 Open Access Article
Open Access ArticleApplication of surface selective site-directed crystallization in a visual assay of DNA†
Jinrong
Chen
ab,
Ruwen
Xie
a,
Rui
Liu
 b,
Lishang
Liu
b,
Lishang
Liu
 *a and
Shusheng
Zhang
*a and
Shusheng
Zhang
 *a
*a
aShandong Province Key Laboratory of Detection Technology of Tumor Markers, Linyi University, Linyi 276005, China. E-mail: liulishanghao@126.com; shushzhang@126.com
bDepartment of Biotechnology, College of Engineering, The University of Suwon, Hwaseong 18323, Korea
First published on 17th October 2024
Abstract
Visual analysis methods have received widespread attention due to their simplicity, economy, and intuitive results. In this work, a visual DNA quantitative analysis method based on surface selective site-directed crystallization (SSSC) was developed. Firstly, we explored the formation of calcium carbonate crystals with unique polymorphism induced by the surface of functionalized glass slides with different groups; among them, the calcite induced by the –COOH functional group has a uniform shape, larger size, and even distribution, so it serves as a signal promoter. In contrast, due to the –N(CH3)3 group acting as a signal inhibitory molecule by inhibiting crystallization, the signal molecule is captured through DNA hybridization, and the crystallization reaction is performed. The calcite growing on the DNA site is visible to the naked eye, and the DNA molecules hybridized on the surface of the glass slide are further quantified. The detection limit of this proposed visual method is 0.1 fM, and only a smartphone is needed to complete basic quantification. This work provides a basis for research into the use of single crystals as digital readouts in the field of DNA analysis, with the advantages of being simple and economical and requiring minimal equipment.
Introduction
Nucleic acid detection has become a key technology in areas ranging from disease diagnosis to environmental assays.1–3 In the past decade, numerous detection platforms based on colorimetric, fluorescent,4–6 Raman,7 magnetic,8,9 motional10 or electrochemical transducers11 have been developed to convert nucleic acid hybridization events into various kinds of signals. However, it remains challenging to quantify DNA molecules without costly and sophisticated instruments. Lateral flow assays have been translated from dedicated laboratory instruments into simple paper-based systems, with limited sensitivity and poor quantitative performance.2 Visible counting strategies have received considerable attention; however, amplification-free and femtomolar sensitivity assays were only achieved by single-molecule fluorescence detection, which requires a sophisticated optical system.4Digital counting strategies were developed but suffered from low sampling efficiency, complicated fabrication, and expensive equipment.12 A variety of probes have been utilized for visible counting strategies such as bubbles, beads, and microorganisms.13–15 Zhou et al. utilized T7 phages (size: ∼5 μm) assembled with gold nanoparticles (GNPs) in a ‘one-to-one’ manner as visible probes to count the virus numbers with the naked eye.16 They also attempted to quantify antigen molecules by counting gold nanoparticles (size: ∼80 nm) assisted by scanning electron microscopy (SEM) manipulation.17 Shlyapnikov et al. presented a microarray-based assay of DNA fragments using magnetic beads (size: ∼1 μm) as visible signals to quantify the DNA with an electrophoresis apparatus.18 Nam et al. utilized polyethyleneimine (PEI)-reduced Cu particles (size: ∼100 nm) on GNPs as an amplified signal to quantify DNA molecules.19 Tekin et al. tried to label protein molecules with micro-sized magnetic beads (size: ∼480 nm) based on dipole–dipole-assisted interaction.20 Tam et al. attempted to increase the size of AuNPs in situ after labeling protein molecules.21 Wang et al. designed a method to capture the target proteins using antigen-labeled paramagnetic microspheres, which were further combined with PtNPs to produce quantifiable microbubbles (size: >35 μm) in response to H2O2.22 Transferring the invisible biomolecule information into visual probes, which are directly countable, is the most cost-effective method. However, it's challenging to directly label biomolecules with visible particles.
Surface-induced selective crystallization has been studied and applied in drug separation and purification, polymorphism discrimination, and rare earth element separation.23 In template-induced nucleation, crystal growth shifts molecular recognition to the nucleation stage. In our previous work, the selective crystallization of CaCO3 at specific sites was analyzed by quartz crystal microbalance (QCM).24,25 The bulk crystals generated on the surface of the QCM sensor act as amplifiers, clearly improving the sensitivity of the mass sensor. Crystals can be quantitatively controlled by tightly controlling the ratio of inhibitor and promoter groups. In fact, the reduction of silver ions to silver in the work of Zhou et al. is similar.1 Group-induced crystals have high potential for application as quantitative molecular indicators.
To demonstrate this capability, in this work, we propose a DNA hybridization assay method based on surface selective site-directed crystallization (SSSC) as shown in Scheme 1. First, the selective crystallization of different forms of CaCO3 crystals is realized on the surface of functionalized glass slides, and –N(CH3)3 is determined to be the signal inhibitory group and –COOH as the signal promoter. Capture DNA (cDNA) is introduced on the surface of the glass slide. When the target DNA (tDNA) is present, it can hybridize with the DNA labeled by the signal promoter (pDNA) and promote the site-specific crystallization of calcite. The target molecules are visualized and detected by observing the crystals, while quantitative detection is achieved by counting calcite crystals using simple optics and smartphones. Therefore, the DNA hybridization strategy proves the selective crystallization and in situ crystallization ability of crystals as quantitative analysis probes. The method is simple to operate and does not require major instruments, and the results are visible to the naked eye. This work uses single crystals as quantitative signals for analysis and detection, providing new ideas for analysis and research workers.
Materials and methods
Reagents and equipment
Bacillus anthrax (BA) human immunodeficiency virus (HIV) and hepatitis B virus (HBV) DNA oligonucleotides were from Sangon Biotech, and they are listed in Table S1.† Glass slides (25 mm × 75 mm) were from Thermo Scientific. HS(CH2)15COOH, HS(CH2)11OH, HS(CH2)11CH3, and HS(CH2)11NH2 were purchased from Sigma-Aldrich. HS(CH2)11N(CH3)3Cl was purchased from Prochimia. CaCl2·2H2O, (NH4)2CO3, tris(2-carboxyethyl) phosphine hydrochloride (TCEP) and bovine serum albumin (BSA) were purchased from Aladdin. Absolute ethanol, sulfuric acid, ammonia solution, and dimethyl sulfoxide were purchased from Shanghai Chemical Reagent Co. Ltd. Hydrogen peroxide and acetic acid were purchased from Sinopharm Chemical Reagent Co. Ltd. 3-(2-Aminoethylamino)propyltrimethoxysilane (EDA) was purchased from Alfa Aesar Co. Succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC) was purchased from Shanghai MERYER Co. All aqueous solutions were prepared using ultrapure water (Milli-Q, Millipore).CaCO3 crystals were characterized by SEM (JSM-7610F), Raman (Renishaw), X-ray diffraction, and optical microscopy (ECLIPSE Ni-U). Confocal images were obtained using a Nikon 6CR609WPQV. A 20× to 200× universal tipscope was purchased from TMALL. A 20× to 400× tipscope was purchased from Jing Dong.
Preparation of glass glides
Glass slides were first treated with a freshly made piranha solution (hydrogen peroxide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) concentrated sulfuric acid = 3
concentrated sulfuric acid = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) at 90 °C for 2 h and thoroughly rinsed with deionized water. The glass slides were allowed to react with EDA in an acetic acid solution (EDA
1, v/v) at 90 °C for 2 h and thoroughly rinsed with deionized water. The glass slides were allowed to react with EDA in an acetic acid solution (EDA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mM acetic acid = 1
1 mM acetic acid = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, v/v) at room temperature for 30 min, followed by washing three times in deionized water, drying with nitrogen, and then baking at 120 °C for 30 min. The glass slides were immersed in a 5 mM DMSO
100, v/v) at room temperature for 30 min, followed by washing three times in deionized water, drying with nitrogen, and then baking at 120 °C for 30 min. The glass slides were immersed in a 5 mM DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethanol (2
ethanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v) solution of a bifunctional coupling reagent, SMCC, overnight and then were washed in ethanol and water, and dried with nitrogen. Functional groups were formed on glass slides (Fig. S1†) by exposing the surfaces to a 10 mM solution of thiol in ethanol for 12 hours, followed by washing with ethanol and drying with nitrogen gas.
3, v/v) solution of a bifunctional coupling reagent, SMCC, overnight and then were washed in ethanol and water, and dried with nitrogen. Functional groups were formed on glass slides (Fig. S1†) by exposing the surfaces to a 10 mM solution of thiol in ethanol for 12 hours, followed by washing with ethanol and drying with nitrogen gas.
Preparation of the hybridization sandwich
The cDNA-immobilized glass slides were obtained by spotting 5 μL of 1 nM cDNA solution (buffer: 0.02 M PBS, 0.25 M NaCl, pH 7.4) for 8 hours at 37 °C, followed by blocking with 10 mM HS-C11–N(CH3)3Cl for 12 hours and rinsing three times with a washing buffer (1 × SSC + 0.1% (v/v) SDS). The prepared glass slides were stored at 4 °C. tDNA of varied concentrations (5 μL solution with buffer: 0.02 M PBS, 0.25 M NaCl, pH 7.4) was hybridized with cDNA immobilized on the surface of the slides. Further hybridization with pDNA (5 μL, 1 nM) was carried out. After each step, the slide was washed in a washing buffer and deionized water to remove non-hybridization DNA, and then dried with nitrogen gas.Promoter optimization
SMCC treated glass slides were exposed to a 10 mM thiol solution (HS(CH2)15COOH, HS(CH2)11OH, HS(CH2)11NH2, or HS(CH2)11N(CH3)3Cl) in ethanol for 12 hours, followed by washing with ethanol and drying with nitrogen gas. The prepared glass slides were placed upside down and the crystallization experiment was conducted.Inhibitor optimization
SMCC treated glass slides were first modified with cDNA (100 nM, 10 nM, 1 nM, 100 pM, 10 pM, or 1 pM) and then immersed in an inhibitor solution (10 mM HS(CH2)11CH3, 10 mM HS(CH2)11N(CH3)3Cl, or 1% BSA) for 12 hours, followed by washing in ethanol and drying with nitrogen gas. The prepared glass slides were placed upside down and the crystallization experiment was conducted.Crystallization of CaCO3 on the glass slide surface
The prepared glass slides were placed upside down, emerged in calcium chloride solution, surrounded with solid ammonium carbonate in a beaker, then sealed and reacted at room temperature. Finally, the slides were washed in deionized water and dried with nitrogen gas.The crystals on the glass slides were observed by optical microscopy and SEM, and the morphology, number, and size of the crystals on the glass surface were recorded. The crystals on the glass slides were characterized by Raman and XRD to confirm their polymorph.
Calibration of the SSSC assay
After hybridization, the glass slides were placed upside down in a flask containing 3 mM calcium chloride solution, surrounded with solid ammonium carbonate. The reaction device was sealed and placed under an optical microscope (20×) to observe and record the crystallization process in real time.The crystallized glass slide was photographed on a black background with a smartphone using an attached tipscope (microscope). The crystals were counted using Image J or the “Count Things” application on the phone. The CV of the crystal number against counting area (take the result of 1 pM tDNA as an example) was analyzed.
The specificity of the DNA assay
Solutions of 1 nM target BA sequence, HIV, HVB, and random sequences were tested by the SSSC assay.Results and discussion
Rationale of the SSSC assay method
Templated heterogeneous nucleation is driven by the characteristics of the templates, including topography, surface functionality, and lattice matching between the crystal and substrate, favoring the directional growth of the crystal. In this work, a glass slide was used as a substrate for the perspective disposal device. The site-specific crystallization was carried out on a glass slide surface for perspective disposal devices. Selective crystallization of CaCO3 on self-assembled monolayer (SAM) modified gold surfaces has been researched;26 however, that on glass slides has rarely been reported even though glass slides have commonly been applied in biological imaging.27,28 Therefore, the modification of the glass slide surface was optimized to select the best inhibitor and promoter for the site-specific crystallization and finally to guarantee the uniformity and discreteness of the generated crystals.In the DNA hybridization assay, as shown in Scheme 1, after immobilization of capture DNA (cDNA) on the slide surface, a layer of inhibiting group was introduced to prevent nonspecific crystallization. Upon the hybridization of the target DNA, probe DNA (pDNA) with a promoter group hybridized further as a specific site for crystallization. The in situ crystallization reaction was carried out by placing glass slides inversely in calcium chloride solution with gas diffusion. The inversion of glass slides can avoid nonspecific crystal settling. Therefore, only in the presence of target DNA can the crystals grow and be visualized. Then, the visual detection of the target can be achieved, and its quantification can be further achieved by crystal counting.
Surface induced acceleration of CaCO3 crystallization
CaCO3 crystallizes in three polymorphs: calcite, aragonite, and vaterite. However, the polymorphic control of CaCO3 crystals was not deeply investigated. Studies of templated CaCO3 nucleation on metal SAM surfaces concluded that –COOH, –PO32−, and –OH groups were active in facilitating nucleation, while –N(CH3)3+ and –CH3 inhibited nucleation. The type of nucleation of CaCO3 induced by –COOH and –OH was oriented, while that induced by –PO32− was random.29 As –NH2 is also a common label in DNA sensing, to select the best crystal candidate as a visual probe for site-specific CaCO3 nucleation, we tested glass slides modified with –COOH, –NH2, –OH functional groups and analyzed the polymorph, morphology, and size distribution of the generated crystals. The morphological results were shown by SEM (Fig. 1) and their polymorphism was demonstrated by XRD and Raman (Fig. S2 and S3†) and the optical microscopy images in Fig. S4.† The polymorphic fraction and coefficient of variation (CV) of size distribution are summarized in Table 1. Rhombohedral and pumpkin-shaped crystals with random size were observed on the –NH2 terminated slide surface and proved to be calcite and vaterite, respectively, with a CV of the crystal size of ∼60%.| Surface group | Proportion of calcite | Proportion of vaterite | Number of crystals | Crystal size (μm) | CV of crystal size |
|---|---|---|---|---|---|
| –NH2 | 86% | 14% | ∼80 | 5–30 | 60% |
| –COOH | 100% | 0 | ∼120 | 10–25 | 19% |
| –OH (tc: 1 h) | 8% | 92% | ∼80 | 5–15 | 43% |
| –OH (tc: 6 h) | 16% | 84% | ∼70 | 10–20 | 32% |
| –OH (tc: 12 h) | 15% | 85% | ∼60 | 5–20 | 30% |
| –OH (tc: 24 h) | 11% | 89% | ∼50 | 5–50 | 32% |
Cubic calcite crystals were exclusively obtained on the –COOH terminated surface. Their strong and sharp XRD peaks indexed to the (104) plane (Fig. S2†) confirmed the excellent crystallization of calcites. All the calcite crystals on the –COOH terminated surface were found to be “standing” on the slide surface with a uniform orientation. Significantly, the calcite crystals were discretely distributed with a unified cubic morphology and unified size (coefficient of variation (CV) ∼ 19%), and even with a unified orientation. The negatively charged –COOH group has been reported to facilitate the crystallization of calcite and influence its growth direction.30 The main driving force for the orientation of calcite on the –COOH terminated surface is believed to be the strong interaction between the deprotonate carboxyl group and calcium ions, providing a high local supersaturation microenvironment.
The crystals generated on the –OH modified glass were more complicated. Crystals with random shapes were observed and were dominant at different crystallization times: hexagonal plate, pumpkin, hexagonal lotus, and flower. They were demonstrated to be vaterite crystals with characteristic XRD peaks of the (110), (112), (114), (104), (300), and (224) planes (Fig. S2†). The calcite crystals on the –COOH terminated surface with a unified morphology, size and orientation showed satisfactory characteristics and were believed to be an ideal candidate as a visual probe. Therefore the –COOH group was determined as the promoter group for site-specific crystallization in visual detection of DNA.
Surface inhibition of determination of CaCO3 crystallization
For DNA hybridization on the substrate surface, non-specific adsorption is always prevented by further surface modification after cDNA immobilization. Functional groups such as –N(CH3)3, –CH3 and –OH and bovine serum albumin (BSA) have also been reported to work as blockers of non-specific nucleic acid, protein, and other biomolecules.29,31 In this work, the blocking of both non-specific nucleic acid and unwanted crystals is critical. The SEM pictures in Fig. 1 showed that the –OH group induced vaterite crystal growth; thus, the –OH group is not a suitable inhibitor candidate. –N(CH3)3, –CH3, and BSA were further modified on a cDNA immobilized slide surface and then a crystallization experiment was introduced on the slide surface. The results are shown in Fig. 2; the –N(CH3)3 group had the best inhibitory effect probably due to repulsive forces against Ca2+ ions, while –CH3 and BSA did not inhibit the crystallization on the cDNA immobilized slide surface. No crystal was found on the –N(CH3)3 terminated surface even with a high cDNA concentration (10 nM). Therefore, the –N(CH3)3 group was selected and used as an inhibitor on the slide surface preventing non-specific nucleation.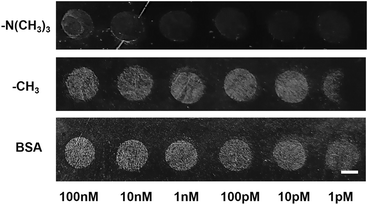 | ||
| Fig. 2 Inhibition effect of –N(CH3)3, –CH3 and BSA on surface crystallization cDNA concentration: 100 nM, 10 nM, 1 nM, 100 pM, 10 pM, and 1 pM. Scale bar: 2 mm. | ||
In addition, QCM was used to verify whether fixing the inhibitory group and increasing the concentration of the carboxyl group could achieve the regulation of the crystal yield (Fig. S5†). The experimental results show that with the increase of carboxyl group concentration, the number of crystals increases and the QCM signal decreases, and the concentration of the carboxyl group is linearly related to the crystal quality signal. This result proves that precise crystal control can be achieved by controlling the amounts of functional groups.
Application of selective crystallization on DNA hybridized sites
Using the DNA sandwich hybridization shown in Scheme 1, –N(CH3)3 as the crystallization inhibitor and COOH as the crystallization promoter, the feasibility of the SSSC DNA assay method was tested. As designed, if no target exists, surface nucleation of CaCO3 would be inhibited by the –N(CH3)3 group, and a clear surface would be observed; if target DNA exists, the –COOH group on pDNA would trigger the site-specific crystallization and a certain amount of calcite crystals would be observed.The inhibitive group –N(CH3)3 and the promoter group –COOH in this design scheme are competitive and balancing in the crystallization process. To achieve the best DNA hybridization differentiation within a certain range, we used Bacillus anthracis DNA analysis as a model and optimized the experimental conditions for the SSSC method. The temperature of the DNA hybridization reaction on the glass sheet, the concentration and the time of the crystallization reaction were optimized. According to the results (Fig. S6–S8†), the DNA hybridization temperature in the subsequent SSSC DNA detection method was determined to be 45 °C, the Ca2+ concentration and crystallization time were 3 mM Ca2+ and 1 h, and the target DNA concentration ranged from 1 nm to 1 fM. At the same time, we also used fluorescence labelled DNA to verify the feasibility of the hybridization reaction on the glass slide. As shown in Fig. S9,† laser confocal microscopy can photograph fluorescently labelled DNA modified on the glass slide, which proves that the hybridization reaction on the glass slide is successful, but this result can only detect DNA of 1 μM.
The site-specific crystallization process on the DNA (100 fM) hybridized surface was observed in situ using optical microscopy (Fig. 3a). Before nucleation took place, the slide surface was clear. As time went by, tiny dots started to appear at the 25th min, and then crystals were found to grow at the same spots from the 30th min to 60th min, only with size increasing. The real time site-specific crystallization was recorded in a video (Video S1 in the ESI†). Together with the crystal size plot over time in Fig. 3b, it was clear that new crystals were not generated after the 36th min, and only an increase in the size of existing crystals was observed. These results demonstrated that the crystals are generated by preferred site-specific heterogeneous nucleation, not by random growth, nor by the attachment of crystals from the bulk solution. The size of the crystals is increased gradually to ∼40 μm in an hour. According to this result, as visual signals, calcites can readily be observed and quantified.
The cooperation of the inhibitor and promoter guaranteed the selective nucleation of crystals. The SEM pictures of crystals with increasing DNA concentration are shown in Fig. 4. These pictures and XRD analysis patterns (Fig. S10†) again confirmed that the CaCO3 crystals obtained are discrete and uniform in size and morphology, indicating that the nucleation is highly restricted. The binding of Ca2+ ions to –COOH groups can increase the nucleation rate of CaCO3 crystals by increasing the local supersaturation ratio of CaCO3 molecules. The –COOH here works as a “spear” to catch Ca2+ ions, and it also plays a key role in the regulation of polymorphism, morphology, size, and direction of the crystal. The obtained calcite crystals show perfect discreteness and can be easily enumerated. These properties make calcite crystals ideal quantitative probes, so it is feasible to transfer the detection of DNA hybridization by calcite crystal counting schemes.
Calibration of the crystal amount induced by DNA hybridization
The results of the analysis of different concentrations of tDNA are shown in Fig. 5a. Next, crystals of DNA sites were quantified using a smartphone. A universal tipscope was attached to a smartphone to take photos (Fig. 5b and c and S11†). The pictures in Fig. 5d were sent to Image J to count the number of calcite crystals. To explore the effect of the sampling area on the counting results, the coefficient of variation (CV) of the number of crystals is plotted against the area in Fig. S12.† As the detection area increased, the CV decreased, so we counted the crystals in the entire puncta area to reduce sampling error. To achieve quantitative analysis more easily, the smartphone app “Count Things” was also used to count the crystals. Examples of smartphone application interfaces are shown in Fig. S13.† Similar enumeration results were obtained using ImageJ and “Count Things” (Fig. S14†). The numbers of crystals were counted over three repeats of results of experiments and found to be positively correlated with the concentration of target DNA, as shown in Fig. 5e. The linear regression follows the equation Y = 122.5X + 1739.4 with an R2 of 0.98. The linear range of the SSSC DNA assay method is from 1 nM to 10 fM, and the limit of detection (LOD) is calculated to be 0.1 fM.Finally, we investigated the specificity and reproducibility of the method. To evaluate the reproducibility of our method, we analyzed the CV of the calcite number in the SSSC DNA assay. It is summarized in Table S2,† with an average value as small as 11%, indicating that the reproducibility of this method is good. The specificity of our approach was investigated using several different DNA sequences (Fig. 6). It could be seen that 1 nm BA DNA and mixed sequences can produce strong signals, while the other DNA sequences (HIV, HVB, and random sequence) cannot show significant signals. This indicates that the method has good specificity.
 | ||
| Fig. 6 The specificity of the SSSC DNA assay method (the concentration of sequences: 1 nM, mean ± standard deviation; n = 3). | ||
Conclusions
In conclusion, this study established a visual DNA quantification method by surface-selective site-specific crystallization and counting calcite crystals. DNA site-induced calcite crystals have been shown for the first time to serve as signal probes to amplify the microscopic world. The crystals obtained were all calcite, and the uniformity of their polymorphic forms and growth directions suggested that they were induced specifically by hybridization of the target DNA rather than randomly.The single calcite crystals crystallized at the site are uniform in size, shape, and distribution. Compared with the reported visible probes such as magnetic beads, bacteriophages, copper particles, and bubbles, calcite crystals have the advantages of low cost and high stability, making them ideal visual digital probes. Compared with previously reported studies also based on visual particle counting (Table 2), the calcite crystal counting method proposed in this work demonstrated a wide linear range without the aid of any professional instruments. Notably, quantitative analysis can be achieved using a smartphone in conjunction with a portable microscope. This method has the advantages of simple operation and no need for amplification, which opens a new route for the design of visual sensors.
| Detection method | Target | Signal | Particle size | Linear range | LOD | Instrument | Ref. |
|---|---|---|---|---|---|---|---|
| Magnetic bead surface coverage assay | Protein | Magnetic beads | 2.8 μm | 1–1 × 108 pg mL−1 | 1 pg mL−1 | Required | 20 |
| Phage-mediated counting | miRNA | Phages | ∼1 μm | 0.2–0.01 fM | 0.003–0.005 fM | Required | 16 |
| Multiplex microarray-based assay | DNA | Magnetic beads | 1 μm | 1000–0.1 fM | 0.1 fM | Required | 18 |
| Simple microbubbling digital assay | Protein | Microbubbles | 35–100 μm | 0.06–1 pg mL−1 | 0.06 pg mL−1 | Not required | 22 |
| Dropcast single-molecule assays | Protein | Paramagnetic beads | 2.7 μm | 10–3 – 102 fM | 0.0192 fM | Required | 12 |
| Pre-equilibrium digital ELISA | Protein | Magnetic beads | ∼4 μm | 40–1000 pg mL−1 | 25.9 pg mL−1 | Required | 32 |
| SSCV | DNA | Calcite | 20–40 μm | 1 nM–1 fM | 0.1 fM | Not required | This work |
Data availability
The data supporting this study's findings are available from the corresponding author upon reasonable request.Author contributions
The manuscript was written through the contributions of all authors. All authors have approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21675076, 81502585, 22076073).Notes and references
- L. Yan, J. Zhou, Y. Zheng, A. S. Gamson, B. T. Roembke, S. Nakayama and H. O. Sintim, Mol. BioSyst., 2014, 10, 970–1003 RSC.
- M. Zhang, J. Ye, J. S. He, F. Zhang, J. Ping, C. Qian and J. Wu, Anal. Chim. Acta, 2020, 1099, 1–15 CrossRef CAS.
- L. Zhang, B. Ding, Q. Chen, Q. Feng, L. Lin and J. Sun, TrAC, Trends Anal. Chem., 2017, 94, 106–116 CrossRef CAS.
- M. Di Antonio, A. Ponjavic, A. Radzevicius, R. T. Ranasinghe, M. Catalano, X. Zhang, J. Shen, L. M. Needham, S. F. Lee, D. Klenerman and S. Balasubramanian, Nat. Chem., 2020, 12, 832–837 CrossRef CAS PubMed.
- J. H. Hwang, S. Park, J. Son, J. W. Park and J. M. Nam, Nano Lett., 2021, 21, 2132–2140 CrossRef CAS.
- A. S. Law, L. C. Lee, K. K. Lo and V. W. Yam, J. Am. Chem. Soc., 2021, 143, 5396–5405 CrossRef CAS PubMed.
- F. Gao, J. Lei and H. Ju, Anal. Chem., 2013, 85, 11788–11793 CrossRef CAS.
- Z. Zhou, Y. Zhang, M. Guo, K. Huang and W. Xu, Biosens. Bioelectron., 2020, 167, 112475 CrossRef CAS.
- M. Bao, E. Jensen, Y. Chang, G. Korensky and K. Du, ACS Appl. Mater. Interfaces, 2020, 12, 43435–43443 CrossRef CAS.
- X. Yu, Y. Li, J. Wu and H. Ju, Anal. Chem., 2014, 86, 4501–4507 CrossRef CAS PubMed.
- E. O. Blair and D. K. Corrigan, Biosens. Bioelectron., 2019, 134, 57–67 CrossRef CAS PubMed.
- C. Wu, P. M. Garden and D. R. Walt, J. Am. Chem. Soc., 2020, 142, 12314–12323 CrossRef CAS.
- M. G. M. Xiong Ding, K. Yin, K. Kadimisetty and C. Liu, Anal. Chem., 2018, 91, 655–672 CrossRef.
- C. M. S. Hsin-I Peng, K. E. Leach, T. D. Krauss and B. L. Miller, ACS Nano, 2009, 3, 2265–2273 CrossRef.
- H. Xu, J. Shen, C. T. Yang, B. Thierry, Y. Zhu, C. B. Mao and X. Zhou, Mater. Today Adv., 2021, 10, 100122 CrossRef CAS.
- X. Zhou, P. Cao, Y. Zhu, W. Lu, N. Gu and C. Mao, Nat. Mater., 2015, 14, 1058–1064 CrossRef CAS.
- X. Zhou, C. T. Yang, Q. Xu, Z. Lou, Z. Xu, B. Thierry and N. Gu, ACS Appl. Mater. Interfaces, 2019, 11, 6769–6776 CrossRef CAS.
- Y. M. Shlyapnikov, E. A. Malakhova and E. A. Shlyapnikova, Anal. Chem., 2019, 91, 11209–11214 CrossRef CAS PubMed.
- J. H. Kim, J. E. Park, M. Lin, S. Kim, G. H. Kim, S. Park, G. Ko and J. M. Nam, Adv. Mater., 2017, 29, 1702945 CrossRef.
- H. C. Tekin and M. Cornaglia, Lab Chip, 2013, 13, 1053–1059 RSC.
- S. C. Tam, Y. H. Cheng, C. N. Lok, H. Y. Au-Yeung, W. X. Ni, X. L. Wei and K. M. Ng, Analyst, 2020, 145, 6237–6242 RSC.
- H. Chen, Z. Li, L. Zhang, P. Sawaya, J. Shi and P. Wang, Angew. Chem., Int. Ed., 2019, 58, 13922–13928 CrossRef CAS PubMed.
- F. Artusio and R. Pisano, Int. J. Pharm., 2018, 547, 190–208 CrossRef CAS PubMed.
- L. S. Liu, C. Wu and S. Zhang, Anal. Chem., 2017, 89, 4309 CrossRef CAS PubMed.
- C. Wu, Z. Sun and L. S. Liu, Analyst, 2017, 142, 2547–2551 RSC.
- H. Deng, X. C. Shen, X. M. Wang and C. Du, Front. Mater. Sci., 2013, 7, 62–68 CrossRef.
- A. M. Travaille, L. Kaptijn, P. Verwer, B. Hulsken, J. A. A. W. Elemans, R. J. M. Nolte and H. van Kempen, J. Am. Chem. Soc., 2003, 125, 11571–11577 CrossRef CAS PubMed.
- H. Deng, X. M. Wang, C. Du, X. C. Shen and F.-Z. Cui, CrystEngComm, 2012, 14, 6647–6653 RSC.
- J. Aizenberg, A. J. Black and G. M. Whitesides, J. Am. Chem. Soc., 1999, 121, 4500–4509 CrossRef CAS.
- J. Aizenberg, Adv. Mater., 2004, 16, 1295–1302 CrossRef CAS.
- J. A. Y. J. Han, J. Am. Chem. Soc., 2003, 125, 4032–4033 CrossRef PubMed.
- Y. Song, E. Sandford, Y. Tian, Q. Yin, A. Kozminski, S. Su, T. Cai, Y. Ye, M. Chung and R. Lindstrom, The Journal of the American Society of Hematology, 2021, 137, 1591–1602 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00149d |
| This journal is © The Royal Society of Chemistry 2024 |





