Open Access Article
Open Access ArticleDetection of surfactants using a hydrophobic natural deep eutectic solvent and smartphone†
Vagner Bezerra
dos Santos
*ab,
Lucas B.
Ayres
 b,
Helayne Santos
de Sousa
a,
Carlos D.
Garcia
b,
Helayne Santos
de Sousa
a,
Carlos D.
Garcia
 *b and
Willian
Toito Suarez
*b and
Willian
Toito Suarez
 c
c
aLIA3 (Laboratório de Instrumentação e Automação em Analítica Aplicada), Universidade Federal de Pernambuco, Recife, PE, Brazil. E-mail: vagner.bsantos@ufpe.br
bDepartment of Chemistry, Clemson University, 211 S. Palmetto Blvd, Clemson, SC 29634, USA. E-mail: cdgarci@clemson.edu
cDepartamento de Química, Universidade Federal de Viçosa, Viçosa, MG, Brazil
First published on 24th July 2024
Abstract
We report on the advantages of a green method to detect surfactants in environmental water samples. The approach is based on the use of a hydrophobic natural deep eutectic solvent (NADES) to extract the complexes formed by the surfactants and methylene blue. The concentration of the surfactant is then determined by measuring the color intensity in the organic phase using a smartphone. Under optimized conditions, an aliquot of 3 mL of the NADES was mixed with 15 mL of water, and then allowed to settle (to enable the separation of the two phases) for 5 min. The procedure allowed quantification of sodium dodecyl sulfate (SDS), as a proxy for alkyl surfactants in the range from 0.010 mg L−1 to 0.600 mg L−1, with a detection limit of 2.0 μg L−1. Besides being a simple alternative to the traditional method (which requires chloroform and a spectrophotometer), the proposed approach offers low waste generation, low power-consumption, and fast analysis time, and is fully compatible with the plastic supplies (e.g. cuvettes, pipettes, tips, etc.) typically used for on-site analysis. The applicability of the approach was demonstrated by measuring various surface water samples and the overall green score of the method was calculated to be 96%.
1. Introduction
Surfactants are a widespread class of chemical compounds1 with pivotal technological applications2 and have widespread use across various products such as pesticides,3 gasoline,4 detergents,5 shampoos,6 cosmetics,7 foods,8 and some pharmaceuticals.9 Given their extensive use, surfactants are commonly present in wastewater and natural aquatic environments, raising significant environmental concerns,10 as these compounds can disrupt aquatic life11 and contribute to broader ecological imbalances.10,12 Indeed, the presence of anionic surfactants at concentrations higher than 2 mg L−1 in water can increase the biochemical oxygen demand and ultimately affect the survival of animals and plants.13 Some studies have reported that surfactant concentrations in the 5–15 mg L−1 range can cause the death of around 50% of larvae fish, fish, and crustaceans.14,15 According to Hammer et al.15 not only the concentration but also the structure of the surfactant is critical, as there is an increase in ecotoxicity by a factor of 4.5 for each additional hydrocarbon present in the alkyl chain of the surfactant – an aspect that is correlated with the surfactant's affinity for cellular membranes and its ability to inhibit enzymes.16 As expected, high concentrations of surfactants in drinking water can also negatively impact human health,17 highlighting the importance of developing robust analytical methods to detect them.Several methods have been reported for the quantification of surfactants in environmental waters, including those based on high-performance liquid chromatography and capillary electrophoresis coupled with UV-vis detection12,18 and gas chromatography coupled with mass spectrometry.18 In addition to fluoresecence,19 UV-vis spectrophotometry has been used due to its low cost and simplicity.18,20–22 However, this method (often referred to as the methylene blue active substance assay, MBAS21–23) typically requires a large volume of chloroform (up to 100 mL per sample) during the extraction step, representing a serious risk due to its high toxicity.20,21 Among other alternatives, the use of deep eutectic solvents (DESs)24–26 seems to be one of the most convenient avenues to remove chloroform.16 DESs are formed by a mixture of two or three solid compounds that often lead to a stable liquid at room temperature27,28 and that present a wide range of physico-chemical properties (density, viscosity, polarity, etc.). As a subclass of DESs, natural deep eutectic solvents (NADESs) are formed with components of natural origin and represent one of the most attractive options to replace traditional organic solvents due to their low vapor pressure and low toxicity.29,30
Aiming to take advantage of the extraordinary properties of NADESs towards the extraction of analytes,31 this report describes the development and application of a hydrophobic NADES for the extraction of surfactants in water. Using a recently developed algorithm,32 several NADESs were developed and assessed to develop a methodology to quantify surfactants from environmental waters, a process that was then coupled with in situ detection using digital imaging. To the best of our knowledge, this is the first report describing this combination. Besides meeting sensitivity requirements to comply with current regulations, this new method also meets several aspects of green chemistry33 and provides the greenest alternative to the traditional methodology.20,31,34
2. Materials and methods
2.1. Reagents and solutions
All reagents used for the experiments were of analytical grade and used as received. All the solutions were prepared using deionized water (>18.0 MΩ cm) from a Millipore Milli-Q system (USA). Hydrated methylene blue was acquired from MP Biomedics (USA), sulfuric acid from BDH (USA), and chloroform from Thermo-Fisher Scientific (USA). Dodecanoic acid, menthol, eucalyptol, borneol, and polyethylene glycol were purchased from Sigma Aldrich (USA). Other compounds used include sodium chloride (Bayer, USA), nickel chloride (Mallinckrodt, USA), copper chloride (Sigma Aldrich, USA), calcium carbonate (Grenrel Storage, USA), magnesium sulfate (Mallinckrodt, USA), aluminum sulfate (Fisher Science, USA), bismuth nitrate (Alfa Aesar, USA), iron(III) chloride (Alfa Aesar, USA), iron(II) sulfate (Acros Organics, USA), potassium phosphate (Sigma-Aldrich, USA), potassium bromide (Alfa Aesar, USA), sodium dihydrogen phosphate monohydrate (Fisher Scientific, USA), sodium octyl sulfate (Sigma, USA), sodium dodecyl sulfate (Sigma-Aldrich, USA), sodium dodecylbenzene sulfonate (Sigma-Aldrich, USA), sodium deoxycholate (Sigma-Aldrich, USA) and cetyltrimethylammonium bromide (Sigma-Aldrich, USA). Methylene blue solutions (0.5 g L−1) were prepared by dissolving 0.0500 g of hydrated methylene blue in the presence of 2.8300 g of sodium dihydrogen phosphate monohydrate and 680 μL of sulfuric acid in deionized water, resulting in a 100 mL solution.21,22 Stock solutions of sodium dodecyl sulfate (13 mg L−1, used as the model surfactant representative of the LAS class)21 were prepared in ultrapure water. Solutions for the MBAS test were prepared according to the reference method described by the American Public Health Association (APHA),22 with the described modifications implemented to avoid the use of chloroform.20–22 In addition, the proposed methodology allowed for reducing the volume of reagents and solutions by 1/1000, as only 100 μL were used per analysis.22 It is also important to note that particular care was taken to avoid the use of surfactants in all glassware/utensils, which were thoroughly rinsed with DI water prior to their use.2.2. Environmental water samples
Surface water samples (2 L, collected at a depth of less than 1 m) were collected at different locations in Clemson, SC using brand-new polypropylene bottles. Following a previously reported process,21 the bottles were thoroughly rinsed, labeled, and filled by immersion, and then kept at 4 °C until use. Before the analysis, the samples were filtered (0.7 μm) to separate any particulate matter suspended in the water. A map with the sample collection sites is provided in Fig. SI 1.†2.3. UV-vis spectrophotometry method
The reference MBAS method was used to detect anionic surfactants in water samples. This method is based on the complexation of the surfactants using methylene blue, followed by extraction with chloroform, acid rinsing (3×) to remove interferences, and detection using UV-vis spectrophotometry at 652 nm.20,22,35 In order to provide a reasonable comparison, the spectrophotometric analysis was performed by using 3 mL of either the NADES or chloroform.2.4. Generation and synthesis of hydrophobic NADESs
NADESs are formed by a combination of hydrogen bond donors (HBDs) and hydrogen bond acceptors (HBAs), leading to stable and viscous liquids at room temperature.36 A recently-described approach37 was used to predict the formation of a group of NADESs with hydrophobic characteristics,32 greatly streamlining the selection process.38 Briefly, a Python script was first used to generate random mixtures of HBDs and HBAs that varied in terms of the stoichiometric ratio (1 to 5), number of components (1 to 5), and chemical structure (n = 198). Then, the probability of formation (p) for each mixture was predicted.37 In this analysis, we focused our attention on mixtures displaying a p > 0.85, which were more likely to lead to a liquid solvent, stable at 25 °C for at least two weeks. Since we were interested in obtaining hydrophobic NADESs, only the most stable mixtures, involving components displaying log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P higher than 0.1, were considered for the experimental validation. This step was critical as it ensured that the generated NADESs would display the appropriate polarity to interact with the methylene blue–surfactant complex, enabling the extraction from the aqueous phase. Also, while some of the NADESs generated by the algorithm could have been also obtained by surveying the literature,17,31,38–42 our approach allowed a systematic and more rational selection of the candidates. Thus, selected binary mixtures (with high probability of formation and hydrophobic characteristics) were prepared by mixing (5 min) and heating (85 °C) until a clear liquid was formed. More information and the composition of the selected NADESs are included in section 3.1 and Table 1 (vide infra).
P higher than 0.1, were considered for the experimental validation. This step was critical as it ensured that the generated NADESs would display the appropriate polarity to interact with the methylene blue–surfactant complex, enabling the extraction from the aqueous phase. Also, while some of the NADESs generated by the algorithm could have been also obtained by surveying the literature,17,31,38–42 our approach allowed a systematic and more rational selection of the candidates. Thus, selected binary mixtures (with high probability of formation and hydrophobic characteristics) were prepared by mixing (5 min) and heating (85 °C) until a clear liquid was formed. More information and the composition of the selected NADESs are included in section 3.1 and Table 1 (vide infra).
| NADES | Component (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) P) |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (average & standard deviation) P (average & standard deviation) |
Notes |
|---|---|---|---|
| #1 | Dodecanoic acid/propylene glycol | 1.6 ± 3.6 | Not stable in water |
| #2 | Menthol/eucalyptol | 2.7 ± 0.3 | Extraction yield: 53% |
| #3 | Menthol/borneol | 2.8 ± 0.2 | High viscosity,43 turbid phase formed when mixed with water samples |
| #4 | Menthol/dodecanoic acid | 3.6 ± 0.8 | Extraction yield: 84% |
2.5. Digital image-based (DIB) method
Aiming to develop a green and point-of-need method to measure surfactants in environmental samples, a smartphone was utilized. The utility of this approach has been previously demonstrated for the detection of biogenic amines,44 copper,45 furfural,46 peroxides,47 and LAS.21 The main advantages of the DIB method include simplicity, analysis time, cost, power consumption, and portability.21,48–50 Here, the DIB method was used to measure the color intensity of a 100 μL sample dispensed on a polystyrene 92-well plate. To control the illumination and thus increase the accuracy of the reading,45,49 an ad hoc chamber (13 cm × 6 cm × 10 cm) was built using black acrylic. Besides including a small hole to place the smartphone's camera (12MP, Samsung Galaxy S22), the box also integrated a white LED (powered by a 5 V battery)51 wrapped in Teflon tape. This strategy enabled not only controlling the light intensity but also providing a more homogeneous distribution of the light inside the chamber.21,23,49 Analysis of the images was performed using ImageJ52 and the free application ColorGrab. In all cases, values reported correspond to the average and standard deviation of the color intensity (R channel) obtained for at least 3 images of 500 × 500 pixels. Similar approaches have been reported in the literature, stating that other color channels (G & B) should not be used because they are not correlated with the complementary color of the ion-pair formed.16,21,23 As a summary of the overall approach, Fig. 1 schematically describes the proposed flowchart. | ||
| Fig. 1 Schematic representation of the method used to detect LAS in environmental water samples using the NADES and digital image method. | ||
2.6. Analytical figures of merit
The proposed methodology was used to develop the corresponding calibration curves and to calculate the linear range, limit of detection (LOD = 3σ/slope, n = 6), and limit of quantification (LOQ = 10σ/slope, n = 6). The repeatability of the method was assessed by measuring the color development of a standard (0.3 mg L−1, n = 4) during three consecutive days and calculating the relative standard deviation (RSD).21,45,46 The recovery was evaluated by comparing the response before and after spiking the environmental water samples at two different levels (0.16 mg L−1 and 0.35 mg L−1).21,45,46 The response of potential interferences was determined using common ions present in environmental water at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 ratio (surfactant
100 ratio (surfactant![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) interference) and at a surfactant concentration of 0.3 mg L−1. The relative error was used to estimate the percentage of interference. The accuracy of the DIB method was calculated by analyzing the same samples by UV-vis spectrophotometry and comparing the results using the F-test and the T-test at a 95% confidence level (n = 3).
interference) and at a surfactant concentration of 0.3 mg L−1. The relative error was used to estimate the percentage of interference. The accuracy of the DIB method was calculated by analyzing the same samples by UV-vis spectrophotometry and comparing the results using the F-test and the T-test at a 95% confidence level (n = 3).
3. Results and discussion
The following paragraphs describe the experiments performed to design and select the hydrophobic NADES, determine the efficiency of the extraction using the NADES as a function of the sample volume and extraction time, calculate the analytical figures of merit, and demonstrate the applicability of the method for the analysis of real samples. Considering its widespread use,13,19 SDS was selected as a model, anionic surfactant.3.1. Synthesis of the hydrophobic NADES
As described in section 2.6, we applied a previously described approach32 to generate a group of hydrophobic NADESs. Briefly, the process started by creating random combinations (n = 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) of HBDs and HBAs. Considering that the selected NADES should not only be safer than chloroform (log
000) of HBDs and HBAs. Considering that the selected NADES should not only be safer than chloroform (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 2.3) but also offer a competitive performance, only mixtures featuring non-toxic compounds and hydrophobic components (log
P = 2.3) but also offer a competitive performance, only mixtures featuring non-toxic compounds and hydrophobic components (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 0.1)36 were considered. Then, the probability of formation of these mixtures was assessed by our transformer-based model, streamlining the subsequent experimental efforts. As can be observed in Fig. 2A, and in agreement with previous results,32 a decreasing distribution in the probability of formation was obtained. In other words, most of the mixtures generated by random combinations are not likely to form a stable NADES, leading only to a few (n = 40) mixtures with a probability of formation higher than 0.85. Out of those, the most frequent components leading to these mixtures include borneol (log
P > 0.1)36 were considered. Then, the probability of formation of these mixtures was assessed by our transformer-based model, streamlining the subsequent experimental efforts. As can be observed in Fig. 2A, and in agreement with previous results,32 a decreasing distribution in the probability of formation was obtained. In other words, most of the mixtures generated by random combinations are not likely to form a stable NADES, leading only to a few (n = 40) mixtures with a probability of formation higher than 0.85. Out of those, the most frequent components leading to these mixtures include borneol (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 2.7), diethylethanolamine (log
P = 2.7), diethylethanolamine (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 0.3), camphor (log
P = 0.3), camphor (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 2.2), hexanediol (log
P = 2.2), hexanediol (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.4), propylene glycol (log
P = 1.4), propylene glycol (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = −0.9), menthol (log
P = −0.9), menthol (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 3.0), octanoic acid (log
P = 3.0), octanoic acid (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 3.0), and dodecanoic acid (log
P = 3.0), and dodecanoic acid (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 4.2), as shown in Fig. 2B.
P = 4.2), as shown in Fig. 2B.
Out of those, the binary mixtures included in Table 1 were selected and synthesized in the laboratory by mild heating (80 °C/5 min) under constant stirring. It is important to note that while all of those NADESs were stable (remained as transparent and liquid mixtures for at least a week at room temperature), only NADESs #2–#4 were hydrophobic enough to be used in the extraction process. The limited stability of NADES #1 in water could be attributed to the large disparity in the polarity of the components, which in turn, allows water to disrupt the hydrogen bond network of the NADES.36,53 As a side note, it is worth mentioning that unlike CHCl3 (1.49 g mL−1),22 NADES #4 features a density (0.91 g mL−1)38 that is lower than that of water, thus simplifying its removal from the extraction mixture.
3.2. Effect of NADES composition on the extraction of the methylene blue–surfactant complex
In its traditional format, the methylene blue active substance (MBAS) test is based on the formation of an ion-pair complex by the cationic methylene blue (pKa = 3.8, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 2.2) and the anionic surfactant (SDS for example, log
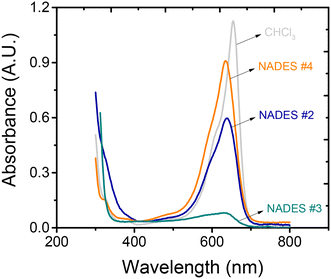
P = 2.2) and the anionic surfactant (SDS for example, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.6), which is then extracted from the aqueous solution using chloroform. In the last step, the concentration of the complex is measured by UV-visible spectrophotometry (see representative spectrum in Fig. 3). In the absence of anionic surfactants, the amount of methylene blue transferred to the non-polar organic phase is negligible.38 Considering the toxicological aspects of CHCl3,54 the next goal of the project was to assess the performance of the proposed NADESs (#2–#4) as alternative extraction solvents. Following previous reports,41 the efficiency of the extraction process using the proposed NADES was compared to that of CHCl3, which was used as a reference (100%).
P = 1.6), which is then extracted from the aqueous solution using chloroform. In the last step, the concentration of the complex is measured by UV-visible spectrophotometry (see representative spectrum in Fig. 3). In the absence of anionic surfactants, the amount of methylene blue transferred to the non-polar organic phase is negligible.38 Considering the toxicological aspects of CHCl3,54 the next goal of the project was to assess the performance of the proposed NADESs (#2–#4) as alternative extraction solvents. Following previous reports,41 the efficiency of the extraction process using the proposed NADES was compared to that of CHCl3, which was used as a reference (100%).
As can be observed in Fig. 3, only NADESs #2 and #4 allowed extraction of the methylene blue–SDS complex from the aqueous phase, although with significantly different yields. The lowest yield was obtained with NADES #2 (53%), followed by NADES #4 (84%). These results were attributed to a combination of the hydrophobicity of these solvents (see average log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of their components in Table 1), the structure of the components forming the NADES, and the specific stoichiometric ratios (1
P values of their components in Table 1), the structure of the components forming the NADES, and the specific stoichiometric ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) selected for these experiments.43 NADES #3 provided a significantly lower extraction efficiency (<7%) and quickly (<30 min) became turbid after being mixed with the aqueous solution containing SDS. Based on these results, NADES #4 was selected as the optimum solvent for the extraction. This decision was also supported by the high hydrophobic character of dodecanoic acid (water solubility 0.15 mg g−1 at 20 °C), leading to a NADES that is not only very stable29,34,40,42 but also immiscible with water.34,38 Moreover, this hydrophobic NADES can interact with various analytes via van der Waals forces, making it suitable for various extraction processes.25,29,31,34,38,39
1) selected for these experiments.43 NADES #3 provided a significantly lower extraction efficiency (<7%) and quickly (<30 min) became turbid after being mixed with the aqueous solution containing SDS. Based on these results, NADES #4 was selected as the optimum solvent for the extraction. This decision was also supported by the high hydrophobic character of dodecanoic acid (water solubility 0.15 mg g−1 at 20 °C), leading to a NADES that is not only very stable29,34,40,42 but also immiscible with water.34,38 Moreover, this hydrophobic NADES can interact with various analytes via van der Waals forces, making it suitable for various extraction processes.25,29,31,34,38,39
3.3. Effect of sample volume and extraction time
In order to determine the optimum conditions for the extraction of the methylene blue–surfactant complex, the effect of the sample volume and time used for the extraction was then investigated. Previous literature reports specifically draw attention to the importance of these variables.38,55 Aiming to simplify the data acquisition, these experiments were followed using the proposed digital image-based (DIB) approach (as described in section 2.5), monitoring the extraction from the water samples (5–250 mL) containing 0.3 mg L−1 of SDS, during an interval of 1–60 min. These ranges were selected from preliminary experiments that considered not only the performance of the analysis but also the practical aspects of the test, to be implemented for on-site detection.As can be observed in Fig. 4A, and in line with previous reports,38,55,56 both variables had a significant effect on the extraction efficiency. That said, the effects were more evident when sample volumes >50 mL and extraction times >30 min were used. As expected, and similar to the reports involving CHCl3,22 the extraction yield was worst when 1 min was used, across the entire range of volumes tested. For instance, when the sample volume was 250 mL and 1 min extraction was used, the process allowed extraction of only 1/10th of the amount removed at 60 min (Fig. 4B). Such a short extraction time would also need to be carefully monitored to minimize errors, as any inconsistency in the extraction time would lead to a large difference in the amount extracted. As also shown in Fig. 4B, such differences were negligible when lower sample volumes were used (Y-scale was set to cover the 0–255 range, which corresponds to the possible range of the RGB scale). From these results, it was also evident that a higher sensitivity could be obtained if the method tolerates/requires larger sample volumes (i.e. 250 mL) and/or longer extraction times (i.e. 60 min). However, considering that such experimental conditions would not align with the principles of green chemistry (amount of waste, power consumption, analysis time), the lowest possible sample volume (15 mL) and extraction time (5 min) were selected as optimum and used for all remaining experiments.
3.4. Analytical figures of merit
To critically assess the analytical performance of the proposed approach, the corresponding calibration curve was obtained. These experiments were performed using the optimum conditions, as previously discussed (3 mL of NADES #4, sample volume: 15 mL, extraction time: 5 min) and the DIB approach. As it can be observed in Fig. 5, the signal intensity (inversely proportional to color intensity) decreased proportionally with respect to the concentration of surfactants in the aqueous phase. In this case, a linear range from 0.010 to 0.600 mg L−1 (R2 = 0.996) was obtained, leading to a competitive limit of detection (LoD = 0.002 mg L−1) and limit of quantification (LoQ = 0.008 mg L−1). It is worth mentioning that the analytical range covers the requirement of the US-EPA for water samples (0.2–0.5 mg L−1)57 and that the LoD of the proposed approach is lower than both the value previously reported using chloroform (0.006 mg L−1)21 and those of other methods reported in the literature.16,23 The inter-day variability of the method, assessed by the standard deviation of the color intensity of a standard (0.30 mg L−1, n = 4) for three consecutive days, was lower than 6%. As also shown in Fig. 5, the presence of the methylene blue–surfactant complex was also evident to the naked eye, potentially enabling a minimally trained analyst to perform the analysis with a printed reference instead of a smartphone.In order to deploy the proposed methodology to quantify the presence of surfactants in surface water, it is critical to evaluate the response of other surfactants. For these experiments, samples containing 0.30 mg L−1 of SDS, sodium octyl sulfate (SOS), sodium dodecylbenzene sulfonate (SDBS), sodium deoxycholate (SDOCh, a non-linear surfactant), or cetyltrimethylammonium bromide (CTAB, a cationic surfactant) were prepared and tested using the optimized conditions for the analysis. As shown in Fig. 6, only a small variation (1 ± 3%) was observed for SDBS with respect to the solution containing SDS. These results can be attributed to the similarities in the chemical structure of the two surfactants. In contrast, SOS and SDOCh lead to significantly lower signals (65 ± 2% and 46 ± 1%, respectively), a finding that was attributed to the lower hydrophobicity of these surfactants.58,59 As expected, only a marginal response (1 ± 1% of the signal obtained for SDS 0.3 g mL−1) was obtained for the cationic surfactant CTAB, which can't form a complex with methylene blue and partition into the organic phase. As a control, the sample containing CTAB was also analyzed using the extraction with CHCl3, yielding similar results (data not shown).
The effect of different cations and anions was also evaluated, at a concentration 100× higher than the surfactants.16,21,23 As can be observed in Table SI 1,† with the exception of Fe3+, only marginal responses (<14%) were observed for all the selected ions. Indeed, the presence of Fe3+ led to a decrease in the signal intensity by 71%, a finding that is in line with the high affinity between this ion and SDS,60–63 which competes with methylene blue. A recovery analysis, performed to determine the efficiency of the extraction process, yielded results in the 84% to 108% range (Table SI 2†), which was considered adequate for the proposed application.
3.5. Analysis of the environmental water samples
In order to demonstrate the broad applicability of the proposed methodology, a number of environmental water samples were collected and analyzed using the optimized conditions. The collection points of the samples are included in Fig. SI 1.† In order to validate these results, we also analyzed the same samples using the CHCl3-based reference method. As can be seen in Table SI 2,† all water samples analyzed presented a surfactant concentration lower than the thresholds set by the US-EPA (0.2–0.5 mg L−1),57 suggesting the success of the stormwater management plans. The results in Table SI 3† also show that no statistical difference (for a confidence level of 95%) was found between the results obtained by both methods, thus supporting the applicability of the proposed approach. It was also noted that the proposed method also rendered higher accuracy than the traditional methodology based on UV-vis spectrophotometry, although a much larger study would be required to support the broad validity of such findings.3.6. Green metrics
Considering that the proposed method was developed to provide a more environmentally benign and safer alternative to the traditional test (using CHCl3), the proposed approach was evaluated using the “Analytical GREEnness Calculator” proposed by Pena-Pereira et al.33 This calculator is based on 12 principles of green chemistry, each of them expressed on a 0–1 scale. Based on the use of the NADES as a non-toxic solvent, requiring only 100 μL per analysis, using a smartphone as a detector for in situ analysis, able to measure 92 samples in only 5 min, without any pretreatment (except simple filtration), the method presented a score of 0.96 (see Fig. 7A). The only threat of the method was the use of MB, which can affect plants and animals if released.64 Notably, the use of the smartphone as the detection device does not generate a threat or penalty, because it is considered an electronic device that the analyst will carry, regardless of the test to be performed. On the other hand, the classic method using chloroform and UV-vis spectrophotometry presented a score of 0.49, mainly attributed to the large amount of chloroform (up to 100 mL) used, limited portability, and low throughput (see Fig. 7B).4. Conclusions
We report on the advantages of a green method to detect surfactants in water, based on the use of a hydrophobic NADES (menthol:dodecanoic acid) and smartphone detection. Under optimized conditions, an aliquot of 3 mL of the NADES was mixed with 15 mL of water for 5 min, then allowed to settle (to enable the separation of the two phases), leading to a linear range from 0.010 mg L−1 to 0.60 mg L−1. The analytical performance of the method was successfully evaluated considering the broad applicability of the approach to common surfactants, the low contribution of interferences, the high recovery values, and the applicability to real samples. Besides enabling the replacement of chloroform, the method can be applied for in situ analysis and allowed a significant reduction of the waste negated, leading to a green score of 0.96. All things considered, the proposed methodology represents one of the simplest and most convenient routes to quantify surfactants in environmental water samples.Data availability
Data reported in this manuscript will be made available upon reasonable requests to the corresponding authors.Author contributions
Vagner Bezerra dos Santos: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing – original draft, and writing – review & editing. Lucas B. Ayres: data curation, formal analysis, investigation, methodology, software, writing – original draft, writing – review & editing. Helayne Santos: conceptualization, data curation, investigation, methodology, writing – original draft, writing – review & editing. Carlos D. Garcia: conceptualization, data curation, funding acquisition, investigation, methodology, project administration, resources, software, supervision, visualization, writing – original draft, writing – review & editing. Willian T Suarez: conceptualization, data curation, funding acquisition, project administration, resources, visualization, and writing – original draft.Conflicts of interest
Authors declare no conflict of interest.Acknowledgements
The authors would like to acknowledge the Fundação de Amparo à Ciência e Tecnologia de Pernambuco (FACEPE) (grants APQ-0942-1.06/22 and APQ-0413-1.06/21), the Conselho Nacional de Ciência e Tecnologia (CNPQ) (grants CNPQ 421147/2018-0, 200421/2023-9, and 308422/2023-6) and CAPES for scholarship. Financial support from the Department of Chemistry at Clemson University and the South Carolina Department of Agriculture ACRE Competitive Grant Program is also acknowledged.References
- S. M. Shaban, J. Kang and D.-H. Kim, Compos. Commun., 2020, 22, 100537 CrossRef.
- T. F. Tadros, Applied Surfactants: Principles and Applications, Wiley, 2006 Search PubMed.
- M. J. L. Castro, C. Ojeda and A. F. Cirelli, Environ. Chem. Lett., 2014, 12, 85–95 CrossRef CAS.
- L. L. Schramm and L. L. Schramm, Surfactants: Fundamentals and Applications in the Petroleum Industry, Cambridge University Press, 2000 Search PubMed.
- Y. Yu, J. Zhao and A. E. Bayly, Chin. J. Chem. Eng., 2008, 16, 517–527 CrossRef CAS.
- P. A. Cornwell, Int. J. Cosmet. Sci., 2018, 40, 16–30 CrossRef CAS PubMed.
- S. Wolfrum, J. Marcus, D. Touraud and W. Kunz, Adv. Colloid Interface Sci., 2016, 236, 28–42 CrossRef CAS PubMed.
- I. Kralova and J. Sjöblom, J. Dispersion Sci. Technol., 2009, 30, 1363–1383 CrossRef CAS.
- M. Y. de Freitas Araújo Reis, R. I. de Araújo Rêgo, B. P. Rocha, G. G. Guedes, Í. M. de Medeiros Ramalho, A. L. de Medeiros Cavalcanti, G. P. Guimarães and B. P. G. de Lima Damasceno, Curr. Pharm. Des., 2021, 27, 4300–4314 CrossRef PubMed.
- J. Arora, A. Ranjan, A. Chauhan, R. Biswas, V. D. Rajput, S. Sushkova, S. Mandzhieva, T. Minkina and T. Jindal, J. Appl. Microbiol., 2022, 133, 1229–1244 CrossRef CAS PubMed.
- K. Jardak, P. Drogui and R. Daghrir, Environ. Sci. Pollut. Res., 2016, 23, 3195–3216 CrossRef CAS PubMed.
- M. Jackson, C. Eadsforth, D. Schowanek, T. Delfosse, A. Riddle and N. Budgen, Environ. Toxicol. Chem., 2016, 35, 1077–1086 CrossRef CAS PubMed.
- G. Jena, K. Dutta and A. Daverey, Chemosphere, 2023, 341, 140082 CrossRef CAS PubMed.
- I. Effendi, S. Nedi, Ellizal, Nursyirwani, Feliatra, Fikar, Tanjung, R. Pakpahan and Pratama, IOP Conf. Ser.: Earth Environ. Sci., 2017, 97, 012030 Search PubMed.
- J. Hammer, A. M. Tukker, J. F. Postma, J. J. H. Haftka, J. L. M. Hermens, P. de Voogt and M. H. S. Kraak, Bull. Environ. Contam. Toxicol., 2018, 101, 99–104 CrossRef CAS PubMed.
- F. N. Feiteira, L. G. T. dos Reis, W. F. Pacheco and R. J. Cassella, Microchem. J., 2015, 119, 44–50 CrossRef CAS.
- G. Salomon and F. Giordano-Labadie, Eur. J. Dermatol., 2022, 32, 677–681 CrossRef CAS PubMed.
- S. O. Badmus, H. K. Amusa, T. A. Oyehan and T. A. Saleh, Environ. Sci. Pollut. Res., 2021, 28, 62085–62104 CrossRef CAS PubMed.
- A. L. Kim, E. V. Musin, A. V. Dubrovskii and S. A. Tikhonenko, Sci. Rep., 2022, 12, 232 CrossRef CAS PubMed.
- M. Koga, Y. Yamamichi, Y. Nomoto, M. Irie, T. Tanimura and T. Yoshinaga, Anal. Sci., 1999, 15, 563–568 CrossRef CAS.
- H. S. de Sousa, R. Arruda-Santos, E. Zanardi-Lamardo, W. T. Suarez, J. L. de Oliveira, R. A. Farias and V. Bezerra dos Santos, Anal. Methods, 2024, 16, 2009–2018 RSC.
- A. W. W. A. American Public Health Association and Water Environment Federation, Standard methods for the examination of water and wastewater, APHA Press, Washington DC, 24th edn, 2024 Search PubMed.
- M. J. A. Lima, C. F. Nascimento and F. R. P. Rocha, Anal. Methods, 2017, 9, 2220–2225 RSC.
- A. Mannu, M. Blangetti, S. Baldino and C. Prandi, Materials, 2021, 14, 2494 CrossRef CAS PubMed.
- D. Li, Front. Plant Sci., 2022, 13, 1004332 CrossRef PubMed.
- A. García-Roldán, L. Piriou and P. Jauregi, Front. Plant Sci., 2023, 13, 1072592 CrossRef PubMed.
- T. Umecky, A. Goto, N. Hayashi and K. Eguchi, ACS Omega, 2023, 8, 14694–14698 CrossRef CAS PubMed.
- Y. Liu, N. Deak, Z. Wang, H. Yu, L. Hameleers, E. Jurak, P. J. Deuss and K. Barta, Nat. Commun., 2021, 12, 5424 CrossRef CAS PubMed.
- F. Mohd Fuad, M. Mohd Nadzir and A. Harun Kamaruddin, J. Mol. Liq., 2021, 339, 116923 CrossRef CAS.
- D. Rente, A. Paiva and A. R. Duarte, Molecules, 2021, 26, 2336 CrossRef CAS PubMed.
- Z.-H. Cai, J.-D. Wang, L. Liu, L.-D. Ruan, Q. Gu, X.-Y. Yan, L.-N. Fu, P.-Q. Zhao, S. Zhang and Y.-J. Fu, Chem. Eng. J., 2023, 457, 141333 CrossRef CAS.
- L. B. Ayres, F. J. V. Gomez, M. F. Silva, J. R. Linton and C. D. Garcia, Sci. Rep., 2024, 14, 2715 CrossRef CAS PubMed.
- F. Pena-Pereira, W. Wojnowski and M. Tobiszewski, Anal. Chem., 2020, 92, 10076–10082 CrossRef CAS PubMed.
- D. J. G. P. van Osch, L. F. Zubeir, A. van den Bruinhorst, M. A. A. Rocha and M. C. Kroon, Green Chem., 2015, 17, 4518–4521 RSC.
- L. S. Clesceri, A. E. Greenberg and A. D. Eaton, Standard Methods for the Examination of Water and Wastewater, APHA American Public Health Association, 20th edn, 1998 Search PubMed.
- L. B. Ayres, G. Weavil, M. Alhoubani, B. G. S. Guinati and C. D. Garcia, J. Mol. Liq., 2023, 389, 122891 CrossRef CAS.
- L. B. Ayres, F. J. V. Gomez, J. R. Linton, M. F. Silva and C. D. Garcia, Anal. Chim. Acta, 2021, 1161, 338403 CrossRef CAS PubMed.
- M. Díaz-Álvarez and A. Martín-Esteban, Adv. Sample Prep., 2022, 4, 100047 CrossRef.
- Y. Fan, H. Luo, C. Zhu, W. Li, D. Wu and H. Wu, Sep. Purif. Technol., 2021, 275, 119112 CrossRef CAS.
- M. Devi, R. Moral, S. Thakuria, A. Mitra and S. Paul, ACS Omega, 2023, 8, 9702–9728 CrossRef CAS PubMed.
- T. Křížek, M. Bursová, R. Horsley, M. Kuchař, P. Tůma, R. Čabala and T. Hložek, J. Cleaner Prod., 2018, 193, 391–396 CrossRef.
- F. Oliveira, E. Silva, A. Matias, J. M. Silva, R. L. Reis and A. R. C. Duarte, Eur. J. Pharm. Sci., 2023, 182, 106368 CrossRef CAS PubMed.
- M. M. Abdallah, S. Müller, A. González de Castilla, P. Gurikov, A. A. Matias, M. D. Bronze and N. Fernández, Molecules, 2021, 26, 1801 CrossRef CAS PubMed.
- V. B. d. Santos, E. F. S. Campos, J. P. B. de Almeida, W. T. Suarez, C. R. S. Oliveira and S. C. B. de Oliveira, Microchem. J., 2023, 189, 108508 CrossRef.
- M. V. Maia, W. T. Suarez, V. B. dos Santos and J. P. B. de Almeida, Microchem. J., 2022, 179, 107500 CrossRef CAS.
- M. de O. K. Franco, W. J. Cardoso, C. B. Vilanculo, V. B. dos Santos, J. P. B. de Almeida, L. F. Capitán-Vallvey and W. T. Suarez, Anal. Methods, 2023, 15, 2300–2308 RSC.
- E. Vidal, C. E. Domini, D. C. Whitehead and C. D. Garcia, Sens. Diagn., 2022, 1, 496–503 RSC.
- S. Soares, L. C. Nunes, W. R. Melchert and F. R. P. Rocha, Microchem. J., 2020, 152, 104273 CrossRef CAS.
- J. P. B. de Almeida, V. B. dos Santos, G. A. do Nascimento, W. T. Suarez, W. M. de Azevedo, A. F. Ferreira and M. V. Maia, Anal. Methods, 2022, 14, 2631–2641 RSC.
- E. Vidal, A. S. Lorenzetti, C. D. Garcia and C. E. Domini, Anal. Chim. Acta, 2021, 1151, 338249 CrossRef CAS PubMed.
- V. B. d. Santos, E. K. N. da Silva, L. M. A. de Oliveira and W. T. Suarez, Food Chem., 2019, 285, 340–346 CrossRef PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- Y. Dai, G.-J. Witkamp, R. Verpoorte and Y. H. Choi, Food Chem., 2015, 187, 14–19 CrossRef CAS PubMed.
- M. Shariati-Rad and F. Fattahi, Anal. Chim. Acta, 2020, 1100, 208–214 CrossRef CAS PubMed.
- A. B. Abdullahi, S. Ismail, U. Alshana and N. Ertaş, J. Food Compos. Anal., 2023, 119, 105263 CrossRef CAS.
- M. Li, C. Rao, C. Wang, X. Cui and Y. Xiong, Sep. Purif. Technol., 2024, 341, 126882 CrossRef CAS.
- U.S. Environmental Protection Agency, 2024.
- C. D. García, B. M. Dressen, A. Henderson and C. S. Henry, Electrophoresis, 2005, 26, 703–709 CrossRef PubMed.
- M. F. Mora, C. E. Giacomelli and C. D. Garcia, Anal. Chem., 2007, 79, 6675–6681 CrossRef CAS PubMed.
- F. I. Talens-Alesson, S. T. Hall, N. P. Hankins and B. J. Azzopardi, Colloids Surf., A, 2002, 204, 85–91 CrossRef CAS.
- F. Kellou-Kerkouche, A. Benchettara and S.-E. Amara, J. Mater., 2013, 2013, 903712 Search PubMed.
- E. A. Streltsova, E. A. Hromysheva and A. F. Tymchuk, Adsorpt. Sci. Technol., 2002, 20, 757–765 CrossRef CAS.
- L. V. Jian-xiao, W. Dong and Z. Ji-ti, J. Dispersion Sci. Technol., 2006, 27, 1073–1077 CrossRef.
- P. O. Oladoye, T. O. Ajiboye, E. O. Omotola and O. J. Oyewola, Results Eng., 2022, 16, 100678 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00196f |
| This journal is © The Royal Society of Chemistry 2024 |