Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Peroxidase-mimicking Prussian blue nanoparticles versus HRP for high colorimetric detection of miRNA-141 in competitive RNA–RNA systems†
Maliana
El Aamri
,
Hasna
Mohammadi
and
Aziz
Amine
 *
*
Faculty of Sciences and Techniques, Laboratory of Process Engineering and Environment, Chemical Analysis and Biosensors Group, Hassan II University of Casablanca, P.A 146, Mohammedia, Morocco. E-mail: azizamine@yahoo.fr
First published on 17th October 2024
Abstract
Rapid and efficient early-stage tumor detection is crucial in cancer diagnostics. Recent research indicates that microRNA-141 expression levels serve as a predictive biomarker for prostate cancer cell count in the human body. In this study, we developed an original competitive system for miRNA-141 detection using Prussian blue nanoparticles (PBNPs), comparing it with a horseradish peroxidase (HRP)-based competitive system for the same target. The competitive system involved miRNA-141 and biotin-miRNA-141 on a magnetic bead-modified capture probe specific to miRNA-141. The synthesized PBNPs were conjugated to avidin, resulting in the formation of avidin–PBNPs. These conjugates were used as a substitute for streptavidin–HRP. The peroxidase-like activity of PBNPs catalyzed the colorimetric substrate (3,3′,5,5′-tetramethylbenzidine), producing a distinct blue color measured at 630 nm. Under optimal conditions, both PBNPs and HRP-based systems exhibited a linear response to miRNA-141 concentrations (50 pM to 300 pM and 80 pM to 500 pM, respectively). Among the two systems investigated in this study, the PBNPs-based bio-assay demonstrated exceptional sensitivity, achieving a remarkably low LOD of 0.61 pM and an analysis time of 32 minutes. These biosensors successfully determined miRNA-141 levels in spiked human serum.
Introduction
Globally, cancer remains a pervasive health challenge, standing as a prominent cause of mortality.1–3 Prostate cancer, particularly affecting men, ranks among the most prevalent forms of cancer worldwide.4,5 Early detection is crucial for unlocking practical, affordable, and high-potential therapeutic interventions.6,7 In this quest, the emergence of novel biomarkers like microRNAs (miRNAs) offers a promising path for advancing cancer diagnosis and understanding.8,9 Over 1200 miRNAs have been identified,10 among which miR-141 is detected at elevated levels in the blood of patients with metastatic prostate cancer.11–13miRNAs, short noncoding RNAs of 19–23 nucleotides, play a vital role in post-transcriptional gene regulation.14 They are diagnostic biomarkers of cell differentiation, proliferation, and apoptosis.15 However, miRNAs feature subsize, ultralow expression and high sequence homology, leading to huge detection challenges.16,17 Although traditional analytic methods including Northern blotting microarrays and reverse-transcription quantitative polymerase chain reaction (RT-PCR) can achieve miRNA assay, they are expensive, time-consuming, and need complicated instruments. So, developing rapid, sensitive, and selective methods for miRNA detection is of great significance.18
Biosensors have emerged as promising tools to overcome these microRNA detection challenges.19 Colorimetric biosensors, in particular, are notable for their simplicity, cost-effectiveness, and ease of interpretation.16,20,21 Colorimetric assays provide a direct visual readout, eliminating the need for complex instruments. They offer rapid and sensitive microRNA quantification, making them ideal for diagnostics and research, without requiring specialized expertise or additional steps.
Enzymes are traditionally the most catalytic components in competitive biosensor systems designed for miRNA detection, catalyzing substrate reactions to produce measurable signals.22 Moreover, ferrocene and methylene blue are commonly integrated into competitive systems. Kutluk et al. innovatively developed an electrochemical biosensor for nanomolar miRNA-197 detection, leveraging a competitive DNA–RNA hybridization approach with glucose oxidase as a catalytic tool.23 In separate studies, Vargas et al. and Zouari et al. devised horseradish peroxidase (HRP)-based competitive RNA–RNA hybridization methods to detect miRNA-21 at nanomolar and femtomolar levels, respectively.22,24
Enzymes often have certain drawbacks, including sensitivity to environmental factors such as pH, temperature, and chemical interference, limiting their stability and reliability in diagnostics. Moreover, producing and purifying natural enzymes can be costly and time-consuming. To address these challenges, researchers are exploring nanozymes, nanomaterials exhibiting enzyme-like catalytic activities akin to natural enzymes.25,26 Nanozymes offer advantages such as enhanced stability, resilience to harsh conditions, and simplified synthesis and modification processes.27 Various nanomaterials, including gold nanoparticles (AuNPs),28 magnetic nanoparticles (Fe3O4NPs),29 cerium oxide nanoparticles (CeO2NPs),30 V2O5 nanowires,31 Co3O4 nanoparticles,32 copper nanoparticles (CuNPs),16 and Prussian blue nanoparticles (PBNPs),33 have been identified with diverse enzyme-mimicking activities (oxidase, peroxidase, catalase, phosphatase), expanding their utility in diagnostics.
PBNPs, characterized by the chemical formula Fe4[Fe(CN)6]3, consist of alternating ferric and ferrous ions coordinated with cyanides.34 The robust peroxidase-like activity exhibited by these artificial enzymes is attributed to the presence of both Fe3+ and Fe2+ in the core of PBNPs.35 This characteristic highlights PBNPs potential as effective and adaptable catalysts and places them as strong candidates to replace natural peroxidase in a biosensor-based competitive system for miRNA detection.
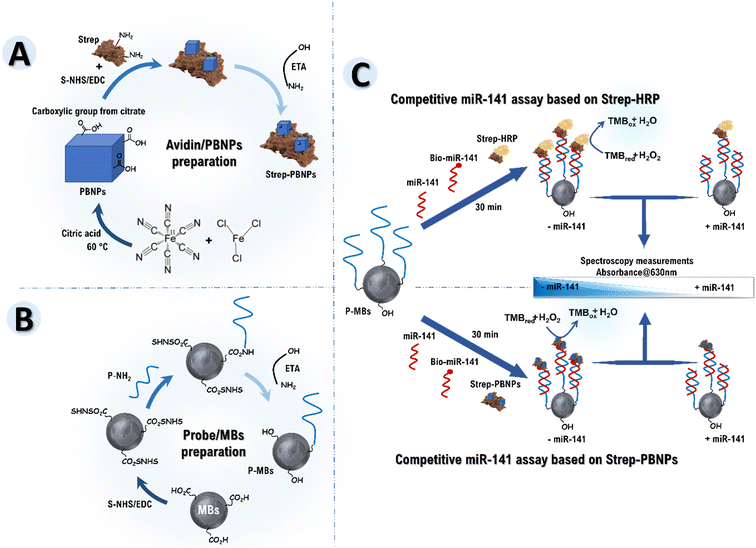
In this work, an innovative colorimetric PBNPs-based competitive biosensor for miRNA-141 detection was developed and compared to an identical system that uses natural HRP. For this, both biosensors were prepared in the same way. A capture probe DNA (P) was immobilized on carboxylated magnetic beads (MBs), enabling amplified immobilization due to the 3D surface of the MBs. In the subsequent step, miRNA-141, biotin-labeled miRNA-141, and avidin–PBNPs conjugates prepared due to the activation of the carboxylated function of citric acid on the surface of PBNPs by EDC/S-NHS, were added in a single step to the formed P/MBs. A competition ensued between miRNA-141 and biotin-labeled miRNA-141 for hybridization with the capture probe. Subsequently, avidin–PBNPs were linked to the biotin of biotin-labeled miRNA-141. Subsequently, 3,3′,5,5′-tetramethylbenzidine (TMB)/H2O2 was introduced as a chromogenic substrate, and the peroxidase-like activity of PBNPs catalyzed the oxidation reaction of TMB, resulting in a distinct blue color. The absorbance of this color at 630 nm was measured, allowing precise quantification and analysis of the detected microRNA-141. On the other hand, for the enzyme-based competitive biosensor, streptavidin–HRP was added instead of prepared avidin–PBNPs conjugates and the competition and detection were performed using the PBNPs-based developed competitive biosensor.
Experimental
Apparatuses
The UV-visible absorption spectra within the wavelength range of 400–800 nm were measured using a narrow bandwidth of 1 nm, and the absorbance of the colorimetric product was measured at 630 nm using an SPECTROstar Omega (BMG Labtech). Data analysis was conducted using MARS version 4.2. Origin 2024 software was utilized for data analysis, artwork, and graphing.Eppendorf high-speed micro-centrifuge DLAB (DLAB Scientific Co., Ltd., China) was used for nanoparticles separation.
Reagents and solutions
Carboxylated magnetic beads (10 mg mL−1) were purchased from Thermos Fisher Scientific (IL, USA). 3,3′,5,5′-Tetramethylbenzidine (TMB), hydrogen peroxide 30% (H2O2) (N-morpholino)ethanesulfonic acid (MES), ethanolamine, N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), sulfo-N-hydroxysuccinimide (S-NHS), artificial human serum and MgCl2 were purchased from Merck (Germany). Tween-20 were obtained from VWR Life Science AMRESCO, Ireland. Potassium hexacyanoferrate(II) (K4[Fe(CN)6]), iron(III) chloride hexahydrate (FeCl3·6H2O), and citric acid were bought from Sigma-Aldrich, Germany. The chemicals employed in the preparation of the phosphate buffer saline solution (0.01 M PBS containing 2.7 mM KCl and 137 mM NaCl, with a pH of 7.4) were procured from Merck in Darmstadt, Germany. All reagents utilized were of a high analytical quality. Unless specified otherwise, all experiments were carried out under room temperature (RT) conditions.Eurogentec provided the lyophilized powder form of HPLC-purified oligonucleotides. To prepare stock solutions, 100 μM concentrations were prepared, divided into small aliquots, and stored at −20 °C. Table 1 provides detailed information regarding the oligonucleotide sequences.
| Nucleic acid | Sequence (5′–3′) |
|---|---|
| Target microRNA-141 | UAA CAC UGU CUG GUA AAG AUG G |
| Probe (NH2-P) | NH2-(CH2)6-AAACCA TCT TTA CCA GAC AGT GTT A |
| Biotin-microRNA-141 | Biotin-UAA CAC UGU CUG GUA AAG AUG G |
| microRNA-21 (non-complementary target) | UAGCUUAUCAGACUGAUGUUGA |
| microRNA-125a (non-complementary target) | UCCCUGAGACCCUUUAACCUGUGA |
| microRNA-146a (non-complementary target) | UGAGAACUGAAUUCCAUGGGUU |
| microRNA-155 (non-complementary target) | UUAAUGCUAAUCGUGAUAGGGGUU |
| microRNA-222 (non-complementary target) | AGCUACAUCUGGCUACUGGGUCUC |
Synthesis of Prussian blue nanoparticles (PBNPs)
PBNPs with sizes ranging from 15 to 24 nm were synthesized according to our previous works36 with minor modifications. Glassware was thoroughly cleaned with aqua regia, washed with distilled water, and dried before synthesis. PBNPs were prepared in a one-step synthesis: a heated aqueous mixture, which already contained 25 mM of citric acid and 1 mM of FeCl3·6H2O, was swiftly combined with a solution composed of 1 mM K4[Fe(CN)6] which in turn is prepared in 25 mM citric acid. The mixture was stirred for 30 min at 60 °C, resulting in a drastic color change to dark blue, indicative of PBNPs formation. The blue dispersion was then cooled to room temperature under continuous stirring and stored at 4 °C until use.Preparation of avidin–PBNPs conjugates
The conjugates of avidin–PBNP were synthesized following this procedure:36 1 mL of synthesized PBNPs dispersion underwent successive centrifuge cycles (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm/30 min) in distilled water, and the blue pellet was dissolved in 0.01 M phosphate buffer (PB, pH 7.4). The conjugation of avidin with PBNPs relied on the EDC/S-NHS coupling method. In brief, 4 mg of EDC and 8 mg of S-NHS, prepared in 50 mM MES buffer (pH 6), were added to 1 mL of PBNPs to activate the surface of citric acid-coated PBNPs before avidin grafting. The activation step was maintained for 1 h at 37 °C. Subsequently, 1 mL of avidin (1 mg mL−1) was introduced into the solution and incubated for 2 h at 37 °C. To block the activating sites of the Prussian blue nanoparticles, 1 mM ethanolamine was utilized. After centrifugation cycles (13
000 rpm/30 min) in distilled water, and the blue pellet was dissolved in 0.01 M phosphate buffer (PB, pH 7.4). The conjugation of avidin with PBNPs relied on the EDC/S-NHS coupling method. In brief, 4 mg of EDC and 8 mg of S-NHS, prepared in 50 mM MES buffer (pH 6), were added to 1 mL of PBNPs to activate the surface of citric acid-coated PBNPs before avidin grafting. The activation step was maintained for 1 h at 37 °C. Subsequently, 1 mL of avidin (1 mg mL−1) was introduced into the solution and incubated for 2 h at 37 °C. To block the activating sites of the Prussian blue nanoparticles, 1 mM ethanolamine was utilized. After centrifugation cycles (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm/10 min), the resulting avidin–PBNP bioconjugates were stored in PBS and kept at 4 °C until use.
000 rpm/10 min), the resulting avidin–PBNP bioconjugates were stored in PBS and kept at 4 °C until use.
Aminated probe immobilization on carboxylated MBs
The aminated DNA capture probe was immobilized using our previously established method.17 HO2C-MBs were activated with EDC/S-NHS. To do this, 2 μL of washed HO2C-MBs (10 mg mL−1) were mixed with 25 μL each of EDC (10 mg mL−1) and S-NHS (10 mg mL−1) in cold MES buffer (50 mM, pH = 6). The mixture was incubated at room temperature with a slight tilt rotation for 30 min.Using NHS-activated MBs, the aminated probe (P-NH2) had to be immobilized in the second phase. This was achieved by incubating the mixture at room temperature for 30 min after adding 50 μL of P-NH2 to the activated MBs. Subsequently, 50 μL of 1 mM ethanolamine in PBS 10 mM (pH = 7.4) was added, and the mixture was incubated at room temperature for 15 min. The MBs underwent three washes with 50 mM MES solution (pH = 6) and magnetic separation following each step.
MicroRNA-141 detection in a competitive system via avidin–PBNPs
The hybridization was carried out in a competitive system: a 50 μL mixture, including 20 μL of a fixed concentration of biotin-miRNA-141 (competitive target), 10 μL of varying concentration of the target miRNA-141 and 20 μL of a fixed amount of avidin–PBNPs, was added to the prepared probe/MBs. The mixture was incubated for 30 min at room temperature. Subsequently, recuperated MBs were washed twice with 0.01 M PBS, pH 7.4, containing 0.01% of Tween 20. The competitive hybridization was performed in 0.01 M PBS, pH 7.4.The detection procedure was performed by adding 70 μL of PBS (10 mM), 30 μL of TMB solution at a concentration of 40 mM, and 50 μL of 25 mM H2O2 solution to the recuperated MBs. The mixture was then incubated for 2 minutes at room temperature. A blue color was obtained due to the peroxidase like-activity of the PBNPs attached to the competitive miRNA-141. In the end, the absorbance of the obtained color was measured at 630 nm.
Detection of microRNA-141 in a competitive system via streptavidin–HRP
The detection of microRNA-141 in a competitive system via streptavidin–HRP was performed following the same protocol described for avidin–PBNPs, with the substitution of avidin–PBNPs with streptavidin–HRP. The experimental steps remained consistent, ensuring comparability between the two detection methods.Real sample
Both developed approaches (PBNPs-based competitive biosensor and HRP-based competitive biosensor) were evaluated for detecting microRNA-141 in commercial artificial human serum samples representing complex matrices. To prepare the samples, the serum was diluted 5 times with PBS buffer (10 mM, pH = 7.4) and then spiked with varying concentrations of microRNA-141. The detection procedure for each approach was carried out following the steps described above.Results and discussion
Peroxidase like activity of PBNPs and their modification with avidin
A very repeatable approach37 was used to synthesize PBNPs, ensuring remarkable stability in dispersion. This process includes mixing equimolar aqueous solutions of FeCl3 and K4[Fe(CN)6] in the presence of citric acid. As shown in Fig. 1A (curve d) and the insert photo (Eppendorf d), a distinct navy blue color solution was obtained at a high concentration of pure PBNPs, which had a maximum absorption peak at 704 nm, suggesting that ferric ions (Fe2+/Fe3+) co-precipitated. In addition to aiding in the nucleation of FeCl3 and K4[Fe(CN)6], the choice of citric acid as a capping agent ensured excellent dispersion of the PBNPs in the working solution, as described in a previous study.36To generate a measured signal. The peroxidase-like activity of PBNPs was investigated by assessing their catalytic role in the oxidation of TMB, a chromogenic peroxidase substrate, in the presence of H2O2. As depicted in Fig. 1A, the presence of PBNPs resulted in the oxidation of TMB by H2O2, forming a blue charge transfer complex known as 3,3′,5,5′-tetramethylbenzidine diimine (TMBDI) with maximal absorption at 630 nm (curve c and Eppendorf c in the insert photo). Crucially, the distinct colors associated with pure PBNPs and TMBDI were corroborated by their maximum absorption peaks. On the other hand, no discernible blue color characteristic of the produced TMBDI was evident in the absence of H2O2 or PBNPs (curves a and b), affirming that the catalytic role of PBNPs in the oxidation of TMB (in the presence of H2O2) was responsible for the observed blue TMBDI. PBNPs demonstrated a catalytic property, functioning as nanozymes that mimic the behavior of natural enzymes. This establishes their potential utility as artificial peroxidases.
The PBNPs were conjugated to avidin (protein) by activating the carboxylated citric acid function on the PBNPs surface by EDC/S-NHS as described in Scheme 1A, forming avidin–PBNP conjugate. In Fig. 1B, curves a and b separately showed the UV-vis spectra of aqueous suspensions of the PBNPs and avidin–PBNP conjugate, respectively. As shown in the spectra, a slight shift in the absorption peak was observed from 704 to 716 nm, which can be attributed to the successful binding of avidin to the PBNPs. Additionally, the stability of the protein–PBNPs composites was studied in our previous work,36 demonstrating excellent stability over 4 weeks, maintaining more than 94% of the initial response.
PBNPs versus HRP based competitive MicroRNA-141 bio-assays
This study develops and compares two competitive biosensors designed for detecting miRNA-141 in competition with biotin-labeled microRNA-141. Specifically, the biosensors include an HRP-based competitive system and a PBNPs-based competitive system. This comparative analysis aims to delineate the respective strengths of each biosensor and, consequently, the advantages of employing PBNPs over HRP in a competitive detection system.To initiate the process for both biosensors, an aminated capture probe named P (P-NH2) was immobilized on commercially available HO2C-MBs, forming P–MBs using EDC/S-NHS chemistry. Following this, ethanolamine was introduced to effectively block the non-specific binding sites on the magnetic beads (Scheme 1B). P/MBs are ready-to-use for microRNA-141 detection.
In the subsequent step, for the HRP-based competitive biosensor, miRNA-141, biotin-labeled miRNA-141, and streptavidin–HRP were added in a single step to the formed P/MBs (Scheme 1C, track a). A competition ensued between miRNA-141 and biotin-labeled miRNA-141 for hybridization with the capture probe. Subsequently, streptavidin–HRP was linked to the biotin of biotin-labeled miRNA-141. On the other hand, for the PBNPs-based competitive biosensor, miRNA-141, biotin-labeled miRNA-141, and synthesized avidin–PBNPs were simultaneously added to the formed P/MBs (Scheme 1C, track b). A similar competition occurred between miRNA-141 and biotin-labeled miRNA-141 for hybridization with the capture probe, followed by linking avidin–PBNPs to the biotin of biotin-labeled miRNA-141.
Subsequently, TMB/H2O2 was introduced as a chromogenic peroxidase substrate, and either HRP (Scheme 1C, track a) or PBNPs (Scheme 1C, track b) leveraged their peroxidase-like activity to catalyze the oxidation reaction of TMB. This catalytic process resulted in the development of a distinct blue color, attributed to the peroxidase activity of HRP or the peroxidase-like activity of PBNPs. Finally, the absorbance of the resultant color at 630 nm was measured, enabling the quantification and analysis of the detected microRNA-141.
In the absence of the miRNA-141 target, the capture probe in both biosensors forms a hybrid with biotin-labeled miRNA-141, resulting in the catalysis of TMB/H2O2 by strep–HRP or avidin–PBNPs linked to biotin and leading to a high absorbance at 630 nm. Conversely, in the presence of miRNA-141 target, its hybridization with the capture probe minimizes the presence of biotin-labeled miRNA-141 on the surface of MBs. This reduction in biotin-labeled miRNA-141 leads to minimal catalysis of TMB/H2O2 due to the absence of strep–HRP or avidin–PBNPs on the MBs' surface. The observed correlation between microRNA-141 concentration and the decreased absorbance of the blue color confirms the effectiveness of our detection methods.
To understand the behavior of HRP and PBNPs in each biosensor (HRP-based competitive biosensor and PBNPs-based competitive biosensor), both systems were hybridized with a mixture containing various concentrations of biotin-labeled miRNA-141 (ranging from 0 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pM) along with strep–HRP or synthesized avidin–PBNPs. TMB/H2O2 was subsequently added to each system; the results are depicted in Fig. 2. In Fig. 2A and B, both systems exhibited similar trends in response to the hybridization of the capture probe. As the concentration of biotin-labeled miRNA-141 increased, the absorbance of the produced blue color increased until reaching 500 pM of biotin-miRNA-141, where the absorbance stabilized. This indicates saturation at 500 pM of biotin-miRNA-141 in both biosensors.
000 pM) along with strep–HRP or synthesized avidin–PBNPs. TMB/H2O2 was subsequently added to each system; the results are depicted in Fig. 2. In Fig. 2A and B, both systems exhibited similar trends in response to the hybridization of the capture probe. As the concentration of biotin-labeled miRNA-141 increased, the absorbance of the produced blue color increased until reaching 500 pM of biotin-miRNA-141, where the absorbance stabilized. This indicates saturation at 500 pM of biotin-miRNA-141 in both biosensors.
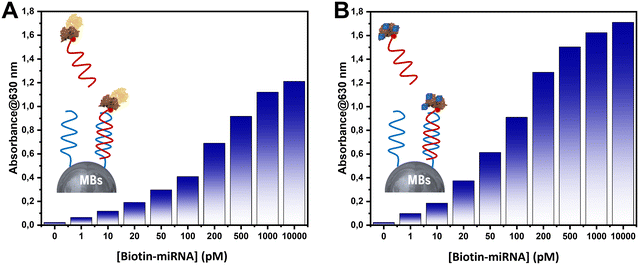 | ||
| Fig. 2 Absorbance measured for various concentrations of biotin-miRNA-141 in the developed systems using (A) strep–HRP and (B) avidin–PBNPs. | ||
In contrast, the HRP-based system reached a maximum absorbance of 1 at 500 pM of biotin-miRNA-141. However, for the PBNPs-based system, a maximum absorbance was achieved with only 100 pM of biotin-miRNA-141. These results highlight that, under our experimental conditions, the PBNPs-based biosensor can detect miRNA-141 at lower concentrations than the HRP-based biosensor in a competitive system under our optimized experimental conditions.
Optimization of the competitive microRNA-141 bio-assays
To ensure a fair comparison between both biosensors, various parameters were optimized. These parameters included, as showed in Table 2, the concentration of the capture probe, the concentration of biotin-labeled miRNA-141 for both systems, the concentration of strep–HRP, and the amount of synthesized avidin–PBNPs conjugate.| Parameter | Tested values | Optimum value |
|---|---|---|
| [Probe] (nM) | 0.1; 1; 10; 100 | 1 |
| Volume (strep–HRP)/volume (PBS) (μL μL−1) | 1/2000; 1/2500; 1/1000; 1/500 | 1/1000 |
| [Bio-miRNA-141] for HRP-based biosensor (nM) | 0.1; 0.2; 0.5; 1 | 0.5 |
| Volume (avidin–PBNPs)/volume (PBS) (μL μL−1) | 1/10; 1/50; 1/100; 1/1000 | 1/100 |
| [Bio-miRNA-141] for PBNPs-based biosensor (nM) | 0.02; 0.05; 0.1; 0.2 | 0.1 |
The optimal condition was determined by discerning a significant difference between the absence and presence of the miRNA-141 target (Fig. S1†). Initially, the concentration of the capture probe for both approaches was optimized. As illustrated in Fig. S1-A,† a concentration of 1 nM for the capture probe was identified as the optimal condition for subsequent work. After that, in optimizing the HRP-based competitive system for miRNA-141 detection, the concentrations of biotin-labeled miRNA-141 (Fig. S1-B†) and strep–HRP (Fig. S1-C†) were systematically adjusted. A significant difference between the absence and presence of the target was achieved using 0.5 nM of biotin-labeled miRNA-141 and 1 μL of (1 mg mL−1) strep–HRP per 1000 μL of PBS.
Finally, optimizing of the PBNPs-based competitive biosensor for miRNA-141 detection involved testing various concentrations of biotin-labeled miRNA-141 and varying amounts of synthesized avidin–PBNPs conjugates. As depicted in Fig. S1-D and E,† the optimal conditions were determined to be 0.1 nM of biotin-labeled miRNA-141 and 1 μL of synthesized avidin–PBNPs per 100 μL of PBS.
Analytical performances of the developed bio-assays
The HRP-based competitive biosensor and PBNPs-based competitive biosensor for miRNA-141 detection demonstrate, as described in Fig. 3, a linear relationship between the logarithm of miRNA-141 concentration for both approaches, spanning different ranges: from 80 pM to 500 pM for the HRP-based competitive biosensor and from 50 pM to 300 pM for the PBNPs-based competitive biosensor. Moreover, the detection limits for these biosensors were estimated to be 2 pM using HRP and 0.61 pM using PBNPs as a peroxidase like activity. The linear regression equation for the proposed method with HRP based competitive system was determined as Y = −0.82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log miRNA-141 (pM) + 2.44, with a high correlation coefficient of R2 = 0.97. However, the linear regression equation for the method-based PBNPs in a competitive system was Y = −1.05
log miRNA-141 (pM) + 2.44, with a high correlation coefficient of R2 = 0.97. However, the linear regression equation for the method-based PBNPs in a competitive system was Y = −1.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log miRNA-141 (pM) + 2.64 with a correlation coefficient of R2 = 0.94. These results demonstrate the remarkable sensitivity of the PBNPs-based competitive biosensor employed in this study, based on the peroxidase-like activity of the PNNPs, enabling highly sensitive microRNA-141 detection. Furthermore, the PBNPs-based biosensor demonstrates quicker detection, requiring only 32 minutes compared to the 45 minutes needed for the HRP-based biosensor. This time efficiency underscores the practical advantages of the PBNPs-based competitive biosensor.
log miRNA-141 (pM) + 2.64 with a correlation coefficient of R2 = 0.94. These results demonstrate the remarkable sensitivity of the PBNPs-based competitive biosensor employed in this study, based on the peroxidase-like activity of the PNNPs, enabling highly sensitive microRNA-141 detection. Furthermore, the PBNPs-based biosensor demonstrates quicker detection, requiring only 32 minutes compared to the 45 minutes needed for the HRP-based biosensor. This time efficiency underscores the practical advantages of the PBNPs-based competitive biosensor.
The PBNPs-based competitive biosensor, with its superior sensitivity and reduced detection time, presents a promising approach for highly sensitive microRNA-141 detection in a competitive system.
Selectivity and repeatability
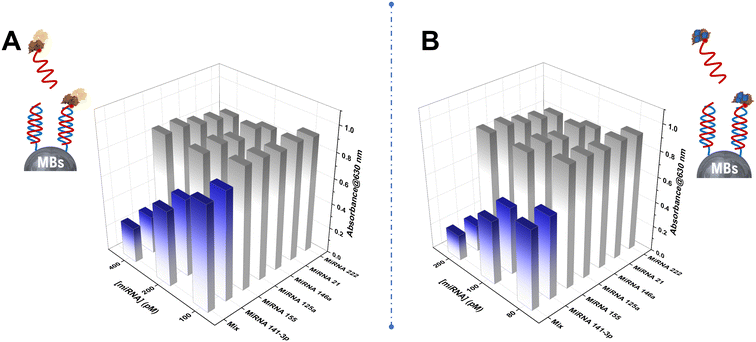
The selectivity of both developed bio-assays was assessed by detecting microRNA-141, various noncomplementary (NC) strands (microRNA-21, microRNA-146a, microRNA-222, microRNA-155, and microRNA-125a), and a mixture of different concentrations using the developed approaches instead of the microRNA-141 target. As illustrated in Fig. 4, the absorbance of both colorimetric assays towards the perfectly matched target miRNA-141 was significantly higher than that for the non-complementary target RNAs, underscoring the methods' high sensitivity specifically towards miRNA-141 detection. Consequently, the developed HRP and PBNPs-based competitive bio-assay exhibit excellent selectivity, offering a reliable and efficient platform for the quantitative detection of miRNA-141 with potential utility in clinical diagnostics.The repeatability of the biosensor-based avidin–PBNPs was assessed by performing ten independent measurements (n = 10) of miRNA-141 at a concentration of 100 pM. The results yielded a mean absorbance of 0.490 ± 0.022 leading to a coefficient of variation of 4.48%. These findings highlight the biosensor's excellent repeatability, demonstrating its reliability for consistent miRNA-141 detection under the optimum conditions.
Serum sample
To investigate the possibility of both proposed systems in a complex matrix, commercial artificial human serum was used to spike target miRNA-141 standards with varied concentrations, diluted with PBS, then evaluated by the HRP and PBNPs-based competitive biosensor. The analytical results for the spiked miRNA are shown in Fig. S2† and Table 3. Recoveries between 98.6% and 106.6% for enzyme-based bio-assay and between 90.6% and 111.5% for nanozyme-based bio-assay (ranging from 100pM to 300 pM) were obtained. It is demonstrated that both developed assays, which were not compromised in serum, could provide a potential analytical tool in real biological samples.| Spiked [miR-141] (pM) | Founded [miR-141] (pM) | Recovery % | RSD % |
|---|---|---|---|
| 100 | 110 | 110 | 6.1 |
| 200 | 223 | 111.5 | 4.21 |
| 300 | 272 | 90.6 | 5.33 |
Conclusion
In conclusion, our study has successfully developed an innovative nanozymatic-based competitive bio-assay utilizing peroxidase-mimicking Prussian blue nanoparticles for the picomolar-level colorimetric detection of microRNA-141. This novel approach offers significant advantages in terms of sensitivity, short analysis time, and a remarkably low detection limit, making it highly suitable for clinical applications. The PBNPs show promising potential for utilization in competitive bio-assay due to these favorable characteristics. Furthermore, the ability of this method to effectively detect a low concentration of miRNA-141 biomarker in artificial human serum samples at 100 pM demonstrates its practicality and potential for real-world diagnostic scenarios. Meanwhile, developed enzyme-based competitive bio-assays possess an established protocol and might exhibit higher precision. The benefits of PBNPs, particularly in challenging working conditions, position them as a compelling alternative for the colorimetric detection of miRNAs in competitive RNA–RNA systems.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
This research was funded by the Moroccan Ministry of Higher Education, Scientific Research and Innovation and the OCP Foundation “APRD research program 2021”. The authors acknowledge the Moroccan Ministry of Higher Education, Scientific Research and Innovation and the OCP Foundation “APRD research program 2021”.References
- F. Bray, M. Laversanne, E. Weiderpass and I. Soerjomataram, The Ever-increasing Importance of Cancer as a Leading Cause of Premature Death Worldwide, Cancer, 2021, 127(16), 3029–3030 CrossRef.
- E. Mahase, Cancer overtakes CVD to become the leading cause of death in high-income countries, BMJ, 2019, 366, l5368 CrossRef.
- R. L. Siegel, K. D. Miller, H. E. Fuchs and A. Jemal, Cancer Statistics, 2021, Ca-Cancer J. Clin., 2021, 71(1), 7–33 CrossRef.
- G. Carioli, P. Bertuccio, P. Boffetta, F. Levi, C. La Vecchia, E. Negri and M. Malvezzi, European Cancer Mortality Predictions for the Year 2020 with a Focus on Prostate Cancer, Ann. Oncol., 2020, 31(5), 650–658 CrossRef CAS.
- F. De Silva and J. Alcorn, A Tale of Two Cancers: A Current Concise Overview of Breast and Prostate Cancer, Cancers, 2022, 14(12), 2954 CrossRef CAS.
- N. Bargahi, S. Ghasemali, S. Jahandar-Lashaki and A. Nazari, Recent Advances for Cancer Detection and Treatment by Microfluidic Technology, Review and Update, Biol. Proced. Online, 2022, 24(1), 1–20 CrossRef.
- D. Crosby, S. Bhatia, K. M. Brindle, L. M. Coussens, C. Dive, M. Emberton, S. Esener, R. C. Fitzgerald, S. S. Gambhir and P. Kuhn, Early Detection of Cancer, Science, 2022, 375(6586), eaay9040 CrossRef CAS.
- N. Arrighetti and G. L. Beretta, Mirnas as Therapeutic Tools and Biomarkers for Prostate Cancer, Pharmaceutics, 2021, 13(3), 380 CrossRef CAS.
- S. Zhang, C. Liu, X. Zou, X. Geng, X. Zhou, X. Fan, D. Zhu, H. Zhang and W. Zhu, MicroRNA Panel in Serum Reveals Novel Diagnostic Biomarkers for Prostate Cancer, PeerJ, 2021, 9, e11441 CrossRef.
- H. V. Tran and B. Piro, Recent Trends in Application of Nanomaterials for the Development of Electrochemical microRNA Biosensors, Microchim. Acta, 2021, 188(4), 128 CrossRef CAS.
- P. Porzycki, E. Ciszkowicz, M. Semik and M. Tyrka, Combination of Three miRNA(miR-141, miR-21, and miR-375) as Potential Diagnostic Tool for Prostate Cancer Recognition, Int. Urol. Nephrol., 2018, 50, 1619–1626 CrossRef CAS.
- Z. Li, Y.-Y. Ma, J. Wang, X.-F. Zeng, R. Li, W. Kang and X.-K. Hao, Exosomal microRNA-141 Is Upregulated in the Serum of Prostate Cancer Patients, OncoTargets Ther., 2015, 139–148 CAS.
- J. Xiao, A.-Y. Gong, A. N. Eischeid, D. Chen, C. Deng, C. Y. Young and X.-M. Chen, miR-141 Modulates Androgen Receptor Transcriptional Activity in Human Prostate Cancer Cells through Targeting the Small Heterodimer Partner Protein, Prostate, 2012, 72(14), 1514–1522 CrossRef CAS.
- Y.-X. Chen, K.-J. Huang and K.-X. Niu, Recent Advances in Signal Amplification Strategy Based on Oligonucleotide and Nanomaterials for microRNA Detection-a Review, Biosens. Bioelectron., 2018, 99, 612–624 CrossRef CAS.
- Y. Zhang, P. Yang and X.-F. Wang, Microenvironmental Regulation of Cancer Metastasis by miRNAs, Trends Cell Biol., 2014, 24(3), 153–160 CrossRef CAS.
- M. El Aamri, H. Mohammadi and A. Amine, A Highly Sensitive Colorimetric DNA Sensor for MicroRNA-155 Detection: Leveraging the Peroxidase-like Activity of Copper Nanoparticles in a Double Amplification Strategy, Microchim. Acta, 2024, 191(1), 32 CrossRef CAS.
- M. El Aamri, R. Zayani, S. Baachaoui, H. Mohammadi, A. Amine and N. Raouafi, An Ingenious Double-Amplified Fluorometric Genosensor Based on Hybridization Chain Reaction and Enzymatic Signal for the Sensitive Detection of piRNA–651, Sens. Actuators, B, 2024, 399, 134749 CrossRef CAS.
- A. Markou, E. G. Tsaroucha, L. Kaklamanis, M. Fotinou, V. Georgoulias and E. S. Lianidou, Prognostic Value of Mature microRNA-21 and microRNA-205 Overexpression in Non–Small Cell Lung Cancer by Quantitative Real-Time RT-PCR, Clin. Chem., 2008, 54(10), 1696–1704 CrossRef CAS.
- M. El Aamri, H. Mohammadi and A. Amine, Development of a Novel Electrochemical Sensor Based on Functionalized Carbon Black for the Detection of Guanine Released from DNA Hydrolysis, Electroanalysis, 2023, 35(1), e202100613 CrossRef CAS.
- M. E. Aamri, H. Mohammadi and A. Amine, Paper-Based Colorimetric Detection of miRNA-21 Using Pre-Activated Nylon Membrane and Peroxidase-Mimetic Activity of Cysteamine-Capped Gold Nanoparticles, Biosensors, 2023, 13(1), 74, DOI:10.3390/bios13010074.
- M. El Aamri, Y. Khalki, H. Mohammadi and A. Amine, Development of an Innovative Colorimetric DNA Biosensor Based on Sugar Measurement, Biosensors, 2023, 13(9), 853 CrossRef CAS.
- M. Zouari, S. Campuzano, J. M. Pingarrón and N. Raouafi, Competitive RNA-RNA Hybridization-Based Integrated Nanostructured-Disposable Electrode for Highly Sensitive Determination of miRNAs in Cancer Cells, Biosens. Bioelectron., 2017, 91, 40–45 CrossRef CAS.
- H. Kutluk, R. Bruch, G. A. Urban and C. Dincer, Impact of Assay Format on miRNA Sensing: Electrochemical Microfluidic Biosensor for miRNA-197 Detection, Biosens. Bioelectron., 2020, 148, 111824 CrossRef CAS.
- E. Vargas, E. Povedano, V. R.-V. Montiel, R. M. Torrente-Rodríguez, M. Zouari, J. J. Montoya, N. Raouafi, S. Campuzano and J. M. Pingarrón, Single-Step Incubation Determination of miRNAs in Cancer Cells Using an Amperometric Biosensor Based on Competitive Hybridization onto Magnetic Beads, Sensors, 2018, 18(3), 863 CrossRef.
- H. Wang, K. Wan and X. Shi, Recent Advances in Nanozyme Research, Adv. Mater., 2019, 31(45), 1805368 CrossRef CAS.
- A. Robert and B. Meunier, How to Define a Nanozyme, ACS Nano, 2022, 16(5), 6956–6959 CrossRef CAS.
- J. Wu, W. Lv, Q. Yang, H. Li and F. Li, Label-Free Homogeneous Electrochemical Detection of MicroRNA Based on Target-Induced Anti-Shielding against the Catalytic Activity of Two-Dimension Nanozyme, Biosens. Bioelectron., 2021, 171, 112707 CrossRef CAS.
- Y. Li, C. Zhang, Y. He, J. Gao, W. Li, L. Cheng, F. Sun, P. Xia and Q. Wang, A Generic and Non-Enzymatic Electrochemical Biosensor Integrated Molecular Beacon-like Catalyzed Hairpin Assembly Circuit with MOF@ Au@ G-Triplex/Hemin Nanozyme for Ultrasensitive Detection of miR-721, Biosens. Bioelectron., 2022, 203, 114051 CrossRef CAS.
- L. Tian, J. Qi, O. Oderinde, C. Yao, W. Song and Y. Wang, Planar Intercalated Copper(II) Complex Molecule as Small Molecule Enzyme Mimic Combined with Fe3O4 Nanozyme for Bienzyme Synergistic Catalysis Applied to the microRNA Biosensor, Biosens. Bioelectron., 2018, 110, 110–117 CrossRef CAS.
- Y. Liu, Y. Zheng, D. Ding and R. Guo, Switching Peroxidase-Mimic Activity of Protein Stabilized Platinum Nanozymes by Sulfide Ions: Substrate Dependence, Mechanism, and Detection, Langmuir, 2017, 33(48), 13811–13820 CrossRef CAS.
- Y. Huang, Z. Liu, C. Liu, E. Ju, Y. Zhang, J. Ren and X. Qu, Self-assembly of Multi-nanozymes to Mimic an Intracellular Antioxidant Defense System, Am. Ethnol., 2016, 128(23), 6758–6762 Search PubMed.
- J. Golchin, K. Golchin, N. Alidadian, S. Ghaderi, S. Eslamkhah, M. Eslamkhah and A. Akbarzadeh, Nanozyme Applications in Biology and Medicine: An Overview, Artif. Cells, Nanomed., Biotechnol., 2017, 45(6), 1069–1076 CrossRef CAS.
- Z. Farka, V. Čunderlová, V. Horáčková, M. Pastucha, Z. Mikušová, A. Hlaváček and P. Skládal, Prussian Blue Nanoparticles as a Catalytic Label in a Sandwich Nanozyme-Linked Immunosorbent Assay, Anal. Chem., 2018, 90(3), 2348–2354, DOI:10.1021/acs.analchem.7b04883.
- J. Estelrich and M. A. Busquets, Prussian Blue: A Nanozyme with Versatile Catalytic Properties, Int. J. Mol. Sci., 2021, 22(11), 5993 CrossRef CAS.
- M. A. Komkova, E. E. Karyakina and A. A. Karyakin, Catalytically Synthesized Prussian Blue Nanoparticles Defeating Natural Enzyme Peroxidase, J. Am. Chem. Soc., 2018, 140(36), 11302–11307 CrossRef CAS.
- N. Seddaoui, R. Attaallah and A. Amine, Development of an Optical Immunoassay Based on Peroxidase-Mimicking Prussian Blue Nanoparticles and a Label-Free Electrochemical Immunosensor for Accurate and Sensitive Quantification of Milk Species Adulteration, Microchim. Acta, 2022, 189(5), 209 CrossRef CAS.
- M. Gautam, K. Poudel, C. S. Yong and J. O. Kim, Prussian Blue Nanoparticles: Synthesis, Surface Modification, and Application in Cancer Treatment, Int. J. Pharm., 2018, 549(1–2), 31–49 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00187g |
| This journal is © The Royal Society of Chemistry 2024 |