Open Access Article
Open Access ArticleColorimetric nano-biosensor for low-resource settings: insulin as a model biomarker
Zia ul Quasim
Syed
,
Sathya
Samaraweera
,
Zhuo
Wang
and
Sadagopan
Krishnan
 *
*
Department of Chemistry, Oklahoma State University, Stillwater, Oklahoma 74078, USA. E-mail: gopan.krishnan@okstate.edu
First published on 23rd August 2024
Abstract
Biomarkers provide critical molecular insights into diseases and abnormal conditions. However, detecting them at ultra-low concentrations is a challenge, particularly in areas with limited resources and access to sophisticated instruments. Our research is primarily focused on mitigating this challenge. In this report, we introduce a colorimetric immunosensor for detecting insulin, an essential hormone biomarker that regulates glucose metabolism, at picomolar concentrations using citrate-functionalized magnetic particles. This immunosensor utilizes a two-antibody sandwich immunoassay: one antibody is covalently conjugated to the nanoparticles to capture and isolate the target marker, while the other is labeled with horseradish peroxidase for colorimetric detection of insulin. We conducted comparative analyses of insulin detection in buffer, saliva, and serum samples, offering valuable analytical insights. Our findings indicate a detection limit of 10 pM, with dynamic ranges of 10 pM to 1 nM, 10 pM to 10 nM, and 50 pM to 1 nM for insulin detection in buffer solution, 2-fold diluted serum, and 20-fold diluted artificial saliva, respectively. We demonstrate the application of the color immunosensor to type 1 diabetes and healthy human serum samples. For human saliva analysis, the detection limit needs to be improved in our future studies. Overall, our study enhances biomarker analysis in biofluids through an equipment-free colorimetric method, which is particularly relevant for point-of-need applications.
1. Introduction
Nanoparticle-based colorimetric sensors offer a straightforward, sensitive, and user-friendly approach to detection, requiring minimal sample preparation and no sophisticated equipment.1 A sensitive and selective enzymatic assay based on the peroxidase-like activity of magnetic greigite (Fe3S4) utilized sulfur vacancies to function as charge trapping and short-range scattering centers by introducing localized gap states. This helps the adsorption of peroxide on the Fe3S4 surface, facilitating glucose detection in serum with a linear range of 0.5–150 μM and a detection limit as low as 0.1 μM.2 António et al. reported a rapid assay for detecting C-reactive protein using citrate-capped gold nanoparticles.3 Platinum nanoparticles for the detection of cardiac troponin have also been reported.4 Other analytes such as cancer cells, proteins, and bacteria have also been detected utilizing magnetic, platinum–gold, and copper nanoparticles, they were able to detect as few as 2.17 × 102 colony forming unit per ml (CFU ml−1) of Listeria monocytogenes bacteria that cause foodborne illness, 10 cells per mL of cancer cells and 1.95 pg mL−1 of alpha-fetoprotein.5–7 Furthermore, a recent study has explored the use of graphene oxide/gold nanoparticles/Triton X-100 nanocomposites for insulin detection. While promising, this approach lacks selectivity.8 The inherent visual nature of colorimetric sensing allows for quick, convenient measurements, making it particularly suitable for point-of-care (POC) tests in remote settings.Magnetic particle-based colorimetric immunosensors have garnered increased attention during the pandemic era due to their significant advantages such as negligible interference with biological matrix,9 lower signal-to-noise ratio, highly stable physical properties, faster separation, and low cost.10 Utilizing magnetic particles (MPs) to immobilize biomolecules such as antibodies, proteins, enzymes, and nucleotides enables the simple, rapid, cost-effective, and efficient capture of target biomolecules from complex real sample matrices.11 These biosensing strategies are not only biocompatible and environmentally friendly but also cost-effective. Additionally, MP-based sensors offer reduced interference because biological samples typically exhibit little to no magnetic signals.12 The high surface-to-volume ratio, ease of dispersibility, and ability to conjugate with a variety of target-specific receptor molecules make MPs highly effective for sensitive detection in minimally processed samples.13–15 Magnetic particles have been versatile in developing assays to detect CA125 antigen,16 hepatitis B antigen,17 and various other analytes.
Compared to the millimolar concentrations of blood glucose, insulin, a hormone regulating glucose, is present at picomolar levels in fasting serum samples of both normal individuals and those with juvenile diabetes (type 1).18 In patients with insulin-resistant diabetes, known as type 2 diabetes, fasting insulin levels typically exceed 70 pM.19 While HbA1C tests are the most common clinical method for diagnosing diabetes, frequent monitoring of insulin is also crucial, particularly for individuals with malignant insulinoma, which can lead to frequent hypoglycemic episodes,20 in cases of pancreatic disorders or damage, helping diagnose impaired insulin production. Monitoring insulin levels is also necessary for understanding unexplained factitious hypoglycemia in non-diabetic individuals,21 assisting athletes with type 1 diabetes in achieving peak performance,22 and in the management of artificial pancreatic systems.
In immunoassays, using selective antibodies for target proteins ensures high specificity in detecting desired molecules,23 such as insulin, which serves as the model protein, hormone, or polypeptide in the present report. A sandwich-type electrochemical immunoassay has been reported for detecting serum insulin with a detection limit of 50 fM24 and in saliva samples with a detection limit of 41 pM.25 Liu et al. demonstrated a fluorescence quenching method for insulin detection with a detection limit of 0.048 U mL−1.26 While effective, this approach requires multiple DNA probes, increasing cost and complexity. G-Quadruplex systems and aptamer-based assays, known for their sensitivity due to multiple binding sites, can detect various analytes. Despite their potential, they pose design challenges.27 Chemiluminescence has been used with a detection limit of 1.6 pM but involves multiple steps.28 Interferometric reflectance spectroscopy offers label-free detection of biological molecules, yet it is costly.29 Chromatography-based techniques, while documented for insulin detection, often require lengthy preparation and running times, making them less ideal for immediate application needs. Given these limitations, especially in resource-constrained or remote settings, there is a critical need for new methods that are user-friendly, cost-effective, and independent of complex instrumentation to detect molecules at ultra-low concentrations.30
In this study, we compared the performance of a magnetic-particle-based colorimetric insulin sandwich immunosensor for detecting ultra-low picomolar concentrations of insulin (Scheme 1). Tests were conducted in a simple buffer solution and then expanded to 2-times diluted serum, 20-times diluted artificial saliva, and human salivary samples. Our goal was to achieve minimal dilution while maintaining sensor functionality, successfully enabled in a 2-fold diluted serum. However, greater dilution was necessary for saliva due to its frothy, viscous nature and to improve signal differentiation from the control saliva (no-insulin) samples at lower concentrations, as detailed in the Results and discussion section. The color intensity of the assay correlates with insulin concentration, providing a reliable, cost-effective means to monitor insulin levels, particularly beneficial for point-of-care applications in resource-limited settings.
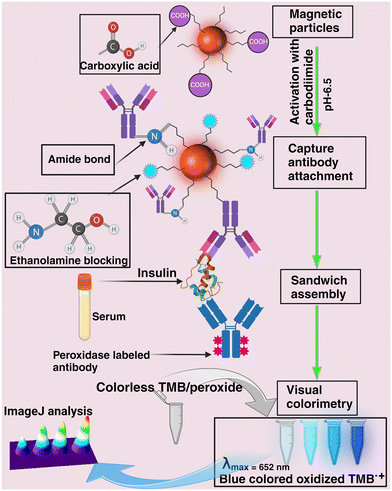 | ||
| Scheme 1 Conceptual schematic illustration of the colorimetric immunosensor developed in this report for insulin detection in biofluids. | ||
2. Experimental section
2.1. Materials
Fluid MAG-CT (200 nm) citrate-functionalized magnetic particles (MPs) were acquired from Chemicell GmbH (Berlin, Germany). We purchased N-hydroxysuccinimide (NHS), polyethylene glycol sorbitan monolaurate (TWEEN® 20), 2-aminoethanol (ethanolamine), disodium hydrogen phosphate (Na2HPO4), 2-(N-morpholino) ethane sulfonic acid hydrate (MES hydrate), a monoclonal anti-insulin antibody produced in mouse, insulin antigen, and human serum from Sigma-Aldrich (St. Louis, MO). Enhanced K-Blue® TMB (3,3′,5,5′-tetramethylbenzidine) substrate was obtained from Neogen® (Lexington, KY). Sodium chloride (NaCl), potassium chloride (KCl), and sodium hydroxide (NaOH) were sourced from Millipore Sigma (Burlington, MA). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and potassium dihydrogen phosphate (KH2PO4) were purchased from Thermo Fisher Scientific (Waltham, MA). DNase-free water was procured from Fisher Bioreagents. The insulin rabbit anti-human polyclonal (HRP) detection antibody was acquired from LS Bio (Shirley, MA). Artificial saliva (sodium carboxy methyl cellulose, potassium phosphate monobasic, potassium chloride, potassium phosphate dibasic, magnesium chloride hexahydrate, calcium chloride dehydrates, methyl-p-hydroxybenzoate) for medical and dental research was purchased from Pickering Laboratories (Mountain View, CA). SpotReady-16 gold spotted glass microarray chips (1 mm × 18 mm × 18 mm) were obtained from Platypus technologies, LLC (Madison, WI). Finally, a magnetic rack (6.73′′ × 2.24′′ × 2.13′′) for 100–250 microliter Eppendorf tubes (16 tubes) was obtained from Sergi Lab Supplies (Seattle, WA).2.2. Methods
UV-vis spectrophotometer was Varian Cary 100 Bio, Varian Inc., CA. The hydrodynamic size and zeta potential of the particles were measured using a ZetaPALS potential analyzer (Brookhaven Instruments Corporation, Holtsville, NY, USA), providing insight into particle stability and interaction. Measurements were conducted at a 90° angle, with the sample diluted 5-fold in 10 mM PBS (pH 7.4, 25 °C). To validate the color sensor results with increasing insulin concentration, we used the SPRimager-II array instrument (Horizon model, GWC Technologies, Madison, WI, USA), which operates at room temperature and uses a fixed SPRi source wavelength of 800 nm. Reflectivity changes at an optimally selected angle indicating surface modification or molecular binding events were recorded. A charge-coupled device (CCD) camera captured these changes as variations in pixel intensity. All experiments were performed in accordance with the Guidelines “Occupational Safety and Health Standards, OSHA Bloodborne Pathogen Standards, U. S. Department of Labor” and approved by the ethics committee at Oklahoma State University. Informed consent was obtained from human participants in this study.2.3. Colorimetric sandwich immunoassay
In this study, magnetic particles (MPs) with a hydrodynamic size of 200 nm were selected due to their rapid magnetic separation in biofluids, a result of enhanced magnetophoretic mobility.31 The choice of 200 nm particles is due to their relatively lower surface area compared to particles smaller than 100 nm. This reduced surface area diminishes the contact area for the intrinsic peroxidase activity of the MPs, consequently lowering the background color in control samples that are not spiked with insulin. Each Eppendorf tube contained 12.5 μL of a 25 mg mL−1 citrate carboxylated MP (–COOH, sodium salt) suspension (0.31 mg, ∼7 × 1010 magnetic particles). To activate the –COOH groups, we added 50 μL of a freshly prepared solution containing 0.4 M EDC and 0.1 M NHS in MES buffer (pH 6.5) and incubated for 15 minutes at room temperature, converting them into reactive N-succinimidyl ester groups. We used a strong neodymium magnetic rack for efficient separation of the –COOH-activated MPs from the bulk unreacted EDC/NHS solution and washed them twice with 10 mM phosphate-buffered saline (PBS). These groups are known to form stable amide bonds with the amine groups on the lysine residues of the anti-insulin monoclonal antibody.18 To accomplish this antibody conjugation, we incubated the MPs with the anti-insulin monoclonal antibody (50 μg mL−1, 50 μL) for 30 minutes under ice-cold conditions (in an ice-box) to support the solution antibody protein stability while conjugating to the particles. After the immobilization, we magnetically separated the antibody-attached MPs and washed them three times with 10 mM phosphate-buffered saline (PBS) to remove unbound antibodies with intermittent magnetic separation at each wash. To block any remaining active chemical sites on the –COOH-activated MPs and thus reduce non-specific binding in subsequent assay steps, we added 50 μL of 2.5 M ethanolamine to the MP-antibody conjugate and incubated it for 15 minutes. Finally, we performed a magnetic separation and washed the ethanolamine-blocked conjugates once with 200 μL of 10 mM PBS.Various concentrations of insulin solutions were prepared in 10 mM PBS buffer, 2-times diluted insulin-free healthy human serum, or 20-times diluted commercial artificial saliva. These solutions were incubated with the ethanolamine-blocked, insulin-antibody-modified MPs for 30 minutes, followed by washing with PBS buffer and intermittent magnetic separation to remove any unbound substances. Next, 50 μL of a 1 μg mL−1 horseradish peroxidase-labeled detection insulin antibody solution in PBS (pH 7.4) was added. This mixture was incubated for another 30 minutes in the ice-cold box with occasional stirring to allow binding to the insulin molecules on the monoclonal antibody MPs, forming a sandwich immuno-assembly. After washing the immunocomplex three times with PBS buffer, we added 20 μL of enhanced K-blue TMB substrate solution (containing 3,3′,5,5′-tetramethylbenzidine, and hydrogen peroxide) and incubated for 25–30 seconds to allow color development. The total assay time is ∼2 h. The blue color intensity, indicative of the bound insulin concentration, was generated from the peroxidase-labeled detection antibody-catalyzed TMB enzymatic reaction. Finally, we magnetically separated the insulin immuno-assembly MPs and measured the absorbance at 652 nm using a UV-vis spectrophotometer to quantify the insulin levels. For visual documentation, photos of the sample tubes were captured using an iPhone 12 camera. Control samples included buffer, two-times diluted serum, and 20-times diluted artificial saliva.
2.4. ImageJ analysis
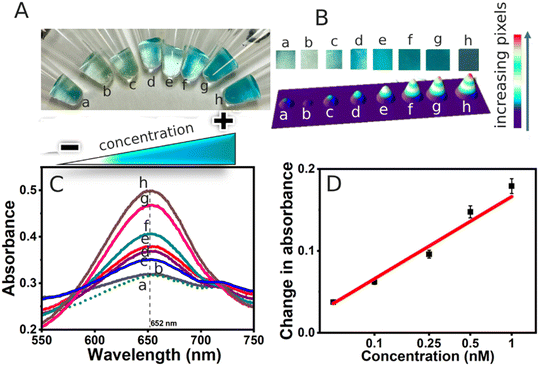
A smartphone camera captured images of the colorimetric change occurring in the sample Eppendorf tubes at the end of the assay. Then, the images corresponding to each insulin concentration were cropped into similar-sized snips. The pixel intensity increase with insulin concentration is represented using the ImageJ software, along with the 3D representation to easily visualize the color intensity changes that correlate with the UV-vis absorbance increase of the TMB oxidation catalyzed by the HRP-labeled detection antibody bound to the magnetic particle-capture antibody-insulin-immunoassembly.3. Results and discussion
3.1. Hydrodynamic size
Magnetic particles and magnetic particle-bound insulin-antibody conjugates were characterized by measuring their size distribution in a liquid environment using dynamic light scattering (DLS), accounting for Brownian motion and hydrodynamic interactions.32 The measured hydrodynamic size of the as-received, unconjugated MPs from Chemicell Inc. was 202 ± 20 nm, which agrees with the manufacturer's specified average size of 200 nm. Following the immobilization of the capture monoclonal anti-insulin antibody onto the MPs, the hydrodynamic size increased to 372 ± 45 nm (N = 3 replicates). This increase is from the antibody immobilization and associated intermolecular and/or interparticle aggregation when attached to the nanoparticle surfaces, as noted in prior studies.33,34 The size distribution was quantified using the polydispersity index (PDI), a dimensionless value ranging from 0 to 1, where values less than 0.1 indicate well-dispersed particles.35 The citrate–carboxylic acid functionalized MPs exhibited a PDI of 0.186, which decreased to 0.009 after the capture insulin-antibody was attached. This indicates a narrower size distribution of the aggregated antibody-MP conjugate compared to the original particles.3.2. Zeta (ζ) potential
Initially, the carboxyl-functionalized magnetic particles exhibited a negative ζ potential of −36.47 ± 0.13 mV, attributed to the citrate (–COOH) surface groups. After the covalent immobilization of activated –COOH groups with the amine groups of lysine residues on the capture antibody to form stable amide bonds, the ζ potential shifted to a less negative value of −15.27 ± 0.19 mV (N = 5 replicates). This shift towards a less negative zeta potential is indicative of the neutralization of the surface negative charge of the MPs due to antibody attachment and subsequent ethanolamine blocking. The remaining net negative charge is likely due to the negative charges on the antibody at pH 7.4, given its isoelectric point (pI) of 6.0–6.5,36 as well as from any residual free citrate groups of the MPs.3.3. Assay in buffer (10 mM PBS, pH 7.4)
The assay results indicate a clear correlation between increasing insulin concentrations (ranging from 10 pM to 10 nM) in the buffer and an increase in color intensity, as shown in Fig. 1(A–D). Visually, color intensities for concentrations from 100 pM to 10 nM were notably distinct compared to the lower 10 pM concentrations, which closely resembled the background color of the buffer, a result of the intrinsic peroxidase activity of MPs. A UV-vis spectral analysis was performed to validate the visual observations. The maximum absorbance at 652 nm corresponds to the formation of the oxidized TMB compound facilitated by the enzymatic activity of the HRP-labeled detection antibody in the sandwich structure. The chromogenic activity of TMB is a result of its two easily oxidizable amino groups of benzidine, which are enhanced due to its sensitivity to HRP.37 In the presence of the horseradish peroxidase/H2O2 system, TMB forms a blue one-electron oxidation product, perpetuating a redox cycle that facilitates the color development characteristic of this assay.38 The sensor demonstrated a robust linear, logarithmic response across the range of 10 pM to 1 nM.3.4. Detection in two times diluted serum
The serum is a complex matrix containing approximately 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different proteins, spanning a wide range of concentrations and molecular weights, which can significantly influence the analytical response of sensors.39 Detecting insulin in serum is particularly challenging due to its low molecular weight (∼6 kDa). Our sandwich assay method addresses this issue by employing two specific antibodies to enhance selectivity. Additionally, to minimize background interference, we diluted the serum 2-fold with 10 mM PBS. In our serum analysis, there was a discernible increase in color intensity corresponding to the rise in spiked insulin concentration from 10 pM to 10 nM (Fig. 2A for visual color changes and Fig. 2B for 3D representation), as compared to the background color of the serum (no-insulin), which is attributable to the intrinsic peroxidase activity of the MPs (Fig. 2C, curve a). Gao et al. have shown that the peroxidase-like activity of MPs increases with increasing temperature, lowering pH (more acidic), and high molar hydrogen peroxide (H2O2) concentrations.40 MPs alone require a high peroxide concentration of 530 mM and a reaction time of over 100 seconds38 to exhibit significant peroxidase activity. To counteract this non-enzymatic background activity, our assay was conducted under ice-cold conditions using a 100 mM peroxide concentration at a physiological pH of 7.4. These measures effectively mitigated the intrinsic peroxidase activity of MPs. Our optimized conditions achieved a linear detection range of 10 pM to 10 nM in 2 times diluted serum (Fig. 2C, curves b–f). While previous studies have utilized 10 to 100-fold serum dilution to enhance sensor response by diminishing nonspecific signals from the complex matrix,8 such extensive dilution can decrease the analyte protein concentration in actual samples, potentially compromising the sensor's ability to detect and overall assay performance. Application of the designed colorimetric sensor serum calibration plot (Fig. 2D) to a type 1 diabetes patient and healthy human serum samples and correlation of the results with an enzyme-linked immunosorbent assay (ELISA) run by an independent laboratory located on our campus (The University Health Services) inferred good agreement at a 95% confidence level (Fig. 2E). The percentage recovery of the ELISA kit was reported as 91%, and the correlation with the present colorimetric sensor infers a similar level of accuracy between the two methods.
000 different proteins, spanning a wide range of concentrations and molecular weights, which can significantly influence the analytical response of sensors.39 Detecting insulin in serum is particularly challenging due to its low molecular weight (∼6 kDa). Our sandwich assay method addresses this issue by employing two specific antibodies to enhance selectivity. Additionally, to minimize background interference, we diluted the serum 2-fold with 10 mM PBS. In our serum analysis, there was a discernible increase in color intensity corresponding to the rise in spiked insulin concentration from 10 pM to 10 nM (Fig. 2A for visual color changes and Fig. 2B for 3D representation), as compared to the background color of the serum (no-insulin), which is attributable to the intrinsic peroxidase activity of the MPs (Fig. 2C, curve a). Gao et al. have shown that the peroxidase-like activity of MPs increases with increasing temperature, lowering pH (more acidic), and high molar hydrogen peroxide (H2O2) concentrations.40 MPs alone require a high peroxide concentration of 530 mM and a reaction time of over 100 seconds38 to exhibit significant peroxidase activity. To counteract this non-enzymatic background activity, our assay was conducted under ice-cold conditions using a 100 mM peroxide concentration at a physiological pH of 7.4. These measures effectively mitigated the intrinsic peroxidase activity of MPs. Our optimized conditions achieved a linear detection range of 10 pM to 10 nM in 2 times diluted serum (Fig. 2C, curves b–f). While previous studies have utilized 10 to 100-fold serum dilution to enhance sensor response by diminishing nonspecific signals from the complex matrix,8 such extensive dilution can decrease the analyte protein concentration in actual samples, potentially compromising the sensor's ability to detect and overall assay performance. Application of the designed colorimetric sensor serum calibration plot (Fig. 2D) to a type 1 diabetes patient and healthy human serum samples and correlation of the results with an enzyme-linked immunosorbent assay (ELISA) run by an independent laboratory located on our campus (The University Health Services) inferred good agreement at a 95% confidence level (Fig. 2E). The percentage recovery of the ELISA kit was reported as 91%, and the correlation with the present colorimetric sensor infers a similar level of accuracy between the two methods.
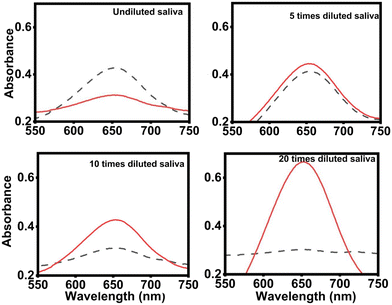
3.5. Detection in 20-times diluted artificial saliva
Fabre et al. reported a 92% confidence interval for the correlation between salivary and serum insulin levels, highlighting saliva as a valuable biofluid for non-invasive detection.41 However, detecting insulin in the saliva is challenging due to its approximately 10-fold lower concentration compared to serum and the complex nature of saliva's composition, which can introduce biological noise and affect the assay's signal response.38,39 To identify the optimal dilution that minimizes background noise and maximizes signal response to spiked insulin, we conducted a series of tests using different dilutions of commercially available artificial saliva (Pickering Laboratories), each spiked with 1 nM insulin.In Fig. 3, the highest signal response relative to the control was noted for the 20-times diluted artificial saliva matrix. The undiluted saliva's background color was more pronounced at lower dilutions than that of the spiked sample response, indicating significant interference. The absorbance changes in 5-times diluted artificial saliva were minimal. Consequently, we chose a 20-times dilution for the saliva matrix to reduce the impact of interfering agents effectively. This approach, however, also means a corresponding dilution of the analyte, impacting the detection of insulin in real saliva samples, which is our future objective to solve.
In Fig. 4 (A for visual color changes and B for 3D representation), we observed a linear response in concentrations ranging from 50 pM to 1 nM (Fig. 4C and D). The intensity for the sample spiked with 10 pM was nearly indistinguishable from the 20-times diluted saliva samples (no spiked insulin). Visually, the intensities at 10 pM and 1 pM appeared lower or comparable to the 20-times diluted saliva samples with no spiked insulin. UV-visible spectral analysis further confirmed that the absorbance at 10 pM closely resembled that of the insulin-unspiked saliva, and the absorbance at 1 pM was equivalent to that of the artificial saliva.
Detecting insulin in human saliva is complex due to its constituents like water, salts, mucus, enzymes, and proteins. Additionally, the viscosity of saliva complicates sample homogenization, leading to frothing even after diligent mixing. In our study, we spiked 20 times diluted, non-fasting, healthy human saliva samples with three different concentrations of insulin. Saliva samples from fasting diabetic patients and non-fasting healthy individuals were collected in 5 mL glass vials and refrigerated at 2–8 °C. Prior to experimentation, samples were taken out of refrigeration, diluted 20-times in PBS, and spiked with insulin. In Fig. 5 (A – type-1 diabetic fasting saliva and B for the 20-times diluted human healthy saliva, and Fig. 5C and D are the respective 3D representations), we noted a significant background signal in the human saliva samples (Fig. 5E and F) compared to the less complex artificial saliva made of salts (Fig. 4C). We achieved detection at 1 nM and 10 nM concentrations above the insulin unspiked saliva control for both the real human saliva and artificial salivary matrix (Fig. 5G).
Optimizing sensor performance and sensitivity typically involves refining various components, such as the type of material and bio-recognizing elements. However, one of the most critical yet often overlooked factors is the background color of the sample matrix. This can substantially affect the sensor's analytical parameters and its ability to detect the analyte effectively. For instance, in our study, the comparative detection limits shown in Table 1 indicate better performance for insulin spiked in a buffer. This is likely due to fewer obstructions for insulin binding to the capture antibody in the buffer, which lacks the complex biological components and viscosity present in serum and saliva that can amplify background signals. Additionally, our sensor design relies on direct modification of magnetic particles, avoiding complex co-materials or hybrid materials that might otherwise increase the background peroxidase-like color from TMB.
| Matrix | LOD 3 × σ/S42 | Linear range (log scale) |
|---|---|---|
| 10 mM PBS | 10 pM | 10 pM–1 nM |
| 2-times diluted serum | 10 pM | 10 pM–10 nM |
| 20-times diluted artificial saliva | 10 pM | 50 pM–1 nM |
3.6. Validation using surface plasmon resonance imaging (SPRi) analysis
To validate the colorimetric results, we employed surface plasmon resonance imaging (SPRi), a label-free, real-time optical biosensing technique.43 This method is particularly adept at monitoring molecular interactions on a metallic sensor surface populated with oscillating electrons known as plasmons.44,45The experimental setup included a 5 nm thin gold layer on a glass surface, with incident light interacting with surface-bound molecules via an evanescent wave. SPR gold chips were placed with the colorimetric insulin sensor solutions, and reflectivity changes were measured against a buffer background.42 As the concentration of spiked insulin increased in the immunoassay solution, a corresponding increase in pixel intensity was noted. Data acquisition from the SPRi difference images was conducted using the Digital Optics V++ software, and the 3D representation of the data was generated using ImageJ software (Fig. 6). These results agreed with the findings obtained from UV-vis spectrometry.
We have compared our sensor with other colorimetric sensors in Table 2. One such sensor uses a G-quadruplex insulin assembly coupled with DNA nanotubes and nanoparticles for picomolar detection of insulin. It's important to note the complexity and time intensity of this method, which requires around 5 hours to complete, with multiple stages including an 80-minute incubation in a thermocycler, a 60-minute ligation period, and additional steps totaling 2 hours and 20 minutes. Additionally, the assay demands specialized techniques like electrophoresis for nanotube separation and gel extraction columns for isolating MNP-coupled DNA nanotubes. Consequently, the process is laborious and requires the skills of trained professionals.46
| Approach | Sample matrix | Limit of detection | Liner range | Ref. |
|---|---|---|---|---|
| 28.7 μIU = 1 ng mL−1 = 172 pM.47 1 μIU = 6 pM. | ||||
| Colorimetric detection of insulin in human serum using graphene oxide/gold nanoparticles/Triton X-100 (GO/AuNPs/TX-100) nanocomposite | Serum (10-fold) | 17.2 pM (0.1 ng mL−1) | 344 pM–52 nM (2–300 ng mL−1) | 8 |
| Colorimetric glucose assay based on magnetic particles having pseudo-peroxidase activity and immobilized glucose oxidase | Plasma | 0.13 mM | 1–24 mM (18–432 mg dL−1) | 48 |
| Colorimetric detection of hydrogen peroxide and glucose using the magnetic mesoporous silica nanoparticles | PBS buffer | 4.0 μM | 10–100 μM | 49 |
| Facile preparation of peroxidase-like core–shell nanorods and application as a platform for colorimetric determination of glucose, insulin, and glucose/insulin ratio | Serum (100-fold) | — | 0.84–6.47 pM (0.014–1.08 μIU mL−1) | 50 |
| A sensitive colorimetric assay using insulin G-quadruplex aptamer arrays on DNA nanotubes coupled with magnetic nanoparticles | Serum (2-fold) | 2.33 pM (0.39 μIU mL−1) | 9.4–600 pM (1.56–100 μIU mL−1) | 46 |
| Colorimetric nano-biosensor for low-resource settings: insulin as a model biomarker | PBS buffer serum (2-fold) | 10 pM | 10 pM–1 nM | This work |
| Artificial saliva (20-fold) | 10 pM | 10 pM–10 nM | ||
| 10 pM | 50 pM–1 nM | |||
Colorimetric sensors developed on peroxidase activity of magnetic particles by Martinkova et al. has good selectivity and stability, however, the detection limit is affected by the interference of the plasma matrix.48 Wang et al. developed a sensor based on core–shell material and demonstrated the detection of glucose detection with good reproducibility, however the limitation of this method is that study was conducted in PBS buffer.49 Huang et al. developed a more straightforward ratiometric assay capable of differentiating between type 1 and type 2 diabetes mellitus, which is also cost-effective. Their method uses core–shell nanorods based on GNR@Au2S/AuAgS/CuS exhibiting peroxidase-like activity for colorimetric signal generation. This assay involves glucose oxidation by glucose oxidase (GOx), producing hydrogen peroxide as a byproduct, which then oxidizes TMB, leading to a visible color change. However, it should be noted that this method requires a 100-fold dilution of serum samples.50 Additionally, a simpler assay using graphene oxide/gold nanoparticle nanocomposites modified with Triton X-100 was able to detect insulin levels consistently over five days, with intra-day and inter-day sample variations of 2.7% and 4.8%, respectively. This approach, which utilizes straightforward spectrometric analysis, is notable for its simplicity, although the long-term stability of the sensor was not evaluated.8 Despite this, such methods represent rapid and sensitive platforms, employing innovative strategies like self-assembling nanocomposites for insulin detection in human serum.
We recognize that the inherent peroxidase activity of the magnetic particles presents a limitation in our sensor, leading to background interference. However, we have demonstrated that this activity can be mitigated. By limiting the detection step reaction to a brief 30 seconds, we ensure that the HRP-driven oxidation of TMB by hydrogen peroxide occurs more rapidly than the non-enzymatic peroxidase-like activity of the MPs, which originates from the iron centers. Additionally, conducting the detection reaction at 4 °C and maintaining a low hydrogen peroxide concentration further minimizes the non-enzymatic catalytic activity of the MPs, thereby reducing the background signal attributable to non-enzymatic TMB oxidation.40
Table 3 presents a comparative account of the developed colorimetric sensor with a representative set of commercial ELISA kits. The commercial colorimetric ELISA kits use different labeling methods, such as HRP, alkaline phosphatase (ALP), and high-affinity HRP–streptavidin–biotin, costing between $450 to $730 per kit. The Mercodia ELISA kit employs an HRP-labeled sandwich antibody platform similar to ours, but its detection limit and linear range are not as good. However, it offers high specificity and minimal cross-reactivity with other proteins in the matrix. Enzo Lifesciences kit also utilizes a sandwich antibody approach with a biotinylated sandwich antibody, which is conjugated to the streptavidin–HRP label for colorimetric detection. It requires a similar sample dilution as ours for the serum and plasma samples, but again, our assay outperforms it in terms of detection limit and assay time. Abcam's kit, though pricier, offers a comparable detection limit and dilution factor for serum samples, but the longer assay time of 5 h may be a drawback. Novus Biologicals and ALPCO kits have excellent detection limits and similar assay time as ours, with good specificity due to the use of two monoclonal antibodies. However, the required sample dilution is not well defined in the manual, and it is up to the users to determine the optimal sample dilution. Our assay is comparable to commercially available kits in terms of assay time, dilution factor, and linear ranges. Compared to the commercial methods, our approach offers advantages such as cost-effectiveness that can be useful in resource-limited settings.
| Approach/name | Sample matrix | Instrument | Limit of detection | Liner range | Time of assay | Ref./company |
|---|---|---|---|---|---|---|
| Mercodia rat insulin ELISA (HRP-label) | Serum, EDTA plasma, and cell culture medium | Required (OD) at 450 nm | 20 nM | 0.02 μM−1.0 μM | 2 h 30 min | Mercodia |
| Immunoreactive insulin in diabetes mellitus patient sera detected by ultrasensitive ELISA with thio–NAD cycling (ALP-label) | 10-fold diluted serum with Tris-buffered saline (TBS) | 2.28 pM | 0–48 pM | Overnight | 51 | |
| Insulin ELISA kit catalog #: ENZ-KIT141-0001 (biotin–streptavidin–HRP label) | Serum, plasma, and tissue culture media | Required (OD) at 450 nm | 15.5 nM | 15.6–500 nM | 3 h | Enzo Lifesciences |
| Human insulin ELISA kit catalog: NBP2-60077 (HRP-label) | Serum, plasma, and tissue culture media | Required (OD) at 450 nm | 12 pM | 60–359 pM | ∼2 h | Novus Biologicals |
| Insulin ELISA catalog number: 80-INSHU-E01.1, E10.1 (label not specified) | Plasma, serum | Required (OD) at 450 nm | 2.4 pM | 18–1198 pM | ∼2 h | ALPCO |
| Human insulin ELISA kit catalog: ab100578 (biotin–streptavidin–HRP label) | Serum, plasma, and cell culture supernatant | Required (OD) at 450 nm | 24 pM | 28–1798 pM | 5 h | Abcam |
| Colorimetric nano-biosensor for low-resource settings: insulin as a model biomarker (sandwich-HRP-label based) | PBS buffer serum (2-fold) | OD at 652 nm and visual detection | 10 pM | 10 pM–1 nM | ∼2 h | This work |
| Artificial saliva (20-fold) | 10 pM | 10 pM–10 nM | ||||
| 10 pM | 50 pM–1 nM |
3.7. Functional stability of the sensor
The functional stability of the sensor is a critical factor in determining its longevity, reliability, and repeatability over time. Stability refers to the sensor's ability to maintain consistent performance and provide accurate readings over extended periods. It encompasses resistance to physical and chemical changes, such as degradation of sensor components, loss of bioactivity in biological elements, or drift in signal response due to environmental factors. Ensuring functional stability means that the sensor can be reliably used in various settings, maintaining accuracy and precision without frequent recalibration or replacement. This is especially important in clinical settings and remote sensing, where consistent and dependable readings are essential for patient diagnosis and continuous monitoring applications.The antibodies are covalently bound to the MPs via stable amide bonds formed with free amine groups on lysine residues. Despite the stability of these bonds, proteins can denature when exposed to water or temperature fluctuations.52 To evaluate the durability of the sensor, we prepared 32 individual vials of the capture antibody-MP conjugates and stored them at 2–8 °C. Over a period of 8 days, new vials were taken from storage for each assay to assess their insulin detection response. Although the conjugates settled at the bottom of the Eppendorf tubes, they were readily resuspended with gentle agitation. We spiked each assay with 1 nM insulin in 10 mM PBS and noted that the control buffer sample (no-insulin) consistently showed a lower response than the spiked samples throughout the 8 days (Fig. 7). The average absorbance response of the spiked samples was consistently higher than that of the buffer samples containing the MP-capture antibody conjugate (no spiked insulin), indicating maintained functional stability over the tested duration in buffer solutions. Evaluating the sensor stability in spiked serum and salivary solutions forms a future objective of our research.
4. Conclusions
The ability to quickly and easily detect biomolecules using a visual sensor is highly advantageous in resource-limited and remote settings. Our developed colorimetric sensor effectively measured spiked insulin across various matrices, including buffer, serum, and saliva. It demonstrated good functional stability and reproducibility, as evidenced by consistent performance over an 8-day storage period and in replicate measurements. The sensor can be applied for insulin measurements in real serum samples and can differentiate between type I and healthy human samples. Independent validation using absorbance spectroscopy and surface plasmon resonance imaging confirmed the reliability of the sensor results. This validation underscores the potential use of the sensor as a robust and reliable tool for insulin detection and indicates its broader applicability for various molecular targets in real samples.Data availability
The article presents the direct absorbance spectra from the instrument presented using Origin Software. Visual color pictures were captured on an iPhone and presented with ImageJ for color intensity representations.All acquired raw data, including the direct absorbance spectra and visual color pictures, are available for reference at the authors' laboratory at Oklahoma State University.
Author contributions
Sadagopan Krishnan: writing – review & editing, writing – supervision, resources, project administration, funding acquisition, formal analysis, and conceptualization. Zia Syed: original draft, visualization, validation, software, methodology, investigation, formal analysis, data curation. Sathya Samaraweera: original draft, visualization, validation, software, methodology, investigation, formal analysis, data curation. Zhuo Wang: visualization, validation, software, methodology, investigation, formal analysis, data curation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The research results discussed in this publication were made possible in total or in part by funding through the award for project number HR21-013 from the Oklahoma Center for the Advancement of Science and Technology. We thank Dr. Todd Green, M.D., and Ms. Allison Harris from the Oklahoma State University Health Services for their generous support in performing the independent ELISA analysis of type 1 diabetes and healthy serum samples.References
- V. S. A. Piriya, P. Joseph, S. C. G. K. Daniel, S. Lakshmanan, T. Kinoshita and S. Muthusamy, Colorimetric Sensors for Rapid Detection of Various Analytes, Mater. Sci. Eng., C, 2017, 78, 1231–1245, DOI:10.1016/j.msec.2017.05.018.
- W. Liu, J. Tian, C. Mao, Z. Wang, J. Liu, R. A. Dahlgren, L. Zhang and X. Wang, Sulfur Vacancy Promoted Peroxidase-like Activity of Magnetic Greigite (Fe3S4) for Colorimetric Detection of Serum Glucose, Anal. Chim. Acta, 2020, 1127, 246–255, DOI:10.1016/j.aca.2020.06.056.
- M. António, R. Ferreira, R. Vitorino and A. L. Daniel-da-Silva, A Simple Aptamer-Based Colorimetric Assay for Rapid Detection of C-Reactive Protein Using Gold Nanoparticles, Talanta, 2020, 214, 120868, DOI:10.1016/j.talanta.2020.120868.
- S. Chen, Z. Yu, Y. Wang, J. Tang, Y. Zeng, X. Liu and D. Tang, Block-Polymer-Restricted Sub-Nanometer Pt Nanoclusters Nanozyme-Enhanced Immunoassay for Monitoring of Cardiac Troponin I, Anal. Chem., 2023, 95(38), 14494–14501, DOI:10.1021/acs.analchem.3c03249.
- S. Alhogail, G. A. R. Y. Suaifan and M. Zourob, Rapid Colorimetric Sensing Platform for the Detection of Listeria Monocytogenes Foodborne Pathogen, Biosens. Bioelectron., 2016, 86, 1061–1066, DOI:10.1016/j.bios.2016.07.043.
- K. Wang, D. Fan, Y. Liu and E. Wang, Highly Sensitive and Specific Colorimetric Detection of Cancer Cells via Dual-Aptamer Target Binding Strategy, Biosens. Bioelectron., 2015, 73, 1–6, DOI:10.1016/j.bios.2015.05.044.
- C. Chen, Y. Liu, Z. Zheng, G. Zhou, X. Ji, H. Wang and Z. He, A New Colorimetric Platform for Ultrasensitive Detection of Protein and Cancer Cells Based on the Assembly of Nucleic Acids and Proteins, Anal. Chim. Acta, 2015, 880, 1–7, DOI:10.1016/j.aca.2015.05.010.
- M. Mirsalari and S. Elhami, Colorimetric Detection of Insulin in Human Serum Using GO/AuNPs/TX-100 Nanocomposite, Spectrochim. Acta, Part A, 2020, 240, 118617, DOI:10.1016/j.saa.2020.118617.
- Z. Altintas, S. S. Kallempudi, U. Sezerman and Y. Gurbuz, A Novel Magnetic Particle-Modified Electrochemical Sensor for Immunosensor Applications, Sens. Actuators, B, 2012, 174, 187–194, DOI:10.1016/j.snb.2012.08.052.
- R. Oropesa-Nuñez, T. Z. G. de la Torre, H. Stopfel, P. Svedlindh, M. Strömberg and K. Gunnarsson, Insights into the Formation of DNA–Magnetic Nanoparticle Hybrid Structures: Correlations between Morphological Characterization and Output from Magnetic Biosensor Measurements, ACS Sens., 2020, 5(11), 3510–3519, DOI:10.1021/acssensors.0c01623.
- C. Tang, Z. He, H. Liu, Y. Xu, H. Huang, G. Yang, Z. Xiao, S. Li, H. Liu, Y. Deng, Z. Chen, H. Chen and N. He, Application of Magnetic Nanoparticles in Nucleic Acid Detection, J. Nanobiotechnol., 2020, 18(1), 62, DOI:10.1186/s12951-020-00613-6.
- X. Zhang, D. B. Reeves, I. M. Perreard, W. C. Kett, K. E. Griswold, B. Gimi and J. B. Weaver, Molecular Sensing with Magnetic Nanoparticles Using Magnetic Spectroscopy of Nanoparticle Brownian Motion, Biosens. Bioelectron., 2013, 50, 441–446, DOI:10.1016/j.bios.2013.06.049.
- H. Shao, C. Min, D. Issadore, M. Liong, T.-J. Yoon, R. Weissleder and H. Lee, Magnetic Nanoparticles and MicroNMR for Diagnostic Applications, Theranostics, 2012, 2(1), 55–65, DOI:10.7150/thno.3465.
- M. V. Yigit, A. Moore and Z. Medarova, Magnetic Nanoparticles for Cancer Diagnosis and Therapy, Pharm. Res., 2012, 29(5), 1180–1188, DOI:10.1007/s11095-012-0679-7.
- P. Farinha, J. M. P. Coelho, C. P. Reis and M. M. Gaspar, A Comprehensive Updated Review on Magnetic Nanoparticles in Diagnostics, Nanomaterials, 2021, 11(12), 3432, DOI:10.3390/nano11123432.
- D. Tang, B. Su, J. Tang, J. Ren and G. Chen, Nanoparticle-Based Sandwich Electrochemical Immunoassay for Carbohydrate Antigen 125 with Signal Enhancement Using Enzyme-Coated Nanometer-Sized Enzyme-Doped Silica Beads, Anal. Chem., 2010, 82(4), 1527–1534, DOI:10.1021/ac902768f.
- Y. Hu, R. Chen, M. Chen, J. An, M. Luo, Y. Lyu, N. Hu, W. Guo, W. Li and Y. Liu, Magnetic Separation and Enzymatic Catalysis Conjugated Colorimetric Immunosensor for Hepatitis B Surface Antigen Detection, Microchem. J., 2021, 168, 106155, DOI:10.1016/j.microc.2021.106155.
- E. Vargas, E. Povedano, S. Krishnan, H. Teymourian, F. Tehrani, S. Campuzano, E. Dassau and J. Wang, Simultaneous Cortisol/Insulin Microchip Detection Using Dual Enzyme Tagging, Biosens. Bioelectron., 2020, 167, 112512, DOI:10.1016/j.bios.2020.112512.
- K. Lian, H. Feng, S. Liu, K. Wang, Q. Liu, L. Deng, G. Wang, Y. Chen and G. Liu, Insulin Quantification towards Early Diagnosis of Prediabetes/Diabetes, Biosens. Bioelectron., 2022, 203, 114029, DOI:10.1016/j.bios.2022.114029.
- T. Okabayashi, Diagnosis and Management of Insulinoma, World J. Gastroenterol., 2013, 19(6), 829, DOI:10.3748/wjg.v19.i6.829.
- E. Leighton, C. A. Sainsbury and G. C. Jones, A Practical Review of C-Peptide Testing in Diabetes, Diabetes Ther., 2017, 8(3), 475–487, DOI:10.1007/s13300-017-0265-4.
- M. C. Riddell, S. N. Scott, P. A. Fournier, S. R. Colberg, I. W. Gallen, O. Moser, C. Stettler, J. E. Yardley, D. P. Zaharieva, P. Adolfsson and R. M. Bracken, The Competitive Athlete with Type 1 Diabetes, Diabetologia, 2020, 63(8), 1475–1490, DOI:10.1007/s00125-020-05183-8.
- I. Kikkas, R. Mallone, N. Tubiana-Rufi, D. Chevenne, J. C. Carel, C. Créminon, H. Volland, C. Boitard and N. Morel, A Simple and Fast Non-Radioactive Bridging Immunoassay for Insulin Autoantibodies, PLoS One, 2013, 8(7), e69021, DOI:10.1371/journal.pone.0069021.
- H. Sun, S. Wu, X. Zhou, M. Zhao, H. Wu, R. Luo and S. Ding, Electrochemical Sandwich Immunoassay for Insulin Detection Based on the Use of Gold Nanoparticle-Modified MoS2 Nanosheets and the Hybridization Chain Reaction, Microchim. Acta, 2019, 186(1), 6, DOI:10.1007/s00604-018-3124-8.
- E. Vargas, H. Teymourian, F. Tehrani, E. Eksin, E. Sánchez-Tirado, P. Warren, A. Erdem, E. Dassau and J. Wang, Enzymatic/Immunoassay Dual-Biomarker Sensing Chip: Towards Decentralized Insulin/Glucose Detection, Angew. Chem., Int. Ed., 2019, 58(19), 6376–6379, DOI:10.1002/anie.201902664.
- C. Liu, J. Han, J. Zhang and J. Du, Novel Detection Platform for Insulin Based on Dual-Cycle Signal Amplification by Exonuclease III, Talanta, 2019, 199, 596–602, DOI:10.1016/j.talanta.2019.03.013.
- Q. Cao, Y. Li, E. Freisinger, P. Z. Qin, R. K. O. Sigel and Z.-W. Mao, G-Quadruplex DNA Targeted Metal Complexes Acting as Potential Anticancer Drugs, Inorg. Chem. Front., 2017, 4(1), 10–32, 10.1039/C6QI00300A.
- Y. Sun, Y. Lin, W. Sun, R. Han, C. Luo, X. Wang and Q. Wei, A Highly Selective and Sensitive Detection of Insulin with Chemiluminescence Biosensor Based on Aptamer and Oligonucleotide-AuNPs Functionalized Nanosilica @ Graphene Oxide Aerogel, Anal. Chim. Acta, 2019, 1089, 152–164, DOI:10.1016/j.aca.2019.09.004.
- R. Chhasatia, M. J. Sweetman, B. Prieto-Simon and N. H. Voelcker, Performance Optimisation of Porous Silicon Rugate Filter Biosensor for the Detection of Insulin, Sens. Actuators, B, 2018, 273, 1313–1322, DOI:10.1016/j.snb.2018.07.021.
- Y. Shen, W. Prinyawiwatkul and Z. Xu, Insulin: A Review of Analytical Methods, Analyst, 2019, 144(14), 4139–4148, 10.1039/C9AN00112C.
- K. Witte, K. Müller, C. Grüttner, F. Westphal and C. Johansson, Particle Size- and Concentration-Dependent Separation of Magnetic Nanoparticles, J. Magn. Magn. Mater., 2017, 427, 320–324, DOI:10.1016/j.jmmm.2016.11.006.
- J. Lim, S. P. Yeap, H. X. Che and S. C. Low, Characterization of Magnetic Nanoparticle by Dynamic Light Scattering, Nanoscale Res. Lett., 2013, 8(1), 381, DOI:10.1186/1556-276X-8-381.
- J. E. Smith, K. E. Sapsford, W. Tan and F. S. Ligler, Optimization of Antibody-Conjugated Magnetic Nanoparticles for Target Preconcentration and Immunoassays, Anal. Biochem., 2011, 410(1), 124–132, DOI:10.1016/j.ab.2010.11.005.
- R. Khan, E. Ahmad, M. Zaman, A. Qadeer and G. Rabbani, Nanoparticles in Relation to Peptide and Protein Aggregation, Int. J. Nanomed., 2014, 899, DOI:10.2147/IJN.S54171.
- M. Danaei, M. Dehghankhold, S. Ataei, F. Hasanzadeh Davarani, R. Javanmard, A. Dokhani, S. Khorasani and M. Mozafari, Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems, Pharmaceutics, 2018, 10(2), 57, DOI:10.3390/pharmaceutics10020057.
- A. Söderberg, B. Sahaf, A. Holmgren and A. Rosén, Monoclonal Antibodies to Human Thioredoxin Reductase, Biochem. Biophys. Res. Commun., 1998, 249(1), 86–89, DOI:10.1006/bbrc.1998.9053.
- X. Zhang, Q. Yang, Y. Lang, X. Jiang and P. Wu, Rationale of 3,3′,5,5′-Tetramethylbenzidine as the Chromogenic Substrate in Colorimetric Analysis, Anal. Chem., 2020, 92(18), 12400–12406, DOI:10.1021/acs.analchem.0c02149.
- P. D. Josephy, T. Eling and R. P. Mason, The Horseradish Peroxidase-Catalyzed Oxidation of 3,5,3′,5′-Tetramethylbenzidine. Free Radical and Charge-Transfer Complex Intermediates, J. Biol. Chem., 1982, 257(7), 3669–3675, DOI:10.1016/S0021-9258(18)34832-4.
- M. De, S. Rana, H. Akpinar, O. R. Miranda, R. R. Arvizo, U. H. F. Bunz and V. M. Rotello, Sensing of Proteins in Human Serum Using Conjugates of Nanoparticles and Green Fluorescent Protein, Nat. Chem., 2009, 1(6), 461–465, DOI:10.1038/nchem.334.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Intrinsic Peroxidase-like Activity of Ferromagnetic Nanoparticles, Nat. Nanotechnol., 2007, 2(9), 577–583, DOI:10.1038/nnano.2007.260.
- B. Fabre, G. Maccallini, A. Oneto, D. Gonzalez, V. Hirschler, C. Aranda and G. Berg, Measurement of Fasting Salivary Insulin and Its Relationship with Serum Insulin in Children, Endocr. Connect., 2012, 1(2), 58–61, DOI:10.1530/EC-12-0024.
- A. Shrivastava and V. Gupta, Methods for the Determination of Limit of Detection and Limit of Quantitation of the Analytical Methods, Chron. Young Sci., 2011, 2(1), 21, DOI:10.4103/2229-5186.79345.
- Z. Wang, Z. Syed, Z. H. Al Mubarak, L. Lehoczky, C. Rodenbaugh, R. A. Bunce and S. Krishnan, Integrating Colorimetry with Surface Sensitive Transducers: Advancing Molecular Diagnostics in Biofluids, Sens. Actuators, B, 2024, 419, 136361, DOI:10.1016/j.snb.2024.136361.
- Z. H. Al Mubarak, G. Premaratne, A. Dharmaratne, F. Mohammadparast, M. Andiappan and S. Krishnan, Plasmonic Nucleotide Hybridization Chip for Attomolar Detection: Localized Gold and Tagged Core/Shell Nanomaterials, Lab Chip, 2020, 20(4), 717–721, 10.1039/C9LC01150A.
- G. Premaratne, A. C. Dharmaratne, Z. H. Al Mubarak, F. Mohammadparast, M. Andiappan and S. Krishnan, Multiplexed Surface Plasmon Imaging of Serum Biomolecules: Fe3O4@Au Core/Shell Nanoparticles with Plasmonic Simulation Insights, Sens. Actuators, B, 2019, 299, 126956, DOI:10.1016/j.snb.2019.126956.
- A. Rafati, A. Zarrabi, S. Abediankenari, M. Aarabi and P. Gill, Sensitive Colorimetric Assay Using Insulin G-Quadruplex Aptamer Arrays on DNA Nanotubes Coupled with Magnetic Nanoparticles, R. Soc. Open Sci., 2018, 5(3), 171835, DOI:10.1098/rsos.171835.
- K. Lian, H. Feng, S. Liu, K. Wang, Q. Liu, L. Deng, G. Wang, Y. Chen and G. Liu, Insulin Quantification towards Early Diagnosis of Prediabetes/Diabetes, Biosens. Bioelectron., 2022, 203, 114029, DOI:10.1016/j.bios.2022.114029.
- P. Martinkova, R. Opatrilova, P. Kruzliak, I. Styriak and M. Pohanka, Colorimetric Glucose Assay Based on Magnetic Particles Having Pseudo-Peroxidase Activity and Immobilized Glucose Oxidase, Mol. Biotechnol., 2016, 58(5), 373–380, DOI:10.1007/s12033-016-9936-z.
- Y. Wang, B. Zhou, S. Wu, K. Wang and X. He, Colorimetric Detection of Hydrogen Peroxide and Glucose Using the Magnetic Mesoporous Silica Nanoparticles, Talanta, 2015, 134, 712–717, DOI:10.1016/j.talanta.2014.12.013.
- F. Tan, Z. Wang, Y. Yang, X. Xie, X. Hua, X. Yang and H. Huang, Facile Preparation of Peroxidase-like Core-Shell Nanorods and Application as Platform for Colorimetric Determination of Glucose, Insulin and Glucose/Insulin Ratio, Talanta, 2019, 204, 285–293, DOI:10.1016/j.talanta.2019.06.005.
- E. Ito, M. Kaneda, H. Kodama, M. Morikawa, M. Tai, K. Aoki, S. Watabe, K. Nakaishi, S. Hashida, S. Tada, N. Kuroda, H. Imachi, K. Murao, M. Yamashita, T. Yoshimura and T. Miura, Immunoreactive Insulin in Diabetes Mellitus Patient Sera Detected by Ultrasensitive ELISA with Thio-NAD Cycling, BioTechniques, 2015, 59(6), 359–367, DOI:10.2144/000114355.
- D. Porter and F. Vollrath, The Role of Kinetics of Water and Amide Bonding in Protein Stability, Soft Matter, 2008, 4(2), 328–336, 10.1039/B713972A.
| This journal is © The Royal Society of Chemistry 2024 |