Open Access Article
Open Access ArticleVitalOne™: a point-of-care platform for rapid, comprehensive, central-lab quality blood testing†
Mounir A.
Koussa
 *,
Miguel
Barreiros‡
,
Paul Said
Ehrlich Perez‡
,
Sae Rin
Jean‡
,
Taehyung Chris
Lee‡
,
Ross
MacLeod‡
,
Aaron
Witham‡
,
Geeta
Bhat
,
Todd
Campbell
,
Sergio
Lizano
,
Marjorie
Toth
,
Amrita
Venkateswaran
,
Don
Yang
,
Nishat
Zaman
,
Wisam
Alfaqheri
,
Afshan
Ardalan
,
Luis
Barbosa
,
Mehran
Behrouzi
,
Vitali
Borisenko
*,
Miguel
Barreiros‡
,
Paul Said
Ehrlich Perez‡
,
Sae Rin
Jean‡
,
Taehyung Chris
Lee‡
,
Ross
MacLeod‡
,
Aaron
Witham‡
,
Geeta
Bhat
,
Todd
Campbell
,
Sergio
Lizano
,
Marjorie
Toth
,
Amrita
Venkateswaran
,
Don
Yang
,
Nishat
Zaman
,
Wisam
Alfaqheri
,
Afshan
Ardalan
,
Luis
Barbosa
,
Mehran
Behrouzi
,
Vitali
Borisenko
 ,
Rohit
Chand
,
Karyn S.
Ho
,
Praveen
Kumar
,
Mate
Lengyel
,
Wei
Luo
,
Fahim
Masum
,
Laura
Piñeros
,
Akhil Rajagopal
Kozhipuram
,
Sergey
Sanders
,
David
Santos
,
Vasu
Nadella
,
Farnoud
Kazemzadeh
and
Iman
Khodadad
,
Rohit
Chand
,
Karyn S.
Ho
,
Praveen
Kumar
,
Mate
Lengyel
,
Wei
Luo
,
Fahim
Masum
,
Laura
Piñeros
,
Akhil Rajagopal
Kozhipuram
,
Sergey
Sanders
,
David
Santos
,
Vasu
Nadella
,
Farnoud
Kazemzadeh
and
Iman
Khodadad
Vital Biosciences, Unit 1 - 3165 Unity Drive, Mississauga, Ontario, Canada L5L 4L4. E-mail: mounir@vitalbio.com
First published on 11th November 2024
Abstract
Diagnostic tools are fundamental to informed healthcare decisions, but the current state of in vitro diagnostics presents barriers to providing timely access to a wide menu of tests. This paper discusses a method for overcoming these challenges with a novel and miniaturized point-of-care (PoC) solution, the VitalOne. While many PoC solutions have limited test menus tailored for specific scenarios, Vital Bio introduces a comprehensive test menu in a compact PoC format that uses centrifugal microfluidic workflows to deliver quantitative results across three modalities that have traditionally required three separate instruments. VitalOne has combined three modalities into a single instrument: hematology, clinical chemistry, and immunoassay. This breadth of central-lab quality results covers a wide variety of use cases and helps to eliminate the send-out gap that impedes the adoption of PoC technologies. This paper provides a comprehensive and transparent view of both our assay performance data and underlying methods of operation. By comparing the VitalOne system with established benchmarks commercialized devices including Roche cobas® c701, Sysmex 140, Beckman DxH® 500, Beckman AU® 480, and Beckman Access® 2 instrument systems, we demonstrate consistent assay performance, resilience to interference, and performance that is compliant with Clinical Laboratory Improvement Amendments (CLIA) total allowable error standards. Additionally, this article elucidates our system's operational intricacies and showcases performance data from a diverse set of analytes.
Introduction
Diagnostics play a pivotal role in healthcare, underpinning the majority of clinical decision-making processes.1–6 However, the current in vitro diagnostic landscape poses challenges in ensuring timely access to these tools, leading to suboptimal patient care.7 The traditional approach requires large-scale equipment operated by highly-trained personnel, restricting their use to centralized labs.3,8 For decades, this has often required patients to visit secondary sites for blood draws and in most cases, the sample is then shipped to the centralized tertiary site for assessment. It's noteworthy that nearly a third of all electronic lab orders are unfilled, a culmination of factors including patient non-adherence to lab testing requisitions and logistical challenges associated with patients securing time for an additional appointment during business hours and obtaining a means of transportation to a draw site.9–12These barriers lead to lengthy delays, missed diagnoses, missed opportunities for intervention, and ultimately poor patient outcomes.3,13 Healthcare professionals are also forced to spend much of their time tracing lab orders to patient encounter notes and reaching out to patients to coordinate follow ups.9 Limited access and frequency also make it difficult for people to take early preventative action with their health.
There is substantial potential in efficiently detecting and leveraging well-established biomarkers when and where they are needed most. Widespread adoption of point-of-care diagnostic solutions is held back by limited test menus that cater primarily to specific use cases such as emergency or operating room scenarios.14–16 While there are plenty of PoC devices on the market, none provide a comprehensive offering (Table 1). There are devices on the market that perform a single panel such as PixCell's HemoScreen™, Sight Dx's Sight OLO®, Hemocue® WBC DIFF (hematology), Abaxis's Piccolo®, Abbott's Afinion™ and Cholestech™ platforms, Thermo Scientific's Indiko™ Clinical Chemistry Analyzer, Horiba's Pentra™ C400 (clinical chemistry), and Genalyte's Maverick™ (immunoassay). Genalyte has recognized the value of multimodal testing solutions and has commercialized services in which a trained technician operates the Maverick system along with separate PoC clinical chemistry analyzer and hematology analyzers made by other companies. Other platforms such as LumiraDx™ and Abbott's i-STAT® can operate on multiple modalities (immunoassay and clinical chemistry) but can only measure a small number of analytes at a time (1–3 for LumiraDx, 1–11 for i-STAT) on cartridges that are run serially. These solutions do little to address the “send out gap”, left open when requisitions cannot be fully covered at the point of care. Lack of breadth in PoC test menus and results in entire orders being sent out for analysis because otherwise a separate sample still needs to be drawn and processed before shipping.
In contrast, our approach with the VitalOne offers a comprehensive, multi-modality test menu in a compact PoC form factor designed to provide central-lab quality results.§ When constructing our menu, we aimed to cover 100% of tests ordered for over 90% of patients in primary care settings, closing the send out gap for the vast majority of patients. With all results ready in 20 minutes, healthcare providers can also discuss the results with their patients at their first visit for immediate intervention. The VitalOne can be run with as little as 110 μL of whole blood for a CBC with a 5-part differential, or with less than 500 μL of whole blood for a panel of over 50 results spanning hematology, clinical chemistry, and immunoassays. The small sample requirement makes the VitalOne more amenable to a PoC setting where capillary blood can be used without needing a trained phlebotomist on site for a venous collection.
Several multidisciplinary design challenges needed to be overcome to realize this vision, synthesizing many individual components to enable the parallel, automated operation of dozens of assays that have fundamentally different lab protocols and require various levels of sensitivity. We succeeded in using centrifugal microfluidics to unify three distinct modalities into a common hardware architecture by fully reimagining the current testing paradigm, which until now required multiple instruments for analysis.
This architecture also makes the VitalOne significantly more reliable, economical, and easy to use than a solution that miniaturizes three disparate hardware architectures into a single unit. The VitalOne system was designed for minimally-skilled users to operate with limited risk of user error, and to this end all sample preparation steps are automated. The platform places sample preparation, precision metering, and calibration into the design of the microfluidic consumable; as a result, the system is more robust, and the system is designed to avoid the need for daily controls and weekly calibration by skilled technicians; this would significantly reduce the personnel cost for operation. Hundreds of built-in quality control checks have also been incorporated to ensure that only trusted results are included in the report to prevent possible patient harm from run errors.
All of this was made possible by starting with reagent formulations that are well-established assay methodologies but customizing the reagents to enable them to operate within our unique architecture that confers significant advantages to these tests. The clinical chemistry subsystem was developed by re-mapping canonical protocols, allowing over 25 tests to run in parallel with a single actuation protocol. The immunoassay subsystem uses magnetic capture beads and proprietary optical reporters for rapid and sensitive assessment with low background in a variety of biological matrices. The hematology subsystem uses image-based analysis to match the performance of flow cytometry-based analyzers without requiring frequent maintenance or skilled calibration. The reaction chemistries for several assays were also adjusted to meet design criteria for freeze-dried reagents and optical readout.
The VitalOne consumable features modular assay lanes that allow all assay types within a single modality to be run together on a single disc, sharing one electromechanical actuation protocol. For example, one-reagent, two-reagent, endpoint, and kinetic assays have all been done before, but not together with a shared workflow. With the VitalOne, users no longer need to think about individual panels to run. They can instead think about the entire menu together, and not have to make choices about sample preparation or which instrument to use. The time to results also stays constant, even when introducing more lanes for additional tests. And while the system runs all chemistries on every run, final results are only reported for requisitioned tests.
Through intentional design, the VitalOne distinguishes itself from prior platforms and sets itself up for successful widespread adoption through the combination of many benefits. Its unified architecture yields reliable and low-cost hardware and consumables. It is easy to use without skilled calibration or sample preparation, allowing minimally-trained users to get quality-checked, accurate results every time. The VitalOne has been designed to be installed with no special infrastructure plumbing or water purification. The system simply requires a standard power outlet and an internet connection greatly lowering barriers to entry. Fast turnaround times were a key design criterion to enable seamless clinical integration with a comprehensive menu that not only closes care gaps, but can also be used with existing reimbursement codes.
With the work presented in this publication, our objective at Vital Bio is to establish a high degree of transparency with the scientific community and the diagnostics industry. By openly sharing both our detailed assay characterization data and the underlying mode of operation of our system, we aim to promote a transparent and open culture.
Experimental
The following is a description of the VitalOne device and microfluidic consumable, the scientific and clinical considerations that went into their design, and the methodologies that underlie their mode of operation.Device description
The VitalOne system is a modular desktop instrument developed to produce laboratory-grade (e.g., CLIA certified) diagnostic results in 20 minutes, directly at the point of care (Fig. 1). The system's architecture is the outcome of design choices aimed at achieving consistent results, high reliability, and simplified user experience without compromising the accuracy or the quality of the results. | ||
| Fig. 1 VitalOne exterior view (left) and interior schematic (right) showing the system design and placement of subsystem modules. | ||
The VitalOne is a versatile platform designed to analyze a broad spectrum of biomarkers, including ions, small molecules, proteins, and cells. To accommodate the diverse methods required to measure these distinct types of analytes, the VitalOne segregates them into three separate subsystem modules that all operate on a unified architecture of centrifugal microfluidics. The VitalOne comprises three distinct assay subsystems tailored for specific analytical methods.
Three primary methods underpin its operation:
a. Clinical Chemistry – consisting of two sub-modalities.
1. Homogeneous liquid reactions with colorimetric readouts.
2. Membrane-liquid interface with colorimetric readouts for electrolytes.
b. Immunoassay – solid–liquid heterogeneous reactions with fluorescence output.
c. Hematology – blood cell staining paired with image-based measurements using fluorescence and darkfield microscopy.
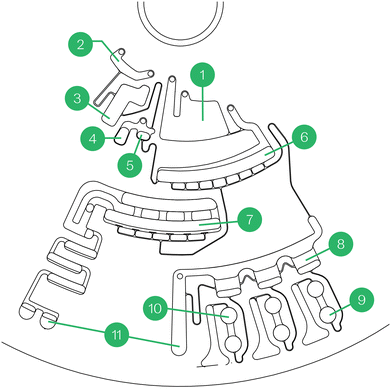
To achieve this, each subsystem has a consistent core of a motor, spindle, and temperature control (Fig. 2). The differences in functionality are enabled by: (1) subsystem-specific components that enable various readout or sample manipulation (e.g., absorbance/fluorescence readers, microscopy units, sonicators, and magnets); and (2) the incorporation of distinct discs each having unique microfluidic structures powered by centrifugal microfluidics. These discs host lyophilized reagents in a bead form called lyospheres and are equipped with channels and chambers for sample and reagent processing. Centrifugal forces aliquot and guide liquids through these channels to complete the required analytical assays.
Consumable description
The VitalOne consumable consists of two main components: a disc pack and a support pack (Fig. 3). The consumable has been designed for room-temperature storage and ease of use while ensuring precise and accurate diagnostic results. | ||
| Fig. 3 VitalOne consumables include a disc pack and support pack that are pre-loaded with all the components required to execute a full assay panel. | ||
System operation
To operate the system, the input drawer is first loaded with a disc pack, which houses the three discs needed for the three modules, and a support pack containing the tip-pack and reagent-pack strip. Following this, tubes containing the samples slated for analysis are placed into the drawer; the system accepts both capillary microtainers and traditional venous vacutainers.The remaining sample preparation and assay workflow steps are automated and to not require additional user input. The instrument mixes the sample within the tubes. This mixing is performed by the pipette head (microtainers) or by inverting the tubes on a tube rocker built into the instrument (vacutainers). The instrument is equipped to de-cap the loaded sample tubes, after inversion if applicable. Having automated de-capping and mixing eliminates both the risk of user exposure to the sample and variability resulting from manual sample mixing.
A gripper within the VitalOne instrument picks up the assay discs sequentially, and positions each of them into their respective modules. Then, a pipette head within the VitalOne instrument picks up a disposable tip and transfers the liquid reagents (primarily buffers and diluents as described in detail below) and the samples into the appropriate microfluidic structures on each disc. Once these discs are populated, they undergo a programmed rotational motion, paired with other operations and optical measurements. After the run is complete, each disc is returned to the disc tray, rendering the disc tray and support pack ready for removal. The processed samples remain contained within the discs allowing for easy disposal. To finish the process, the machine re-caps the sample tubes, ensuring that they can be safely removed without direct contact with the samples.
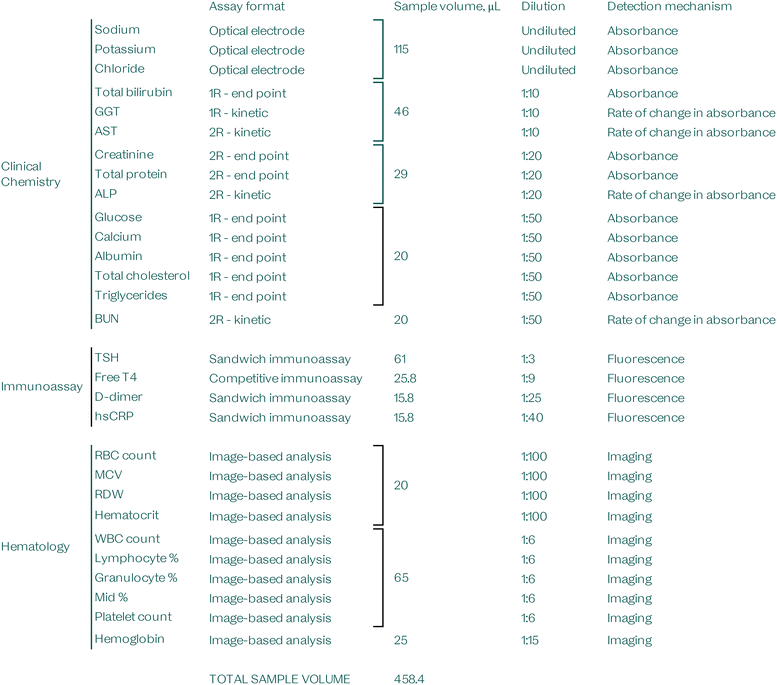
Assay menu and sample volume
The VitalOne is designed to support standard vacutainers used for venous blood collection as well as capillary microtainers; the system was designed to require low total sample volume to be compatible with capillary sample collection. Table 2 shows the sample volumes required for dispensing by the pipette head and disposable tip to ensure complete filling. The VitalOne is designed to perform a complete blood count (CBC) with as little as 110 μL of whole blood and a comprehensive panel with a full array of immunoassays and clinical chemistry assays, returning over 50 results with as little as 500 μL of whole blood. Smaller panels of tests that represent routine panels for quarterly chronic-disease management visits with over 25 results can be returned with just 300 μL of sample.Time-to-results (TTR)
The VitalOne system was designed with TTR as a core requirement. When running the Clinical Chemistry panel alone (full comprehensive metabolic panel (CMP), lipid panel, hemoglobin A1c (HbA1c), and GGT) the system can return these results within 15 minutes of the user initiating a test. Since all analytes on each modular lane for a given subsystem can be run in parallel using a shared actuation routine, the full panel of hematology, clinical chemistry, and immunoassays can be executed with a the TTR of only 20 minutes, regardless of how many tests are requisitioned.Microfluidic disc description
The following sections describe the microfluidic disc designs for each VitalOne subsystem and their operation. Please see the ESI† for annotated videos of the microfluidic workflows.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 1
20, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) and either a 1-reagent (1R) reaction or 2-reagent (2R) reactions that produces a change in absorbance. The system reads the absorbance at each well across all wavelengths and software is then used to select appropriate endpoint or kinetic read windows for each assay.
50) and either a 1-reagent (1R) reaction or 2-reagent (2R) reactions that produces a change in absorbance. The system reads the absorbance at each well across all wavelengths and software is then used to select appropriate endpoint or kinetic read windows for each assay.
The design of each clinical chemistry sample dilution is composed of similar structures, tuned for the dilution factor and required volume of the diluted sample, and designed to match the target rotation protocol. The disc design includes some overhead to ensure correct metering for each overflow.
The sample separation structure/element (Fig. 4, structure 3) is designed so that under rotation, whole blood blocks a vent port inside the chamber, partially compressing the air volume inside it, allowing for the use of pneumatic force to prime the siphon when the speed is changed. This structure is used both as a plasma separation chamber, where blood is spun for long periods of time, and as a functional valve operated by speed changes, thereby allowing the extraction of cell-free plasma.
The dilution structure (structures 6–7) also performs multiple functions: buffer metering, valving to control liquid motion (plasma metering and diluted sample), and mixing of buffer and plasma/sample. All of this is achieved by combining an unvented chamber in order to use centrifugal force and the buffer-diluted sample to compress the air inside which acts as a ballast.
Two unique reaction structures were developed to be used in a modular way where all detection wells are located at the same radius and both 1R and 2R structures are designed to be operated at the same speeds; because these modular lanes share a common actuation routine, they can be combined onto a common disc, but separate development discs for 1R and 2R assays were used in this study and are shown in Fig. 4 (2R) and the electronic supplementary information (1R). In the 1R disc design (ESI†), structure 10 from the 2R design (Fig. 4) is removed because a second reagent bead is not required; the surrounding siphon structures are modified to fill the available space.
The microfluidic designs enable reaction volume metering, reagent resuspension, reagent homogenization, and assay readout while minimizing dead volume (non-contributing to signal/readout). Each reaction structure is composed of an unvented region which includes a detection well and a vented region which is designed for accurate metering of reaction volume. All the reaction structures for the same dilution factor are arranged side by side with an inverted-V-shaped connecting channel between them. The last reaction structure is connected to an overflow chamber through a siphon channel with a U-shaped portion. The connecting channels are designed in a way to enable active pneumatic mixing inside the reaction chamber without contamination between reactions nor loss of volume to the overflow chamber.
Relative to clinical chemistry, the same sample separation element is tuned to allow for a larger volume, and an additional overflow chamber region was designed within the sample plasma separation chamber to increase precision of metered plasma volume, removing the need for subsequent metering. The extracted plasma volume is defined by the radial position of the overflow radius and the outlet channel entrance.
The second element used is a reagent resuspension element with both unvented and vented chambers where lyophilized buffer beads are reconstituted and resulting liquid is homogenized by changing the rotation speed. The outlet of this element is a siphon which works as a speed-defined valve and connects to the detection structures.
One of the detection elements is designed to allow for precise sample metering and avoid contamination to downstream chambers. This is achieved by using a similar structure to the sample separation element but tuned for a low volume.
The electrolyte detection element is a simple shallow chamber defined by the minimum volume required for detection and manufacturing tolerances of the ion-selective optical electrodes (optodes).
This design enables the incorporation of dozens of assays on a single disc with a single workflow while enabling each to have unique reaction conditions.
This disc also has unique features like the ability to have start-and-stop workflows and specialized structures to enable magnetic pulldown and sonication-assisted resuspension of capture and reporter nanoparticles.
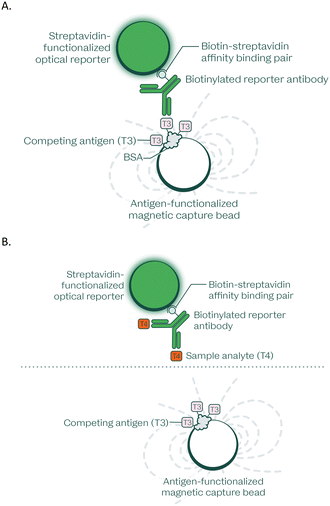
The microfluidic structures have been designed to accommodate sandwich assays (e.g., high-sensitivity C-reactive protein (hsCRP)), competitive assays (e.g., free thyroxine (free T4)), and assays requiring releasing solutions (e.g., vitamin D). All immunoassay subtypes are actuated by a single rotational protocol, and their dry reagents are unitized and pre-loaded onto the disc as lyospheres (in structures 10–11).
Notably, the resulting signal in sandwich immunoassays is proportional to the amount of analyte in the sample, whereas the signal in the competitive immunoassays is inversely proportional to the amount of analyte in the sample.
The workflow for a sandwich immunoassay is as follows: After plasma separation and plasma dilution; 45 μL of diluted sample is injected into the reaction chamber which contains two lyospheres. One lyosphere contains the capture-antibody coated magnetic nanoparticles and the other contains biotinylated reporter antibody. Because biotinylated antibodies are used, the system was designed to be robust to biotin interference (results from interference study shown in Table 5). This mixture is homogenized by rotating the disc to align each reaction chamber with a sonicating probe that makes contact with the flexible capping layer side of the reaction chamber and homogenizes its contents. After a 5-minute incubation the disc is rotated to align the reaction chambers with magnets that attract and pellet the magnetic nanoparticles. A subsequent rotation protocol sweeps the magnetic particles into an appendix (structure 12) enabling a centrifugally driven flushing of the fluid in the chamber without losing magnetic nanoparticles. This enables high-efficiency pellet washing, greatly increasing robustness to endogenous and exogenous interferents and reducing assay background. A third lyosphere containing our proprietary reporter particles (structure 10), each containing hundreds of quantum dots, are then reconstituted and loaded into the reaction chamber (structure 11) with a centrifugation step. The structures are aligned once more to the sonicator and the pellet of magnetic nanoparticles is then resuspended in the presence of the reconstituted reporter particles. The mixture is then incubated for another 3 minutes before repeating the magnetic pulldown and wash procedures. The final step is resuspension of the particles in buffer and rotating the disc to align each reaction structure with a fluorescence reader for signal capture.
The workflow for a competitive immunoassay is as follows: After plasma separation and plasma dilution, 45 μL of diluted sample is injected into the reaction chamber which contains two lyospheres. One lyosphere contains magnetic nanoparticles coated with a competitive binding agent (molecules identical or similar in nature to the analyte of interest that will compete with the endogenous analyte for epitope binding sites on the antibody). The other lyosphere contains biotinylated reporter antibody. Homogenization, pelleting, and washing steps are performed with the same workflow and at the same time as the sandwich assays.
Rather than pneumatic mixing using high-speed centrifugation balanced with an air ballast, the reaction chamber of the hematology disc is designed with large, rounded structures to enable gentle mixing while preserving cellular morphology (Fig. 7). This combined with the low-RPM inertial mixing (LRIM) protocol ensures that the diluted sample is well homogenized and thoroughly mixed with the reagents. The implementation of geometric valves in series with capillary pressure valves enables a high degree of robustness to changes in the hydrophilicity of the plastic.
The hematology cartridge also employs imaging chambers tuned to each analyte's concentration and density. To keep the instrumentation simple and reliable, all the microscopic imaging is done with the same optical setup.
The cartridge design is split into three independent structures with similar elements: (1) white blood cells (WBC) and platelets (PLT); (2) hemoglobin (HGB); (3) RBC. Each structure is composed of similar buffer and sample metering elements, with an inertial mixing structure and an imaging chamber adjusted to the target analytes. All imaging chambers (Fig. 7, structures labeled 4) are at the same radius meaning all imaging can happen simply by rotating the disc over the imaging sensor. On each structure the dilution factor is adjusted to the required volume of diluted sample for the imaging chamber and matched to the target rotation protocol.
Lyophilized reagent bead synthesis
To make our reagents room-temperature-stable, they are freeze dried into unitized lyospheres so that a single bead can be placed inside the appropriate chamber of a sealed microfluidic consumable for future reconstitution with a neat patient sample or one diluted with water or buffer. This eliminates the need for precision dispensing of powders or liquids in the assembly line. This work is carried out in a clean and dry room with a particle count maintained under 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 PPM and relative humidity maintained under 8%.
000 PPM and relative humidity maintained under 8%.
User interface
In addition to the above-mentioned features that minimise the number of user steps, the system has been equipped with a 12 inch full-color touch screen display capable of displaying rich video of all sample and consumable loading and unloading steps to ensure even a minimally trained operator would be able to operate the instrument.Assay methodology
For the purposes of this study, individual subsystem test benches were isolated as modular units and used for data acquisition with all the same on-board hardware as the fully integrated VitalOne units. In some cases to increase throughput, modular microfluidic lanes were also isolated onto development discs to test various analytes individually. The hardware and microfluidic geometries were preserved.Clinical chemistry
The clinical chemistry subsystem performs automated spectrophotometric measurements on the reaction wells, which are positioned on the outer radius of the disc in the path of the on-board absorbance readers (Fig. 4, structure 9 and Fig. 5, structure 6). The reaction well positions are defined by their angular positions relative to a homing feature on the disc. The data stream is parsed using a high-resolution encoder (0.1 degree resolution) that allows tracking of the angular displacement of the motor during each rotation. These features enable continuous readout data at all wavelengths on the path of the reaction wells without pausing rotation. The continuous readout capability is a particular advantage for kinetic assays where multiple measurements must be taken over time.The analyte concentrations are correlated with absorption changes at target wavelengths. These reactions are either of an end point type or a kinetic type and the reactions may require multiple reagents. These assays share fluidic structures and are grouped by sample dilutions.
Electrolytes and HbA1c are measured in an independent fluidic structure on the same disc. Electrolytes are measured by bringing undiluted plasma into contact with ion-selective optical membranes. The HbA1c structure contains a lysis structure to lyse and dilute whole blood as input for the reaction.
In this study, the clinical chemistry subsystem was used to analyze lithium-heparin anticoagulated whole blood samples. Once the sample and required buffers were dispensed on disc, the clinical chemistry subsystem started the sample preparation steps within which it performed a plasma separation step. Next, the chemistry disc meters the separated plasma and aliquoted it to five groups of assays (as described below) each containing a number of measurands and their associated reagents. It should be noted that while the assay groups share plasma separation and dilution steps, measurands are measured based on individual assays and each have their independent reagents, reaction and measurement wells. After the instrument loaded blood and buffer onto the disc, all fluidic manipulation and transfers were carried out using inertial and centrifugal forces based on a predefined protocol.
The clinical chemistry disc was split into various modular lanes to accommodate six assay types: two-reagent (2R) - end point; 2R - kinetic; one-reagent (1R) - end point; 1R - kinetic; calculated; three-reagent (3R) - end point; and optical electrode. Each assay type may share microfluidic elements for sample preparation, followed by splitting into aliquots for the introduction of assay-specific reagents. Where required, modular lanes were isolated to allow individual analytes to be quantified separately on development discs; the same geometries can be combined onto an integrated disc for parallel operation.
2R - end point
2R - kinetic
1R - end point
1R - kinetic
Optical electrode
Immunoassay
The immunoassay subsystem performs automated quantification of proteins and small molecules of interest in plasma generated from lithium-heparin whole blood samples.18,19In this study, the immunoassay subsystem was used for an automated sample preparation workflow within which it performed a plasma separation step on disc. Then, the immunoassay disc metered the separated plasma and started the reactions in each of the assay lanes. Two classes of immunoassays can be performed in parallel: sandwich assays for larger proteins (C-reactive protein, thyroid stimulating hormone, and D-dimer; Scheme 1) and competitive assays for smaller proteins and molecules (free thyroxine; Scheme 2). Other than instrument loading of blood and buffer onto the disc, all fluidic manipulation and transfers were done using inertial and centrifugal forces based on a predefined protocol that is shared between both assay classes.
The protocol combined the prepared sample with monoclonal antibody-coupled magnetic particles for capture, and subsequently with a reporter antibody-reporter signalling particle for fluorescent detection.
The capture and reporter particles were conjugated to the capturing antibodies either directly or indirectly through streptavidin–biotin complex formation. Through magnetic pulldown and washing steps, background and potential interferents in the sample matrix were replaced with a stable and clean matrix that enhances kinetics to accelerate the time to results.
The immunocomplexes present at the end point were resuspended and quantified by fluorescence readout. Similar to the clinical chemistry subsystem, the immunoassay reaction wells are positioned on the outer radius of the disc in the path of the on-board fluorescence readers (Fig. 6, structure 11). The reaction well positions are defined by their angular positions relative to a homing feature on the disc. The data stream is parsed using a high-resolution encoder (0.1 degree resolution) that allows tracking of the angular displacement of the motor during each rotation. These features enable continuous readout data at all positions on the path of the reaction wells without pausing rotation.
Hematology
Blood components were quantified within the hematology subsystem using image-based analysis.20 The hematology subsystem processes K2EDTA anticoagulated whole blood samples for quantitative, multi-parameter, automated in vitro diagnostic analysis. The entire assay reaction occurs inside the microfluidic hematology disc.In this study, the hematology subsystem was used to automate the sample preparation steps on disc, within which it metered and diluted the sample to the designed specifications by directing the sample and diluents through fluidic paths. The diluted sample was then mixed with appropriate tagging reagents and the mixture was then loaded into an imaging chamber for blood component analysis and quantification by an inverted fluorescent microscope which measured fluorescence and scattering properties of the cell structures. All fluidic manipulation and transfers were done using inertial and centrifugal forces based on a predefined protocol.
After deposition of blood and buffer on the consumable disc, the following workflow was carried out by the numbered structures on hematology disc (Fig. 7) for RBC, WBC, PLT, HCT, and HGB assay.
Here we provide a more detailed description of each assay protocol for the measurands on the hematology panel:
Experimental results
In this study, we characterized the performance of the VitalOne with a panel of analytes that spans across all three assay modalities – hematology, clinical chemistry, and immunoassay – selecting a representative subset of analytes that would challenge every aspect of our platform's intended capabilities. We completed an in-depth quantitative analysis of complete blood count with 3-part differential, a diverse set of clinical chemistry assays that includes each assay type (1R, 2R, kinetic, end point, electrolyte), and selected sandwich and competitive immunoassays as proof of concept.This is our first demonstration that all steps of our microfluidic workflow and on-board readout are possible on disc, including sample preparation, plasma separation, sample dilution, reagent mixing and incubation, multi-spectral cell imaging, and optical readout at several thousand RPM. These capabilities are the foundation for our immediate roadmap to an expanded menu that will cover 50+ of the most requisitioned assays in primary care, leaving space for future expansion for more specialized test menus.
Assay measuring range, precision, and linearity
To obtain quantitative measurements on the VitalOne platform, a 5–8 point calibration curve was generated for each analyte across the desired assay measuring range (AMR) using commercially available validation kits on three separate days, resulting in a master calibration curve using the same dilution as the samples to be measured. These curves were used to assess the limit of blank (LoB), limit of detection (LoD), and limit of quantitation (LoQ) for each analyte; they were also used to empirically estimate AMR (Table 3).These results confirm that we were successful in tuning assay protocols and sample dilutions to achieve target AMR that span across physiologically relevant concentrations, defined by medical decision levels and reference intervals for each analyte.
Precision was assessed using 4 samples and 3–5 blank replicates, tested at 2 timepoints (AM and PM) on each of 3 days by a single operator. The reported precision values were calculated as the average of the coefficient of variation (ratio of the standard deviation to the mean) related as a % for each sample.
To verify linear performance across physiologically relevant levels, contrived samples representing 3–6 concentrations of each analyte were tested spanning the relevant AMR.
Linearity is one of the most important characteristics for evaluating assay accuracy, as it establishes the reportable range. Linear regression was used to give the line of best fit (Fig. 8–10). These studies were carried out on disc for hematology and clinical chemistry, and using liquid reagents on an automated pipetting robot customized with a magnetic pulldown station for immunoassays.
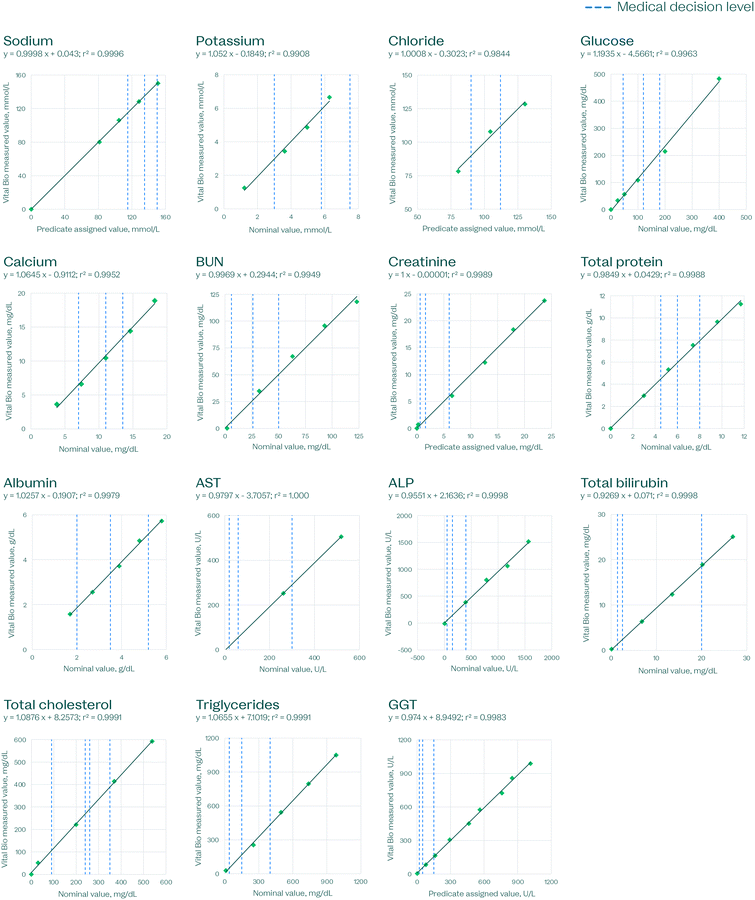 | ||
| Fig. 8 Clinical chemistry linearity on the VitalOne platform. Each data point is based on the average of 3–5 replicates per level. | ||
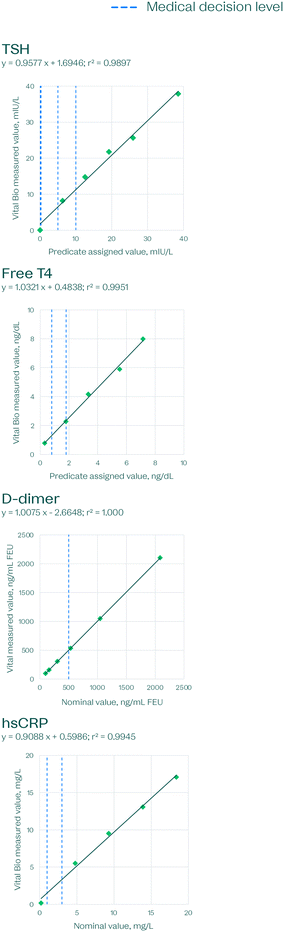 | ||
| Fig. 9 Immunoassay linearity on the VitalOne platform. Each data point is based on the average of 3 replicates per level. | ||
 | ||
| Fig. 10 Hematology linearity on the VitalOne platform. Each data point is based on the average of 3 replicates per level. | ||
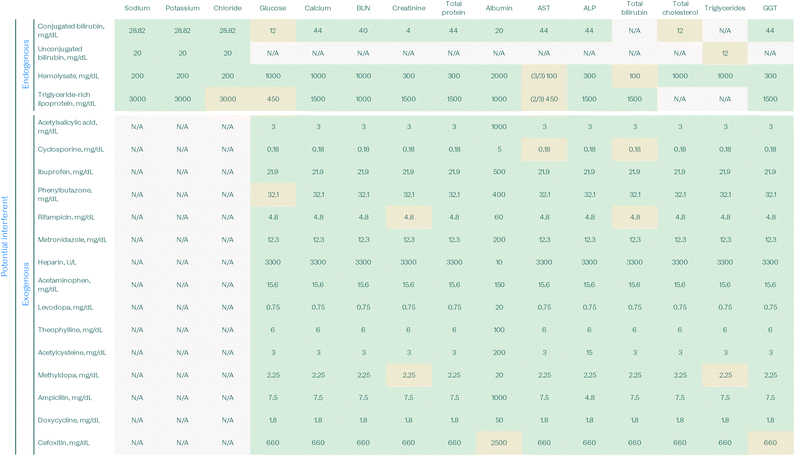
Interference testing
To check for sensitivity to potential interferences that may be present in patient samples, endogenous and exogenous interference testing was performed following Clinical & Laboratory Standards Institute (CLSI) guidelines.32 Samples were spiked with appropriate dilutions of potential interferents using ASSURANCE kits obtained from Sun Diagnostics. Assay performance in the presence of potential interferents is considered unaffected when results fall within 10% percent recovery of controls without interferent.For endogenous interference testing, hemolysis, icterus, and lipemia (HIL) interference was investigated to empirically establish optimized HIL index thresholds for the purposes of reporting HIL interference to advise clinicians on the interpretation of laboratory results in the instructions for use (IFU) in cases where hemolysis, icterus, or lipemia are present. Several potential exogenous interferents, such as common over-the-counter medications and antibiotics, were also tested.
Tables 4–6 summarize the interference findings. When marked in green, the concentration listed represents the highest concentration tested where the assay is not sensitive to the potential interferent. The values marked in tan denote concentrations where the assay result is affected by the potential interferent, but is either at a low level of interference or is a known interferent in predicate central laboratory instruments and will therefore be listed as a potential interferent in the IFU.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 sample dilution was used to assess interference for total bilirubin; the interference study will be repeated with a 1
50 sample dilution was used to assess interference for total bilirubin; the interference study will be repeated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 sample dilution to match the protocol used otherwise used in prototype verification. This table is based on 3–4 technical replicates per test
10 sample dilution to match the protocol used otherwise used in prototype verification. This table is based on 3–4 technical replicates per test
Robustness testing
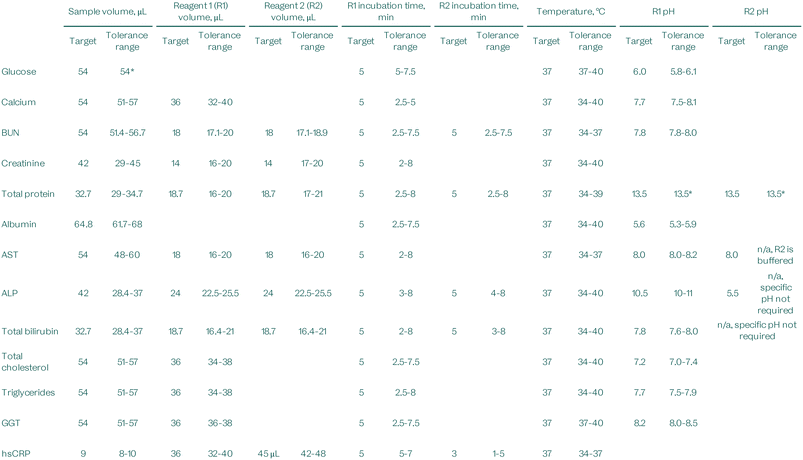
Robustness testing was carried out to mimic the extremes of variances in volumes, times, or temperatures that may be encountered when running an assay protocol. This testing was carried out using liquid reagents so that each parameter could be controlled independently. These results help determine the tolerance ranges over which the system is robust enough to deliver accurate results.Parameters such as sample volume (±5 and 10%), reagent volume (±5 and 10%), incubation time (±2.5 and 3 minutes), incubation temperature (±3 °C), and assay reagent pH (±0.3 pH units) were varied from the standard assay protocol to identify the point of failure for that parameter (Table 7). If a parameter failed at the standard testing range, a smaller range was tested to attempt to establish the tolerance range. Values marked with an asterisk (*) have tight requirements and further investigation needed to determine the relevant tolerance ranges.
These results are used to determine the maximum range that the system can deviate from the target parameters and the patient results can be trusted. Engineering strategies and microfluidic disc manufacturing tolerances are developed to keep them well within these ranges, and internal control systems will monitor for deviations beyond tolerance limits so that results can be suppressed.
Method comparison
One of the key aims of the VitalOne platform is to match the performance of instruments used in centralized clinical labs, bringing the same high-quality results to the point of care in a fraction of the turnaround time.To compare the results from the VitalOne platform against predicate instruments, we applied our technology to blood samples collected from consenting donors under an IRB-approved protocol, and residual samples sourced from Labcorp, Discovery Life Science, and Precision for Medicine. A minimum of 25–30 well-characterised patient samples were tested using both the VitalOne platform and a predicate instrument; Roche cobas c701, Beckman AU480, Beckman DxH 500, Beckman Access 2, and Sysmex 140 were used in this study and are widely used in clinical laboratories around the world. The resulting comparator data are also summarized in a table in the electronic supplementary information.
Although an effort was made to acquire samples that span the AMR, these are natural patient samples, and it was not always possible to obtain samples far outside the reference interval at the more uncommon medical decision levels. The matched samples were analyzed together on site to minimize error arising from sample transportation and storage.
Accepting the results from each predicate instrument as the ground truth for the analyte concentrations in each sample, the results from the VitalOne platform were plotted against the predicate and compared to the total allowable error limits established by Ricós33 (used for hemoglobin, WBC count, MCV, lymphocyte %, mid %, granulocyte %) or CLIA34 (used for all other assays) (Fig. 11–13). The target is for 95% of results to fall within total allowable error.
 | ||
| Fig. 11 Clinical chemistry method comparison, demonstrating the performance of the VitalOne platform compared to predicate devices (Roche cobas c701 and Beckman AU480) that are commonly used in centralized lab facilities. Each data point represents a single measurement. The sodium and AST data sets each contain an outlier (marked with an x) that were excluded from the subsequent linear regression. A correction was applied to compensate for a −3.6% bias observed in the GGT dataset, a −2.7% bias observed in the albumin dataset, and a −0.145 mmol L−1 bias observed in the potassium dataset. Total allowable error shown established by CLIA.34 | ||
 | ||
| Fig. 12 Immunoassay method comparison, demonstrating the performance of the VitalOne platform compared to predicate devices (Beckman access 2, Beckman AU480 and Sysmex 140) that are commonly used in centralized lab facilities. Each data point represents a single measurement. A correction was applied to compensate for a −7.8% bias observed in the hsCRP dataset. Total allowable error shown established by CLIA.34 | ||
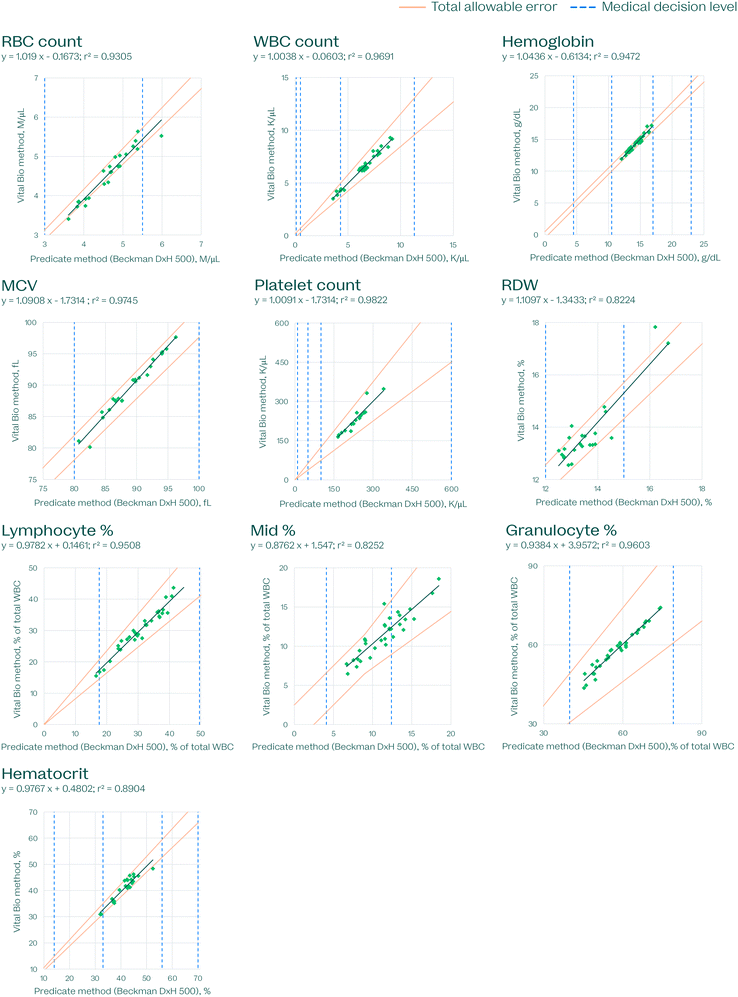 | ||
| Fig. 13 Hematology method comparison, demonstrating the performance of the VitalOne platform compared to a predicate device (Beckman DxH 500) that is commonly used in centralized lab facilities. Each data point represents a single measurement. A correction was applied to compensate for biases observed in the following datasets: WBC count, −11.42% bias; RBC count, +14.26% bias. Total allowable error shown established by by Ricós33 (hemoglobin, WBC count, MCV, lymphocyte %, mid %, granulocyte %) or CLIA34 (used for all other assays). | ||
We applied a correction factor to compensate for systematic biases to the comparators in the following datasets: GGT, −3.6% bias; albumin, −2.7% bias; potassium, −0.145 mmol L−1 bias; hsCRP -7.8% bias; WBC count, −11.42% bias; RBC count, +14.26% bias. These correction factors were determined by the median % bias and were subtracted from the Vital Bio measured values (Fig. 11–13).
Fully integrated user testing
In addition to the thorough analytical performance characterizations undertaken in a controlled laboratory environment, we set up the fully integrated VitalOne instrument for field testing at sites in New York and San Francisco to assess the usability and reliability of the system when run outside of a lab by a minimally trained user. For this pilot study, an integrated consumable was manufactured and assembled to deliver 35 results in parallel within 26 minutes, including assays across all three VitalOne subsystems.The following results were automatically generated after each run was complete:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) globulin ratio, creatinine, eGFR.
globulin ratio, creatinine, eGFR.
The system was operated by a phlebotomist who was trained by being shown how to operate the system once and allowed to practice using the onscreen instructions 2–3 more times. The fully integrated system was operated via the touch screen and graphical user interface (GUI). The operator had a 100% success rate in loading the samples and the consumables properly. The system delivered 35 results in over 95% of the runs. A single run had a microfluidic failure that was detected by the automated on-board quality control systems resulting in 29 results being reported. Real-time quality control systems on the VitalOne automatically detect failures. If a failure is detected the system will either resolve the failure or suppress the subset of impacted patient results ensuring that no erroneous information is reported.
This real-time quality control ensures that the VitalOne will be suitable for use in both CLIA-waived and moderate complex settings.
An annotated video of the internal operation of the fully integrated device is included in the ESI.†
Discussion
To demonstrate the capability of a point-of-care diagnostics platform to perform a comprehensive menu, it is critical to perform an extensive testing regimen. We did that by sourcing thousands of samples to develop our technology and optimize assay conditions, culminating in this method comparison test of the VitalOne system side-by-side with established benchmarks in the field. The primary objective was to ascertain whether our methodologies, which merge well-established chemical principles with innovations in robotics, optics, and microfluidics, could yield an easy to use, economical, and quick turn-around PoC instrument without sacrificing accuracy or precision.Consistency in assay performance is paramount, particularly across different medical decision levels. Our findings indicate that irrespective of the reagent batch in use, our assays consistently deliver reliable results. This uniformity is an indication of the robustness of our methods and the reproducibility of our system. Furthermore, our assays have demonstrated that their analytical measurement ranges encompass clinically relevant ranges. This accomplishment mirrors the capabilities of the predicate instruments we benchmarked against, solidifying our position as a comparable alternative with vastly improved turnaround time.
Assays in a clinical setting are invariably subjected to many variables, including potential interference from endogenous and exogenous agents and preanalytical errors. Our rigorous validation processes confirmed that our assays possess a commendable degree of robustness against these potentially interfering agents. The resilience observed in our system closely matches, and in certain parameters, such as biotin interference, even surpasses that of the predicate instruments.
Our method comparison studies, pivotal in establishing our performance in the existing diagnostics landscape, were conducted against Roche cobas c701, Sysmex 140, Beckman DxH 500, Beckman AU480, and Beckman Access 2 predicate instruments. The data exhibited a strong correlation with the benchmarked instruments. More significantly, the results consistently adhered to the stringent standards set by CLIA's total allowable error limits, reinforcing the reliability and clinical relevance of our diagnostic system. The data presented here clearly demonstrate the ability of our system to reliably measure these analytes in a general clinical setting. Additional studies to demonstrate the performance of the system in specific clinics or hospital units are currently being planned.
This publication serves a dual purpose. Firstly, it delineates our system's mode of operation, breaking down the scientific and engineering principles and methodologies employed. Secondly, we present performance data from a significant set of representative analytes, establishing the broad capability of our diagnostic system.
While the system offers significant advantages over central-lab instruments and point-of-care systems with smaller test menus, it does have a limitation in sample throughput. The system is designed to process one sample at a time, built around a model where 0.5 to 2 instruments are allocated per physician. This configuration envisions a world where lab testing is seamlessly integrated into the patient care journey. Consequently, while hospitals and central labs might find the system appealing as a backup or for specific use cases, it is not designed to meet the high patient sample throughput provided by central lab instrumentation.
This study serves as a broad demonstration of the quantitative capabilities of the VitalOne in vitro diagnostic system, but more testing is still required to clear the bar to regulatory approval and clinical use. As a culmination of the development work performed on thousands of patient samples, we applied our technology to 25–30 natural blood samples per analyte in this study. More work needs to be done to ensure that we both increase our sample size and cover a greater range of physiologically relevant concentrations at either end of the extremes by actively sourcing samples from selected patient groups.
It is also imperative for the reader to appreciate that the scope of our system extends beyond the menu demonstrated in this particular publication. The system is in late-stage development to further improve performance. Our ambitions and ongoing design improvements are geared towards enabling the system to execute a more expansive range of tests. When constructing our menu, we aimed to cover 100% of tests ordered for over 90% of patients in primary care settings, closing the send out gap for the vast majority of patients, and several more tests will be required to accomplish this goal. Among the tests we are incorporating are:
• A complete blood count inclusive of a 5-part differential.
• A comprehensive metabolic panel (CMP) – and it is crucial to underscore that this is a genuine CMP, complete with electrolytes and total CO2 – diverging from the sometimes-seen point-of-care “CMP” which lacks these critical analytes.
• A lipid panel, aimed at providing insights into cardiovascular health.
• HbA1c, crucial for monitoring long-term glucose management.
• GGT, an enzyme indicative of liver function and potential damage.
• A robust panel of 9 immunoassays, expanding the diagnostics capability into inflammatory markers, cardiac markers, thyroid markers, vitamins, endocrine markers, cancer markers and more.
• Over time, we plan to expand to common hormone markers, urinalysis, and other emerging analytes with high diagnostic value.
This more expansive menu would require approximately 650 μL of whole blood split across lithium-heparin and EDTA sample tubes.
The analytes presented here cover representative assays of all types in our final intended menu giving confidence in the ability for the same system to expand to the full menu described above as part of our commitment to pushing the boundaries of point-of-care diagnostics and providing clinicians with a comprehensive tool that aids in more informed decision-making and ultimately better patient outcomes.
Conclusions
In this study, we have demonstrated the capability of the VitalOne platform, a point-of-care diagnostics system, to perform a wide array of assays with precision and accuracy comparable to centralized laboratory instruments. Our findings underscore the system's versatility, reinforcing its potential as a transformative force in diagnostic testing. Importantly, this paper not only presents the performance of the system but also delves into the underlying scientific and engineering principles that enable this high level of functionality.Our combination of well-established chemical principles with innovations in robotics, optics, and microfluidics results in a diagnostic system that not only matches the performance of traditional laboratory equipment but also offers significant advantages in terms of usability, cost-efficiency, and rapid turnaround time. The detailed explanation of these principles provides a comprehensive understanding of how the VitalOne platform achieves its exceptional consistency across various medical decision levels, ensuring reliable and clinically relevant results regardless of the reagent batch used.
Furthermore, our method comparison studies, conducted against established benchmarks in the field such as Roche cobas c701, Sysmex 140, Beckman DxH 500, Beckman AU480, and Beckman Access 2 instruments, revealed a strong correlation, adhering to the stringent standards set by CLIA's total allowable error limits. This substantiates the clinical relevance of our diagnostic system, while the thorough discussion of the scientific and engineering underpinnings of our platform emphasizes the rigor and transparency, and innovation of our approach.
In summary, the VitalOne platform represents a significant advancement in point-of-care diagnostics, offering a comprehensive, accurate, and efficient alternative to traditional laboratory testing. This innovation is poised to transform the way clinicians approach diagnostics, enabling more informed decision-making and ultimately enhancing patient outcomes. The detailed exposition of both the performance and the foundational principles of the platform underscores its scientific and practical merit, making a compelling case for its broad adoption in healthcare settings.
Data availability
The method comparison data that support the findings of this study are available within the ESI.†Author contributions
Mounir A. Koussa: conceptualization, funding acquisition, methodology, project administration, supervision, writing – original draft, writing – review & editing. Miguel Barreiros: investigation, methodology, supervision, visualization, writing – original draft. Paul Said Ehrlich Perez: investigation, methodology, supervision. Sae Rin Jean: methodology, project administration, supervision, writing – review & editing. Taehyung Chris Lee: investigation, methodology, supervision. Ross MacLeod: software, supervision. Aaron Witham: formal analysis, investigation, methodology, supervision. Geeta Bhat: conceptualization, methodology, project administration, supervision, writing – review & editing. Todd Campbell: conceptualization, project administration, supervision. Sergio Lizano: conceptualization, methodology, project administration, supervision. Marjorie Toth: conceptualization, project administration, supervision. Amrita Venkateswaran: data curation, formal analysis, investigation, methodology, supervision. Don Yang: investigation, methodology, supervision. Nishat Zaman: formal analysis, investigation, methodology, supervision. Wisam Alfaqheri: investigation. Afshan Ardalan: data curation, formal analysis, investigation, methodology. Luis Barbosa: investigation. Mehran Behrouzi: investigation. Vitali Borisenko: investigation, methodology. Rohit Chand: data curation, formal analysis, investigation. Karyn S. Ho: formal analysis, funding acquisition, visualization, writing – original draft, writing – review & editing. Praveen Kumar: data curation, formal analysis, investigation. Mate Lengyel: investigation. Wei Luo: data curation, formal analysis, investigation. Fahim Masum: data curation, investigation. Laura Piñeros: data curation, investigation. Akhil Rajagopal Kozhipuram: investigation, methodology, visualization. Sergey Sanders: investigation. David Santos: investigation. Vasu Nadella: conceptualization, funding acquisition, writing – review & editing. Farnoud Kazemzadeh: conceptualization, funding acquisition, methodology, project administration, supervision, writing – review & editing. Iman Khodadad: conceptualization, funding acquisition, methodology, project administration, supervision, writing – review & editing.Conflicts of interest
All authors are paid employees who hold stock options at Vital Bio.Acknowledgements
The work was supported by the National Research Council of Canada Industrial Research Assistance Program (NRC IRAP) project numbers 951901, 965186, and 1000908, as well as by Innovative Solutions Canada project number 988343. The authors would like to thank Vital Bio's Scientific Advisory Board members Dr. Susan Evans and Dr. Nader Rifai for their thoughtful reviews of our manuscript. Eric Mahoney, Erden Ertorer, Imrul MD Kayes, Claire Noël, Aaron Campbell have also made significant contributions in supporting this work with the design and construction of the sub-system modules that has enabled the readouts. The authors would also like to thank every Vital Bio employee, past and present, whose contributions enabled us to build our common vision for the VitalOne.References
- U.-P. Rohr, C. Binder, T. Dieterle, F. Giusti, C. G. M. Messina, E. Toerien, H. Moch and H. H. Schäfer, The Value of In Vitro Diagnostic Testing in Medical Practice: A Status Report, PLoS One, 2016, 11, e0149856 CrossRef PubMed.
- C. Binder, M. Schmid, T. Dieterle and H. H. Schäfer, Costs and benefits of diagnostic testing: four ways to improve patient care by purposive use of in vitro diagnostics, Swiss Med. Wkly., 2017, 147, w14546, DOI:10.4414/smw.2017.14546.
- M. P. McRae, K. S. Rajsri, T. M. Alcorn and J. T. McDevitt, Smart Diagnostics: Combining Artificial Intelligence and In Vitro Diagnostics, Sensors, 2022, 22(17), 6355, DOI:10.3390/s22176355.
- G. Giuseppe and M. Plebani, The add value of laboratory diagnostics: the many reasons why decision-makers should actually care, J. Lab. Precis. Med., 2017, 2, 100, DOI:10.21037/jlpm.2017.12.07.
- G. Lippi and M. Plebani, Cost, profitability and value of laboratory diagnostics: In God we trust, all others bring data, J. Lab. Med., 2019, 43(1), 1–3, DOI:10.1515/labmed-2018-0321.
- G. Lippi, The irreplaceable value of laboratory diagnostics: Four recent tests that have revolutionized clinical practice, Electron. J. Int. Fed. Clin. Chem. Lab. Med., 2019, 30(1), 7–13 CAS.
- L. T. Car, N. Papachristou, A. Bull, A. Majeed, J. Gallagher, M. El-Khatib, P. Aylin, I. Rudan, R. Atun, J. Car and C. Vincent, Clinician-identified problems and solutions for delayed diagnosis in primary care: A PRIORITIZE study, BMC Fam. Pract., 2016, 17, 131, DOI:10.1186/s12875-016-0530-z.
- S.-M. Yang, S. Lv, W. Zhang and Y. Cui, Microfluidic point-of-care (POC) devices in early diagnosis: A review of opportunities and challenges, Sensors, 2022, 22, 1620 CrossRef CAS PubMed.
- E. A. Boohaker, R. E. Ward, J. E. Uman and B. D. McCarthy, Patient notification and follow-up of abnormal test results: A physician survey, Arch. Intern. Med., 1996, 156(3), 327–331, DOI:10.1001/archinte.156.3.327.
- The post-Theranos world, Nat. Biotechnol., 2022, 40, 139 CrossRef.
- I. Litchfield, L. Bentham, A. Hill, R. J. McManus, R. Lilford and S. Greenfield, Routine failures in the process for blood testing and the communication of results to patients in primary care in the UK: a qualitative exploration of patient and provider perspectives, BMJ Qual. Saf., 2015, 24(11), 681–690, DOI:10.1136/bmjqs-2014-003690.
- N. Ramsay, T. Johnson and T. Badrick, Investigating Patient Adherence with Pathology Testing in Primary Care and How Point of Care Testing Can Improve It, Point Care, 2016, 15(4), 144–151, DOI:10.1097/POC.0000000000000110.
- M. Jimenez-Barragan, M. Rodriguez-Oliva, C. Sanchez-Mora, C. Navarro-Bustos, S. Fuentes-Cantero, S. Martin-Perez, J. M. Garrido-Castilla, L. Undabeytia-Lopez, A. Luque-Cid, J. de Miguel-Melendez and A. Leon-Justel, Emergency severity level-3 patient flow based on point-of-care testing improves patient outcomes, Clin. Chim. Acta, 2021, 523, 144–151, DOI:10.1016/j.cca.2021.09.011.
- C. M. Pandey, S. Augustine, S. Kumar, S. Kumar, S. Nara, S. Srivastava and B. D. Malhotra, Biotechnol. J., 2018, 13 Search PubMed.
- S. Sachdeva, R. W. Davis and A. K. Saha, Front. Bioeng. Biotechnol., 2021, 8 Search PubMed.
- S. Sharma, J. Zapatero-Rodríguez, P. Estrela and R. O'Kennedy, Biosensors, 2015, 5 Search PubMed.
- M. Bruchez, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Semiconductor nanocrystals as fluorescent biological labels, Science, 1979, 281(5385), 2013–2016, DOI:10.1126/science.281.5385.2013.
- M. A. Koussa, Kinetic modulation for magnetic analyte detection, US Pat., US20230324416A1, 2022 Search PubMed.
- A. Ward, A. Chandrasekaran, D. Chen, C. Blanchard, P. Garden, B. Demarco, J. Forman, M. A. Koussa and L. Caldwell, Magnetic particle-based immunoassay and methods of using the same, US Pat., US20210389313A1, 2023 Search PubMed.
- I. Khodadad, A. Wong and F. Kazemzadeh, Imaging biological tissue or other subjects, US Pat., US10966611, 2019 Search PubMed.
- P. Fossati, L. Prencipe and G. Berti, Enzymic creatinine assay: A new colorimetric method based on hydrogen peroxide measurement, Clin. Chem., 1983, 29(8), 1494–1496, DOI:10.1093/clinchem/29.8.1494.
- T. Weichselbaum, An accurate and rapid method for the determination of proteins in small amounts of blood serum and plasma, Am. J. Clin. Pathol., 1946, 10, 40–49, DOI:10.1093/ajcp/16.3_ts.40.
- A. Karmen, A note on the spectrometric assay of glutamic-oxalacetic transaminase in human blood serum, J. Clin. Investig., 1955, 34(1), 131–133 CrossRef CAS PubMed.
- D. D. Van Slyke and G. E. Cullen, A permanent preparation of urease, and its use in the determination of urea, J. Biol. Chem., 1914, 19(2), 211, DOI:10.1016/s0021-9258(18)88302-8.
- H. A. Assink, B. G. Blijenberg, G. J. M. Boerma and B. Leijnse, The Introduction of Bromocresol Purple for the Determination of Serum Albumin on SMAC and ACA, and the Standardization Procedure, Clin. Chem. Lab. Med., 1984, 22(10), 685–692, DOI:10.1515/cclm.1984.22.10.685.
- T. Ueno, S. Hirayama, M. Sugihara and T. Miida, The bromocresol green assay, but not the modified bromocresol purple assay, overestimates the serum albumin concentration in nephrotic syndrome through reaction with α2-macroglobulin, Ann. Clin. Biochem., 2016, 53(Pt 1), 97–105, DOI:10.1177/0004563215574350.
- J. W. Janssen and A. R. Helbing, Arsenazo III: An improvement of the routine calcium determination in serum, Eur. J. Clin. Chem. Clin. Biochem., 1991, 29(3), 197–201 CAS.
- D. Shin, K. P. Yoon, S. K. Kwon and C. G. Kang, Determination of Total Calcium in Serum with Arsenazo III Method, Ann. Lab. Med., 1994, 14(1), 12–19 Search PubMed.
- W. E. Morf, K. Seiler, P. R. Sørensen and W. Simon, in Ion-Selective Electrodes, 1989 Search PubMed.
- K. Wang, K. Seiler, W. E. Morf, U. E. Spichiger, W. Simon, E. Lindner and E. Pungor, Characterization of Potassium-Selective Optode Membranes Based on Neutral Ionophores and Application in Human Blood Plasma, Anal. Sci., 1990, 6(5), 715–720, DOI:10.2116/analsci.6.715.
- S. T. Avecilla, S. M. Marionneaux, T. D. Leiva, J. A. Tonon, V. T. Chan, C. Moung, R. C. Meagher and P. Maslak, Comparison of manual hematocrit determinations versus automated methods for hematopoietic progenitor cell apheresis products, Transfusion, 2015, 56(2), 528–532, DOI:10.1111/trf.13346.
- Clinical and Laboratory Standards Institute (CLSI), Interference testing in clinical chemistry, 3rd edn, 2018.
- C. Ricós, V. Alvarez, F. Cava, J. V. García-Lario, A. Hernández, C. V. Jiménez, J. Minchinela, C. Perich and M. Simón, Current databases on biological variation: pros, cons and progress, Scand. J. Clin. Lab. Invest., 1999, 7, 491, DOI:10.1080/00365519950185229.
- CLIA, CLIA Proficiency Testing criteria, Westgard Online.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00105b |
| ‡ MB, PSEP, SRJ, TCL, RM, and AW are equal contributors to this work. |
| § The VitalOne has not yet been evaluated for regulatory clearance, nor is it currently available for purchase, but it is currently in late-stage development. |
| This journal is © The Royal Society of Chemistry 2024 |