, filamentous structures as they expand. After division, the length of the daughter cells is chosen such that the combined length of the two daughter cells is equal to the length of the mother cell.
We incorporate the Hertzian theory of elastic contact through repulsive forces to describe mechanical interactions between cells within close proximity.38 Each nonmotile bacterial cell in the colony follows over-damped dynamics described by the following equations of motion:
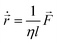 | (3) |
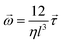 | (4) |
![[F with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/i_char_0046_20d1.gif) and
and ![[small tau, Greek, vector]](https://www.rsc.org/images/entities/i_char_e1ca.gif) represent the corresponding linear forces and torques acting on the cell. The interaction force between two spherocylinders is approximated by considering the force between two spheres arranged along the principal axis of the rods at their minimum distances. When the closest distance between neighboring spherocylinders is r, resulting in an overlap h = d0 − r, the magnitude of the force is given by F = Ed01/2h3/2, where E is a parameter that quantifies the strength of the repulsive interaction and is proportional to the cell's elastic modulus. This force expression in prior literature consistently yielded results when modeling rod-shaped bacterial colonies.7,35 In our simulations, we use a finite value of E (Table 1), allowing for some degree of cell deformation.10,35,37,39 The expression for torque calculation on the i’th cell by the j’th cell in close proximity is given by
represent the corresponding linear forces and torques acting on the cell. The interaction force between two spherocylinders is approximated by considering the force between two spheres arranged along the principal axis of the rods at their minimum distances. When the closest distance between neighboring spherocylinders is r, resulting in an overlap h = d0 − r, the magnitude of the force is given by F = Ed01/2h3/2, where E is a parameter that quantifies the strength of the repulsive interaction and is proportional to the cell's elastic modulus. This force expression in prior literature consistently yielded results when modeling rod-shaped bacterial colonies.7,35 In our simulations, we use a finite value of E (Table 1), allowing for some degree of cell deformation.10,35,37,39 The expression for torque calculation on the i’th cell by the j’th cell in close proximity is given by 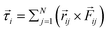 . The distance between the center of mass of the i’th cell and the point of contact between two cells is denoted by
. The distance between the center of mass of the i’th cell and the point of contact between two cells is denoted by ![[r with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/i_char_0072_20d1.gif) ij, while the force between them is represented by
ij, while the force between them is represented by ![[F with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/i_char_0046_20d1.gif) ij. In addition to the direct mechanical cell–cell interactions, there exists competition for local nutrients, which introduces an indirect interaction mediated by the environment.
ij. In addition to the direct mechanical cell–cell interactions, there exists competition for local nutrients, which introduces an indirect interaction mediated by the environment.
| Parameters | Symbols | Simulations (units) |
|---|---|---|
| Box size | L x × Ly | 200 × 400 μm2 |
| Critical length | l c | 3.0 μm |
| Diameter of cells | d 0 | 1.0 μm |
| Linear growth rate | ϕ | 1.5 μm h−1 |
| Cell-division rate | k div | 0.1 h−1 |
| Elastic modulus of alive cells | E | 3 × 105 Pa |
| Friction coefficient | η | 200 Pa h |
| Initial nutrient concentration | C 0 | 10–400 fg μm−3 |
| Nutrient consumption rate | k c | 6.0 h−1 |
| Diffusion rate of nutrient | D | 10–400 μm2 h−1 |
To perform our agent-based simulations, we consider a two-dimensional box with periodic boundary conditions. The initial setup involves the inoculation of a small number (N = 300) of bacterial cells of equal proportion of blue and red colored cells (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with the same aspect ratio and random orientations along a one-dimensional (1D) line connecting two points (0,Ly/2) and (Lx,Ly/2) in a thin strip approximately lmax in the y-direction. For the time evolution of the equations of motion (eqn (3) and (4)), we utilize the Euler technique. To solve the nutrient diffusion described by eqn (1), we employ a central finite-difference scheme. Both the nutrient diffusion equation and the equation of motion are solved numerically while considering periodic boundary conditions. This comprehensive approach allows us to study the mechanical interactions between cells, nutrient competition, and other dynamics in a simulated environment, providing insights into colony growth on a hard agar plate.
1) with the same aspect ratio and random orientations along a one-dimensional (1D) line connecting two points (0,Ly/2) and (Lx,Ly/2) in a thin strip approximately lmax in the y-direction. For the time evolution of the equations of motion (eqn (3) and (4)), we utilize the Euler technique. To solve the nutrient diffusion described by eqn (1), we employ a central finite-difference scheme. Both the nutrient diffusion equation and the equation of motion are solved numerically while considering periodic boundary conditions. This comprehensive approach allows us to study the mechanical interactions between cells, nutrient competition, and other dynamics in a simulated environment, providing insights into colony growth on a hard agar plate.
This comprehensive agent-based model allows us to simulate bacterial colony growth on a hard agar plate while considering nutrient availability, cell growth, division, and orientation dynamics, all of which are crucial factors in understanding the behavior of bacterial colonies in complex environments.
3 Results and discussion
The current study focuses on the spatiotemporal dynamics, morphology and emergent ordering in a mixture of two visually distinguishable (red and blue) rod-shaped bacterial cells, identical in their growth and mechanical aspects except for the presence of the inherent demographic noise. We begin our study by simulating a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of red and blue cells from a line inoculation as described in the Model and method section. Bacteria growing in a Petri dish-like setting in the presence of a fixed amount of initially sourced nutrient forming colonies of varied morphologies were reported in the existing literature.40–45 These studies unravel the influence of resource limitations as the primary reason underlying such morphological outcomes, further classified in the literature as diffusion-limited aggregation, Eden-like, concentric-ring-like, homogeneous spreading, and dense branching morphology.46–48
1 mixture of red and blue cells from a line inoculation as described in the Model and method section. Bacteria growing in a Petri dish-like setting in the presence of a fixed amount of initially sourced nutrient forming colonies of varied morphologies were reported in the existing literature.40–45 These studies unravel the influence of resource limitations as the primary reason underlying such morphological outcomes, further classified in the literature as diffusion-limited aggregation, Eden-like, concentric-ring-like, homogeneous spreading, and dense branching morphology.46–48
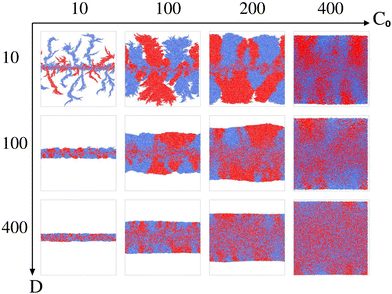
To explore the role of resource limitation in bacterial spreading dynamics consisting of two visually distinct bacterial cells, we consider two parameters: the initial nutrient concentration (C0) and the nutrient diffusivity (D). We carried out simulations by varying the parameters C0 and D and observed emergence of a diverse range of spatial morphologies in expanding colonies as shown in Fig. 1. The simulated results show similar colony morphologies following the transitions from fingerlike to branching to smooth front on increasing nutrient concentration and diffusion value as predicted in the earlier study for a single nonmotile species.10 However, the initially well-mixed bacterial cells colored as blue and red, undergo a significant spatial segregation and sectoring in a growing colony when the local nutrient availability reduces for low values of C0 and D. The simulated colony images (Fig. 1) visually suggest a substantial influence of resource scarcity on both the instability at the colony front and the spatial segregation or demixing as the colony develops. Building upon these visual observations, we proceed to systematically investigate the impact of resource limitation on front instability, cell size distribution, nematic ordering, and the emergence of demixing and sectoring in a growing colony of two rod-shaped bacterial cells.
3.1 Colony morphology at the fronts
To quantify variations in the colony morphologies, we introduce the roughness parameter denoted as σf. This parameter measures the ensemble-averaged standard deviation of the height of the colony front and is calculated as follows: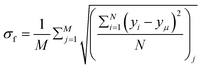 .10 Here, M = 16 represents the number of simulations for which we calculate the average roughness, and N is the number of bins into which we divide the simulation box along the x-direction. The bin size is chosen to be equal to the minimal length of the cell (ld = 1 μm). For each bin i, we determine the corresponding maximum colony front height, denoted as yi. The term yμ represents the mean front height, calculated as the average of yi values across all bins. σf allows us to quantitatively assess the roughness of the colony periphery and provide valuable insights into the morphological variations observed in our simulations.
.10 Here, M = 16 represents the number of simulations for which we calculate the average roughness, and N is the number of bins into which we divide the simulation box along the x-direction. The bin size is chosen to be equal to the minimal length of the cell (ld = 1 μm). For each bin i, we determine the corresponding maximum colony front height, denoted as yi. The term yμ represents the mean front height, calculated as the average of yi values across all bins. σf allows us to quantitatively assess the roughness of the colony periphery and provide valuable insights into the morphological variations observed in our simulations.
The roughness calculation results shown in Fig. 2 are consistent with the spectrum of colony morphology obtained from our simulations. As shown in Fig. 2(a), for a constant diffusion coefficient, D = 10 while increasing C0 will result in a decrease in the roughness and an increase in the smoothness of the colony's front. This follows the transition in colony morphology from fingerlike to branch to smooth front. Similarly, increasing nutrient diffusivity while keeping C0 = 100 fixed causes smoothening of the colony fronts, which is in agreement with a decreasing value of roughness on increasing diffusion value depicted in Fig. 2(b).
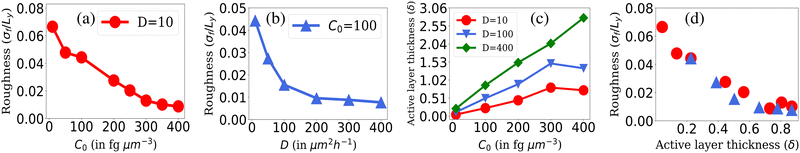 | ||
| Fig. 2 Roughness of the colony fronts. The normalized standard deviation of the front height (roughness σf/Ly) where Ly = 400 is shown with respect to (a) different values of C0 s at fixed D = 10 and (b) for different values of nutrient diffusion coefficients, D at fixed C0 = 100. (c) The active layer thickness, δ is shown for different C0 values for fixed values of D = 10, 100, and 400 and (d) for different values of δ corresponding to the C0 and D values of Fig. 2(a) and (b). Standard error bars are comparable to the marker size of all plots. | ||
The observed phenomenon where lower nutrient availability leads to a rougher colony front compared to higher nutrient conditions can be explained by the dynamics of bacterial growth and resource utilization in our model.
• Lower nutrient availability and roughness: when nutrients are scarce, bacterial cells cannot multiply indefinitely due to limited resources. As a result, in regions where nutrients become depleted, the colony growth halts, leading to the formation of less dense colonies. The cells in these regions might not have the resources to grow and divide rapidly, resulting in irregular and rough colony surfaces.
• Higher nutrient availability and smoothness: conversely, when there is an excess of nutrients, bacterial cells can proliferate more rapidly. This abundance of resources allows the colony to expand and become populated with a larger number of bacteria. As a result, the colony front is smoother because the cells can efficiently utilize the available nutrients to support their growth and division, resulting in a denser and smoother colony structure.
• Effect of nutrient diffusion: the nutrient diffusion coefficient (D) also plays a role in colony morphology. A higher diffusion coefficient allows nutrients to penetrate deeper into the colony. In this scenario, the colony does not necessarily expand to a larger size because the nutrient uptake by cells is concentrated in a smaller region due to the efficient diffusion of nutrients. This concentration of bacterial growth results in a localized, dense, and smooth colony in a given area.
Indeed, the observed roughness or smoothness of the bacterial colony front is a consequence of local nutrient availability and diffusion dynamics within the model. These instabilities occur because individual cells experience stochastic fluctuations in their growth and division due to the discrete nature of the particles. Consequently, some cells may outcompete others for the limited resources, leading to uneven growth at the colony front. This uneven growth results in the formation of irregularities and roughness in the colony's leading edge, a phenomenon observed in both single and multi-species bacterial colonies.
However, the colony morphology at the front is not solely dependent on the concentration or diffusion value but both C0 and D jointly participate in deciding the morphology of the expanding colony. This combined influence is determined by a dimensionless parameter defined as the active layer thickness (δ), as investigated in previous literature13 considering spherical bacterial cells. To calculate δ in our model with rod-like bacterial cells, we start by non-dimensionalizing the diffusion eqn (1). We achieve this by normalizing the substrate concentration using the initial concentration, denoted as 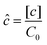 . Next, we normalize the area of the i-th cell by the area of a cell with the critical length (Ac = πr02 + 2r0lc), represented as
. Next, we normalize the area of the i-th cell by the area of a cell with the critical length (Ac = πr02 + 2r0lc), represented as 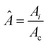 . Finally, we scale the spatial coordinates by the height of the diffusion boundary layer
. Finally, we scale the spatial coordinates by the height of the diffusion boundary layer ![[h with combining tilde]](https://www.rsc.org/images/entities/i_char_0068_0303.gif) . The square root of the coefficient of the Laplacian of ĉ in the dimensionless diffusion eqn (2) at steady state is termed the active layer thickness, denoted as δ (eqn (3)).
. The square root of the coefficient of the Laplacian of ĉ in the dimensionless diffusion eqn (2) at steady state is termed the active layer thickness, denoted as δ (eqn (3)).
Importantly, δ is a dimensionless number, and a crucial estimator of describing the resource limitation within a growing colony.
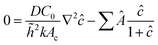 | (5) |
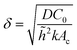 | (6) |
As illustrated in Fig. 2(c), when we maintain a fixed diffusion coefficient (D) value and increase the initial nutrient concentration (C0), we observe a higher active layer δ. Conversely, lower C0 results in lower δ values. Similarly, if we vary the diffusion coefficient (D) while keeping the initial nutrient concentration (C0) constant, an increase in the D value leads to an increase in δ. These results align with our understanding of the active layer thickness.
The active layer thickness (δ) indeed provides valuable insight into the portion of the population actively participating in the growth of the colony. A larger δ suggests that a greater number of cells are participating in colony expansion, resulting in a denser and smoother colony. Conversely, a smaller δ value indicates that fewer cells at the colony's edge are participating in growth, leading to front instabilities as it expands. To establish the relationship between δ and the front roughness (σf), within the colony, we illustrated in Fig. 2(d) the roughness against δ values obtained from diverse simulations. In this representation, the red circle markers correspond to δ and roughness (σ/Ly) values derived from simulations aligning with the data in Fig. 2(a), where D = 10 is held constant while varying the C0 values. For each (D = 10,C0) pair in Fig. 2(a), we computed the roughness (σ/Ly) and δ, subsequently depicted with red markers in Fig. 2(d). Likewise, the blue markers in Fig. 2(d) represent the (σ/Ly) and δ values computed from simulations associated with the data in Fig. 2(b), where C0 = 100 is fixed, and various D values are considered. For each (D,C0 = 100) pair in Fig. 2(b), we determined the roughness (σ/Ly) and δ, presented with blue markers in Fig. 2(d). The result implies that the roughness of the colony front is higher for lower active layer thickness. This establishes the connection between δ and σf and sheds light on how changes in nutrient availability (controlled by C0) and diffusion (controlled by D) affect the active layer thickness and colony front roughness in a compact way.
3.2 Sectoring and demixing
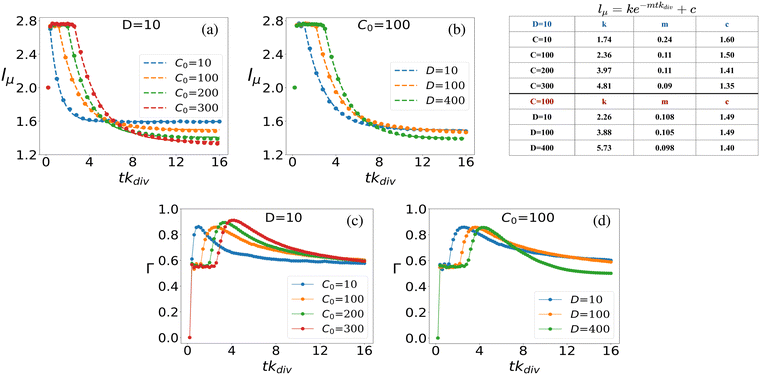
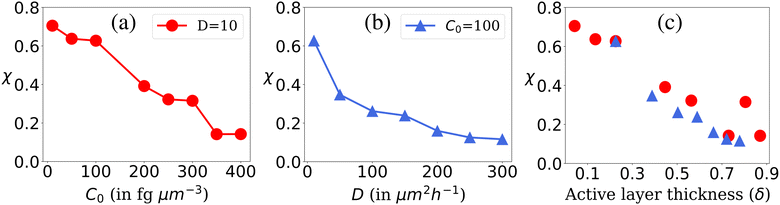
The availability of resources and nutrient diffusion play pivotal roles in determining population fluctuations within bacterial colonies. Higher resource availability and greater nutrient diffusivity tend to stabilize populations, while resource scarcity and limited diffusion can lead to more significant population fluctuations and the dominance of specific genotypes within the colony. The spatial demixing and sectoring due to the resource limitation stems from population fluctuation in the active layer. To quantitatively assess the population fluctuations within the bacterial colony as the resource levels vary, we calculate the ensemble standard deviation of the population ratio between the two bacterial types 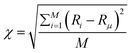 , when the colony reaches full development where Ri is the ratio of the population of bacteria type one and bacteria type two of the i-th simulation of the ensemble and Rμ is the mean population ratio of the ensemble and M = 16 denotes the number of simulations. In environments with high nutrient concentrations, the ratio approaches 1 for each simulation in the ensemble, resulting in a low standard deviation, indicating minimal population fluctuations. Conversely, in low-nutrient environments, the ratio fluctuates significantly around 1, leading to a higher standard deviation and increased population fluctuations. Fig. 3(a) supports that lower nutrient concentrations lead to higher population fluctuations, while Fig. 3(b) shows that higher diffusion values reduce fluctuations by allowing deeper nutrient penetration. Population fluctuations are influenced by both nutrient concentration and diffusion. A smaller δ indicates fewer cells actively contributing to colony growth, leading to higher fluctuations. Conversely, higher δ values involve more cells, reducing fluctuations. Fig. 3(c) demonstrates that smaller δ values result in increased population fluctuations, regardless of whether nutrient concentration or diffusion is altered. Higher δ values lead to more stable populations by involving more cells in colony growth.
, when the colony reaches full development where Ri is the ratio of the population of bacteria type one and bacteria type two of the i-th simulation of the ensemble and Rμ is the mean population ratio of the ensemble and M = 16 denotes the number of simulations. In environments with high nutrient concentrations, the ratio approaches 1 for each simulation in the ensemble, resulting in a low standard deviation, indicating minimal population fluctuations. Conversely, in low-nutrient environments, the ratio fluctuates significantly around 1, leading to a higher standard deviation and increased population fluctuations. Fig. 3(a) supports that lower nutrient concentrations lead to higher population fluctuations, while Fig. 3(b) shows that higher diffusion values reduce fluctuations by allowing deeper nutrient penetration. Population fluctuations are influenced by both nutrient concentration and diffusion. A smaller δ indicates fewer cells actively contributing to colony growth, leading to higher fluctuations. Conversely, higher δ values involve more cells, reducing fluctuations. Fig. 3(c) demonstrates that smaller δ values result in increased population fluctuations, regardless of whether nutrient concentration or diffusion is altered. Higher δ values lead to more stable populations by involving more cells in colony growth.
 | ||
| Fig. 3 Population fluctuation representing the ensemble standard deviation of the population ratio (χ) of the two bacterial types. Plot of χ for (a) different C0 values at fixed D = 10, (b) different D values at fixed C0 = 100 and (c) for different values of δ corresponding to C0 and D values of Fig. 3(a) and (b). | ||
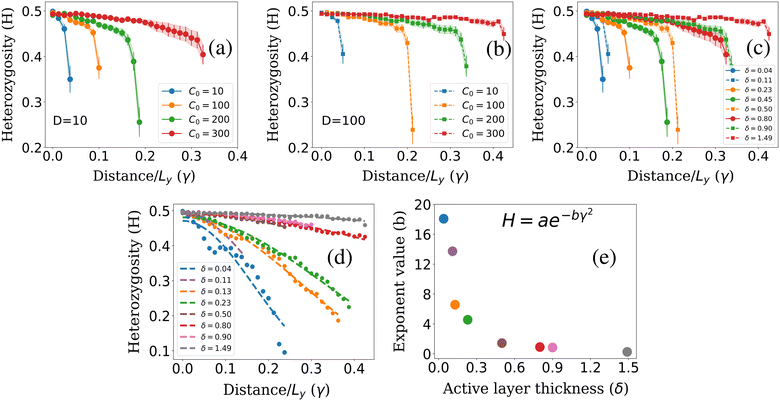
To calculate colony heterozygosity, we discretize the simulation box into xi and yi bins, each comparable in size to lc. We track colony expansion along the y-axis from the inoculum (0, Ly/2) to (Lx,Ly/2), counting red and blue cells within each xi bin. These counts are averaged over the colony and normalized along colony expansion as Nr(y) and Nb(y). Heterozygosity (H), ensemble-averaged over M simulations, is computed as: 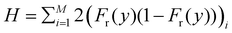 . Here, Fr(y) = Nr(y)/(Nr(y) + Nb(y)). This approach, based on Mitri et al. (2016),12 quantifies colony genetic diversity during expansion. When H = 0.5, it indicates that the colony remains well-mixed in a region and retains its genetic diversity. Conversely, when H = 0 along the colony's expansion, it signifies that the colony has fully demixed and lost its genetic diversity. In our simulations, initially well-mixed bacterial colonies had an H value of ∼0.5. However, as these colonies expanded under varying conditions of nutrient concentration (C0) and diffusion coefficient (D), diversification events occurred within the colony. The ensemble average of heterozygosity, as illustrated in Fig. 4(a)–(c) as a function of scaled distance γ (distance scaled by dividing it with Ly) at fixed scaled time, i.e. tkdiv = 16, showcases these dynamic processes. Fig. 4(a) and (b) depict the variation of H with respect to C0 for two different nutrient diffusivities: D = 10 and D = 100.
. Here, Fr(y) = Nr(y)/(Nr(y) + Nb(y)). This approach, based on Mitri et al. (2016),12 quantifies colony genetic diversity during expansion. When H = 0.5, it indicates that the colony remains well-mixed in a region and retains its genetic diversity. Conversely, when H = 0 along the colony's expansion, it signifies that the colony has fully demixed and lost its genetic diversity. In our simulations, initially well-mixed bacterial colonies had an H value of ∼0.5. However, as these colonies expanded under varying conditions of nutrient concentration (C0) and diffusion coefficient (D), diversification events occurred within the colony. The ensemble average of heterozygosity, as illustrated in Fig. 4(a)–(c) as a function of scaled distance γ (distance scaled by dividing it with Ly) at fixed scaled time, i.e. tkdiv = 16, showcases these dynamic processes. Fig. 4(a) and (b) depict the variation of H with respect to C0 for two different nutrient diffusivities: D = 10 and D = 100.
 | ||
| Fig. 4 Heterozygosity (H) plots for different initial nutrient concentrations C0 for fixed nutrient diffusion coefficients: (a) D = 10, (b) D = 100. (c) Plot of heterozygosity for different values of δ corresponding to C0 and D values of Fig. 4(a) and (b) at scaled time tkdiv = 16. (d) Fitted curve for different δ values at steady state. (e) Exponent (b) for different δ corresponding to Fig. 4(d). | ||
However, it's important to note that δ, which measures the active population participating in colony growth, provides further insight into the spatial dynamics of colony diversification. The relationship between colony diversification, represented by H, and δ is crucial. To verify this relationship, we calculate the respective δ values corresponding to simulations with different C0 and D. We then plot H vs. distance for different δ values, as shown in Fig. 4(c). The results confirm that lower δ values lead to more rapid colony demixing compared to higher δ values. Furthermore, wider error bars observed at lower δ values indicate increased noise when the active layer of cells is smaller. In environments with limited resources (lower δ), only a few cells at the colony's edge have access to these resources. Consequently, the colony expands with a common lineage and loses diversity more rapidly than it does under conditions of higher δ values. In summary, the relationship between δ and heterozygosity (H) verifies that a lower δ value correlates with a faster loss of colony diversity. This insight highlights the importance of δ in understanding how genetic diversity is retained or lost as a colony expands.
We further explored the dynamics of colony diversity loss in space by establishing a mathematical function to describe it. For different δ values, when the colony fully settled, we plotted the H value against γ and found that the data fitted well with a suitable curve described by the function: H = ae−bγ2 Here, a is approximately 0.5, and b is the exponent value. This curve provides insights into how rapidly a colony will lose its diversity spatially, especially for lower δ values, as shown in Fig. 4(d). In Fig. 4(e), we plotted the exponent value (b) for different δ values. It becomes evident that for lower δ values, the exponent value is higher, signifying that the colony will lose its diversity spatially more quickly. Conversely, for higher δ values, the exponent has a lower value, indicating that the colony will retain its diversity spatially for a more extended period. It is worth noting that for lower δ values, the data appears more scattered around the fitted curve due to higher noise in the system, which can be observed in Fig. 4(d). This analysis provides valuable insights into how various factors, such as δ, influence the spatial dynamics of colony diversity loss.
One plausible justification for the exponential form of the heterozygosity fitting curve, H = ae−bγ2, is rooted in the impact of resource availability on cell proliferation. In lower-nutrient environments, a scarcity of resources allows one cell lineage to dominate growth, division, and expansion due to chance. Consequently, cells from this lineage increase selectively at the colony front, while others decrease. This dominance results in a rapid increase in the exposed genotype due to favorable conditions for division into daughter cells, influencing the colony's genetic diversity. In nutrient-rich environments, both cell types access resources equally as the colony grows, leading to an even distribution of genotypes and maintaining genetic diversity throughout the spatial region. Consequently, H, representing diversity, remains close to 0.5 in environments abundant in resources (b → 0). This consistency indicates a colony's continual mix within higher resource availability. Conversely, in lower nutrient environments, cells encounter diverse opportunities for proliferation. Scarce resources favor one cell lineage's exposure to growth and division, resulting in selective expansion at the colony's front. This selective exposure leads to a rapid increase in specific genotypes exposed to nutrients, impacting heterozygosity, denoted by H = ae−bγ2. A higher exponential term (b) accelerates the colony's loss of genetic diversity (H = ae−bγ2), leading to a reduction in H as the colony expands spatially. This outcome aligns with our findings in lower nutrient environments, indicating a faster spatial loss of genetic diversity and demixing as γ increases.
3.3 Cell length variation and nematic ordering in growing bacterial colonies
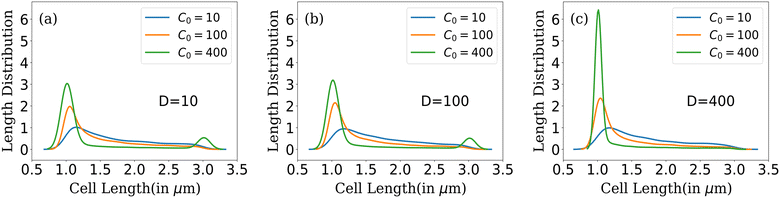 | ||
| Fig. 5 Cell length distribution of bacteria for different C0 at fixed nutrient diffusion coefficients: (a) D = 10, (b) D = 100 and (c) D = 400. | ||
In conditions of lower resource availability (C0 = 10, 100) and (D = 10, 100), the distribution depicts a colony with diverse cell lengths. This diversity arises due to resource scarcity, which limits cell growth, leading cells to cease growth in regions where nutrients are no longer available. This results in a varied and diverse distribution of bacterial cells throughout the colony. As nutrient concentration increases, we find that the distribution displays less diverse cell lengths for C0 = 400 (Fig. 5(a) and (b)). The distribution tends towards zero between the maximum and minimum cell lengths but does not completely reach zero for higher nutrient concentrations. In this scenario, the colony exhibits a higher number of cells at two specific conditions: during cell division, when cell length is at its maximum and an even higher probability of finding cells just after division (when parent cells divide into daughters). However, with higher available resources and a quick replenishment of resources (C0 = 400 and D = 400) as depicted in Fig. 5(c), most cells rapidly reach the division condition, exhaust the available nutrients, and divide into daughters, resulting in a prominent peak at the minimum cell length. At this stage, only a few cells can elongate to the critical length for division, leading to the elimination of the second peak.
In summary, colonies with lower nutrient concentrations display a wide range of bacterial cell lengths, reflecting the challenges posed by limited nutrient accessibility. In contrast, higher nutrient concentrations promote rapid growth and division, resulting in a peak around conditions favorable for bacterial proliferation. Moreover, increasing the diffusion value facilitates better nutrient penetration within the colony, leading to a concentration of bacterial lengths around conditions conducive to growth following cell division. This analysis provides insights into the complex interplay between nutrient availability, cell growth, and length distribution within bacterial colonies as they expand.
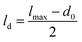 , where lmax represents the value decided by the distribution where the cell will divide. Since most cells divide around the lc = 3 threshold, if we consider lmax = lc = 3, then ld = 1. Consequently, after the division of a parent cell into two daughter cells, the mean length of the colony significantly decreases. Therefore, with time, as nutrients in the bacterial colony shift bacteria towards division, subsequent cell divisions lead to a decrease in the average cell length of the colony. Notably, the decline in lμ takes longer for colonies with higher C0 values. This phenomenon occurs due to the increased nutrient availability and penetration, which favor cell growth. However, as the colony expands and time progresses, the average cell length starts to decrease due to limited nutrient availability for cells deeper within the colony and the onset of cell division. Similarly, in Fig. 6(b), where C0 is fixed at 100 and D varies, a similar trend is observed. Lower D values cause lμ to start decreasing earlier, while higher D values result in colonies that take longer to decay. Notably, at longer times, the mean length of the colony attains higher values for colonies having lower C0. This behavior is attributed to the intricate interaction between resource limitation and the stochastic nature of bacterial division within the colonies.
, where lmax represents the value decided by the distribution where the cell will divide. Since most cells divide around the lc = 3 threshold, if we consider lmax = lc = 3, then ld = 1. Consequently, after the division of a parent cell into two daughter cells, the mean length of the colony significantly decreases. Therefore, with time, as nutrients in the bacterial colony shift bacteria towards division, subsequent cell divisions lead to a decrease in the average cell length of the colony. Notably, the decline in lμ takes longer for colonies with higher C0 values. This phenomenon occurs due to the increased nutrient availability and penetration, which favor cell growth. However, as the colony expands and time progresses, the average cell length starts to decrease due to limited nutrient availability for cells deeper within the colony and the onset of cell division. Similarly, in Fig. 6(b), where C0 is fixed at 100 and D varies, a similar trend is observed. Lower D values cause lμ to start decreasing earlier, while higher D values result in colonies that take longer to decay. Notably, at longer times, the mean length of the colony attains higher values for colonies having lower C0. This behavior is attributed to the intricate interaction between resource limitation and the stochastic nature of bacterial division within the colonies.
The dynamics of mean cell length within the bacterial colony, as depicted in Fig. 6(a) and (b), can be accurately described by exponential decay curves of the form lμ = ke−mtkdiv + c. Here, the parameters k, m, and c are fitted to the data to provide the best representation of the decay dynamics. This exponential decay behavior is consistent for both scenarios: when D is fixed at 10 and C0 varies (Fig. 6(a)), and when C0 is fixed at 100 and D varies (Fig. 6(b)). For each curve plotted in Fig. 6(a) and (b), the corresponding fitted parameters are provided in the inset table in Fig. 6. These parameters capture the characteristics of the exponential decay and provide quantitative insights into the dynamics of mean cell length as the colony evolves over time, under varying nutrient conditions and diffusion coefficients.
In what follows, we further delve into exploring the fluctuations in mean-length of the rod-shaped bacteria within the growing colony. In Fig. 6(c), we present the ensemble average standard deviation of cell length within the bacterial colony for various initial nutrient concentrations (C0) at a fixed nutrient diffusion coefficient (D = 10). The plot depicts how the standard deviation of cell length changes over time, with error bars included to represent the uncertainty in the data (note that the error bars are small compared to the data points). When analyzing the temporal dynamics of the standard deviation of cell length within the bacterial colony, we can observe specific patterns. At the initial time point (tkdiv = 0), before the colony commences its growth, all colonies exhibit the same characteristic: cells possess identical lengths (l = 2), resulting in a standard deviation of zero (Γ = 0), regardless of the initial nutrient concentration (C0) values. As nutrients become available to the bacterial colony, a sudden surge in cell length variability occurs, leading to a noticeable peak in the deviation of cell length over time. This peak represents the period of rapid cell growth and division, which leads to significant variations in cell length. Subsequently, the fluctuations in cell length start to diminish as the colony matures, and Γ stabilizes. The time it takes for the fluctuations to peak and then stabilize depends on the initial nutrient concentration (C0). Interestingly, colonies with higher initial nutrient concentrations (C0) exhibit a delay in the peak of heightened fluctuation and take more time to stabilize compared to colonies with lower initial concentrations. This phenomenon is attributed to the fact that higher nutrient concentrations enable a larger proportion of the bacterial population to be in conditions favorable for growth and division, leading to prolonged fluctuations before stabilization. However, regardless of the initial nutrient concentration, all colonies eventually reach a constant value of Γ which signifies that as the colony settles, the variability in cell lengths becomes consistent and less pronounced. Similar trends are observed when varying the diffusion coefficient (D) for a fixed initial nutrient concentration (C0 = 100) in Fig. 6(d), where higher diffusion values delay the peak of fluctuation and prolong the stabilization period before convergence to a constant Γ value.
The time evolution of cell length fluctuations exhibits a response similar to an impulse response function. The Γ value experiences a sharp increase in a short time, resembling the initial peak of an impulse response, and subsequently, it gradually decays to reach a stable and constant value. In colonies that started growing with lower initial nutrient concentrations, the fluctuation impulse peak is shorter in duration, and Γ asymptotically approaches a constant value more rapidly. This behavior is consistent with the fact that lower nutrient concentrations result in more stringent growth conditions and, consequently, shorter periods of heightened fluctuation before reaching a stable state.
Given that our study involves modeling bacterial colonies in 2D as spherocylinders, we have utilized an established formulation from Barci et al.50 to describe ordering in a two-dimensional nematic. To quantify the extent of alignment within the colony, we employ a two-dimensional nematic order parameter, defined as: 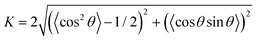 . Here, θ denotes the angle between a cell's long axis and with some reference axis. The angular brackets signify averaging over all cells. This formulation is consistent with the methodology presented in ref. 51 and has been employed in a prior study52 to explore the role of growth rate in the orientational alignment of E. coli in a slit. The resulting nematic order value (K) ranges from 0 to 1. A value of K = 1 indicates a high degree of order, signifying that bacterial cells within the colony are uniformly aligned with the reference axis, while K = 0 implies a complete absence of order.
. Here, θ denotes the angle between a cell's long axis and with some reference axis. The angular brackets signify averaging over all cells. This formulation is consistent with the methodology presented in ref. 51 and has been employed in a prior study52 to explore the role of growth rate in the orientational alignment of E. coli in a slit. The resulting nematic order value (K) ranges from 0 to 1. A value of K = 1 indicates a high degree of order, signifying that bacterial cells within the colony are uniformly aligned with the reference axis, while K = 0 implies a complete absence of order.
Using the scalar order parameter formula, we evaluated the ordering of rod-shaped bacteria within the colony, employing horizontal and vertical axes as reference directions for different D and C0 values, as depicted in Fig. S1 (ESI†). Our findings suggest that the rod-shaped cells within the colony lack global order, regardless of the chosen reference direction. However, upon closer inspection, patches of ordered cells become apparent within the colony, particularly in relatively small domain sizes. This prompts us to investigate the dependency of cell ordering on domain size more closely. Consequently, we focus on determining the local ordering within a vicinity defined by the rcut = r/d0, where d0 represents the diameter of a cell, r represents the radial distance and rcut is a dimensionless distance analogous parameter.
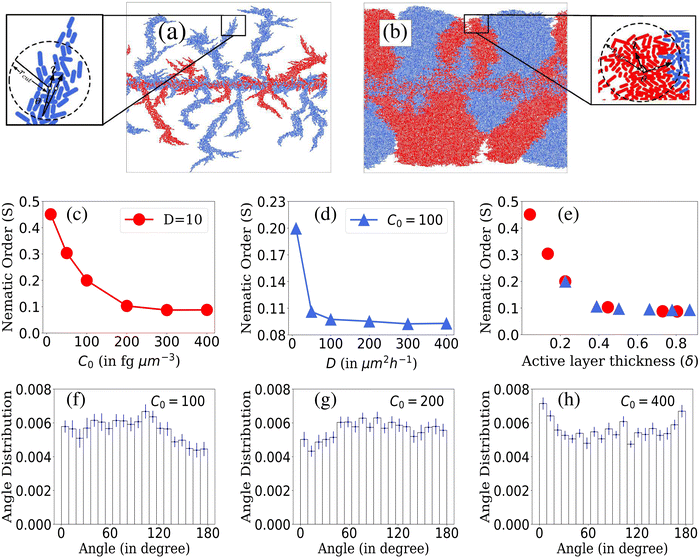
To calculate the local order, we perform neighbor searching around each cell, using a cutoff distance of rcut = 10/d0. For a visual demonstration, in Fig. 7(a) and (b), a zoomed-in section of the simulated colony outcomes is presented for (D = 10, C0 = 10) and (D = 10, C0 = 200), respectively, illustrating the arrangement of neighboring cells around the ith cell within the colony's frontal region. The ith cell is marked by the index ‘i’ at the arrowhead, while the arrow along the rod cells represents the orientation vector of each rod-shaped cell. The angle θ depicts the relationship between the ith cell and its neighboring rod cell. Within this zoomed-in region, depicted in Fig. 7(a) and (b), a circular boundary delineates the search region around the ith cell, denoted by a radius of rcut. The nematicity is then computed using the formula mentioned above, and the average is taken over all cells within the colony. Finally, to obtain an ensemble average of the nematic order, we perform this calculation across M simulations. The ensemble-averaged nematic order (S) is given by: 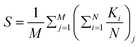 . Here, N represents the total number of cells in a simulation, and M = 16 denotes the number of simulations over which the nematicity is calculated. In Fig. 7(a), observed at lower concentration, there's a qualitative validation of longer cells’ prevalence, indicating a more ordered arrangement of rod cells within the colony. However, in the case of higher concentration as depicted in Fig. 7(b), the colony appears denser with a higher presence of shorter cells, resulting in a comparatively less ordered system than the lower concentration scenario.
. Here, N represents the total number of cells in a simulation, and M = 16 denotes the number of simulations over which the nematicity is calculated. In Fig. 7(a), observed at lower concentration, there's a qualitative validation of longer cells’ prevalence, indicating a more ordered arrangement of rod cells within the colony. However, in the case of higher concentration as depicted in Fig. 7(b), the colony appears denser with a higher presence of shorter cells, resulting in a comparatively less ordered system than the lower concentration scenario.
In Fig. 7(c)–(e), we present the local nematic order of bacterial cells within a colony when it reached a steady state. In cases where D is held constant and C0 varies as shown in Fig. 7(c), colonies with lower C0 values exhibit greater nematic order than those with higher C0 values. This phenomenon is attributed to the wider range of bacterial lengths within colonies growing with lower initial nutrient conditions. Increased length diversity tends to foster greater alignment and ordering within the system. On the other hand, for colonies with fixed C0 and varying D, nematic order is more prominent at lower D values. Higher D values result in denser colonies that lack significant ordering, as shown in Fig. 7(d). A large number of densely packed cells of smaller lengths show random orientation within the colony for higher D, which limits the extent of alignment among bacteria. These observations align with the variation in the nematic order parameter concerning the active layer thickness (δ), as depicted in Fig. 7(e). Notably, higher values of δ correlate with lower orientational order within colonies.
We delved deeper into the impact of domain size on local nematic order. In Fig. S2(a) and (b) (ESI†), we observe a decline in local nematic order with increasing domain size (rcut). Notably, for a fixed rcut value, the system exhibits higher order when nutrient concentration and diffusivity are lower. The domain size of rcut = 10/d0, highlighted in Fig. S2(a) and (b) (ESI†), elucidates these trends. Interestingly, in environments with limited resources, greater ordering is consistently observed across varying domain sizes, contrasting with environments featuring higher resource availability. Additionally, as domain size (rcut) increases, the system transitions from perfect alignment, observed in very small domains, to increased disorder. Moreover, for higher C0 and D values, the decline in ordering accelerates with increasing domain size, eventually stabilizing, indicating a level of stability in colony order. This stability may arise from the random alignment of additional cells within the region, sustaining overall colony order.
In low-nutrient environments with lower (C0, D) values, colonies tend to form finger-like structures due to limited resources, resulting in less densely packed colonies spatially. This environment promotes the presence of longer cells with a wider length distribution. Previous studies have suggested that longer rod-shaped particles tend to progress towards a nematic phase.53 Conversely, in higher-nutrient settings with higher (C0, D) values, colonies tend to have a higher number of shorter cells, leading to denser colony formations. The increased density of shorter cells seemingly conflicts with mechanical interactions, causing them to orient randomly in space and resulting in lower ordering. The observed ordering in bacterial colonies in our studies seems to be influenced by the interplay between cell density and length distribution, which varies based on the C0 and D values.
In what follows, to decipher the alignment of interfacial cells at the colony periphery, we generated ensemble-averaged histograms showing the normalized angle distribution across various nutrient concentrations while maintaining a constant nutrient diffusivity (D = 10), illustrated in Fig. 7(f)–(h). The presented data suggest that due to the discrete nature of cells and inherent stochasticity, drawing definitive conclusions about the alignment (parallel, perpendicular, or random) of cells at the colony front is challenging. It appears that the alignment tends towards randomness. Obtaining a comprehensive understanding of the ordering dynamics of rod-shaped cells within the colony might necessitate a broader sampling of the colony cells. Relying solely on boundary cells may not sufficiently capture the complete ordering behavior of the system. Subsequently, we explore the angular distribution of all cells within the colony. The heat map representations of colony cell orientations for (D = 10, C0 = 10) and (D = 10, C0 = 200) in Fig. S3(a) and (b) (ESI†) qualitatively illustrate how adjacent cells within the colony form patches exhibiting a common angle orientation, indicated by the consistent color representation. Furthermore, normalized histogram plots of colony cell angle distributions for (D = 10, C0 = 10) and (D = 10, C0 = 200) in Fig. S3(c) and (d) (ESI†) demonstrate that the colony cells do not significantly conform to any preferred common angle ordering. However, in the case of low resource availability, the cells make smaller angles with respect to the director ranging from 30 to 100 degrees. Furthermore, in Fig. S4(a) and (b) (ESI†), we calculate the spatial orientational autocorrelation function54 to quantify the range over which cells share a common orientation. Colonies with higher nutrient concentration and diffusivity exhibit a rapid decrease in correlation values, indicating a disorderly state over shorter ranges compared to colonies with lower C0 and D values. For further details regarding calculating the orientational autocorrelation, please refer to the ESI.†
To this end, it is worth mentioning that, in a similar context of nonmotile bacterial colonies, a recent study by Schwarzendahl et al.55 identified +1/2 and −1/2 topological defects.56–58 Their methodology involved computations of the nematic tensorial orientation field, mapping it to continuum fields using a smoothing function and identifying these defects through integration and clustering algorithms. However, it is important to note that a comprehensive investigation of active-nematics, including the specific context of these topological defects, requires a detailed study beyond the current scope of our research. In our current focus, we aim to delve deeper into understanding the connection between resource availability and the orientational order that may emerge within the bacterial colony.
In summary, these findings shed light on the complex interplay between nutrient concentration, diffusion, and nematic order in bacterial colonies, offering valuable knowledge about the alignment and organization of bacterial cells in dynamic, evolving environments.
4 Concluding remarks
Understanding the dynamics of bacterial colonies is fundamental for unraveling the complexity of microbial ecosystems. In this comprehensive study, we employ a model featuring two visually distinct types of rod-shaped bacteria that are otherwise identical, with resource-related parameters, initial nutrient concentration (C0), and nutrient diffusion coefficient (D), as our primary variables of interest. These parameters collectively govern the resource availability to colony cells and, consequently, shape the colony's behavior. Notably, we find that resource availability plays a pivotal role in regulating colony morphology, genetic diversity, and microscopic ordering. Our investigation encompasses diverse facets of colony dynamics:• Broadening the research focus: while our model is built upon previous work,7,10 our study focuses on a wider question. Specifically, we explore how resource limitations (nutrient concentration and diffusivity) not only impact front instabilities but also contribute to spatial patterning and sectoring when two neutral bacterial strains coexist and grow together, sharing identical growth and mechanical properties but differing genetically. These phenomena are primarily driven by population fluctuations and the intricate interplay of resource availability.
• Introduction of a dimensionless parameter: our study introduces a novel dimensionless parameter, δ, reflecting the active layer thickness of a growing colony. This parameter plays a crucial role in quantifying the intricate interplay of resource availability and actively influences colony front characteristics. Lower δ values are associated with less dense and rougher colony fronts, contributing to a deeper understanding of how resource limitations shape bacterial colonies.
• Population fluctuations and genetic diversity: the incorporation of a novel population fluctuation parameter (χ) allows us to observe pronounced fluctuations in cell population ratios, particularly in low-resource conditions. Lower δ values are linked to higher population fluctuations, increasing the likelihood of one cell lineage dominating due to nutrient scarcity. Heterozygosity analyses reveal that colonies with lower δ values lose genetic diversity more rapidly as they expand.
• Temporal dynamics of colony and cell length: our study delves into the temporal dynamics of colony mean length and cell length fluctuations, uncovering diverse length distributions within colonies, especially in lower resource environments. Initial disturbances upon nutrient exposure gradually subside as colonies expand. This model reveals exponential decay patterns in colony mean length, with lower initial nutrient concentrations resulting in faster decay. Cell length fluctuations exhibit impulse-like responses before stabilizing, providing insights into the dynamic behavior of bacterial colonies over time.
• Impact on colony microstructure (nematic ordering in colonies): colonies in lower nutrient conditions exhibit higher nematic ordering, highlighting the impact of resource availability on colony microstructure. This study establishes a robust relationship between δ and nematicity, emphasizing the role of resource contributions from both C0 and D in shaping the structural organization of bacterial colonies.
Our findings deepen our understanding of microbial ecosystems, illustrating the multifaceted nature of bacterial colony dynamics. The influence of resource availability, governed by C0 and D reflected as active layer thickness, emerges as a pivotal factor shaping colony behavior. Additionally, our approach reveals previously unexplored relationships between genetic diversity, population and cell length fluctuations, nematic ordering and resource availability. This comprehensive exploration underscores the multifaceted nature of bacterial colony dynamics and the necessity of considering multiple parameters in modeling and interpreting their behavior.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the Indian Institute of Science Education and Research Thiruvananthapuram (IISERTVM), India for providing the support of computing resources. Additionally, we extend our appreciation for the allocation of high-performance computing time at the Padmanabha Cluster, IISERTVM, India. PG acknowledges the Start-up Research grant (SRG/2022/000043) by the Science and Engineering Research Board (SERB), India. RK gratefully acknowledges SERB, India for the partial financial support. Lastly, KM gratefully acknowledges MOE, India for PMRF-ID-0803052.Notes and references
- S. Arnaouteli, N. C. Bamford, N. R. Stanley-Wall and Á. T. Kovács, Nat. Rev. Microbiol., 2021, 19, 600–614 CrossRef CAS PubMed
.
- K. Sauer, P. Stoodley, D. M. Goeres, L. Hall-Stoodley, M. Burmølle, P. S. Stewart and T. Bjarnsholt, Nat. Rev. Microbiol., 2022, 20, 608–620 CrossRef CAS PubMed
.
- G. El Tekle and W. S. Garrett, Nat. Rev. Cancer, 2023, 23, 600–618 CrossRef CAS PubMed
.
- T. Roose, S. J. Chapman and P. K. Maini, SIAM Rev., 2007, 49, 179–208 CrossRef
.
- M. J. Wade, J. Harmand, B. Benyahia, T. Bouchez, S. Chaillou, B. Cloez, J.-J. Godon, B. M. Boudjemaa, A. Rapaport and T. Sari,
et al.
, Ecol. Modell., 2016, 321, 64–74 CrossRef
.
- A. Mitchell and J. Wimpenny, J. Appl. Microbiol., 1997, 83, 76–84 CrossRef CAS PubMed
.
- F. Farrell, O. Hallatschek, D. Marenduzzo and B. Waclaw, Phys. Rev. Lett., 2013, 111, 168101 CrossRef CAS PubMed
.
- C. Giverso, M. Verani and P. Ciarletta, J. R. Soc., Interface, 2015, 12, 20141290 CrossRef PubMed
.
- H. Shimada, T. Ikeda, J.-I. Wakita, H. Itoh, S. Kurosu, F. Hiramatsu, M. Nakatsuchi, Y. Yamazaki, T. Matsuyama and M. Matsushita, J. Phys. Soc. Jpn., 2004, 73, 1082–1089 CrossRef CAS
.
- N. Rana, P. Ghosh and P. Perlekar, Phys. Rev. E, 2017, 96, 052403 CrossRef PubMed
.
- P. Ghosh and H. Levine, Phys. Rev. E, 2017, 96, 052404 CrossRef PubMed
.
- S. Mitri, E. Clarke and K. R. Foster, ISME J., 2016, 10, 1471–1482 CrossRef CAS PubMed
.
- C. D. Nadell, K. R. Foster and J. B. Xavier, PLoS Comput. Biol., 2010, 6, e1000716 CrossRef PubMed
.
- K. P. Koutsoumanis and A. Lianou, Appl. Environ. Microbiol., 2013, 79, 2294–2301 CrossRef CAS PubMed
.
- B. von Bronk, S. A. Schaffer, A. Götz and M. Opitz, PLoS Biol., 2017, 15, e2001457 CrossRef PubMed
.
- M. Kaern, T. C. Elston, W. J. Blake and J. J. Collins, Nat. Rev. Genet., 2005, 6, 451–464 CrossRef CAS PubMed
.
- J. W. Wimpenny and R. Colasanti, FEMS Microbiol. Ecol., 1997, 22, 1–16 CrossRef CAS
.
- K. Nagarajan, C. Ni and T. Lu, ACS Synth. Biol., 2022, 11, 3564–3574 CrossRef CAS PubMed
.
- X. Zhang, X. Wang, K. Nie, M. Li and Q. Sun, Phys. Biol., 2016, 13, 046002 CrossRef PubMed
.
- Z. You, D. J. Pearce, A. Sengupta and L. Giomi, Phys. Rev. Lett., 2019, 123, 178001 CrossRef CAS PubMed
.
- K. Cremin, S. J. Duxbury, J. Rosko and O. S. Soyer, Interface Focus, 2023, 13, 20220062 CrossRef PubMed
.
- L. Eigentler, F. A. Davidson and N. R. Stanley-Wall, Open Biol., 2022, 12, 220194 CrossRef CAS PubMed
.
- W. Liu, T. A. Tokuyasu, X. Fu and C. Liu, Curr. Opin. Microbiol., 2021, 63, 109–116 CrossRef CAS PubMed
.
- A. E. Blanchard and T. Lu, BMC Syst. Biol., 2015, 9, 1–13 CrossRef PubMed
.
- J. Van Gestel, F. J. Weissing, O. P. Kuipers and A. T. Kovács, ISME J., 2014, 8, 2069–2079 CrossRef CAS PubMed
.
- V. Bucci, C. D. Nadell and J. B. Xavier, Am. Nat., 2011, 178, E162–E173 CrossRef PubMed
.
- I. Frost, W. P. Smith, S. Mitri, A. S. Millan, Y. Davit, J. M. Osborne, J. M. Pitt-Francis, R. C. MacLean and K. R. Foster, ISME J., 2018, 12, 1582–1593 CrossRef CAS PubMed
.
- W. Kim, F. Racimo, J. Schluter, S. B. Levy and K. R. Foster, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E1639–E1647 CAS
.
- W. P. Smith, Y. Davit, J. M. Osborne, W. Kim, K. R. Foster and J. M. Pitt-Francis, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E280–E286 CrossRef CAS PubMed
.
- S. Estrela and S. P. Brown, PLoS Comput. Biol., 2018, 14, e1006179 CrossRef PubMed
.
- F. Goldschmidt, R. R. Regoes and D. R. Johnson, ISME J., 2018, 12, 136–144 CrossRef CAS PubMed
.
- F. Goldschmidt, L. Caduff and D. R. Johnson, ISME J., 2021, 15, 2415–2426 CrossRef PubMed
.
- J. Monod, Annu. Rev. Microbiol., 1949, 3, 371–394 CrossRef CAS
.
-
B. Chezeau and C. Vial, Biohydrogen, Elsevier, 2019, pp. 445–483 Search PubMed
.
- P. Ghosh, J. Mondal, E. Ben-Jacob and H. Levine, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E2166–E2173 CrossRef CAS PubMed
.
- W. Donachie, K. Begg and M. Vicente, Nature, 1976, 264, 328–333 CrossRef CAS PubMed
.
- P. Bera, A. Wasim and P. Ghosh, Soft Matter, 2023, 19, 1034–1045 RSC
.
- D. Volfson, S. Cookson, J. Hasty and L. S. Tsimring, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15346–15351 CrossRef PubMed
.
- P. Bera, A. Wasim, J. Mondal and P. Ghosh, Soft Matter, 2021, 17, 7322–7331 RSC
.
- E. Ben-Jacob, I. Cohen and D. L. Gutnick, Annu. Rev. Microbiol., 1998, 52, 779–806 CrossRef CAS PubMed
.
- P. Deng, L. de Vargas Roditi, D. Van Ditmarsch and J. B. Xavier, New J. Phys., 2014, 16, 015006 CrossRef PubMed
.
- E. Ben-Jacob, Contemp. Phys., 1997, 38, 205–241 CrossRef CAS
.
- J. A. Shapiro, BioEssays, 1995, 17, 597–607 CrossRef CAS PubMed
.
- N. Verstraeten, K. Braeken, B. Debkumari, M. Fauvart, J. Fransaer, J. Vermant and J. Michiels, Trends Microbiol., 2008, 16, 496–506 CrossRef CAS PubMed
.
- J.-i Wakita, H. Itoh, T. Matsuyama and M. Matsushita, J. Phys. Soc. Jpn., 1997, 66, 67–72 CrossRef CAS
.
- J.-i Wakita, K. Komatsu, A. Nakahara, T. Matsuyama and M. Matsushita, J. Phys. Soc. Jpn., 1994, 63, 1205–1211 CrossRef CAS
.
- T. Sams, K. Sneppen, M. H. Jensen, C. Ellegaard, B. E. Christensen and U. Thrane, Phys. Rev. Lett., 1997, 79, 313 CrossRef CAS
.
- K. Kawasaki, A. Mochizuki, M. Matsushita, T. Umeda and N. Shigesada, J. Theor.
Biol., 1997, 188, 177–185 CrossRef CAS PubMed
.
- F. D. Farrell, M. Gralka, O. Hallatschek and B. Waclaw, J. R. Soc., Interface, 2017, 14, 20170073 CrossRef PubMed
.
- D. G. Barci and D. A. Stariolo, Phys. Rev. B: Condens. Matter Mater. Phys., 2009, 79, 075437 CrossRef
.
- H. Chaté, F. Ginelli and R. Montagne, Phys. Rev. Lett., 2006, 96, 180602 CrossRef PubMed
.
- J. Sheats, B. Sclavi, M. Cosentino Lagomarsino, P. Cicuta and K. D. Dorfman, R. Soc. Open Sci., 2017, 4, 170463 CrossRef PubMed
.
- V. Narayan, N. Menon and S. Ramaswamy, J. Stat. Mech.: Theory Exp., 2006, 2006, P01005 Search PubMed
.
- G. Duclos, S. Garcia, H. Yevick and P. Silberzan, Soft Matter, 2014, 10, 2346–2353 RSC
.
- F. J. Schwarzendahl and D. A. Beller, Front. Phys., 2022, 10, 940980 CrossRef
.
- D. Dell’Arciprete, M. L. Blow, A. T. Brown, F. D. Farrell, J. S. Lintuvuori, A. F. McVey, D. Marenduzzo and W. C. Poon, Nat. Commun., 2018, 9, 4190 CrossRef PubMed
.
- R. C. Coelho, N. A. Araújo and M. M. T. da Gama, Soft Matter, 2022, 18, 7642–7653 RSC
.
- K. Copenhagen, R. Alert, N. S. Wingreen and J. W. Shaevitz, Nat. Phys., 2021, 17, 211–215 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sm00066h |
| This journal is © The Royal Society of Chemistry 2024 |