Open Access Article
Open Access ArticleA FokI-driven signal amplification platform for the simultaneous detection of multiple viral RNA pathogens†
Juan R.
Tejedor
 *abcd,
Annalisa
Roberti
*abcd,
Annalisa
Roberti
 ab,
Cristina
Mangas
bc,
Marta E.
Álvarez-Argüelles
ab,
Cristina
Mangas
bc,
Marta E.
Álvarez-Argüelles
 be,
Susana
Rojo-Alba
be,
Susana
Rojo-Alba
 be,
José A.
Boga
be,
José A.
Boga
 be,
Agustín F.
Fernández
be,
Agustín F.
Fernández
 abcd,
Santiago
Melón
abcd,
Santiago
Melón
 be,
Mercedes
Rodríguez
be,
Mercedes
Rodríguez
 be and
Mario F.
Fraga
be and
Mario F.
Fraga
 *abcd
*abcd
aNanomaterials and Nanotechnology Research Center (CINN-CSIC), El Entrego, 33940, Spain. E-mail: jr.tejedor@cinn.es; mffraga@cinn.es; Tel: +34 985733644
bHealth Research Institute of Asturias (ISPA), Avenida del Hospital Universitario s/n, 33011 Oviedo, Spain
cUniversity Institute of Oncology (IUOPA), University of Oviedo, Oviedo, 33006, Spain
dCenter for Biomedical Network Research on Rare Diseases (CIBERER), Madrid, 28029, Spain
eCentral University Hospital of Asturias (HUCA) Oviedo, 33011, Spain
First published on 14th February 2024
Abstract
Accurate signal quantification is a critical parameter for precise pathogen diagnosis. Furthermore, the development of novel diagnostic methods amenable for portable or scalable purposes may facilitate the real-time surveillance of emerging pathogenic threats. Here, we extrapolated the use of a FokI-driven signal amplification approach, powered by the nickase activity of the FokI restriction endonuclease, to enhance the signal detection of multiple respiratory viral pathogens relevant to human health. This approach utilises a set of dumbbell-like fluorescent oligonucleotides against three families of human single-stranded RNA respiratory viruses (Coronaviridae, Paramyxoviridae, and Pneumoviridae), including human betacoronaviruses (OC43/HKU1), human betacoronavirus SARS-CoV-2, human parainfluenza viruses (HPIV1/HPIV3/HPIV2/HPIV4), and human metapneumovirus (HMPV)/respiratory syncytial virus (HRSV). FokI-assisted digestion of these dumbbell-like fluorescent probes in the presence of their respective viral targets enhanced their detectable signal in a highly specific manner. In addition, this technique exhibited high multiplex potential to detect multiple viral targets in a single assay. A molecular coupling between the FokI-assisted reaction and the isothermal rolling circle amplification technique significantly improved the limit of detection in samples from infected patients, ensuring adequate specificity and sensitivity. These results highlight the diagnostic potential of this methodology, representing a cost-effective alternative for the identification of respiratory viral pathogens of interest to the public healthcare system.
Introduction
Human respiratory viruses account for a total of 17.2 billion cases of upper tract infections per year (ref. 1) and pose a serious threat to national healthcare systems worldwide. The recent pandemic caused by the severe acute respiratory syndrome coronavirus (SARS-CoV-2) has highlighted the potential threats associated with these respiratory viruses,2 but other viral infectious agents, including rhinovirus, influenza, coronavirus, parainfluenza, and pneumovirus, also significantly impact healthcare resource utilisation, leading to a financial burden on society.3–7 In particular, infections caused by viruses associated with the Coronaviridae family, such as OC43 and HKU1, which are single-stranded, positive-sense RNA viruses (ssRNA+), have been linked to the downregulation of relevant genes involved in the innate immune response, contributing to 15–30% of cases of common colds in adults.8 On the other hand, members of the Paramyxoviridae family, including human respiroviruses (HPIV1/HPIV3) and rubulaviruses (HPIV2/HPIV4), are negative-sense ssRNA viruses principally involved in acute lower respiratory infections (ALRI) in childhood.9 The estimated incidence of parainfluenza infections is approximately 13% of all ALRI cases, with a significant burden of parainfluenza in ALRI morbidity and mortality among young children.10 Members of the Pneumoviridae family, which encompasses the negative-sense ssRNA viruses human metapneumovirus (HMPV) and syncytial respiratory virus (HRSV), are also major contributors to respiratory infections in infants.11,12 Since severe and life-threatening lower respiratory infections can occur in elderly individuals, infants, and immunocompromised patients following infection with any of these aforementioned viral pathogens, early detection of these infectious agents and their accurate interpretation at the epidemiological level, are crucial steps to combat their adverse effects on global public health.The quantitative real-time reverse transcription polymerase reaction (RT-PCR) technique stands as a gold standard in microbiology laboratories.13 However, while this technology excels in terms of specificity and sensitivity, its reliance on specialised equipment calls for alternative approaches that can be scaled up to enable real-time surveillance of specific diseases, as has been proposed in recent years.14,15 Among them, nucleic acid amplification techniques (NAAT) based on loop-mediated isothermal amplification (LAMP),16,17 recombinase polymerase amplification (RPA),18,19 or their combination with CRISPR/Cas systems20,21 have been successfully used to diagnose COVID-19 patients during the recent SARS-CoV-2 pandemic. In addition, other isothermal amplification approaches, such as rolling circle amplification (RCA)22 or multiple displacement amplification (MDA),23 show promise for detecting viral targets in the absence of additional reverse transcription steps. In this vein, we have recently developed a method that combines a signal amplification strategy mediated by the action of the FokI restriction endonuclease coupled to RCA technology, enhancing the detection of SARS-CoV-2 in COVID-19 patients.24 The present work further expands the catalogue of potential human respiratory virus targets and focuses on the development of a multiplex assay for the rapid and accurate detection of various viral families, including Coronaviridae (OC43/HKU1 and SARS-CoV-2), Paramyxoviridae (HPIV1, HPIV3, HPIV2 and HPIV4) and Pneumoviridae (HMPV, HRSV).
Experimental section
Design of synthetic oligonucleotides
The FASTA sequences corresponding to multiple human respiratory viruses (HKU1, OC43, 229E, NL63, SARS-CoV-2, HPIV1, HPIV3, HPIV2, HPIV4, HMPV and HRSV) were retrieved from NCBI (accession numbers NC_006577.2, NC_006213.1, NC_002645.1, NC_005831.2, NC_045512.2, NC_003461.1, NC_001796.2, NC_003443.1, NC_021928.1, NC_039199.1 and NC_001781.1 respectively). Multiple sequence alignment (MSA) was conducted using the Clustal Omega software,25 and the visualization of the MSA was performed using the Jalview software.26 For the design of fluorescent dumbbell-like oligonucleotides targeting these viral entities, specific or shared target regions across multiple viruses were selected. In addition, these oligonucleotides contained a FokI restriction site within their duplex sequence and were characterized to include a 6 bp stem structure in the dumbbell-like oligonucleotide, preventing self-cleavage by FokI before the hybridisation step. A total of four custom dumbbell-like oligonucleotides were designed targeting the human betacoronaviruses OC43 and HKU1 (O01), SARS-CoV-2 (O05), human parainfluenza viruses (HPIV1, HPIV3, HPIV2 and HPIV4, O09) and human pneumoviruses (HMPV and HRSV, O13). These sequences encompassed a common hairpin region containing a unique FokI restriction site and were labelled with a 6-FAM fluorophore at their 5′ end and BHQ-1 quencher at their 3′ end, respectively. In addition, for multiplex purposes, another dumbbell-like oligonucleotide targeting human pneumoviruses (O17) was labelled with a HEX fluorophore and BHQ-1 quencher at their 5′ and 3′ ends, respectively. Padlock probes (O04, O08, O12, O16) were designed to optimize hybridisation with their viral targets while avoiding internal secondary structures, and these probes included a phosphate group at their 5′ end for subsequent ligation purposes. The structures of dumbbell-like oligonucleotides and padlock probes were confirmed in silico using the RNAstructure software.27 Short synthetic RNAs and DNAs corresponding to the aforementioned viral targets (30 nt length) were used to establish the single or multiplex FokI-assisted signal amplification conditions. All oligonucleotides used in this study were synthetized by Metabion (Germany) and are listed in Table S1.†FokI-assisted signal amplification assays
All experiments were conducted at 37 °C in a final reaction volume of 20 μl. Conventional hybridisation assays were performed using 2 μl of NEB 10× Cutsmart buffer (NEB #B7204), 100 nM of fluorescent dumbbell-like oligonucleotides targeting different viral targets, and decreasing concentrations of either target DNA, target RNA (5 nM, 1 nM, 500 pM, 100 pM, 50pM, 0 M) or control DNA/RNA (5 nM) for each respective reaction. In addition to the conventional hybridisation protocol, FokI-assisted signal amplification assays were performed by supplementing the reaction with 5 units of FokI RE (NEB, #R0109S). Fluorescence intensity measurements for these experiments were recorded using a StepOnePlus™ real-time PCR system (Applied Biosystems). The excitation and emission wavelengths (488 nm and 520 nm, respectively) were set to detect the FAM channel, and the reactions were monitored for 60 cycles (each cycle 1 min at 37 °C). For the multiplexed reactions, a similar experimental setup was adopted. However, these reactions simultaneously included 100 nM of fluorescent dumbbell-like oligonucleotides (O05 and O17) targeting SARS-CoV-2 and pneumovirus, labelled with FAM and HEX fluorophores, respectively. The excitation and emission wavelengths were configured to simultaneously detect FAM and VIC channels (488 nm excitation, 520 nm emission, and 529 nm excitation, 549 nm emission, respectively). It is noted that the HEX dye has a similar excitation/emission spectrum to the VIC dye.Molecular coupling between FokI-assisted signal amplification and RCA
The hybridisation of padlock probes with their corresponding synthetic RNA molecules and the subsequent padlock probe circularization step were conducted in a reaction volume of 8 μl. The concentrations of the reaction components were as follows: 0.8 μl of NEB 10× SplintR ligase reaction buffer, 5 units of SplintR ligase (NEB, #M0375S), 10 nM of padlock probe oligonucleotides and variable amounts of each target RNA molecule (10 fmol, 1 fmol, 100 amol, 10 amol, 1 amol, 100 zmol or 0 mol), with the exception of the control RNA condition, which was consistently assayed at 10 fmol. This reaction mixture was incubated for 5 min at room temperature, and then 12 μl of the RCA premix was added to the resulting reaction. The concentrations and amounts of the RCA premix components were: 2 μl of NEB 10× Cutsmart buffer, 500 uM dATP, dGTP, dCTP and dTTP (Promega, #U1240), 40 nM of the universal rolling circle primer (O20), 0.01 units of pyrophosphatase inorganic from E. coli (NEB, #M0361S), 0.5 μl of QualiPhi DNA polymerase (4Basebio, #510100), 100 nM of 5′ FAM and 3′ BHQ-1 labelled dumbbell-like oligonucleotides, and 5 units of FokI RE, resulting in a final reaction volume of 20 μl. Fluorescence intensity in the FAM channel was recorded over 60 cycles (each cycle 1 min at 37 °C) using a StepOnePlus™ real-time PCR system, as previously indicated. A similar set of experiments was conducted for multiplexed reactions, where both padlock probes (O08 and O16, 10 nM each) and both fluorescent dumbbell-like oligonucleotides (O05 and O17, 100 nM each) targeting SARS-CoV-2 and pneumovirus were simultaneously incorporated. The excitation and emission wavelengths were configured to detect FAM and VIC channels simultaneously, as previously indicated. An additional 3-step multiplex reaction simultaneously incorporating all padlock probes (O04, O08, O12 and O16, 10 nM each) and each of the fluorescent dumbbell-like oligonucleotides (100 nM) was performed in the context of the FAM channel to evaluate the specific detection of a given viral target. For all experiments, fluorescence measurements were recorded using a StepOnePlus™ real-time PCR system.Sample collection and viral detection in human samples
A total of 20 nasopharyngeal swab samples were collected from patients infected by multiple coronavirus or pneumovirus species at the Hospital Universitario Central de Asturias (HUCA). All experiments were conducted in accordance with the Declaration of Helsinki, as revised in 2013, and with the approval of the research ethics committee of the Principality of Asturias (ref 2020.309). Informed consents were not required, as Organic Law 3/2018, of 5 December (Seventeenth Additional Provision, 2b), on Data Protection and Guarantee of Digital Rights provides, in relation to the processing of health data, that health authorities and public institutions with public health control responsibilities may conduct scientific research without the consent of the data subject in situations of exceptional relevance and seriousness for public health, such as actions arising from the COVID19 pandemic. RNA isolation was carried out using a MagNA pure 96 system (Roche Diagnostics) following the manufacturer's recommendations. Initial diagnosis was established by personnel at the virology unit at HUCA. An additional viral detection assay was performed using a coupled RCA – FokI-assisted signal amplification method, as previously described in the previous section, with slight modifications: 5 μl of purified RNA was incubated in the presence of a cocktail of padlock probes targeting betacorona, SARS-CoV-2, parainfluenza, or pneumoviruses (O04, O08, O12, and O16, 10 nM each) during the padlock circularization step, as previously detailed. Subsequently, 12 μl of the RCA premix was added to the reaction, excluding FokI and the dumbbell-like oligonucleotides. This reaction was incubated for up to 60 minutes, and the resultant reaction was divided into 4 equal samples. Each sample was assayed for 60 minutes with FokI, along with a specific 5′ FAM and 3′ BHQ-1 labelled dumbbell-like oligonucleotide targeting either betacorona, SARS-CoV-2, parainfluenza, or pneumovirus.To account for potential RNA degradation over time, an additional virus detection step was conducted using an in-house real-time (RT)-PCR approach. Viral genomes were amplified using Superscript III (Invitrogen, #12574026), random primers and oligodT as per the manufacturer's recommendations. SYBR green RT-qPCR detection was performed using primers targeting betacoronaviruses HKU1 or OC43,28 SARS-CoV-2 specific primers,29 and a primer pair designed against a comprehensive dataset of human pneumovirus.30 This involved the use of a 2× SYBRgreen mastermix (Applied Biosystems, #A25742), 400 nM forward and reverse primers, and 1 μl of RT product from the previous step. Amplifications and data analysis were carried out using a StepOnePlus™ real-time PCR system under the following conditions: hot start at 95 °C for 10 min, followed by 45 cycles at 95 °C for 30 s and 60 °C for 1 min. Cycle thresholds were inferred using a threshold line of 0.1 for all conditions analysed.
Data normalization and statistical analyses
For all experiments involving FokI-assisted signal amplification, multicomponent raw data were downloaded from the instrument and used for downstream analyses. Signal intensities for the different conditions were normalized firstly to their corresponding background signal at time 0 (intra-sample normalization), and subsequently to the signal of the non-template control (inter-sample normalization), and this normalization was executed through an iterative subtraction in each cycle, following the methodology described by Tejedor and colleagues.24 The limit of detection (LOD) for each reaction was calculated as LOD = 3.3Sy/slope, where Sy represents the standard error of the predicted y-value for each x in the regression and slope represents the slope value (a) of the calibration plot y = ax + b.Multiplex detection in the context of patient's samples was conducted similarly. However, for each patient, a z-score was computed for each independent dumbbell-like fluorescent oligonucleotide targeting a specific virus at 60 min timepoint. This calculation took in consideration the information observed for the remaining signals. The maximum z-score was considered after the normalization procedure, and a sample was classified as positive if the z-score exceeded 1.2. Sensitivities and specificities were calculated using the R Bioconductor package caret (6.0–90), considering the predictions derived from the FokI-assisted signal amplification protocol, and the observations validated using the PCR approach, which served as the gold standard. Data representation and subsequent statistical analyses were carried out using the R programming language (v.4.0.3).
Results
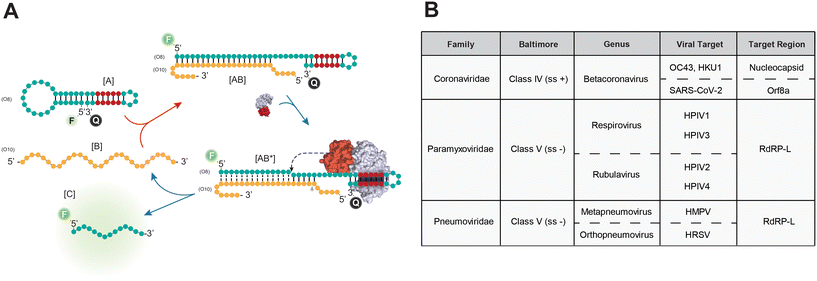
A FokI-assisted digestion platform for enhanced detection of ssRNA viruses
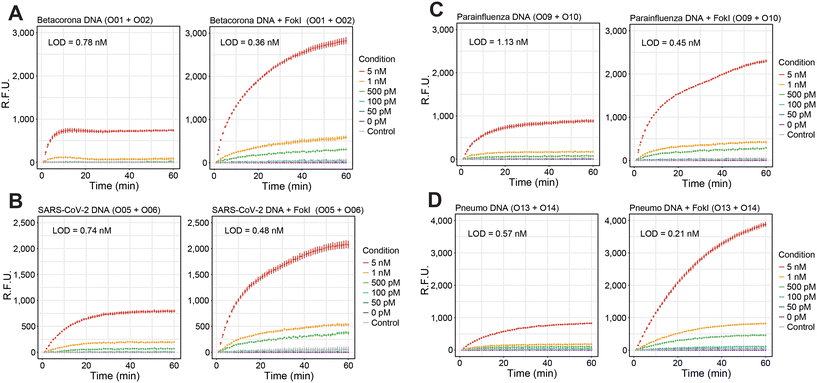
Recent studies have demonstrated the potential of FokI-assisted digestion of fluorescent dumbbell-like oligonucleotides in enhancing the detection of nucleic acid targets,24,31 particularly in the context of SARS-CoV-2 infection.24 This working principle relies on a FokI-assisted reaction (Fig. 1A) using a combination of fluorescent dye and quencher molecules at the 5′ and 3′ ends of the dumbbell-like structure [A]. In the absence of any complementary target molecule, the fluorescence of the dumbbell-like oligonucleotide is suppressed due to a Förster resonance energy transfer (FRET) resulting from the proximity of the fluorescent dye and the quencher molecule. The presence of the target sequence [B] initiates a conventional hybridisation assay, where one molecule of nucleic acid substrate can hybridize with one molecule of the dumbbell-like oligonucleotide, generating a heteroduplex substrate [AB]. This process results in the release of the fluorescent signal from the reporter oligonucleotide. The FokI-assisted digestion system, which combines the presence of dumbbell-like structures and native FokI restriction endonuclease, augments the detection of nucleic acids through an asymmetric cleavage facilitated by FokI, which allows for the rapid release of the fluorescent portion of the dumbbell-like oligonucleotide [C] and enables the recirculation of the target molecule [B] for further rounds of digestion.The efficacy of the proposed signal amplification method was evaluated in a panel of human single-stranded RNA respiratory viruses corresponding to the families Coronaviridae, Paramyxoviridae and Pneumoviridae (Fig. 1B). A set of 4 fluorescent dumbbell-like oligonucleotides, labelled with 6-FAM and BHQ1 at their 5′ and 3′ ends, respectively, was designed to enable the combined detection of human betacoronaviruses (OC43/HKU1), human betacoronavirus SARS-CoV-2, human parainfluenza viruses (HPIV1/HPIV3/HPIV2/HPIV4), and human metapneumovirus (HMPV)/respiratory syncytial virus (HRSV), respectively (Fig. 1B and S1†). The performance of the FokI-assisted signal amplification strategy was evaluated by real-time fluorescence measurements in the presence of different DNA or RNA target concentrations, comparing it with both a conventional hybridisation assay and the proposed FokI-assisted signal amplification method (Fig. 2 and S2†). The latter strategy exhibited enhanced fluorescence detection in all scenarios when compared to conventional hybridisation assays. Although the performance of the reaction was superior in the presence of ssDNA substrates (Fig. 2, average LOD conventional assay = 0.85 nM, average LOD FokI-assisted assay = 0.37 nM), the improved detection of ssRNA molecules was also evident in the FokI-assisted signal amplification assay (Fig. 2, average LOD conventional assay = 0.92 nM, average LOD FokI-assisted assay = 0.65 nM).
To test the specificity and the multiplex capabilities of the FokI-assisted digestion method, a series of experiments were conducted using a simultaneous combination of fluorescently labelled dumbbell-like oligonucleotides targeting the Orf8a region of SARS-CoV-2 and the common RdRP-L region shared between the HMPV and HRSV pneumoviruses (Fig. S3A†). These oligonucleotides were labelled with different fluorophores (FAM or HEX, respectively) at their 5′ end, while maintaining the BHQ1 quencher and their 3′ end to prevent signal interference and enable the detection of different target molecules within a single fluorescence channel. Enhanced detection of either SARS-CoV-2 or pneumoviruses was observed exclusively in the presence of the corresponding DNA (Fig. S3B†) or RNA (Fig. S3C†) target molecules and their respective fluorescent channels (FAM – SARS-CoV-2 or VIC – pneumoviruses). Notably, detection of other sequences or unrelated control substrates was absent under these experimental conditions within their corresponding fluorescent channels. This indicates efficient fluorescence release by the reporter dumbbell-like oligonucleotides in their respective emission spectra, illustrating the highly specific signal amplification mediated by FokI-assisted digestion for the target sequence of interest.
Molecular coupling between FokI-assisted signal amplification and RCA enhances ssRNA target detection
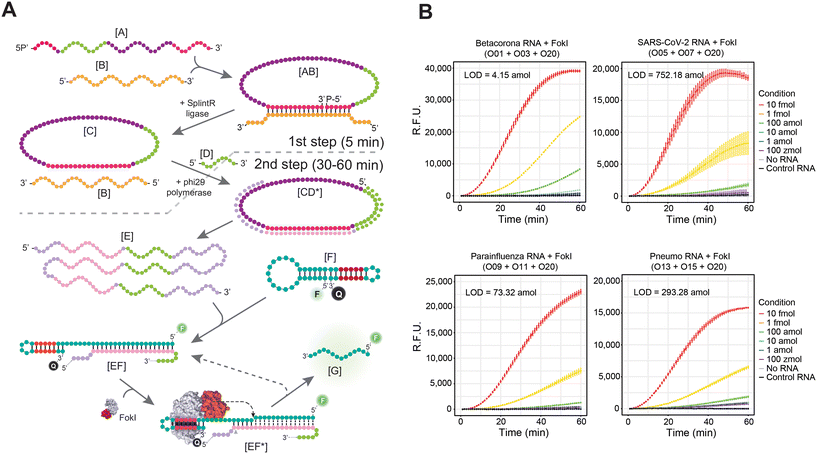
We have recently demonstrated that the LOD of this technique can be further enhanced by simultaneously coupling the FokI-assisted signal amplification reaction with a classical isothermal nucleic acid amplification method, such as the RCA technique.24 This approach depends on amplifying circular DNA targets by means of the high processivity and strand displacement capabilities of the Phi29 DNA polymerase,32 and the proposed reaction involves the use of padlock probes [A] and a nucleic acid target [B] (Fig. 3A). When the padlock probe hybridizes with the target molecule, it adopts a circular structure [AB], and through the catalytic activity of a DNA ligase enzyme, it undergoes complete circularization by ligating its splinted 5′ and 3′ ends [C]. The resulting single stranded, circularized DNA template can be indefinitely amplified in the presence of a complementary RCA primer [D], dNTPs, and the DNA polymerase Phi29, thereby increasing the number of target molecules in the reaction mixture [E]. The simultaneous molecular coupling of the FokI-assisted signal amplification assay improves the detection results obtained in classical RCA approaches, as these target molecules [B, E] can hybridize with the custom-designed dumbbell-structures [F] and release the fluorescent signal of the reporter molecule. In addition, the asymmetric cleavage reaction mediated by FokI enhances the excision of the fluorescent portion of the probe [G] and enables the potential reuse of the target sequence [B, E] for further rounds of digestion, resulting in an improved detection signal of the reaction.The efficacy of the molecular coupling between FokI-assisted digestion and the RCA reaction was tested at short incubation times (<60 min) in the context of human betacoronaviruses (OC43/HKU1), human betacoronavirus SARS-CoV-2, human parainfluenza viruses (HPIV1/HPIV3/HPIV2/HPIV4), and human metapneumovirus (HMPV)/respiratory syncytial virus (HRSV) respectively (Fig. 3B). This molecular coupling enhanced the detection limit in all scenarios compared to the single FokI-assisted signal amplification approach, but extent of the signal amplification was sequence dependent, being the combination of padlock probe/dumbbell-like oligonucleotide designed against human betacoronaviruses (OC43/HKU1) the most effective in terms of target detection (LOD betacoronavirus = 4.15 amol; LOD SARS-CoV-2 = 752.18 amol, LOD parainfluenza = 73.32 amol; LOD pneumo = 293.28 amol). Notably, the simultaneous use of padlock probes and fluorescently labelled dumbbell-like oligonucleotides against the Orf8a region of SARS-CoV-2 and the common RdRP-L region between the HMPV and HRSV pneumoviruses (FAM or HEX respectively) allowed multiplexed detection of these viral targets in a single reaction tube (Fig. S4A†). In the context of DNA targets, enhanced detection of either SARS-CoV-2 or pneumoviruses was achieved only in the presence of their specific sequences (Fig. S4B†) and their corresponding fluorescent channels (FAM – SARS-CoV-2 or VIC – pneumoviruses), demonstrating the high specificity of this approach. However, the detection of RNA sequences from these viruses, especially in the context of the VIC channel (Fig. S4B†), was noisy and less specific, despite detecting the correct cognate sequence in each case analysed.
A multiplex padlock ligation step allows the detection of multiple viral targets in a single combined assay
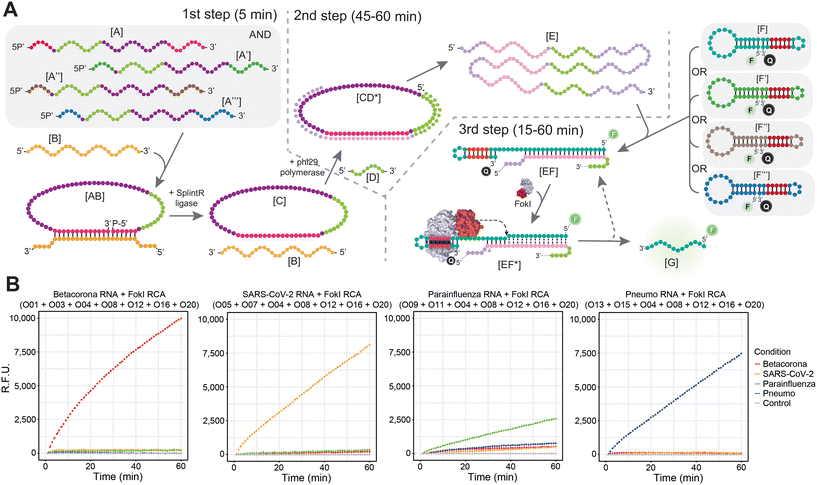
To overcome the limitations arising from the concurrent presence of combined padlock probes and multiple fluorescently labelled dumbbell-like oligonucleotides in a simultaneous amplification reaction, particularly in the context of RNA targets, a different multiplex strategy was implemented (Fig. 4). Incorporating multiple padlock probes in the initial step of the reaction might facilitate the detection of multiple viral targets, each expected to hybridize exclusively to its corresponding padlock probe during the subsequent ligation step (Fig. 4A). The resulting circularized DNA template can undergo indefinite amplified in the context of the RCA reaction, as described in the previous section. However, rather than conducting a simultaneous molecular coupling between the FokI-assisted signal amplification reaction and the RCA step, the resulting sample is divided into several distinct FokI-mediated signal amplification assays in separate tubes, each tube containing its specific fluorescent dumbbell-like oligonucleotide probe designed against the cognate viral RNA sequence. Theoretically, following the completion of the uncoupled RCA and the FokI-assisted signal amplification step, each dumbbell-like oligonucleotide should amplify the fluorescent signal corresponding to its respective molecular target (Fig. 4A). This was demonstrated through a combined reaction involving multiple padlock probes against human betacoronaviruses (OC43/HKU1), human betacoronavirus SARS-CoV-2, human parainfluenza viruses (HPIV1/HPIV3/HPIV2/HPIV4), and human metapneumovirus (HMPV)/respiratory syncytial virus (HRSV) alongside specific fluorescent dumbbell-like oligonucleotides (FAM) in the context of each specific viral RNA target condition (Fig. 4B). The exclusive detection of each corresponding RNA target was achieved in the presence of their respective dumbbell-like oligonucleotides, but no detection occurred in the presence of other dumbbell-like oligonucleotides designed for different viral targets, thereby confirming the high specificity of the proposed multiplexed padlock step. This strategy presents a simple and cost-effective method for detecting viral targets, since a single multiplexed padlock RCA amplification assay is required per patient, and the resulting amplification product can be divided into multiple FokI-assisted signal amplification reactions driven by different dumbbell-like oligonucleotides against distinct viral RNA targets of interest, without necessitating additional fluorescence measurement requirements.Detection of multiple viral targets in real-world samples
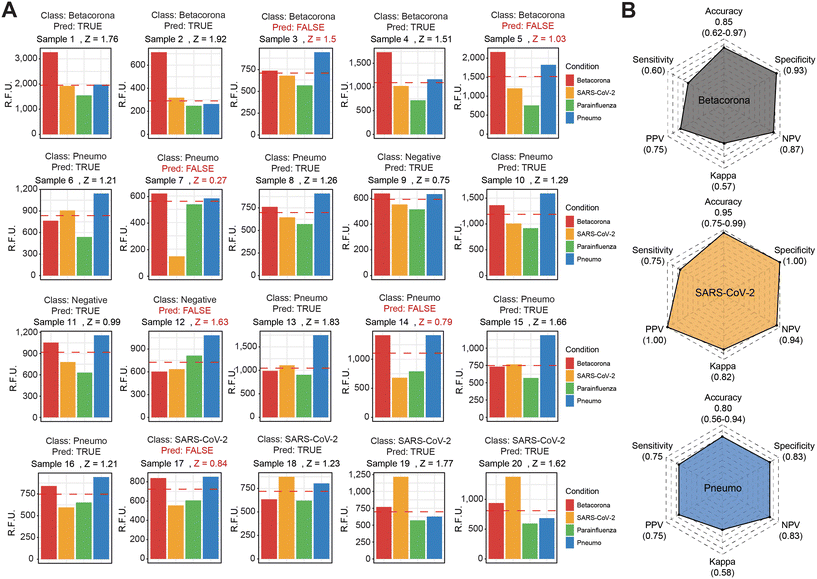
Considering the enhanced signal detection in the context of multiple dumbbell-like oligonucleotides and their corresponding viral molecules in a test tube, we further investigated whether this molecular coupling between FokI-assisted signal amplification and RCA could serve as a diagnostic method for detecting multiple viral targets in RNA isolated from human nasopharyngeal swabs diagnosed at the Hospital Universitario Central de Asturias. We selected a cohort of 20 patients diagnosed with various viral infections (5 betacoronaviruses, 4 SARS-CoV-2 and 8 pneumoviruses), including 3 control samples with no detection of any of these targets in the samples of interest (Fig. 5 and S5†). The use of a 3-step, coupled reaction between FokI-assisted signal amplification and multiplexed RCA, which involved a simultaneous analysis of multiple dumbbell-like oligonucleotides, revealed significant differences in the signal intensity of particular viral species depending on the sample of interest at reaction times of 60 minutes (Fig. 5A). The accuracy, sensitivity and specificity of the multiplexed FokI-assisted signal amplification reaction, compared to the gold standard RT-PCR approach, for the different dumbbell-like oligonucleotides tested were, respectively: 0.85, 0.60 and 0.93 for betacoronaviruses; 0.95, 0.75 and 1 for SARS-CoV-2 infected samples; and 0.80, 0.75 and 0.83 for human pneumoviruses (Fig. 5B). These results demonstrate the feasibility of using this method for detecting multiple viral targets, and offers a cost-effective, time-saving, and specific diagnostic strategy for monitoring human respiratory viral infection rates in the population of interest.Discussion
In recent years, the number of diagnostic methods for the early diagnosis of human diseases has dramatically increased, largely motivated by the needs highlighted during the COVID-19 pandemic.33,34 In this regard, the development of new diagnostic methodologies as alternatives to the PCR technique has led to the implementation of diagnostic tests centred on the use of new tools based on LAMP, RPA, or CRISPR/Cas9, among others.35–37 Some of these methodologies are also highly portable to point-of-care (POC) systems,38,39 which could prove effective in responding to critical situations, whether in remote locations or with limited equipment, or when facing reagent shortages associated with the PCR technique.40 To address these limitations and expand the catalogue of available diagnostic methodologies, this work focused on implementing recent advances observed in the context of signal amplification strategies24 with classical nucleic acid amplification technologies such as the RCA technique. Our findings demonstrate that the combination of a FokI-assisted signal amplification strategy with RCA improves the limit of detection of viral pathogens in human samples, ensuring adequate specificity and sensitivity, thereby reinforcing the diagnostic potential of this approach for single-stranded RNA viruses. In addition, this detection can be performed in a multiplexed manner, allowing the screening of multiple human respiratory viruses using a single assay, considerably reducing the processing cost per sample.Despite the progress shown in this study, this work has some limitations. One of the main limitations is the sample size of the validation cohort of real-world samples, from which the specificity and sensitivity values of the work have been inferred using a small number of subjects. In this respect, analysis in larger cohorts will serve to determine more precisely the extent of these improvements in the context of other diagnostic systems. Furthermore, despite the obvious improvements introduced by the coupling of the FokI-assisted signal amplification reaction and RCA, the observed limit of detection values do not outperform recently established methods focusing on exponential amplification methods, such as LAMP or PCR itself.41 Being aware of these limitations, we propose that the coupling of the FokI-assisted signal amplification reaction and RCA could serve as a complementary alternative for potential multiplexing in relevant clinical settings, including human respiratory virus diseases. The panel designed in this work could be complemented with new molecular beacons designed against additional viruses in the RCA multiplexing step, resulting in a rapid and cost-effective solution for the diagnosis of human respiratory diseases of concern.
Conclusions
This work demonstrates that FokI-assisted signal amplification enhances the detection of viral RNAs from multiple human respiratory viruses compared to conventional oligonucleotide hybridisation approaches. Furthermore, we observed that the molecular coupling between FokI-assisted signal amplification and the RCA method further improves the detection limit of these technologies. In addition, the multiplexing of padlock probes emerges as a viable approach for identifying specific respiratory viruses in human samples with adequate sensitivity and specificity. This cost-effective methodology could serve as a diagnostic alternative for the simultaneous detection of multiple human respiratory viruses, holding significant potential for public health applications.Author contributions
Conceptualization: J. R. T. and M. F. F.; data curation: J. R. T., A. R. and M. R.; formal analysis: J. R. T., A. R., M. E. A. A., M. R., J. A. B., S. R. A., A. F. F. and S. M.; funding acquisition: J. R. T., A. F. F and M. F. F.; investigation: J. R. T., A. R., M. E. A. A., M. R., J. A. B., and S. R. A.; methodology: J. R. T. and A. R.; project administration: C. M.; supervision: J. R. T., S. M. and M. F. F.; validation: J. R. T., M. E. A. A., M. R., J. A. B., S. R. A. and S. M.; visualization: J. R. T.; writing – original draft: J. R. T., A. F. F. and M. F. F.; writing – review and editing: J. R. T., A. R., A. F. F. and M. F. F. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare the following competing financial interest(s): a patent application (EP22382308) has been filed on the use of the FokI-assisted signal amplification system for the detection of human respiratory viral pathogens (J. R. T., A. F. F., and M. F. F.).Acknowledgements
The authors would like to thank Jennifer Kefauver for manuscript editing. This work received support from the European Commission – NextGenerationEU, through CSIC's Global Health Platform (PTI Salud Global), and the Spanish Ministry of Science and Innovation under the Recovery, Transformation, and Resilience Plan (GL2021-03-39 and GL2021-03-40). ISCIII provided funding (COV00624 to J. R. T. and M. F. F., PI21/01067 to M. F. F.), along with support from CSIC (202020E092 to M. F. F.), the PCTI from the Asturias Government co-funded by 2018-2022/FEDER (IDI/2021/000077 to M. F. F.), and the IUOPA. J. R. T. has an RyC contract from the Spanish Ministry of Science and Innovation (RYC2021-031799-I). A. R. receives support from the PTI Salud Global.References
- X. Jin, J. Ren, R. Li, Y. Gao, H. Zhang and J. Li,
et al., Global burden of upper respiratory infections in 204 countries and territories, from 1990 to 2019, eClinicalMedicine, 2021, 37, 100986 CrossRef PubMed
.
- M. Jit, A. Ananthakrishnan, M. McKee, O. J. Wouters, P. Beutels and Y. Teerawattananon, Multi-country collaboration in responding to global infectious disease threats: lessons for Europe from the COVID-19 pandemic, Lancet Reg. Health Eur., 2021, 9, 100221 CrossRef PubMed
.
- A. M. Near, J. Tse, Y. Young-Xu, D. K. Hong and C. M. Reyes, Burden of influenza hospitalization among high-risk groups in the United States, BMC Health Serv. Res., 2022, 22(1), 1209 CrossRef PubMed
.
- J. S. Nguyen-Van-Tam, M. O'Leary, E. T. Martin, E. Heijnen, B. Callendret and R. Fleischhackl,
et al., Burden of respiratory syncytial virus infection in older and high-risk adults: a systematic review and meta-analysis of the evidence from developed countries, Eur. Respir. Rev., 2022, 31(166), 220105 CrossRef PubMed
.
- K. C. Halabi, M. S. Stockwell, L. Alba, C. Vargas, C. Reed and L. Saiman,
et al., Clinical and socioeconomic burden of rhinoviruses/enteroviruses in the community, Influenza Other Respir. Viruses, 2022, 16(5), 891–896 CrossRef CAS PubMed
.
- H. Bolek, L. Ozisik, Z. Caliskan and M. D. Tanriover, Clinical outcomes and economic burden of seasonal influenza and other respiratory virus infections in hospitalized adults, J. Med. Virol., 2022, 95(1), 28153 CrossRef PubMed
.
- R. K. Zimmerman, G. K. Balasubramani, H. E. A. D'Agostino, L. Clarke, M. Yassin and D. B. Middleton,
et al., Population-based hospitalization burden estimates for respiratory viruses, 2015-2019, Influenza Other Respir. Viruses, 2022, 16(6), 1133–1140 CrossRef PubMed
.
-
D. X. Liu, J. Q. Liang and T. S. Fung, Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae), Encyclopedia of Virology, Elsevier, 2021, pp. 428–440 Search PubMed
.
- K. J. Henrickson, Parainfluenza viruses, Clin. Microbiol. Rev., 2003, 16(2), 242–264 CrossRef CAS PubMed
.
- X. Wang, Y. Li, M. Deloria-Knoll, S. A. Madhi, C. Cohen and V. L. Arguelles,
et al., Global burden of acute lower respiratory infection associated with human parainfluenza virus in children younger than 5 years for 2018: a systematic review and meta-analysis, Lancet Global Health, 2021, 9(8), e1077–e1087 CrossRef CAS PubMed
.
- C. Griffiths, S. J. Drews and D. J. Marchant, Respiratory Syncytial Virus: Infection, Detection, and New Options for Prevention and Treatment, Clin. Microbiol. Rev., 2017, 30(1), 277–319 CrossRef CAS
.
- X. Wang, Y. Li, M. Deloria-Knoll, S. A. Madhi, C. Cohen and A. Ali,
et al., Global burden of acute lower respiratory infection associated with human metapneumovirus in children under 5 years in 2018: a systematic review and modelling study, Lancet Global Health, 2021, 9(1), e33–e43 CrossRef CAS PubMed
.
- I. M. Mackay, K. E. Arden and A. Nitsche, Real-time PCR in virology, Nucleic Acids Res., 2002, 30(6), 1292–1305 CrossRef CAS PubMed
.
- C. C. Ginocchio, Life beyond PCR: alternative target amplification technologies for the diagnosis of infectious diseases, part I, Clin. Microbiol. Newsl., 2004, 26(16), 121–128 CrossRef PubMed
.
- M. Fakruddin, K. S. B. Mannan, A. Chowdhury, R. M. Mazumdar, M. N. Hossain and S. Islam,
et al., Nucleic acid amplification: Alternative methods of polymerase chain reaction, J. Pharm. BioAllied Sci., 2013, 5(4), 245–252 CrossRef PubMed
.
- V. L. Dao Thi, K. Herbst, K. Boerner, M. Meurer, L. P. Kremer and D. Kirrmaier,
et al., A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples, Sci. Transl. Med., 2020, 12(556), eabc7075 CrossRef CAS PubMed
.
- S. P. Kidd, D. Burns, B. Armson, A. D. Beggs, E. L. A. Howson and A. Williams,
et al., Reverse-Transcription Loop-Mediated Isothermal Amplification Has High Accuracy for Detecting Severe Acute Respiratory Syndrome Coronavirus 2 in Saliva and Nasopharyngeal/Oropharyngeal Swabs from Asymptomatic and Symptomatic Individuals, J. Mol. Diagn., 2022, 24(4), 320–336 CrossRef CAS PubMed
.
- L. G. Liang, M. J. Zhu, R. He, D. R. Shi, R. Luo and J. Ji,
et al., Development of a multi-recombinase polymerase amplification assay for rapid identification of COVID-19, influenza A and B, J. Med. Virol., 2022, 95(1), e28139 CrossRef PubMed
.
- A. A. El Wahed, P. Patel, M. Maier, C. Pietsch, D. Rüster and S. Böhlken-Fascher,
et al., Suitcase Lab for Rapid Detection of SARS-CoV-2 Based on Recombinase Polymerase Amplification Assay, Anal. Chem., 2021, 93(4), 2627–2634 CrossRef CAS
.
- M. Z. Ahmed, P. Badani, R. Reddy and G. Mishra, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas Advancement in Molecular Diagnostics and Signal Readout Approaches, J. Mol. Diagn., 2021, 23(11), 1433–1442 CrossRef CAS PubMed
.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita and J. Singh,
et al., CRISPR-Cas12-based detection of SARS-CoV-2, Nat. Biotechnol., 2020, 38(7), 870–874 CrossRef CAS PubMed
.
- M. G. Mohsen and E. T. Kool, The Discovery of Rolling Circle Amplification and Rolling Circle Transcription, Acc. Chem. Res., 2016, 49(11), 2540–2550 CrossRef CAS PubMed
.
- L. D. W. Luu, M. Payne, X. Zhang, L. Luo and R. Lan, Development and comparison of novel multiple cross displacement amplification (MCDA) assays with other nucleic acid amplification methods for SARS-CoV-2 detection, Sci. Rep., 2021, 11(1), 1873 CrossRef CAS PubMed
.
- J. R. Tejedor, G. Martín, A. Roberti, C. Mangas, P. Santamarina-Ojeda and R. Fernández Pérez,
et al., Enhanced Detection of Viral RNA Species Using FokI-Assisted Digestion of DNA Duplexes and DNA/RNA Hybrids, Anal. Chem., 2022, 94(18), 6760–6770 CrossRef CAS PubMed
.
- F. Sievers, A. Wilm, D. Dineen, T. J. Gibson, K. Karplus and W. Li,
et al., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega, Mol. Syst. Biol., 2011, 7, 539 CrossRef PubMed
.
- A. M. Waterhouse, J. B. Procter, D. M. A. Martin, M. Clamp and G. J. Barton, Jalview Version 2--a multiple sequence alignment editor and analysis workbench, Bioinformatics, 2009, 25(9), 1189–1191 CrossRef CAS PubMed
.
- J. S. Reuter and D. H. Mathews, RNAstructure: software for RNA secondary structure prediction and analysis, BMC Bioinf., 2010, 11, 129 CrossRef
.
- M. Zhao, Y. Xu, D. Zhang, G. Li, H. Gao and X. Zeng,
et al., Establishment and evaluation of a quadruple quantitative real-time PCR assay for simultaneous detection of human coronavirus subtypes, Virol. J., 2022, 19(1), 67 CrossRef CAS PubMed
.
- J. D. Pearson, D. Trcka, S. Lu, S. J. Hyduk, M. Jen and M. M. Aynaud,
et al., Comparison of SARS-CoV-2 indirect and direct RT-qPCR detection methods, Virol. J., 2021, 18(1), 99 CrossRef CAS PubMed
.
- S. van Boheemen, T. M. Bestebroer, J. H. Verhagen, A. D. M. E. Osterhaus, S. D. Pas and S. Herfst,
et al., A family-wide RT-PCR assay for detection of paramyxoviruses and application to a large-scale surveillance study, PLoS One, 2012, 7(4), e34961 CrossRef CAS PubMed
.
- Y. Weizmann, Z. Cheglakov, V. Pavlov and I. Willner, An autonomous fueled machine that replicates catalytic nucleic acid templates for the amplified optical analysis of DNA, Nat. Protoc., 2006, 1(2), 554–558 CrossRef CAS PubMed
.
- L. Blanco, A. Bernad, J. M. Lázaro, G. Martín, C. Garmendia and M. Salas, Highly efficient DNA synthesis by the phage phi 29 DNA
polymerase. Symmetrical mode of DNA replication, J. Biol. Chem., 1989, 264(15), 8935–8940 CrossRef CAS
.
- Q. Song, X. Sun, Z. Dai, Y. Gao, X. Gong and B. Zhou,
et al., Point-of-care testing detection methods for COVID-19, Lab Chip, 2021, 21(9), 1634–1660 RSC
.
- T. Dong, M. Wang, J. Liu, P. Ma, S. Pang and W. Liu,
et al., Diagnostics and analysis of SARS-CoV-2: current status, recent advances, challenges and perspectives, Chem. Sci., 2023, 14(23), 6149–6206 RSC
.
- H. Jayakody, G. Kiddle, S. Perera, L. Tisi and H. S. Leese, Molecular diagnostics in the era of COVID-19, Anal. Methods, 2021, 13(34), 3744–3763 RSC
.
- M. C. Marqués, R. Ruiz, R. Montagud-Martínez, R. Márquez-Costa, S. Albert and P. Domingo-Calap,
et al., CRISPR-Cas12a-Based Detection of SARS-CoV-2 Harboring the E484K Mutation, ACS Synth. Biol., 2021, 10(12), 3595–3599 CrossRef PubMed
.
- Y. Zhu, W. Jiang, R. Chen, J. Jouha, Q. Wang and L. Wu,
et al., A post-pandemic perspective: Evolution of SARS-CoV-2 early detection, TrAC, Trends Anal. Chem., 2023, 117458 Search PubMed
.
- M. Shang, J. Guo and J. Guo, Point-of-care testing of infectious diseases: recent advances, Sens. Diagn., 2023, 2(5), 1123–1144 RSC
.
- G. Ruiz-Vega, M. Soler, M. C. Estevez, P. Ramirez-Priego, M. D. Pazos and M. A. Noriega,
et al., Rapid and direct quantification of the SARS-CoV-2 virus with an ultrasensitive nanobody-based photonic nanosensor, Sens. Diagn., 2022, 1(5), 983–993 RSC
.
- M. N. Esbin, O. N. Whitney, S. Chong, A. Maurer, X. Darzacq and R. Tjian, Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection, RNA, 2020, 26(7), 771–783 CrossRef CAS
.
- Y. Mardian, H. Kosasih, M. Karyana, A. Neal and C. Y. Lau, Review of Current COVID-19 Diagnostics and Opportunities for Further Development, Front. Med., 2021, 8, 615099 CrossRef
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00316g |
| This journal is © The Royal Society of Chemistry 2024 |