Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorescence coding techniques for RNA detection
Junren
Wang
,
Qin
Xiang
*,
Haifeng
Dong
 * and
Xueji
Zhang
* and
Xueji
Zhang
Shenzhen Key Laboratory for Nano-Biosensing Technology, Research Center for Biosensor and Nanotheranostic, Guangdong Key Laboratory of Biomedical Measurements and Ultrasound Imaging, School of Biomedical Engineering, Shenzhen University Medical School, Shenzhen University, Shenzhen 518060, China. E-mail: qinxiang@szu.edu.cn; hfdong@ustb.edu.cn
First published on 16th April 2024
Abstract
Fluorescence imaging along with a wide range of fluorescence detection methodologies enables the visualization of macromolecular dynamics in clinical diagnostics and fundamental biomedical research. However, multiplexed fluorescence-based detection has hitherto been largely limited by the overlap of the emission spectra of fluorophores, which only with four or five fluorochrome dyes enable simultaneous use. Fluorescent barcodes with high resolution and high throughput have emerged as a versatile biosensor platform for multianalyte analysis via fine control of fluorescence proportional types, spatial geometries, specific fluorescence properties, and time dimensions. Fluorescence imaging of RNA enables the visualization of physiological and pathological processes at the subcellular level, which is highly important for biomedical diagnostics and therapies and has made tremendous progress toward understanding the complexity of biological systems. In this review, we provide the design and development of fluorescence coding techniques for multiplexed bioanalysis detection and then discuss their applications in RNA biosensors. Additionally, associated challenges in the field and potential solutions will be discussed. Overall, we hope this review will inspire researchers and pave the way for the future design of fluorescent biosensors.
1. Introduction
Conducting numerous high-throughput studies in intricate biological systems is crucial for biological research and medical diagnostics.1 Fluorescence imaging is the predominant technique for the in situ detection of cells and intracellular compounds.2,3 Fluorescence imaging is a method of visualizing and examining organisms or materials using fluorescent markers. It has several benefits including exceptional sensitivity, precise resolution, and non-destructive properties. This approach finds extensive application in several areas such as biomedical research, cell biology, and molecular biology.4–6 As technology advances, fluorescence imaging techniques such as multiphoton fluorescence imaging7 and super-resolution fluorescence imaging8,9 are continuously being enhanced and refined. These advancements offer greater potential for scientific inquiry and medical diagnostics. However, the presence of spectral overlap among fluorescent dyes can make the differentiation of fluorescence signals of distinct dyes difficult when many dyes are used to identify various molecules or cells. This can lead to the mixing of information or incorrect classification.10In order to address this issue, the concept of fluorescence coding has been suggested as a means of encoding various chemicals or organisms by utilizing different fluorescence colors or intensities. This approach has the potential to greatly enhance the ability to detect or image many targets simultaneously. Fluorescence coding is a versatile technique that can be employed to mark proteins for monitoring their whereabouts and mobility within cells or tissues.11 Additionally, it can be used to construct fluorescent probes capable of identifying the existence and function of biomolecules.12 Moreover, fluorescence coding enables the tracking and examination of cellular behaviors and interactions.13 In summary, fluorescence coding has an extensive array of applications in biological detection. The present fluorescence codes used for multiple detections mostly consist of ratio-type codes, spatial geometry codes, fluorescence property codes, and time dimension codes.
Ribonucleic acid (RNA) is a crucial molecule involved in the process of gene expression. It serves as a transporter of genetic information during transcription and translation. RNA has been identified as a potential biomarker for several illnesses, including early cancer identification and screening.14 RNA can be classified into several categories including mRNA,15 tRNA,16 microRNA (miRNA),17 and small-molecule RNA. Messenger RNA (mRNA), a type of RNA that is not double-stranded, regulates protein production and carries genetic information.18 Tumor-associated mRNAs are frequently used as specific indicators to assess the local or systemic migration of tumor cells, and any change in their expression levels is directly associated with the onset and progression of tumors.19,20 Approximately 30% of genes are directly regulated by miRNAs, which are a group of small, non-coding RNAs. They inhibit the production of proteins via regulating the post-transcriptional stage of mRNA degradation.21 Research has demonstrated that miRNAs play a significant role in a wide range of biological processes such as apoptosis, migration, aging, differentiation, and proliferation. Additionally, they have been demonstrated to contribute to the development of several illnesses such as diabetes,22 leukemia,23 and cancer.24
RNA biomarkers25,26 have higher sensitivity and specificity, and RNA detection holds significant therapeutic promise. However, several challenges need to be overcome before its application to real samples. To effectively detect RNA, it is necessary to employ techniques with high specificity. This is because RNA has a low molecular weight and a high degree of similarity within the same family. Furthermore, the assay must have sensitivity and accuracy as RNA is found in both healthy and malignant cells at low levels, with the primary distinction being the varying amounts produced. Furthermore, it is evident that multiplexed assays are essential, as relying solely on the dysregulation of a single RNA for illness diagnosis is not appropriate. This is due to the high expression of certain RNAs in various types of disease-causing cells.27,28
This article provides an overview of the current research on fluorescence coding for multiplexed detection. It discusses the application of fluorescence coding for multiplexed detection, in particular RNA detection. Additionally, it evaluates the obstacles and opportunities associated with this technique.
2. Fluorescence coding technique
Fluorescent markers,29 compounds that absorb light from the surrounding environment and emit fluorescence at specific wavelengths, are the main components used in fluorescence imaging. Fluorescent dyes,30 fluorescent proteins,31,32 and similar substances are commonly employed as fluorescent markers. In the 1960s, researchers began investigating the application of fluorescent materials for interpreting biological data.33 Fluorescent materials have the ability to stimulate and emit energy, resulting in the production of visible light due to their luminescence properties. Fluorescent materials are ideal for labeling purposes. Fluorescence imaging can be used to conduct studies on the distribution and metabolism of fluorescent markers in animals. This technique allows for the tracking of tumor growth and metastasis in live mice.34,35 Additionally, it can be utilized to observe the localization and distribution of particular proteins within cells36 and explore the physiological processes of cells.Fluorescence imaging serves as the fundamental basis for fluorescence encoding technology.37–39 Spectral overlap is a phenomenon that occurs when numerous fluorescent dyes are employed simultaneously. This poses a challenge in precisely measuring the matching fluorescence intensity, thereby affecting the precision of detection outcomes. Consequently, the utilization of fluorescence imaging technology is limited to the simultaneous employment of just four to five distinct types of fluorescent dyes. In addition, varying excitation spectra require the use of separate exciters, resulting in increased costs and complexity of the apparatus. Fluorescence coding technology has been developed to solve these problems. It is the most commonly used coding technology because it enables easy visualization of barcodes, detection signal localization, and reading. This technology primarily utilizes the distinct characteristics of the coding original, such as fluorescence intensity, spatial geometry, and temporal lifetime, to fulfill the requirement of multiple detection and achieve various coding methods. The fluorescence coding approach is extensively employed in biomedical research to achieve more precise labeling of cells,40 proteins41 and DNA,42 among other substances. As technology has progressed, the use of fluorescence coding has gradually increased. In the 1980s, researchers initiated an inquiry into the application of fluorescent particles for coding. Fluorescent particles have the ability to mark and monitor items, such as identifying tumor markers in biomedical research.43,44 With the advancement of nanotechnology, fluorescence coding has shown significant improvement in recent years. Researchers investigated the application of nanoparticles for fluorescence coding.45 Nanoparticles possess reduced dimensions and enhanced fluorescence characteristics, facilitating the utilization of more intricate and accurate encoding techniques. The nano-fluorescence coding technology has diverse applications in the domains of biology, materials science, and information storage.46–48 Fluorescence coding primarily emerged from the investigation and utilization of fluorescent materials. Due to technological advancements, fluorescence coding technology has undergone constant development and improvement, offering new prospects for research and application in diverse domains.
3. Classification of fluorescence codes
3.1 Fluorescence coding based on proportion
Proportional fluorescence coding is a strategy that was discovered early on and is now frequently employed. This technique employs various proportions of fluorescent dyes to represent distinct biomolecules or cells. By measuring the intensity of different fluorescence signals, it can identify and quantify the presence and concentration of target biomolecules or cells. This enables the simultaneous detection of multiple biomolecules or cells, enhancing the efficiency and throughput of the assay.49 Proportional fluorescence coding has numerous benefits in the field of biological detection. First, it has the capability to simultaneously detect multiple target biomolecules or cells, enabling efficient high-throughput detection. Second, it can achieve various encodings by modifying the concentration of different ratios of fluorescent dyes, offering great flexibility and adaptability.50–52In 2005, Luo's team used Y-DNA to construct dendritic DNA (DL-DNA) nanostructures, which demonstrated the superiority of DNA nanostructures. The DL-DNA was employed as a carrier for fluorescent dyes in Alexa Fluor 488 (G) and BODIPY 630/650 (R), to create fluorescence intensity-encoded nano-barcodes. This allowed for the precise control of the ratio between the two types of dyes (Fig. 1A).61 Initially, Y-DNA was synthesized and labeled with fluorescent markers. Then, using sticky ends, five different combinations of fluorescent barcodes were created: 4G1R, 2G1R, 1G1R, 1G2R, and 1G4R. These barcodes were used to detect various pathogen DNAs through fluorescence microscopy and spot blotting. This technique enables the simultaneous detection of multiple pathogens using flow cytometers, which are typically limited to detecting only two colors. In 2006, the main focus was on preparing DL-DNA by solid-phase and liquid-phase methods. Additionally, the multivalent anisotropy of DL-DNA was used to create fluorescence coding approaches for detecting various diseases simultaneously and quickly.62 Subsequently, three Y-DNA donor molecules and one X-DNA acceptor molecule were employed to amplify and identify the signals of various diseases by encoding three fluorescence ratios, namely 1G1R, 1G3R, and 4G, utilizing quantum dots (QDs) of red (R) and green (G) hues.63 Nevertheless, the aforementioned approaches are susceptible to aggregation-induced fluorescence bursts and have limited encoding capability.
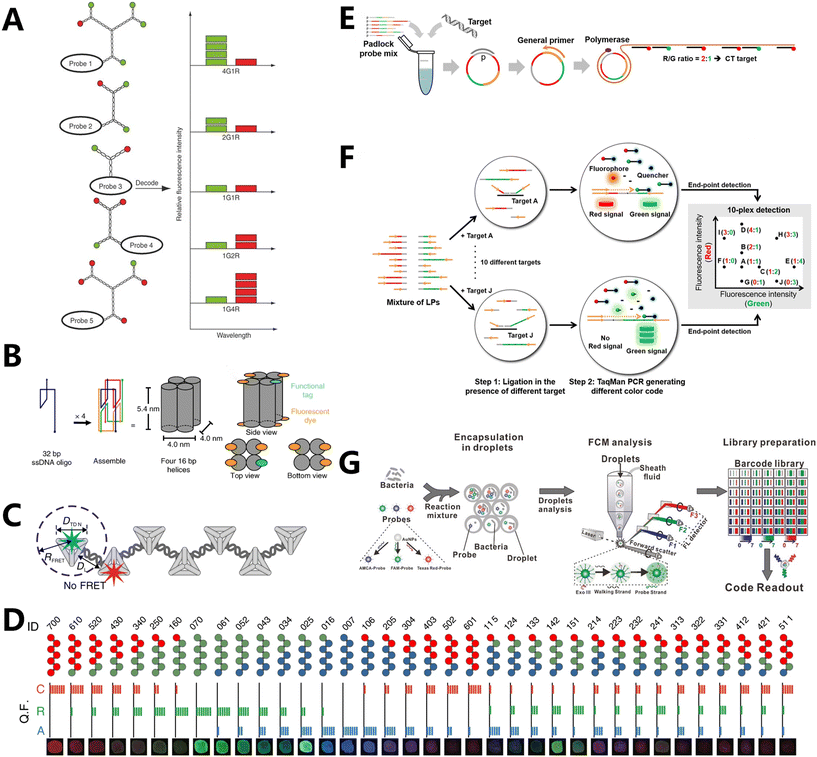 | ||
| Fig. 1 Fluorescence coding based on proportion. (A) Schematic illustration of barcode decoding. The nanobarcodes 4G1R, 2G1R, 1G1R, 1G2R and 1G4R were decoded based on the ratio of fluorescence intensity.61 (B) Design and assembly of DNA FluoroCubes.71 (C) Schematic illustration of the FDF structure with spatial control of fluorophores.72 (D) Thirty-six FDF barcodes encoded with different fluorophore combinations.72 (E) Schematic illustration of sequence-encoded amplicon (SeqEA) for multiplexed imaging of individual RNAs in single cells.74 (F) LiNC PCR is performed using two steps.76 (G) Schematic of the SDwalker-Drop platform for the super-multiplex bacterial phenotype analysis.79 | ||
In 2006, Rothemund showcased innovative research in the field of DNA origami technology.64 DNA origami technology is a method that harnesses the inherent self-assembly capability of DNA molecules to construct structures and devices at the nanoscale. The process relies on the fundamental concept of base complementary pairing between DNA molecules, allowing for the spontaneous formation of desired structures when specified conditions are met through the design of suitable DNA sequences.65,66 The technology of DNA origami is characterized by its exceptional accuracy and controllability, enabling meticulous assembly at the nanoscale.67 Additionally, it possesses a high degree of self-assembly efficacy and rapid self-assembly velocity, enabling the formation of intricate structures within a short timeframe.68 Furthermore, the DNA origami technology has superior biocompatibility and biostability, which make it suitable for implementation in live organisms.69 When comparing small Y-DNA molecules to DNA origami technology, it is observed that DNA origami technology prevents fluorescence resonance energy transfer (FRET) due to its larger size. Additionally, it allows for the precise placement of fluorophores at specific positions and their assembly into predetermined fluorescence-aligned structures. This significantly enhances the capacity for fluorescence encoding. Jungmann and his colleagues used DNA origami to construct fluorescent probes smaller than 100 nm in size. These probes have adjustable optical properties, and were constructed by combining 24 DNA double-helix structures into a two-dimensional (2D) rectangular DNA origami with a dimension of 90 × 60 nm2. The origami structures were loaded with varying amounts of three different fluorescent dyes (ATTO 647N, Cy3, and ATTO 488).70 By employing three distinct fluorescent dyes and conducting trials at five varying intensity levels, researchers obtained a total of 53 − 1 = 124 fluorescent probes. While the encoding capacity is significantly enhanced, the large-scale production of DNA origami measuring around 100 nm often necessitates numerous single-stranded DNAs. The complexity of the strand design and synthesis process imposes limitations on its practical use. Larger DNA nanoparticles are not suitable for detecting and imaging at the cellular level.
FluoroCubes, which are only 6 nm in size, have been recently prepared by DNA engineering, and can be customized with six fluorescent moieties (Fig. 1B).71 This method enables the efficient imaging of individual molecules, along with a significant 54-fold extension in fluorescence duration and a substantial 43-fold enhancement in photon emission. Fan et al. have used Cayley dendritic fractal DNA frameworks (FDFs) to develop fluorescence intensity-encoded barcodes (Fig. 1C).72 The Cayley dendritic framework structure was initially fabricated by building DNA tetrahedra with a side length of 20 bp. Subsequently, the n-node structure (n = 53) could be achieved using only 16 strands. By accurately attaching three fluorescent motifs (Alexa 488, Cy5, and ROX) to the FDFs to create the F2,3 structure, they not only prevented fluorescence energy resonance transfer, which reduces fluorescence crosstalk, but also produced 36 coding barcodes with distinct fluorescence colors and intensities (Fig. 1D). This significantly reduces the quantity of DNA strands and diminishes the complexity and workload associated with coding, in contrast to the prior method of using large-sized DNA origami. At the same time, it enhances the capabilities of recognizing and imaging many functions in living cells.
Lee et al. applied RCA to produce biotin surface-modified nanospheres with many functionalities to generate barcodes for cancer cell detection.73 The nanospheres were subsequently linked to different proportions of red and green QDs via biotin–streptavidin interactions. Wang et al. used lock-in probes to accomplish proportionate fluorescence encoding with RCA (Fig. 1E).74 In particular, lock-in probes were used to attach two probes with different fluorophores by incorporating a specific number of binding sites to target nucleic acids. The study describes the design of six lock-in probes that can be recycled. These probes were created using two separate fluorescent dyes, namely red (R) and green (G), and each probe has a unique R/G ratio. After performing nucleic acid amplification, these hybridized probes were used to identify the gene sequences of six infectious diseases, thereby expanding the method's potential to simultaneously detect several diseases.
Furthermore, the PCR has been utilized for encoding, in addition to the RCA approach. The reaction is a molecular biology technique used to amplify specific DNA fragments in a controlled laboratory setting. Due to its exceptional specificity and sensitivity, it is widely utilized in various domains such as gene amplification and medical diagnostics. This technique allows for the amplification of several target fragments from minuscule DNA samples.75 Wang et al. developed a novel fluorescence-based scheme called Ligation-eNabled fluorescence-Coding PCR (LiNC PCR) using PCR technology. This scheme significantly enhances the multiplexing capability of standard PCR analysis (Fig. 1F).76 In this approach, TaqMan probes were inserted using probe locations that corresponded to red (R) and green (G) fluorescence signals. The probes were built with ten R/G ratios, namely, 1/1, 2/1, 1/2, 4/1, 1/4, 1/0, 0/1, 3/3, 3/0, and 0/3. The binding sites provide distinctive fluorescence signals during the PCR procedure, enabling clear and unequivocal identification of the amplified targets. Consequently, the detection performance and ability to simultaneously identify several ovarian cancer epigenetic indicators were considerably enhanced by using only two universal TaqMan probes.
For example, Nie's team developed a technique to combine several fluorescent colors by incorporating red, green, and blue QDs into polymer microspheres at varying levels of fluorescence intensity.77 Similarly, Xu's team used mesoporous beads of similar sizes to microcarriers in order to develop a barcode library with a large encoding capacity.78 Subsequently, they combined different proportions of red QDs in the outermost layer and enclosed varying proportions of fluorescein isothiocyanate (FITC) isomer I green dye within the mesopores. Ultimately, they employed ten specific tumor markers as exemplar targets to demonstrate the suitability of this multiplexed bioassay platform. Based on a similar idea, Pei et al. developed a detection method called SDwalker-Drop, which utilizes stochastic DNA walkers in droplets. This technique combines DNA walker technology with coded combinations of fluorescent dyes to enable simultaneous identification and multiplex detection of pathogens (Fig. 1G).79 The paper utilized a mixture of three dyes (AMCA, FAM, and TR) with distinct spectral properties, along with eight intensity levels ranging from 0 to 7. These dyes were immobilized in thiol-labeled oligonucleotide gold nanoparticles, which functioned as spherical nucleic acid probes (SNPs). Theoretically, this system could achieve a coding capacity of 83 − 1 = 511. Through experimentation, researchers successfully obtained 20 patterns for the detection and identification of bacterial phenotypes. The process of enclosing bacteria and probes within droplets that are 50 picometers in size, and the enzymatic cascade reaction of SDwalker for signal transduction and amplification caused by bacteria, significantly enhances the potential of droplet-based microfluidics to detect numerous bacterial species simultaneously. Nevertheless, this method remains unsuitable for identifying and seeing chemicals within living cells.
Klymchenko et al. addressed this issue by creating fluorescent polymer nanoparticle barcodes that are suitable for both in vitro and in vivo cellular imaging.80 Researchers used a biodegradable poly(lactic-co-glycolic acid) polymer mixed with three colors of cyanine dye in different ratios to form homogeneous 40 nm-sized RGB (red, green, and blue) barcodes. These barcodes serve the dual purpose of distinguishing between 13 different cell populations, and can be passed down through multiple cell generations. Consequently, they enable the tracking of tumor cells in live animals and facilitate cell tracing in developing embryos. Hu's team used fluorescent proteins conjugated to magnetic beads to generate encoded microspheres. These microspheres were subsequently deployed to immobilize capture probes on their surface, enabling the detection of target oligonucleotides within the pathogen.81 Recently, the technique of DNA-induced fusions has been employed to carry out numerous randomized fusions of six different populations of cargo liposomes. A total of 566 distinct fusion sequences were detected using various ratios of chromophore-encoded liposome populations, as viewed by total internal reflection microscopy. Machine learning enables the categorization of enormous amounts of data, which make it a valuable tool for high-throughput drug screening, biosynthesis, and epitope mapping.82
3.2 Fluorescence coding based on spatial geometry
The use of fluorescence emission color-based coding is often problematic in detection devices due to its susceptibility to photobleaching and the need for several lasers to match different dyes with varying excitation spectra, which adds to the complexity. However, spatial geometry-based fluorescence coding allows for the identification of many targets utilizing a reduced number of fluorescent dyes. Spatial geometric fluorescence coding is a scalable technique that allows for the adjustment of the number of tiny areas to accommodate various detection samples. The fundamental technique entails partitioning the surface of an item into several small regions and applying fluorescent dyes, either of the same or different hues, to each individual region. Subsequently, the dyes are encoded based on their placements and colors, resulting in a system that is exceptionally resilient to errors due to the ability to selectively stimulate and detect fluorescent dyes' hues.The improved capacity to precisely locate and adjust DNA origami technology has opened up new possibilities for using fluorescence coding methods. Yin's team utilized DNA origami techniques to create submicron fluorescent barcodes with geometric encoding. The encoding mostly involves accurately positioning fluorescent dyes on the surface in a specific spatial geometry (Fig. 2A).86 Fluorescent labeling was applied to three specific sections of a six-helix bundle of DNA nanorods, which had a length of roughly 800 nm. The labeled portions were spaced at intervals of 450 nm and 270 nm, respectively. This facilitated the differentiation of the asymmetrical patterns during the process of decoding using fluorescence microscopy. The three sections were identified using the fluorophores “blue” (B, Alexa Fluor 488), “red” (R, Alexa Fluor 647), and “green” (G, Cy3). The BRG barcode obtained is distinct from the GRB barcode because of the fluorophore labeling of the three areas. Consequently, the three fluorophores that have different spectrum properties can be utilized to generate a total of 33 = 27 unique barcodes. Subsequently, advancements were made in total internal reflection fluorescence microscopy, resulting in the ability to accurately identify 216 different barcodes (Fig. 2B). This breakthrough showcased barcodes with increased spatial information density and achieved a super-resolution of 40 nm. Wei et al. constructed DNA origami nanorods consisting of five 10-helix bundles. Each bundle had a length of approximately 250 nm and contained both fluorescent and non-fluorescent groups (Fig. 2C).87 The five monomers unite to create a 1.25 μm DNA barcode. Each 10-helix bundle can exist in two states: “on” (1) or “off” (0). When there are two or more “on” monomers in a row in the barcode, they cannot be told apart and appear as a longer “on” monomer. However, the presence of “off” monomers creates a distinction between them. This study demonstrates the utilization of these barcodes for both multiplexed single-molecule analysis and the identification of synthetic single-stranded DNA target sequences. The spatial geometric encoding capability of barcodes is improved by including fluorescent moieties of different colors. This enables the barcodes to be encoded in a spatially geometric manner, even in scenarios where there is only one color of fluorescence, as well as in cases where there are fluctuations in the intensity of fluorescence in individual units.
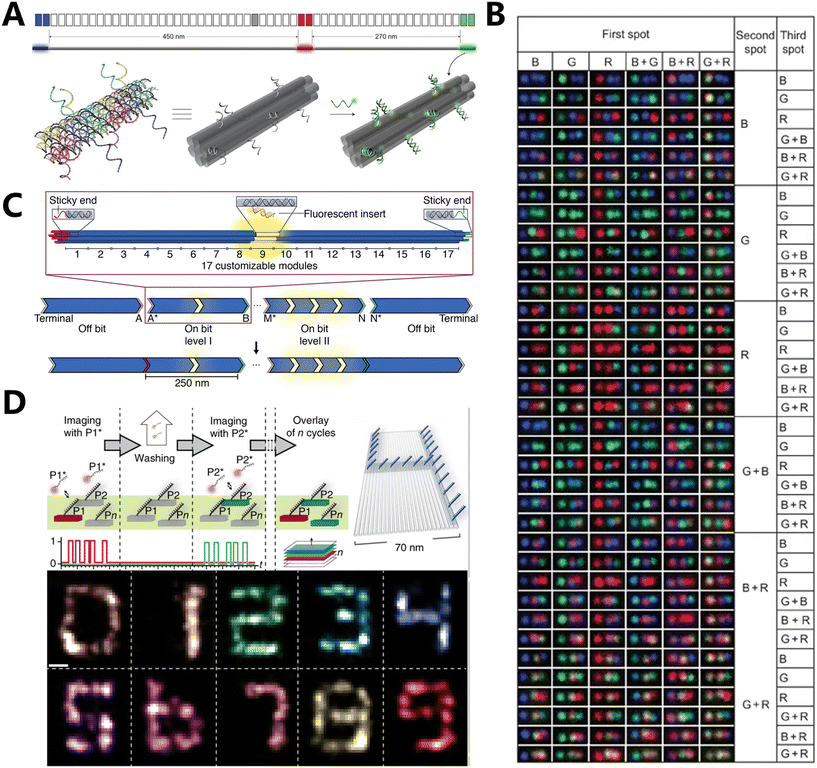 | ||
| Fig. 2 Fluorescence coding based on spatial geometry. (A) Three-dimensional illustrations to show the details of one fluorescently labelled zone.86 (B) Representative TIRF for 216 barcode species.86 (C) Design schematic for monochromatic fluorescent barcodes hierarchically assembled from modular DNA origami nanorods.87 (D) Exchange-PAINT schematic showing sequential imaging of multiple targets using imager strands labeled with the same fluorophore.88 | ||
Ganji et al. have recently employed DNA-PAINT along with the DNA nanostructure design to simultaneously detect and analyze individual dinucleotides using DNA nanotechnology. This allowed them to evaluate the energy associated with the stacking of bases at the single-molecule level.92 DNA-PAINT enables the direct measurement of the binding kinetics between fluorophore-labeled oligonucleotides and their corresponding strands of DNA origami nanostructures. The imager strands' binding kinetics are compared to the stacked and unstacked conformations of their terminal nucleotides in a single experiment using multiplexed imaging of patterned origami structures. The variation in individual base stacking energies resulted in a much wide range of stability in the residence duration during the examination of single-molecule data. Based on the observed stability of dwell duration, a distinctive grid pattern was created using five rectangular DNA origami structures: box, L, U, C, and H shapes. As a result, the structures were able to be easily differentiated in DNA-PAINT imaging, and an experiment was conducted to show that they are suitable for simultaneous super-resolution imaging. In addition to this, Jungmann et al. used a modified version of DNA-PAINT called Exchange-PAINT, which was combined with orthogonal DNA barcode imaging and wash cycles. This enabled them to achieve sequential target multiplexing and imaging through sequences, resulting in improved resolution. They referred to this technique as resolution enhancement by sequential imaging (RESI).93 This study confirms that RESI is capable of conducting intramolecular imaging in intact cells, thereby connecting the realms of super-resolution microscopy and structural biology research.
3.3 Fluorescence coding based on fluorescence properties
Although spatial geometric fluorescence encoding can enhance detection resolution and enable numerous detections using a reduced number of fluorescent moieties, it necessitates the fixing of fluorescent dyes in specific spatial places, which increases the complexity of design and synthesis. Furthermore, the encoding capacity remains restricted as a result of the object's dimensions, rendering it inappropriate for detection settings that need encoding with high capacity. It is also unsuitable for analyzing single-cell samples if the object's size is too large. Xie et al. employed the inherent characteristics of fluorescent groups, in particular fluorescence anisotropy (FA) and fluorescence polarization (FP),94 to encode and address these issues (Fig. 3A).95 The purpose of FA-based encoding in polystyrene microspheres was to encode and multiplex five DNAs by converting them into encoding signals. This was achieved by utilizing FRET, which causes a decrease in steady-state FA as the dye loading increases. Additionally, a combination of dyes with varying intrinsic FA values leads to different FA values at the same fluorescence intensity level. Meanwhile, Zhang's team has created a new type of biosensor called fluorescence anisotropy reporter (FLARE), which uses a technique called homochromatic FRET to measure the interaction between two fluorescent proteins. The biosensor detects changes in the conformation of molecular switches by analyzing the loss of polarization of emitted light by fluorescence polarization microscopy to observe the FA of the sensor. The biosensor is used to monitor specific biochemical activities (Fig. 3B).96 Research uses the monochromatic sensor's ability to integrate multiple signals and provides quantitative measurements utilizing various color variations. This enables the simultaneous imaging of three active reporter genes in a single cell, demonstrating multiplexed capabilities.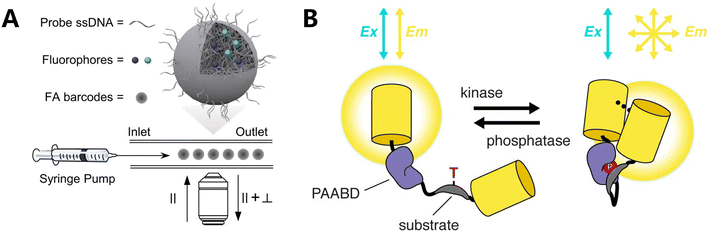 | ||
| Fig. 3 Fluorescence coding based on fluorescence properties. (A) Illustration of FA-encoded microspheres encapsulating lipophilic fluorescent labels inside PS microspheres for multiplexed biochemical detection on an FA imaging platform. The surface of the microspheres was covalently modified with DNA probes.95 (B) Schematic of a kinase activity FLARE.96 | ||
3.4 Fluorescence coding based on the time dimension
Although fluorescence-based encoding enables cell detection and precise size control, it is limited by issues such as excessive background signals, intensity-signal artifacts, and the possibility of false-positive results. Therefore, the advancement of technologies that allow for the regulation of luminescence sequences and the use of micro- and nanomaterials with extended fluorescence lifetimes has led to the emergence of new possibilities for high-throughput detection. Additionally, these materials enhance the detecting signal range.Reif et al. created a temporal reporter framework comprising fluorescent moiety reporter strands and DNA devices.100 Upon attaching the reporter strand to the DNA device, a momentary fluorescent spot becomes visible when observed using a microscope. In the study, the fluorescent intensity traces that are captured over a sufficient duration of time are referred to as temporal DNA barcodes. This is because the random intensity trajectories vary for each DNA device, enabling the distinction of individual molecules. Meanwhile, they introduce a method for barcoding single molecules that does not require amplification and relies on time-based signals (Fig. 4A).101 This is achieved by using reporter gene strands colored with dyes, which temporarily connect to specific nucleic acid strands and produce signals with different intensities over time. The DNA strands are designed to produce distinct temporal signals that can be used for molecular-scale identification.
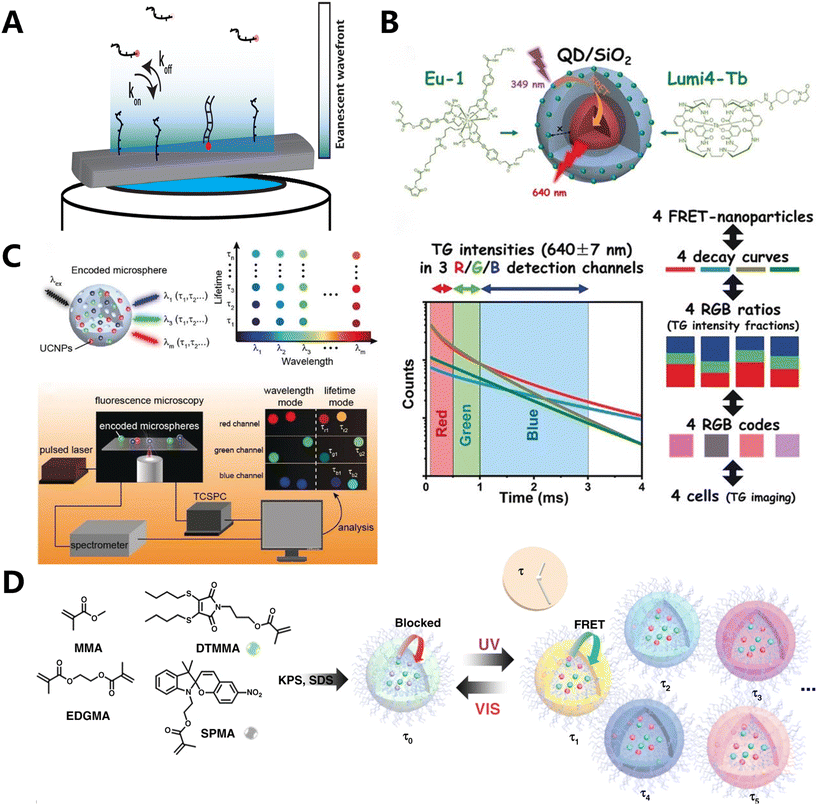 | ||
| Fig. 4 Fluorescence coding based on the time dimension. (A) Overview of the temporal barcoding framework.101 (B) A prototypical system for FRET lifetime encoding with individual nanoparticles.104 (C) Manipulating the luminescence emission color and decay lifetimes of UCNPs (designated as τλ-UCNPs) to create 2D barcodes.107 (D) Fabrication of the photo-switchable polymeric nanogels.108 | ||
Based on the aforementioned fluorescence characteristics, Hildebrandt et al. identified and characterized two distinct types of QDs coated with a layer of SiO2 and encased by two different rare earth complexes (Tb and Eu). The thickness of the SiO2 shell layer varied between 6 nm and 12 nm (Fig. 4B).104 Conventional organic dyes are often used because they are affordable, come in a wide range of options, and have a high quantum yield. However, their application is hampered by their broad luminescence spectrum and less persistent fluorescence, which makes them prone to photobleaching. QDs105,106 outperform traditional fluorescent dyes in several aspects, such as enhanced photostability, increased fluorescence intensity, and the ability to generate various fluorescence hues using the same wavelength by adjusting their size or composition. The text witnessed four separate photoluminescence (PL) decays due to FRET from Tb or Eu donors to QD acceptors. The discovery of these decays occurred in three distinct time-detection windows employing time-gated (TG) luminescence intensity monitoring. Consequently, the attachment of Eu or Tb complexes to QDs with two different SiO2 coating thicknesses resulted in the production of four distinct QD luminescence decays within three detection windows labeled as red (R), green (G), and blue (B). Each of these decays exhibited distinct variations in light intensity. Consequently, unique RGB codes were created for each of the four nanomaterials, taking into account the different levels of intensity seen in these three detection windows. The ability to identify and track several cells was demonstrated in this manner.
Despite the advancements made in utilizing QDs for fluorescence lifetime encoding, these materials still exhibit very brief lives ranging from 10 to 100 nanoseconds, which endow them with the potential for heightened signal interference. Consequently, there is a preference for rare earth metal compounds that exhibit luminescence durations ranging from microseconds to milliseconds and are encoded utilizing FRET based on UCNP. Zhang's team systematically design a methodology for manipulating the luminescence emission color and decay lifetimes of UCNPs (referred to as τλ-UCNPs) (Fig. 4C).107 By adjusting the shell thickness and controlling the FRET, the luminescence intensity of these particles can be modified. This allows for the encoding of the τλ-UCNPs based on their emission color and luminescence decay lifetime. Subsequently, they generated microspheres encoded with lifespan wavelength binary information using τλ-UCNPs and porous polystyrene (PS) microspheres. This unique color-lifetime encoding approach was used to evaluate nine subtypes of human papillomavirus (HPV) in patient samples. The encoding capacity of this strategy is improved by three orders of magnitude compared to the standard strategy. This demonstrates the strategy's capacity for efficient and rapid identification of biological samples.
Irrespective of the concentration of the material and the technique used for measurement, employing fluorescence lifetimes as readouts produces more precise and consistent results compared to conventional fluorescent barcodes. Nevertheless, the absence of methods to enable temporal and spatial control over the encoding process has limited the application of fluorescence lifetimes in this domain. Although there has been significant advancement in using semiconductor QDs and lanthanide complexes for encoding, achieving real-time adjustment of fluorescence lifetimes remains a challenge due to the poisonous nature of QDs.
O'Reilly et al. synthesized a two-component switching nanogel by utilizing spiropyran (SP) monomers and methacrylate maleimide dithiomaleimides (DTMs) (Fig. 4D).108 DTMs, being non-toxic and capable of modifying substituents, can be utilized to alter the fluorescence properties. By combining methyl methacrylate (MMA) as the hydrophobic matrix and ethylene glycol dimethacrylate (EGDMA) as the cross-linking agent, crosslinked nanogels are synthesized. These nanogels have different ratios of DTMMA and SPMA. Upon exposure to light, a photoisomerization-induced FRET process occurs. This enables the real-time adjustment of fluorescence lifespan by the manipulation of light. A variety of nanogels with a similar surface chemistry but wider dynamic lifetimes were used for encoding and counting different systems, after the successful implementation of dynamic photocontrol of fluorescence durations.
4. Application of fluorescence coding technology in RNA detection
RNA molecules have a low molecular weight, exhibit large variability in their abundance and play a crucial role in guiding protein synthesis within cells. Recent studies have shown that RNA expression differs between normal cells and those cells affected by various diseases, which makes it a valuable biomarker for diagnosing disorders. Studying RNA expression and control can contribute to tumor diagnosis and treatment by improving the understanding of different tumor mechanisms.4.1 Proportional type-based fluorescence coding in RNA detection
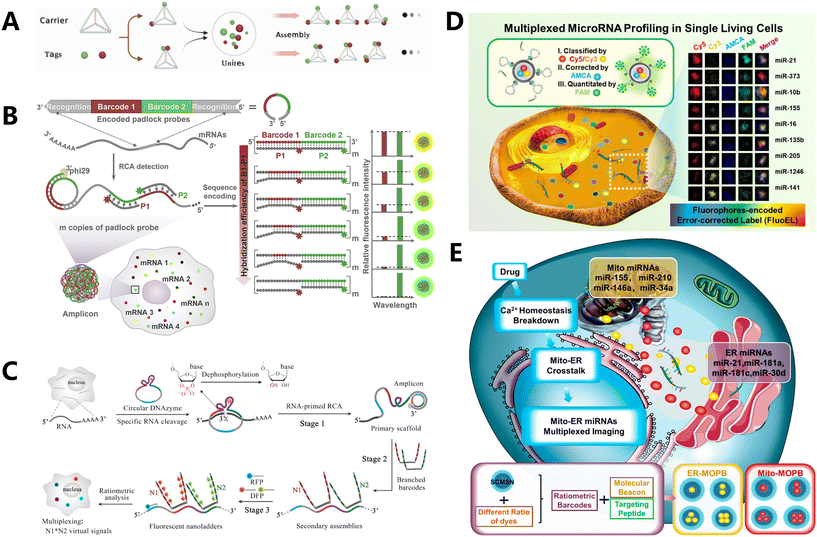 | ||
| Fig. 5 Proportional type-based fluorescence coding in RNA detection. (A) Construction procedure and in vitro multiplexed sensing application of Tetra-FICT.109 (B) Schematic illustration of sequence-encoded amplicon (SeqEA) for multiplexed imaging of individual RNAs in single cells.110 (C) Overview of hierarchical DNA branch assembly-encoded fluorescent nanoladders for single-cell transcript imaging.111 (D) Schematic of FluoELs for multiplexed miRNA detection.113 (E) Scheme of subcellular miRNAs' multiplexed detection using ratiometric barcodes in living cells.114 | ||
Most fluorescence-encoded microspheres currently available in the market range in size from 2 to 100 micrometers, which poses challenges in their detection within living cells. Furthermore, biomolecules are arranged in a random manner within the internal space of the cell. The complex environment within the living cell can easily lead to uneven refraction of fluorescence, resulting in a divergence of the detection signal. Our team developed a technique called fluorophore-encoded error-corrected labels (FluoELs), which is a form of fluorescence labeling coding with error correction. This technology allows for accurate examination of marker molecules in living cells (Fig. 5D).113 Living cellular miRNAs can be identified using FluoELs, which consist of cell-safe silica nanoparticles (MSN) as the structural framework and varying proportions of coding bi-fluorescent fluorophores (Cy5 and Cy3) contained in the core. In this study, a fixed amount of AMCA fluorescence was used to correct the quantitative bias caused by refractive fluctuations in living cells. After fabricating the fluorescent coding spheres, a 20 nm silica shell layer was added to the outer surface. This layer allowed for the loading of more molecular beacon (MB) detection probes with the right size and low cytotoxicity. Additionally, the structure prevented FRET between the signal recognition component of MB probes and the coding dye, enabling specific detection of miRNAs. This method allows for accurate quantification and in situ detection of nine specific miRNAs in breast cancer cells. It also permits the examination of variations within individual cells and the quantitative assessment of miRNA levels in living cells. Using this information, we proceeded to develop multiplexed organelles portrait barcodes (MOPB), an innovative fluorescence coding method that utilizes heterochromatic dye-encoded barcodes to identify miRNAs in different locations of organelles within living cells (Fig. 5E).114 The shell–core mesoporous silica nanoparticle (SFMSN) was loaded with varying proportions of AMCA, Cy3, and Cy5 dyes, ensuring good biosafety. The surface of these nanoparticles was then modified with organelle targeting peptide (TP) and MB detection probes, resulting in the formation of MOPBs. The paper presented a model of the endoplasmic reticulum (ER) and mitochondria (Mito). Four distinct ratios of Cy3/AMCA dyes were employed to create SFMSN-encoding spheres. These spheres were then utilized to generate ER-MOPB, which targets the ER, and Mito-MOPB, which targets the Mito. ER-TP and Mito-TP were combined with the corresponding MOPBs with MB probes that covalently bind and react with target miRNA. Intracellular miRNAs were quantified by measuring the fluorescence intensity of FAM on MB. This allows for the accurate identification of miRNAs from eight distinct organelles, facilitating the discovery of biomarkers within the subcellular milieu. Similarly, He et al. suggested utilizing apolipoproteins as carriers to create fluorescence-encoded microspheres. These microspheres were loaded with two different fluorophores (fluorescein and rhodamine B) in seven different amount ratios. The purpose was to detect and visualize seven specific mRNA targets in situ.115 Chen's team constructed the core of the sensor using UCNPs, which are nanoparticles that obtained three fluorescence-encoded probes by embedding Cy3 and Cy5 in a proportional manner into poly-L-lysine-packaged UCNPs (PLL-UCNPs). Each barcode in the sensor simultaneously packaged the same number of AMCA fluorophores for error correction.116 Finally, they acquired in situ pictures of the three separate miRNAs.
4.2 Fluorescence coding based on spatial geometry in RNA detection
Dai's group created a method for miRNA profiling that is extremely specific to sequences and allows for multiplexing (Fig. 6C).120 This method utilizes DNA nanostructures and repeating probe binding, which enables super-resolution geometric barcoding. In order to trap and render the relevant miRNAs immobile, they initially functionalized origami structures that were only partially complementary to the target miRNA sequences (13–17 nT) on a glass surface. The unpaired region that miRNAs capture functions as a binding site for DNA-PAINT technology imaging. This binding is detected and identified through the temporary pairing of complementary imaging strands that are labeled with fluorescent markers. The work employed a square lattice configuration, with a 20 nm gap between each of the four convenient markers and eight anchoring strands, to attain a high level of multiplexing capability. This configuration enabled the simultaneous identification and quantification of eight different miRNAs.
4.3 Fluorescence coding based on fluorescence properties in RNA detection
FA has great potential for imaging biomolecules and performing multiplexed analysis since it can efficiently distinguish fluorophores that have similar emission spectra. Regrettably, the extensive use of this substance in the field of biology and biotechnology has been impeded by its susceptibility to alterations in the environment.Li et al. achieved precise control over the FA level and stability of a fluorophore in complex situations by attaching fluorescent motifs at various locations on the DNA tetrahedron (Fig. 7A).121 First, the researchers created a DNA tetrahedral structure to control the FA properties of the fluorophore, known as FA DNA frameworks (FAFs). They devised three methods to position the fluorophore Alexa 647: first, by attaching it to the edge of the structure, second, by attaching it to the 4th or 11th nucleotide protruding from the 13th nucleotide on the structure, and third, by placing it at the 11th nucleotide on the structure, extending inward from the 8th nucleotide (Fig. 7B). They discovered that the shorter overhanging structure resulted in higher FA. Furthermore, the fluorophore positioned within the structure exhibited higher FA compared to when it was positioned outside the structure. This suggests that different positions on the structure can influence the movement of the fluorophore, leading to variations in FA. By merging alterations in FA with two distinct fluorescent hues of fluorophores, they achieved barcodes including eight unique pseudo-colors that can be used for miRNA analysis and cell labeling.
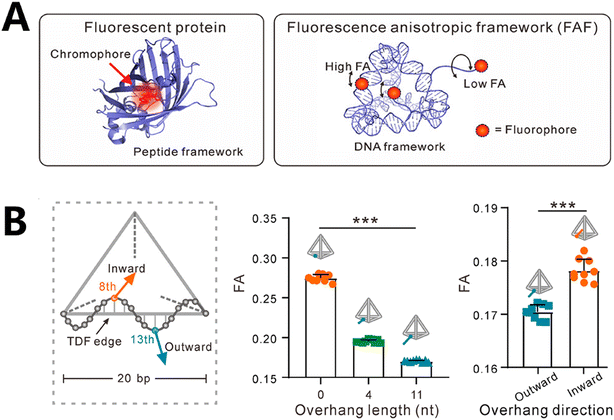 | ||
| Fig. 7 Fluorescence coding based on fluorescence properties in RNA detection. (A) Schematic of the peptide-like framework structure accommodating fluorophores with controllable FA features.121 (B) Regulation of FA by adjusting the fluorophores' placement on FAFs.121 | ||
4.4 Fluorescence coding based on the time dimension in RNA detection
Sun et al. employed fluorescent lifetime imaging of nanoflares to specifically detect endogenous mRNAs (Fig. 8C).123 The researchers generated nanoflares by forming AuNP–DNA complexes and attaching Cy5-modified oligonucleotides to gold nanoparticles, they observed a significant decrease in fluorescence lifespan. In the presence of the target mRNA, the recognition sequence on the AuNP would form a hybrid with the target, resulting in an extension of the fluorescence lifetime. The suitability of this approach for identifying endogenous mRNAs was assessed using MDA-MB-231 cells. Fluorescence lifetimes can be accurately determined by studying the fluorescence decay trails in cells, which allows for the detection of target mRNAs in cells. This presents novel possibilities for the detection of a wide range of supplementary biomolecules.
5. Summary and outlook
Fluorescence coding has emerged as a crucially pivotal technique in biomedical detection due to its versatility, sensitivity, and multiplexing capabilities. This methodology involves tagging molecules with fluorescent labels based on the fine control of fluorescence scaling type, spatial geometry, specific fluorescence properties, and time dimensions, allowing for their detection and quantification by fluorescence microscopy or spectroscopy. Based on its high throughput, high spatiotemporal resolution, and real-time monitoring, fluorescence coding has shown great potential in the tracing of pathophysiological microenvironment variations. Developing a fluorescence coding technique for RNA detection in anomalous microenvironments offers numerous advantages in the diagnosis and tracing of diverse pathophysiological processes that have revolutionized the field of molecular biology and biomedicine.Fluorescence coding allows for the simultaneous detection of multiple RNA targets within the same sample, attributing to encoding fluorescent motifs in multiple ways, the multiplexing capability of which is invaluable for studying complex biological processes. By employing different fluorescent labels with distinct emission spectra, different proportions, and spatial geometry, and using nanomaterials, researchers can detect multiple RNAs simultaneously, distinguish between various RNA species, and monitor their dynamics in real time, providing insights into cellular responses to stimuli or disease states.
However, there are several challenges in RNA detection by fluorescence coding techniques. Low signal intensity and high background noise in physiological environment can obscure the detection of low-abundance RNA targets, limiting the utility of fluorescence coding techniques for sensitive biomarker detection and quantification. To solve the problem, multiplexed fluorescence imaging strategy based on multicolor encoded hybridization chain reaction amplifiers can improve the sensitivity of fluorescence coding techniques, enabling the detection of low-abundance RNA targets with improved signal-to-noise ratios. Furthermore, fluorescence coding can be combined with advanced imaging techniques such as fluorescence in situ hybridization (FISH) or single-molecule imaging, to visualize RNA molecules with high spatial resolution. It allows researchers to study RNA localization, trafficking, and dynamics within subcellular compartments or complex tissue environments, providing mechanistic insights into RNA-mediated biological processes.
In summary, fluorescence coding has become an indispensable tool for RNA detection in biomedical research, offering sensitivity, multiplexing capabilities, and compatibility with high-throughput screening platforms. By harnessing the power of fluorescence coding technology, researchers can elucidate the roles of RNA molecules in health and disease conditions, paving the way for the development of novel diagnostics and therapeutic interventions.
Author contributions
Junren Wang: investigation, and writing – original draft, review and editing. Qin Xiang: writing – review and editing, and supervision. Haifeng Dong: project administration, funding acquisition, and writing – review and editing. Xueji Zhang: resources, project administration, and supervision.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
The work was supported by Special Foundation for State Major Research Program of China (Grant 2022YFB3207202), National Natural Science Foundation of China (Grant 22374102), Shenzhen Medical Research Fund (Grant D2301011), Guangdong Province Pearl River Team (2021ZT09C289), Shenzhen Key Laboratory for Nano-Biosensing Technology (ZDSYS20210112161400001). We thank Instrumental Analysis Center of Shenzhen University for the assistance with the Electron Microscope/Nuclear Magnetic Resonance spectroscope/Mass Spectrometer/Confocal Microscope/Small Animal Diagnostics Imaging/Material Characterization technical support.References
- R. Ren, S. L. Cai, X. N. Fang, X. Y. Wang, Z. Zhang, M. Damiani, C. Hudlerova, A. Rosa, J. Hope, N. J. Cook, P. Gorelkin, A. Erofeev, P. Novak, A. Badhan, M. Crone, P. Freemont, G. P. Taylor, L. H. Tang, C. Edwards, A. Shevchuk, P. Cherepanov, Z. F. Luo, W. H. Tan, Y. Korchev, A. P. Ivanov and J. B. Edel, Nat. Commun., 2023, 14, 7362 CrossRef CAS PubMed.
- T. R. Mempel, M. J. Pittet, K. Khazaie, W. Weninger, R. Weissleder, H. von Boehmer and U. H. von Andrian, Immunity, 2006, 25, 129–141 CrossRef CAS PubMed.
- A. Raj, P. van den Bogaard, S. A. Rifkin, A. van Oudenaarden and S. Tyagi, Nat. Methods, 2008, 5, 877–879 CrossRef CAS PubMed.
- W. Q. Zhang, B. J. Du, M. H. Gao and C. H. Tung, ACS Nano, 2021, 15, 16442–16451 CrossRef CAS PubMed.
- S. Wang, W. X. Ren, J. T. Hou, M. Won, J. An, X. Y. Chen, J. Shu and J. S. Kim, Chem. Soc. Rev., 2021, 50, 8887–8902 RSC.
- D. H. Li, R. S. Gamage, A. G. Oliver, N. L. Patel, S. Muhammad Usama, J. D. Kalen, M. J. Schnermann and B. D. Smith, Angew. Chem., Int. Ed., 2023, 62, e202305062 CrossRef CAS PubMed.
- D. Li, X. Q. Deng, Z. R. Xu, D. L. Wang, G. X. Xu, P. Y. Zhang, P. Qiu, W. X. Xie, D. Wang, B. Z. Tang and K. Wang, Adv. Funct. Mater., 2023, 33, 2303967 CrossRef CAS.
- B. Huang, M. Bates and X. W. Zhuang, Annu. Rev. Biochem., 2009, 78, 993–1016 CrossRef CAS PubMed.
- L. Wei, Z. Chen, L. Shi, R. Long, A. V. Anzalone, L. Zhang, F. Hu, R. Yuste, V. W. Cornish and W. Min, Nature, 2017, 544, 465–470 CrossRef CAS PubMed.
- F. H. Hu, C. Zeng, R. Long, Y. P. Miao, L. Wei, Q. Z. Xu and W. Min, Nat. Methods, 2018, 15, 194–200 CrossRef CAS PubMed.
- T. Konry, R. B. Hayman and D. R. Walt, Anal. Chem., 2009, 81, 5777–5782 CrossRef CAS PubMed.
- C. C. Li, H. Y. Chen, X. Luo, J. Hu and C. Y. Zhang, Chem. Sci., 2021, 12, 12407–12418 RSC.
- Z. Cao, L. Wang, R. Liu, S. Lin, F. Wu and J. Liu, Mater. Today Bio, 2022, 15, 100311 CrossRef CAS PubMed.
- G. J. Goodall and V. O. Wickramasinghe, Nat. Rev. Cancer, 2020, 21, 22–36 CrossRef PubMed.
- C. Buccitelli and M. Selbach, Nat. Rev. Genet., 2020, 21, 630–644 CrossRef CAS PubMed.
- S. Q. Huang, B. Sun, Z. P. Xiong, Y. Shu, H. H. Zhou, W. Zhang, J. Xiong and Q. Li, J. Exp. Clin. Cancer Res., 2018, 37, 1–11 CrossRef CAS PubMed.
- H. F. Dong, J. P. Lei, L. Ding, Y. Q. Wen, H. X. Ju and X. J. Zhang, Chem. Rev., 2013, 113, 6207–6233 CrossRef CAS PubMed.
- G. Faure, A. Y. Ogurtsov, S. A. Shabalina and E. V. Koonin, Nucleic Acids Res., 2016, 44, 10898–10911 CrossRef CAS PubMed.
- F. Q. Li, X. Q. Zhang, W. Ho, M. P. Tang, Z. Y. Li, L. Bu and X. Y. Xu, Nat. Commun., 2023, 14, 4223 CrossRef CAS PubMed.
- S. L. Cao, J. R. Wang, S. X. Ji, P. Yang, Y. Y. Dai, S. Guo, M. D. Montierth, J. P. Shen, X. Zhao, J. X. Chen, J. J. Lee, P. A. Guerrero, N. Spetsieris, N. Engedal, S. Taavitsainen, K. X. Yu, J. Livingstone, V. Bhandari, S. M. Hubert, N. C. Daw, P. A. Futreal, E. Efstathiou, B. Lim, A. Viale, J. J. Zhang, M. Nykter, B. A. Czerniak, P. H. Brown, C. Swanton, P. Msaouel, A. Maitra, S. Kopetz, P. Campbell, T. P. Speed, P. C. Boutros, H. T. Zhu, A. Urbanucci, J. Demeulemeester, P. Van Loo and W. Y. Wang, Nat. Biotechnol., 2022, 40, 1624–1633 CrossRef CAS PubMed.
- N. Y. Wang, J. Zheng, Z. Chen, Y. Liu, B. Dura, M. Kwak, J. Xavier Ferrucio, Y. C. Lu, M. M. Zhang, C. Roden, J. J. Cheng, D. S. Krause, Y. Ding, R. Fan and J. Lu, Nat. Commun., 2019, 10, 95 CrossRef PubMed.
- R. L. Liu, C. L. Liu, X. Z. He, P. Sun, B. Zhang, H. R. Yang, W. Y. Shi and Q. G. Ruan, Nat. Commun., 2022, 13, 3545 CrossRef CAS PubMed.
- R. Bhayadia, K. Krowiorz, N. Haetscher, R. Jammal, S. Emmrich, A. Obulkasim, J. Fiedler, A. Schwarzer, A. Rouhi, M. Heuser, S. Wingert, S. Bothur, K. Döhner, T. Mätzig, M. Ng, D. Reinhardt, H. Döhner, C. M. Zwaan, M. V. D. H. Eibrink, D. Heckl, M. Fornerod, T. Thum, R. K. Humphries, M. A. Rieger, F. Kuchenbauer and J.-H. Klusmann, J. Clin. Oncol., 2018, 36, 1007–1016 CrossRef CAS PubMed.
- A. Dhawan, J. G. Scott, A. L. Harris and F. M. Buffa, Nat. Commun., 2018, 9, 5228 CrossRef CAS PubMed.
- M. N. Islam, M. K. Masud, M. H. Haque, M. S. A. Hossain, Y. Yamauchi, N. T. Nguyen and M. J. A. Shiddiky, Small Methods, 2017, 1, 1700131 CrossRef.
- C. G. Sheng, J. Zhao, Z. H. Di, Y. Y. Huang, Y. L. Zhao and L. L. Li, Nat. Biomed. Eng., 2022, 6, 1074–1084 CrossRef CAS PubMed.
- X. C. Lu, K. Y. S. Kong and P. J. Unrau, Chem. Soc. Rev., 2023, 52, 4071–4098 RSC.
- T. Jet, G. Gines, Y. Rondelez and V. Taly, Chem. Soc. Rev., 2021, 50, 4141–4161 RSC.
- P. H. Ma, X. X. Jia, Y. Y. He, J. H. Tao, Q. Wang and C.-I. Wei, Trends Food Sci. Technol., 2024, 143, 104310 CrossRef CAS.
- S. Zeng, X. S. Liu, Y. S. Kafuti, H. Kim, J. Y. Wang, X. J. Peng, H. D. Li and J. Yoon, Chem. Soc. Rev., 2023, 52, 5607–5651 RSC.
- M. Hirano, R. Ando, S. Shimozono, M. Sugiyama, N. Takeda, H. Kurokawa, R. Deguchi, K. Endo, K. Haga, R. Takai-Todaka, S. Inaura, Y. Matsumura, H. Hama, Y. Okada, T. Fujiwara, T. Morimoto, K. Katayama and A. Miyawaki, Nat. Biotechnol., 2022, 40, 1132–1142 CrossRef CAS PubMed.
- B. C. Campbell, M. G. Paez-Segala, L. L. Looger, G. A. Petsko and C. F. Liu, Nat. Methods, 2022, 19, 1612–1621 CrossRef CAS PubMed.
- W. E. Yasso, Sedimentology, 1966, 6, 287–301 CrossRef.
- Y. Zhang, Q. S. Wang, T. Ma, D. W. Zhu, T. J. Liu and F. Lv, J. Controlled Release, 2020, 328, 127–140 CrossRef CAS PubMed.
- X. Gao, Y. Cui, R. M. Levenson, L. W. Chung and S. Nie, Nat. Biotechnol., 2004, 22, 969–976 CrossRef CAS PubMed.
- S. Shikha, X. Zheng and Y. Zhang, Nano-Micro Lett., 2018, 10, 31 CrossRef PubMed.
- A. M. Milosevic, L. Rodriguez-Lorenzo, S. Balog, C. A. Monnier, A. Petri-Fink and B. Rothen-Rutishauser, Angew. Chem., Int. Ed., 2017, 56, 13382–13386 CrossRef CAS PubMed.
- Y. J. Xie, Z. Z. Tong, T. L. Xia, J. C. Worch, J. Y. Rho, A. P. Dove and R. K. O'Reilly, Adv. Mater., 2023, 36, 2308154 CrossRef PubMed.
- Z. C. Liang, C. L. Hao, C. Chen, W. Ma, M. Z. Sun, L. G. Xu, C. L. Xu and H. Kuang, Adv. Mater., 2021, 34, 2107449 CrossRef PubMed.
- L. Rodriguez Lorenzo, K. Fytianos, F. Blank, C. von Garnier, B. Rothen Rutishauser and A. Petri Fink, Small, 2014, 10, 1341–1350 CrossRef CAS PubMed.
- T. Konry, R. B. Hayman and D. R. Walt, Anal. Chem., 2009, 81, 5777–5782 CrossRef CAS PubMed.
- J. Z. Song, Q. Yang, F. T. Lv, L. B. Liu and S. Wang, ACS Appl. Mater. Interfaces, 2012, 4, 2885–2890 CrossRef CAS PubMed.
- Y. K. Leng, W. J. Wu, L. Li, K. Lin, K. Sun, X. Y. Chen and W. W. Li, Adv. Funct. Mater., 2016, 26, 7581–7589 CrossRef CAS.
- L. Martín Banderas, A. Rodríguez Gil, Á. Cebolla, S. Chávez, T. Berdún Álvarez, J. M. Fernandez Garcia, M. Flores Mosquera and A. M. Gañán Calvo, Adv. Mater., 2006, 18, 559–564 CrossRef.
- M. Liong, J. Lu, M. Kovochich, T. Xia, S. G. Ruehm, A. E. Nel, F. Tamanoi and J. I. Zink, ACS Nano, 2008, 2, 889–896 CrossRef CAS PubMed.
- H. Y. Xia, Y. G. Ding, J. J. Gong, A. Lilienkampf, K. Xie and M. Bradley, ACS Appl. Mater. Interfaces, 2022, 14, 27107–27117 CrossRef CAS PubMed.
- J. Y. Zhang, S. Y. Qin, S. N. Zhang, C. Sun, Y. X. Ren, L. Y. Zhang, J. L. Liu, J. M. Xiao, W. Hu, H. Yang and D. K. Yang, Adv. Mater., 2023, 36, 2305872 CrossRef PubMed.
- H. W. Ji, H. H. Chen, T. Z. Mao, L. F. Zhao, K. Zhang, M. J. Yang, X. B. Zhou, Q. Wang, L. Wu and Y. L. Qin, Adv. Opt. Mater., 2023, 11, 2300991 CrossRef CAS.
- M. H. Lee, J. S. Kim and J. L. Sessler, Chem. Soc. Rev., 2015, 44, 4185–4191 RSC.
- S.-H. Park, N. Kwon, J.-H. Lee, J. Yoon and I. Shin, Chem. Soc. Rev., 2020, 49, 143–179 RSC.
- J. Q. Chang, Y. Zhang, Y. Li, Z. W. Han, F. Tian, C. Liu, Q. Feng, Y. G. Wang, J. S. Sun and L. Zhang, Small Methods, 2020, 5, 2001047 CrossRef PubMed.
- N. Makukhin, V. Tretyachenko, J. Moskovitz and J. Míšek, Angew. Chem., Int. Ed., 2016, 55, 12727–12730 CrossRef CAS PubMed.
- N. C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- J. D. Watson and F. H. C. Crick, Nature, 1953, 171, 737–738 CrossRef CAS PubMed.
- Y. Hu and C. M. Niemeyer, Adv. Mater., 2019, 31, e1806294 CrossRef PubMed.
- C. M. Niemeyer, Curr. Opin. Chem. Biol., 2000, 4, 609–618 CrossRef CAS PubMed.
- R. Meyer, S. Giselbrecht, B. E. Rapp, M. Hirtz and C. M. Niemeyer, Curr. Opin. Chem. Biol., 2014, 18, 8–15 CrossRef CAS PubMed.
- H. Pei, R. Sha, X. Wang, M. Zheng, C. Fan, J. W. Canary and N. C. Seeman, J. Am. Chem. Soc., 2019, 141, 11923–11928 CrossRef CAS PubMed.
- D. Huang, K. Patel, S. Perez-Garrido, J. F. Marshall and M. Palma, ACS Nano, 2019, 13, 728–736 CrossRef CAS PubMed.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2019, 119, 6459–6506 CrossRef CAS PubMed.
- Y. Li, Y. T. Cu and D. Luo, Nat. Biotechnol., 2005, 23, 885–889 CrossRef CAS PubMed.
- S. H. Um, J. B. Lee, S. Y. Kwon, Y. Li and D. Luo, Nat. Protoc., 2006, 1, 995–1000 CrossRef CAS PubMed.
- J. B. Lee, Y. H. Roh, S. H. Um, H. Funabashi, W. Cheng, J. J. Cha, P. Kiatwuthinon, D. A. Muller and D. Luo, Nat. Nanotechnol., 2009, 4, 430–436 CrossRef CAS PubMed.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- L. M. Smith, Nature, 2006, 440, 283–284 CrossRef CAS PubMed.
- A. V. Pinheiro, D. Han, W. M. Shih and H. Yan, Nat. Nanotechnol., 2011, 6, 763–772 CrossRef CAS PubMed.
- Y. Ke, L. L. Ong, W. M. Shih and P. Yin, Science, 2012, 338, 1177–1183 CrossRef CAS PubMed.
- B. Wei, M. Dai and P. Yin, Nature, 2012, 485, 623–626 CrossRef CAS PubMed.
- S. Li, Q. Jiang, S. Liu, Y. Zhang, Y. Tian, C. Song, J. Wang, Y. Zou, G. J. Anderson, J. Y. Han, Y. Chang, Y. Liu, C. Zhang, L. Chen, G. Zhou, G. Nie, H. Yan, B. Ding and Y. Zhao, Nat. Biotechnol., 2018, 36, 258–264 CrossRef CAS.
- J. B. Woehrstein, M. T. Strauss, L. L. Ong, B. Wei, D. Y. Zhang, R. Jungmann and P. Yin, Sci. Adv., 2017, 3, e1602128 CrossRef PubMed.
- S. Niekamp, N. Stuurman and R. D. Vale, Nat. Methods, 2020, 17, 437–441 CrossRef CAS PubMed.
- J. Li, J. Dai, S. Jiang, M. Xie, T. Zhai, L. Guo, S. Cao, S. Xing, Z. Qu, Y. Zhao, F. Wang, Y. Yang, L. Liu, X. Zuo, L. Wang, H. Yan and C. Fan, Nat. Commun., 2020, 11, 2185 CrossRef CAS PubMed.
- S. Han, J. S. Lee and J. B. Lee, Nanoscale, 2017, 9, 14094–14102 RSC.
- Y. Zhang, L. Chen, K. Hsieh and T. H. Wang, Anal. Chem., 2018, 90, 12180–12186 CrossRef CAS PubMed.
- C. Ramakers, J. M. Ruijter, R. H. Deprez and A. F. Moorman, Neurosci. Lett., 2003, 339, 62–66 CrossRef CAS PubMed.
- J. S. Park, T. Pisanic, Y. Zhang and T. H. Wang, Anal. Chem., 2021, 93, 2351–2358 CrossRef CAS PubMed.
- M. Y. Han, X. H. Gao, J. Z. Su and S. M. Nie, Nat. Biotechnol., 2001, 19, 631–635 CrossRef CAS PubMed.
- Y. Wang, C. Chen, J. He, Y. M. Cao, X. X. Fang, X. M. Chi, J. W. Yi, J. C. Wu, Q. S. Guo, H. Masoomi, C. Z. Wu, J. Ye, H. C. Gu and H. Xu, Small, 2021, 17, 2100315 CrossRef CAS PubMed.
- M. Xiao, K. Zou, L. Li, L. Wang, Y. Tian, C. Fan and H. Pei, Angew. Chem., Int. Ed., 2019, 58, 15448–15454 CrossRef CAS PubMed.
- B. Andreiuk, A. Reisch, M. Lindecker, G. Follain, N. Peyriéras, J. G. Goetz and A. S. Klymchenko, Small, 2017, 13, 1701582 CrossRef PubMed.
- B. J. Zheng, Y. Q. Qin, Y. Q. Xiang, A. Guo, Y. Y. Chen and Y. G. Hu, Anal. Chem., 2023, 95, 6542–6549 CrossRef CAS PubMed.
- M. G. Malle, P. M. G. Loffler, S. S. Bohr, M. B. Sletfjerding, N. A. Risgaard, S. B. Jensen, M. Zhang, P. Hedegard, S. Vogel and N. S. Hatzakis, Nat. Chem., 2022, 14, 558–565 CrossRef CAS PubMed.
- M. J. Dejneka, A. Streltsov, S. Pal, A. G. Frutos, C. L. Powell, K. Yost, P. K. Yuen, U. Müller and J. Lahiri, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 389–393 CrossRef CAS.
- Y. H. Zhang, L. X. Zhang, R. R. Deng, J. Tian, Y. Zong, D. Y. Jin and X. G. Liu, J. Am. Chem. Soc., 2014, 136, 4893–4896 CrossRef CAS PubMed.
- X. Zeng, S. K. Vanga, E. T. Poh, Y. Shi, C. H. Sow, A. A. Bettiol and X. G. Liu, Adv. Opt. Mater., 2020, 8, 2001168 CrossRef CAS.
- C. Lin, R. Jungmann, A. M. Leifer, C. Li, D. Levner, G. M. Church, W. M. Shih and P. Yin, Nat. Chem., 2012, 4, 832–839 CrossRef CAS PubMed.
- V. Pan, W. Wang, I. Heaven, T. Bai, Y. Cheng, C. Chen, Y. Ke and B. Wei, ACS Nano, 2021, 15, 15892–15901 CrossRef CAS PubMed.
- R. Jungmann, M. S. Avendano, J. B. Woehrstein, M. Dai, W. M. Shih and P. Yin, Nat. Methods, 2014, 11, 313–318 CrossRef CAS PubMed.
- D. A. Helmerich, M. Budiarta, D. Taban, S. Doose, G. Beliu and M. Sauer, Adv. Mater., 2023, 36, 2310104 CrossRef PubMed.
- F. Schueder, J. Lara-Gutierrez, B. J. Beliveau, S. K. Saka, H. M. Sasaki, J. B. Woehrstein, M. T. Strauss, H. Grabmayr, P. Yin and R. Jungmann, Nat. Commun., 2017, 8, 2090 CrossRef PubMed.
- R. Iinuma, Y. Ke, R. Jungmann, T. Schlichthaerle, J. B. Woehrstein and P. Yin, Science, 2014, 344, 65–69 CrossRef CAS PubMed.
- A. Banerjee, M. Anand, S. Kalita and M. Ganji, Nat. Nanotechnol., 2023, 18, 1474–1482 CrossRef CAS PubMed.
- S. C. M. Reinhardt, L. A. Masullo, I. Baudrexel, P. R. Steen, R. Kowalewski, A. S. Eklund, S. Strauss, E. M. Unterauer, T. Schlichthaerle, M. T. Strauss, C. Klein and R. Jungmann, Nature, 2023, 617, 711–716 CrossRef CAS PubMed.
- C. Vinegoni, P. F. Feruglio, I. Gryczynski, R. Mazitschek and R. Weissleder, Adv. Drug Delivery Rev., 2019, 151, 262–288 CrossRef PubMed.
- W. Y. Huang, Y. Cheng, J. Y. Zhai, Y. M. Qin, W. A. Zhang and X. J. Xie, Analyst, 2023, 148, 4406–4413 RSC.
- B. L. Ross, B. Tenner, M. L. Markwardt, A. Zviman, G. Shi, J. P. Kerr, N. E. Snell, J. J. McFarland, J. R. Mauban, C. W. Ward, M. A. Rizzo and J. Zhang, eLife, 2018, 7, e35458 CrossRef PubMed.
- X. Shaulli, R. Rivas-Barbosa, M. J. Bergman, C. Zhang, N. Gnan, F. Scheffold and E. Zaccarelli, ACS Nano, 2023, 17, 2067–2078 CrossRef CAS PubMed.
- W. Y. Wu, S. H. Luo, C. Y. Fan, T. J. Yang, S. W. Zhang, W. X. Meng, T. Xu, W. Ji and L. S. Gu, Light: Sci. Appl., 2023, 12, 9 CrossRef CAS PubMed.
- E. Perego, S. Zappone, F. Castagnetti, D. Mariani, E. Vitiello, J. Rupert, E. Zacco, G. G. Tartaglia, I. Bozzoni, E. Slenders and G. Vicidomini, Nat. Commun., 2023, 14, 8224 CrossRef CAS PubMed.
- S. Shah, A. K. Dubey and J. Reif, ACS Synth. Biol., 2019, 8, 1100–1111 CrossRef CAS PubMed.
- S. Shah, A. K. Dubey and J. Reif, Nano Lett., 2019, 19, 2668–2673 CrossRef CAS PubMed.
- F. Cole, J. Zähringer, J. Bohlen, T. Schröder, F. Steiner, M. Pfeiffer, P. Schüler, F. D. Stefani and P. Tinnefeld, Nat. Photonics, 2024, 1–7, DOI:10.1038/s41566-024-01384-4.
- A. Martin and P. Rivera-Fuentes, Nat. Chem., 2023, 16, 28–35 CrossRef PubMed.
- C. Chen, L. Ao, Y. T. Wu, V. Cifliku, M. Cardoso Dos Santos, E. Bourrier, M. Delbianco, D. Parker, J. M. Zwier, L. Huang and N. Hildebrandt, Angew. Chem., Int. Ed., 2018, 57, 13686–13690 CrossRef CAS PubMed.
- C. M. Green, J. Spangler, K. Susumu, D. A. Stenger, I. L. Medintz and S. A. Díaz, ACS Nano, 2022, 16, 20693–20704 CrossRef CAS PubMed.
- Y. X. Ma, G. B. Mao, W. R. Huang, G. Q. Wu, W. Yin, X. H. Ji, Z. S. Deng, Z. M. Cai, X.-E. Zhang, Z. He and Z. Q. Cui, J. Am. Chem. Soc., 2019, 141, 13454–13458 CrossRef CAS PubMed.
- L. Zhou, Y. Fan, R. Wang, X. Li, L. Fan and F. Zhang, Angew. Chem., Int. Ed., 2018, 57, 12824–12829 CrossRef CAS PubMed.
- Y. Xie, M. C. Arno, J. T. Husband, M. Torrent-Sucarrat and R. K. O'Reilly, Nat. Commun., 2020, 11, 2460 CrossRef CAS PubMed.
- X. S. Zhao, Y. Xu, Z. T. Chen, C. R. Tang and X. Q. Mi, Biosens. Bioelectron., 2024, 248, 115994 CrossRef CAS PubMed.
- R. J. Deng, K. X. Zhang, L. D. Wang, X. J. Ren, Y. P. Sun and J. H. Li, Chem, 2018, 4, 1373–1386 CAS.
- X. Cao, F. Chen, J. Xue, Y. Zhao, M. Bai and Y. Zhao, Nucleic Acids Res., 2023, 51, e13 CrossRef CAS PubMed.
- X. J. Qu, H. J. Jin, Y. Q. Liu and Q. J. Sun, Anal. Chem., 2018, 90, 3482–3489 CrossRef CAS PubMed.
- W. Wei, W. Dai, F. Yang, H. Lu, K. Zhang, Y. Xing, X. Meng, L. Zhou, Y. Zhang, Q. Yang, Y. Cheng and H. Dong, Angew. Chem., Int. Ed., 2022, 61, e202116909 CrossRef CAS PubMed.
- W. Wei, H. Lu, W. Dai, X. Zheng and H. Dong, ACS Nano, 2022, 16, 20329–20339 CrossRef CAS PubMed.
- Y. Wang, Y. Xiong, Y. Duan, K. Shi, C. Su, L. Ding, J. Wang and L. He, ACS Sens., 2023, 8, 1918–1928 CrossRef CAS PubMed.
- Y. Wen, W. Y. Liu, J. H. Wang, Y. L. Yu and S. Chen, Anal. Chem., 2023, 95, 12152–12160 CrossRef CAS PubMed.
- J. Lee, P. W. Bisso, R. L. Srinivas, J. J. Kim, A. J. Swiston and P. S. Doyle, Nat. Methods, 2014, 13, 524–529 CrossRef CAS PubMed.
- S. Jung, J. Kim, D. J. Lee, E. H. Oh, H. Lim, K. P. Kim, N. Choi, T. S. Kim and S. K. Kim, Sci. Rep., 2016, 6, 22975 CrossRef CAS PubMed.
- N. Juthani and P. S. Doyle, Analyst, 2020, 145, 5134–5140 RSC.
- W. Xu, P. Yin and M. Dai, Angew. Chem., Int. Ed., 2018, 57, 14075–14079 CrossRef CAS PubMed.
- Q. Huang, B. Chen, J. Shen, L. Liu, J. Li, J. Shi, Q. Li, X. Zuo, L. Wang, C. Fan and J. Li, J. Am. Chem. Soc., 2021, 143, 10735–10742 CrossRef CAS PubMed.
- O. K. Wade, J. B. Woehrstein, P. C. Nickels, S. Strauss, F. Stehr, J. Stein, F. Schueder, M. T. Strauss, M. Ganji, J. Schnitzbauer, H. Grabmayr, P. Yin, P. Schwille and R. Jungmann, Nano Lett., 2019, 19, 2641–2646 CrossRef CAS PubMed.
- J. Shi, M. Zhou, A. H. Gong, Q. J. Li, Q. Wu, G. J. Cheng, M. Y. Yang and Y. C. Sun, Anal. Chem., 2016, 88, 1979–1983 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |