Open Access Article
Open Access ArticleAutomated electrosynthesis reaction mining with multimodal large language models (MLLMs)†
Shi Xuan
Leong
 ab,
Sergio
Pablo-García
ab,
Sergio
Pablo-García
 acd,
Zijian
Zhang
cd and
Alán
Aspuru-Guzik
acd,
Zijian
Zhang
cd and
Alán
Aspuru-Guzik
 *acdefgh
*acdefgh
aDepartment of Chemistry, University of Toronto, Lash Miller Chemical Laboratories, 80 St. George Street, ON M5S 3H6, Toronto, Canada. E-mail: alan@aspuru.com
bDivision of Chemistry and Biological Chemistry, School of Chemistry, Chemical Engineering and Biotechnology, Nanyang Technological University, 21 Nanyang Link, Singapore 637371
cDepartment of Computer Science, University of Toronto, Sandford Fleming Building, 10 King's College Road, ON M5S 3G4, Toronto, Canada
dVector Institute for Artificial Intelligence, 661 University Ave. Suite 710, ON M5G 1M1, Toronto, Canada
eAcceleration Consortium, 80 St. George St., M5S 3H6, Toronto, Canada
fDepartment of Materials Science & Engineering, University of Toronto, 184 College St., M5S 3E4, Toronto, Canada
gDepartment of Chemical Engineering & Applied Chemistry, University of Toronto, 200 College St., ON M5S 3E5, Toronto, Canada
hLebovic Fellow, Canadian Institute for Advanced Research (CIFAR), 661 University Ave., M5G 1M1, Toronto, Canada
First published on 9th October 2024
Abstract
Leveraging the chemical data available in legacy formats such as publications and patents is a significant challenge for the community. Automated reaction mining offers a promising solution to unleash this knowledge into a learnable digital form and therefore help expedite materials and reaction discovery. However, existing reaction mining toolkits are limited to single input modalities (text or images) and cannot effectively integrate heterogeneous data that is scattered across text, tables, and figures. In this work, we go beyond single input modalities and explore multimodal large language models (MLLMs) for the analysis of diverse data inputs for automated electrosynthesis reaction mining. We compiled a test dataset of 65 articles (MERMES-T24 set) and employed it to benchmark five prominent MLLMs against two critical tasks: (i) reaction diagram parsing and (ii) resolving cross-modality data interdependencies. The frontrunner MLLM achieved ≥96% accuracy in both tasks, with the strategic integration of single-shot visual prompts and image pre-processing techniques. We integrate this capability into a toolkit named MERMES (multimodal reaction mining pipeline for electrosynthesis). Our toolkit functions as an end-to-end MLLM-powered pipeline that integrates article retrieval, information extraction and multimodal analysis for streamlining and automating knowledge extraction. This work lays the groundwork for the increased utilization of MLLMs to accelerate the digitization of chemistry knowledge for data-driven research.
Introduction
Despite today's increasingly data-driven scientific landscape, most of the past chemical knowledge remains locked in one way or another, using legacy data formats such as the hypertext markup language (HTML) and portable document format (PDF), or hidden behind paywalls. One way forward is the automated data mining from these “locked” scientific data stemming from publications and patents. If fully automated, this task would be essential to help construct databases which can be leveraged as collective knowledge to significantly expedite materials and reaction discovery and uncover fundamental governing chemistries.1–13 Despite advancements in specialized text mining or diagram parsing tools,14–18 extracting accurate and comprehensive experimental data records remains challenging due to at least two main hurdles: (i) key chemical and/or materials information is usually scattered across various data modalities within both the main text and ESI,† including tables, figures/schemes, and textual descriptions and (ii) the user-desired data is typically overwhelmed by a substantial amount of extraneous content, i.e., “data flooding”. These two challenges underscore the need for robust yet flexible data mining workflows capable of multimodal analysis, while efficiently filtering and processing the specialized data at hand.Recently, large language models (LLMs) such as Claude,19,20 Gemini,21 GPT22 and LLaMA23 among others24,25 have demonstrated immense potential in text-based knowledge extraction from scientific publications, using a combination of prompt engineering, model fine-tuning and in-context learning techniques.3,22,26–40 Compared to traditional natural language processing (NLP) methods that rely heavily on rule-based syntax or dictionary-matching, LLM-powered literature mining exhibits higher adaptability and generalizability due to the natural language processing, conversational capabilities, and general-purpose versatility of LLMs. Nonetheless, the integration of visual information poses an additional layer of complexity. The emergent development of multimodal large language models (MLLMs), with the ability to receive and process multiple data types including images, text, language, audio, and other heterogeneity, offers a more well-rounded task-solver.41–44 These MLLMs have reported promising accuracies in various vision-language multimodal tasks such as image captioning and visual question-answering in generic everyday context, across different evaluation benchmarks. The next logical question for us arises: can state-of-the-art MLLMs effectively process and integrate textual and visual inputs from scientific literature for specialized, domain-specific tasks such as reaction mining?
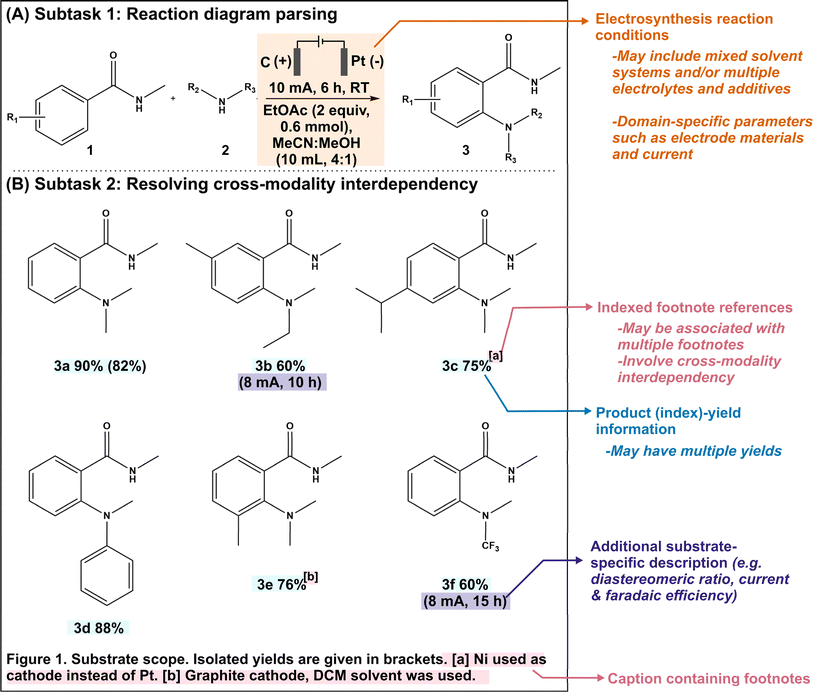
In this study, we demonstrate that MLLMs exhibit multimodal cognition capabilities that are suitable for chemical applications. We focus on two essential subtasks in automated reaction mining. The first subtask involves parsing reaction conditions into categorical data (10 different categories), which is a requirement due to the conventional use of graphical representations to summarize novel reactions (Fig. 1A). The second subtask involves resolving cross-modality interdependencies, where reference labels within figure images are defined elsewhere in the text (Fig. 1B). This capability is also required since substrate-specific variations in reaction conditions are typically conveyed through such index cross-references. Being able to identify and connect these distributed pieces of information together is thus crucial to maintain data coherency and integrity during the automated mining process. Using 65 literature articles reporting novel organic electrosynthesis reactions as our proof-of-concept test dataset, we evaluated the zero-shot performances of five prominent MLLMs, namely GPT-4V,22 Gemini Pro,21 Claude 3,19 InternVL45 and LLaVA.46,47 The resulting frontrunner model (GPT-4V) was selected for further performance refinement through strategic incorporation of single-shot visual prompts and automated image cropping. Finally, we developed an integrated tool called MERMES, a Multimodal Reaction Mining pipeline for ElectroSynthesis, which is an end-to-end automated workflow that effectively leverages multimodal information from scientific publications for automated reaction mining. MERMES features three sequential modules: an article retrieval module to download HTML webpages from the publisher, an information extraction module to extract all image–caption pairs from incoming literature articles and identify the relevant figure images, and a multimodal information analysis module to extract the key chemical information from the filtered data subset. We envisage that these MLLM-based automated reaction mining workflows play an integral role towards the complete digitization of chemistry knowledge to facilitate data-driven research.
Methodology
Study scope and overview of knowledge retrieval tasks
This work focuses on mining organic electrosynthesis reactions. The recent renaissance of this field as an efficient and sustainable alternative to traditional chemical syntheses has led to the growth of scientific literature on novel electrosynthesis reactions.48–50 To expedite the electrification of industrially-relevant organic reactions by enabling data-driven investigations of various parameter–property relationships such as yields and selectivities across the high-dimensional parameter and compound spaces, it is necessary to create a unified and structured electrosynthesis reaction database and leverage existing literature as past collective knowledge.To evaluate the multimodal cognition ability of the MLLMs to assist in knowledge retrieval from scientific literature, we investigated two essential subtasks within automated electrosynthesis reaction mining. The first subtask consists of the extraction of reaction conditions from reaction schemes (Fig. 1A). This is a specialized task which requires domain knowledge and context-awareness to interpret symbolic representations, such as electrode polarity indicators, and to ensure accurate role assignment, such as distinguishing between an electrolyte and solvent. The second subtask is that of resolving cross-modality data interdependencies (Fig. 1B), where reference labels within figure images are defined elsewhere in the text. This capability is important since substrate-specific variations in reaction conditions are typically conveyed through footnote cross-references. Being able to identify and connect these distributed pieces of information is essential to maintain the coherency and integrity of the mined data. We would like to clarify that the subtask of image-to-SMILES translation is beyond the scope of this work because there are numerous available rule-based and deep-learning based toolkits including DECIMER, SwinOCSR and MolNexTR with high reported accuracy rates.51–53
Evaluation dataset (MERMES-T24) construction
To benchmark our selected models, we manually curated a test dataset comprising 65 literature articles reporting new organic electrosynthesis reactions, published across 16 peer-reviewed journals to ensure diversity of writing and graphic presentation styles (full DOI list in Table S1†). We named this dataset MERMES-T24 for further reference by other researchers. Only articles that reported on batch reactions in undivided cells were considered within the scope of this study. The papers, published between February 2017 and December 2023, were downloaded in HTML format, and the image–caption pairs were compiled individually. Out of the 475 image–caption pairs, we identified 87 pairs that were relevant to our defined tasks (i.e. reaction diagram schemes depicting standard electrosynthesis conditions and/or demonstrating cross-modality interdependency).Multimodal large language model (MLLM) selection
We selected five state-of-the-art MLLMs for preliminary evaluation of their zero-shot performances in our specified tasks, namely GPT-4V (gpt-4-vision-preview),22 Gemini Pro (gemini-1.0-pro),21 Claude 3 (Claude 3 Opus),19 InternVL (InternVL-chat-v1.2-chinese-plus)45 and LLaVA (LLaVA-v1.6-34b).46,47 All five MLLMs are autoregressive language models based on the transformer architecture. We chose these models because they previously demonstrated comparable performances on multiple text-image multimodal evaluation tasks including DocVQA,54 ChartQA,55 AI2D56 and MME.57 In addition, researchers can access these tools through various application programming interfaces (APIs) or by downloading their models, which include pre-trained weights. This accessibility simplifies their integration into our final automated reaction mining pipeline.Results and discussion
Subtask 1: electrosynthesis reaction diagram parsing
The formal prompt for subtask 1 consists of three main parts: (i) instructions to identify and categorize as the following 10 reaction parameters, namely the anode, cathode, electrolytes/additives, amounts of electrolytes/additives, solvents, amounts of solvents, current, duration, air/inert atmosphere, and temperature, (ii) succinct contextual information, and (iii) single-shot visual examples (Fig. 2). The latter two parts serve to pre-condition the MLLM for our domain-specific task. In addition, the prompt also instructs the MLLM to include any other related reaction parameter that does not fall under the predefined categories in a separate “Others” column, to accommodate more complex reactions. To minimize hallucinations, we have adopted prompt engineering strategies that have been previously reported.29,58 Within this framework, the models are directed to set any specific parameter to “N.R. (not reported)” if they cannot find the relevant information in the supplied image and/or caption.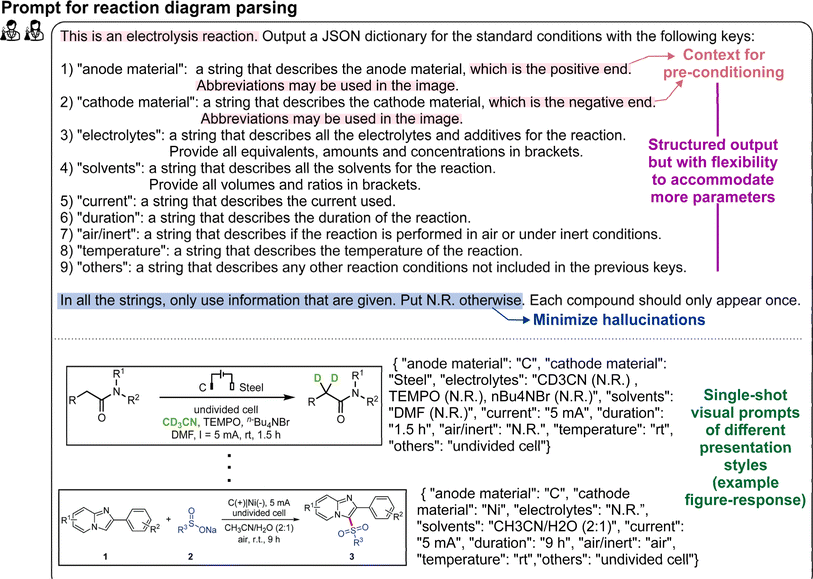 | ||
| Fig. 2 [Single-shot visual prompting for electrosynthesis reaction diagram parsing] The full prompt used in this subtask for electrosynthesis reaction diagram parsing is provided. The example images are adapted with permission from ref. 59 and ref. 60, with permission from the Royal Society of Chemistry. | ||
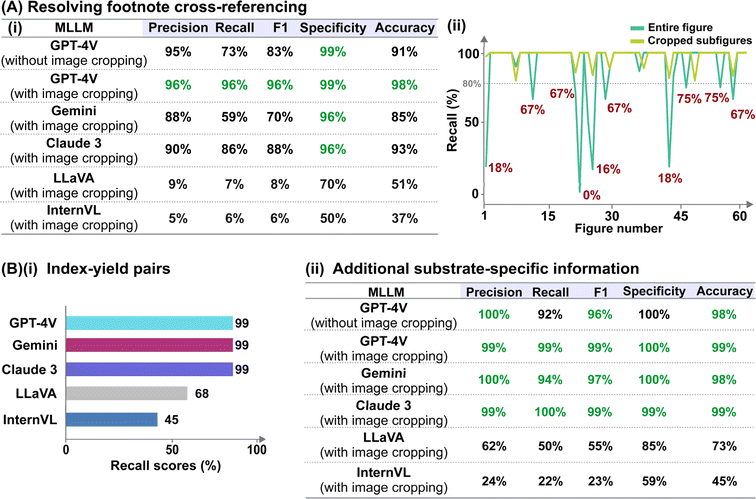
We analysed the results of reaction diagram parsing using two evaluation metrics with different tolerance levels: (i) hard match evaluation requires the correct matching of reaction parameters with their intended role, while (ii) soft match evaluation requires only correct identification of the parameter, regardless of its role. For instance, an anode material annotated as the cathode, or solvents classified as electrolytes/additives are considered incorrect under hard match evaluation but correct under soft match evaluation. Accurate hard matches for the anode and cathode material are especially critical for automated electrosynthesis reaction mining because swapping the electrode polarity would influence the actual yield and selectivity. In addition, it is important to note that we do not accommodate partial answers for reactions involving mixed solvent systems and/or multiple electrolytes/additives across both evaluation metrics. In other words, the identified parameter is only considered “correct” when all reported solvents/electrolytes/additives are present.
The outcomes of our preliminary evaluation of the zero-shot performances of the five MLLMs, summarized in Fig. 3A, reveal that GPT-4V surpasses the others in both hard and soft match evaluations (detailed discussion in ESI Note 1; supplementary evaluation metrics provided in Tables S2–S5†). Given its strong performance, we chose GPT-4V for further refinement. The model's hard match accuracy was lowest for the identification of solvents and electrode materials (both cathode and anode), with scores of 83% and 85% respectively (Fig. 3A). In the case of solvents, misclassification primarily arises from mis-labelling solvents as electrolytes, in which the model achieved excellent 100% soft-match accuracy (Fig. 3A). For electrode classification, we deconvoluted the model's performance based on the presentation style of the electrode systems, categorized into four categories based on whether the anode and cathode materials are explicitly indicated by their respective polarities or inferred schematically from circuit symbols (Fig. 3B-i). While the model achieved 100% accuracy for the first two styles, its performance dropped to 75–78% and 70% for styles 3 and 4, respectively (Fig. 3B-iii). We hypothesize that these two styles are poorly predicted due to their ambiguous representations, which necessitate prior understanding of the commonly employed chemical nomenclature. In this regard, we demonstrate the effectiveness of single-shot visual prompting in boosting the “chemical-awareness” of the MLLM for improved adaptability to specialized image mining tasks. By providing single-shot examples of a representative reaction diagram from each presentation style before analysing the actual dataset, we observed notable improvements to 100% and 89% for the latter two styles, boosting the overall hard match electrode accuracy to 99% (Fig. 3B-ii and iii; Tables S6 and S7†). In addition, hard match accuracies for all other parameter categories also reached near-unity (Fig. 3B-ii). Importantly, the model is robust against more intricate scenarios, such as those involving mixed solvent systems and/or multiple electrolytes and additives (present in 57% of investigated reaction diagrams), or in instances where multiple units such as ratio, % mol, mmol and molarity are utilized (present in all investigated reaction diagrams).
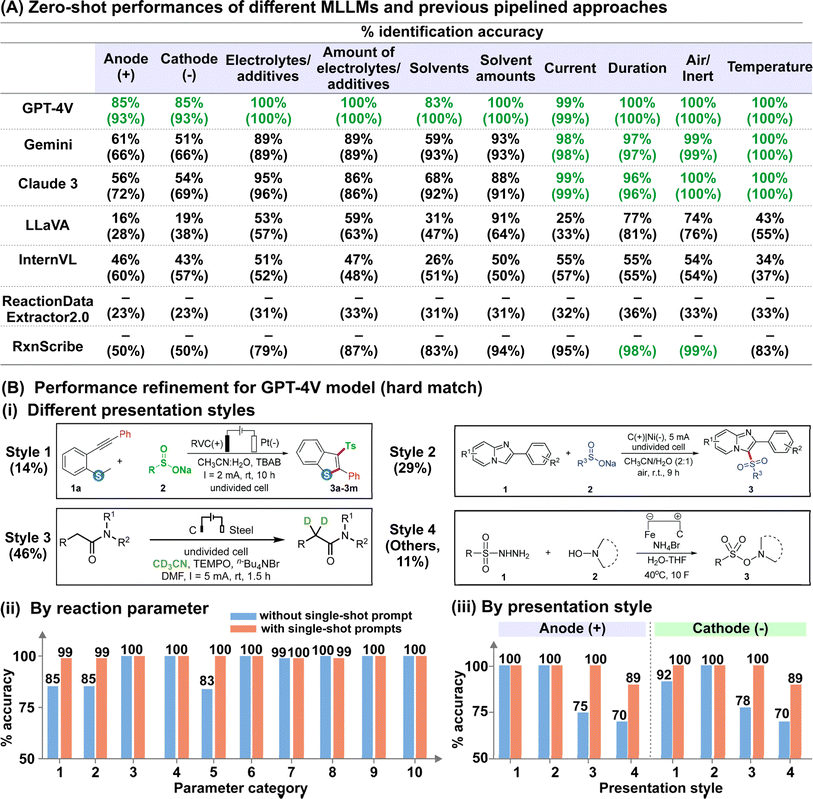 | ||
| Fig. 3 [Performance evaluation of electrosynthesis reaction diagram parsing] (A) % hard match (soft match) identification accuracy for each parameter using different MLLMs and two specialized reaction diagram parsing toolkits previously reported, as benchmarks. For the benchmark models, only the soft match scores are tabulated because the different categories are not specified. The highest scores for each category are in green. (B) Evaluation of the effectiveness of single-shot visual prompts. (i) Examples of different reaction diagram presentation styles. The % distribution of each style within our dataset is included in brackets. The example images are adapted with permission from ref. 61 (style 1), ref. 59 (style 2), ref. 60 (style 3), and ref. 62 (style 4), with permission from the Royal Society of Chemistry. (ii) % of correctly identified for each reaction parameter category with and without the integration of single-shot visual prompts. Categories 1–10 refer to anode, cathode, electrolytes/additives, amounts of electrolytes/additives, solvents, amounts of solvents, current, duration, air/inert atmosphere, and temperature, respectively. (iii) % of correctly identified anode and cathode material for each presentation style under hard match evaluation. | ||
To further evaluate our MLLM workflow, we benchmarked its performance against ReactionDataExtractor2.0 (ref. 14) and RxnScribe,18 two established deep-learning based toolkits for reaction diagram parsing. Notably, the frontrunner MLLM demonstrated superior performance in interpreting electrosynthesis reaction diagrams, which the aforementioned toolkits struggled to parse accurately. Although both ReactionDataExtractor2.0 and RxnScribe are designed for single-line chemical reaction diagrams resembling those in our study, they achieved soft match accuracies ranging from 23–36% to 50–99% on our test dataset, respectively (Fig. 3A). The poor performance of ReactionDataExtractor2.0 across all categories is due to its failure at identifying the regions with reaction conditions for most of the reaction diagrams, despite accurately identifying the location and direction of the reaction arrows (ESI Note 2†). On the other hand, we observe that although RxnScribe can accurately identify standard parameters such as duration and temperature with >90% accuracy, domain-specific information such as the electrode materials and the electrolytes are poorly identified, whereby RxnScribe makes mistakes such as missing one or both electrode materials (ESI Note 2†). Since ReactionDataExtractor2.0 and RxnScribe are specialized toolkits designed at recognizing organic reaction schemes, this limits their ability to extrapolate beyond familiar data, making them less effective at interpreting annotations specific to electrosynthesis reactions in our MERMES-T24 dataset, which are outside their design focus. This thus emphasizes the key advantage of general-purpose models, which have the capability to interpret symbolic cues with in-context learning that is straightforward to implement at low training costs and eliminates the need for rewriting programs or retraining models each time the target chemical application changes.
Subtask 2: resolving cross-modality interdependency
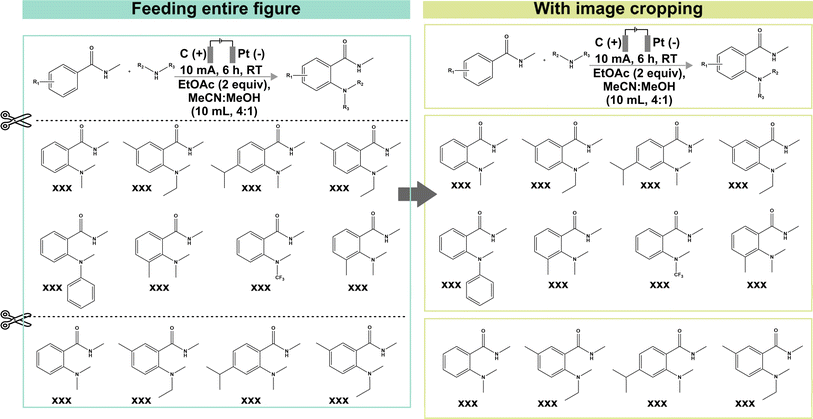
The formal prompt for subtask 2 contains succinct contextual information to pre-condition the MLLM to recognize the correlations between superscript letters (found in the figure) and footnote references (found in the caption) (full prompt in Fig. S1†). In addition, the prompt also instructs the MLLM to identify substrate-specific information including the corresponding yields (as index–yield pairs) and other additional details that are related to the investigated reaction (as a separate “Others” column). These additional details can vary in terms of contents, ranging from diastereomeric and product ratios to reaction durations. From the initial data subset, we further identify image–caption pairs that demonstrate such cross-modality interdependency (74 in total). Each model prediction was assigned as follows: true positive for correct identification of index–yield or footnote references; false positive for incorrect assignment or redundant information; true negative for correct identification of compounds without reported yields or footnote references; and false negative for missing information. It is worth noting that certain compounds may be associated with multiple yields and/or footnote references, which further increases the task complexity. In these cases, we do not accommodate partial answers and the prediction is only assigned as a “true positive” when all associated yields and/or footnote references are correctly identified.Instead of providing the raw images, we cropped each figure into smaller subfigures using an in-house automated image cropper code released as part of MERMES before collectively passing these subfigures into the model to extract the information (Fig. 4). Using GPT-4V as the test model, we demonstrate that incorporating image cropping as an additional preprocessing step prior to multimodal analysis of substrate scope diagrams effectively improved the overall recall of footnote cross-references from 73% to 96%, while maintaining excellent precision and specificity at ≥96% (Fig. 5A-i; ESI Note 3†). This is because the model is prone to overlooking footnote cross-references (i.e. higher false negative counts) when the lines of information within the figure are densely packed, such as in the case of Markush structures with varying R substituents, where individual recall scores of each figure can fall below 20% (Fig. 5A-ii and S4; Table S8†). The incorporation of image cropping is thus useful to circumvent information overload or neglect, with only minor trade-offs in terms of current service cost (∼$0.039 USD versus ∼$0.034 USD per figure with and without image cropping for GPT-4V), and execution time (∼33.7 seconds per figure versus ∼328.6 seconds per figure with and without image cropping for GPT-4V). Comparing across different MLLMs, GPT-4V outperforms the other models by 6–90% and 10–90% for precision and recall, respectively, attaining an excellent F1 score of 96% (Fig. 5A-i; Tables S9–S13†). In particular, InternVL and LLaVA often fail to identify the presence of superscript letters, resulting in high false negative counts. In comparison, Gemini and Claude 3 can accurately identify the presence of superscript letters in the figures and match them with the corresponding footnote references in the caption; however, they struggle in instances with multiple superscript letters associated with the same product.
As for the identification of substrate-specific information, GPT-4V, Gemini and Claude 3 achieve excellent performances, attaining 99% recall of the index–yield pairs, and high overall precision, recall and specificity of ≥94% at identifying additional substrate-specific information (Fig. 5B; Tables S14–S20†). In comparison, LLaVA and InternVL often make mistakes such as incorrect identification of diastereomeric ratios, e.g. reporting as “1.2.1” instead of “1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.1” or failure to identify the presence of any substrate-specific information. They also struggle to interpret index labels denoted by double alphabet letters (e.g., 3ab, 3ac, 2az) (Tables S21–S24†). At the same time, we observe that they are more prone to hallucinations and would provide false information such as giving additional index–yield pairs beyond the provided substrate scope.
2.1” or failure to identify the presence of any substrate-specific information. They also struggle to interpret index labels denoted by double alphabet letters (e.g., 3ab, 3ac, 2az) (Tables S21–S24†). At the same time, we observe that they are more prone to hallucinations and would provide false information such as giving additional index–yield pairs beyond the provided substrate scope.
MERMES: multimodal reaction mining pipeline for electrosynthesis
Leveraging our findings, we developed an end-to-end workflow towards automated reaction mining. Modelling the cognitive process of a human scientist during literature reading, our workflow features three sequential modules to perform the following: article retrieval, information extraction and multimodal analysis. Our automated workflow is designed to process articles in HTML format, which offers standardized, machine-readable document structure to facilitate automated data extraction. In brief, the article retrieval module initiates the job by downloading content and high-resolution images from the URLs of the articles (Step 1 in Fig. 6). Next, the information extraction module extracts the image–caption pairs and identifies those of relevance to our defined task via MLLM, guided by user-provided natural language prompts (Step 2 in Fig. 6). The filtered image–caption pairs are directed to the final multimodal analysis module, where the pertinent chemical information is extracted by natural language prompts (Step 3 in Fig. 6). When analysing the figures, we adopt the similar strategy to crop the figure into smaller subfigures parts to ensure the performance of the MLLM. We note that our framework can be easily extended to process other data contents in the article. Our future work will include extending the pipeline to efficiently mine other document formats, such as PDF, which remains challenging for machines to parse and sort the data. The full code is available in GitHub: https://www.github.com/aspuru-guzik-group/MERMES.Conclusions
We demonstrate that multimodal large language models are capable of chemistry-relevant multimodal cognition skills to interpret and assimilate electrosynthesis information from scientific publications. Our findings reveal that the strategic integration of text-based preconditioning prompts and single-shot visual prompts for in-context learning enables MLLMs to grasp domain-specific concepts for accurate parameter identification and role assignment, achieving ≥99% accuracy across 10 different parameter categories for the frontrunner MLLM. At the same time, they can resolve cross-modality data interdependency with excellent F1 scores of ≥96%, while offering enough flexibility to handle disparate amounts of information to extract additional, non-standardized details not consistently reported across all figures. This level of accuracy significantly surpasses the 80% benchmark for manual data extraction by humans.63 We developed a toolkit (MERMES) for carrying out these tasks, serving as a multimodal pipeline for automated reaction mining from scientific publications. Moving forward, we will equip MERMES with additional capabilities such as image-to-SMILES translation and utilize MERMES to create a comprehensive electrosynthesis reaction database as an invaluable data resource for numerous potential downstream applications from computer-aided reaction discovery and optimization, reaction prediction and other predictive tasks. From a broader perspective, by exemplifying the potential of harnessing multimodal large language models in (electro)chemical information processing, we envisage that MLLM-based reaction mining workflows such as MERMES will open new opportunities for data-driven research across numerous domains.Data availability
The source code of our automated workflow extraction code, MERMES, together with additional data mining scripts, can be found in our Github repository (https://www.github.com/aspuru-guzik-group/MERMES). To improve the reproducibility of this work, a frozen version of the repository has been uploaded to Zenodo (https://doi.org/10.5281/zenodo.12713560).64 The prompts used in this work, together with the raw responses from the tested MLLM models are compiled and uploaded to Zenodo (https://doi.org/10.5281/zenodo.12701834) for data transparency and reproducibility.65Author contributions
S. X. L., conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization & writing – original draft preparation, review & editing. S. P.-G., conceptualization, formal analysis, methodology, supervision & writing – review & editing. Z. Z. conceptualization, methodology, software & writing – review & editing. A. A.-G., conceptualization, funding acquisition, resources, project administration, supervision & writing – review & editing.Conflicts of interest
A. A.-G is a founder of Kebotix, Inc., Axiomatic, Inc., and Intrepid Labs, Inc.Acknowledgements
S. X. L. acknowledges support from Nanyang Technological University, Singapore and the Ministry of Education, Singapore for the Overseas Postdoctoral Fellowship. S. P.-G. acknowledges that this material is based upon work supported by the U.S. Department of Energy, Office of Science, Subaward by the University of Minnesota, Project title: Development of Machine Learning and Molecular Simulation Approaches to Accelerate the Discovery of Porous Materials for Energy-Relevant Applications under Award Number DE-SC0023454. A. A.-G. acknowledges support from the CIFAR, the Canada 150 Research Chairs Program as well as Anders G. Frøseth. This research is part of the University of Toronto's Acceleration Consortium, which receives funding from the Canada First Research Excellence Fund (CFREF).References
- H. Wang, T. Fu, Y. Du, W. Gao, K. Huang, Z. Liu, P. Chandak, S. Liu, P. Van Katwyk, A. Deac, A. Anandkumar, K. Bergen, C. P. Gomes, S. Ho, P. Kohli, J. Lasenby, J. Leskovec, T.-Y. Liu, A. Manrai, D. Marks, B. Ramsundar, L. Song, J. Sun, J. Tang, P. Veličković, M. Welling, L. Zhang, C. W. Coley, Y. Bengio and M. Zitnik, Scientific discovery in the age of artificial intelligence, Nature, 2023, 620, 47–60 CrossRef CAS PubMed.
- O. Kononova, H. Huo, T. He, Z. Rong, T. Botari, W. Sun, V. Tshitoyan and G. Ceder, Text-mined dataset of inorganic materials synthesis recipes, Sci. Data, 2019, 6, 203 CrossRef PubMed.
- J. Dagdelen, A. Dunn, S. Lee, N. Walker, A. S. Rosen, G. Ceder, K. A. Persson and A. Jain, Structured information extraction from scientific text with large language models, Nat. Commun., 2024, 15, 1418 CrossRef CAS PubMed.
- Z. Jensen, E. Kim, S. Kwon, T. Z. H. Gani, Y. Román-Leshkov, M. Moliner, A. Corma and E. Olivetti, A Machine Learning Approach to Zeolite Synthesis Enabled by Automatic Literature Data Extraction, ACS Cent. Sci., 2019, 5, 892–899 CrossRef CAS PubMed.
- M. Ansari and S. M. Moosavi, Agent-Based Learning of Materials Datasets from Scientific Literature, arXiv, 2023, preprint, arXiv:2312.11690, DOI: DOI:10.48550/arXiv.2312.11690.
- A. B. Georgescu, P. Ren, A. R. Toland, S. Zhang, K. D. Miller, D. W. Apley, E. A. Olivetti, N. Wagner and J. M. Rondinelli, Database, Features, and Machine Learning Model to Identify Thermally Driven Metal–Insulator Transition Compounds, Chem. Mater., 2021, 33, 5591–5605 CrossRef CAS.
- C. Karpovich, Z. Jensen, V. Venugopal and E. Olivetti, Inorganic Synthesis Reaction Condition Prediction with Generative Machine Learning, arXiv, 2021, preprint, arXiv:2112.09612, DOI: DOI:10.48550/arXiv.2112.09612.
- J. E. Saal, A. O. Oliynyk and B. Meredig, Machine Learning in Materials Discovery: Confirmed Predictions and Their Underlying Approaches, Annu. Rev. Mater. Res., 2020, 50, 49–69 CrossRef CAS.
- H. Huo, C. J. Bartel, T. He, A. Trewartha, A. Dunn, B. Ouyang, A. Jain and G. Ceder, Machine-Learning Rationalization and Prediction of Solid-State Synthesis Conditions, Chem. Mater., 2022, 34, 7323–7336 CrossRef CAS PubMed.
- E. Kim, K. Huang, A. Saunders, A. McCallum, G. Ceder and E. Olivetti, Materials Synthesis Insights from Scientific Literature via Text Extraction and Machine Learning, Chem. Mater., 2017, 29, 9436–9444 CrossRef CAS.
- Y. Luo, S. Bag, O. Zaremba, A. Cierpka, J. Andreo, S. Wuttke, P. Friederich and M. Tsotsalas, MOF Synthesis Prediction Enabled by Automatic Data Mining and Machine Learning, Angew. Chem., Int. Ed., 2022, 61, e202200242 CrossRef CAS PubMed.
- O. Kononova, T. He, H. Huo, A. Trewartha, E. A. Olivetti and G. Ceder, Opportunities and challenges of text mining in materials research, iScience, 2021, 24, 102155 CrossRef PubMed.
- T. He, W. Sun, H. Huo, O. Kononova, Z. Rong, V. Tshitoyan, T. Botari and G. Ceder, Similarity of Precursors in Solid-State Synthesis as Text-Mined from Scientific Literature, Chem. Mater., 2020, 32, 7861–7873 CrossRef CAS.
- D. Wilary and J. M. Cole, ReactionDataExtractor 2.0: A Deep Learning Approach for Data Extraction from Chemical Reaction Schemes, J. Chem. Inf. Model., 2023, 63, 6053–6067 CrossRef CAS PubMed.
- E. Beard and J. M. Cole, ChemSchematicResolver: A Toolkit to Decode 2D Chemical Diagrams with Labels and R-Groups into Annotated Chemical Named Entities, J. Chem. Inf. Model., 2020, 60, 2059–2072 CrossRef CAS PubMed.
- A. Tharatipyakul, S. Numnark, D. Wichadakul and S. Ingsriswang, ChemEx: information extraction system for chemical data curation, BMC Bioinf., 2012, 13, S9 CrossRef PubMed.
- S. H. M. Mehr, M. Craven, A. I. Leonov, G. Keenan and L. Cronin, A universal system for digitization and automatic execution of the chemical synthesis literature, Science, 2020, 370, 101–108 CrossRef CAS PubMed.
- Y. Qian, J. Guo, Z. Tu, C. W. Coley and R. Barzilay, RxnScribe: A Sequence Generation Model for Reaction Diagram Parsing, J. Chem. Inf. Model., 2023, 63, 4030–4041 CrossRef CAS PubMed.
- Anthropic, The Claude 3 Model Family: Opus, Sonnet, Haiku, 2024, https://www-cdn.anthropic.com/de8ba9b01c9ab7cbabf5c33b80b7bbc618857627/Model_Card_Claude_3.pdf.
- Anthropic, Model Card and Evaluations for Claude Models, 2023, https://www-cdn.anthropic.com/files/4zrzovbb/website/bd2a28d2535bfb0494cc8e2a3bf135d2e7523226.pdf.
- Gemini Team et al., Gemini: A Family of Highly Capable Multimodal Models, arXiv, 2023, preprint, arXiv:2312.11805, DOI: DOI:10.48550/arXiv.2312.11805.
- OpenAI et al., GPT-4 Technical Report, arXiv, 2023, preprint, arXiv:2303.08774, DOI: DOI:10.48550/arXiv.2303.08774.
- H. Touvron, T. Lavril, G. Izacard, X. Martinet, M.-A. Lachaux, T. Lacroix, B. Rozière, N. Goyal, E. Hambro, F. Azhar, A. Rodriguez, A. Joulin, E. Grave and G. Lample, LLaMA: Open and Efficient Foundation Language Models, arXiv, 2023, preprint, arXiv:2302.13971, DOI: DOI:10.48550/arXiv.2302.13971.
- J. Devlin, M.-W. Chang, K. Lee and K. Toutanova, BERT: Pre-Training of Deep Bidirectional Transformers for Language Understanding, arXiv, 2019, preprint, arXiv:1810.04805, DOI: DOI:10.48550/arXiv.1810.04805.
- R. Anil et al., PaLM 2 Technical Report, arXiv, 2023, preprint, arXiv:2305.10403, DOI: DOI:10.48550/arXiv.2305.10403.
- M. Thway, A. K. Y. Low, S. Khetan, H. Dai, J. Recatala-Gomez, A. P. Chen and K. Hippalgaonkar, Harnessing GPT-3.5 for text parsing in solid-state synthesis – case study of ternary chalcogenides, Digital Discovery, 2024, 3, 328–336 RSC.
- W. Zhang, Q. Wang, X. Kong, J. Xiong, S. Ni, D. Cao, B. Niu, M. Chen, R. Zhang, Y. Wang, L. Zhang, X. Li, Z. Xiong, Q. Shi, F. Cheng, Z. Fu and M. Zheng, Fine-Tuning ChatGPT Achieves State-of-the-Art Performance for Chemical Text Mining, ChemRxiv, 2023, preprint, DOI: DOI:10.26434/chemrxiv-2023-k7ct5.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, ChatGPT Chemistry Assistant for Text Mining and the Prediction of MOF Synthesis, J. Am. Chem. Soc., 2023, 145, 18048–18062 CrossRef CAS PubMed.
- Z. Zheng, Z. Rong, N. Rampal, C. Borgs, J. T. Chayes and O. M. Yaghi, A GPT-4 Reticular Chemist for Guiding MOF Discovery, Angew. Chem., Int. Ed., 2023, 62, e202311983 CrossRef CAS PubMed.
- Y. Zhu, P. Zhang, C. Zhang, Y. Chen, B. Xie, Z. Dou, Z. Liu and J.-R. Wen, INTERS: Unlocking the Power of Large Language Models in Search with Instruction Tuning, arXiv, 2024, preprint, arXiv:2401.06532, DOI: DOI:10.48550/arXiv.2401.06532.
- M. P. Polak and D. Morgan, Extracting accurate materials data from research papers with conversational language models and prompt engineering, Nat. Commun., 2024, 15, 1569 CrossRef CAS PubMed.
- K. M. Jablonka, Q. Ai, A. Al-Feghali, S. Badhwar, J. D. Bocarsly, A. M. Bran, S. Bringuier, L. C. Brinson, K. Choudhary, D. Circi, S. Cox, W. A. De Jong, M. L. Evans, N. Gastellu, J. Genzling, M. V. Gil, A. K. Gupta, Z. Hong, A. Imran, S. Kruschwitz, A. Labarre, J. Lála, T. Liu, S. Ma, S. Majumdar, G. W. Merz, N. Moitessier, E. Moubarak, B. Mouriño, B. Pelkie, M. Pieler, M. C. Ramos, B. Ranković, S. G. Rodriques, J. N. Sanders, P. Schwaller, M. Schwarting, J. Shi, B. Smit, B. E. Smith, J. Van Herck, C. Völker, L. Ward, S. Warren, B. Weiser, S. Zhang, X. Zhang, G. A. Zia, A. Scourtas, K. J. Schmidt, I. Foster, A. D. White and B. Blaiszik, 14 examples of how LLMs can transform materials science and chemistry: a reflection on a large language model hackathon, Digital Discovery, 2023, 2, 1233–1250 RSC.
- L. Patiny and G. Godin, Automatic extraction of FAIR data from publications using LLM, ChemRxiv, 2023, preprint, DOI: DOI:10.26434/chemrxiv-2023-05v1b-v2.
- S. Liu, J. Wang, Y. Yang, C. Wang, L. Liu, H. Guo and C. Xiao, ChatGPT-Powered Conversational Drug Editing Using Retrieval and Domain Feedback, arXiv, 2023, preprint, arXiv:2305.18090, DOI: DOI:10.48550/arXiv.2305.18090.
- Z. Xie, X. Evangelopoulos, Ö. H. Omar, A. Troisi, A. I. Cooper and L. Chen, Fine-tuning GPT-3 for machine learning electronic and functional properties of organic molecules, Chem. Sci., 2024, 15, 500–510 RSC.
- L. Chen, M. Zaharia and J. Zou, How is ChatGPT's behavior changing over time?arXiv, 2023, preprint, arXiv:2307.09009, DOI: DOI:10.48550/arXiv.2307.09009.
- Y. Zhou, A. I. Muresanu, Z. Han, K. Paster, S. Pitis, H. Chan and J. Ba, Large Language Models Are Human-Level Prompt Engineers, arXiv, 2023, preprint, arXiv:2211.01910, DOI: DOI:10.48550/arXiv.2211.01910.
- G. Yona, R. Aharoni and M. Geva, Narrowing the Knowledge Evaluation Gap: Open-Domain Question Answering with Multi-Granularity Answers, arXiv, 2024, preprint, arXiv:2401.04695, DOI: DOI:10.48550/arXiv.2401.04695.
- A. G. Parameswaran, S. Shankar, P. Asawa, N. Jain and Y. Wang, Revisiting Prompt Engineering via Declarative Crowdsourcing, arXiv, 2023, preprint, arXiv:2308.03854, DOI: DOI:10.48550/arXiv.2308.03854.
- D. Vidhani and M. Mariappan, Optimizing Human–AI Collaboration in Chemistry: A Case Study on Enhancing Generative AI Responses through Prompt Engineering, Chemistry, 2024, 6, 723–737 CrossRef.
- C. Wu, J. Lei, Q. Zheng, W. Zhao, W. Lin, X. Zhang, X. Zhou, Z. Zhao, Y. Zhang, Y. Wang and W. Xie, Can GPT-4V(Ision) Serve Medical Applications? Case Studies on GPT-4V for Multimodal Medical Diagnosis, arXiv, 2023, preprint, arXiv:2310.09909, DOI: DOI:10.48550/arXiv.2310.09909.
- Y. Shi, D. Peng, W. Liao, Z. Lin, X. Chen, C. Liu, Y. Zhang and L. Jin, Exploring OCR Capabilities of GPT-4V(Ision) : A Quantitative and In-Depth Evaluation, arXiv, 2023, preprint, arXiv:2310.16809, DOI: DOI:10.48550/arXiv.2310.16809.
- S. Yin, C. Fu, S. Zhao, K. Li, X. Sun, T. Xu and E. Chen, A Survey on Multimodal Large Language Models, arXiv, 2023, preprint, arXiv:2306.13549, DOI: DOI:10.48550/arXiv.2306.13549.
- Z. Zheng, Z. He, O. Khattab, N. Rampal, M. A. Zaharia, C. Borgs, J. T. Chayes and O. M. Yaghi, Image and data mining in reticular chemistry powered by GPT-4V, Digital Discovery, 2024, 3, 491–501 RSC.
- Z. Chen, J. Wu, W. Wang, W. Su, G. Chen, S. Xing, M. Zhong, Q. Zhang, X. Zhu, L. Lu, B. Li, P. Luo, T. Lu, Y. Qiao and J. Dai, InternVL: Scaling up Vision Foundation Models and Aligning for Generic Visual-Linguistic Tasks, arXiv, 2024, preprint, arXiv:2312.14238, DOI: DOI:10.48550/arXiv.2312.14238.
- H. Liu, C. Li, Q. Wu and Y. J. Lee, Visual Instruction Tuning, arXiv, 2023, preprint, arXiv:2304.08485, DOI: DOI:10.48550/arXiv.2304.08485.
- H. Liu, C. Li, Y. Li and Y. J. Lee, Improved Baselines with Visual Instruction Tuning, arXiv, 2023, preprint, arXiv:2310.03744, DOI: DOI:10.48550/arXiv.2310.03744.
- C. Kingston, M. D. Palkowitz, Y. Takahira, J. C. Vantourout, B. K. Peters, Y. Kawamata and P. S. Baran, A Survival Guide for the “Electro-curious”, Acc. Chem. Res., 2020, 53, 72–83 CrossRef CAS PubMed.
- C. Zhu, N. W. J. Ang, T. H. Meyer, Y. Qiu and L. Ackermann, Organic Electrochemistry: Molecular Syntheses with Potential, ACS Cent. Sci., 2021, 7, 415–431 CrossRef CAS PubMed.
- D. Pollok and S. R. Waldvogel, Electro-organic synthesis – a 21 st century technique, Chem. Sci., 2020, 11, 12386–12400 RSC.
- Y. Chen, C. T. Leung, Y. Huang, J. Sun, H. Chen and H. Gao, MolNexTR: A Generalized Deep Learning Model for Molecular Image Recognition, arXiv, 2024, preprint, arXiv:2403.03691, DOI: DOI:10.48550/arXiv.2403.03691.
- K. Rajan, H. O. Brinkhaus, M. I. Agea, A. Zielesny and C. Steinbeck, DECIMER.ai: an open platform for automated optical chemical structure identification, segmentation and recognition in scientific publications, Nat. Commun., 2023, 14, 1–18 Search PubMed.
- Z. Xu, J. Li, Z. Yang, S. Li and H. Li, SwinOCSR: end-to-end optical chemical structure recognition using a Swin Transformer, J. Cheminf., 2022, 14, 1–13 Search PubMed.
- M. Mathew, D. Karatzas and C. V. Jawahar, DocVQA: A Dataset for VQA on Document Images, arXiv, 2021, preprint, arXiv:2007.00398, DOI: DOI:10.48550/arXiv.2007.00398.
- A. Masry, D. X. Long, J. Q. Tan, S. Joty and E. Hoque, ChartQA: A Benchmark for Question Answering about Charts with Visual and Logical Reasoning, arXiv, 2022, preprint, arXiv:2203.10244, DOI: DOI:10.48550/arXiv.2203.10244.
- A. Kembhavi, M. Salvato, E. Kolve, M. Seo, H. Hajishirzi and A. Farhadi, Computer Vision – ECCV 2016, Springer, Cham, 2016, pp. 235–251 Search PubMed.
- C. Fu, P. Chen, Y. Shen, Y. Qin, M. Zhang, X. Lin, J. Yang, X. Zheng, K. Li, X. Sun, Y. Wu and R. Ji, MME: A Comprehensive Evaluation Benchmark for Multimodal Large Language Models, arXiv, 2023, preprint, arXiv:2306.13394, DOI: DOI:10.48550/arXiv.2306.13394.
- K. Hatakeyama-Sato, N. Yamane, Y. Igarashi, Y. Nabae and T. Hayakawa, Prompt engineering of GPT-4 for chemical research: what can/cannot be done?ChemRxiv, 2023, preprint, DOI: DOI:10.26434/chemrxiv-2023-s1x5p.
- L. Han, M. Huang, Y. Li, J. Zhang, Y. Zhu, J. K. Kim and Y. Wu, An electrolyte- and catalyst-free electrooxidative sulfonylation of imidazo[1,2- a ]pyridines, Org. Chem. Front., 2021, 8, 3110–3117 RSC.
- S. Ning, C. Wu, L. Zheng, M. Liu, Y. Zhang, X. Che and J. Xiang, Electrochemical α-deuteration of amides, Green Chem., 2023, 25, 9993–9997 RSC.
- M.-M. Zhang, Y. Sun, W.-W. Wang, K.-K. Chen, W.-C. Yang and L. Wang, Electrochemical synthesis of sulfonated benzothiophenes using 2-alkynylthioanisoles and sodium sulfinates, Org. Biomol. Chem., 2021, 19, 3844–3849 RSC.
- A. O. Terent’ev, O. M. Mulina, V. D. Parshin, V. A. Kokorekin and G. I. Nikishin, Electrochemically induced oxidative S–O coupling: synthesis of sulfonates from sulfonyl hydrazides and N -hydroxyimides or N -hydroxybenzotriazoles, Org. Biomol. Chem., 2019, 17, 3482–3488 RSC.
- S. Huang and J. M. Cole, A database of battery materials auto-generated using ChemDataExtractor, Sci. Data, 2020, 7, 260 CrossRef PubMed.
- Z. Zhang and S. X. LeongAspuru-Guzik-Group/MERMES: First Release [code], 2024, DOI:10.5281/zenodo.12713560.
- S. X. Leong, Automated Electrosynthesis Reaction Mining with Multimodal Large Language Models - Raw Data and Prompts [datasets], 2024, DOI:10.5281/zenodo.12701834.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc04630g |
| This journal is © The Royal Society of Chemistry 2024 |