Open Access Article
Open Access ArticlepH-responsive i-motif-conjugated nanoparticles for MRI analysis†
Kristine Y.
Ma
 ab,
Mireia
Perera-Gonzalez
ab,
Mireia
Perera-Gonzalez
 b,
Nicole I.
Langlois
b,
Nicole I.
Langlois
 c,
Owen M.
Alzubi
a,
Joseph D.
Guimond
c,
Owen M.
Alzubi
a,
Joseph D.
Guimond
 a,
Chris A.
Flask
a,
Chris A.
Flask
 d and
Heather A.
Clark
d and
Heather A.
Clark
 *a
*a
aSchool of Biological and Health Systems Engineering, Arizona State University, Tempe AZ, USA. E-mail: Heather.Ann.Clark@asu.edu
bDept. of Bioengineering, Northeastern University, Boston MA, USA
cDept. of Chemistry and Chemical Biology, Northeastern University, Boston MA, USA
dDepts. of Radiology, Biomedical Engineering, and Pediatrics, Case Western Reserve University, Cleveland OH, USA
First published on 6th March 2024
Abstract
Gadolinium (Gd)-based contrast agents (CAs) are widely used to enhance anatomical details in magnetic resonance imaging (MRI). Significant research has expanded the field of CAs into bioresponsive CAs by modulating the signal to image and monitor biochemical processes, such as pH. In this work, we introduce the modular, dynamic actuation mechanism of DNA-based nanostructures as a new way to modulate the MRI signal based on the rotational correlation time, τR. We combined a pH-responsive oligonucleotide (i-motif) and a clinical standard CA (Gd-DOTA) to develop a pH-responsive MRI CA. The i-motif folds into a quadruplex under acidic conditions and was incorporated onto gold nanoparticles (iM-GNP) to achieve increased relaxivity, r1, compared to the unbound i-motif. In vitro, iM-GNP resulted in a significant increase in r1 over a decreasing pH range (7.5–4.5) with a calculated pKa = 5.88 ± 0.01 and a 16.7% change per 0.1 pH unit. In comparison, a control CA with a non-responsive DNA strand (T33-GNP) did not show a significant change in r1 over the same pH range. The iM-GNP was further evaluated in 20% human serum and demonstrated a 28.14 ± 11.2% increase in signal from neutral pH to acidic pH. This approach paves a path for novel programmable, dynamic DNA-based complexes for τR-modulated bioresponsive MRI CAs.
Introduction
Magnetic resonance imaging (MRI) offers multiple advantages as an anatomical imaging technique including deep tissue penetration, minimal invasiveness, and 3D scanning capabilities. Traditionally, paramagnetic metal-based contrast agents (CAs) have been used to improve imaging resolution for enhanced structural and functional details. Recent advances in the field have led to the development of bioresponsive MRI CAs, which offer new strategies for achieving biomarker detection and molecular imaging1 by responding to a physiological trigger. To achieve bioresponsive modulation of MRI signals (“relaxivity”), several strategies can be used to tune the properties of paramagnetic metal-based CAs. One popular approach is to alter the hydration state (q), which refers to the number of water molecules directly coordinated to the metal center of a coordinate complex.2 Some examples of q-modulated MRI CAs include different chelate complexes that respond to enzymatic cleaveage,3,4 cations (e.g., zinc,5 calcium,6–8 copper,9–11 and mercury12), pH,13 and oxidation reactions.14,15 While CAs featuring q-modulation have paved the way for bioresponsive MR imaging, issues with thermodynamic instability,16 metal displacement by coordinating anions in vivo,17 and synthetic inefficiency (i.e. laborious purification procedures) present challenges for the development of new CAs for MR imaging in complex systems. Additional methods to modulate the signal of a CA without depending on q-modulation have the potential to unlock new avenues for bioresponsive imaging.Enhancement of relaxivity can be alternatively attained by increasing the molecular weight of a CA by incorporating paramagnetic metal chelates onto a nanosized platform. This increase in CA size slows down the rotation of CA described by the rotational correlation time, τR.18 In our previous works, we have demonstrated this phenomenon by developing nanoplatforms that are simultaneously modulated by q and τR parameters. We paired q-modulated pH-responsive CAs with enzymes on a nanoparticle to demonstrate acetylcholine imaging in a rat brain.19 The pH-sensing mechanism of this CA, Gd(NP-DO3A),20 was dependent on changing the protonation state of the nitrophenol arm of the ligand, which consequently changes q (q = 1 vs. q = 2). Using the same pH-responsive Gd-based CA, we developed a DNA-dendrimer-based MRI CA that demonstrated a significant increase in relaxivity for use in quantitative pH measurement.21
Building from our previous works, we engineered a pH-responsive CA based on τR-modulation that is independent of q-modulation. We paired the FDA-approved Gd-DOTA (Dotarem®) CA with a stimuli-responsive DNA i-motif oligonucleotide. The i-motif is one of the simplest dynamic DNA structures that can be programmed to reversibly respond to a specific stimulus, in this case – pH.22 In acidic environments, hemiprotonation of cytidine bases in the i-motif sequence induces a strand conformational change through folding, forming a series of intercalated cytidine–cytidine+ base pairs in a quadruplex secondary structure.23 This folding process increases the rigidity of the complex, thus slowing the τR relative to the unfolded linear structure, resulting in an increase in relaxivity (r1). The rate of water exchange between the inner-sphere and bulk solvent, kex = 1/τM, is another parameter that can be modulated for designing bioresponsive CAs. While modulating τM is a complex process that is beyond the scope of this work, the i-motif folding mechanism can potentially impact water access to the CA and thus affect the MRI signal as well.
In this work, we coupled Gd-DOTA onto an i-motif sequence24 for a novel pH-responsive DNA-based CA design. The i-motif coupled with Gd-DOTA was loaded onto a gold nanoparticle (GNP) to expand the size of the complex and further increase MRI signals by slowing τR. The GNP and size were chosen as a platform to load the pH-responsive CA as it has been widely reported in the literature with established protocols for loading DNA,25,26 and for use as a biosensor platform in vivo.27 We demonstrated that our pH-responsive i-motif conjugated GNP (iM-GNP) can increase MRI signals (r1) through a bioresponsive mechanism, compared to a non-responsive control composed of a 33 nt repeating thymine oligonucleotide (T33-GNP) which does not fold in response to pH changes. Over traditional “on–off” MRI responsive systems, our approach features reversibility, tunability and high sensitivity over a physiologically-relevant pH range. Our approach is of special interest for paving a path for engineering novel applications of programmable dynamic DNA-based complexes for non q-modulated bioresponsive CAs for MR imaging.
Experimental methods
Gd-DOTA & oligonucleotide coupling preparation
Custom amine/thiol-functionalized single-strand DNA oligonucleotides were purchased from MilliporeSigma, with sequences reported in Table S1.† The concentration of DNA was determined via the absorbance at 260 nm, ε = 282.8 mM−1 cm−1 for the i-motif strand and ε = 267.9 mM−1 cm−1 for the control T33 strand. Gd-DOTA was conjugated onto the DNA oligonucleotide via thioamide coupling on the amine terminal end of the DNA oligonucleotide. Gd–p-SCN-Bn-DOTA (2.46 mg, 3 μmol, macrocyclics) and the amine/thiol-functionalized DNA oligonucleotide (0.02 μmol) were dissolved in 0.5 mL of 0.1 M carbonate buffer and incubated for 20 h at room temperature. DNA–Gd-DOTA conjugates were purified with ethanol precipitation followed by an Illustra NAP-10 column (GE Health) to remove excess Gd–p-SCN-Bn-DOTA. The conjugates were eluted in nanopure water and then lyophilized overnight (Labconco).Analysis of Gd-DOTA coupling to DNA by HPLC
Chromatographic analyses were performed using an Agilent 1260 Infinity II HPLC System, equipped with a vial sampler, quaternary pump, wide-range diode array detector, and ChemStation software. A method for the separation of free DNA from Gd-DOTA conjugates was developed using ion-paired reversed phase chromatography on a Zorbax EclipsePlus C18 column (4.6 mm × 250 mm, 5 μm, Agilent). Mobile phase A was 50 mM triethylammonium acetate (TEAA, pH 7.0), with acetonitrile as mobile phase B. Samples of unpurified DNA–Gd-DOTA conjugates were reduced with 0.1 M tris(2-carboxyethyl)phosphine (TCEP) for 1 h, diluted in the mobile phase, and then analyzed at a flow rate of 0.65 mL min−1 using a gradient elution method (10–30% B over 20 min). Unmodified DNA samples were diluted in the mobile phase and analyzed using the same gradient method for comparison. The online absorbance signal for the DNA was monitored at 260 nm. The retention times of the i-motif strand, iM–Gd-DOTA, the T33 strand, and T33–Gd-DOTA were 9.15 min, 9.44 min, 9.81 min, and 10.26 min, respectively. Residual Gd–p-SCN-Bn-DOTA present in the samples exhibited peaks with retention times of 9.85 min, 10.58 min, 11.15 min, 11.80 min, and 11.98 min. Disulfide dimers present in the non-reduced, unreacted DNA samples exhibited peaks with retention times of 10.05 min and 10.60 min for the i-motif and T33 strands.Preparation of Gd-DOTA oligonucleotide functionalized gold nanoparticles
Oligo-functionalized gold nanoparticles (GNPs) (10 nm) were prepared according to literature procedures.25 Briefly, the Gd-DOTA-conjugated i-motif and T33 oligos (0.02 μmol) were resuspended in a solution of 0.1 M tris(2-carboxyethyl)phosphine (TCEP) (5.73 mg, 20 μmol) and 0.1 M phosphate buffer (PB) at pH 8 to reduce the disulfide bonds at the terminal thiol ends of the oligonucleotides. After 1 h in the reducing environment, the freshly deprotected oligos (4 nmol) were diluted in nanopure water (250 μL) and then added to 1 mL of gold colloid (10 nM, Sigma-Aldrich) in an Eppendorf tube. The solution was shaken for 2 h prior to salt stabilization. iM-GNP and T33-GNP were buffered to a pH of 7.2 with a final concentration of 10 mM PB and 0.01% SDS. After 30 min equilibration, samples were salt aged with NaCl solution, slowly increasing the NaCl concentration to 1.0 M over a 4 h period. GNPs were sonicated during this salt aging process. Particles were shaken overnight to yield fully functionalized GNPs. Removal of any unbound oligonucleotides was performed by washing through sequential centrifugation steps (10 kDa MWCO, 4000 × g, 20 °C), supernatant removal, and resuspension (0.1% SDS in water). This procedure was repeated four times. To determine the concentration of each GNP sample, we used the reported extinction coefficient at Abs = 530 nm for 10 nm particles: ε = 1.06 × 108 M−1 cm−1.pH titration (1.4 T NMR)
iM-GNP and T33-GNP (30 nM) were dispersed in 20 mM MES/HEPPS/HEPES at pH = 9. The pH titration was carried out by adding 1–3 μL increments of 0.1 M HCl. With each addition, the solution was vigorously vortexed, the pH was measured using a calibrated pH meter (Orion Star™), and T1 measurements were obtained using a 1.4 T Bruker minispec mq60 NMR analyzer (60 MHz, Bruker Inc., Billerica, MA) at 37 °C. The relaxivity (r1) at each point was calculated with the GNP concentration. The gain was set to 56 dB and T1 delays of 6–8000 ms were used. Four T1 replicate measurements were performed, with the last three being averaged to stabilize the temperature effect on the measurement. T1 values were measured in the pH range of 4.5–7.5. Relaxivity values were obtained by using eqn (1) and plotted against pH, fitted to a dose–response curve.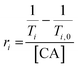 | (1) |
Relaxivity analysis (3.0 T MRI)
The T1 maps were calculated on a voxel-by-voxel basis by three-parameter fitting of eqn (2) to the signal intensity images at each TR.| Mz(TR) = Mo·(1 − e−TR/T1) + C | (2) |
Regions of interest (ROI) were manually defined from the MR signal intensity images for each capillary tube. The resulting T1 values were averaged over each ROI. Relaxivity values (ri) were obtained from eqn (2), where T1 = longitudinal relaxation of the CA in buffer.
Cell viability by MTT assay
Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide reduction (MTT) assay. HEK293T cells were seeded in 96-well plates at a density of 2.5 × 104 cells in 100 μL of cell media per well. After 24 h incubation (37 °C, 5% CO2), cells were treated with water-soluble iM-GNP by adding 50 μL of various concentrations for final working concentrations (0 nM to 20 nM). After 24 h exposure and incubation, media in wells were carefully replaced with 200 μL of MTT–DMEM (0.2 mg mL−1). Cells were incubated for 3 h and media were carefully replaced with 100 μL of dimethyl sulfoxide (DMSO). After 15 min of shaking at room temperature, the absorbance was measured at 540 nm with a BioTek Synergy H1 microplate reader. The viability was assessed by a percentage with respect to controls (set as 100%).Cell viability by NR assay
The cell viability was determined using the neutral red (NR) uptake assay. HEK293T cells were seeded in 96-well plates at a density of 2.5 × 104 cells in 100 μL of cell media per well. After 24 h incubation (37 °C, 5% CO2), cells were treated with water-soluble iM-GNP by adding 50 μL of various concentrations for final working concentrations (0 nM to 20 nM). After 24 h exposure and incubation, media in wells were carefully replaced with 100 μL neutral red medium (50 μg mL−1). Cells were incubated for 2 h. Media were carefully replaced with 150 μL of phosphate buffered saline (PBS) and centrifuged (1000 × g, 20 °C) for 10 min. Media were carefully removed, and the cell pellet was resuspended in 150 μL of neutral red destain solution (50% ethanol, 1% glacial acetic acid). After 10 min of shaking at room temperature, the absorbance was measured at 540 nm with a BioTek Synergy H1 microplate reader.Cell viability analysis
The viability was assessed by a percentage with respect to controls (0 nM, set as 100%) depicted by eqn (3). Values were corrected with blanks. The statistical significance was assessed by one-way analysis of variance (ANOVA).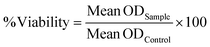 | (3) |
Results & discussion
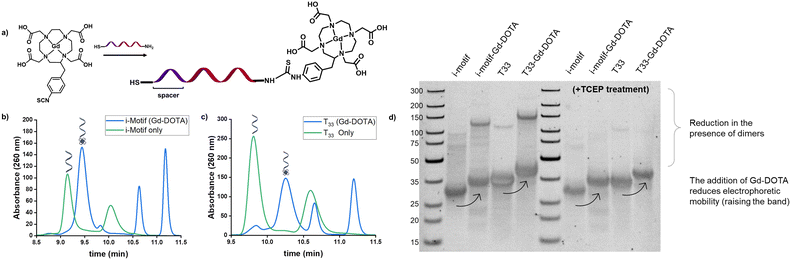
To prepare our CAs, Gd-DOTA was conjugated onto the DNA i-motif sequence, purified, and validated before loading onto GNPs. By using commercially-available Gd-DOTA, the need for chelating Gd as a separate step during synthesis is eliminated, thus preventing non-specific interactions of Gd3+ to the DNA.28 To tailor the sensor for actuation in a relevant dynamic range, we chose a reported i-motif sequence which undergoes a change in folding from 5 to 95% between pH 6.26 and 5.9.24 This range is useful in the study of disease states such as metastatic tumors (pH 6.1–6.4 (ref. 29)) or tissue acidosis (pH 6.0–7.0 (ref. 30)). A 12 nt length spacer of thymine bases was chosen to maintain an ∼4 nm distance between the GNP surface and the i-motif strand to provide flexibility and freedom31 for the i-motif to fold and unfold by minimizing electrostatic repulsions with the GNP surface. We also prepared a control group using repeating thymine residues with the same number of base pairs (T33) as the i-motif with the spacer. The 5′ end of each oligo was amine-modified for coupling to Gd-DOTA, and the 3′ end was thiol-modified for GNP attachment. With a commonly used primary amine reaction, we coupled the isothiocyanate-functionalized Gd-DOTA with the amine end of the DNA through a thioamide linkage (Fig. 1a). Upon purification and reduction of the disulfide bonds via TCEP, successful coupling of the Gd-DOTA was validated by ion-paired reverse-phase liquid chromatography, as evidenced by shifts to later retention times compared to the unmodified oligonucleotide strands (Fig. 1b and c). Peaks from the unmodified i-motif/T33 strands were not observed in the chromatograms after the modifications, indicating a high reaction conversion (DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gd-DOTA ≈ 1
Gd-DOTA ≈ 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio). Residual Gd–p-SCN-Bn-DOTA in the reacted samples contributed to additional peaks in the chromatogram as shown in Fig. S1.† Additionally, in the non-reduced reference DNA strands, a secondary peak was observed at later retention times that is consistent with the presence of disulfide dimers compared to the reduced single strands. These results were further validated through gel electrophoresis (Fig. 2d) where it can be observed that a reduced electrophoretic mobility is associated with Gd-DOTA modification on the strands and that TCEP treatment significantly reduced the observation of disulfide dimers present in the samples.
1 ratio). Residual Gd–p-SCN-Bn-DOTA in the reacted samples contributed to additional peaks in the chromatogram as shown in Fig. S1.† Additionally, in the non-reduced reference DNA strands, a secondary peak was observed at later retention times that is consistent with the presence of disulfide dimers compared to the reduced single strands. These results were further validated through gel electrophoresis (Fig. 2d) where it can be observed that a reduced electrophoretic mobility is associated with Gd-DOTA modification on the strands and that TCEP treatment significantly reduced the observation of disulfide dimers present in the samples.
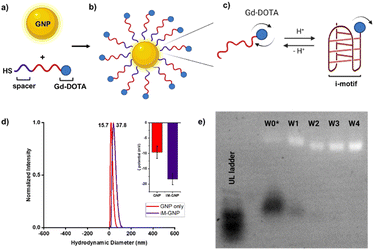 | ||
| Fig. 2 Scheme of pH-responsive iM-GNP. a) Thiolated oligonucleotides were loaded onto GNP after TCEP treatment b) iM-GNP in the fully expanded form at basic pH. c) Folding of the i-motif oligonucleotides occurs in the presence of increased H+. d) DLS analysis shows that the average size of unmodified commercial 10 nm GNP was 15.7 nm, while iM-GNP had a larger average size of 37.8 nm. Inset shows the ζ potential of the unmodified GNP = −9.7 ± 2.0 mV and iM-GNP = −18.4 ± 1.8 mV. e) Electrophoretic mobility of iM-GNP (2.0% agarose gel, 1× TAE). The first lane (from left to right) corresponds to the ultralow ladder. W0* is the iM-GNP reaction solution before washing. W1, W2, W3, and W4 represent the iM-GNP concentrate after 1, 2, 3, and 4 washes, respectively. It appears that after at least 3 washes, there is no free DNA (black bands) and the iM-GNP sensor (white bands) is no longer changing the size/charge due to non-covalently bound DNA. This is further validated by ICP-MS analysis (Fig. S3†). | ||
The Gd-DOTA-conjugated i-motif and T33 control were loaded onto GNPs following established protocols25,26 for maximal DNA loading per GNP (Fig. 2a–c). Salt aging decreases the electrostatic interaction between neighboring oligonucleotides and thus improves the loading efficiency of the DNA on the surface of the GNP. To remove the unbound DNA, sequential centrifugation, supernatant removal, and resuspension (0.01% SDS in water) were performed and repeated. The unbound DNA in the sample is completely removed after at least three washes as confirmed by both agarose gel electrophoresis32 (Fig. 2e) and ICP-MS (Fig. S3†). The zeta potential of the GNPs increased in magnitude after DNA loading (unmodified GNP = −9.7 ± 2.0 mV and iM-GNP = −18.4 ± 1.8 mV), which indicated successful gold–thiol linkage and DNA immobilization onto the GNP (Fig. 2d). This increase in negative surface charge is consistent with observations in previous reports for DNA immobilization on the surface of GNPs.33 The gadolinium content was estimated using ICP analysis, yielding 123.5 ± 30.4 units of oligo–Gd-DOTA per GNP. DLS was used to measure the average diameter of iM-GNP (37.8 nm) in comparison to unmodified GNPs (15.7 nm) (Fig. 2d). The increase in hydrodynamic size of the NP can be attributed to the immobilization of the DNA on the surface, further corroborating successful functionalization to yield the iM-GNP (Fig. S2†).
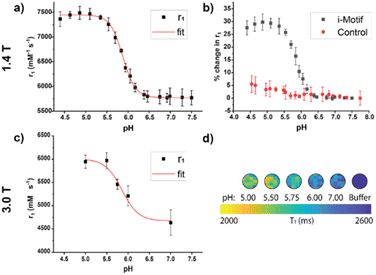
We characterized the pH-responsive properties of the iM-GNP and the control, T33-GNP, using both the benchtop 1.4 T NMR and the preclinical 3.0 T MRI scanner. From the 1.4 T NMR at 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C, the relaxivity profiles of iM-GNP as a function of pH were obtained between pH 4.50 and 7.50 with a calculated pKa of 5.88 ± 0.01 and a 16.7% change per 0.10 pH unit (Fig. 3a). These measured values are lower than the reported pKa of the i-motif strand (6.26 ± 0.01 at 25
°C, the relaxivity profiles of iM-GNP as a function of pH were obtained between pH 4.50 and 7.50 with a calculated pKa of 5.88 ± 0.01 and a 16.7% change per 0.10 pH unit (Fig. 3a). These measured values are lower than the reported pKa of the i-motif strand (6.26 ± 0.01 at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C).24 We hypothesize that this slight shift in observed pKa can be attributed to factors including modification of the sequence with additional thymine bases as a spacer, the coupling of the DNA with Gd-DOTA as the reporter, and differences in buffer and instrument conditions. The per metal-center magnetic relaxivity measured at 1.4 T for iM-GNP was approximated by accounting for the ICP-MS measured 123 Gd-DOTA sites per GNP: acidic (pH = 5) r1 = 60.7 ± 1.1 mM−1 s−1 and neutral (pH = 7) r1 = 47.1 ± 1.7 mM−1 s−1 (n = 3). The per metal center relaxivities resulted in significantly higher values compared to the standard Gd-DOTA agent (∼3.11 mM−1 s−1 at 1.4 T).34 When comparing the percent change in r1 between iM-GNP and T33-GNP, there is a clear increase in signal change for iM-GNP (Fig. 3b). The iM-GNP signal enhancement is also significantly greater than the Gd-DOTA conjugated i-motif strand alone, which exhibited a per-metal center relaxivity of 48.8 mM−1 s−1 at pH 5 (Fig. S5†).
°C).24 We hypothesize that this slight shift in observed pKa can be attributed to factors including modification of the sequence with additional thymine bases as a spacer, the coupling of the DNA with Gd-DOTA as the reporter, and differences in buffer and instrument conditions. The per metal-center magnetic relaxivity measured at 1.4 T for iM-GNP was approximated by accounting for the ICP-MS measured 123 Gd-DOTA sites per GNP: acidic (pH = 5) r1 = 60.7 ± 1.1 mM−1 s−1 and neutral (pH = 7) r1 = 47.1 ± 1.7 mM−1 s−1 (n = 3). The per metal center relaxivities resulted in significantly higher values compared to the standard Gd-DOTA agent (∼3.11 mM−1 s−1 at 1.4 T).34 When comparing the percent change in r1 between iM-GNP and T33-GNP, there is a clear increase in signal change for iM-GNP (Fig. 3b). The iM-GNP signal enhancement is also significantly greater than the Gd-DOTA conjugated i-motif strand alone, which exhibited a per-metal center relaxivity of 48.8 mM−1 s−1 at pH 5 (Fig. S5†).
In addition to τR, the folding of the i-motif can contribute to reduced water activity due to an ordered layer of water on the surface caused by nanoconfinement conditions.35 The restricted space makes the water molecules more ordered than in the bulk solution affecting τM and thus affecting relaxivity.35 Water molecules in enclosed environments have a considerably different diffusion than in the bulk, which leads to increased relaxivities.36 Previous works showed that introducing steric constraints on the water-binding site37,38 increases the water exchange rate and thus contributes to increased relaxivity. Under confined conditions, the strong outer sphere relaxation from the i-motif could be the influencing τM properties of the Gd-DOTA. Understanding the degree of impact of τM from folding and unfolding of the i-motif coupled with the MR CA is outside the scope of this work but could be a subject of future studies. For the control, T33-GNP, there was no significant change in r1in vitro at 1.4 T over the pH range of 4.50–7.50 (Fig. S4†). When comparing the percent change in r1 between iM-GNP and T33-GNP, there is a clear increase in signal change for iM-GNP (Fig. 3b).
The change in MR response was also characterized using the 3.0 T MRI. We expected that the relaxivity profiles from 3.0 T would exhibit similar trends to the data obtained from 1.4 T, with a slight downward intensity shift in the pH-relaxivity curve due to the increase in magnetic field strength.39 We prepared 30 nM iM-GNP (n = 3) in solutions of increasing pH buffers (pH 5.00, 5.50, 5.75, 6.00, and 7.00) to capture the change in relaxivity as a function of pH. Using an in-house developed MRI testbed for small volume analysis (Fig. S6†), we observed a significant change in signal between pH 5.0 and 7.0, Δ1/T1 28.6 ± 7.4% with a calculated pKa of 5.86 ± 0.14 and a 12.7% change per 0.10 pH unit (Fig. 3c). The raw plots for the 3.0 T MRI signal intensity over repetition time were used to calculate the iM-GNP T1 values and are reported in (Fig. S7†).
To demonstrate the suitability of the application of iM-GNP in complex biological environments, we prepared 20% human serum (HS) in pH 4.99 buffer and pH 8.79 buffer. There was a measurable signal change between the basic pH condition and the acidic pH condition of 28.14 ± 11.2% (n = 2). Our results suggest that the i-motif can fold not only in vitro under well-controlled conditions, but also in a complex medium. Previous reports show that various media conditions (i.e., water, plasma, blood) do not impact the r1 signal of the Gd-based CA significantly,41 and that the folding capabilities of the i-motif are not hindered in complex in vivo environments (i.e., pH-responsive nanomachine42).
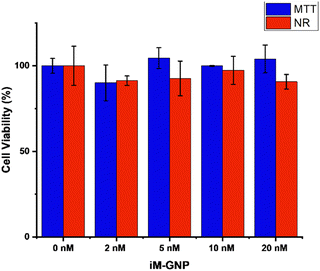
We evaluated the potential toxicity of the iM-GNP with two different in vitro cell viability assays. MTT and NR assays were applied to iM-GNP treated HEK293T cells to examine biocompatibility and safety. Both assays are commonly used for investigating the cell viability of drugs, chemical exposure, and nanoparticles.40 Our ANOVA analysis from MTT (p = 0.14) and NR (p = 0.57) assays depict no significant differences (p > 0.05) between the control and different iM-GNP concentration treatment groups (Fig. 4), indicating the iM-GNP's biocompatibility.
A significant advantage in using the i-motif as the MR signal modulating element is the ease of tunability of the i-motif response range by making changes in the DNA sequence. The i-motif based system can be modified by using various cytosine derivatives and by extending the length of the C–C+ strands to tune the pH range by +0.14 and −0.22 pH units in addition to the folding kinetics.24
Future work could address several research avenues that would improve the quantitative aspects of this study. First, there is an observed drop in signal for both the acidic and neutral group when measuring the iM-GNP in 20% HS over an extended period of time at 37 °C (Fig. S8†). Enzymatic destabilization from digestion by nucleases present in HS is likely the source of DNA structural instability, causing the i-motif to lose function overtime. Future adaptations of this work could investigate ways to stabilize the i-motif against degradation by introducing alternative materials (e.g. PS-DNA, LNA, other XNAs).43
Second, future studies could investigate the optimal balance between CA size and CA homogeneity (i.e. same number of Gd–CA per DNA nanostructure21) without compromising the potential for accurate pH quantification. This is because accurate pH quantification for this study is hindered by the inhomogeneity of Gd attachments to each nanoparticle complex; however, the use of GNPs enabled dense packing of Gd-DOTA onto a single platform to maximize the signal. Third, potential studies can also include quantifying pH independent of CA concentration, by using a second non-pH-responsive CA with the same concentration in tandem with the iM–CA, as in our previously reported work.21 This second CA could be: 1) a T2-modulated CA (using dual contrast MR fingerprinting44); 2) a 19F or 13C based CA (using multi-nuclear MRI);45 and/or 3) a radiolabeled agent (using PET/MRI).46 In summary, the desirable characteristics of the pH-responsive i-motif-conjugated NPs for MR analysis enable another step towards translation of more bioresponsive CAs into future in vivo studies.
Conclusions
We have presented a τR-modulated bioresponsive CA based on the actuation mechanism of the i-motif for imaging pH using MRI. The pH-responsiveness was demonstrated by 1.4 T relaxometer analysis with a measured pKa of 5.88 ± 0.01 and a 16.7% change per 0.10 pH unit, with similar results to 3.0 T MRI analysis. As a platform, the iM-GNP demonstrated robust MR signals with a 1.4 T measured per-metal center relaxivity of r1 = 60.7 ± 1.1 mM−1 s−1 at acidic pH, a 20-fold increase compared to Gd-DOTA alone. This MR CA demonstrated a significant change in signal in vitro compared to the non-responsive T33-GNP control particles. Upon analysis of both particles in 20% HS under acidic and neutral conditions, the iM-GNP demonstrated a significant change in signal of 28.14 ± 11.2%. The iM-GNP's mechanism is not only effective in a complex medium, but also shows potential for biocompatibility as indicated by cellular assays. By utilizing the rotational motion of a stimuli-responsive DNA-based i-motif, combined with an FDA approved CA, this biocompatible platform presents an unparalleled opportunity to develop bioresponsive CAs for in vivo translation with ease of synthesis and tunability.Author contributions
We strongly encourage authors to include author contributions and recommend using CRediT for standardised contribution descriptions. Please refer to our general author guidelines for more information about authorship.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Dr. Guoxin Rong at the Institute for Chemical Imaging of Living Systems (RRID: SCR_022681) at Northeastern University for MRI imaging support. The authors would like to thank Dr. Isen Andrew C. Calderon for assistance in carrying out preliminary studies, for helpful discussions regarding experimental design and data analysis. This work was supported by the US National Institute of Health, NINDS (1UF1NS107713-01).Notes and references
- B. B. Bartelle, A. Barandov and A. Jasanoff, Neuroscience, 2016, 36, 4139–4148 CrossRef CAS PubMed.
- M. P. Lowe, D. Parker, O. Reany, S. Aime, M. Botta, G. Castellano, E. Gianolio and R. Pagliarin, J. Am. Chem. Soc., 2001, 123, 7601–7609 CrossRef CAS PubMed.
- R. A. Moats, S. E. Fraser and T. J. Meade, Angew. Chem., Int. Ed. Engl., 1997, 36, 726–728 CrossRef CAS.
- A. Y. Louie, M. M. Hüber, E. T. Ahrens, U. Rothbächer, R. Moats, R. E. Jacobs, S. E. Fraser and T. J. Meade, Nat. Biotechnol., 2000, 18, 321–325 CrossRef CAS PubMed.
- J. L. Major, G. Parigi, C. Luchinat and T. J. Meade, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13881–13886 CrossRef CAS PubMed.
- A. Mishra, P. Fousková, G. Angelovski, E. Balogh, A. K. Mishra, N. K. Logothetis and É. Tóth, Inorg. Chem., 2008, 1370–1381 CrossRef CAS PubMed.
- A. Mishra, N. K. Logothetis and D. Parker, Chem. – Eur. J., 2011, 17, 1529–1537 CrossRef CAS PubMed.
- G. Angelovski, P. Fouskova, I. Mamedov, S. Canals, E. Toth and N. K. Logothetis, ChemBioChem, 2008, 9, 1729–1734 CrossRef CAS PubMed.
- E. L. Que and C. J. Chang, J. Am. Chem. Soc., 2006, 128, 15942–15943 CrossRef CAS PubMed.
- E. L. Que, E. Gianolio, S. L. Baker, A. P. Wong, S. Aime and C. J. Chang, J. Am. Chem. Soc., 2009, 131, 8527–8536 CrossRef CAS PubMed.
- W.-S. Li, J. Luo and Z.-N. Chen, Dalton Trans., 2011, 40, 484–488 RSC.
- M. Andrews, A. J. Amoroso, L. P. Harding and S. J. Pope, Responsive, Dalton Trans., 2010, 39, 3407–3411 RSC.
- S. Zhang, K. Wu and A. D. Sherry, Angew. Chem., Int. Ed., 1999, 38, 3192–3194 CrossRef CAS PubMed.
- C. Tu, R. Nagao and A. Y. Louie, Angew. Chem., 2009, 121, 6669–6673 CrossRef.
- C. Tu, E. A. Osborne and A. Y. Louie, Tetrahedron, 2009, 65, 1241–1246 CrossRef CAS PubMed.
- M. Polasek and P. Caravan, Inorg. Chem., 2013, 52, 4084–4096 CrossRef CAS PubMed.
- J. Wahsner, E. M. Gale, A. Rodríguez-Rodríguez and P. Caravan, Chem. Rev., 2019, 119, 957–1057 CrossRef CAS PubMed.
- P. Caravan, C. T. Farrar, L. Frullano and R. Uppal, Contrast Media Mol. Imaging, 2009, 4, 89–100 CrossRef CAS PubMed.
- Y. Luo, E. H. Kim, C. A. Flask and H. A. Clark, ACS Nano, 2018, 12, 5761–5773 CrossRef CAS PubMed.
- M. Woods, G. E. Kiefer, S. Bott, A. Castillo-Muzquiz, C. Eshelbrenner, L. Michaudet, K. McMillan, S. D. Mudigunda, D. Ogrin and G. Tircsó, J. Am. Chem. Soc., 2004, 126, 9248–9256 CrossRef CAS PubMed.
- H. Seo, K. Y. Ma, E. E. Tuttle, I. A. C. Calderon, A. D. Buskermolen, C. A. Flask and H. A. Clark, ACS Sens., 2021, 6, 727–732 CrossRef CAS PubMed.
- K. Gehring, J.-L. Leroy and M. Guéron, Nature, 1993, 363, 561–565 CrossRef CAS PubMed.
- Y. Zhao, Z.-X. Zeng, Z.-Y. Kan, Y.-H. Hao and Z. Tan, ChemBioChem, 2005, 6, 1957–1960 CrossRef CAS PubMed.
- L. Lannes, S. Halder, Y. Krishnan and H. Schwalbe, ChemBioChem, 2015, 16, 1647–1656 CrossRef CAS PubMed.
- H. D. Hill, J. E. Millstone, M. J. Banholzer and C. A. Mirkin, ACS Nano, 2009, 3, 418–424 CrossRef CAS PubMed.
- S. J. Hurst, A. K. R. Lytton-Jean and C. A. Mirkin, Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes, Anal. Chem., 2006, 78, 8313–8318 CrossRef CAS PubMed.
- Y. Li, H. J. Schluesener and S. Xu, Gold Bull., 2010, 43, 29–41 CrossRef CAS.
- O. Edogun, N. H. Nguyen and M. Halim, Anal. Bioanal. Chem., 2016, 408, 4121–4131 CrossRef CAS PubMed.
- M. Anderson, A. Moshnikova, D. M. Engelman, Y. K. Reshetnyak and O. A. Andreev, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 8177–8181 CrossRef CAS PubMed.
- F. E. Díaz, E. Dantas and J. Geffner, Mediators Inflammation, 2018, 2018, 1218297 Search PubMed.
- J.-H. Oh and J.-S. Lee, Anal. Chem., 2011, 83, 7364–7370 CrossRef CAS PubMed.
- D. Zanchet, C. M. Micheel, W. J. Parak, D. Gerion, S. C. Williams and A. P. Alivisatos, J. Phys. Chem. B, 2002, 106, 11758–11763 CrossRef CAS.
- H. Yang, Z. Chen, L. Zhang, W.-Y. Yung, K. C.-F. Leung, H. Y. E. Chan and C. H. J. Choi, Small, 2016, 12, 5178–5189 CrossRef CAS PubMed.
- X. Wu, A. C. Dawsey, B. N. Siriwardena-Mahanama, M. J. Allen and T. J. Williams, J. Fluorine Chem., 2014, 168, 177–183 CrossRef CAS PubMed.
- J. Huang, S. Gambietz and B. Saccà, Small, 2022, 2202253 Search PubMed.
- K. Malzahn, S. Ebert, I. Schlegel, O. Neudert, M. Wagner, G. Schütz, A. Ide, F. Roohi, K. Münnemann, D. Crespy and K. Landfester, Adv. Healthcare Mater., 2016, 5, 567–574 CrossRef CAS PubMed.
- C. H. Huang and A. Tsourkas, Curr. Top. Med. Chem., 2013, 13, 411–421 CrossRef CAS PubMed.
- S. Avedano, L. Tei, A. Lombardi, G. B. Giovenzana, S. Aime, D. Longo and M. Botta, Chem. Commun., 2007, 4726–4728 RSC.
- G. E. Hagberg and K. Scheffler, Contrast Media Mol. Imaging, 2013, 8, 456–465 CrossRef CAS PubMed.
- K. P. Steckiewicz, E. Barcinska, A. Malankowska, A. Zauszkiewicz-Pawlak, G. Nowaczyk, A. Zaleska-Medynska and I. Inkielewicz-Stepniak, J. Mater. Sci.: Mater. Med., 2019, 30, 22 CrossRef PubMed.
- M. Rohrer, H. Bauer, J. Mintorovitch, M. Requardt and H.-J. Weinmann, Invest. Radiol., 2005, 40, 715–724 CrossRef PubMed.
- S. Surana, J. M. Bhat, S. P. Koushika and Y. Krishnan, Nat. Commun., 2011, 2, 340 CrossRef PubMed.
- N. I. Langlois, K. Y. Ma and H. A. Clark, Appl. Phys. Rev., 2023, 10, 011304 CAS.
- C. E. Anderson, S. B. Donnola, Y. Jiang, J. Batesole, R. Darrah, M. L. Drumm, S. M. Brady-Kalnay, N. F. Steinmetz, X. Yu, M. A. Griswold and C. A. Flask, Sci. Rep., 2017, 7, 8431 CrossRef PubMed.
- G. Gambino, T. Gambino, R. Pohmann and G. Angelovski, Chem. Commun., 2020, 56, 3492–3495 RSC.
- G. Thomas, J. Boudon, L. Maurizi, M. Moreau, P. Walker, I. Severin, A. Oudot, C. Goze, S. Poty, J.-M. Vrigneaud, F. Demoisson, F. Denat, F. Brunotte and N. Millot, ACS Omega, 2019, 4, 2637–2648 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00285c |
| This journal is © The Royal Society of Chemistry 2024 |