Open Access Article
Open Access ArticleDecoding the role of mycobacterial lipid remodelling and membrane dynamics in antibiotic tolerance†
Anjana P.
Menon
abc,
Tzong-Hsien
Lee
bc,
Marie-Isabel
Aguilar
 *bc and
Shobhna
Kapoor
*bc and
Shobhna
Kapoor
 *ab
*ab
aDepartment of Chemistry, Indian Institute of Technology Bombay, Mumbai 400076, India. E-mail: shobhnakapoor@chem.iitb.ac.in
bIITB-Monash Research Academy, Indian Institute of Technology Bombay, Mumbai 400076, India. E-mail: mibel.aguilar@monash.edu
cDepartment of Biochemistry & Molecular Biology, Monash University, Clayton, VIC 3800, Australia
First published on 29th October 2024
Abstract
Current treatments for tuberculosis primarily target Mycobacterium tuberculosis (Mtb) infections, often neglecting the emerging issue of latent tuberculosis infection (LTBI) which are characterized by reduced susceptibility to antibiotics. The bacterium undergoes multiple adaptations during dormancy within host granulomas, leading to the development of antibiotic-tolerant strains. The mycobacterial membrane plays a crucial role in drug permeability, and this study aims to characterize membrane lipid deviations during dormancy through extensive lipidomic analysis of bacteria cultivated in distinct media and growth stages. The results revealed that specific lipids localize in different regions of the membrane envelope, allowing the bacterium to adapt to granuloma conditions. These lipid modifications were then correlated with the biophysical properties of the mycomembrane, which may affect interactions with antibiotics. Overall, our findings offer a deeper understanding of the bacterial adaptations during dormancy, highlighting the role of lipids in modulating membrane behaviour and drug permeability, ultimately providing the groundwork for the development of more effective treatments tailored to combat latent infections.
Introduction
Tuberculosis (TB) remains a formidable global health challenge and a leading cause of death worldwide, irrespective of HIV co-infection.1. The ability of Mycobacterium tuberculosis (Mtb) to inhibit phagosome-lysosome fusion and survive within granulomas facilitates the emergence of persistent cells, leading to latent tuberculosis infection (LTBI).2 Recent upticks in TB cases3 underscore the urgency for improved treatment regimens that act via different mechanisms compared to current therapies that predominantly target replicating Mycobacterium during early infection phases, yielding suboptimal results for LTBI.4Mycobacterium exhibits a complex membrane envelope, featuring a unique outer membrane composed of a non-covalently bound outer leaflet and a peptidoglycan-linked inner leaflet.5 Chemical and microscopic evidence confirms that the envelope consists of a inner membrane separated by a periplasmic region, surrounded by a cell wall comprising peptidoglycan covalently linked to arabinogalactan esterified by mycolic acids, forming the inner leaflet of the outer membrane.6 The outer leaflet of the outer membrane comprises mycolic acids and glycolipid derivatives,7 while the inner membrane mainly contains glycolipid derivatives such as phosphatidylinositol mannosides (PIM), and lipoarabinomannan (LAM).8 Prior research has indicated that membrane lipids play a crucial role in determining the permeability of drug molecules across the membrane envelope, thereby influencing their concentration within the cytoplasm.9 While various factors contribute to drug resistance, membrane permeability is a significant factor.10,11 Moreover, studies have reported remodelling of the mycobacterial lipidome and membrane properties following rifampicin treatment or within the host microenvironment.12 Therefore, a comprehensive understanding of the spatial and compositional distributions of lipids and their dynamics within the mycobacterial membrane under different conditions is essential for effective drug design or uptake.
In this work, we first establish membrane systems representing the early and late infection stages of Mtb, aligning with the growth phases of the Mycobacterium surrogate, Mycobacterium smegmatis (Msm).13 We demonstrate these systems to exhibit varying tolerance to a structurally diverse class of antibiotics, confirming the efficacy of these models in representing the respective infection stages. We then conducted a comprehensive lipidomic analysis of lipids extracted from Msm cultivated under these stages, revealing highly specific modifications in the lipidome of both the inner and outer cell envelope layers, which correlated with variable drug tolerance. Finally, we investigated the structural and functional implications of growth stage-induced lipid remodelling on mycobacterial membrane dynamics and its impact on antibiotic partitioning.
Our findings collectively demonstrate how pathogens adapt to diverse conditions by modifying their lipidome across various parameters, including fatty acid unsaturation, chain length, head group, and molecular geometry of the lipids. These alterations, though sometimes subtle, significantly influence membrane biophysical properties such as packing, fluidity, order, and permeability, consequently impacting diffusion, lipid/protein localization and clustering, and the partitioning of external molecules, including drugs. Therefore, a multiparametric analysis of molecular changes under varying conditions facilitates a clearer understanding of the factors governing antibiotic tolerance in mycobacterial species and may foster the discovery of effective strategies to combat antibiotic tolerance.
Results and discussion
Bacterial stress-adaptation upon carbon-depletion leads to drug tolerance in dormant bacteria
In a healthy individual, primary infection of Mtb is followed by a period of latency or dormancy within granulomas, until the immune system is compromised.14 During this stage, the mycobacterial membrane, serving as the first point of contact with the host microenvironment, is likely to adapt to foster bacterial survival. Thus, examining membrane characteristics during early and latent growth states is imperative.15Msm growth stages are comparable to different infection stages of Mtb16 and thus could be used to infer bacterial characteristics with the infection stage. We tracked Msm growth, noting the onset of the exponential (log) phase at ∼0.03 OD600, extending to 1.0 OD600 (Fig. 1A). The transition phase concluded at ∼1.5 OD600, initiating the stationary phase. The absence of tyloxapol did not hinder cell growth (Fig. 1B); however, its inclusion facilitated the dispersion of cell aggregates, enhancing cellular aeration.17 Within 5 h of culture, cell doubling (from an initial culture density of 0.05 OD600 to 0.4 OD600) resulted in reduction of glycerol availability to ∼25% per cell (Fig. 1C). Furthermore, after 10 h of culture, i.e., >1.0 OD600, 50% of the glycerol was consumed, diminishing glycerol availability to less than ∼2% per cell, promising complete dormancy above 3.0 OD600, which in the absence of tyloxapol mirrors a model for the late growth stage. Consequently, we selected bacterial culture densities of 0.8 OD600 and 3.0 OD600 growth, respectively, to represent early and late growth/infection stages.
To validate the membrane systems, we evaluated the drug susceptibility of various antibiotics with distinct mechanisms18–21 against Msm (Fig. 1D). The IC50 values of antibiotics notably increased at the late growth stage by 2–10 fold (moxifloxacin < rifabutin = amikacin < clarithromycin = ∼2 < 4 < 10), indicating diminished drug permeation and/or efficacy leading to a resistant phenotype (Fig. 1E). This observation is consistent with prior research demonstrating reduced potency of diverse antibiotic classes against dormant bacteria.22 Consequently, these growth stage bacterial systems could serve as suitable models for elucidating membrane characteristics during drug tolerance and bacterial growth.
Lipidomic analysis of mycobacterial cell envelope lipid layers
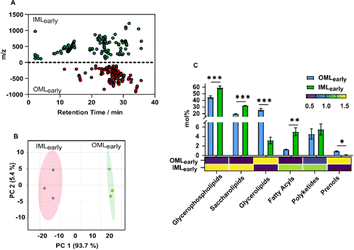
The compositional variations in the mycobacterial cell envelope layers were analysed through global analysis of lipids extracted from the outer (OML) and inner (IML) membrane layers of Msm at the early infection stage: denoted as OMLearly and IMLearly. The lack of isotopically labelled mycobacterial lipid standards prompted us to use reserpine, which is otherwise not present in the lipidome, and thus, all mass spectroscopy data reflect relative concentrations and changes. We identified 275 compounds appearing at the respective retention times (±0.1 s), exhibiting a 5-fold change in distribution across both the IML and OML (Fig. 2A). Compound identification and subsequent principal component analysis (PCA) demonstrated significant variation among lipid classes, with the most abundant class of mycobacterial lipids (glycerophospholipids) displaying at least a 30-fold difference across both layers (Fig. 2B). This confirms the scope to study lipid diversity across the cell wall.Next, mycobacterial lipids were classified into six major lipid classes based on LipidMaps23 (Fig. 2C, ESI excel Tables S1 and S2†). The predominant lipids identified were glycerophospholipids (GPhLs) (44.9% in OMLearly and 59.3% in IMLearly) and saccharolipids (ScLs) (19.7% in OMLearly and 32.1% in IMLearly), which together constituted 90% of the lipids in IMLearly. The high abundance of GPhLs, which are the primary membrane-forming lipids,24 is consistent with their crucial role in both lipid layers. ScLs in the outer membrane are known to facilitate infection and enable evasion of the late stages of phagocytosis,25 while ScLs in the inner membranes promote membrane permeability. The remaining lipid classes in IMLearly included fatty acyls (FAcs) (5%), glycerolipids (GL) (3.2%), polyketides (PK) (5.5%), and prenols (PR) (0.14%). In OMLearly, GL (25.7%), which serve as the major storage lipid in mycobacteria, were found in similar abundance to ScLs. The abundance of PK (4.6%) was similar in both OMLearly and IMLearly. As Msm is a non-virulent strain, the biomarkers for bacterial virulence,26 were observed in relatively lower concentrations across the bilayer. The distribution of FAcs in OMLearly (1.3%) was approximately four times lower than in IMLearly, reflecting their important role in maintaining cell integrity and biofilm formation.27 The presence of surfactants and the growth conditions may contribute to the overall reduced biosynthesis of FAcs. Conversely, PR lipids, primarily involved as carrier lipids in the biosynthesis of the cell wall,28 were minimally present in OMLearly (0.9%), though in higher amounts than in IMLearly.
A multi-parametric analysis of lipids was conducted focusing on the molecular geometry (conical/cylindrical/inverted-conical), degree of unsaturation, and acyl chain length (Fig. 3A–D). First, we observed a higher abundance of conical and inverted-conical shaped lipids in IMLearly compared to OMLearly (Fig. 3A). The ratio of inverted-conical/conical29 was higher in OMLearly (ratio = 3) than in IMLearly (ratio = 2.75), potentially rendering OMLearly relatively unstable in the bilayer configuration. Additionally, IMLearly exhibited a greater overall abundance of unsaturated lipids compared to OMLearly (Fig. 3B), suggesting a more fluid membrane bilayer in IMLearly. However, upon further classification of the degree of unsaturation and acyl chain lengths across different lipid conformations in each extract, the majority of polyunsaturated lipids in IMLearly were found to be in an inverted-conical shape (=43.37 mol%) (Fig. 3C), potentially resulting in a compact lipid bilayer30 when combined with long acyl chains (Fig. 3D). The equal abundance of saturated lipids (=13.90 mol%) in the inverted-conical conformation and short-chained lipids (=17.26 mol%) further supports this. Monounsaturated lipids with long-chains in the cylindrical conformation conform to a more global conical lipid configuration in IMLearly to accommodate membrane proteins.31 Moreover, the abundance of unsaturation in conical lipids does not significantly alter membrane packing. In contrast, for OMLearly, the saturated lipid population (=25.50 mol%) was higher in the inverted-conical configuration, indicating the presence of long acyl chains linking into the inner leaflet (covalently linked peptidoglycan-associated lipid leaflet). Moreover, the abundant unsaturated lipids (=27.26 mol%) in the cylindrical configuration, along with shorter chain lengths and a combination of existing conical configurations, compensated for a high proportion of inverted-conical configurations, resulting in compactness for surface attaching molecules.
In IMLearly, long-chained lipids (>30) increased while shorter-chained lipids (<30) decreased across all geometries compared to OMLearly. The enhanced hydrophobic and van der Waals interactions of long acyl chains facilitate more effective packing, supporting higher membrane ordering/low fluidity in IMLearly, as previously reported.32 This suggests that IMLearly accumulates more conical lipids, promoting membrane curvature compared to OMLearly, aligning with the recent discovery of the inner membrane's functional role in vesiculation in Gram-negative bacteria.33
The major components (by abundance) of conical lipids in IMLearly (Fig. 3E) were CL (cardiolipins), PA (phosphatidic acid) (GPhL); FA (branched fatty acids), FE (fatty acid esters) (FAc). For inverted conical geometry, LPA (lyso-PA) and AcPIMs (acyl-phosphatidylinositol mannosides) (GPhL); GPLs (glycopeptidolipids), DAT (diacyl trehalose) (ScL); and mycobactins (PK) (Fig. S1†) were key components. In contrast, in OMLearly, PE (phosphatidylethanolamine) and LPE (lyso-PE) (GPhL) and DAT (diacyltrehalose) (ScL) were the abundant subclasses for conical and inverted-conical shaped lipids, respectively (Fig. 3E). MAG (monoacyl glycerol) (GL) in OMLearly, and PI (phosphatidylinositol) (GPhL) and linear PK (PK) in IMLearly showed a substantial increase.
PA is the common precursor34 for the synthesis of all the lipids present in the mycobacterial cell wall; this justifies its abundance during the early growth stage. CL's multifunctionality within the IML is well-documented, primarily stabilizing SecYEG-mediated protein translocons across the inner membranes,35 and contributing to the function of the type VII secretion system (T7SS).36 Thus, the abundance of CL in the early growth stages, especially in IMLearly emerges as critical. PI (phosphatidylinositol) forms the precursor37 for the PIMs and AcPIMs. These PI derivatives are essential for the bacterium to persist and replicate inside the macrophages, in addition to providing structural integrity. Among ScLs and in the overall lipid subclasses, GPLs ranks the highest in abundance, and plays a major role in modulating membrane permeability.38 Trehalose derivatives (sulfolipids (SL), trehalose monomycolate (TMM), trehalose dimycolate (TDM), and di-, tri-, poly-acyl trehalose (DAT, TAT, PAT)) have properties to activate macrophages to initiate infection.39 Its failure to segregate into the OML regions and accumulate more in the IML regions would indicate the reduced infectious nature of Msm. The FAs and FEs contribute to the biosynthesis of ScLs and other FAcs, and hence they exhibit high abundance during the early growth stages. Furthermore, due to the continuous replication state of the bacterium during early infection, there is the least accumulation of complex storage lipids (di- and tri-acyl glycerol (DAG and TAG)) but high accumulation of MAG (monoacyl glycerol).40
Comparative lipidome changes in cell envelope layers with the growth stage
The dormancy of bacteria can profoundly affect their survival and pathogenesis, and confer drug resistance.41 To elucidate the contribution of the membrane lipid composition in these relationships, we analysed lipidome modifications using the above workflow in each Msm membrane layer under early and late growth conditions, denoted as I/OMLearly and I/OMLlate, respectively. Of note, in addition to changes in O.D., the absence of tyloxapol in late-stage bacteria could also impact the lipidome, but is reported to not alter the bacterial metabolome including fatty acids, rendering it a better additive for bacterial culturing.42Global analysis revealed the modulation of at least 85 lipids in the OML and 244 lipids in the IML, demonstrating at least a 5-fold upregulation or downregulation in the late growth stage (Fig. S2A†). PCA indicated significant variation, with a 30-fold change in the GPhL class and a 5-fold in the ScL class across growth stages in the OML and IML, respectively (Fig. S2B†). In the late growth stage, overall, there was an increase in ScLs and a decrease in PR in both the OML and IML (Fig. 4A, 5A, ESI excel Tables S1 and S2†). Additionally, OMLlate lipids exhibited an increased abundance of FAc, while PK were decreased compared to that in OMLearly. Multi-parametric analysis revealed OMLlate to have an increased abundance of conical (PE (GPhL); FA (FAc)) and a decreased abundance of inverted conical (LPA (GPhL), LPE (GPhL); DAT (ScL); mycobactin (PK)) lipids (Fig. 4B, ESI excel Tables S1 and S2†). DAG (conical) and LPE and GPL (inverted conical) showed opposite trends whereby the overall change in inverted-conical lipids was minimized. Very little change was observed in cylindrical lipids in all lipid categories.
In IMLlate, in addition to ScLs and PR, a decrease in GPhLs was seen compared to its early growth counterpart (Fig. 5A, ESI excel Tables S1 and S2†). Furthermore, overall, no change in the conformations of lipids was seen; however, a decrease in PA (GPhL) and FE (FAc) for conical lipids, alongside an increase in CL (GPhL), possibly to maintain the relative abundance of conical lipids, was observed (Fig. 5B, ESI excel Tables S1 and S2†). The overall reduction of inverted-conical lipids (LPE (GPhL), LPG (GPhL), and GPL (ScL)) was compensated for by the increase in LPA (GPhL), PIM (GPhL) and DAT (ScL). No significant change was observed in cylindrical lipids, expect for a 2 mol% decrease in PI (GPhL).
The significant increase in FAc levels in OMLlate, could mark the formation of biofilms during dormancy that could facilitate its survival within the macrophages,41 and it remained unchanged in IMLlate. The decreased levels of PK and PR could underline a subsided virulence with OMLlate and cell wall metabolism in IMLlate respectively at dormancy. Furthermore, the ratio of inverted-conical lipids to conical lipids decreased during the late stage, indicating a higher propensity for vesiculation by the OML layer.43 Alternatively, this could suggest that the growth stage may regulate the mechanisms of vesicle production (OMV, IMV vs. I-OMV) in Gram-negative bacteria. Next, the specific modulation of charged, neutral and bulky lipids in each layer with the growth stage is likely to impact interaction with antibiotics, thereby fine tuning their membrane partitioning. This could alter bacterial antibiotic tolerance and subsequently contribute to drug resistance.
The increased levels of FAs in the OMLlate could also help in bacterial survival during the late growth stage. The OML also demonstrated a higher accumulation of membrane-associated mycobactins (Fig. S3†) than in the IML for improved iron-acquisition from the host during infection.44 The iron-limitation in the environment is known to enhance virulence of the cells whereby it can avail a targeted amount of iron from the host.45 The decrease in the levels of the mycobactins at the late growth stage can be correlated with the possibly enhanced virulence. The increase in exochelins in IMLlate on the other hand, even in the absence of mycobactins, can still facilitate iron-acquisition,46 thus restoring metabolic functioning of the cell. Furthermore, a ubiquinone increase is observed under anoxic conditions resulting from the limited oxygen supply during biofilm formation at the late infection stage.47 PR are involved in the catabolism of the peptidoglycan-arabinogalactan framework of the bacterial membrane,48 and its complete absence in the late growth state could induce alteration in the catabolic pathway of the mycobacterial cell wall.
Correlation of membrane biophysics with growth-induced lipidome rewiring
We then investigated the biophysical properties of Msm membranes generated using lipids extracted from each layer under different conditions (denoted as I/OMLearly/late) and then correlated the properties with the changes in the growth stage-induced lipidome and drug tolerance. First, membrane ordering in the head group region was assessed using Laurdan generalized polarization (GP).49 Both OMLearly and IMLearly exhibited reduced GP values compared to their late stage counterparts at lower temperatures (Fig. 6A and B) indicating reduced ordering within the head group regions or higher fluidity. The high abundance of inverted-conical to conical lipids at the early growth stage (Fig. 6C and D) underlines the increased disorder within the membrane head groups by disrupting the membrane packing. In addition to the geometrical configuration of these lipids, the head group moieties can also influence membrane biophysics. A higher abundance of serovar in GPLs (Fig. 6E) in IMLearly, also contributes to higher disorder in the head-group region compared to the OML at various temperatures. Similarly, mannosylation in PIMs (Fig. 6F) may contribute to lower ordering in both I/OMLearly compared to their late stage counterparts. OMLlate exhibited a higher proportion of PE (Fig. 6G) which mitigates electrostatic repulsions, thus preserving membrane order. At higher temperatures, both IMLlate and OMLlate showed pronounced disorder (Fig. 6A and B). This phenomenon is attributed to the cumulative influence of large head group lipids (including PIMs, GPLs, trehalose, sulfolipids, and PKs) that are more abundant in OMLlate and IMLlate (Fig. 6H) compared to their early stage counterparts; large head groups foster electrostatic repulsion thereby promoting increased membrane hydration.50Next, membrane fluidity was evaluated in all systems in a depth-dependent manner using the anisotropy of TMA-DPH (head-group region) and DPH (hydrophobic chain region) fluorescent probes. At physiological temperature (35 °C), higher membrane fluidity (low anisotropy) was observed in the hydrophobic chain region for all OML and IML membranes compared to the head-group region (Fig. 7A). For IMLlate, fluidity at this site increased at higher temperatures (>35 °C, Fig. 7A), with less effect on TMA-DPH (interfacial region fluidity). In contrast, for OMLlate, membrane fluidity decreased at high temperatures (45 °C) for both head and acyl chain regions, warranting further examination. Unsaturation or kinks are factors that regulate membrane fluidity.51 However, the observed prevalence of polyunsaturated lipids followed the order OMLlate > IMLearly > OMLearly > IMLlate (approximately 40 > 39>36 > 26 mol%) (Fig. 7B). This suggests that other factors such as lipid chain length and branching could account for high and low fluidity in IMLlate and OMLlate membranes, respectively. In line with this, we observed a higher abundance of C20–25 acyl chain lengths and long-chain lipids (>C60) in both OMLlate and IMLlate, along with C35 (only for IMLlate). The elevated abundance of C20–C25 lipids during the late stage renders the bilayer more fluid in the DPH region due to weaker van der Waals interactions (Fig. 7C). However, in OMLlate, the presence of long-chained lipids and poly-unsaturation reverses this effect, thereby promoting membrane ordering (Fig. 7C). This increase in membrane order is evident with increasing temperature in OMLlate. Furthermore, lipid chains in the OML can adopt W or U configurations leading to low fluidity in the interfacial region due to conformationally ordered packing of the acyl chain configurations.52
Lipidome changes and membrane biophysical attributes govern antibiotic interactions
Drug tolerance towards various antibiotics critically depends on the molecular and biophysical properties of bacterial lipid membranes.53 To understand how the observed drug tolerance in intact bacteria at the late growth stage (Fig. 1B) is likely influenced by the altered lipidome culminating in low drug permeation, we investigated the interactions of the antibiotic rifabutin with Msm model membranes developed from different growth stages. The partition coefficient (Kp or log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D) of the drug, determined by using the third derivative of the absorbance of rifabutin as a function of wavelength at various lipid concentrations,54 followed the trend IMLlate > IMLearly = OMLearly > OMLlate (Fig. 8A and E). Rifabutin interacts both electrostatically and via hydrophobic interactions with membranes,55,56 with the membrane surface charge being a predominant factor governing its selective partitioning. Thus, assessment of the zeta potential (Fig. 8B) and the cumulative negative charge of the lipids (Fig. 8C), suggests an enhanced partitioning of rifabutin within the IML. High negative charges lead to lose membrane packing and enhanced drug partitioning. In contrast, the elevated presence of PE (Fig. 6G) and diminished abundance of CL (Fig. 8D) rendered OMLlate less receptive to rifabutin, resulting in lower Kp (Fig. 8E). Conversely, IMLlate exhibited the highest abundance of CL, followed by IMLearly (Fig. 8D) and the lowest abundance of PE (Fig. 6G), potentially favouring rifabutin uptake. Our data indicate that with dormancy, the retention of CL increases in IMLlate but decreases in OMLlate, validating the importance of CL in the cytoplasmic region and onset of drug tolerance across all antibiotic classes, irrespective of their mode of entry or mechanism of action. Enhanced membrane fluidity in IMLlate facilitates higher uptake of rifabutin compared to early stage and OML systems.57 The variable drug partitioning in the IML with growth stages could also stem from a reduced abundance of GPL serovars and lack of mannosylation in PIMs at the late growth stage.
D) of the drug, determined by using the third derivative of the absorbance of rifabutin as a function of wavelength at various lipid concentrations,54 followed the trend IMLlate > IMLearly = OMLearly > OMLlate (Fig. 8A and E). Rifabutin interacts both electrostatically and via hydrophobic interactions with membranes,55,56 with the membrane surface charge being a predominant factor governing its selective partitioning. Thus, assessment of the zeta potential (Fig. 8B) and the cumulative negative charge of the lipids (Fig. 8C), suggests an enhanced partitioning of rifabutin within the IML. High negative charges lead to lose membrane packing and enhanced drug partitioning. In contrast, the elevated presence of PE (Fig. 6G) and diminished abundance of CL (Fig. 8D) rendered OMLlate less receptive to rifabutin, resulting in lower Kp (Fig. 8E). Conversely, IMLlate exhibited the highest abundance of CL, followed by IMLearly (Fig. 8D) and the lowest abundance of PE (Fig. 6G), potentially favouring rifabutin uptake. Our data indicate that with dormancy, the retention of CL increases in IMLlate but decreases in OMLlate, validating the importance of CL in the cytoplasmic region and onset of drug tolerance across all antibiotic classes, irrespective of their mode of entry or mechanism of action. Enhanced membrane fluidity in IMLlate facilitates higher uptake of rifabutin compared to early stage and OML systems.57 The variable drug partitioning in the IML with growth stages could also stem from a reduced abundance of GPL serovars and lack of mannosylation in PIMs at the late growth stage.
Next, partitioning of rifabutin mitigated the temperature-induced increase in the membrane fluidity likely affecting drug localization and specific intermolecular interactions, thereby altering the hydration network of the lipid bilayer surface (Fig. S4†). Rifabutin-induced quenching of TMA-DPH (head-group region) or DPH (acyl chain region) probes serves as an indirect predictor of drug localization within the membrane bilayer. OMLlate demonstrated a substantial decrease in quenching in the acyl chain region, while that in the interfacial region increased marginally (Fig. S5†). On the other hand, IMLlate did not affect the quenching in the interfacial region but was significantly reduced in the acyl chain region. Collectively, these data imply that the drug's probable location along the membrane bilayer depth shifts towards the interfacial region in both layers during the late growth stage reflecting modulated hydrophobic and electrostatic interactions with the altered lipidome.
Collectively, these findings indicate the existence of a drug partitioning gradient dependent on the bacterial cell envelope layer and growth stage, wherein the OML acts as the rate limiting barrier for effective drug uptake and subsequent intracellular concentration. Importantly under late growth conditions the disparity between the drug uptake (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D) in the IML vs. OML further increases. Correlating these with the drug activity in intact bacteria (Fig. 1B) suggests an intricate interplay of lipidome changes within each layer that regulate the drug uptake and resistance phenotype. These factors should be consulted while designing novel chemotypes, for accessing the activity of antibiotics in treating drug resistance strains and for determining the treatment regimens contingent upon the growth stage.
D) in the IML vs. OML further increases. Correlating these with the drug activity in intact bacteria (Fig. 1B) suggests an intricate interplay of lipidome changes within each layer that regulate the drug uptake and resistance phenotype. These factors should be consulted while designing novel chemotypes, for accessing the activity of antibiotics in treating drug resistance strains and for determining the treatment regimens contingent upon the growth stage.
Conclusion
Mycobacterium species are linked to persistence, variable drug tolerance and virulence.58 One key component of mycobacteria is their unique cell membrane architecture with spatially resolved inner and outer membrane layers with distinct lipid repertoires. Furthermore, mycobacterial membranes are now known to structurally and compositionally adapt to drug exposure, within the host microenvironment, under stressed conditions and possibly in the dormant growth stage. This is likely to impact the intracellular concentrations of drugs by modulating their passive diffusion through the membrane layers under variable conditions. Therefore, characterisation of multiple properties of the mycobacterial membrane lipidome and membrane dynamics, especially with the growth/infection stage is crucial for the development and rationalization of effective treatment strategies.In this work, we demonstrate bacterial membrane systems representing early and late growth stages, with the latter showing high drug tolerance (increased IC50) to various antibiotics. We revealed distinctive changes in the lipidome of the inner and outer membrane layers of mycobacteria under these growth states. The global lipid changes aid in categorizing lipid classes based on chain length, poly-unsaturation, charge and molecular geometry. Membranes derived from these lipid extracts demonstrate how specific lipid changes regulate membrane ordering/fluidity, influencing antibiotic interaction and partitioning. Taken together, the results demonstrate that the mycobacterial outer membrane is pivotal in controlling drug-partitioning, especially during the late growth stage, driven by upregulation of long-chained lipids, reduced abundance of charged lipids, a higher level of GLP serovars, and a high ratio of inverted-conical/conical lipids, all contributing to low membrane fluidity or high ordering. Conversely, the inner membrane, especially in the late growth stage, shows the most effective drug uptake due to increased levels of charged lipids, cardiolipin, decreased GPL serovar levels, and accumulation of lipids with large head groups, resulting in high membrane fluidity and low packing.
This work reveals functional insights into specific lipidome changes in both the inner and outer cell membrane layers in synergy to bacterial survival and drug resistance. Also the work suggests that an effective anti-infective strategy to tackle dormant/persistent mycobacteria should combine (a) outer membrane layer disrupting agents along with antibiotics to re-sensitise the resistant bacteria, (b) a liposomal antibiotic formulation containing short chained synthetic mycobacterial outer membrane lipids to mitigate the membrane ordering effect of long-chained lipids induced at the late growth stage and (c) cardiolipin-fused outer membrane liposomal antibiotic formulations.
Data availability
Data are available in the article and ESI† and can be provided by the authors upon request.Author contributions
The experiments were performed by A. P. M. S. K., T. H. L. and M. A. supervised the project. S. K. and A. P. M. designed the project. The paper was written by S. K. and A. P. M. and revised by M. A. and T. H. L. All authors contributed to data analysis, interpreted the data, and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from DST-SERB (EMR/2016/005414 and WEA/2020/000032). Central UV-vis, fluorescence and mass spectroscopy facilities at the Department of Chemistry, IIT Bombay are gratefully acknowledged. MIA acknowledges the support of the National Health and Medical Research Council Project grant APP1142750 and Ideas Grant ID2011990.References
- Global tuberculosis report 2023, Geneva: World Health Organization, 2023, Licence: CC BY-NC-SA 3.0 IGO Search PubMed.
- R. L. Kinsella, D. X. Zhu, G. A. Harrison, A. E. Mayer Bridwell, J. Prusa and S. M. Chavez, et al., Perspectives and Advances in the Understanding of Tuberculosis, Annu. Rev. Pathol.: Mech. Dis., 2021, 16, 377–408 CrossRef CAS.
- K. R. Schildknecht, Tuberculosis—United States, 2022, MMWR Morbidity and Mortality Weekly Report, 2023, vol. 72 Search PubMed.
- S. H. Lee, Diagnosis and treatment of latent tuberculosis infection, Tuberc. Respir. Dis., 2015, 78(2), 56–63 CrossRef.
- P. Adhyapak, A. T. Srivatsav, M. Mishra, A. Singh, R. Narayan and S. Kapoor, Dynamical organization of compositionally distinct inner and outer membrane lipids of mycobacteria, Biophys. J., 2020, 118(6), 1279–1291 CrossRef CAS PubMed.
- M. Daffé and H. Marrakchi, Unraveling the structure of the mycobacterial envelope, Microbiol. Spectrum, 2019, 7(4), 7 Search PubMed.
- C. L. Dulberger, E. J. Rubin and C. C. Boutte, The mycobacterial cell envelope—a moving target, Nat. Rev. Microbiol., 2020, 18(1), 47–59 CrossRef CAS.
- M. Jackson, The mycobacterial cell envelope—lipids, Cold Spring Harbor Perspect. Med., 2014, 4(10), a021105 CrossRef PubMed.
- A. P. D. W. Menon, T. H. Lee, M. I. Aguilar, M. Duan and S. Kapoor, Mutually Exclusive Interactions of Rifabutin with Spatially Distinct Mycobacterial Cell Envelope Membrane Layers Offer Insights into Membrane-Centric Therapy of Infectious Diseases, ACS Bio Med Chem Au, 2022, 2(4), 395–408 CrossRef CAS PubMed.
- T.-H. Lee, V. Hofferek, F. Separovic, G. E. Reid and M.-I. Aguilar, The role of bacterial lipid diversity and membrane properties in modulating antimicrobial peptide activity and drug resistance, Curr. Opin. Chem. Biol., 2019, 52, 85–92 CrossRef CAS PubMed.
- T.-H. Lee, P. Charchar, F. Separovic, G. E. Reid, I. Yarovsky and M.-I. Aguilar, The intricate link between membrane lipid structure and composition and membrane structural properties in bacterial membranes, Chem. Sci., 2024, 15(10), 3408–3427 RSC.
- M. Mishra, A. D. Gupta, R. Dadhich, M. N. Ahmad, A. Dasgupta and S. Chopra, et al., Mycobacterial lipid-derived immunomodulatory drug-liposome conjugate eradicates endosome-localized mycobacteria, J. Ophthalmol. Clin. Res., 2023, 360, 578–590 CAS.
- N. Lelovic, K. Mitachi, J. Yang, M. R. Lemieux, Y. Ji and M. Kurosu, Application of Mycobacterium smegmatis as a surrogate to evaluate drug leads against Mycobacterium tuberculosis, J. Antibiot., 2020, 73(11), 780–789 CrossRef CAS.
- A. R. Sholeye, A. A. Williams, D. T. Loots, A. M. Tutu van Furth, M. van der Kuip and S. Mason, Tuberculous granuloma: emerging insights from proteomics and metabolomics, Front. Neurol., 2022, 13, 804838 CrossRef PubMed.
- C. Peetla, S. Vijayaraghavalu and V. Labhasetwar, Biophysics of cell membrane lipids in cancer drug resistance: Implications for drug transport and drug delivery with nanoparticles, Adv. Drug Delivery Rev., 2013, 65(13–14), 1686–1698 CrossRef CAS PubMed.
- V. Chaturvedi, N. Dwivedi, R. P. Tripathi and S. Sinha, Evaluation of Mycobacterium smegmatis as a possible surrogate screen for selecting molecules active against multi-drug resistant Mycobacterium tuberculosis, J. Gen. Appl. Microbiol., 2007, 53(6), 333–337 CrossRef CAS.
- L. Herman, S. C. De Smedt and K. Raemdonck, Pulmonary surfactant as a versatile biomaterial to fight COVID-19, J. Controlled Release, 2022, 342, 170–188 CrossRef CAS PubMed.
- M. S. Ramirez and M. E. Tolmasky, Amikacin: uses, resistance, and prospects for inhibition, Molecules, 2017, 22(12), 2267 CrossRef.
- I. I. Salem, Clarithromycin. Analytical Profiles of Drug Substances and Excipients, Elsevier, 1996, vol. 24, p. 45–85 Search PubMed.
- H. Mosaei and N. Zenkin, Inhibition of RNA polymerase by rifampicin and rifamycin-like molecules, EcoSal Plus, 2020, 9(1) DOI:10.1128/ecosalplus.ESP-0017-2019.
- P. F. Chan, V. Srikannathasan, J. Huang, H. Cui, A. P. Fosberry and M. Gu, et al., Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin, Nat. Commun., 2015, 6(1), 10048 CrossRef CAS PubMed.
- J. L. Khawbung, D. Nath and S. Chakraborty, Drug resistant Tuberculosis: A review, Comp. Immunol., Microbiol. Infect. Dis., 2021, 74, 101574 CrossRef CAS.
- E. Layre, R. Al-Mubarak, J. T. Belisle and D. B. Moody, Mycobacterial lipidomics, Molecular genetics of Mycobacteria, 2014, pp. 341–360 Search PubMed.
- I. M. López-Lara and O. Geiger, Bacterial lipid diversity, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2017, 1862(11), 1287–1299 CrossRef.
- G. Etienne, C. Villeneuve, H. Billman-Jacobe, C. Astarie-Dequeker, M.-A. Dupont and M. Daffe, The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis, Microbiology, 2002, 148(10), 3089–3100 CrossRef CAS.
- C. Hertweck, The biosynthetic logic of polyketide diversity, Angew. Chem., Int. Ed., 2009, 48(26), 4688–4716 CrossRef CAS.
- J. Yao and C. O. Rock, Exogenous fatty acid metabolism in bacteria, Biochimie, 2017, 141, 30–39 CrossRef CAS.
- J. Baddiley, Lipid intermediates in the biosynthesis of bacterial cell-wall components, Portland Press Ltd, 1973 Search PubMed.
- V. Kumar, Complementary molecular shapes and additivity of the packing parameter of lipids, Proc. Natl. Acad. Sci. U. S. A., 1991, 88(2), 444–448 CrossRef CAS.
- P. Janmey and P. K. Kinnunen, Biophysical properties of lipids and dynamic membranes, Trends Cell Biol., 2006, 16(10), 538–546 CrossRef CAS PubMed.
- T. G. Pomorski, T. Nylander and M. Cárdenas, Model cell membranes: Discerning lipid and protein contributions in shaping the cell, Adv. Colloid Interface Sci., 2014, 205, 207–220 CrossRef CAS.
- K. J. Seu, L. R. Cambrea, R. M. Everly and J. S. Hovis, Influence of lipid chemistry on membrane fluidity: tail and headgroup interactions, Biophys. J., 2006, 91(10), 3727–3735 CrossRef CAS PubMed.
- N. Mozaheb and M.-P. Mingeot-Leclercq, Membrane vesicle production as a bacterial defense against stress, Front. Microbiol., 2020, 11, 600221 CrossRef PubMed.
- S. K. Angala, A. Carreras-Gonzalez, E. Huc-Claustre, I. Anso, D. Kaur and V. Jones, et al., Acylation of glycerolipids in mycobacteria, Nat. Commun., 2023, 14(1), 6694 CrossRef CAS PubMed.
- V. A. Gold, A. Robson, H. Bao, T. Romantsov, F. Duong and I. Collinson, The action of cardiolipin on the bacterial translocon, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(22), 10044–10049 CrossRef CAS PubMed.
- N. Sengupta, S. Padmanaban and S. Dutta, Cryo-EM reveals the membrane-binding phenomenon of EspB, a virulence factor of the mycobacterial type VII secretion system, J. Biol. Chem., 2023, 299(4), 104589 CrossRef CAS PubMed.
- M. Gilleron, M. Jackson, J. Nigou and G. Puzo, Structure, Biosynthesis, and Activities of the Phosphatidyl-myo-Inositol-Based Lipoglycans, The mycobacterial cell envelope, 2008, pp. 75–105 Search PubMed.
- L. Pang, X. Tian, W. Pan and J. Xie, Structure and function of mycobacterium glycopeptidolipids from comparative genomics perspective, J. Cell. Biochem., 2013, 114(8), 1705–1713 CrossRef CAS PubMed.
- A. A. Khan, Synthesis and Biological Evaluation of Trehalose Glycolipids, Open Access Te Herenga Waka-Victoria University of Wellington, 2012 Search PubMed.
- G. C. Burdge and P. C. Calder, Introduction to fatty acids and lipids. Intravenous Lipid Emulsions, 2015, 112, 1–16 Search PubMed.
- S. Lipworth, R. Hammond, V. Baron, Y. Hu, A. Coates and S. Gillespie, Defining dormancy in mycobacterial disease, Tuberculosis, 2016, 99, 131–142 CrossRef CAS PubMed.
- M. Opperman, R.-D. Pietersen, M. van Reenen, D. Beukes, B. Baker and I. du Preez, The effect of Tyloxapol on the metabolome of Mycobacterium tuberculosis, J. Microbiol. Methods, 2024, 107028 CrossRef CAS PubMed.
- T. Nagakubo, N. Nomura and M. Toyofuku, Cracking open bacterial membrane vesicles, Front. Microbiol., 2020, 10, 509337 Search PubMed.
- K. Patel, S. Butala, T. Khan, V. Suvarna, A. Sherje and B. Dravyakar, Mycobacterial siderophore: a review on chemistry and biology of siderophore and its potential as a target for tuberculosis, Eur. J. Med. Chem., 2018, 157, 783–790 CrossRef CAS.
- G. M. Rodriguez, N. Sharma, A. Biswas and N. Sharma, The iron response of mycobacterium tuberculosis and its implications for tuberculosis pathogenesis and novel therapeutics, Front. Cell. Infect. Microbiol., 2022, 12, 876667 CrossRef CAS PubMed.
- L. D. Horwitz and M. A. Horwitz, The exochelins of pathogenic mycobacteria: unique, highly potent, lipid-and water-soluble hexadentate iron chelators with multiple potential therapeutic uses, Antioxid. Redox Signaling, 2014, 21(16), 2246–2261 CrossRef CAS PubMed.
- S. S. Abby, K. Kazemzadeh, C. Vragniau, L. Pelosi and F. Pierrel, Advances in bacterial pathways for the biosynthesis of ubiquinone, Biochim. Biophys. Acta, Bioenerg., 2020, 1861(11), 148259 CrossRef CAS PubMed.
- P. K. Crellin, C.-Y. Luo and Y. S. Morita, Metabolism of plasma membrane lipids in mycobacteria and corynebacteria, Lipid Metab., 2013, 6, 119–148 Search PubMed.
- S. A. Sánchez, M. Tricerri, G. Gunther and E. Gratton, Laurdan generalized polarization: from cuvette to microscope, Modern research and educational topics in microscopy, 2007, 2, 1007–1014 Search PubMed.
- R. Dadhich, A. Singh, A. P. Menon, M. Mishra, C. Athul and S. Kapoor, Biophysical characterization of mycobacterial model membranes and their interaction with rifabutin: towards lipid-guided drug screening in tuberculosis, Biochim. Biophys. Acta, Biomembr.|, 2019, 1861(6), 1213–1227 Search PubMed.
- T.-A. Hagve, Effects of unsaturated fatty acids on cell membrane functions, Scand. J. Clin. Lab. Invest., 1988, 48(5), 381–388 CrossRef CAS PubMed.
- W. Groenewald, M. S. Baird, J. A. Verschoor, D. E. Minnikin and A. K. Croft, Differential spontaneous folding of mycolic acids from Mycobacterium tuberculosis, Chem. Phys. Lipids, 2014, 180, 15–22 Search PubMed.
- M.-P. Mingeot-Leclercq and J.-L. Décout, Bacterial lipid membranes as promising targets to fight antimicrobial resistance, molecular foundations and illustration through the renewal of aminoglycoside antibiotics and emergence of amphiphilic aminoglycosides, MedChemComm, 2016, 7(4), 586–611 RSC.
- M. Pinheiro, M. Arede, C. Nunes, J. M. Caio, C. Moiteiro and M. Lucio, et al., Differential Interactions of Rifabutin with Human and Bacterial Membranes: Implication for Its Therapeutic and Toxic Effects, J. Med. Chem., 2013, 56(2), 417–426 CrossRef CAS PubMed.
- M. Pinheiro, C. Pereira-Leite, M. Arêde, C. Nunes, J. M. Caio and C. Moiteiro, et al., Evaluation of the Structure–Activity Relationship of Rifabutin and Analogs: A Drug–Membrane Study, ChemPhysChem, 2013, 14(12), 2808–2816 CrossRef CAS PubMed.
- M. Pinheiro, M. Arêde, C. Nunes, J. M. Caio, C. Moiteiro and M. Lúcio, et al., Differential interactions of rifabutin with human and bacterial membranes: implication for its therapeutic and toxic effects, J. Med. Chem., 2013, 56(2), 417–426 Search PubMed.
- T. Prasad, A. Chandra, C. K. Mukhopadhyay and R. Prasad, Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells, Antimicrob. Agents Chemother., 2006, 50(11), 3597–3606 CrossRef CAS PubMed.
- M. McGrath, N. Gey van Pittius, P. Van Helden, R. Warren and D. Warner, Mutation rate and the emergence of drug resistance in Mycobacterium tuberculosis, J. Antimicrob. Chemother., 2014, 69(2), 292–302 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc06618a |
| This journal is © The Royal Society of Chemistry 2024 |