Open Access Article
Open Access ArticleElectrochemical immunomagnetic assay for interleukin-6 detection in human plasma
Grace
Buckey
 ,
Olivia E.
Owens
,
Olivia E.
Owens
 ,
Hannah A.
Richards
,
Hannah A.
Richards
 and
David E.
Cliffel
and
David E.
Cliffel
 *
*
Department of Chemistry, Vanderbilt University, 7330 Stevenson Center, VU Station B351822, Nashville, TN 37235-1822, USA. E-mail: d.cliffel@vanderbilt.edu
First published on 8th May 2024
Abstract
An electrochemical immunoassay for interleukin-6 (IL-6) was developed based on IL-6 capture using magnetic beads and electrochemical signal production using horseradish peroxidase/tetramethylbenzidine. We achieved IL-6 detection from the 50–1000 pg mL−1 range, which is a physiologically relevant IL-6 range for a variety of biological systems. The sandwich assay performed well in phosphate buffered solution as well as in cellular media and human plasma spiked with IL-6, and decreased time to IL-6 concentration readout to approximately one hour. There is also future potential to apply this assay to real-time point-of-care human disease diagnostics.
Interleukin-6 (IL-6) is a multifunctional cytokine that plays a significant role in the immune system and inflammation. It is typically produced by monocytes and macrophages in response to an injury or an infection,1 but when abnormally elevated it becomes problematic in many situations. IL-6 is responsible for recruiting the enzymes that degrade the extracellular matrix of the fetal membrane, leading to preterm birth (PTB).2,3 By detecting IL-6 in models of PTB, we hope to provide further insight into the mechanisms underlying PTB. IL-6 is also involved in musculoskeletal infection. The damage caused by infection generates measurable increases of IL-6 in blood and can indicate musculoskeletal infection severity.4 Short response time of the sensor is especially important in this application, as timely diagnosis and treatment can significantly improve patient outcomes. Endometriosis is a chronic inflammatory disease that affects millions worldwide and yet remains poorly understood.5,6 Numerous studies have examined the role of IL-6 in endometriosis, reporting relationships of IL-6 to infertility5 and growth regulation of endometriotic lesions4 or using IL-6 as a biomarker for endometriosis diagnosis.7 Overall, the detection of IL-6 in endometriosis studies remains vitally important in better understanding endometriosis. Lastly, IL-6 can also be used as an indicator of COVID-19 severity and survival, and it is especially important to detect it early in disease progression for fast diagnosis of patients with a higher risk of disease deterioration.8–10
Enzyme-linked immunosorbent assays (ELISA) are the standard technique for IL-6 detection. They achieve low detection limits but are also expensive, time-consuming, and require trained personnel.1 Traditional ELISAs take greater than three hours to run and conversations with Vanderbilt medical colleagues suggest that typical hospital turnaround times for IL-6 concentration readings span 1–2 days. In comparison, electrochemical immunoassays achieve excellent specificity, sensitivity, wide linear ranges, quick readout, and low cost due to strong antibody–antigen interactions and the electrochemical instrumentation.11 Antibodies are used to capture analyte molecules12 while enzymes are used as labels combined with a substrate that the enzyme converts into an electrochemically detectable product. These enzyme labels can be attached to a secondary antibody or the analyte in order to form sandwich or competitive assays, respectively. Electrochemical detection of the enzymatic product correlates to the amount of enzyme in solution and also the amount of analyte. This further improves the detection limits and sensitivity.
In this work, poly-horseradish peroxidase (poly-HRP) labels the secondary antibody to form an antibody sandwich with the target analyte. As a result, the amount of poly-HRP is directly proportional to the amount of analyte present. Similar reports have used enzymatic labels such poly-HRP, alkaline phosphatase (ALP), and glucose oxidase (GO). Poly-HRP has served as a signal amplifying enzymatic label for an electrochemical magnetoimmunosensor to detect IL-6 levels present in saliva and urine samples.13 ALP has been used to trigger Prussian blue nanoparticle formation, correlating to the electrochemical redout for zearalenone detection.14 More recently, GO was used as an electrochemical label with scanning electrochemical microscopy to identify human growth hormone.15 The use of nanoparticles has been investigated as an alternative to enzymatic labeling. Some examples include using platinum nanoparticles to catalyze chemical signal transduction between palladium nanowires and H2 to develop a gas-based electrochemical biosensor16 and using gold nanoparticles with ferricyanide to detect SARS-CoV-2 nucleocapsid protein in a smartphone based electrochemical immunoassay.17
Horse radish peroxidase (HRP) is a commonly used enzyme in electrochemical immunoassays due to its reactivity with a large number of substrates.18–22 The substrate is generally H2O2 combined with an electrochemical mediator to improve conversion efficiency and examples include hydroquinone,19o-phenylenediamine,20 or tetramethylbenzidine (TMB).18,22,23 TMB is a particularly common choice for HRP substrate, due to its superior performance and sensitivity compared to other HRP substrates and its photothermal and colorimetric nature.11 In the presence of hydrogen peroxide, HRP oxidizes TMB to TMB diimine which can be electrochemically detected via reduction of TMB diimine back to TMB. Previous work suggests the use of TMB for a photothermal-thermoelectric coupled immunoassay for the electrochemical detection of temperature changes from oxidized TMB.24 Likewise, a TMB system was employed to initiate a cascade of enzymatic-like reactions for a smartphone based neural network-assisted multimodal immunoassay for acute myocardial monitoring.25 The highly reproducible TMB system, in combination with nanomaterials, makes the HRP/TMB enzyme-substrate complex desirable for the development of the electrochemical immunomagnetic assay in the present study.
Previous groups have developed IL-6 sensors using HRP. However, literature examples range from two hours19,20 to two days.18 While one report was able to achieve more rapid IL-6 detection, the linear range of the sensor is not mentioned,22 which is a vital determinant of sensor performance for the intended use. Finally, another group reports a sensor fabrication method and verifies proof of concept using IL-6 and HRP/TMB23 but this sensor had a lengthy and complex sensor fabrication procedure while we desired a straightforward sensor fabrication.
Nanomaterials have been combined with electrochemical immunoassays in order to improve their performance.18 The use of nanomaterials such as nanoparticles, nanowires, graphene, and carbon nanotubes, can enhance the surface area of the sensor which decreases detection limits, allowing for sensitive detection of low-level biomarkers. Magnetic nanoparticles are a commonly used nanomaterial, also used to increase the sensor surface area. In addition to being easy to use, magnetic nanoparticles separate complex samples from the electrochemical detection which helps to minimize biofouling.12
Magnetic beads exhibit proven advantages for electrochemical immunoassays. Recent investigations demonstrate the use of magnetic beads for the development of a nucleic acid-based magnetic potentiometric aptasensing platform to indirectly detect prostate-specific antigen.26 Similarly, for carcinoembryonic antigen detection, magnetic beads were used to fabricate a photoelectrochemical immunosensing probe.27 Magnetic beads have even been integrated into disposable electrochemical immunoassays for use in clinical settings.28 They are a notable tool for the versatile design and fabrication of many electrochemical immunoassays.
In this work, we combine magnetic beads with the HRP/TMB system in order to detect IL-6 in the 50–1000 pg mL−1 range, which is physiologically relevant IL-6 range for a variety of biological systems. We adapted the assay protocol from Del Rio et al.23 for our final assay design, but used magnetic beads for capture antibody attachment in order to simplify the sensor fabrication and improve the readout time. Due to the complex environment of each of the biological systems of interest, we tested our IL-6 sensor in a variety of media. Our sandwich assay performed well in PBS as well as in complex solutions of cellular media and plasma spiked with IL-6. Overall, our sensor demonstrated good performance in biologically complex environments and shortened the time required for IL-6 concentration readings.
Experimental
Materials
Recombinant Human Interleukin-6 (IL-6, Mw = 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 718 g mol−1), was purchased (Recombinant Human, Carrier Free) from R&D Systems. Recombinant Human Matrix Metalloproteinase-9 (MMP-9, Mw = 93 kDa) and Recombinant Human Matrix Metalloproteinase-3 (MMP-3, Mw = 54 kDa) were purchased from Sigma Aldrich, USA. KCl (Certified ACS), MgCl2, NaCl, Trizma base, 10× phosphate buffer solution, IL-6 Human matched antibody pair (CHC1263), Dynabeads antibody coupling kit (14311D), and Dulbecco's Modified Eagle Media (DMEM) were obtained from Thermo Fisher Scientific. The following chemicals were used as received without additional purification and were obtained from Sigma Aldrich (St. Louis, MO, USA): 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate system for ELISA (T0440). Normal patient pooled plasma was obtained from Jonathan Schoenecker's lab and spiked with IL-6 for assay calibration in human plasma. All experiments were performed in accordance with the NIH and Vanderbilt IRB Guidelines, and approved by the ethics committee at Vanderbilt University. De-identified human plasma was obtained with informed consent from human donors.
718 g mol−1), was purchased (Recombinant Human, Carrier Free) from R&D Systems. Recombinant Human Matrix Metalloproteinase-9 (MMP-9, Mw = 93 kDa) and Recombinant Human Matrix Metalloproteinase-3 (MMP-3, Mw = 54 kDa) were purchased from Sigma Aldrich, USA. KCl (Certified ACS), MgCl2, NaCl, Trizma base, 10× phosphate buffer solution, IL-6 Human matched antibody pair (CHC1263), Dynabeads antibody coupling kit (14311D), and Dulbecco's Modified Eagle Media (DMEM) were obtained from Thermo Fisher Scientific. The following chemicals were used as received without additional purification and were obtained from Sigma Aldrich (St. Louis, MO, USA): 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate system for ELISA (T0440). Normal patient pooled plasma was obtained from Jonathan Schoenecker's lab and spiked with IL-6 for assay calibration in human plasma. All experiments were performed in accordance with the NIH and Vanderbilt IRB Guidelines, and approved by the ethics committee at Vanderbilt University. De-identified human plasma was obtained with informed consent from human donors.
Instrumentation
Electrochemical measurements were performed using a CHI 1440 4-Channel Potentiostat (CH Instruments, Austin, TX). Carbon screen-printed electrodes (CSPE) from Pine Research Instrumentation, Inc. with a 2 mm diameter carbon working electrode, a Ag/AgCl reference electrode, and a carbon counter electrode were used for electrochemical interrogation. All experiments were completed on a bare unactivated CSPE.Methods
Capture antibodies conjugation to epoxy-coated magnetic beads
IL-6 antibodies were conjugated to epoxy-coated magnetic beads using the manufacturers protocol and buffers (C1, C2, HB, LB, and SB). Epoxy-coated magnetic beads were washed with 1 mL of C1 buffer, mixed, placed on magnet, and the supernatant was removed. 149 μL of C1 buffer, 0.9 μL of 100 mg mL−1 IL-6 antibody, and 150 μL of C2 buffer were combined, mixed with beads, and allowed to incubate at 37 °C for 16–24 h while mixing. After mixing, the solution was placed on magnet and the supernatant was removed. 800 μL of HB buffer was added, mixed, placed on magnet, and supernatant was removed. This was repeated with LB buffer once, and SB buffer four times. Then 800 μL of SB buffer was mixed at room temperature for 15 minutes before placing on magnet and removing supernatant. The beads were resuspended in 300 μL SB to create a 10 mg mL−1 stock and stored at 4 °C until use. We found that aliquoting the beads into 10 μL aliquots and stored ready to be used in assays improved the replicability of the assays. Beads were typically only stored for up to one month before use; however, the product manual cites stability in solution for at least one year.PolyHRP/TMB Sandwich assay procedure
10 μL of magnetic bead stock were used per assay and mixed with 50 μL of IL-6 and 2 μL of biotinylated detection antibody per assay. In order to prevent agglomeration, the biotinylated detection antibody was diluted in buffer containing 0.1% Tween-20. Then, the mixture was placed on mixer at room temperature for 30 minutes. After mixing, the mixture was placed on magnet stand and the supernatant was removed with a transfer pipette after one minute. Three washes were performed to reduce nonspecific binding but also to ensure minimal bead loss. The washes were performed by resuspending beads in 1 mL of 1× PBS 0.01% Tween-20, placing on magnet stand for one minute, rinsing out pipette tip, waiting one minute, and removing supernatant with transfer pipette. This was done twice with 1× PBS with 0.01% Tween-20. One wash was done with PBS that did not contain Tween-20. After the third wash, 15 μL of 4 μg mL−1 streptavidin poly-HRP was added to the beads and the mixture was placed on the mixer for 5 minutes. The poly-HRP enzyme was conjugated to the detection antibody via streptavidin-biotin coupling. After streptavidin polyHRP incubation, the mixture was placed on the magnet stand and supernatant was removed with transfer pipette after one minute. The three washes were repeated as before, with two washes with 1× PBS with 0.01% Tween-20 and one wash with PBS containing no Tween-20. Finally, 100 μL of TMB was added to the beads, mixed several times until beads are resuspended, and placed on mixer at room temperature for 5 min. The mixture was removed from mixer with 2 min remaining, then placed on magnet stand with 1.5 min remaining, the supernatant was removed at 0.5 min remaining and dropcasted onto a bared unactivated CSPE. At 0 minutes remaining, electrochemical reduction was performed using chronoamperometry for 20 seconds at −0.1 V. We integrated the current to collect total charge passed in 20 seconds to plot with the IL-6 calibrant concentration.IL-6 solutions were measured in PBS, DMEM, and human plasma. Many measurements were made in PBS to optimize the system and verify its repeatability. Repeatability for DMEM and human plasma were tested at least three times each.
Results and discussion
Scheme 1 displays the electrochemical immunoassay steps based on using magnetic beads and polymeric HRP. Chronoamperometry was used to detect TMB diimine reduction. Then, the current of the chronoamperometry curve was integrated to get the total charge, which is plotted with the calibrant concentration. In chronoamperometry, the current in the initial milliseconds is largely controlled by charging current. Since nonfaradaic current affects the later parts of the current-time curve less, using the total charge passed produces better signal-to-noise ratios. Furthermore, chronocoulometric data is cleaner as integration smooths the random noise from the current-time curves. As a result, we report charge vs. concentration for IL-6 calibrations.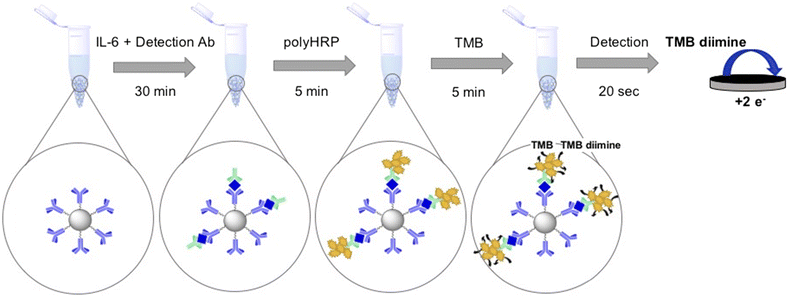 | ||
| Scheme 1 Sandwich assay protocol. The time to run one assay is approximately an hour including wash time. | ||
We first verified replicability of the TMB reduction using chronoamperometry. In order to verify that the reduction of oxidized TMB was sufficiently replicable across different CSPEs, assay size was increased by 6 times and incubated with 50 ng mL−1 IL-6-HRP then split six ways before incubation with TMB. The six TMB solutions were electrochemically reduced using chronoamperometry (Fig. 1). These assays demonstrated sufficient replicability, with a percent difference of 8.3% between the largest and smallest charges integrated over 20 seconds. Thus, it was determined that the HRP/TMB enzyme/substrate system was appropriate for moving forward with assay optimization and calibration.
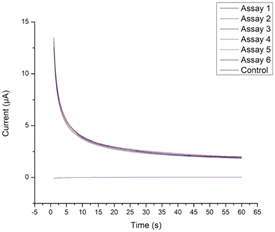 | ||
| Fig. 1 TMB reduction across six CPSEs demonstrated sufficient replicability with a percent difference of 8.3% between the largest and smallest charges. | ||
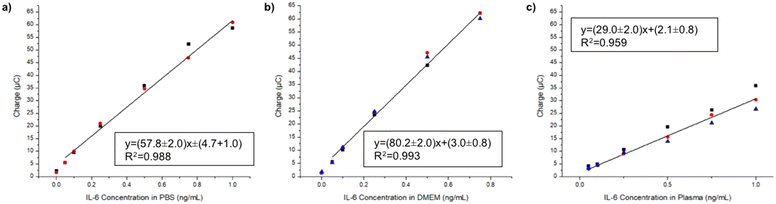
Next, we calibrated the sensor in PBS. Two calibrations from 0.05–1 ng mL−1 IL-6 in PBS were run on two separate days (Fig. 2a). They exhibited good day to day replicability, with the largest percent difference of 11.0% occurring at 0.75 ng mL−1. These calibrations also demonstrated excellent linearity, with an R2 of 0.988. In order to verify the performance of the assay in complex media, we tested three calibration replicates from 0.05–1 ng mL−1 IL-6 in DMEM cellular media (Fig. 2b). One replicate was run on one day and the other two were run on another. They exhibited desirable day to day replicability, with the largest percent difference of 10.6%. This calibration also exhibited excellent linearity, with an R2 of 0.993. Next, we ran triplicate calibrations in plasma (Fig. 2c) which showed an increase in variability, as expected. The sensitivity of this calibration was much less compared to calibrations in other media. We suspect that this is due to a combination of the batch of detection antibody and the matrix. Future work includes applying the IL-6 sandwich immunoassay as a potential diagnostic assay in a wide variety of biological samples.
Conclusions
The polyHRP/TMB system combined with magnetic beads for IL-6 capture allowed for the detection of IL-6 in the physiologically relevant concentration range. The sandwich assay demonstrated excellent linearity and replicability and achieved detection in 50–1000 pg mL−1 range. Overall, the assay exhibited good performance in biologically complex solutions including cellular media and human plasma and shortened the time required for IL-6 concentration readout. Replicability, linearity, and assay time were all improved compared to similar previously published works. Additionally, the simplicity of the assay procedure further improves upon previously reported designs. Future work involves validating the assay's measurements with conventional ELISAs and applying this immunoassay to a wide variety of biological systems as a potential diagnostic.Author contributions
Conceptualization, G. B., O. E. O., H. A. R., and D. E. C.; methodology, G. B. and O. E. O.; validation, G. B.; formal analysis, G. B.; investigation, G. B.; writing–original draft preparation, G. B.; writing–review and editing, G. B., O. E. O., H. A. R., and D. E. C.; visualization, G. B.; supervision, G. B. and D. E. C.; project administration, G. B. and D. E. C.; funding acquisition, D. E. C.; resources, D. E. C.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This research was funded by the National Institutes of Health, grant number 1R01HD102752, and by the United States Environmental Protection Agency, grant numbers 83950401 and 83950101. G. B., O. E. O., and H. A. R. were funded by The Training Program in Environmental Toxicology NIH T32 ES007028. We would like to thank Dr. Andrea Locke for her encouragement and help with editing and Dr. Jonathan Schoenecker and Dr. Stephanie N. Moore-Lotridge for supplying us with human plasma.Notes and references
- L. McCrae, W. Ting and M. Howlader, Biosens. Bioelectron., 2023, 13, 100288 CAS
.
- R. Goldenberg, J. Culhane, J. Iams and R. Romero, Lancet, 2008, 371(9606), 75–84 CrossRef PubMed
.
- A. Flores-Pliego, M. Espejel-Nuñez, N. Castillo-Castrejon, N. Meraz-Cruz, J. Beltran-Montoya, V. Zaga-Clavellina, S. Nava-Salazar, M. Sanchez-Martinez, F. Vadillo-Ortega and G. Estrada-Gutierrez, PLoS ONE, 2015 Search PubMed.
- A. Bergqvist, C. Bruse, M. Carlberg and K. Carlström, Fertil. Steril., 2001, 75(3), 489–495 CrossRef CAS PubMed
.
- G. G. Incognito, F. Di Guardo, F. A. Gulino, F. Genovese, D. Benvenuto, C. Lello and M. Palumbo, Int. J. Fertil. Steril., 2023, 17(4), 226–230 CAS
.
- V. Stephens, J. Rumph, S. Ameli, K. Bruner-Tran and K. Osteen, Front. Physiol., 2022, 12, 807685 CrossRef PubMed
.
- J. Jiang, Z. Jiang and M. Xue, Gynecol. Endocrinol., 2019, 35(7), 571–575 CrossRef CAS PubMed
.
- D. Del Valle, S. Kim-Schulze, H. H. Huang, N. Beckmann, S. Nirenberg, B. Wang, Y. Lavin, T. Swartz, D. Madduri, A. Stock, T. Marron, H. Xie, M. Patel, K. Tuballes, O. Oekelen, A. Rahman, P. Kovatch, J. Aberg, E. Schadt, S. Jagannath, M. Mazumdar, A. Charney, A. Firpo-Betancourt, D. Mendu, J. Jhang, D. Reich, K. Sigel, C. Cordon-Cardo, M. Feldmann, S. Parekh, M. Merad and S. Cnjatic, Nat. Med., 2020, 26, 1636–1643 CrossRef CAS
.
- X. Liu, H. Wang, S. Shi and J. Xiao, Postgrad. Med. J., 2022, 98(1165), 871–879 CrossRef
.
- H. Huan, O. Mab, C. Lic, R. Liua, L. Zhaoc, W. Wangd, P. Zhanga, X. Liue, G. Goaf, F. Liug, Y. Jiangh, X. Chengi, C. Zhuaand and X. Yuchen, Emerging Microbes Infect., 2020, 9, 1123–1130 CrossRef
.
- G. Volpe, R. Draisci, G. Palleschi and D. Compagnone, Analyst, 1998, 123, 1303–1307 RSC
.
- F. Bettazzi, A. Natale, E. Torres and I. Palchetti, Sensors, 2018, 18(9), 2965 CrossRef PubMed
.
- I. Ojeda, M. Moreno-Guzman, A. Gonzalez-Cortes, P. Yanez-Sedeno and J. M. Pingarron, Anal. Bioanal. Chem., 2014, 406(25), 6363–6371 CrossRef CAS
.
- C. Shang, Y. Li, Q. Zhang, S. Tang, X. Tang, H. Ren, P. Hu, S. Lu, P. Li and Y. Zhao, Sens. Actuators, 2022, 358, 131525 CrossRef CAS
.
- I. Morkvenaite-Vilkonciene, A. Kisieliute, W. Nogala, A. Popov, B. Brasiunas, M. Kamarauskas, A. Ramanavicius, S. Linfield and A. Ramanaviciene, Electrochim.
Acta, 2023, 463, 142790 CrossRef CAS
.
- S. Lv, K. Zhang, L. Zhu, D. Tang, R. Niessner and D. Knopp, Anal. Chem., 2019, 91(18), 12055–120662 CrossRef CAS PubMed
.
- R. Zheng and D. Tang, Talanta, 2020, 219, 121215 CrossRef PubMed
.
- C. Zhang, D. Shi, X. Li and J. Yuan, Talanta, 2022, 240, 123173 CrossRef CAS PubMed
.
- R. Malhotra, V. Patel, J. Vaqué, J. Gutkind and J. Rusling, Anal. Chem., 2010, 82(8), 3118–3123 CrossRef CAS PubMed
.
- G. Wang, H. Huang, G. Zhang, X. Zhang, B. Fang and L. Wang, Langmuir, 2010, 27, 1224–1231 CrossRef
.
- G. Wang, H. Huang, B. Wang, X. Zhang and L. Wang, Chem. Commun., 2012, 48, 720–722 RSC
.
- S. Punj, D. Sidhu, D. Bhattacharya, M. Wang and P. K. Wong, IEEE Open J. Nanotechnol., 2020, 1, 31–37 Search PubMed
.
- J. Del Rio, O. Henry, P. Jolly and D. Ingber, Nat. Nanotechnol., 2019, 14, 1143–1149 CrossRef PubMed
.
- L. Huang, J. Chen, Z. Yu and D. Tang, Anal. Chem., 2020, 92(3), 2809–2814 CrossRef CAS PubMed
.
- Z. Yu, H. Gong, M. Li and D. Tang, Biosens. Bioelectron., 2022, 218, 114751 CrossRef CAS PubMed
.
- S. Tian, L. Huang, Y. Gao, Z. Yu and D. Tang, Sens. Diagn., 2023, 2(3), 707–713 RSC
.
- R. Zeng, M. Qiu, Q. Wan, Z. Huang, X. Liu, D. Tang and D. Knopp, Anal. Chem., 2022, 94(43), 15155–15161 CrossRef CAS PubMed
.
- A. Molinero-Fernandez, M. Moreno-Guzman, M. A. Lopez and A. Escarpa, Biosensors, 2020, 10(6), 66 CrossRef CAS
.
| This journal is © The Royal Society of Chemistry 2024 |