Open Access Article
Open Access ArticleNon-fibril amyloid aggregation at the air/water interface: self-adaptive pathway resulting in a 2D Janus nanofilm†
Hao
Ren‡
a,
Huan
Chen‡
b,
Yu
Kang‡
c,
Wei
Liu
a,
Yongchun
Liu
a,
Fei
Tao
a,
Shuting
Miao
a,
Yingying
Zhang
a,
Qian
Liu
d,
Mingdong
Dong
 d,
Yonggang
Liu
d,
Yonggang
Liu
 *c,
Bing
Liu
*b and
Peng
Yang
*c,
Bing
Liu
*b and
Peng
Yang
 *a
*a
aKey Laboratory of Applied Surface and Colloid Chemistry, Ministry of Education, Xi'an Key Laboratory of Polymeric Soft Matter, International Joint Research Center on Functional Fiber and Soft Smart Textile, School of Chemistry and Chemical Engineering, Shaanxi Normal University, Xi'an 710119, China. E-mail: yangpeng@snnu.edu.cn
bFirst Affiliated Hospital, School of Medicine, Xi'an Jiaotong University, Xi'an, 710061, China. E-mail: bliu2018@xjtu.edu.cn
cState Key Laboratory of Polymer Physics and Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, 130022, China. E-mail: yonggang@ciac.ac.cn
dInterdisciplinary Nanoscience Center (iNANO), Aarhus University, Aarhus C, Denmark
First published on 7th May 2024
Abstract
The amyloid states of proteins are implicated in several neurodegenerative diseases and bioadhesion processes. However, the classical amyloid fibrillization mechanism fails to adequately explain the formation of polymorphic aggregates and their adhesion to various surfaces. Herein, we report a non-fibril amyloid aggregation pathway, with disulfide-bond-reduced lysozyme (R-Lyz) as a model protein under quasi-physiological conditions. Very different from classical fibrillization, this pathway begins with the air–water interface (AWI) accelerated oligomerization of unfolded full-length protein, resulting in unique plate-like oligomers with self-adaptive ability, which can adjust their conformations to match various interfaces such as the asymmetric AWI and amyloid-protein film surface. The pathway enables a stepwise packing of the plate-like oligomers into a 2D Janus nanofilm, exhibiting a divergent distribution of hydrophilic/hydrophobic residues on opposite sides of the nanofilm. The resulting Janus nanofilm possesses a top-level Young's modulus (8.3 ± 0.6 GPa) among amyloid-based materials and exhibits adhesive strength two times higher (145 ± 81 kPa) than that of barnacle cement. Furthermore, we found that such an interface-directed pathway exists in several amyloidogenic proteins with a similar self-adaptive 2D-aggregation process, including bovine serum albumin, insulin, fibrinogen, hemoglobin, lactoferrin, and ovalbumin. Thus, our findings on the non-fibril self-adaptive mechanism for amyloid aggregation may shed light on polymorphic amyloid assembly and their adhesions through an alternative pathway.
1 Introduction
An amyloid is a particular form of protein in nature, consisting of a universal cross-β-structure from misfolded protein.1 It is not only related to human neurodegenerative diseases,2 but is also involved in the bioprocess of various organisms, such as interfacial adhesion of E. coli and barnacles,3in vivo enamel growth,4 and life origin.5 In the past few decades, considerable progress has been made in in vitro aggregation of amyloidogenic proteins/peptides, indicating that the kinetics of fibrillation follows the oligomer–protofibril–fibril pathway, with a long lag phase (∼days) of oligomerization (Scheme 1a).2,6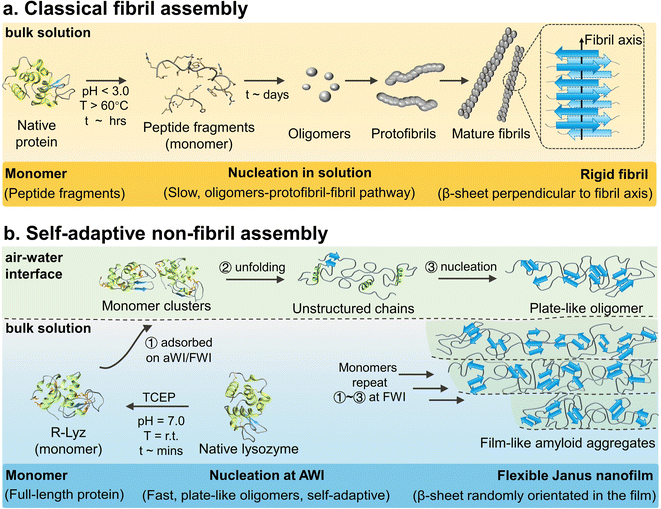 | ||
| Scheme 1 Schematic illustration of the aggregation mechanism. (a) Classical lysozyme amyloid fibril2,6 and (b) self-adaptive assembly of R-Lyz at the AWI. | ||
Although extensive research has been conducted on interface-accelerated fibrillation,7–10 the classical pathway fails to explain various bio-processes. First, in vitro synthesis of amyloid fibrils mainly involves peptide fragments as monomers.7 However, in nature, organisms such as microbes and barnacles can easily produce amyloid aggregates from full-length proteins (e.g., CsgA 13k, Cp19k and even Cp100k) under mild biological conditions to achieve adhesion and fulfill biological functions.8 The difference of monomer type may imply undetectable aspects in the amyloid transformation pathway (e.g., elusive function of interfaces). Second, it is hard for the classical fibrillation pathway to explain the morphological diversity of amyloid aggregates observed in nature. For instance, various non-fibril structures and hierarchical organization at multiple scales are frequently observed in many cases, such as self-assembled protein membranes,11 flake/sponge-like aggregates,12 and network structures.13 The non-fibril pathway may be the underlying cause for the diversity of morphological structures. Third, without notably considering the surface activity and adaptivity of solution species on surfaces, the classical fibrillation mechanism could not explain well in vivo amyloid adhesion at interfaces.6,14 In general, micron-scale amyloid fibrils showed a low interfacial activity for high rigidity, which is not favorable for supporting the easy adjustment of their conformation and shape to interact with various surfaces. In contrast, adhesion-related proteins in nature, such as the CsgA,15 cement protein (Cp 20k),16 and mussel foot protein (Mfp),17 are all rich in unstructured structures and present robust interfacial adhesion on the surfaces from hard rock to soft tissues by forming amyloid plaques.
The above examples underline some interesting clues but are still difficult to be understood at the molecular level based on the classical fibrillation mechanism.7 Herein, we decipher a non-fibril amyloid aggregation pathway at interfaces with disulfide-bond-reduced lysozyme (R-Lyz) as a model protein.18 Unlike traditional fibrillation under harsh conditions (Scheme 1a), the native lysozyme was unfolded under near-physiological conditions by the disulfide bond reducing agent tris(2-carboxyethyl)phosphine (TCEP) (Scheme 1b). Throughout the assembly process, the lysozyme monomers maintain their primary structure, mimicking the preservation of full-length proteins involved in amyloid-based adhesion in nature. At the AWI of the R-Lyz solution, a non-fibril amyloid nanofilm consisting of plate-like oligomers forms within minutes. The oligomerization step at interfaces occurs more rapidly (∼minutes) compared to the traditional fibrillation pathway (∼days).19 Moreover, unstructured protein chains had high flexibility to adjust to interact with various surfaces, exhibiting self-adaptivity, even on the initially formed protein nanofilm. Consequently, the newly formed plate-like oligomers attached and packed well onto the initial nanofilm, ultimately assembling into a Janus nanofilm with divergent hydrophilic/hydrophobic residues on opposite sides. The internal structure of the film shares the cross-β motif with randomly orientated β-sheet stacking.20 However, the assembly behavior differs from the secondary nucleation in the traditional fibrillation pathway, where oligomers detach from the fibril interface.21 Upon drying, the film exhibits a top-level Young's modulus (8.3 ± 0.6 GPa) in amyloid-based materials, and the adhesion strength is twice that of barnacle cement. This self-adaptive pathway provides a general strategy for fabricating Janus protein films from several amyloidogenic proteins, including bovine serum albumin (BSA), insulin, fibrinogen, hemoglobin, lactoferrin, and ovalbumin. Compared to other bottom-up heterogeneous processes, this self-adaptive assembly strategy produces Janus nanofilms through the assembly of proteins at the heterogeneous interface (AWI), without the need for any external polymer substrate or energy inputs.22,23 This strategy not only provides an opportunity for producing protein-based nanoscale Janus films but also holds promise for applications in various types of surface modification, separation, and wet adhesion. It particularly underscores the crucial role of interfaces in amyloid assembly and adhesion enhancement.24
2 Results and discussion
2.1. The prediction of amyloid assembly behavior of full-length proteins at interfaces by molecular dynamics (MD) simulation
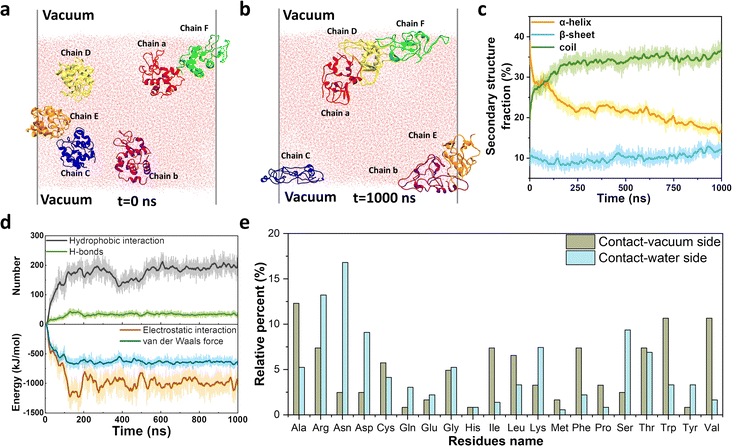
In natural systems, several adhesion-related plaques consist of full-length unstructured/unfolded proteins that interact with various interfaces. Hence, we choose disulfide-bond-reduced lysozyme (R-Lyz), one of the most studied amyloidogenic proteins, as a model protein to predict the assembly behavior of full-chain proteins at the interface.18 Firstly, a vacuum–water interface (VWI) was utilized to mimic the AWI for replica exchange molecular dynamics (REMD) simulation.25 The α-helices of R-Lyz unfolded into the random coils, and part of them further formed β-sheets in the presence of the VWI (Fig. S1a–c†). In contrast, the R-Lyz structure remained relatively unchanged and maintained an α-helix-dominated conformation in water without the VWI (Fig. S1d–f†). The free energy change (ΔG) of R-Lyz at the VWI (−95 kJ mol−1) is significantly lower than in water (−10 kJ mol−1), suggesting that the VWI strongly encourages the R-Lyz molecule to enrich at the VWI (Fig. S1a and d†). Both the radius of gyration (Rg) and the root-mean-square distance (RMSD) of R-Lyz at the VWI increased compared to those in water, indicating accelerated unfolding and the loss of the α-helix structure of R-Lyz facilitated by the interface (Fig. S2†). This unfolding was accompanied by the gradually exposed hydrophobic residues, as indicated by the increase in the solvent accessible surface area (SASA) of R-Lyz at the VWI (Fig. S3†). The exposed hydrophobic regions automatically move to the VWI, further enabling the α-helix to random coil transition, which has higher tendency to form β-sheets because the hydrophobic core regions are easier to encounter in flexible coils.26To further clarify the intermolecular interaction between monomers in amyloid aggregation, we performed an all-atom molecular dynamics (MD) simulation with six randomly orientated R-Lyz in water with the existence of a VWI (Fig. 1a and b). After 1000 ns simulation, all R-Lyz spontaneously moved towards the interface (snapshots in Fig. S4†), accompanied by the loss of the α-helix structure and an increase in flexible coils and β-sheets (Fig. 1c). Statistical analysis of the non-covalent interaction between R-Lyz monomers revealed that hydrophobic interactions were more pronounced than hydrogen bonds (Fig. 1d). In terms of intermolecular energy potential analysis, electrostatic interactions emerged as the dominant driving force for monomer aggregation, surpassing van der Waals forces. Hence, it can be concluded that hydrophobic and electrostatic interactions are the main driving forces for monomer aggregation.
Furthermore, unstructured/unfolded protein chains exhibit self-adaptive behavior, allowing them to adjust the distribution of their residues at interfaces. Hydrophobic and nonpolar residues (e.g., Ala, Val, Leu, Ile, Phe, Pro, and Trp) tend to migrate towards and orient themselves at the VWI, while hydrophilic and positively/negatively charged residues (e.g., Arg, Asn, Asp, Lys, and Ser) tend to face the water (Fig. 1e). The relative percentage of different residue types at the VWI is summarized in Fig. S5,† with 62.3% hydrophobic residues on the vacuum-side, while only 21.8% on the water-side. This remarkable self-adaptivity of unstructured protein chains may contribute to the adhesion sites for amyloid plaques on different surfaces,8,27 as demonstrated by the following experimental evidence.
2.2. Unfolding and interface-accelerated aggregation of full-length proteins
To obtain full-length R-Lyz and validate the simulation findings, we utilized TCEP as a disulfide-bond reducing agent to unfold the native lysozyme.28 After reduction, R-Lyz remained its primary structure and maintained its full chain length, as evidenced by the m/z signal at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 (M + H)+ and 7144 (M + 2H)2+ observed in MALDI-TOF mass spectrometry (Fig. 2a). In contrast, in traditional lysozyme amyloid fibrils obtained under acidic conditions and at high temperatures (70 °C),18 the peptide mainchain was hydrolyzed into small fragments with molecular weight from 1900 to 7500 Da and formed micron-length mature fibrils after 7 days of incubation (Fig. S6†). The small-angle X-ray scattering (SAXS) of the R-Lyz solution reveals the increase in Rg (from 1.37 nm to 1.5 nm) in the Guinier plot (Fig. S7a, inset†) and non-convergence of the Kratky plot (Fig. S7b†), indicating the loss of globular structure and thus an increase in random coil content and chain flexibility.29 The formation of protein aggregates in the R-Lyz solution was then monitored by asymmetric flow field-flow fractionation coupled with multi-angle laser light scattering (AF4-MALLS), with small species eluting first and larger species eluting later from the parabolic channel (Fig. S8†).30 According to the time evolution of the weight average aggregation number (Nw,agg) plot, no aggregates can be detected in the first 3 h of cultivation (Fig. 2b). Then, a few aggregates with Nw,agg ∼16 formed in solution (Fig. 2b–i), followed by an exponential increase of the aggregation number to a steady value of Nw,agg ∼103 at 7–8 h. Coincidentally, a 7 h lag phase for hydrophobic aggregation and β-sheet formation was observed according to the 8-anilinonaphthalene-1-sulfonic acid (ANS) and thioflavin T (ThT) fluorescence stain of R-Lyz in bulk solution (Fig. S9†). The results from AF4-MALLS and fluorescence staining revealed that aggregation in bulk solution is noticeably frustrated, consistent with simulation results (Fig. S1†). After 28 h of cultivation, 84% of R-Lyz was still in the monomeric state, while 15% had aggregated into curly filaments as the dominant morphology (weight average molecular weight, Mw ∼2 × 107 g mol−1) (Fig. S10, S11† and 2b-ii). From a macroscopic aspect, the solution is transparent and would exhibit the Tyndall effect when illuminated with a red laser after 24 h of incubation (Fig. 2b-iii).
280 (M + H)+ and 7144 (M + 2H)2+ observed in MALDI-TOF mass spectrometry (Fig. 2a). In contrast, in traditional lysozyme amyloid fibrils obtained under acidic conditions and at high temperatures (70 °C),18 the peptide mainchain was hydrolyzed into small fragments with molecular weight from 1900 to 7500 Da and formed micron-length mature fibrils after 7 days of incubation (Fig. S6†). The small-angle X-ray scattering (SAXS) of the R-Lyz solution reveals the increase in Rg (from 1.37 nm to 1.5 nm) in the Guinier plot (Fig. S7a, inset†) and non-convergence of the Kratky plot (Fig. S7b†), indicating the loss of globular structure and thus an increase in random coil content and chain flexibility.29 The formation of protein aggregates in the R-Lyz solution was then monitored by asymmetric flow field-flow fractionation coupled with multi-angle laser light scattering (AF4-MALLS), with small species eluting first and larger species eluting later from the parabolic channel (Fig. S8†).30 According to the time evolution of the weight average aggregation number (Nw,agg) plot, no aggregates can be detected in the first 3 h of cultivation (Fig. 2b). Then, a few aggregates with Nw,agg ∼16 formed in solution (Fig. 2b–i), followed by an exponential increase of the aggregation number to a steady value of Nw,agg ∼103 at 7–8 h. Coincidentally, a 7 h lag phase for hydrophobic aggregation and β-sheet formation was observed according to the 8-anilinonaphthalene-1-sulfonic acid (ANS) and thioflavin T (ThT) fluorescence stain of R-Lyz in bulk solution (Fig. S9†). The results from AF4-MALLS and fluorescence staining revealed that aggregation in bulk solution is noticeably frustrated, consistent with simulation results (Fig. S1†). After 28 h of cultivation, 84% of R-Lyz was still in the monomeric state, while 15% had aggregated into curly filaments as the dominant morphology (weight average molecular weight, Mw ∼2 × 107 g mol−1) (Fig. S10, S11† and 2b-ii). From a macroscopic aspect, the solution is transparent and would exhibit the Tyndall effect when illuminated with a red laser after 24 h of incubation (Fig. 2b-iii).
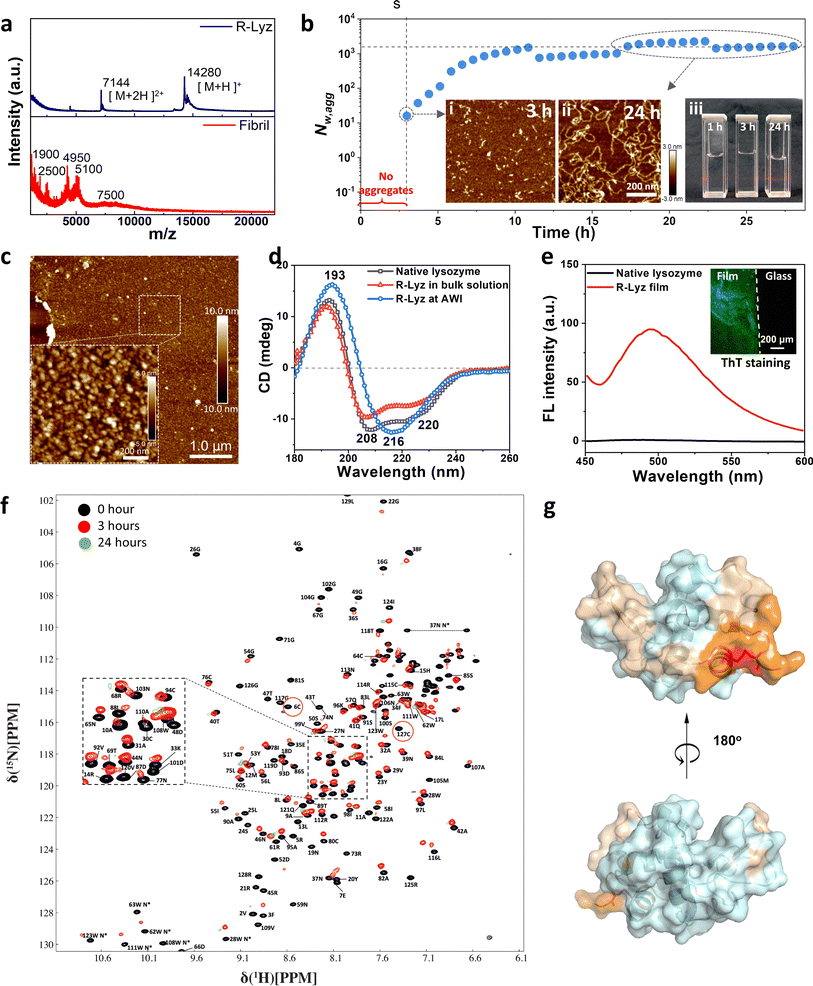 | ||
| Fig. 2 Full-length R-Lyz aggregation in solution and at the AWI. (a) MALDI-TOF spectra of R-Lyz solution and traditional fibril solution; (b) time-dependent Nw,agg of R-Lyz solution; inset: AFM images of the aggregation morphology of R-Lyz after (i) 3 h and (ii) 24 h of reaction; (iii) digital images of the reaction solution after 1, 3 and 24 h of incubation under a red laser; (c) AFM image of the R-Lyz aggregation structure at the AWI after 10 min of incubation; (d) far-UV CD spectrum; (e) fluorescence spectrum of the nanofilm after staining with ThT, inset: dark-field confocal image of the ThT stained nanofilm. (f) The overlay of the HSQC spectra of lysozyme before and after adding TCEP for 0 h (black), 3 h (red) and 24 h (green); (g) the projection of the residue with CSP larger than 0.4 ppm and significant intensity drops of more than 92% on the structure of the lysozyme. Among the colored residues, residues that experienced the peak broadening effect are labelled in Fig. S15.† | ||
However, the aggregation of R-Lyz at the AWI differs much from that in bulk solution. A film-like aggregate was discovered in a short cultivation time (10 min) and was composed of 20–30 nm particles (Fig. 2c), which is in a β-sheet-dominated conformation as proven by the strong negative peak at 216 nm in the far-UV circular dichroism (CD) spectrum (Fig. 2d) and the increase of the infrared absorption peaks at 1624 and 1696 cm−1 in the Fourier transform infrared (FTIR) spectrum (Fig. S12†). The percentage of the β-sheet component increases from 4–7% in the native state to 34–38% at the AWI (Fig. S13†). In contrast, R-Lyz in bulk solution still had the α-helix structure and only contained ∼6% β-sheet component, which is very similar to native lysozyme.31 The fluorescence increase at 484 nm and bright green emission in the confocal image of the ThT stained nanofilm also indicate an amyloid-like inner structure (Fig. 2e and S14†).
The unfolding and aggregation of R-Lyz were analyzed at the residue level using a 1H–15N heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) experiment (Fig. 2f, S15 and S16†). The spectra recorded at 3 h after the addition of TCEP indicated that the aggregation initiated around residues C6–C127 (Fig. 2f, red cycles). Being consistent with the MD results (Fig. 1), the NMR experiments also indicate that the first aggregated residues in bulk solution are primarily composed of neutral and small hydrophobic residues, such as Gly (4, 26, 49, 102, 126), Ala (10, 11, 32, 122), Val (120), Ile(124) and Leu (129), as well as charged residues, such as Arg (5, 45, 125, 128) and Asp (18, 48, 52, 66, 87, 101, 119), according to the cross-peak intensity decrease (Fig. 2f, S15 and S16†). Additionally, the residues experiencing the most significant conformational changes, with chemical shift perturbations (CSP) larger than 0.4 ppm, and the first aggregated residues, with intensity drops of more than 92%, have been highlighted on the structure of the native lysozyme in wheat (Fig. 2g).
The NMR and MD simulation results suggest that the driving forces of unfolded protein aggregation are similar in bulk solution and at the interface. However, the presence of the AWI significantly promotes the nucleation of R-Lyz into oligomers within 10 min. It is not only ∼280 times faster than the oligomerization step of traditional lysozyme fibrillation, which typically has a lag phase of ∼48 h,32 but also ∼20 times faster than the nucleation of full-length R-Lyz in bulk solution, which has a lag phase of ∼3 h (Fig. 2b). These findings suggest a unique surface-induced assembly pathway for unfolded amyloidogenic proteins. Understanding this process holds promise for revealing the mechanisms underlying amyloid-based assembly and adhesion at various surfaces.
2.3. Plate-like oligomer formation at the AWI
The assembly of unfolded full-length proteins at a confined two-dimensional interface may afford great opportunities to biopolymer chains for polymorph development of non-fibril amyloids. Hence, we utilized AFM to investigate the morphology of the original oligomers at the AWI. Plate-like oligomers ∼20–30 nm in width and ∼3–5 nm in height were detected at the AWI (Fig. 3a), composed of the R-Lyz monomer 3–5 nm in width (Fig. 3b and c). If we estimate the aggregation number of the oligomer by the volume of the oligomer and monomer, we can get the Nagg of the plate-like oligomer of around 27–112, which is in the same order as the Nw,agg of oligomers in bulk solution after 3 h of cultivation (Fig. 2b). The plate-like morphology was also proven by synchrotron radiation grazing-incidence small-angle X-ray scattering (SR-GISAXS) (Fig. 3d). The log–log plot of q against I(q) in the middle-q region (0.1–1.0 nm−1) shows I(q) − q−α with α being 2 (Fig. 3e), revealing a plate-like shape of the assembly unit according to the theory.33 The pair distance distribution function (PDDF) curve of the scattering data also proved the plate-like shape of the oligomer (Fig. 3f).34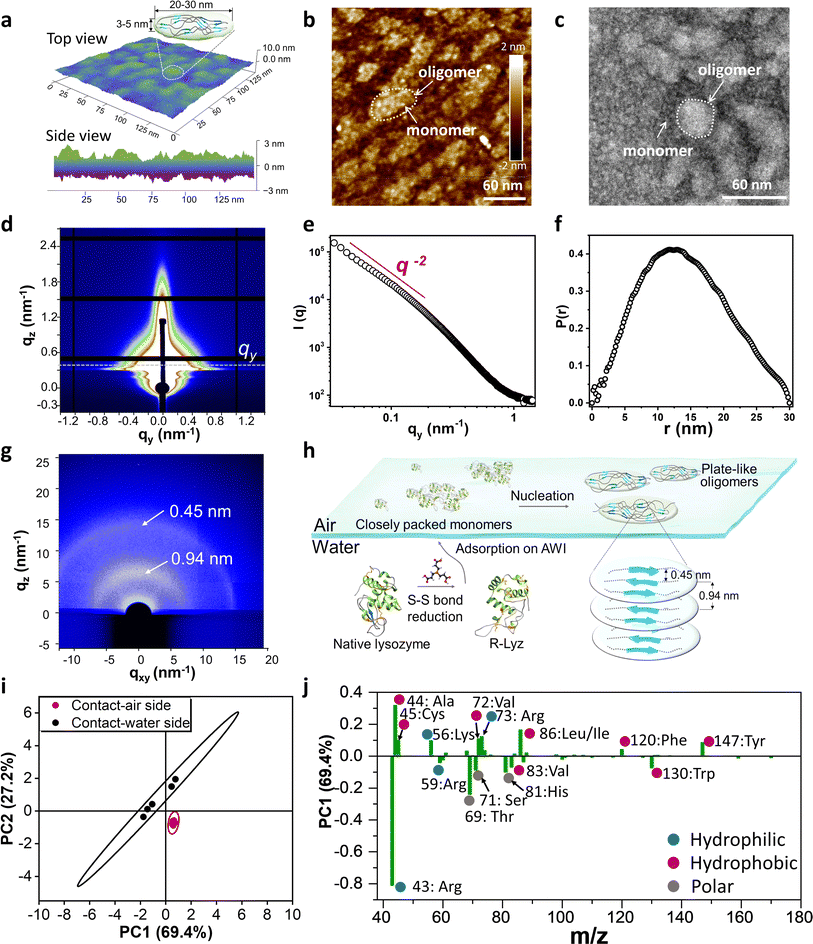 | ||
| Fig. 3 Characterization of plate-like oligomers. (a) AFM 3D-image and the schematic illustration of the plate-like oligomer formed at the AWI; (b) AFM image and (c) TEM image of the monomer and oligomers at the AWI of R-Lyz solution after 5 min of assembly; (d) SR-GISAXS image of the amyloid-like nanofilm at the AWI on a silicon wafer; (e) log–log plot of q against I(q); (f) PDDF plot of I(q); (g) SR-GIWAXS of the nanofilm on a silicon wafer; (h) schematic illustration of plate-like oligomer aggregation at the AWI; (i) the score plot TOF-SIMS data after PCA; (j) the loading plot and assignment of major peaks of PC1 (75.1%) after PCA of TOF-SIMS data (hydrophobic, hydrophilic, and neutral residues were marked with red, blue, and grey points, respectively. Residues' hydrophilic/hydrophilic scores at pH 7 were counted according to Monera et al.37). | ||
The internal structure of the plate-like oligomer was examined using synchrotron radiation grazing-incidence wide-angle X-ray scattering (SR-GIWAXS) (Fig. 3g), where two strong dispersion diffraction rings at q = 6.7 nm−1 and 13.8 nm−1 were observed, corresponding to the inter-β-sheet spacing (∼0.94 nm) and the inter-β-strand spacing (∼0.45 nm) within the protein stacking structure of the film.35 These results indicate that the plate-like oligomers share the cross-β-motif with traditional fibrils.20 Different from the previously reported cross-β diffraction pattern in traditional amyloid fibrils,21 these β-sheet structures in the plate-like oligomers are randomly distributed according to the blurred circular ring of the GIWAXS image. This random arrangement contributes to the flexibility of the film, thereby increasing the contact area and promoting adhesion to different surfaces. The schematic diagram for the assembly of R-Lyz into plate-like oligomers at the AWI is displayed in Fig. 3h. Initially, R-Lyz monomers concentrate at the AWI, followed by the collision of α-helices with random coils.36 This collision further facilitates the spatial alignment of β-sheet core regions into a randomly arranged cross-β-motif, ultimately leading to the significant formation of plate-like oligomers. Furthermore, this plate-like structure 4–5 nm in height and 20–30 nm in width has also been found in Aβ1–42 and the E. coli adhesion layer (Fig. S17†), suggesting that this particular structure may be involved in natural adhesion processes.
As revealed by MD simulation (Fig. 1e), unfolded/unstructured protein chains have high flexibility to adjust their residue distribution at interfaces, which is the critical character for the possible interfacial adhesion of non-fibril assembly. To prove the self-adaptive behavior of unfolding chains at the AWI, we measured the time-of-flight secondary ion mass spectrometry (TOF-SIMS) spectra for the contact-air (CA) and contact-water (CW) sides of the original deposition consisting of plate-like oligomers (Fig. S18†). Principal component analysis (PCA) was utilized to analyze the residual differences of both sides (the spectrum for the contact-air side was selected as a reference spectrum). Fig. 3j shows that the two 95% confidence ellipses are well separated for TOF-SIMS data of the CA-side and CW-side, indicating the different residue exposure of both sides. According to the positively loaded fragments in PC1 (69.4%) (Fig. 3j, where the CA-side was set as the reference spectrum), the contact-water side was predominant in charged and polar amino acid fragments, with m/z 43/59 (Arg), 71 (Ser), 81 (His), and 69 (Thr). In contrast, aromatic amino acid fragments 147 (Tyr) and 120 (Phe), and alkyl amino acid fragments 86 (Ile/Leu), 72 (Val), 45 (Cys) and 44 (Ala) are rich in the contact-air side. This distribution diversity of protein residues is consistent with MD simulations (Fig. 1e), demonstrating the self-adaptive ability of unfolded/unstructured protein chains to adjust their orientation at the molecular level. Combined with non-fibril aggregation driven by a plate-like oligomer, it provides a possible pathway to the polymorph diversity of amyloid-based interfacial adhesion.
2.4. Self-adaptive assembly of plate-like oligomers
As illustrated above, unfolded protein chains aggregating into plate-like oligomers have enough flexibility to adjust their residue distribution at interfaces, which is fundamental to mesoscopic assembly and offers high adhesion potential of the plate-like oligomers on surfaces. In nature, amyloid-mediated adhesion is typically not a monolayer adsorption. For instance, barnacles or E. coli usually produces macroscopic size adhesion plaques on substrates with thickness >1 μm, no matter what the morphologies are.12,38 This suggests that the mechanism of solution species adhering to a surface through multilayer packing is plausible. The process may involve the interfacial adsorption of amyloid species on a bare surface, followed by adhesion to a surface already covered by amyloid aggregates. Thereupon, we employed a quartz crystal microbalance with dissipation monitoring (QCM-D) to track the adsorption of R-Lyz on an Au-coated chip. The adsorption of R-Lyz caused a significant frequency shift of ∼−400 Hz (Fig. 4a), whereas native lysozyme and traditional amyloid fibrils only exhibited a frequency decrease of ∼10–25 Hz (Fig. 4a, inset). Consequently, the adsorbed mass of R-Lyz (7470 ng cm−2) was ∼17 times higher than that of native lysozyme (443 ng cm−2) and ∼24 times higher than that of traditional fibrils (315 ng cm−2) (Fig. 4b and S19†). These results highlight the high surface activity and aggregation tendencies of unfolded protein chains and plate-like oligomers compared to native and fibril species. It is noteworthy that a deceleration in frequency decrease is observed during QCM-D measurement, suggesting a diminished interfacial activity of filaments within the bulk solution compared to monomers and oligomers. For additional evidence, we manually removed the initial film (first 30 min) at the AWI, leading to the formation of a new film consisting of filaments with large interspaces attributable to their reduced self-adaptivity (Fig. S20†).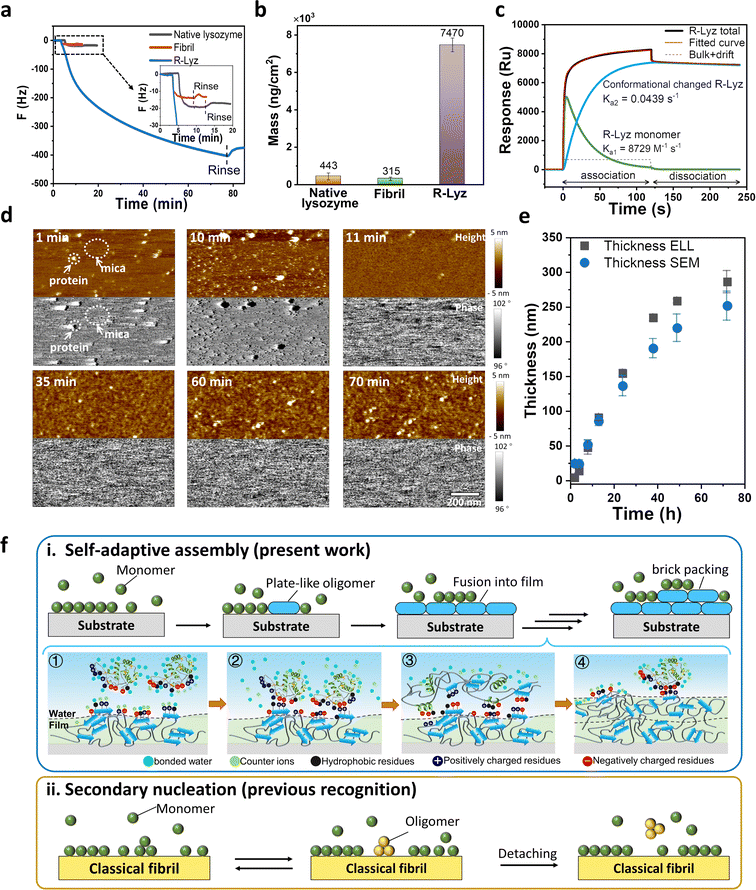 | ||
| Fig. 4 Characterization of self-adaptive assembly. QCM-D analysis of native lysozyme, traditional amyloid fibrils and R-Lyz solution on an Au sensor. (a) Frequency curve; (b) adsorption mass; (c) SPR response curve and two-state reaction fitting of R-Lyz solution; (d) snapshots from the in situ AFM movie of R-Lyz solution on the mica–water interface; (e) thickness of the nanofilm at the AWI against incubation time detected from ellipsometer (ELL) and scanning electron microscope (SEM) cross-section images; (f) schematic comparison of self-adaptive assembly (i) (① R-Lyz approaching the protein film surface; ② entropic gain due to counterion and bonded water release; ③ loss of secondary structure; conformational change and formation of plate-like oligomers; ④ the repeating of processes 1–3) and classical secondary nucleation21) and (ii) of amyloidogenic proteins. | ||
Besides, the ΔD/ΔF curve of the R-Lyz solution experienced a two-step adsorption profile, with a turning point at ∼−30 Hz (Fig. S21†), implying a “soft” layer formed at the initial stage of the assembly, followed by the formation of a relatively rigid layer.39 This two-step profile was further confirmed by surface plasmon resonance (SPR) (Fig. 4c and S22†). The curve is well fitted with a two-state reaction model, involving the rapid adsorption of a “soft” R-Lyz monomer layer on the solid–water interface (SWI) (Ka1 = 8729 M−1 s−1; Kd1 = 0.1792 M−1 s−1), followed by a slow α-helix to β-sheet conformational change (Ka2 = 0.0439 s−1; Kd2 = 2.921 × 10−4 s−1), leading to the irreversible binding of plate oligomers.40,41 The conformational transition was also supported by CD results of solution species and protein nanofilm (Fig. 2d). The results reveal that the plate-like oligomers exhibit a multilayer aggregation behavior and follow a brick-like packing manner, as captured by the in situ AFM recording (Fig. 4d and S23 and Movie S1†). Initially, R-Lyz was rapidly adsorbed onto the mica surface, as indicated by black dots in the phase image representing different viscoelasticities compared to the mica substrate. Subsequently, the entire mica surface was covered by plate-like oligomers (Fig. 4d, 11 min). Instead of forming traditional fibrils, these oligomers fused together to form a flat 2D nanofilm on the mica surface. The phase image shows an identical distribution after the mica surface is fully occupied by proteins. Over the next hour, plate-like oligomers (3–5 nm in height and 20–30 nm in width) were generated on the initial 2D nanofilm (Fig. 4d and S24†). AFM images of the film at the AWI also revealed a closely packed layer of plate-like oligomers with a thickness of ∼4 nm on the initial film (Fig. S25†). Further evidence of multilayer aggregation was provided by the steady increase in film thickness over time (Fig. 4e and S26†). Prolonged incubation at high buffer salt concentration (e.g., 250 mM NaCl) and low TCEP concentration (e.g., 0.5 mM) resulted in an increase in film thickness from ∼5 nm at 2 h to ∼280 nm after 72 h.
Based on the above evidence, it is reasonable to conclude that plate-like oligomers exhibit a step-by-step packing behavior at the interface, and the newly formed plate-like oligomers will not detach from the existing 2D nanofilm surface. Accordingly, we proposed a self-adaptive surface-induced assembly pathway of plate-like oligomers at the AWI, as illustrated in Fig. 4f–i. R-Lyz monomers firstly adsorbed on the substrate surface due to the increase in entropy and caused the loss of secondary structure, which then promoted the encounter of amyloid core regions in unfolded chains and produced plate-like oligomers. As the assembly proceeded, the adjacent plate-like oligomers fused into a flat 2D nanofilm. Further, R-Lyz monomers continued to be adsorbed on the newly formed nanofilm surface owing to the entropic gain from counterion and bonded water release, driven by hydrophobic and electrostatic interactions between protein residues. The adsorbed R-Lyz unfolded under the limitation of chain movements by the interface, followed by the step-wise formation of plate-like oligomers. The β-sheet structures inside and between plate-like oligomers could act as physical interaction points,11,42 allowing the newly formed packing units to adhere firmly on the original nanofilm surface (Fig. 4f–i). It is worth noting that this process is very different from previously reported secondary nucleation of traditional amyloid fibrils, where mature fibrils act as seeds to enhance the heterogeneous nucleation of monomers;14,21 the new oligomers on the fibril surface automatically detached and released into the bulk solution to continue the oligomer–protofibril–mature fibril pathway (Fig. 4f-ii). Considering the unstructured nature of critical proteins of the amyloid adhesion system (e.g., Cp20K, Csg A),15,16 the step-by-step self-adaptive assembly revealed in the present work may be closely related to the possible pathway for the formation of adhesion plaques adopted by organisms.
2.5. Bottom-up fabrication of the Janus nanofilm
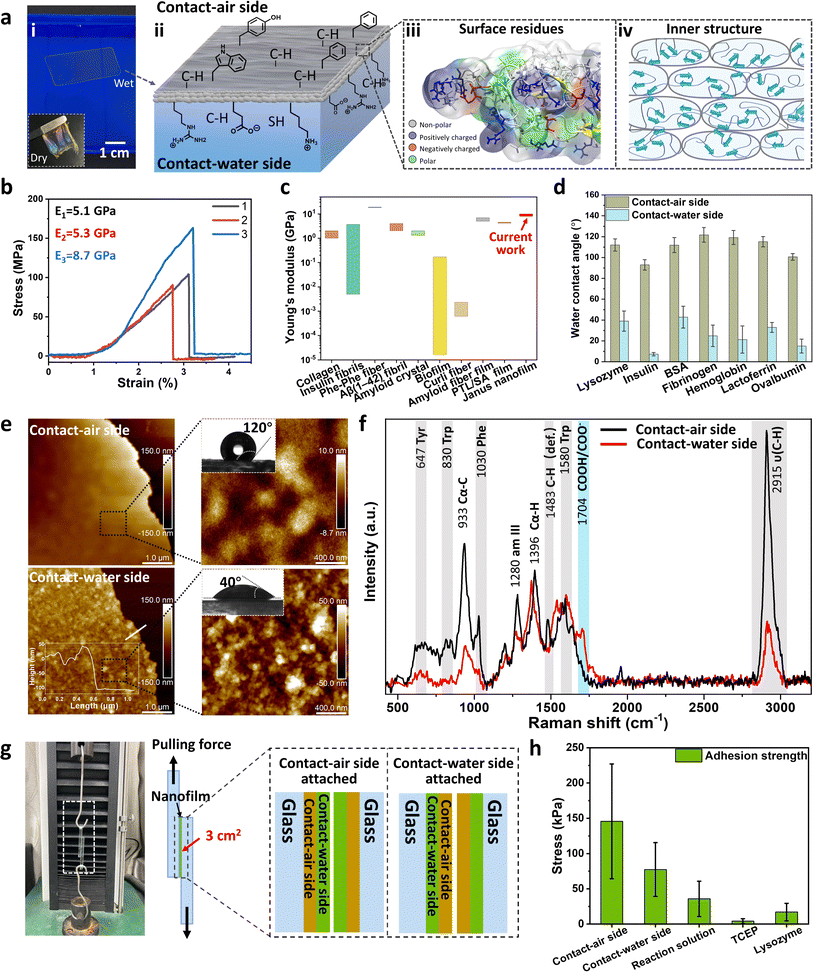
As mentioned above, plate-like oligomers are closely packed following the self-adaptive process. This multilayer stacking of assemble units can increase the strength of the obtained film through multi-noncovalent inter-oligomer interactions.42 We utilized a ∼150 nm nanofilm at the AWI to investigate their mechanical properties (Fig. S27†).43 The nanofilm can be taken out from the water surface with a plastic frame and dried into a free-standing film under ambient conditions (Fig. 5a-i, inset). In addition, the obtained film is very stable in dimethylsulfoxide and vitamin C, and it can partially dissolve in sodium dodecyl sulfate (SDS) after 44 h of soaking due to the dissociation of the inner hydrogen bonding of β-sheets (Fig. S28†). The Young's modulus of the overlapped dry film (∼16 layers, see Fig. S29†) was measured by a tension test, which is 5.1–8.7 GPa from the slope of the stress–strain curve (Fig. 5b). The result is consistent with the nanoindentation results (∼8.3 ± 0.6 GPa, Fig. S30†). It must be noted that although the film consists of randomly orientated β-sheets (Fig. 3g), the Young's modulus of the dried film ranked at the top level among amyloid-based materials with highly ordered cross-β-structures (Fig. 5c),44 which may benefit from the brick-packing of plate-like oligomers (Fig. 5a-iv).Further, based on the high self-adaptivity of unfolded proteins, the newly formed nanofilm exhibited a distinct Janus character in the wet state for several amyloidogenic proteins, where the water contact angle (WCA) of the CA-side is much higher than that of the CW-side (Fig. 5d). Take the film assembled from R-Lyz as an example: the WCA of the CA-side is 120°, and that of the CW-side is 40° (Fig. 5e). Moreover, the morphology of the CA-side appears to be a flat wrinkled surface for the diffusion of protein chains driven by surface tension at the AWI, while the CW-side is rough and constitutes plate-like oligomers ranging from 20 to 30 nm. In addition, the chemical components of the Janus nanofilm have apparent differences on both sides. From surface-enhanced Raman spectroscopy (SERS), the stretching vibration of –CH3 at 2915 cm−1 and –CH3 deformation at 1483 cm−1 are much stronger on the CA-side than on the CW-side. The signals of aromatic rings on the CA-side, such as the vibration peaks of Phe at 1030 cm−1, Trp at 830 cm−1, and Tyr at 647 cm−1, are significantly stronger than those on the CW-side of the film. In contrast, the COOH/COO− signal at 1704 cm−1 is higher at the CW-side, proving that the CW-side is rich in charged groups. In addition, grazing angle (45°) X-ray photoelectron spectroscopy (XPS) of the Janus nanofilm reflects that the hydrophilic components of NH2/NH3+ (at 400.4 eV, 87.1%) and sulfate (at 167.3 eV, 88.4%) on the CW-side are more pronounced than those on the CA-side (Fig. S31†). As the film comprises diverse hydrophilic functional groups, it is prone to swelling upon exposure to water. Consequently, we measured the reflectance spectrum to assess the swelling behavior of the film under variable humid conditions (Fig. S32†). The color of the film on the silicon wafer changes from light cyan at 0% relative humidity (RH) to bright pink at 100% RH, indicating an increase in film thickness due to swelling. Using the Bragg equation, we determined the swelling ratio of the film to be 38.6%, with the film thickness increasing from ∼176 nm to ∼244 nm. Schematic diagrams of the obtained Janus 2D film at the AWI are provided in Fig. 5a-ii and iii, with the CA-side of the film exposed to more hydrophobic residues and the CW-side exposed to a higher content of hydrophilic residues.
The divergent residue distribution was also supported by the different adhesion performances at the opposite sides of the nanofilm, as measured by AFM with a SiO2 spherical probe (Fig. S33†). The adhesion force on the CA-side is two times higher than on the CW-side, suggesting that the excessive hydrophobic residues on the CA-side contributed mainly to the adhesion force on the SiO2 probe surface. Furthermore, this Janus nanofilm can act as the glue to stick two pieces of glass. The internal randomly distributed β-sheet structures provide the film with adequate flexibility in the wet state over traditional amyloid fibrils, which can increase the contact area of possible rough surfaces by adjusting their shape. After drying, stable adhesion is obtained between two glass sheets (Fig. 5g). When the CA-side was attached to the glass surface, the average adhesion strength from shear testing was 145 ± 81 kPa (Fig. 5h), two times higher than the adhesion strength obtained by attaching the CW-side to the glass surface (77 ± 38 kPa). For blank control experiments, sandwiching the TCEP solution between two glasses causes almost no adhesion after drying, while the adhesion strength of the CA-side of the Janus nanofilm is 8.5 and 4.1 times higher than those upon sandwiching native lysozyme or R-Lyz solution between two glass sheets respectively. The results indicate that the unfolding of native protein increases the adhesion strength, and the brick-packing structure of the plate-like oligomers' inner structure further increases it. Compared with natural adhesion species, the adhesion strength of the CA-side is two times higher than that of barnacle cement on a smooth glass surface (68 kPa)45 and ∼103 times higher than that of biofilm on a glass surface (80 ± 22 Pa).46 This indicated that such self-adaptive non-fibril amyloid aggregation has the possibility to create materials that would mimic or even surpass natural bioadhesive systems.
3 Conclusion
In summary, we demonstrate a non-fibril amyloid aggregation pathway at the AWI using full-length amyloidogenic proteins. The unfolded protein triggered by the disulfide bond reducing agent experienced interface-accelerated nucleation within several minutes, while no aggregation occurred in the bulk solution during the same period. The plate-like oligomers, as the first discovered structure in all known protein aggregation systems, exhibit unprecedented self-adaptive properties, where unfolded full-length proteins adjust their residue distribution at interfaces from the AWI to amyloid-film surfaces, which further promotes inter-oligomer interactions by the encounter of the amyloid core regions, hydrophobic region, and opposite charge regions of flexible unfolded chains. Based on this self-adaptive assembly, the plate-like oligomers are closely packed into a 2D Janus nanofilm at the AWI, where inner- and inter-oligomer β-sheet structures randomly distribute, acting as the cross-linking points. This unique brick-like packing structure of plate-like oligomers endows the resultant Janus nanofilm with a top-level Young's modulus (∼8.3 ± 0.6 GPa), which is one of the highest among existing amyloid-based materials, and adhesion strength (145 ± 81 kPa), which is two times higher than that of natural species after drying. Such self-adaptive assembly discovered in R-Lyz has its generality since such features have been further found in other unfolded proteins, such as BSA, insulin, fibrinogen, hemoglobin, lactoferrin, and ovalbumin (Fig. 5d).This work highlights the importance of the interface in the assembly of full-length proteins into a 2D film-like amyloid structure with self-adaptive characteristics. The obtained film aggregates have a high Young's modulus but also exhibit better flexibility and adhesion strength, which provides new insight into the mechanism of polymorph amyloid-based aggregation and adhesion. In light of the universality of self-adaptive assembly and the asymmetry of the AWI, we can easily obtain protein Janus nanofilms, which have never been reported before and are expected to be applied in many fields, such as interface modification, drug separation, biomineralization, and other health-related areas.11,47 Following this clue, the surface behavior of other functional or amyloidogenic proteins/peptides (e.g., α-synuclein, Aβ1–42, CsgA, Cp20K, etc.) at various interfaces in future studies may provide us with a better understanding of the possible pathways as well as the application of non-fibril amyloids.
Data availability
Data available on request from the authors.Author contributions
H. R. and P. Y. initiated the project. H. R. performed material synthesis, AFM, CD, Raman, XPS, QCM-D, SPR, and film adhesion tests; H. C. and B. L. performed 2D-NMR analysis; Y. K. and Y. L. carried out AF4 analysis; F. T. and S. M. performed the synchrotron WAXS/SAXS; Y. L. performed the computer simulations; Y. Z. performed in situ AFM; W. L. carried out ellipsometry and SEM; S. M. performed the mechanical analysis; L. Q. and M. D. performed CD measurements and component analysis. The manuscript was written by H. R. and P. Y. All authors reviewed and approved the manuscript.Conflicts of interest
There is no conflict of interest to report.Acknowledgements
We are grateful for funding from the National Natural Science Foundation of China (No. 21905166, 22375122, 51903147, 21905165), the National Science Fund for Distinguished Young Scholars (52225301), the Fundamental Research Funds for the Central Universities (No. GK202304040, 2020TS092, GK202305001), the Shanghai Synchrotron Radiation Facility (2019-SSRF-PT-011403, 2019-SSRF-PT-011509), and the Open Research Fund of State Key Laboratory of Polymer Physics and Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences (2020-1, 2021-16).References
- R. Riek and D. S. Eisenberg, The activities of amyloids from a structural perspective, Nature, 2016, 539, 227–235 CrossRef PubMed.
- L. Gremer, D. Scholzel, C. Schenk, E. Reinartz, J. Labahn, R. Ravelli, M. Tusche, C. Lopez-Iglesias, W. Hoyer, H. Heise, D. Willbold and G. F. Schroder, Fibril structure of amyloid-β (1-42) by cryo-electron microscopy, Science, 2017, 358, 116–119 CrossRef CAS PubMed.
- C. Liang, Z. Ye, B. Xue, L. Zeng, W. Wu, C. Zhong, Y. Cao, B. Hu and P. B. Messersmith, Self-assembled nanofibers for strong underwater adhesion: the trick of barnacles, ACS Appl. Mater. Interfaces, 2018, 10, 25017–25025 CrossRef CAS PubMed.
- Y. Ha, J. Yang, F. Tao, Q. Wu, Y. Song, H. Wang, X. Zhang and P. Yang, Phase-transited lysozyme as a universal route to bioactive hydroxyapatite crystalline film, Adv. Funct. Mater., 2018, 28, 1704476 CrossRef.
- C. P. J. Maury, Origin of life. Primordial genetics: information transfer in a pre- RNA world based on self-replicating beta-sheet amyloid conformers, J. Theor. Biol., 2015, 382, 292–297 CrossRef CAS PubMed.
- S. M. Butterfield and H. A. Lashuel, Amyloidogenic protein-membrane interactions: mechanistic insight from model systems, Angew. Chem., Int. Ed., 2010, 49, 5628–5654 CrossRef CAS.
- J. Adamcik and R. Mezzenga, Amyloid polymorphism in the protein folding and aggregation energy landscape, Angew. Chem., Int. Ed., 2018, 57, 8370–8382 CrossRef CAS PubMed.
- C. Liang, J. Strickland, Z. Ye, W. Wu, B. Hu and D. Rittschof, Biochemistry of barnacle adhesion: an updated review, Front. Mar. Sci., 2019, 6, 565 CrossRef.
- Q. Lu, Q. Tang, Y. Xiong, G. Qing and T. Sun, Protein/peptide aggregation and amyloidosis on biointerfaces, Materials, 2016, 9, 740 CrossRef PubMed.
- H. Okumura and S. G. Itoh, Molecular dynamics simulations of amyloid-β (16–22) peptide aggregation at air–water interfaces, J. Chem. Phys., 2020, 152, 95101 CrossRef CAS.
- D. Wang, Y. Ha, J. Gu, Q. Li, L. Zhang and P. Yang, 2D protein supramolecular nanofilm with exceptionally large area and emergent functions, Adv. Mater., 2016, 28, 7414–7423 CrossRef CAS.
- D. E. Barlow, G. H. Dickinson, B. Orihuela, J. L. Kulp, D. Rittschof and K. J. Wahl, Characterization of the adhesive plaque of the barnacle Balanus amphitrite: amyloid-like nanofibrils are a major component, Langmuir, 2010, 26, 6549–6556 CrossRef CAS PubMed.
- Q. Ren, A. H. Kwan and M. Sunde, Solution structure and interface-driven self-assembly of NC2, a new member of the Class II hydrophobin proteins, Proteins, 2014, 82, 990–1003 CrossRef CAS.
- S. Campioni, G. Carret, S. Jordens, L. Nicoud, R. Mezzenga and R. Riek, The presence of an air-water interface affects formation and elongation of α-synuclein fibrils, J. Am. Chem. Soc., 2014, 136, 2866–2875 CrossRef CAS PubMed.
- M. L. Evans and M. R. Chapman, Curli biogenesis: order out of disorder, Biochim. Biophys. Acta, Mol. Cell Res., 2014, 1843, 1551–1558 CrossRef CAS.
- H. Mohanram, A. Kumar, C. S. Verma, K. Pervushin and A. Miserez, Three-dimensional structure of Megabalanus rosa cement protein 20 revealed by multi-dimensional NMR and molecular dynamics simulations, Philos. Trans. R. Soc., B, 2019, 374, 20190198 CrossRef CAS PubMed.
- J. H. Waite, Mussel adhesion-essential footwork, J. Exp. Biol., 2017, 220, 517–530 CrossRef PubMed.
- M. A. Williams, J. M. Thornton and J. M. Goodfellow, Modelling protein unfolding: hen egg-white lysozyme, Protein Eng., 1997, 10, 895–903 CrossRef CAS PubMed.
- M. G. Iadanza, M. P. Jackson, E. W. Hewitt, N. A. Ranson and S. E. Radford, A new era for understanding amyloid structures and disease, Nat. Rev. Mol. Cell Biol., 2018, 19, 755–773 CrossRef CAS PubMed.
- J. C. Stroud, C. Liu, P. K. Teng and D. Eisenberg, Toxic fibrillar oligomers of amyloid-β have cross-β structure, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7717–7722 CrossRef CAS PubMed.
- Y. Shen, L. Posavec, S. Bolisetty, F. M. Hilty, G. Nyström, J. Kohlbrecher, M. Hilbe, A. Rossi, J. Baumgartner, M. B. Zimmermann and R. Mezzenga, Amyloid fibril systems reduce, stabilize and deliver bioavailable nanosized iron, Nat. Nanotechnol., 2017, 12, 642–647 CrossRef CAS PubMed.
- M. Lei, H. Liao, S. Wang, H. Zhou, J. Zhu, H. Wan, G. F. Payne, C. Liu and X. Qu, Electro-sorting create heterogeneity: constructing a multifunctional Janus film with integrated compositional and microstructural gradients for guided bone regeneration, Adv. Sci., 2024, 11, 2307606 CrossRef CAS PubMed.
- M. Wu, F. Yang, J. Yang, Q. Zhong, V. Körstgen, P. Yang, P. Müller-Buschbaum and Z. Xu, Lysozyme membranes promoted by hydrophobic substrates for ultrafast and precise organic solvent nanofiltration, Nano Lett., 2020, 20, 8760–8767 CrossRef CAS PubMed.
- R. Qin, Y. Guo, H. Ren, Y. Liu, H. Su, X. Chu, Y. Jin, F. Lu, B. Wang and P. Yang, Instant adhesion of amyloid-like nanofilms with wet surfaces, ACS Cent. Sci., 2022, 8, 705–717 CrossRef CAS PubMed.
- S. G. Itoh, M. Yagi-Utsumi, K. Kato and H. Okumura, Effects of a hydrophilic/hydrophobic interface on amyloid-β peptides studied by molecular dynamics simulations and NMR experiments, J. Phys. Chem. B, 2018, 123, 160–169 CrossRef PubMed.
- J. A. J. Housmans, B. Houben, M. Monge-Morera, D. Asvestas, H. H. Nguyen, G. Tsaka, N. Louros, S. Carpentier, J. A. Delcour, F. Rousseau and J. Schymkowitz, Investigating the sequence determinants of the curling of amyloid fibrils using ovalbumin as a case study, Biomacromolecules, 2022, 23, 3779–3797 CrossRef CAS PubMed.
- A. Kumar, H. Mohanram, J. Li, H. Le Ferrand, C. S. Verma and A. Miserez, Disorder-order interplay of a barnacle cement protein triggered by interactions with calcium and carbonate ions: a molecular dynamics study, Chem. Mater., 2020, 32, 8845–8859 CrossRef CAS.
- J. A. Burns, J. C. Butler and J. Moran, Selective reduction of disulfides by tris (2-carboxyethyl) phosphine, J. Org. Chem., 1991, 56, 2648–2650 CrossRef CAS.
- D. A. Jacques and J. Trewhella, Small-angle scattering for structural biology—expanding the frontier while avoiding the pitfalls, Protein Sci., 2010, 19, 642–657 CrossRef CAS PubMed.
- J. C. Giddings, Field-flow fractionation: analysis of macromolecular, colloidal, and particulate materials, Science, 1993, 260, 1456–1465 CrossRef CAS PubMed.
- A. Micsonai, F. Wien, L. Kernya, Y. Lee, Y. Goto, M. Réfrégiers and J. Kardos, Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E3095–E3103 CrossRef CAS PubMed.
- L. N. Arnaudov and R. de Vries, Thermally induced fibrillar aggregation of hen egg white lysozyme, Biophys. J., 2005, 88, 515–526 CrossRef CAS PubMed.
- H. D. T. Mertens and D. I. Svergun, Structural characterization of proteins and complexes using small-angle X-ray solution scattering, J. Struct. Biol., 2010, 172, 128–141 CrossRef CAS PubMed.
- Y. Lin, Y. Lu, C. Tsao, A. Saeki, J. Li, C. Chen, H. Wang, H. Chen, D. Meng, K. Wu, Y. Yang and K. Wei, Enhancing photovoltaic performance by tuning the domain sizes of a small-molecule acceptor by side-chain-engineered polymer donors, J. Mater. Chem. A, 2019, 7, 3072–3082 RSC.
- D. Eisenberg and M. Jucker, The amyloid state of proteins in human diseases, Cell, 2012, 148, 1188–1203 CrossRef CAS PubMed.
- G. Kim, M. Gurau, J. Kim and P. S. Cremer, Investigations of lysozyme adsorption at the air/water and quartz/water interfaces by vibrational sum frequency spectroscopy, Langmuir, 2002, 18, 2807–2811 CrossRef CAS.
- O. D. Monera, T. J. Sereda, N. E. Zhou, C. M. Kay and R. S. Hodges, Relationship of sidechain hydrophobicity and α-helical propensity on the stability of the single-stranded amphipathic α-helix, J. Pept. Sci., 1995, 1, 319–329 CrossRef CAS PubMed.
- S. Raman, L. Karunamoorthy, M. Doble, R. Kumar and R. Venkatesan, Barnacle adhesion on natural and synthetic substrates: adhesive structure and composition, Int. J. Adhes. Adhes., 2013, 41, 140–143 CrossRef CAS.
- P. J. Molino, M. J. Higgins, P. C. Innis, R. M. I. Kapsa and G. G. Wallace, Fibronectin and bovine serum albumin adsorption and conformational dynamics on inherently conducting polymers: a QCM-D study, Langmuir, 2012, 28, 8433–8445 CrossRef CAS PubMed.
- H. Nagaoka and T. Imae, Analytical investigation of two-step adsorption kinetics on surfaces, J. Colloid Interface Sci., 2003, 264, 335–342 CrossRef CAS PubMed.
- R. Gamsjaeger, A. Johs, A. Gries, H. J. Gruber, C. Romanin, R. Prassl and P. Hinterdorfer, Membrane binding of β2-glycoprotein I can be described by a two-state reaction model: an atomic force microscopy and surface plasmon resonance study, Biochem. J., 2005, 389, 665–673 CrossRef CAS PubMed.
- L. Wang, S. Gopalakrishnan and V. M. Rotello, Tailored functional surfaces using nanoparticle and protein “nanobrick” coatings, Langmuir, 2019, 35, 10993–11006 CrossRef CAS PubMed.
- B. Dai, S. G. Kang, T. Huynh, H. Lei, M. Castelli, J. Hu, Y. Zhang and R. Zhou, Salts drive controllable multilayered upright assembly of amyloid-like peptides at mica/water interface, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 8543–8548 CrossRef CAS PubMed.
- T. P. J. Knowles and R. Mezzenga, Amyloid fibrils as building blocks for natural and artificial functional materials, Adv. Mater., 2016, 28, 6546–6561 CrossRef CAS PubMed.
- D. J. C Crisp, G. Walker, G. A. Young and A. B. Yule, Adhesion and substrate choice in mussels and barnacles, J. Colloid Interface Sci., 1985, 104, 40–50 CrossRef.
- P. C. Y. Lau, J. R. Dutcher, T. J. Beveridge and J. S. Lam, Absolute quantitation of bacterial biofilm adhesion and viscoelasticity by microbead force spectroscopy, Biophys. J., 2009, 96, 2935–2948 CrossRef CAS PubMed.
- Y. Liu, F. Tao, S. Miao and P. Yang, Controlling the structure and function of protein thin films through amyloid-like aggregation, Acc. Chem. Res., 2021, 54, 3016–3027 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures and supplementary data. See DOI: https://doi.org/10.1039/d4sc00560k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |