Open Access Article
Open Access ArticleElectrochemical detection of tumor cells based on proximity labelling-assisted multiple signal amplification†
Guozhang
Zhou‡
ab,
Fei
Zhou‡
a,
Xiaomeng
Yu
ac,
Daiyuan
Zhou
a,
Jiaqi
Wang
a,
Bing
Bo
*d,
Ya
Cao
*ab and
Jing
Zhao
 *a
*a
aCenter for Molecular Recognition and Biosensing, Shanghai Engineering Research Center of Organ Repair, School of Life Sciences, Shanghai University, Shanghai 200444, China. E-mail: conezimint@shu.edu.cn; jingzhao@t.shu.edu.cn
bJoint International Research Laboratory of Biomaterials and Biotechnology in Organ Repair (Ministry of Education), Shanghai University, Shanghai, 200444, China
cState Key Laboratory of Analytical Chemistry for Life Science, School of Life Sciences, Nanjing University, Nanjing 210023, P.R. China
dDepartment of Medical Oncology, Shanghai Pulmonary Hospital & Thoracic Cancer Institute, Tongji University School of Medicine, Shanghai 200433, China. E-mail: bbtice@163.com
First published on 2nd August 2024
Abstract
Malignant tumors are the second leading cause of human deaths worldwide, and early cancer screening and diagnosis can effectively reduce cancer mortality. Herein, we propose a new electrochemical method for the highly sensitive detection of MUC1-positive tumor cells based on proximity labelling-assisted multiple signal amplification. Specifically, a MUC1 aptamer-modified electrode was prepared for capturing MUC1-positive tumor cells, followed by binding of G4-DNA strands to the cells with the aid of a mild reduction reaction. A hemin/G4-DNA complex was then formed and acted as a mimic of horseradish peroxidase, catalysing the proximal labelling of tyramine-modified gold nanoparticles to induce silver-enhanced electrochemical signal amplification. Electrochemical results demonstrated that the method was able to specially identify MUC1-positive tumor cells and generate corresponding electrochemical responses in the range of 100 cells per mL to 1 × 106 cells per mL with a detection limit of 21 cells per mL. Furthermore, the method displayed good stability and anti-interference performance in complex serum environments. Therefore, our work may provide an effective tool to improve the accuracy of cell-based tissue examination and liquid biopsy for early diagnosis of cancers in the future.
Introduction
Cancer is a malignant disease characterized by the abnormal proliferation and dissemination of tumor cells and is one of the primary causes of human deaths worldwide. According to the International Agency for Research on Cancer (IARC), there were 20 million new cases of cancers and 9.74 million cancer-related deaths globally in 2022.1 A 77% rise in new cancer cases is expected to exceed the number of cases beyond 35 million by 2050. Among the different types of cancers, lung cancer is the most commonly diagnosed cancer, accounting for 12.4% of the total number of new cancer cases, which is followed by female breast cancer.1 Although low-income countries have a lower incidence of cancer compared to higher-income countries, these populations always face an increased risk of cancer-related mortality due to inadequate access to timely diagnosis and treatment. Therefore, early detection is essential to improve the cure and survival rates of cancer patients. Tumor cells, containing abundant tumor-related information, are the main cells comprising tumor tissues and have become an important tumor marker to present the molecular characteristics of malignant tumors. Meanwhile, circulating tumor cells (CTCs), which are released into the bloodstream by primary tumors, have attracted increasing attention in liquid biopsy for tumor evaluation during whole disease progression.2–4 Despite this, the number of tumor cells remains quite limited in the early stages of cancer, especially for CTCs, which presents a significant challenge for early detection in cancer diagnosis. Therefore, it is urgently required to develop some highly sensitive and specific approaches to identify and detect tumor cells, which may advance cell-based tissue examination and liquid biopsy.5–7In order to tackle these challenges, the introduction of new analytical technologies boosts the development of cell-based biosensors. Among them, electrochemical technique offers a powerful tool for cancer diagnosis, leveraging its unique features of easy manipulation, low cost, and inherently high sensitivity.8–11 Furthermore, signal amplification coupled with electrochemical technique has evolved into multiple signal amplification strategies, contributing to increased sensitivity for early detection of many cancers.12–14 Tyramine signal amplification (TSA), an enzyme-mediated proximal labelling technique, is an emerging signal amplification method that utilizes horseradish peroxidase (HRP) to catalyze the oxidation of tyramine-based substrates, thereby generating a large number of free radicals directly deposited on the adjacent protein surface.15,16 Thus, TSA serves as the foundation for multiple signal amplification and demonstrates the advantages of good sensitivity, stability, and rapid response in the biomedine and clinical diagnosis, such as the detection of fungi, human chorionic gonadotrophin, and hepatocellular carcinoma cells.17–20
In this work, we are proposing an electrochemical method to sensitively detect tumor cells in combination with the electrochemical aptasensor and TSA-assisted multiple signal amplification. The electrochemical aptasensor specifically recognizes and captures the tumor cells through the selective binding of surface protein and corresponding aptamers. Subsequently, mild reduction facilitates the reduction of the disulfide bond within the membrane proteins of the captured cells,21–23 and generates a large number of reactive thiol groups to bind to G-rich DNA (G4-DNA strands) through Michael addition, which was the first signal amplification; G4-DNA strands interact with hemin to form the hemin/G4-DNA complex as HRP mimics, catalysing TSA-assisted proximal labelling of tyramine-modified gold nanoparticles (Ty–AuNPs) onto the neighbouring proteins at the cell surface, which was the second signal amplification; a large number of the silver atoms were deposited onto AuNPs to generate the amplified electrochemical signal for the quantitative determination, which was the third signal amplification. It is worth mentioning that Ty–AuNPs used in this study can be used to develop robust nanointerfaces for enhancing electrochemical signals, due to excellent biocompatibility, large surface area, and high interfacial reactivity.24–26 Therefore, the proximal labelling-assisted multiple signal amplification significantly enhances the sensitivity of the electrochemical aptasensor for the detection of tumor cells, thereby improving its diagnostic accuracy and precision.
Experimental
Material and reagents
The DMEM medium, fetal bovine serum (FBS), and trypsin (0.25% EDTA) were purchased from Thermo Fisher Scientific. A mammary epithelial cell medium (MEpiCM) was obtained from ScienCell Research Laboratories. Recombinant mucin 1 (MUC-1), carcinoembryonic antigen (CEA), folate receptor (FR), alpha-fetoprotein (AFP), and programmed cell death ligand 1 (PD-L1) were purchased from China National Biotec Group Company Limited. Tris(2-carboxyethyl)phosphine (TCEP), methylcyclohexane (MCH), ascorbic acid, chloroauric acid (HAuCl4), 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), tyramine and silver nitrate (AgNO3) were purchased from Sigma Aldrich. Gold electrodes were purchased from Wuhan Gaoshi Ruilian Technology Company Limited (China). Tween-20 (Tween-20) and Hoechst fluorescent dye 33342 (Hoechst 33342) were purchased from Beijing Solepol Technology Company Limited. All the DNA strands were synthesized by Hippo Biotechnology Company Limited, and their sequences are listed in Table S1.†Cell cultures and preparation of Ty–AuNPs
Breast cancer cells MCF-7 and lung cancer cells A549, and normal human breast cells MCF-10A were obtained from the National Collection of Authenticated Cell Cultures. MCF-7 and A549 cells were cultured in the DMEM medium containing cultures in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS. Human normal mammary epithelial cells MCF-10A cells were cultured using MEpiCM. MUC-1 expression in these cells was analysed by using flow cytometry. AuNPs were prepared by trisodium citrate reduction based on our previous work.27 Afterward, 4 mg mL−1 of tyramine was mixed with AuNPs solution at the volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and incubated overnight. The mixture was then centrifuged at 12
3, and incubated overnight. The mixture was then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 15 min, and the precipitate was re-suspended and stored in the dark at 4 °C.17
000 g for 15 min, and the precipitate was re-suspended and stored in the dark at 4 °C.17
Preparation of aptamer-functionalized electrodes
MUC-1 aptamer-functionalized gold electrode was prepared through Au–S bonds. 10 nM mercapto group-modified MUC-1 aptamer was first incubated with 10 mM TCEP at 4 °C for 1 h, and the mixture was then transferred to the surface of the gold electrode and reacted overnight at 4 °C. After that, MUC-1 aptamer-modified electrode was gently rinsed with PBS, and then treated by using 100 μL of 1 mM MCH solution at 37 °C for 30 min. Finally, the electrode was thoroughly rinsed with PBS and prepared for further use.28Electrochemical measurements
The cells at the desired concentrations were incubated with MUC-1 aptamer-functionalized electrode at 37 °C for 2 h. 1 mM TCEP was used to perform the mild reduction at 37 °C for 30 min, and 200 μL of 100 nM G4-DNA solution was incubated with the cell-enriched electrode at 37 °C for 1 h. After that, 50 μL of 100 μM hemin were added and reacted with G4-DNA strands at 37 °C for 20 min to form the hemin/G4-DNA complex. 100 μL of AuNPs–Ty were added and incubated with the electrode at 37 °C for 1 h for the deposition of tyramine onto the cell surface. Finally, a solution containing 0.5 mM AgNO3 and 0.25 mM ascorbic acid was incubated with the electrode at 37 °C for 7 min. Silver was dissolved in HNO3, and detected using differential pulse voltammetry (DPV) according to our previous work.29,30Results and discussion
The principle of the electrochemical method for the detection of tumor cells
Mucin-1 (MUC-1) is a membrane glycoprotein overexpressed in many cancers, such as breast, lung, and pancreatic cancers.31–33 MUC-1 plays a significant role in tumorigenesis, progression, and treatment response.34–36 Taking MUC1-positive tumor cells as an example, proximal labelling-assisted multiple signal amplification is designed for the electrochemical detection of tumor cells as illustrated in Scheme 1. Specifically, MUC-1 aptamer was self-assembled on the surface of the gold electrode via Au–S bonds and was used to capture MUC1-positive tumor cells through selective recognition and binding of MUC1 protein that was overexpressed at the surface of the tumor cells. Subsequently, the disulfide bonds within the membrane proteins were destroyed through a TCEP-mediated mild reduction reaction, exposing abundant thiol groups for the linking of the maleimide group-modified G4-DNA strands through Michael's addition reaction. After interacting with hemin, the hemin/G4-DNA complex worked as the HRP mimic and catalyzed the oxidation of tyramine to the reactive radicals, and facilitated the deposition of tyramine-modified gold nanoparticles (Ty–AuNPs) through the interaction with the tyrosine residues at the neighbouring membrane proteins. Finally, silver ions were deposited at the surface-enriched AuNPs and led to a greatly enhanced electrochemical signal, realizing highly sensitive detection of MUC1-positive tumor cells. | ||
| Scheme 1 Schematic of the electrochemical detection of MUC-1-positive tumor cells based on the proximal labelling-assisted multiple signal amplification. | ||
Identification of MUC-1 protein at the MUC-1 aptamer-functionalized electrode
MUC-1 aptamer-functionalized electrode was first adopted to investigate the feasibility of the recognition and binding of MUC-1 proteins. Electrochemical impedance spectroscopy (EIS) was conducted to characterize the stepwise reactions occurring at the gold electrode (Fig. 1A). The semicircle portion observed at a higher frequency in a typical Nyquist plot is associated with the electron-transfer-limited process, whereas the linear portion at a lower frequency is associated with the diffusion process. Accordingly, the stepwise modification of the electrode could be investigated by the observation of the semicircle at a higher frequency. No impedance was detected at the bare electrode, but an increased impedance with a larger semicircle was observed after the modification of the gold electrode with MUC-1 aptamers due to its electrostatic repulsion with the negatively charged potassium ferrocyanide/ferrocyanide. A significant increase in impedance value was observed after the incubation with MUC-1 proteins, attributed to the enhanced steric hindrance at the electrode surface following the selective capture of target proteins by using MUC-1 aptamers. Furthermore, the reduction of disulfide bonds within the surface-captured proteins using TCEP enabled the exposure of the thiol groups for covalent binding of the maleimide group-modified G4-DNA chains through the Michael addition reaction, resulting in the increased impedance at the electrode again. Therefore, EIS results first demonstrated the preparation of a MUC-1 aptamer-functionalized electrode for the selective recognition and binding of the target protein, which could be labelled by G4-DNA through mild reduction.Because the formation of the hemin/G4-DNA complex was essential to catalyze the proximal labelling to induce multiple signal amplification, we studied the catalytic activities of the hemin/G4 complex using UV-vis spectroscopy. In general, HRP catalyzes the oxidation of ABTS to ABTS+. In the presence of H2O2, exhibiting a characteristic absorption peak at approximately 420 nm. As shown in Fig. 1B, a substantial increase of the absorption at 420 nm was observed when the hemin/G4-DNA complex was present compared to that with only hemin or G4-DNA, indicating the peroxide-like activity of hemin/G4-DNA. In our work, AuNPs were synthesized through the sodium citrate reduction method and adopted as the nanocarrier for the in situ deposition of silver atoms and aroused silver-enhanced electrochemical signals. The morphological structure of AuNPs were also confirmed by using TEM, and the modification with tyramine was further confirmed by using UV-vis spectroscopy (Fig. 1C and D).
Electrochemical determination of MUC-1 proteins was performed using proximal labelling-assisted multiple signal amplification. Fig. 2A schematically illustrates the principle of the electrochemical detection of MUC-1 proteins. G4-DNA linking to the target protein interacted with hemin, and formed the hemin/G4-DNA complex to catalyze the proximal labelling and subsequently induced the silver-enhanced electrochemical responses. Fig. 2B shows the electrochemical results obtained under different reaction conditions. The peak current obtained with MUC-1 protein was found to significantly increase after proximal labelling-assisted multiple signal amplification compared to that without MUC-1 or without the binding of hemin/G4-DNA. The results were in line with the design of our method. MUC-1 proteins were enriched onto the electrode through the selective interaction with MUC-1 aptamers at the electrode and were linked with G4-DNA probes with the aid of mild reduction. Afterward, the hemin/G4-DNA complex linked to the surface-captured proteins catalyzed proximal labelling-assisted multiple signal amplification, resulting in a greatly enhanced electrochemical response. In contrast, proximal labelling-assisted multiple signal amplification was inhibited by the absence of the target proteins and the catalytic DNA, hemin/G4-DNA complex. Furthermore, the electrochemical method was proven to be highly specific in identifying the target proteins, as the high peak current was obtained only with the addition of the target protein, MUC-1 (Fig. 2C). Overall, the electrochemical results demonstrated the feasibility of the proximal labelling-assisted multiple signal amplification in the construction of an electrochemical aptasensor for the detection of tumor-associated protein, MUC-1.
Electrochemical detection of MUC-1-positive tumor cells
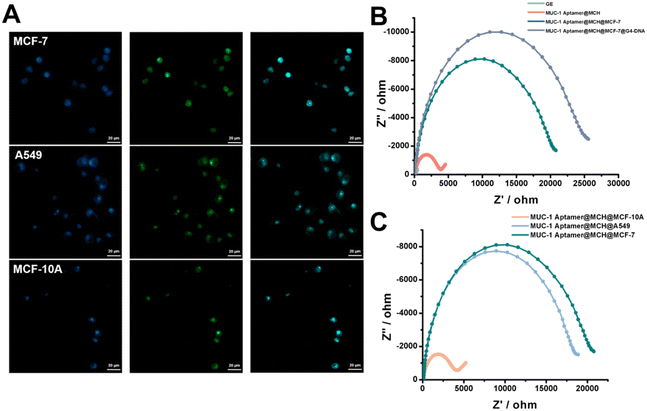
MUC-1 is overexpressed at the surface of the tumor cells and functions as a cell adhesion molecule, promoting tumor cell aggregation, invasion, and metastasis, therefore, MUC-1 is also recognized as an important surface biomarker to identify the tumor cells. To prove the feasibility of the electrochemical detection of MUC-1-positive tumor cells, three cell lines were chosen in our work, including a breast cancer cell line MCF-7, lung cancer cell line A549 and normal breast cell line MCF-10A. The mild reduction-mediated labelling of the cells was first studied by using FAM-modified G4-DNA strands (Fig. 3A). The cells were stained by using the fluorescent dye Hoechst, highlighting the nucleus within the cells, including MCF-7, A549, and MCF-10A cells. Meanwhile, intense green fluorescence was observed to surround the cells after the labelling of FAM-modified G4-DNA probes after the mild reduction. The overlapping images clearly showed the colocalization of both fluorescent signals, indicating the binding of G4-DNA onto the surface of these cells through mild reduction. Flow cytometry was also used to identify MUC-1 expression in these cells, confirming high expression of MUC-1 proteins in the tumor cells (such as MCF-7 and A549 cells) while much lower expression in the MCF-10A cells (Fig. S1†). MUC-1 aptamer-functionalized electrode was employed to capture MUC-1-positive tumor cells. Fig. 3B shows EIS results obtained at the electrode after the stepwise reactions for the capture of MUC-1-positive MCF-7 cells. The impedance values were found to increase with the modification of the electrode surface with MUC-1 aptamers, and the capture of MUC-1-positive MCF-7 in sequence, and a more obvious increase in impedance was observed following the binding of G4-DNA strands through mild reduction, indicating that the surface-captured cells were able to provide more binding sites for the linking of G4-DNA strands compared to the protein. Fig. 3C further shows EIS results obtained at the MUC-1 aptamer-functionalized electrode with the addition of different cells, such as MCF-7, A549 and MCF-10A cells. The impedance value was largely increased with the addition of both MCF-7 and A549 cells, but nearly no changes in the impedance were observed with the addition of MCF-10A cells. Therefore, the results were in line with the MUC-1 expression at different cells and demonstrated the feasibility of the MUC-1 aptamer-functionalized electrode in the identification and binding of MUC-1-positive tumor cells, such as MCF-7 and A549 cells.Proximal labelling-assisted multiple signal amplification was utilized to determine MUC-1-positive tumor cells by taking MCF-7 cells as an example. Fig. 4A shows the electrochemical responses obtained under different reaction conditions. A quite high electrochemical response was observed with MCF-7 cells after proximal labelling-assisted multiple signal amplification, while much lower electrochemical responses were observed at the MUC-1 aptamer-functionalized electrode, and after the incubation with the hemin/G4-DNA complex in the absence of MCF-7 cells, or with MCF-7 cells but without proximal labelling-assisted multiple signal amplification. After the capture of MUC-1-positive MCF-7 cells onto the electrode surface, the hemin/G4-DNA complex was linked to thiol groups exposed at the surface of MCF-7 cells through mild reduction. Accordingly, Ty–AuNPs were deposited onto the cell surface through proximal labelling of the neighbouring proteins, and led to the significantly enhanced electrochemical signal after silver accumulation. However, the electrochemical signal was prohibited when the target cells were absent or the multiple signal amplification procedures were inhibited.
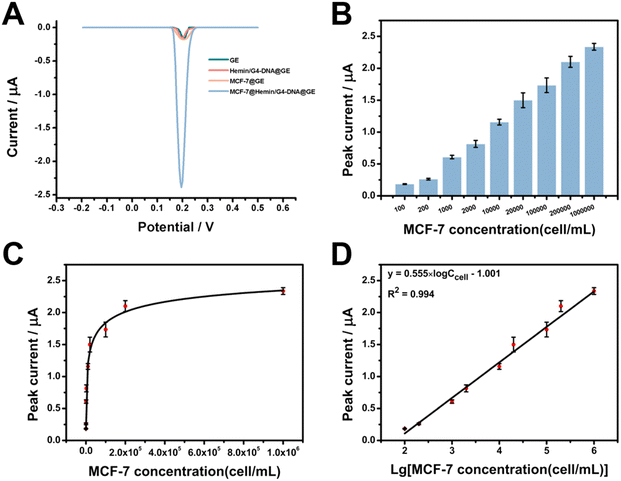
Having verified the feasibility of detecting MUC-1-positive tumor cells, we then optimized several experimental conditions to achieve the best detection performance. As shown in Fig. S2,† the electrochemical signal reached a state of equilibrium when the concentration of MUC1 aptamers used for electrode functionalization was 10 nM, as such the optimal concentration was chosen as 10 nM. As shown in Fig. S3,† the electrochemical signal reached a state of equilibrium when the reaction time for the capture of MCF-7 cells at MUC-1 aptamer-functionalized electrodes was 90 min, so the optimal time was chosen as 90 min. As shown in Fig. S4,† the electrochemical signal reached a state of equilibrium when the reaction time for the linking of G4-DNA strands was 45 min, so the optimal reaction time was chosen as 45 min. As shown in Fig. S5,† the electrochemical signal reached a state of equilibrium when the time for the deposition of Ty–AuNPs was 60 min and the optimal time was chosen as 60 min. Under the optimized condition, the peak currents were found to increase with the concentrations of MCF-7 cells (Fig. 4B). The enhanced concentration of MCF-7 cells provided an increased number of binding sites for the linking of G4-DNA probes and subsequently contributed to enhanced electrochemical signals after the multiple signal amplification procedures. Fig. 4C shows the relationship between the peak currents and the concentrations of MCF-7 cells in the range from 100 to 106 cells per mL, confirming the similar tendency in Fig. 4B. Fig. 4D further revealed a linear relationship between peak currents and the logarithmic value of the concentrations of MCF-7 cells in the range from 100 to 106 cells per mL. The regression equation was I = 0.555 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ccell − 1.001 μA, R2 = 0.994, and the detection of the limit was calculated to be 21 cell per mL at the ratio of signal-to-noise of 3. As shown in Table S2,† our work exhibited a wider or comparable linear range and a lower detection limit compared to the previous works, demonstrating an improved performance of our work toward cell analysis.37–41 In addition, the electrochemical detection at each concentration was repeated at least three times, and the average relative standard deviation was 4.93%, indicating a satisfactory reproducibility of our method. Therefore, the electrochemical results demonstrated the usability of proximal labelling-assisted multiple signal amplification in the electrochemical detection of MUC-1-positive MCF-7 cells with high sensitivity.
Ccell − 1.001 μA, R2 = 0.994, and the detection of the limit was calculated to be 21 cell per mL at the ratio of signal-to-noise of 3. As shown in Table S2,† our work exhibited a wider or comparable linear range and a lower detection limit compared to the previous works, demonstrating an improved performance of our work toward cell analysis.37–41 In addition, the electrochemical detection at each concentration was repeated at least three times, and the average relative standard deviation was 4.93%, indicating a satisfactory reproducibility of our method. Therefore, the electrochemical results demonstrated the usability of proximal labelling-assisted multiple signal amplification in the electrochemical detection of MUC-1-positive MCF-7 cells with high sensitivity.
The specificity of the electrochemical detection of MUC-1-positive tumor cells
The specificity of the electrochemical method was studied with the addition of different cells. As shown in Fig. 5A, the peak currents obtained from MUC-1-positive tumor cells, such as MCF-7 and A549 cells, were much higher than those from MCF-10A, which exhibited low expression of MUC-1 proteins. Therefore, the electrochemical results demonstrated that only MUC-1-positive tumor cells were captured by the MUC-1 aptamer-functionalized electrode through the selective interaction of the surface protein and the corresponding aptamers, and promoted the proximal labelling-assisted multiple signal amplification for the electrochemical detection of the target cells. Furthermore, we studied the potential application of the electrochemical methods in a complex environment using the diluted serum as an example. As shown in Fig. 5B, the peak currents obtained in the PBS were comparable to those obtained in the diluted serum samples with the addition of different concentrations of MCF-7 cells. The recoveries were calculated based on the regression equation, showing the recoveries from 91.2% to 95.5% (Table S3†). Therefore, the electrochemical results demonstrated the good anti-interference performance of the electrochemical method, which may avoid the unspecific adsorption onto the electrode surface even in a complex environment, suggesting the potential use of our method in clinical applications.Conclusions
In conclusion, we report an electrochemical method for the sensitive and specific detection of tumor cells based on the recognition and labelling of the surface protein, such as MUC-1. MUC-1 aptamer-functionalized electrode was constructed to ensure the specificity for the recognition and capture of MUC1-positive cancer cells, such as breast cancer cell MCF-7 and lung cancer cell A549. In the meanwhile, the multiple signal amplification consisted of the mild reduction-mediated labelling of G4-DNA strands, TSA-based proximal labelling of Ty–AuNPs and silver-enhanced electrochemical signal amplification, contributing to enhanced sensitivity for the electrochemical detection. Under the optimized conditions, the electrochemical method demonstrated the improved sensitivity for the detection of MUC-1-positive tumor cells (taking MCF-7 cells as the example) with a limit of the detection as low as 21 cell per mL, and excellent stability and anti-interference capability even in a complex serum environment. Therefore, our method may pave the way for the selective detection of tumor cells with high sensitivity, which may facilitate cell-based tissue examination and liquid biopsy. Furthermore, our methods could be easily extended to detect more types of tumor cells by changing the aptamers to identify different tumor-associated surface proteins in the future.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Guozhang Zhou: methodology, investigation, writing – original draft. Fei Zhou: methodology, investigation, writing – original draft. Xiaomeng Yu: validation, investigation. Daiyuan Zhou: investigation. Jiaqi Wang: investigation. Bing Bo: methodology, writing – review and editing. Ya Cao: methodology, writing – review and editing. Jing Zhao: conceptualization, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81871449, 81972799 and 82202834).Notes and references
- F. Bray, M. Laversanne, H. Sung, J. Ferlay, R. L. Siegel, I. Soerjomataram and A. Jemal, Ca-Cancer J. Clin., 2024, 74, 229–263 CrossRef PubMed.
- F. Vajhadin, S. Ahadian, J. Travas-Sejdic, J. Lee, M. Mazloum-Ardakani, J. Salvador, G. E. Aninwene, P. Bandaru, W. Sun and A. Khademhossieni, Biosens. Bioelectron., 2020, 151, 111984 CrossRef CAS PubMed.
- H. Safarpour, S. Dehghani, R. Nosrati, N. Zebardast, M. Alibolandi, A. Mokhtarzadeh and M. Ramezani, Biosens. Bioelectron., 2019, 148, 111833 CrossRef PubMed.
- A. K. Mattox, C. Bettegowda, S. Zhou, N. Papadopoulos, K. W. Kinzler and B. Vogelstein, Sci. Transl. Med., 2019, 11, 507 Search PubMed.
- D. Lin, L. Shen, M. Luo, K. Zhang, J. Li, Q. Yang, F. Zhu, D. Zhou, S. Zheng, Y. Chen and J. Zhou, Signal Transduction Targeted Ther., 2021, 6, 404 CrossRef CAS PubMed.
- N. Soda, K. Clack and M. J. A. Shiddiky, Sens. Diagn., 2022, 1, 343–375 RSC.
- M. Capuozzo, F. Ferrara, M. Santorsola, A. Zovi and A. Ottaiano, Cell, 2023, 12, 2590 CrossRef CAS PubMed.
- X. Yu, Y. Cao, Y. Zhao, J. Xia, J. Yang, Y. Xu and J. Zhao, Anal. Chem., 2023, 95, 15900–15907 CrossRef CAS PubMed.
- H. Liu, X. Yuan, T. Liu, W. Zhang, H. Dong and Z. Chu, Adv. Healthcare Mater., 2024, 13, 2304355 CrossRef CAS PubMed.
- W. Zhang, R. Wang, F. Luo, P. Wang and Z. Lin, Chin. Chem. Lett., 2020, 31, 589–600 CrossRef CAS.
- S.-S. Shi, X.-J. Li, R.-N. Ma, L. Shang, W. Zhang, H.-Q. Zhao, L.-P. Jia and H.-S. Wang, Lab Chip, 2024, 24, 367–374 RSC.
- Y.-L. Jia, X.-Q. Li, Z.-X. Wang, H. Gao, H.-Y. Chen and J.-J. Xu, Anal. Chem., 2024, 96, 7172–7178 CrossRef CAS PubMed.
- J. Liu, Z. Liu, C. Zhao, Y. Jiao, B. Li, J. Shi, Z. Chen and Z. Zhang, Nanoscale, 2024, 16, 8950–8959 RSC.
- T. Hu, X. Ke, Y. Ou and Y. Lin, Anal. Chem., 2022, 94, 8506–8513 CrossRef CAS PubMed.
- W. Xue, L. Wang, K. Yi, L. Sun, H. Ren and F. Bian, Biosens. Bioelectron., 2024, 255, 116270 CrossRef CAS PubMed.
- W. Chen, Z. Li, W. Cheng, T. Wu, J. Li, X. Li, L. Liu, H. Bai, S. Ding, X. Li and X. Yu, J. Nanobiotechnol., 2021, 19, 450 CrossRef CAS PubMed.
- P. Zuo, F. Gong, Y. Yang, X. Ji and Z. He, Chin. Chem. Lett., 2023, 34, 107259 CrossRef CAS.
- F. Chen, D. Wang, J. Chen, J. Ling, H. Yue, L. Gou and H. Tang, Sens. Actuators, B, 2020, 305, 127472 CrossRef CAS.
- C. Fu, L. Zhang, M. Bao, Y. Zhang, Y. Li, Y. Wu and Y. M. Jung, Analyst, 2022, 147, 5718–5724 RSC.
- Y. Zhang, H. Gu and H. Xu, Sens. Diagn., 2024, 3, 9–27 RSC.
- L. Li, B. Han, Y. Wang, J. Zhao and Y. Cao, Biosens. Bioelectron., 2019, 145, 111714 CrossRef CAS PubMed.
- B. Han, L. Dong, L. Li, L. Sha, Y. Cao and J. Zhao, Sens. Actuators, B, 2020, 325, 128762 CrossRef CAS.
- H. Kim, K. Shin, O. K. Park, D. Choi, H. D. Kim, S. Baik, S. H. Lee, S. H. Kwon, K. J. Yarema, J. Hong, T. Hyeon and N. S. Hwang, J. Am. Chem. Soc., 2018, 140, 1199–1202 CrossRef CAS PubMed.
- J. Xu, Y. Liu, K. J. Huang, Y. Y. Hou, X. Sun and J. Li, Anal. Chem., 2022, 94, 16980–16986 CrossRef CAS PubMed.
- J. Xu, Y. Liu, Y. Li, Y. Liu and K. J. Huang, Anal. Chem., 2023, 95, 13305–13312 CrossRef CAS PubMed.
- D. Tang, J. Shi, Y. Wu, H. Luo, J. Yan, K. J. Huang and X. Tan, Anal. Chem., 2023, 95, 16374–16382 CrossRef CAS PubMed.
- H. Shi, T. Zeng, Q. Liang, J. Yang, R. Chen, S. Wu, N. Duan, J. Zhao, G. Li and Y. Yin, Anal. Chem., 2024, 96, 3662–3671 CrossRef CAS PubMed.
- Y. Tang, Y. Dai, X. Huang, L. Li, B. Han, Y. Cao and J. Zhao, Anal. Chem., 2019, 91, 7531–7537 CrossRef CAS PubMed.
- Y. Cao, L. Zhou, G. Zhou, W. Liu, H. Cui, Y. Cao, X. Zuo and J. Zhao, Biosens. Bioelectron., 2024, 255, 116245 CrossRef CAS PubMed.
- Y. Zhao, Q. Liu, Y. Qin, Y. Cao, J. Zhao, K. Zhang and Y. Cao, ACS Appl. Mater. Interfaces, 2023, 15, 6411–6419 CrossRef CAS PubMed.
- N. Yamashita and D. Kufe, Int. J. Mol. Sci., 2022, 23, 8219 CrossRef CAS PubMed.
- Z. Li, D. Yang, T. Guo and M. Lin, Biomolecules, 2022, 12, 952 CrossRef CAS PubMed.
- E. Atta Manu, K. Bedu-Addo, N. A. Titiloye, C. Ameh-Mensah, F. Opoku and B. M. Duduyemi, J. Oncol., 2020, 2020, 9752952 CAS.
- X. Chen, I. K. Sandrine, M. Yang, J. Tu and X. Yuan, Front. Immunol., 2024, 15, 1356913 CrossRef CAS PubMed.
- X. Wei, Y. Lai, J. Li, L. Qin, Y. Xu, R. Zhao, B. Li, S. Lin, S. Wang, Q. Wu, Q. Liang, M. Peng, F. Yu, Y. Li, X. Zhang, Y. Wu, P. Liu, D. Pei, Y. Yao and P. Li, Onco Targets Ther, 2017, 6, e1284722 Search PubMed.
- Y. Chen, Z. Ye, M. Ma, J. Yang, R. Liu, Y. Zhang, P. Ma and D. Song, Biosens. Bioelectron., 2024, 254, 116241 CrossRef CAS PubMed.
- S. Zhang, Y. Huang, Y. Chen, S. Yan, H. Dai and J. Zhao, Sens. Diagn., 2023, 2, 140–146 RSC.
- C. Wang, X. P. Zhao, F. F. Liu, Y. Chen, X. H. Xia and J. Li, Nano Lett., 2020, 20, 1846–1854 CrossRef CAS PubMed.
- J. Lei, L. Shi, W. Liu, B. Li and Y. Jin, Analyst, 2022, 147, 3219–3224 RSC.
- B. Yuan, L. Guo, K. Yin, X. Wang, Q. Liu, M. He, K. Liu and J. Zhao, Analyst, 2020, 145, 2676–2681 RSC.
- A. Sanati, Y. Esmaeili, M. Khavani, E. Bidram, A. Rahimi, A. Dabiri, M. Rafienia, N. A. Jolfaie, M. R. K. Mofrad, S. H. Javanmard, L. Shariati and A. Zarrabi, Anal. Chim. Acta, 2023, 1252, 341017 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Flow cytometry assay of cell phenotypes; results of optimization experiments; sequences of DNA probes; comparison of currently available methods; detection results in diluted serum samples. See DOI: https://doi.org/10.1039/d4sd00217b |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |