Open Access Article
Open Access ArticleA high-fidelity DNAzyme-assisted CRISPR/Cas13a system with single-nucleotide resolved specificity†
Yunping
Wu
,
Ruigang
Jin
,
Yangyang
Chang
and
Meng
Liu
 *
*
School of Environmental Science and Technology, Key Laboratory of Industrial Ecology and Environmental Engineering (Ministry of Education), Dalian University of Technology, Dalian POCT Laboratory, Dalian, 116024, China. E-mail: mliu@dlut.edu.cn
First published on 2nd April 2024
Abstract
A CRISPR/Cas system represents an innovative tool for developing a new-generation biosensing and diagnostic strategy. However, the off-target issue (i.e., mistaken cleavage of nucleic acid targets and reporters) remains a great challenge for its practical applications. We hypothesize that this issue can be overcome by taking advantage of the site-specific cleavage ability of RNA-cleaving DNAzymes. To test this idea, we propose a DNAzyme Operation Enhances the Specificity of CRISPR/Cas13a strategy (termed DOES-CRISPR) to overcome the problem of relatively poor specificity that is typical of the traditional CRISPR/Cas13a system. The key to the design is that the partial hybridization of the CRISPR RNA (crRNA) with the cleavage fragment of off-target RNA was not able to activate the collateral cleavage activity of Cas13a. We showed that DOES-CRISPR can significantly improve the specificity of traditional CRISPR/Cas13a-based molecular detection by up to ∼43-fold. The broad utility of the strategy is illustrated through engineering three different systems for the detection of microRNAs (miR-17 and let-7e), CYP2C19*17 gene, SARS-Cov-2 variants (Gamma, Delta, and Omicron) and Omicron subtypes (BQ.1 and XBB.1) with single-nucleotide resolved specificity. Finally, clinical evaluation of this assay using 10 patient blood samples demonstrated a clinical sensitivity of 100% and specificity of 100% for genotyping CYP2C19*17, and analyzing 20 throat swab samples provided a diagnostic sensitivity of 95% and specificity of 100% for Omicron detection, and a clinical sensitivity of 92% and specificity of 100% for XBB.1 detection.
Introduction
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) protein system refers to an immune response system originating from archaea and bacteria, which is composed of a Cas effector and CRISPR RNA (crRNA).1–3 Cas12a (type V) and Cas13a (type VI) exhibit the target-activated nonspecific endonuclease activity of single-stranded DNA (ssDNA and Cas12a) or RNA (ssRNA and Cas13a).4–6 Furthermore, the active Cas12a and Cas13a enable the detection of nucleic acid in one system with pM-level sensitivity and rapid response time.7–10 These properties make them ideal candidates for wide-ranging applications in biosensing, clinical diagnosis, and environmental monitoring.11–17 However, the off-target issue (i.e., mistaken cleavage of nucleic acid targets and reporters) remains a great challenge for their practical applications.18,19 This is due largely to the fact that the Cas12a effector is tolerable to mismatches in the protospacer-adjacent motif (PAM)-distal region of the target double-stranded DNA (dsDNA), and exhibits no specificity for ssDNA.4,20 In addition, Cas13a is incapable of discriminating a single-base difference in target RNA.6There are two common approaches for improving the specificity of Cas12a and Cas13a nucleases. One is to engineer crRNA, including the introduction of synthetic mismatch into the spacer domain of crRNA, designing hairpin-spacer crRNA, and modifying crRNA with 2′-O-methyl.21–25 However, the number and location of mismatches in crRNA have to be carefully designed to reduce the off-target effect without sacrificing the cleavage activity of Cas proteins.22,23 Furthermore, the use of hairpin-spacer crRNA and 2′-O-methyl modified crRNA only improves the specificity of the original CRISPR/Cas system by 2- to 3-fold.24,25 The other approach is engineering high-fidelity Cas proteins.26–28 It remains challenging to work with because of the sophisticated protein expression and screening process. Moreover, all these strategies are aimed at optimizing different components of the CRISPR/Cas system without overcoming the fundamental trade-off between the cleavage efficiency and the specificity. Thus, strategies that can significantly improve the specificity are highly desirable for their practical applications, such as biosensing, as they will avoid false positive results.
DNAzymes (also known as deoxyribozymes, DNA enzymes, or catalytic DNAs), are single-stranded DNA molecules with catalytic capabilities that are isolated from random-sequence DNA pools by in vitro selection.29–33 As we will show for the first time in this work, DNAzyme operation can be uniquely exploited to enhance the specificity of the CRISPR/Cas13a system. We term this approach DOES-CRISPR. More specifically, we employ RNA-cleaving DNAzymes (DZs) that precisely cleave the desired off-target RNA that contains any purine–pyrimidine junction. Because Cas13a needs higher complementarity between the crRNAs and the target RNA before it is catalytically activated, this site-specific cleavage of RNA in turn would efficiently block the off-target activity of Cas13a, thus resulting in an increase in specificity. We showed that DOES-CRISPR can significantly improve the specificity of CRISPR/Cas13a-based molecular detection by up to ∼43-fold. Furthermore, we demonstrated the versatility of DOES-CRISPR for the detection of microRNAs (miRNA17 and let7e), CYP2C19*17 gene, SARS-Cov-2 variants (Gamma, Delta, and Omicron), and Omicron subtypes (BQ.1 and XBB.1) with single-nucleotide resolved specificity. Finally, clinical evaluation of this assay using blood samples and throat swab samples suggested the promise of DOES-CRISPR for clinical use.
Results and discussion
Engineering the DOES-CRISPR system
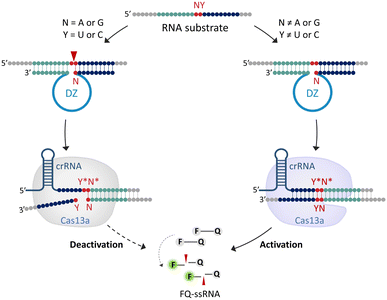
The system features two key components (Fig. 1): (1) 10-23DZs for their ability to efficiently cleave any NY junction in a given RNA substrate (denoted as RS-NY), where N denotes a purine (A or G) and Y is a pyrimidine (U or C); (2) CRISPR/Cas13a for its ability to trigger non-specific collateral cleavage of fluorogenic FQ-ssRNA reporters upon RNA recognition. For the off-target RNA that contains a purine–pyrimidine junction, cleavage occurs after an unpaired purine nucleotide of the RNA that was flanked by oligonucleotides complementary to 10-23DZ (17 or 18 nucleotides). Partial hybridization of the crRNA with the cleavage fragment was not able to activate the collateral cleavage activity of Cas13a. Therefore, the FQ-ssRNA reporters remain intact, resulting in a negligible fluorescence increase. However, on recognition of the full-length RNA target, the crRNA-complementary RNA triggered Cas13a to cleave the FQ-ssRNA reporter substrate. The cleavage of multiple reporters is accompanied by an increase in fluorescence intensity. Our approach, therefore, rationally integrates the site-specific cleavage ability of DZs with the target RNA-triggered collateral cleavage activity of CRISPR/Cas13a. This can therefore overcome the problem of relatively poor specificity that is typical of the traditional CRISPR/Cas13a system.Effect of DNAzyme-mediated off-target RNA cleavage on the specificity of the CRISPR/Cas13 system
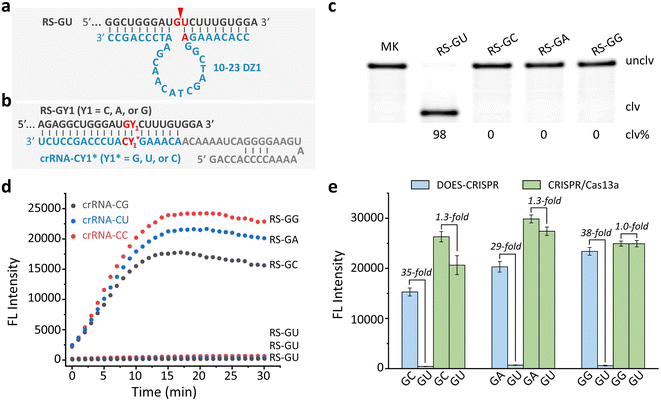
The well-known 10-23DZs have the ability to cleave RS-NY (RS = RNA substrate; N = A or G; Y = U or C), with robust activity for AU, GU and GC sites, but reduced activity for AC sites (Fig. S1).† We estimated that the observed rate constant (kobs) was ∼3 min−1 on the basis of the observation that nearly 90% of the RS-AU (Fig. S2a†), RS-GU (Fig. S2b†), or RS-GC (Fig. S2c†) was cleaved in 1 min. Two conditions must be met for successful DOES-CRISPR. First, 10-23DZs should specifically precisely cleave the desired off-target RNA. To confirm this, we first choose RS-GU as the off-target RNA and RS-GY1 (Y1 = C, A, or G) as the corresponding target RNA. 10-23DZ1 was used to cleave RS-GU (Fig. 2a), and crRNA-CY1* (Y1* = G, U, or C) was designed to bind RS-GY1 through standard Watson–Crick base-pairing (Fig. 2b). As expected, only the RS-GU was cleaved by 10-23DZ1 (98%, Fig. 2c), whereas the RS-GY1 remained intact.Second, CRISPR/Cas13a is activated by full-length RS-GY1, rather than the cleavage fragments of RS-GU to perform trans-cleavage of FQ-ssRNA reporters. An experiment was carried out to examine the time-dependent fluorescence changes of FQ-ssRNA in the presence of crRNA-CY1*/Cas13a, 10-23DZ1, and RS-GY1 (or RS-GU). As shown in Fig. 2d, RS-GY1 was indeed able to activate the crRNA-CY1*/Cas13a system, reflected by the fluorescence enhancement over time. However, no increase in fluorescence was observed when each DOES-CRISPR reaction was tested with off-target RS-GU. We determined the signaling enhancements (i.e., S/B, defined as the fluorescence intensity in the presence of RS-GY1 compared to that in the presence of RS-GU) to be 35, 29, and 38-fold for RS-GC, RS-GA, and RS-GG, respectively (Fig. 2e). In contrast, the original CRISPR/Cas13a system provided an S/B value as high as 1.3-fold for RS-GC and RS-GA.
To demonstrate the general applicability of DOES-CRISPR, we designed the other two off-target RNAs, named RS-AU (Fig. S3a†) and RS-GC (Fig. S3b†), and their corresponding target RNAs, named RS-AY2 (Y2 = C, A, or G) and RS-GY3 (Y3 = U, A, or G). Once again, 10-23DZ1 and 10-23DZ2 were used to cleave RS-AU and RS-GC, respectively. Two crRNAs, denoted as crRNA-UY2* (Y2* = G, U, or C, Fig. S3c†) and crRNA-CY3* (Y3* = A, U, or C, Fig. S3d†) were designed to hybridize with targets RS-AY2 and RS-GY3, respectively. The successful DOES-CRISPR was confirmed experimentally: (1) more than 95% of off-target RNAs can be efficiently cleaved by 10-23DZs (Fig. S3e and f†); (2) DOES-CRISPR exhibited a maximum 14 and 47-fold S/B value for RS-AA (over RS-AU, Fig. S3g†) and RS-GG (over RS-GC, Fig. S3h†), respectively, in comparison with that of 1.6 and 1.3-fold for the CRISPR/Cas13a system. Taken together, these results demonstrated that DNAzyme-mediated cleavage of off-target RNA can robustly enhance the specificity of the CRISPR/Cas13a system.
DOES-CRISPR for microRNA detection
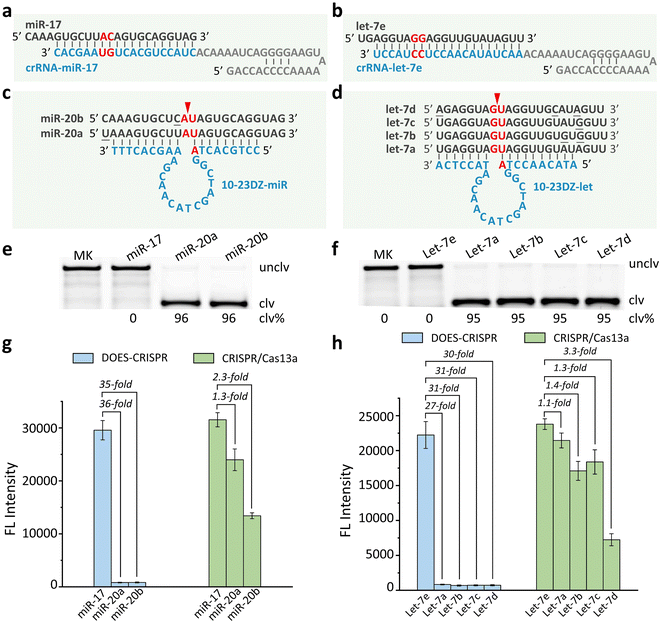
We next investigate the possibility of exploiting DOES-CRISPR for biosensing applications. MicroRNAs (miRNAs), as a class of endogenous non-coding small RNAs, have been regarded as biomarkers for disease diagnosis and cancer screening.34–38 We first choose miR-17 and let-7e as the model miRNA targets. crRNA-miR-17 and crRNA-let-7e were designed to guide Cas13a to bind the corresponding targets (Fig. 3a and b). Two off-target miRNA groups miR-20X (X = a or b) and let-7Z (Z = a, b, c, or d) were used for the specificity test (Fig. 3c and d). 10-23DZ-miR and 10-23DZ-let were able to catalyze the cleavage of AU and GU junctions within the off-targets miR-20X (96%, Fig. 3e) and let-7Z (95%, Fig. 3f), respectively. Note that 10-23DZs can tolerate several mismatches in each of the substrate-binding arms within the 10-23DZ-miR/miR-20X or 10-23DZ-let/let-7Z complex.We performed real-time fluorescence monitoring of the cleavage of FQ-ssRNA reporters by DOES-CRISPR for miRNA detection (Fig. S4†). In the presence of miR-17 (or let-7e), fluorescence intensity increased gradually with reaction time, indicating that miR-17 (or let-7e) can indeed initiate DOES-CRISPR. The limit of detection (LOD, defined as 3σ, σ = standard deviation of the blank samples) for miR-17 and let-7e detection was determined to be 5 pM (Fig. S5a†) and 10 pM (Fig. S5b†), respectively. Besides the high sensitivity, each system also exhibited excellent selectivity for its cog-nate target. No increase in fluorescence was observed when testing with miR-20X and let-7Z. We found that DOES-CRISPR activated by miR-17 reached ∼35-fold improvement compared to miR-20X (Fig. 3g). Our assay also provides a 27- to 31-fold improvement for let-7e compared to the related off-targets let-7Z (Fig. 3h). In contrast, CRISPR/Cas13a was less specific, and displayed only 1.1–3.3-fold improvement for target miRNAs versus off-target miRNAs.
DOES-CRISPR for CYP2C19*17 gene detection
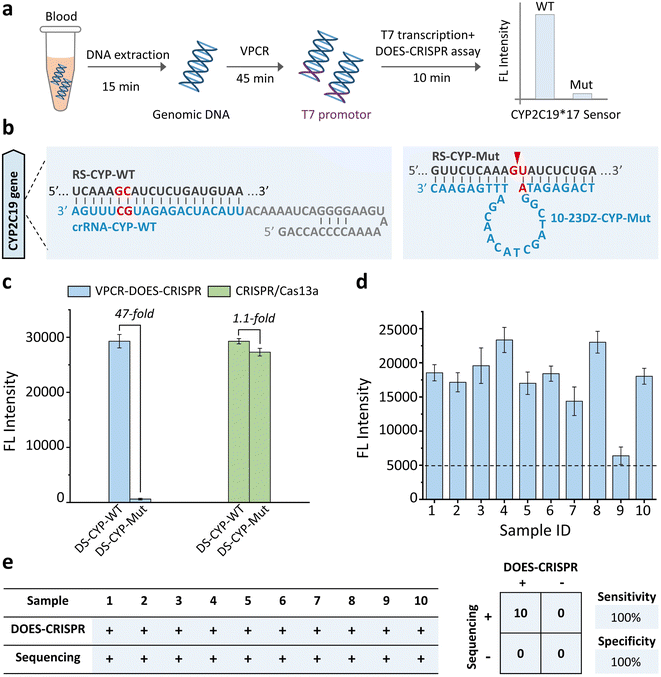
We next investigated whether our DOES-CRISPR assay is capable of genotyping DNA polymorphisms. The CYP2C19*17 gene was used as a DNA target. The polymorphism of the CYP2C19*17 gene (C806T rs12248560) is associated with the ultra-rapid metabolism of clopidogrel, which can increase the inhibitory effect on platelets and the bleeding risk.39 Therefore, specific genotyping of the CYP2C19*17 gene is critical for clopidogrel individualized medication. The overall operation of DOES-CRISPR includes (Fig. 4a) (1) DNA extraction (15 min), (2) V-shape polymerase chain reaction (VPCR) (45 min), and (3) T7 transcription and DOES-CRISPR in one-step reaction (10 min). The wild-type and mutant-type DNA sequences of the CYP2C19*17 gene were denoted as DS-CYP-WT and DS-CYP-Mut, respectively. As shown in Fig. 4b, crRNA-CYP-WT was designed to recognize the corresponding RNA of DS-CYP-WT (named RS-CYP-WT), and 10-23DZ-CYP-Mut was used to cleave GU dinucleotides within the RNA of DS-CYP-Mut (named RS-CYP-Mut). As expected, the immediate cleavage of RS-CYP-Mut (Fig. S6† inset) in turn completely arrested the CRISPR/Cas13a reaction, as no fluorescence enhancement was observed. In sharp contrast, the presence of DS-CYP-WT could generate the corresponding RS-CYP-WT that activates Cas13a for collateral-cleavage activity to produce fluorescence signals (Fig. S6†). Using VPCR-DOES-CRISPR, we can detect DS-CYP-WT at a concentration as low as 10 copies per μL (Fig. S7†). Furthermore, VPCR-DOES-CRISPR exhibited ∼47-fold selectivity for DS-CYP-WT versus DS-CYP-Mut (Fig. 4c).We then evaluate the feasibility of VPCR-DOES-CRISPR toward a set of 10 clinical whole blood samples. The genomic DNA in blood samples was first extracted and amplified by VPCR (Fig. S8†). Using the commonly used clinical threshold of 10 copies per μL to decide the genotype of CYP2C19*17 (Fig. 4d), we achieved a clinical sensitivity of 100% and specificity of 100%. All these 10 blood samples were categorized as wide-type CYP2C19*17, which is consistent with the traditional Sanger sequencing result (Fig. S9† and 4e).
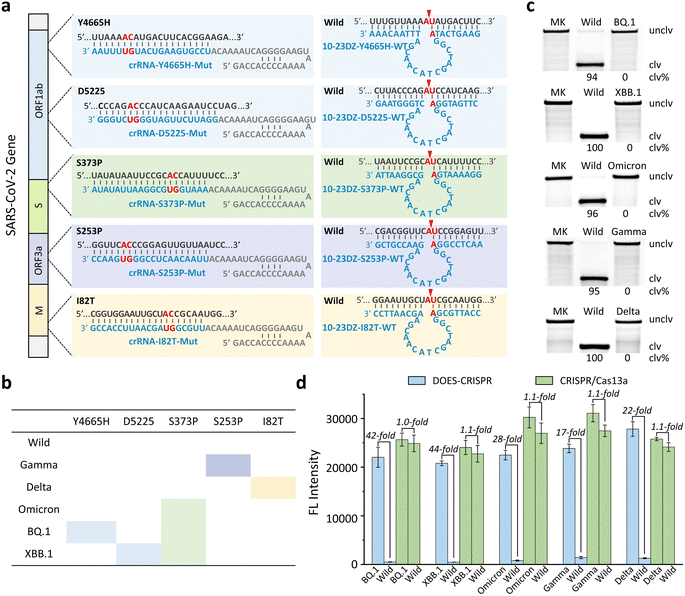
DOES-CRISPR for SARS-CoV-2 variant and subvariant detection
We next challenged our DOES-CRISPR to detect various RNA mutations in the SARS-CoV-2 genome. SARS-CoV-2, a ∼30 kb betacoronavirus, has caused a global pandemic of coronavirus disease.40 In addition, the emergence of SARS-CoV-2 variants and subvariants increased the risk of immune escape, virus transmission, and vaccine ineffectiveness.41–44 The Delta, Gamma, and Omicron variants, listed as variants of concern of SARS-CoV-2, are dominant worldwide.45–47 Furthermore, XBB.1 and BQ.1, as the prevalent subvariants of the Omicron variant, have been recognized as the variants of interest of SARS-CoV-2.48–50 Therefore, it is urgent to develop a highly specific method to genotype these variants and subvariants. Here, five critical single-nucleotide mutations (SNM), including Y4665H, D5225, S373P, S253P, and I82T, were selected as the RNA targets (Fig. 5a). S253P, I82T, and S373P mutations can be used to distinguish the Gamma, Delta, and Omicron variants, respectively. The BQ.1 and XBB.1 subvariants of Omicron can be further identified by using the Y4665H and D5225 mutations, respectively (Fig. 5b).A set of crRNAs, named crRNA-Y4665H-Mut, crRNA-D5225-Mut, crRNA-S373P-Mut, crRNA-S253P-Mut, and crRNA-I82T-Mut, were designed to target each RNA mutation. Five specific 10-23DZs were programmed to recognize and catalyze the cleavage of AU dinucleotides in the corresponding wild-type RNA of SARS-CoV-2, producing the maximum cleavage yields ranging from 94% to 100% (Fig. 5c). We measured the fluorescence responses of DOES-CRISPR and the traditional CRISPR/Cas13a system toward mutated RNAs and their corresponding wild-type RNAs. The LOD of DOES-CRISPR for Gamma (Fig. S10†) and Delta (Fig. S11†) detection is 25 pM, and for Omicron (Fig. S12†), BQ.1 (Fig. S13†), and XBB.1 (Fig. S14†) detection is 10 pM, without the preamplification step. Moreover, DOES-CRISPR exhibited 17- to 44-fold improvement for mutated versus wide-type RNAs (Fig. 5d). All together, these results suggested that our assay has the ability to identify single-nucleotide RNA mutations in the SARS-CoV-2 genome. Our DOES-CRISPR strategy offers higher sensitivity and shorter sample-to-answer time compared with traditional CRISPR-based detection methods for nucleic acid targets (Table S2).†
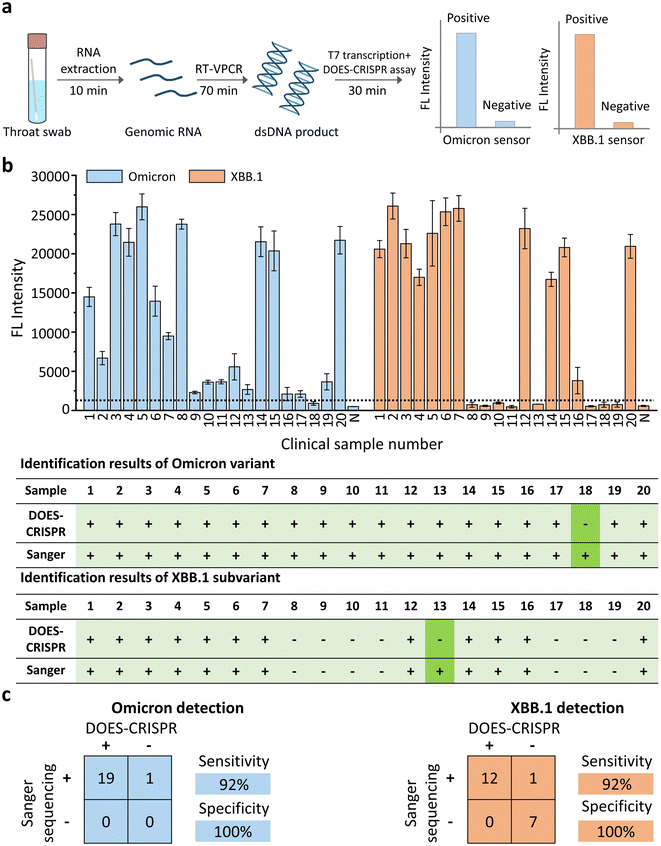
DOES-CRISPR for Omicron variant and XBB.1 subvariant detection in clinical throat swab samples
Finally, we investigated the feasibility of DOES-CRISPR assay for the detection of the Omicron variant and its subvariant XBB.1 using clinical throat swab samples. A total of 20 throat swab samples were collected from persons suspected of SARS-CoV-2 infection. 13 samples were confirmed Omicron-positive and XBB.1-positive, and 7 samples were confirmed Omicron-positive and XBB.1-negative by using the Sanger sequencing results (Fig. S15†). These samples were then detected in parallel by the DOES-CRISPR assay. The overall procedure includes (Fig. 6a) (1) genomic RNA extraction from throat swabs (10 min), (2) reverse-transcription and VPCR (70 min), and (3) T7 transcription and the DOES-CRISPR reaction (30 min). At a signal threshold of 2-fold measured over the background fluorescence signal, we were able to correctly categorize 12 Omicron-positive and XBB.1-positive samples and 7 Omicron-positive and XBB.1-negative samples (Fig. 6b). Therefore, DOES-CRISPR demonstrated a diagnostic sensitivity of 95% and specificity of 100% for Omicron detection, and a clinical sensitivity of 92% and specificity of 100% for XBB.1 detection (Fig. 6c). This result confirmed the potential of DOES-CRISPR as a diagnostic tool for COVID-19 detection in patient samples.Conclusions
In summary, we have demonstrated for the first time that 10-23DZs can be used to fundamentally overcome the off-target issue in the traditional CRISPR/Cas13a system. We term this approach DOES-CRISPR. The approach begins with 10-23DZs that precisely cleave the desired off-target RNA that contains any purine–pyrimidine junction, which in turn would efficiently block the off-target activity of Cas13a because Cas13a needs higher complementarity between the crRNAs and the target RNA. To our knowledge, an integrated DNAzyme recognition-RNA cleavage-CRISPR/Cas13a activation strategy has never been reported before. Significantly, we demonstrated that DOES-CRISPR can significantly improve the specificity of traditional CRISPR/Cas13a-based molecular detection by up to ∼43-fold. Furthermore, this approach can be adopted for detection of wide-ranging targets, including microRNAs (miR-17 and let-7e), CYP2C19*17 gene, SARS-Cov-2 variants (Gamma, Delta, and Omicron), and Omicron subtypes (BQ.1 and XBB.1) with single-nucleotide resolved specificity. Clinical evaluation of a DOES-CRISPR-based CYP2C19*17 sensor using 10 patient blood samples demonstrated a clinical sensitivity of 100% and specificity of 100%. In addition, DOES-CRISPR demonstrated a diagnostic sensitivity of 95% and specificity of 100% for Omicron detection, and a clinical sensitivity of 92% and specificity of 100% for XBB.1 detection when analyzing 20 clinical throat swab samples. Given that a specific 10-23DZ can be easily engineered to contain two binding arms that are complimentary to the target RNA sequence, DOES-CRISPR is suitable for the detection of both mRNA and miRNA targets. With a wide variety of RNA-cleaving DNAzymes currently available for the specific cleavage of various purine–purine, purine–pyrimidine, pyrimidine–pyrimidine and pyrimidine–purine junctions,51,52 we envision that the described strategy will find diverse applications in chemical biology, medical diagnostics, DNA computing, and biosensing. Furthermore, it is conceivable that the same design can be extended to improve the specificity of Cas12a by using DNA-cleaving DNAzymes. Currently, we are also engineering a fully printed paper-based analytical device for point-of-care testing in resource-limited settings.53Data availability
Experimental data is available in the ESI† online.Author contributions
M. L. applied for the funding. M. L. conceived the idea and supervised the project. Y. P. W. performed the experiments and data collection. Y. P. W., R. G. J., Y. Y. C., and M. L. analyzed the data. Y. P. W., R. G. J., and M. L. wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (No. 2023YFC3205804).Notes and references
- L. A. Marraffini and E. J. Sontheimer, Nat. Rev. Genet., 2010, 11, 181–190 CrossRef CAS PubMed.
- R. Barrangou and L. Marraffini, Mol. Cell, 2014, 54, 234–244 CrossRef CAS PubMed.
- G. Knott and J. Doudna, Science, 2018, 361, 866–869 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed.
- O. Abudayyeh, J. Gootenberg, S. Konermann, J. Joung, I. Slaymaker, D. Cox, S. Shmakov, K. Makarova, E. Semenova, L. Minakhin, K. Severinov, A. Regev, E. Lander, E. Koonin and F. Zhang, Science, 2016, 353, aaf5573 CrossRef PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, C. Myhrvold, R. P. Bhattacharyya, J. Livny, A. Regev, E. V. Koonin, D. T. Hung, P. C. Sabeti, J. J. Collins and F. Zhang, Science, 2017, 356, 438–442 CrossRef CAS.
- D. Boehm and C. L. Tsou, Cell, 2020, 184, 323–333 Search PubMed.
- D. Samanta, S. Ebrahimi, N. Ramani and C. Mirkin, J. Am. Chem. Soc., 2022, 144, 16310–16315 CrossRef CAS.
- H. Li, J. Yang, G. Wu, Z. Weng, Y. Song, Y. Zhang, J. Vanegas, L. Avery, Z. Gao, H. Sun, Y. Chen, K. Dieckhaus, X. Gao and Y. Zhang, Angew. Chem., Int. Ed., 2022, 61, e202203826 CrossRef CAS PubMed.
- H. Li, Y. Xie, F. Chen, H. Bai, L. Xiu, X. Zhou, Q. Hu and K. Yin, Chem. Soc. Rev., 2023, 52, 361–382 RSC.
- Y. Li, S. Li, J. Wang and G. Liu, Trends Biotechnol., 2019, 37, 730–743 CrossRef CAS PubMed.
- Y. Tang, L. Gao, W. Feng, C. Guo, Q. Yang, F. Li and X. C. Le, Chem. Soc. Rev., 2021, 50, 11844–11869 RSC.
- W. Feng, A. M. Newbigging, J. Tao, Y. Cao, H. Peng, C. Le, J. Wu, B. Pang, J. Li, D. L. Tyrrell, H. Zhang and X. C. Le, Chem. Sci., 2021, 12, 4683–4698 RSC.
- S. Chen, R. Wang, S. Peng, S. Xie, C. Lei, Y. Huang and Z. Nie, Chem. Sci., 2022, 13, 2011–2020 RSC.
- Y. Chen, Y. Mei and X. Jiang, Chem. Sci., 2021, 12, 4455–4462 RSC.
- Y. Ma, Q. Mou, P. Yan, Z. Yang, Y. Xiong, D. Yan, C. Zhang, X. Zhu and Y. Lu, Chem. Sci., 2021, 12, 11740–11747 RSC.
- Y. Dai, Y. Wu, G. Liu and J. J. Gooding, Angew. Chem., Int. Ed., 2020, 59, 20754–20766 CrossRef CAS.
- J. Tao, D. E. Bauer and R. Chiarle, Nat. Commun., 2023, 14, 212 CrossRef CAS PubMed.
- L. Zhang, H. Rube, C. Vakulskas, M. Behlke, H. Bussemaker and M. Pufall, Nucleic Acids Res., 2020, 48, 5037–5053 CrossRef CAS PubMed.
- D. Swarts, J. Oost and M. Jinek, Mol. Cell, 2017, 66, 221–233 CrossRef CAS.
- T. Zhou, R. Huang, M. Huang, J. Shen, Y. Shan and D. Xing, Adv. Sci., 2020, 7, 1903661 CrossRef CAS.
- C. Myhrvold, C. A. Freije, J. S. Gootenberg, O. O. Abudayyeh, H. C. Metsky, A. F. Durbin, M. J. Kellner, A. L. Tan, L. M. Paul, L. A. Parham, K. F. Garcia, K. G. Barnes, B. Chak, A. Mondini, M. L. Nogueira, S. Isern, S. F. Michael, I. Lorenzana, N. L. Yozwiak, B. L. MacInnis, I. Bosch, L. Gehrke, F. Zhang and P. C. Sabeti, Science, 2018, 360, 444–448 CrossRef CAS.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439–444 CrossRef CAS.
- Y. Ke, S. Huang, B. Ghalandari, S. Li, A. Warden, J. Dang, L. Kang, Y. Zhang, Y. Wang, Y. Sun, J. Wang, D. Cui, X. Zhi and X. Ding, Adv. Sci., 2021, 8, 2003611 CrossRef CAS PubMed.
- Y. Ke, B. Ghalandari, S. Huang, S. Li, C. Huang, X. Zhi, D. Cui and X. Ding, Chem. Sci., 2022, 13, 2050–2061 RSC.
- H. Tong, J. Huang, Q. Xiao, B. He, X. Dong, Y. Liu, X. Yang, D. Han, Z. Wang, X. Wang, W. Ying, R. Zhang, Y. Wei, C. Xu, Y. Zhou, J. Li, M. Cai, Q. Wang, M. Xue and H. Yang, Nat. Biotechnol., 2022, 41, 1–12 Search PubMed.
- B. P. Kleinstiver, A. A. Sousa, R. T. Walton, Y. E. Tak, J. Y. Hsu, K. Clement, M. M. Welch, J. E. Horng, J. Malagon-Lopez, I. Scarfò, M. V. Maus, L. Pinello, M. J. Aryee and J. K. Joung, Nat. Biotechnol., 2019, 37, 276–282 CrossRef CAS PubMed.
- J. Zhou, P. Chen, H. Wang, H. Liu, Y. Li, Y. Zhang, Y. Wu, C. Paek, Z. Sun, J. Lei and L. Yin, Mol. Ther., 2022, 30, 244–255 CrossRef CAS PubMed.
- Y. Wang, Y. Wang, D. Song, X. Sun, Z. Li, J. Y. Chen and H. Yu, Nat. Chem., 2022, 14, 350–359 CrossRef PubMed.
- S. K. Silverman, Acc. Chem. Res., 2009, 42, 1521–1531 CrossRef CAS PubMed.
- E. McConnell, I. Cozma, Q. Mou, J. Brennan, Y. Lu and Y. Li, Chem. Soc. Rev., 2021, 50, 8954–8994 RSC.
- M. Liu, D. Chang and Y. Li, Acc. Chem. Res., 2017, 50, 2273–2283 CrossRef CAS PubMed.
- S. Santoro and G. Joyce, Biochemistry, 1998, 37, 13330–13342 CrossRef CAS.
- D. P. Bartel, Cell, 2004, 116, 281–297 CrossRef CAS.
- G. A. Calin and C. M. Croce, Nat. Rev. Cancer, 2006, 6, 857–866 CrossRef CAS PubMed.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435, 834–838 CrossRef CAS.
- Y. S. Lee and A. Dutta, Annu. Rev. Pathol., 2009, 4, 199–227 CrossRef CAS PubMed.
- H. Dong, J. Lei, L. Ding, Y. Wen, H. Ju and X. Zhang, Chem. Rev., 2013, 113, 6207–6233 CrossRef CAS PubMed.
- S. Sim, C. Risinger, M. L. Dahl, E. Aklillu, M. Christensen, L. Bertilsson and M. Ingelman-Sundberg, Clin. Pharmacol. Ther., 2006, 79, 103–113 CrossRef CAS PubMed.
- B. Hu, H. Guo, P. Zhou and Z. L. Shi, Nat. Rev. Microbiol., 2020, 19, 1–14 Search PubMed.
- W. Garcia-Beltran, E. Lam, K. Denis, A. Nitido, Z. Garcia, B. Hauser, J. Feldman, M. Pavlovic, D. Gregory, M. Poznansky, A. Sigal, A. Schmidt, A. Iafrate, V. Naranbhai and A. Balazs, Cell, 2021, 184, 2372–2383 CrossRef CAS PubMed.
- A. Fontanet, B. Autran, B. Lina, M. P. Kieny, S. S. A. Karim and D. Sridhar, The Lancet, 2021, 397, 952–954 CrossRef CAS PubMed.
- R. Gupta, Nat. Rev. Immunol., 2021, 21, 340–341 CrossRef CAS PubMed.
- D. M. Altmann, R. J. Boyton and R. Beale, Science, 2021, 371, 1103–1104 CrossRef CAS PubMed.
- I. Hadj Hassine, Rev. Med. Virol., 2022, 32, e2313 CrossRef CAS PubMed.
- G. Vaidyanathan, Nature, 2021, 593, 321–322 CrossRef CAS.
- Z. Cui, P. Liu, N. Wang, L. Wang, K. Fan, Q. Zhu, K. Wang, R. Chen, R. Feng, Z. Jia, M. Yang, G. Xu, B. Zhu, W. Fu, T. Chu, L. Feng, Y. Wang, X. Pei, P. Yang, X. S. Xie, L. Cao, Y. Cao and X. Wang, Cell, 2022, 185, 860–871 CrossRef CAS PubMed.
- D. Planas, T. Bruel, I. Staropoli, F. Guivel-Benhassine, F. Porrot, P. Maes, L. Grzelak, M. Prot, S. Mougari, C. Planchais, J. Puech, M. Saliba, R. Sahraoui, F. Femy, N. Morel, J. Dufloo, R. Sanjuán, H. Mouquet, E. André and O. Schwartz, Nat. Commun., 2023, 14, 824 CrossRef CAS PubMed.
- A. Dijokaite-Guraliuc, R. Das, D. Zhou, H. Ginn, C. Liu, H. Duyvesteyn, J. Huo, R. Nutalai, P. Supasa, M. Selvaraj, T. Silva, M. Plowright, T. Newman, H. Hornsby, A. Mentzer, D. Skelly, T. Ritter, N. Temperton, P. Klenerman and G. Screaton, Cell Rep., 2023, 42, 112271 CrossRef CAS PubMed.
- J. Lewnard, V. Hong, J. Kim, S. Shaw, B. Lewin, H. Takhar, M. Lipsitch and S. Tartof, Nat. Commun., 2023, 14, 3854 CrossRef CAS PubMed.
- K. Schlosser, J. Gu, L. Sule and Y. Li, Nucleic Acids Res., 2008, 36, 1472–1481 CrossRef CAS PubMed.
- K. Schlosser, J. Gu, J. Lam and Y. Li, Nucleic Acids Res., 2008, 36, 4768–4777 CrossRef CAS PubMed.
- Y. Zhang, Q. Zhang, F. Cheng, Y. Chang, M. Liu and Y. Li, Chem. Sci., 2021, 12, 8282–8287 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc01501k |
| This journal is © The Royal Society of Chemistry 2024 |