Open Access Article
Open Access ArticleDNAzyme-RCA-based colorimetric and lateral flow dipstick assays for the point-of-care testing of exosomal m5C-miRNA-21†
Hao
Zhang‡
a,
Yue
Tang‡
c,
Yingshun
Zhou
a,
Yiguo
Wang
d,
Haibin
Si
*a,
Lu
Li
*a and
Bo
Tang
 *ab
*ab
aDepartment College of Chemistry, Chemical Engineering and Materials Science, Collaborative Innovation Center of Functionalized Probes for Chemical Imaging, Key Laboratory of Molecular and Nano Probes, Ministry of Education, Shandong Normal University, Jinan, 250014, P. R. China. E-mail: tangb@sdnu.edu.cn; lilu5252@163.com; haibinsi@sdnu.edu.cn
bLaoshan Laboratory, Qingdao 266237, P. R. China
cDepartment of Emergency Medicine, Shandong Provincial Clinical Research Center for Emergency and Critical Care Medicine, Qilu Hospital of Shandong University, Jinan, 250014, P. R. China
dFirst Affiliated Hospital of Shandong First Medical University, Shandong Provincial Qianfoshan Hospital, Jinan, 250014, P. R. China
First published on 22nd May 2024
Abstract
Methylation of microRNAs (miRNAs) is a post-transcriptional modification that affects miRNA activity by altering the specificity of miRNAs to target mRNAs. Abnormal methylation of miRNAs in cancer suggests their potential as a tumor marker. However, the traditional methylated miRNA detection mainly includes mass spectrometry, sequencing and others; complex procedures and reliance on large instruments greatly limit their application in point-of-care testing (POCT). Based on this, we developed DNAzyme-RCA-based gold nanoparticle (AuNP) colorimetric and lateral flow dipstick (LFD) assays to achieve convenient detection of exosomal 5-methylcytosine miRNA-21 (m5C-miRNA-21) for the first time. The two assays achieved specific recognition and linear amplification of m5C-miRNA-21 through the DNAzyme triggered RCA reaction and color output with low background interference through AuNP aggregation induced by base complementary pairing. The lowest concentration of m5C-miRNA-21 visible to the naked eye of the two assays can reach 1 pM and 0.1 pM, respectively. Detection of exosomal m5C-miRNA-21 in clinical blood samples showed that the expression level of m5C-miRNA-21 in colorectal cancer patients was significantly higher than that in healthy individuals. This approach not only demonstrates a new strategy for the detection of colorectal cancer but also provides a reference for the development of novel diagnostic tools for other miRNA methylation-related diseases.
Introduction
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that regulate gene expression by promoting mRNA degradation and/or inhibiting mRNA translation at the post-transcriptional level.1 Studies have shown that miRNAs are the key regulator of fundamental physiological processes, including cell differentiation, proliferation, and apoptosis.2 As a consequence, their expression level is significantly associated with numerous human diseases.3,4 However, the physiological and pathological functions of miRNA are not only correlated with miRNA expression levels.5 Several mechanisms that have been reported to regulate miRNA activity also include single nucleotide polymorphisms, miRNA tailing, editing, and methylation.6–8 In particular, nucleotide methylation modifications within miRNA sequences are critical to their functions, and have received increasing attention. Methylation of miRNAs, as a novel form of post-transcriptional base modification, mainly includes N6-methyladenosine (m6A), 2′-O-methylation, 5-methylcytosine (m5C), and m7G, which can greatly affect miRNA activity by changing the specificity of miRNA to its target mRNA.9–12 Specifically, miRNA methylation has been demonstrated to inhibit the translation of tumor suppressor factors by influencing miRNA interactions with binding proteins, thereby promoting the development of bladder and liver cancers. Moreover, researchers have detected m5C on mature miRNAs, and further demonstrated that methylated miRNAs on cytosine residues inhibited the formation of miRNA/mRNA double strands, leading to the loss of miRNA's cancer-inhibiting function.13,14 In addition, multiple studies have shown that some tumor-associated miRNA methylation levels in the tumor cells of cancer patients are significantly increased compared with healthy individuals. In particular, for some miRNAs that are similarly expressed in tumor and normal tissues, their methylation levels were significantly increased in tumor tissues.15,16 This suggests that methylated miRNAs have the potential to serve as biomarkers to distinguish cancer patients from healthy individuals and provide a valuable tool for studying the initiation and progression of various cancers. Exosomal miRNA is an important form of miRNAs in blood, with good stability and various modification states, and plays a key role in RNA-based intercellular communication.17 By analyzing the expression and modification status of exosomal miRNA, they can be effectively utilized for cancer diagnosis and therapeutic monitoring.18 Therefore, it has been used as a broad non-invasive tumor marker.19–23 The development of an accurate and convenient detection method for exosomal methylated miRNAs is expected to provide a new way for cancer diagnosis and monitoring.Recently, some studies have reported the detection of methylated miRNA; these methods commonly first use antibodies to immunologically capture methylated RNA, followed by sequencing analysis.15,24–27 However, the complex procedures of the above schemes are not friendly for the detection of low content and easily degraded miRNAs. In addition, their high cost and reliance on large instruments have limited the use of methylated miRNAs as markers for point-of-care testing (POCT) of disease. In contrast, colorimetric assay and colorimetric assay based on lateral flow dipsticks (LFDs) have received considerable attention in the field of point-of-care diagnostics due to their low cost, rapid response, and simple visual assessment.28–31 So far, due to the low content of methylated miRNA and the difficulty in identifying specific methylation sites, no mature convenient detection method for methylated miRNA has been reported.
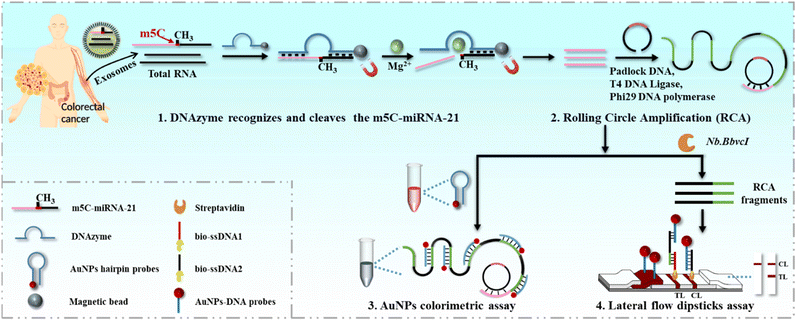
In this study, we developed colorimetric and lateral flow dipstick (LFD) assays based on the DNAzyme triggered rolling circle amplification (DNAzyme-RCA) reaction to achieve the point-of-care testing of exosomal 5-methylcytosine miRNA-21 (m5C-miRNA-21) for the first time. miRNA-21 is known to be highly correlated with the occurrence and development of tumors, and many studies have shown the expression or structural abnormalities of miRNA-21 in a variety of tumors.32,33 In recent years, it has been clearly pointed out that the methylation level of miRNA-21 is significantly different in tumor tissues and normal tissues.34 In this scheme, we modified DNAzyme with methylation recognition ability to specifically recognize and cleave m5C-miRNA-21 in the presence of Mg2+.35 Then, a padlock DNA was designed using one of the cleaved strands of m5C-miRNA-21 as the primer, and the RCA reaction occurred under the action of T4 ligase and DNA polymerase to achieve linear amplification of m5C-miRNA-21. In order to adapt to the application in different scenarios, the RCA products induced by m5C-miRNA-21 were then detected by colorimetric and LFD assays, respectively. (1) In colorimetric assay, AuNPs modified with short nucleic acid strands (AuNP hairpin probes) complementary to the repeat sequences of RCA products were used for coupling the RCA product for signal output. Due to the regular aggregation of AuNPs caused by RCA products, AuNP solution changes from red to blue, and this color change can be quickly recognized by the naked eye, so as to achieve a specific and sensitive detection independent of equipment. (2) In LFD assay, the RCA products are first cut by nicking endonuclease (Nb.BbvcI) into repeating nucleic acid fragments (RCA fragments). Subsequently, these RCA fragments are added onto a conjugate pad of LFDs and combined with AuNPs–DNA probes prepositioned to form RCA fragments/AuNPs–DNA complexes. As the solution flows over the test line (TL), these complexes hybridize with the bio-ssDNA-1 on the TL, resulting in a color band visible to the naked eye due to the presence of AuNPs. We applied DNAzyme-RCA based AuNP colorimetric and LFD assays to detect the exosomal m5C-miRNA-21 in the blood of clinical colorectal cancer patients, and both assays showed good specificity and sensitivity. The results showed that colorectal cancer patients exhibited higher m5C-miRNA-21 expression levels compared to healthy individuals and significant individual heterogeneity. The protocol is expected to be a new strategy and product for tumor POCT diagnosis based on methylated miRNA (Scheme 1).
Results and discussion
Design and validation of the DNAzyme triggered RCA reaction
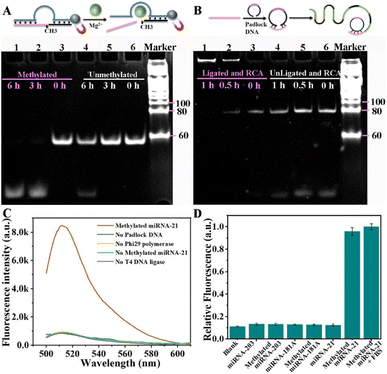
The DNAzyme can recognize specific RNA or DNA sequences and thus exhibit the cleavage ability.36 In this protocol, in order to selectively recognize m5C-miRNA-21 from total RNA, we designed a binding arm that is complementary to the miRNA-21 sequence. The binding arms of miRNA-21 were then fused to the hammerhead ribozyme sequence possessing 5-methylcytosine recognition and cleavage, forming a DNAzyme that facilitates the sequence-specific cleavage of m5C-miRNA-21 in the presence of Mg2+.37 The structural stability of DNAzyme (conformation 1) and DNAzyme/miRNA-21 (conformation 2) was calculated using NUPACK software (Fig. S1†). The Gibbs free energies of conformation 1 and conformation 2 are −14.40 kcal mol−1 and −30.94 kcal mol−1, respectively, indicating that DNAzyme tends to bind miRNA-21. The structural stability of padlock DNA combined with the cleaved strands of m5C-miRNA-21 was also calculated (Fig. S1C†). The Gibbs free energy of the complex was −18.21 kcal mol−1, indicating that the padlock DNA could effectively combine with the cleaved strands of m5C-miRNA-21.The cleavage of DNAzyme and the RCA reaction were confirmed by polyacrylamide gel electrophoresis (PAGE) experiments. The gel electrophoresis image of the cleaved products of DNAzyme is presented in Fig. 1A. As shown in lanes 1–3, the DNAzyme effectively recognizes and cleaves m5C-miRNA-21 within 3 hours. In contrast, unmethylated miRNA-21 (lanes 4–6) remained intact during the same time. The results demonstrate good recognition and cleavage capabilities of the DNAzyme towards m5C-miRNA-21. Successful cyclization of padlock DNA is a prerequisite for RCA amplification. In this scheme, the cleaved strands of m5C-miRNA-21 by DNAzyme were used as bridging primers to achieve cyclic formation of padlock DNA. The gel electrophoresis image of the RCA reaction is presented in Fig. 1B. With increasing amplification time, bands in lanes 1–3 adjacent to sample pores progressively intensified, indicating that the cleavage product successfully triggered the RCA reaction. Conversely, no bands were observed in lanes 4–6 near the sample pores, suggesting an unsuccessful RCA reaction with unmethylated miRNA-21.
The fluorescence experiments based on molecular beacons (MBs) were used to further validate the DNAzyme triggered RCA reaction. Specifically, fluorescent labeled molecular beacons were designed and synthesized, which were complementary to RCA products, and then fluorescence analysis was performed using a fluorescence spectrophotometer. As shown in Fig. 1C, a strong fluorescence signal was observed when padlock DNA, polymerase, ligase, and m5C-miRNA-21 were present. This suggests that DNAzyme successfully identified and cleaved m5C-miRNA-21 to trigger the RCA reaction. To verify the specificity of this protocol, we performed experiments using unmethylated miRNA-21 or other miRNAs as detection targets. As shown in the fluorescence results in Fig. 1D, the fluorescence intensity corresponding to m5C-miRNA-21 was significantly higher than that of other detection targets.
The AuNP colorimetric assay for m5C-miRNA-21
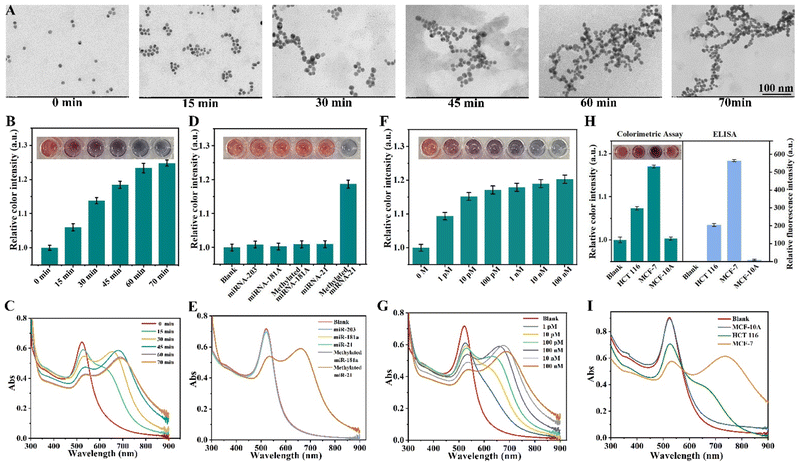
The AuNP colorimetric method is an ideal means of POCT because of its simple operation, low cost and high sensitivity. In this scheme, AuNP hairpin probes that are complementary to the RCA products are used for the signal output of the RCA reaction. In order to verify the connection between AuNPs and RCA products, AuNP hairpin probes incubated with RCA products for different times were examined by transmission electron microscopy (TEM). As shown in Fig. 2A, with the extension of the reaction time, the degree of regular aggregation of AuNPs increases, indicating that more AuNP hairpin probes are connected to the RCA products by complementary base pairing. The aggregation of AuNPs leads to the reduction of the distance between them, resulting in the change of the local surface plasmon resonance (LSPR) effect. This change alters the particles' ability to absorb and scatter light, as shown by a redshift (color change from red to blue, top of Fig. 2B) in the absorption spectrum.38,39 In order to quantify the color of the solution, the solution photos are converted using ImageJ into the color intensity based on RGB mode and calculated according to the weighting formula I = 0.3R + 0.59G + 0.11B. The ratios of the color intensity of the sample to the blank are the results of the colorimetric assay and are determined as the ordinates.40 It can be observed that the color intensity corresponds well to the aggregation of AuNPs, demonstrating a good detection ability independent of the experimental equipment (Fig. 2B, bottom). Then, the above reaction process was further verified using ultraviolet-visible (UV-vis) absorption spectra. In Fig. 2C, with the increase of the reaction time, that is, the increase of the AuNP aggregation degree, the absorption peaks of AuNP solution at 530 nm decrease, while the corresponding absorption peaks at 730 nm gradually increase. The spectral changes reached a stable state after 60 minutes, indicating that the reaction was complete, which was consistent with TEM analysis. The color-changing AuNP colorimetric assay does not require the removal of unconnected AuNPs, effectively avoiding high background color caused by residual AuNPs in the reaction solution, and is expected to have higher sensitivity.To evaluate the specificity of AuNP colorimetric assay for m5C-miRNA-21, miRNA-21 and other methylated/unmethylated miRNAs were also examined. Fig. 2D shows that AuNP solution exhibits a distinct blue color when m5C-miRNA-21 is present, which is consistent with a distinct absorption peak at 730 nm in the UV-vis spectra (Fig. 2E). In contrast, the control sample showed no visually distinguishable color change, and the UV-vis spectra showed that the absorptions were still mainly around 530 nm. The ability of AuNP colorimetric assay to detect different concentrations of m5C-miRNA-21 was verified. As depicted in Fig. 2F and G, within the detection range of 1 pM to 100 nM, the absorption peaks around 530 nm in the UV-vis spectra gradually weaken and new absorption peaks emerge at 730 nm with the increasing concentration of m5C-miRNA-21. The color analysis of the AuNP solution demonstrated a strong concentration correlation within the range of 1 pM to 100 nM.
Furthermore, we employed this method to detect cells with different expression levels of miRNA-21. As shown in Fig. 2H, significant color changes in the AuNP solutions were observed in cell lysates with high (MCF-7) and medium (HCT116) expression levels of miRNA-21, which are also evidenced by UV-vis spectra (Fig. 2I). However, the detection results of the lysate of cells with low miRNA-21 expression levels (MCF-10A) showed only slight variations from the control samples. Analysis of the results showed that the expression levels of m5C-miRNA-21 in these three cell lines were positively correlated with the expression levels of miRNA-21 in the corresponding cells.41,42 In order to further verify the colorimetric assay, m5C-miRNA-21 of three cell lines was simultaneously detected using the ELISA methylation kit as a control. The detection accuracy of the ELISA methylation kit was first verified using the synthesized m5C-miRNA-21 standard at different concentrations (Fig. S4†). In addition, because the ELISA methylation kit does not have the specificity and sensitivity to detect miRNA-21 in actual samples, we improved the ELISA methylation kit using the miRNA-21 extraction strategy in this paper (improved ELISA) to detect m5C-miRNA-21 in three cell lines. As shown in Fig. 2H, the results of colorimetric assay were consistent with the improved ELISA.
The LFD assay for m5C-miRNA-21
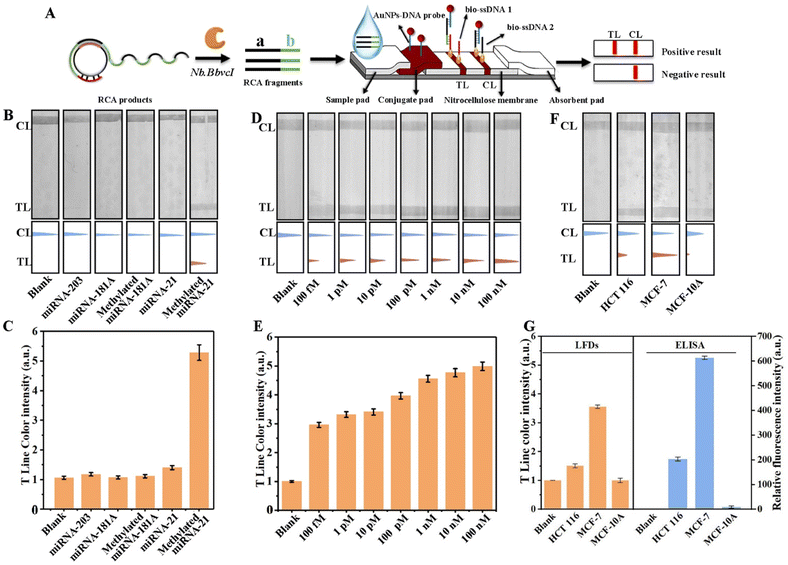
As a common, inexpensive and standardized detection method, LFDs have been widely used in point-of-care testing. They can quickly detect the presence or absence of target analytes without the need for professional personnel or laboratory capacity. Therefore, in order to further improve the practicability of m5C-miRNA-21 detection in this paper, portable LFDs based on DNAzyme-RCA were developed.To meet the needs of LFD assay, the RCA products were initially engineered to be cleaved by Nb.BbvcI, resulting in the generation of RCA fragments with region a and region b (Fig. 3A). Subsequently, in order to realize the identification of RCA fragments, AuNPs–DNA probes that complement with the region a of the RCA fragments are placed on the conjugate pad. At the same time, TL and control line (CL) were pre-modified with biotinylated single-stranded DNA 1 (bio-ssDNA 1) and single-stranded DNA 2 (bio-ssDNA 2), which were complementary to the region b of the RCA fragments and the AuNPs–DNA probes, respectively, and were used to fix the RCA fragment/AuNPs–DNA complexes and the blank AuNPs–DNA probes, respectively. The sequences of all nucleic acid probes have been presented in the ESI Table S1.† In a typical analysis, the complex solution is dropped onto the sample pad and flows to the conjugate pad, where the cleaved RCA fragments are bound to the AuNPs–DNA probes by nucleic acid hybridization. After hybridization, these RCA fragment/AuNPs–DNA complexes flow through the TL and bind to it by pre-modified bio-ssDNA 1. At the same time, excess AuNPs–DNA probes continued to flow and bind to pre-modified bio-ssDNA 2 on CL. Finally, due to the accumulation of AuNPs, the colored bands are visible with the naked eye. To more accurately compare the differences among the samples, the colored bands on the TL are usually quantified. Specifically, the original strip photos are first converted into gray images using ImageJ software, and the gray values of TL are read. Then the ratios of gray values of the sample to blank control are used as LFD assay results.
To optimize the performance of LFDs, four common running buffers (PBS, SSC, DEPC–water, and tris–HCl) were tested, and Fig. S2† shows that 2× SSC exhibited the highest TL/CL ratio, indicating its suitability for detection. m5C-miRNA-21, miRNA-21, and other methylated or unmethylated miRNA were used to evaluate the specificity of the LFDs. As shown in Fig. 3B, TL bands appeared only when m5C-miRNA-21 was present, and no obvious TL bands were found according to blank control and other miRNAs. The CL bands for all detection targets were evident, confirming the normal function of LFDs. Besides, six repeated tests of m5C-miRNA-21 at a concentration of 10 pM were performed, and the consistent, clear bands in Fig. S3A† exhibit the reproducibility of LFD assay. In addition, stability testing at different temperatures (4 °C, 25 °C, and 37 °C) revealed that even after 14 days at room temperature, LFDs maintained their ability to detect m5C-miRNA-21, highlighting their remarkable stability (Fig. S3B†).
The ability of LFD assay for different concentrations of m5C-miRNA-21 was also verified. As shown in Fig. 3D, in the detection range from 1 pM to 100 nM, the TL bands gradually become stronger with the increase of the concentration of m5C-miRNA-21, indicating that the LFDs have good detection capability. The grayscale analysis of TL bands (Fig. 3E) also confirms the detection results. To verify the stability of LFDs for m5C-miRNA-21 detection in a biological system, lysates from MCF-10A, MCF-7, and HCT 116 cells were examined. As shown in Fig. 3F, no obvious TL bands corresponding to m5C-miRNA-21 were observed in MCF-10A lysate, while obvious TL bands were found in lysates of MCF-7 and HCT 116 cells. This difference in m5C-miRNA-21 levels across different cell types aligns with colorimetric measurements. To verify the accuracy of the LFD assay, m5C-miRNA-21 in three cell lines was detected simultaneously using the improved ELISA. As shown in Fig. 3G, the results of LFD assay and the improved ELISA strategy were consistent. In addition, we compared colorimetric and LFD assays in this work with exosomal miRNA detection methods reported in the literature. As shown in Table S2,† colorimetric and LFD assays maintain good sensitivity despite the addition of methylation recognition design.
The point-of-care testing of exosomal m5C-miRNA-21 from clinical blood samples
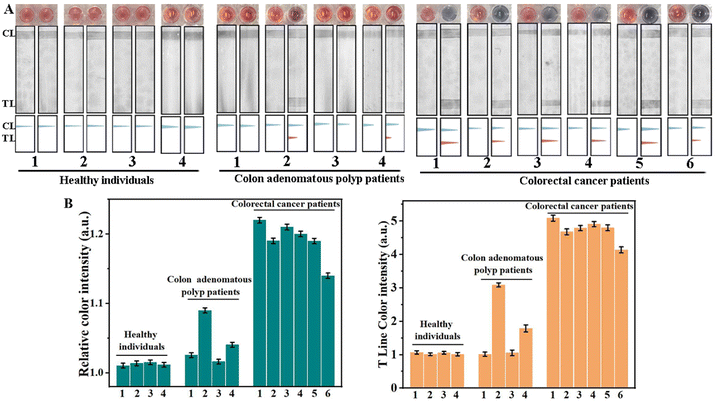
Exosomes in blood derived from colorectal cancer patients were obtained by ultracentrifugation, which were then analyzed using TEM and dynamic light scattering (DLS). As shown in Fig. S5A,† these exosomes presented a typical cup-shaped morphology under a TEM. Additionally, DLS revealed a standard size-distribution profile for the native exosomes, with an average size of 122 nm (Fig. S5B†), aligning with previous reports.43 The exosomes were then broken using the TRIzol reagent, and the resulting solutions were used for AuNP colorimetric and LFD assays respectively. We performed quantitative analysis of exosomal miRNA-21 by the qRT-PCR method (Fig. S5C†), and the concentration of exosomal miRNA-21 in 1 mL of serum calculated from the test result is 0.191 nM (Fig. S5D†). The concentration determined is in line with that reported in the literature.44 This indicated that we have successfully extracted exosomal miRNAs from blood samples. The stability of exosome extraction by ultracentrifugation is the prerequisite for the detection of exosomal miRNA. We used BCA protein concentration assay to detect exosomes extracted from the same cell culture medium. As shown in Fig. S6,† there was little difference in exosomal protein content from three identical cell culture media, indicating stable extraction efficiency of exosomes.We examined exosomal m5C-miRNA-21 in the blood of six colorectal cancer patients, four colon adenomatous polyp patients and four healthy donors, with SSC as the control sample for each assay. Clinicopathological characteristics of all patients and healthy individuals are shown in Table S3.† As shown in Fig. 4, the colorimetric and LFD results of the four healthy donors showed no significant difference compared to those of the control samples. The color intensity analysis of the colorimetric solution and TL bands using ImageJ showed that no m5C-miRNA-21 was detected. Correspondingly, in samples from six clinical colorectal cancer patients, both colorimetric and LFD results indicated the presence of m5C-miRNA-21. In addition, the detection results of different patients showed significant individual heterogeneity in the content of exosomal m5C-miRNA-21 in blood samples, and this individual heterogeneity showed a good correlation between AuNP colorimetric assay and LFD assay. We compared the test results with the clinical stage of colorectal cancer patients and found no statistically significant correlation. Notably, we detected the presence of m5C-miRNA-21 in colon adenomatous polyp patients 2 and 4, although the color intensities in AuNP colorimetric and LFD assays were weaker than that in colorectal cancer patients. According to relevant literature, approximately 80% to 95% of colorectal cancers develop from polyp; colon adenomatous polyps are considered a significant precancerous lesion.45,46 This implies that the scheme may be capable of identifying cancer transformation in advance. To further validate this, we will need to continue monitoring more similar patients in the future. Nevertheless, the AuNP colorimetric and LFD assays based on DNAzyme-RCA developed in this paper still proved to be a point-of-care testing with good specificity and high sensitivity.
Conclusions
In summary, a DNAzyme-RCA-based AuNP colorimetric assay was developed for the first point-of-care testing (POCT) of exosomal m5C-miRNA-21. This assay achieved specific recognition and linear amplification of m5C-miRNA-21 through the DNAzyme triggered RCA reaction, and achieved low background interference color output through RCA reaction induced AuNP aggregation. The lowest concentration of m5C-miRNA-21 visible to the naked eye can reach 1 pM. Furthermore, the DNAzyme-RCA method was developed for LFD assay. The results showed that LFDs could recognize m5C-miRNA-21 with high specificity, and the lowest concentration we could detect was as low as 0.1 pM. The DNAzyme-RCA-based AuNP colorimetric and LFD assays were both used to detect exosomal m5C-miRNA-21 in blood samples derived from colorectal cancer patients. The results showed that the expression level of m5C-miRNA-21 in clinical colorectal cancer patients was significantly higher than that in healthy donors. The AuNP colorimetric and LFD assays based on the DNAzyme-RCA method in this paper are expected to provide new POCT products for colorectal cancer.Ethical statement
All experiments were approved by the ethics committee of the First Affiliated Hospital of Shandong First Medical University & Shandong Provincial Qianfoshan Hospital. The ethical approval number was YXLL-KY2022(039). All methods were performed in accordance with the Declaration of Helsinki and the relevant guidelines and regulations. The admission certificate signed indicates that medical information, biological specimens and examination results of patients may be used for scientific research, and personal identity and privacy will not be disclosed in the research achievement.Data availability
Detailed materials list, experimental procedures and methods, additional data mentioned in the text including Fig. S1–S6 and Tables S1–S3,† and additional references. The raw data is available upon request from the corresponding author.Author contributions
B. Tang, L. Li, H. Si and Y. Tang proposed the idea, designed the experiments, and revised the manuscript. H. Zhang performed experimental work and wrote the manuscript. Y. Zhou helped to perform part of the additional experiment. All authors contributed to the analysis and discussion of the results. All authors contributed to the manuscript descriptions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22134004), Natural Science Foundation of Shandong Province of China (ZR2022MB045 and ZR2023MB050), Taishan Scholars Program of Shandong Province (tsqn.201909077), and Major Science and Technology Innovation Project of Shandong Province (2022SFXGFY01 and 2021ZDSYS09).Notes and references
- J. A. Vidigal and A. Ventura, Trends Cell Biol., 2015, 25, 137–147 CrossRef CAS PubMed
.
- F. Ingenito, G. Roscigno, A. Affinito, S. Nuzzo, I. Scognamiglio, C. Quintavalle and G. Condorelli, Int. J. Mol. Sci., 2019, 20, 4687 CrossRef CAS PubMed
.
- D. Zheng, M. Huo, B. Li, W. Wang, H. Piao, Y. Wang, Z. Zhu, D. Li, T. Wang and K. Liu, Front. Cell Dev. Biol., 2020, 8, 616161 CrossRef PubMed
.
- C. Z. J. Lim, A. Natalia, N. R. Sundah and H. Shao, Adv. Biosyst., 2020, 4, 1900309 CrossRef PubMed
.
- K. I. Kozaki and J. Inazawa, Cancer Sci., 2012, 103, 837–845 CrossRef CAS PubMed
.
- I. Heo, C. Joo, J. Cho, M. Ha, J. Han and V. N. Kim, Mol. Cell, 2008, 32, 276–284 CrossRef CAS PubMed
.
- T. Katoh, Y. Sakaguchi, K. Miyauchi, T. Suzuki, S. Kashiwabara and T. Baba, Genes Dev., 2009, 23, 433–438 CrossRef CAS PubMed
.
- Y. Kawahara, B. Zinshteyn, T. P. Chendrimada, R. Shiekhattar and K. Nishikura, EMBO Rep., 2007, 8, 763–769 CrossRef CAS PubMed
.
- D. Dominissini, S. M. Moshkovitz, S. Schwartz, M. S. Divon, L. Ungar, S. Osenberg, K. Cesarkas, J. J. Hirsch, N. Amariglio, M. Kupiec, R. Sorek and G. Rechavi, Nature, 2012, 485, 201–206 CrossRef CAS PubMed
.
- M. Schaefer, Methods Enzymol., 2015, 560, 297–329 CAS
.
- Q. Dai, S. M. Moshkovitz, D. Han, N. Kol, N. Amariglio, G. Rechavi, D. Dominissini and C. He, Nat. Methods, 2017, 14, 695–698 CrossRef CAS PubMed
.
- L. Pandolfini, I. Barbieri, A. J. Bannister, A. Hendrick, B. Andrews, N. Webster, P. Murat, P. Mach, R. Brandi, S. C. Robson, V. Migliori, A. Alendar, M. d'Onofrio, S. Balasubramanian and T. Kouzarideset, Mol. Cell, 2019, 74, 1278–1290 CrossRef CAS PubMed
.
- M. Shivakumar, S. Han, Y. Lee and D. Kim, BMC Genomics, 2021, 22, 1–12 CrossRef PubMed
.
- J. Liu and K. Jiang, J. Cell. Mol. Med., 2022, 26, 2505–2519 CrossRef CAS PubMed
.
- J. Briand, A. A. Sérandour, A. Nadaradjane, G. Bougras-Cartron, D. Heymann, B. Ory, F. M. Vallette and P. F. Cartron, Mol. Ther.--Nucleic Acids, 2020, 22, 72–83 CrossRef CAS PubMed
.
- Y. Toiyama, Y. Okugawa, K. Tanaka, T. Araki, K. Uchida, A. Hishida, M. Uchino, H. Ikeuchi, S. Hirota, M. Kusunoki, C. R. Boland and A. Goel, Gastroenterology, 2017, 153, 1634–1646 CrossRef CAS PubMed
.
- B. Lin, T. Tian, Y. Lu, D. Liu, M. Huang, L. Zhu, Z. Zhu, Y. Song and C. Yang, Angew. Chem., Int. Ed., 2021, 60, 7582–7586 CrossRef CAS PubMed
.
- Y. Cao, X. Yu, T. Zeng, Z. Fu, Y. Zhao, B. Nie, J. Zhao, Y. Yin and G. Li, J. Am. Chem. Soc., 2022, 144, 13475–13486 CrossRef CAS PubMed
.
- R. Reshke, J. A. Taylor, A. Savard, H. Guo, L. H. Rhym, P. S. Kowalski, M. T. Trung, C. Campbell, W. Little, D. G. Anderson and D. Gibbings, Nat. Biomed. Eng., 2020, 4, 52–68 CrossRef CAS PubMed
.
- J. U. Lee, W. H. Kim, H. S. Lee, K. H. Park and S. J. Sim, Small, 2019, 15, 1804968 CrossRef PubMed
.
- H. Valadi, K. Ekström, A. Bossios, M. Sjöstrand, J. J. Lee and J. O. Lötvall, Nat. Cell Biol., 2007, 9, 654–659 CrossRef CAS PubMed
.
- A. Thind and C. Wilson, J. Extracell. Vesicles, 2016, 5, 31292 CrossRef PubMed
.
- L. X. Zhang, W. Z. Li, R. Parvin, X. C. Wang, Q. H. Fan and F. F. Ye, Adv. Funct. Mater., 2022, 32, 2207879 CrossRef CAS
.
- C. R. Alarcón, H. Lee, H. Goodarzi, N. Halberg and S. F. Tavazoie, Nature, 2015, 519, 482–485 CrossRef PubMed
.
- J. L. Zhang, R. H. Bai, M. Li, H. L. Ye, C. Wu, C. F. Wang, S. P. Li, L. P. Tan, D. M. Mai, G. L. Li, L. Pan, Y. F. Zheng, J. C. Su, Y. Ye, Z. Q. Fu, S. Y. Zheng, Z. X. Zuo, Z. X. Liu, Q. Zhao, X. Che, D. Xie, W. H. Jia, M. S. Zeng, W. Tan, R. F. Chen, R. H. Xu, J. Zheng and D. X. Lin, Nat. Commun., 2019, 10, 1858 CrossRef PubMed
.
- M. Cheray, A. Etcheverry, C. Jacques, R. Pacaud, G. Bougras-Cartron, M. Aubry, F. Denoual, P. Peterlongo, A. Nadaradjane, J. Briand, F. Akcha, D. Heymann, F. M. Vallette, J. Mosser, B. Ory and P. F. Cartron, Mol. Cancer, 2020, 19, 36–51 CrossRef CAS PubMed
.
- T. Ohshiro, M. Konno, A. Asai, Y. Komoto, A. Yamagata, Y. Doki, H. Eguchi, K. Ofusa, M. Taniguchi and H. Ishii, Sci. Rep., 2021, 11, 19304 CrossRef CAS PubMed
.
- E. Xiong, L. Jiang, T. Tian, M. L. Hu, H. H. Yue, M. Q. Huang, W. Lin, Y. Z. Jiang, D. B. Zhu and X. M. Zhou, Angew. Chem., Int. Ed., 2021, 60, 5307–5315 CrossRef CAS PubMed
.
- R. Pandey, D. Chang, M. Smieja, T. Hoare, Y. F. Li and L. Soleymanii, Nat. Chem., 2021, 13, 895–901 CrossRef CAS PubMed
.
- M. M. Ali, R. Silva, D. White, S. Mohammadi, Y. F. Li, A. Capretta and J. D. Brennan, Angew. Chem., Int. Ed., 2022, 134, e202112346 CrossRef
.
- M. M. Ali, M. Wolfe, K. Tram, J. Gu, C. D. M. Filipe, Y.
F. Li and J. D. Brennan, Angew. Chem., Int. Ed., 2019, 131, 10012–10016 CrossRef
.
- Y. Zhou, H. Ren, B. Dai, J. Li, L. Shang, J. Huang and X. Shi, J. Exp. Clin. Cancer Res., 2018, 37, 1–18 CrossRef PubMed
.
- W. Li, X. Huang and D. Bi, Arch. Med. Sci., 2022, 18, 406 CAS
.
- M. Konno, J. Koseki, A. Asai, A. Yamagata, T. Shimamura, D. Motooka, D. Okuzaki, K. Kawamoto, T. Mizushima, H. Eguchi, S. Takiguchi, T. Satoh, K. Mimori, T. Ochiya, Y. Doki, K. Ofusa, M. Mori and H. Ishii, Nat. Commun., 2019, 10, 3888 CrossRef PubMed
.
- C. P. M. Scheitl, M. M. Ghaem, A. K. Lenz and C. Höbartner, Nature, 2020, 587, 663–667 CrossRef CAS PubMed
.
- Q. Q. Hu, Z. X. Tong, A. Yalikong, L. P. Ge, Q. Shi, X. Y. Du, P. Wang, X. Y. Liu, W. Q. Zhan, X. Gao, D. Sun, T. Fu, D. Ye, C. H. Fan, J. Liu, Y. S. Zhong, Y. Z. Jiang and H. Z. Gu, Nat. Chem., 2024, 16, 122–131 CrossRef CAS PubMed
.
- A. Liaqat, M. V. Sednev, C. Stiller and C. Höbartner, Angew. Chem., Int. Ed., 2021, 133, 19206–19210 CrossRef
.
- X. H. Huang, S. Neretina and M. A. El-Sayed, Adv. Mater., 2009, 21, 4880–4910 CrossRef CAS PubMed
.
- R. A. Reynolds, C. A. Mirkin and R. L. Letsinger, J. Am. Chem. Soc., 2000, 122, 3795–3796 CrossRef CAS
.
- J. Y. Yue, L. P. Song, Y. T. Wang, P. Yang, Y. Ma and B. Tang, Anal. Chem., 2022, 94, 14419–14425 CrossRef CAS PubMed
.
- A. Qu, M. Sun, L. Xu, C. Hao, X. Wu, C. Xu, N. A. Kotov and H. Kuang, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 3391–3400 CrossRef CAS PubMed
.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435, 834–838 CrossRef CAS PubMed
.
- M. Esteves, R. Abreu, H. Fernandes, C. Serra-Almeida, P. A. Martins, M. Barão, A. C. Cristóvão, C. Saraiva, R. Ferreira and L. Ferreira, Mol. Ther., 2022, 30, 3176–3192 CrossRef CAS PubMed
.
- F. Causa, A. Aliberti, A. M. Cusano, E. Battista and P. A. Netti, J. Am. Chem. Soc., 2015, 137, 1758–1761 CrossRef CAS PubMed
.
- F. A. Sinicrope, N. Engl. J. Med., 2022, 386, 1547–1558 CrossRef CAS PubMed
.
- N. Keum and E. Giovannucci, Nat. Rev. Gastroenterol. Hepatol., 2019, 16, 713–732 CrossRef PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc02648a |
| ‡ Co-first author with the equally contribution. |
| This journal is © The Royal Society of Chemistry 2024 |