Open Access Article
Open Access ArticleInflachromene ameliorates Parkinson's disease by targeting Nrf2-binding Keap1†
Junhyeong
Yim
 a,
Yoon Soo
Hwang‡
a,
Yoon Soo
Hwang‡
 b,
Jae-Jin
Lee§
c,
Ju Hee
Kim
c,
Jeong Yeob
Baek¶
b,
Jae-Jin
Lee§
c,
Ju Hee
Kim
c,
Jeong Yeob
Baek¶
 d,
Jaeyeong
Jeong||
d,
Young Il
Choi
d,
Jaeyeong
Jeong||
d,
Young Il
Choi
 c,
Byung Kwan
Jin
d and
Seung Bum
Park
c,
Byung Kwan
Jin
d and
Seung Bum
Park
 *abc
*abc
aDepartment of Biophysics and Chemical Biology, Seoul National University, Seoul 08826, Korea. E-mail: sbpark@snu.ac.kr
bCRI Center for Chemical Proteomics, Department of Chemistry, Seoul National University, Seoul 08826, Korea
cSPARK Biopharma, Inc., Seoul, 08791, Korea
dDepartment of Biochemistry & Molecular Biology, School of Medicine, Kyung Hee University, Seoul 02447, Korea
First published on 2nd February 2024
Abstract
Parkinson's disease (PD) is the most common neurodegenerative disease characterized by movement disorder. Despite current therapeutic efforts, PD progression and the loss of dopaminergic neurons in the substantia nigra remain challenging to prevent due to the complex and unclear molecular mechanism involved. We adopted a phenotype-based drug screening approach with neuronal cells to overcome these limitations. In this study, we successfully identified a small molecule with a promising therapeutic effect for PD treatment, called inflachromene (ICM), through our phenotypic screening strategy. Subsequent target identification using fluorescence difference in two-dimensional gel electrophoresis (FITGE) revealed that ICM ameliorates PD by targeting a specific form of Keap1. This interaction led to upregulating various antioxidants, including HO-1, NQO1, and glutathione, ultimately alleviating PD symptoms. Furthermore, ICM exhibited remarkable efficacy in inhibiting the loss of dopaminergic neurons and the activation of astrocytes and microglia, which are critical factors in PD pathology. Our findings suggest that the phenotypic approach employed in this study identified that ICM has potential for PD treatment, offering new hope for more effective therapeutic interventions in the future.
Introduction
Parkinson's disease (PD) is a chronic and progressive neurological disorder1,2 characterized by a range of primary motor symptoms such as bradykinesia,3 tremors,4 and rigidity,5 and accompanied by secondary non-motor symptoms including depression,6 cognitive dysfunction,7 and behavioral disorders.8,9 These motor symptoms arise due to the degeneration of dopaminergic neurons in the substantia nigra, resulting in a reduction in the neurotransmitter dopamine. With the global population aging, the incidence and prevalence of PD are on the rise.1,10 Despite affecting only 1–1.5% of individuals over 60 years old, PD poses a significant economic burden, with the United States alone incurring costs of $51.9 billion in 2017.11 Unfortunately, as of now, there is no cure for PD,12 and available therapies primarily focus on alleviating symptoms by either supplementing dopamine13,14 or inhibiting its clearance.15,16 However, these treatments often trigger common side effects such as nausea, constipation, and confusion. Furthermore, prolonged use of levodopa, a blood–brain barrier-permeable dopamine precursor, can lead to a decrease in efficacy and toxicity, accelerating the death of dopaminergic neurons.14To address the pressing need for novel therapeutics targeting the progression of PD rather than mere symptom relief, our proposed approach involves using a phenotypic assay to overcome the complexities associated with the disease's mechanisms. Our study employed a phenotypic viability recovery assay with neuronal cells exposed to neurotoxins, leading us to a pivotal discovery that ICM effectively inhibits the progression of PD. Further investigation using our novel target identification method unveiled that ICM activates the Nrf2 signaling pathway by directly binding to a specific form of Keap1, thereby mitigating PD progression. This groundbreaking finding suggests a potential avenue for developing novel treatments that target the fundamental causes of PD, thus holding promise for more effective disease management.
Results and discussion
The neuroprotective effect of ICM through Nrf2 activation and the reduction of mitochondrial ROS
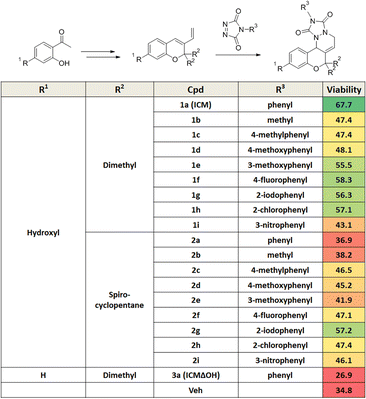
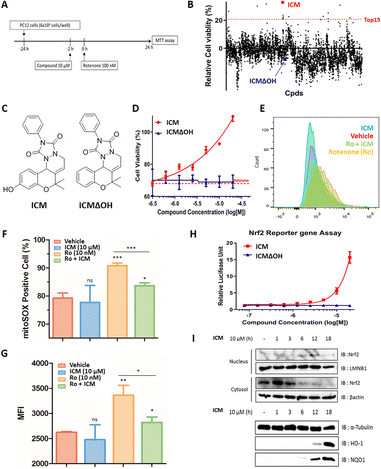
Mitochondrial dysfunction is a prominent characteristic of PD.17,18 Various studies have highlighted the disruptive effects of certain pesticides, like rotenone, on neurons by inhibiting mitochondria complex I, resulting in PD-like symptoms. Due to its ability to penetrate cells independently of the dopamine transporter, rotenone has emerged as a commonly used neurotoxin in in vitro PD models.19–23 To investigate whether pretreatment with a small molecule capable of activating neuroprotective pathways could alleviate neurotoxin-induced cytotoxicity, we screened approximately 4700 compounds from our in-house privileged substructure-based diversity-oriented synthesis (pDOS) library.24,25 The pretreatment of neuron-like PC12 rat pheochromocytoma cells with these compounds, followed by co-treatment with rotenone, enabled the measurement of cell viability after 24 h (Fig. 1A). During this screening campaign, we identified specific hit compounds featuring benzopyran-embedded structures (Fig. S1†). Subsequent preliminary structure–activity relationship studies led to the recognition of ICM as the lead compound,26,27 alongside its negative counterpart, ICMΔOH (Table 1 and Fig. 1B, C). Even though the modification at either R2 dimethyl group or R3 phenyl group did not improve the efficacy of ICM, we identified that the R1 hydroxyl group is the key functional group of ICM and its modification to other functional groups like amine may enhance the activity of ICM.To authenticate the neuroprotective effects of ICM,28,29 we conducted the experiment wherein PC12 cells were exposed to varying doses of ICM or ICMΔOH, followed by co-treatment with rotenone. The results unequivocally confirmed that ICM significantly reinstates cell viability following rotenone-induced cytotoxicity without any toxicity of ICM itself, whereas ICMΔOH fails to yield a similar effect (Fig. 1D and S2†). Additionally, our investigation revealed that rotenone is capable of increasing mitochondrial reactive oxidative stress (ROS), and notably, ICM effectively reduces the levels of mitochondrial ROS under rotenone-treated conditions, bringing them back to basal levels (Fig. 1E–G and S3A, B†).
Considering the pivotal role of Nrf2 as the primary regulator of the cellular oxidative defense mechanism,30–33 we embarked on an investigation to ascertain whether ICM could activate the Nrf2 pathway. Our evaluation of Nrf2 activation involved an Nrf2 reporter gene assay, coupled with the monitoring of Nrf2 translocation through subcellular fractionation and immunoblotting. The Nrf2 reporter gene assay distinctly revealed that ICM activates Nrf2 in a dose- and time-dependent manner, whereas ICMΔOH does not elicit a similar response (Fig. 1H and S4†). Normally, Nrf2 binds to Keap1 dimers and remains in the cytosol at its inactive state. However, upon activation, Nrf2 translocates to the nucleus, assuming its role as a transcription factor. Our observations showed the translocation of Nrf2 from the cytosol to the nucleus after 6 h of ICM treatment, followed by the upregulation of heme-oxygenase 1 (HO-1) and NAD(P)H quinone oxidoreductase-1 (NQO1) after an additional 6 h, indicating that ICM safeguards PC12 cells via the activation of the Nrf2 pathways (Fig. 1I). These findings highlight the potential of ICM in neuroprotective therapies against PD.
FITGE identifies Keap1 as the target protein of ICM
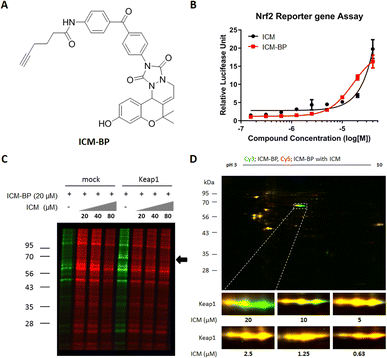
In our effort to pinpoint the target protein of ICM, we utilized the FITGE method,34 which involved the synthesis of photoaffinity probes, ICM-BP and ICMΔOH-BP, incorporating the benzophenone moiety as the photocrosslinker and an alkyne as a handle for click chemistry (Fig. 2A and S7A†). Before deploying ICM-BP and ICMΔOH-BP for FITGE-based target identification, we first evaluated their efficacy on Nrf2 activation via Nrf2 reporter gene assay. The results indicated that ICM-BP activates Nrf2 to a degree comparable to that of ICM, the original compound, whereas ICMΔOH-BP fails to induce a similar response (Fig. 2B and S7B†).Subsequently, these photoaffinity probes were employed for FITGE-based target identification. Initially, HEK293T cells were treated with ICM-BP in the presence or absence of ICM as a competitor. After 30 min of ICM competition, the cells were exposed to 365 nm UV radiation to photocrosslink ICM-BP to its nearby target proteins. The resulting cells were then lysed, and the lysates, with or without ICM competition, were labeled with azide-linked Cy5 or Cy3 fluorescent dyes, respectively, through click reaction. The initial one-dimensional (1D) gel analysis did not reveal any difference in protein bands labeled by ICM-BP in the absence or presence of ICM. However, the overexpression of Keap1 led to the distinct visualization of a band at 69 kDa in the ICM-BP labeling patterns with ICM competition (Fig. 2C and S5A†). The relatively low endogenous expression level of Keap1 rendered it imperceptible without Keap1 overexpression (Fig. S5B†).35,36 Furthermore, two-dimensional (2D) gel analysis demonstrated discrete labeling patterns in a dose-dependent manner upon competition between ICM-BP and ICM (Fig. 2D). The protein spots outcompeted in the 2D gel analysis were further subjected to LC/MS/MS analysis, confirming Keap1 as the target protein of ICM (Table S1†). To clarify the off-target effects of ICM, we also conducted proteome analysis using competition between ICM and its target validation probe, ICM-alkyne, without the benzophenone moiety to eliminate the photocrosslinking activity (Fig. 4A). The outcome of the chemoproteomics study with ICM-lkyne showed that ICM selectively bound to Keap1 except either non-specific proteins, like albumin and hnRNP, or Nrf2-mediated metabolism related proteins, like ENO-1 and FASN (Fig. S6†). Conversely, ICMΔOH-BP exhibited entirely different labeling patterns in live HEK293T cells but not in lysates (Fig. S7C–E†). This emphasizes the importance of conducting target identification studies in live cells and employing appropriate negative probes for accurate assessments. Overall, our FITGE-based approach successfully identified Keap1 as the target protein of ICM, providing insights into the underlying mechanism of ICM's neuroprotective effects through Nrf2 activation and the reduction of mitochondrial ROS.
Nrf2-binding Keap1 is the target of ICM
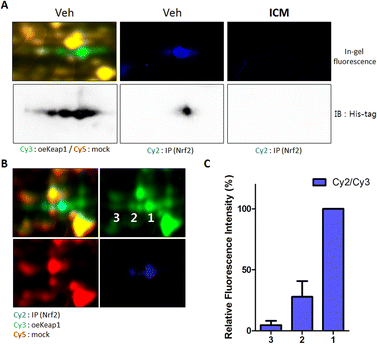
Notably, the three spots labeled by ICM-BP displayed a concentration-dependent competitive response with ICM (Fig. 2D). At 20 μM concentration of ICM, the spot corresponding to Keap1 with a high isoelectric point (pI) value was preferentially outcompeted. As the ICM concentration increased, the pI value of the out-competed protein spots gradually decreased. Given Keap1's known role in interacting with Nrf2 and facilitating Nrf2 degradation through proteasome-mediated ubiquitination,37 we hypothesized that the pI value of Keap1 might be linked to its interaction with Nrf2. To validate the direct binding of Nrf2 with Keap1, we performed a pull-down assay of Nrf2 proteins using an Nrf2-specific antibody. Upon immunoblotting the Nrf2-binding proteome with a Keap1-specific antibody, we observed that Keap1 from a specific spot directly bound to Nrf2 among the three spots; however, this Keap1–Nrf2 interaction was obstructed by ICM treatment (Fig. 3A).To further elucidate the pI values of Nrf2-binding Keap1, we conducted 2D gel analysis of a mixed sample containing Cy2-labeled Nrf2-binding proteome (in cyan), Cy3-labeled Keap1 overexpressed proteome (in green), and Cy5-labeled mock proteome (in red). This analysis revealed that Keap1 with a high pI value was involved in the interaction with Nrf2 (Fig. 3B). The ratio of the entire Keap1 protein to the Nrf2-binding Keap1 protein, as indicated by the fluorescence signals, reinforced the direct binding of Nrf2 with Keap1 having a high pI value (Fig. 3C).
To gain insights into the relationship between Nrf2 and the pI values of Keap1, we delved into the post-translational modifications (PTMs) of Keap1 in the three spots using mass spectrometry (Fig. S8†). Among various PTMs, we specifically observed a significant disparity in cysteine oxidation, which exhibited a correlation with the pI value of Keap1 (Table S2†). As the number of cysteine oxidation events increased, the pI value of Keap1 decreased, suggesting that the potential binding sites of ICM might involve specific cysteine residues, such as Cys77, 151, 241, 288, 297, and 319 of Keap1. These findings strongly support that ICM directly targets Nrf2-binding Keap1, likely through an interaction with specific cysteine residues. The disruption of the Keap1–Nrf2 interaction by ICM could explain the activation of the Nrf2 pathway and the subsequent neuroprotective effects observed in our previous investigations.
Validation of the covalent binding between ICM and Keap1 using the target validation probe ICM-alkyne
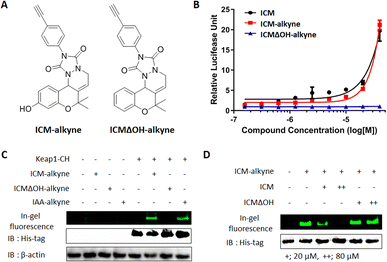
To confirm the covalent binding between ICM and Keap1, we synthesized the target validation probe, ICM-alkyne, without benzophenone to eliminate the photocrosslinking activity, along with a negative probe, ICMΔOH-alkyne (Fig. 4A).38,39 Before employing ICM-alkyne for target validation, we assessed Nrf2 activation using an Nrf2 reporter gene assay. The results confirmed that ICM-alkyne activates Nrf2 to a level comparable to that of ICM, the original compound, while the negative probe did not exhibit this activity (Fig. 4B).Given that electrophiles, like iodoacetamide, covalently bind to the cysteine residues of Keap1, leading to Nrf2 activation,40 we also prepared iodoacetamide with an alkyne (IAA-alkyne) as a positive control probe. With or without transient Keap1 overexpression, HEK293T cells were treated with ICM-alkyne, ICMΔOH-alkyne, and IAA-alkyne for 12 min, followed by click reaction with an azide-linked Cy3 fluorescent dye. 1D gel analysis revealed two distinct bands after ICM-alkyne and IAA-alkyne treatments in cells with Keap1 overexpression, while these bands were absent in cells treated with ICMΔOH-alkyne (Fig. 4C and S9A†), indicating the covalent binding of ICM-alkyne and IAA-alkyne to the overexpressed Keap1 in cells. To further validate the covalent binding of ICM, we compared the labeling patterns of ICM-alkyne in the competition with either ICM or ICMΔOH, in the context of Keap1 overexpression. As shown in Fig. 4D and S9B,† ICM competition resulted in a dose-dependent reduction in the labeling of ICM-alkyne to Keap1, while ICMΔOH did not elicit a similar effect. This confirmed that ICM effectively occupies the binding site of ICM-alkyne on Keap1, hindering the covalent binding of ICM-alkyne to Keap1.
The successful covalent binding of ICM-alkyne to Keap1 reinforced the concept that ICM directly targets Keap1, particularly through its interaction with cysteine residues. This binding event likely contributes to Nrf2 activation and the subsequent neuroprotective effects observed during ICM treatment. These findings enhance our understanding of the molecular mechanism underlying ICM's action and underscore the potential of Keap1 as a promising therapeutic target for the treatment of PD and related neurodegenerative conditions.
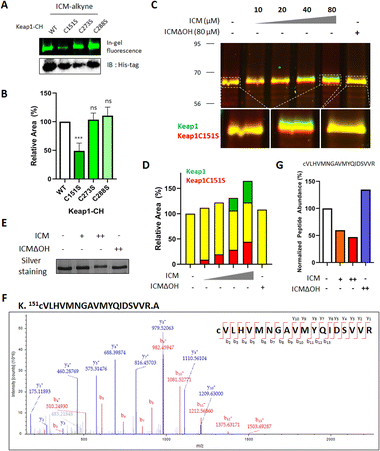
ICM covalently binds to Cys151 of Keap1
Keap1 functions as a vital intracellular sensor in mammals, sensing oxidative stress through three main residues, Cys151, Cys273, and Cys288.41–45 Our observations of the covalent binding between ICM and Keap1, along with the implication of cysteine as the targeted amino acid (Fig. 3, 4, S9 and Table S2†), prompted an investigation to identify the specific cysteine residue on Keap1 responsible for the binding with ICM. To address this, we transfected HEK293T cells with either wild-type (WT) Keap1 or mutants where each of the three major cysteines was substituted by serine (C151S, C273S, and C288S). Subsequently, we treated the resulting cells with ICM-alkyne to evaluate covalent binding and found that the C151S-mutant Keap1 exhibited a notable reduction in covalent labeling with ICM-alkyne (Fig. 5A, B and S10†).To further confirm whether Cys151 of Keap1 was the critical residue for covalent binding with ICM, HEK293T cells transfected with either WT or C151S-mutant Keap1 were treated with ICM and ICMΔOH. While WT Keap1 exhibited a molecular shift in 1D gel electrophoresis upon ICM treatment in a dose-dependent manner, no such shift was observed with ICMΔOH, even at the highest dose (green signals in Fig. 5C and S11†). Conversely, the C151S-mutant Keap1 displayed no molecular shift upon ICM treatment (red signals in Fig. 5C and S11†), confirming that Cys151 of Keap1 is the primary binding site for ICM. As the ICM concentration increased, the green and red signals intensified without merging (Fig. 5D, S11 and S12†).
The subsequent validation using MS/MS analysis confirmed that Cys151 is indeed the key residue of Keap1 responsible for specific binding to ICM. We treated Keap1-overexpressed HEK293T cells with either 10/100 μM ICM or 100 μM ICMΔOH, followed by cell lysis. The ICM-mediated molecular shift of Keap1 was confirmed through 1D gel analysis (Fig. 5E and S13A†), and the Keap1 in the lysates underwent analysis using ultra-performance liquid chromatography coupled with tandem mass spectrometry (LC/MS/MS). The mass analysis results confirmed that ICM treatment led to a dose-dependent reduction in the mass signals of the tryptic peptide of Keap1 containing Cys151, irrespective of cysteine carbamidomethylation. Conversely, ICMΔOH showed no effect (Fig. 5F, G, S13B, C and Table S3†). Despite our efforts, we were unable to identify the mass signals of peptides covalently modified with ICM, probably due to their inefficient ionization. It is also interesting that Cys151-specific covalent labeling of ICM to Keap1 was only observed in live cells, but not in lysates (Fig. S7C–E†), which may result from the unknown metabolic processes of ICM in the live cellular system. Overall, these comprehensive findings unequivocally demonstrate that ICM covalently binds to Cys151 of Keap1, providing a better understanding of the molecular basis for ICM's interaction with Keap1 and its subsequent activation of the Nrf2 pathway.
ICM activates Nrf2, leading to an increase in antioxidants and glutathione via p-p62-mediated Keap1 degradation
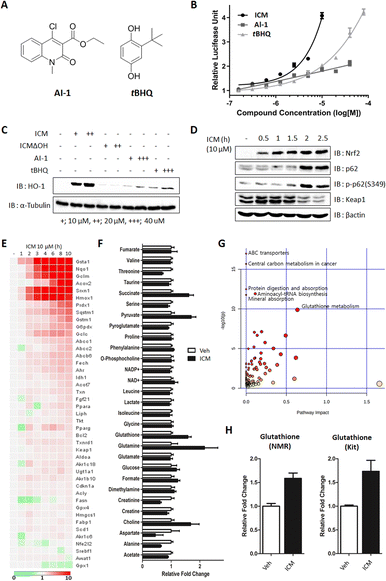
Small molecules, such as AI-1 and tert-butylhydroxylquinone (tBHQ), have been shown to activate Nrf2 and alleviate the symptoms of MPTP-induced parkinsonism by enhancing antioxidant activity, specifically through the cellular upregulation of HO-1 and NQO1 (Fig. 6A).46,47 To monitor the upregulation of antioxidants, we conducted Nrf2 reporter gene assays and immunoblots for ICM, AI-1, and tBHQ. The results indicated that 10 μM AI-1 and tBHQ induced 2.04- and 1.87-fold increases, respectively, while 40 μM AI-1 and tBHQ induced 3.38- and 2.12-fold increases in Nrf2 transcriptional activity compared to the vehicle. Notably, 10 μM ICM showed an even greater 4.08-fold increase in Nrf2 transcriptional activity compared to the vehicle, surpassing the efficacy of AI-1 and tBHQ at the same concentration (Fig. 6B).As the levels of Nrf2-driven antioxidants correlated with Nrf2 transcriptional activity, we further confirmed the increase in HO-1 and NQO1 using immunoblot assays. It was observed that 10 μM ICM induced a greater increase in HO-1 and NQO1, both at the mRNA and protein levels, than 40 μM AI-1 or tBHQ, consistent with the outcomes of the Nrf2 reporter gene assay (Fig. 6C and S14A, B†). Furthermore, 10 μM ICM resulted in a higher induction of most Nrf2-driven genes compared to 40 μM AI-1 or tBHQ (Fig. S15 and S16†).
Nrf2 activation is known to increase both p62 and p-p62 (S349), leading to the autophagic degradation of Keap1.48 As the positive feedback loop, the increase of p62 and p-p62 also activates Nrf2 through Keap1 autophagic degradation. Time-course monitoring of Nrf2, p62, and p-p62 demonstrated that ICM induced Nrf2 activation within 5 min without affecting p62 and p-p62 levels (Fig. S17†). After 2 h of ICM treatment, an increase in both p62 and p-p62 was observed, followed by the subsequent degradation of Keap1, indicating that ICM induces Nrf2 activation and Keap1 degradation through the upregulation of p62 and p-p62 (Fig. 6D).
Considering that Nrf2 activation induces metabolic reprogramming and enhances resistance to various cellular stresses,48 we monitored Nrf2-mediated genes and metabolites following ICM treatment. Among the 43 Nrf2-mediated genes, Hmox1, Nqo1, and glutathione-related genes, including Gsta1, Gclm, and Gstm1, significantly increased after 2 h of ICM treatment (Fig. 6E). An NMR-based metabolomics study also revealed substantial increases in cellular glutathione, succinate, pyruvate, glutamine, and choline among 29 metabolites (Fig. 6F and S18A†). The subsequent kit-based quantitative assay double confirmed a significant increase of glutathione (Fig. 6H). Integrated analysis of transcriptome and metabolome data confirmed that glutathione metabolism is one of the key pathways regulated by ICM (Fig. 6G and S18B, C†). Overall, ICM demonstrated a neuroprotective effect by activating Nrf2 and subsequently increasing cellular antioxidants, including HO-1, NQO1, and glutathione. These findings hold significant promise for the development of potential therapies targeting Nrf2 activation for PD and other oxidative stress-related disorders.
ICM demonstrates a neuroprotective effect in an in vivo PD model
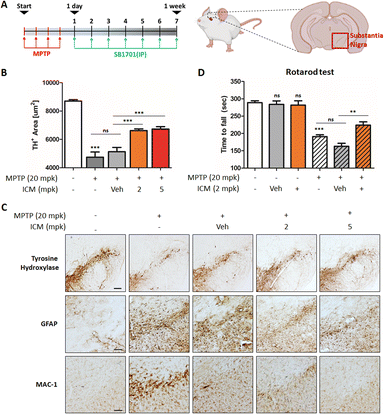
The neurotoxin MPTP is commonly used to induce in vivo PD models, leading to various insults, including oxidative stress and mitochondrial damage, ultimately resulting in the selective death of dopaminergic neurons in the substantia nigra and the activation of microglia.21,23 To assess the neuroprotective potential of ICM, we administered MPTP and ICM via intraperitoneal injection following the schedule depicted in Fig. 7A. Quantification of the positive cells of tyrosine hydroxylase (TH), a marker for dopaminergic neurons that converts tyrosine to levodopa, revealed a significant decrease in dopaminergic neurons induced by MPTP, which was effectively rescued by the treatment with 2 mg kg−1 ICM, in contrast to the vehicle treatment (Fig. 7B). In assessing the activation levels of astrocytes and microglia, we employed glial fibrillary acidic protein (GFAP), as a marker for astrocyte injury and stress, and macrophage-1 antigen (MAC-1) as an adhesion and recognition receptor in microglia. MPTP treatment led to increased activation levels of both astrocytes and microglia, whereas ICM efficiently ameliorated their over-activation (Fig. 7C).MPTP-induced loss of dopaminergic neurons contributed to decreased dopamine release and motor imbalances. To assess motor coordination and balance, we conducted a rotarod test. Mice treated with MPTP exhibited a significant decline in balance performance, which was effectively restored by ICM treatment (Fig. 7D). Nrf2 activation plays a central role in the PD treatment; however, metabolic pathways including glycolysis and fatty acid synthesis may also be associated with ICM efficacy in the in vivo PD model (Fig. 6F, G and S6†). Collectively, these results demonstrate that ICM effectively protects against MPTP-induced dopaminergic neuronal death and alleviates the over-activation of astrocytes and microglia, leading to the restoration of MPTP-induced movement disorders in the in vivo PD model. These findings highlight the therapeutic potential of ICM as a neuroprotective agent in PD and emphasize its promise for further exploration in clinical applications.
Conclusion
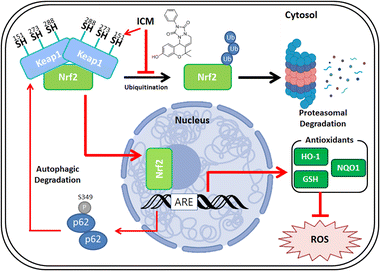
Parkinson's disease (PD) presents a complex neurological challenge, with its progression intricately linked to the aging process.1,2,10 While a cure for PD progression remains elusive,12 available medications like levodopa and MAO-B inhibitors offer only temporary symptom relief.13–16 To tackle these limitations, our phenotypic screening of our in-house 4700-membered pDOS library led to the discovery of ICM, a benzopyran-embedded small molecule demonstrating the restoration of neuronal cell death in in vitro and in vivo neurotoxin-induced PD models. Our novel target ID method FITGE revealed that ICM binds to specific Nrf2-binding Keap1, followed by upregulating Nrf2-driven antioxidants like HO-1, NQO1, and glutathione (Fig. 8). Our findings suggest that while our phenotype-based screening did not directly elucidate the unknown mechanism of PD, the Nrf2-driven upregulation of antioxidants effectively inhibited PD progression. Our results underscore the significance of strong Nrf2 activation and the subsequent increment of antioxidants as the principal axes for PD medication. This discovery holds promise for the development of Nrf2/antioxidant activators as potential therapeutics for PD and various other neurodegenerative diseases.Although ICM showed sufficient selectivity to Keap1 and exceeded the activity of other known Nrf2 activators,46,47 our SAR study did not improve the activity of lCM in this study. However, we identified that the hydroxyl group of ICM plays a crucial role in modulating Keap1 in the living cellular system. Therefore, we are currently pursuing further SAR study to improve the efficacy and pharmacokinetic properties of ICM. Despite these unsolved issues including blood–brain barrier permeability, this study suggests Nrf2 activation via autophagic degradation of Keap1 as a new prominent therapeutic strategy for the treatment of various neurodegenerative diseases.
Data availability
The datasets supporting this article have been uploaded as part of the ESI.†Author contributions
J. Y. and S. B. P. designed the study, prepared the figures, and wrote the manuscript. J. Y. performed the high-throughput screening and molecular biology work. Y. S. H. synthesized and characterized the compounds. J.-J. L., J. H. K. and Y. C. conducted mass-spectrometry analysis. J. Y. B., J. J. and B. K. J. performed in vivo experiments. All authors critically reviewed the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by the Creative Research Initiative Grant (2014R1A3A2030423 to S. B. P.) and Creative Challenging Research Grant (RS-2023-00250887 to J. Y.) through the National Research Foundation of Korea (NRF) funded by the Korean government (Ministry of Science & ICT). We gratefully acknowledge the National Center for Inter-university Research Facilities (NCIRF) for technical support of FACS analysis. We gratefully appreciate Dr Young-Joon Surh Cancer Institute, Seoul National University for kindly providing pARE-luc plasmid for Nrf2 reporter gene assay. Figures were created with BioRender.com.References
- J. Apfeld and W. Fontana, Biology, 2017, 7, 1 CrossRef PubMed.
- M. J. Armstrong and M. S. Okun, JAMA, 2020, 323, 548–560 CrossRef PubMed.
- D. E. Vaillancourt, J. Prodoehl, L. Verhagen Metman, R. A. Bakay and D. M. Corcos, Brain, 2004, 127, 491–504 CrossRef PubMed.
- M. Löhle, A. Storch and H. Reichmann, J. Neural Transm., 2009, 116, 1483–1492 CrossRef PubMed.
- A. A. Moustafa, S. Chakravarthy, J. R. Phillips, A. Gupta, S. Keri, B. Polner, M. J. Frank and M. Jahanshahi, Neurosci. Biobehav. Rev., 2016, 68, 727–740 CrossRef PubMed.
- D. Aarsland, S. Påhlhagen, C. G. Ballard, U. Ehrt and P. Svenningsson, Nat. Rev. Neurol., 2012, 8, 35–47 CrossRef CAS PubMed.
- N. Caballol, M. J. Martí and E. Tolosa, Mov. Disord., 2007, 22, S358–S366 CrossRef PubMed.
- W. Poewe, Eur. J. Neurol., 2008, 15, 14–20 CrossRef PubMed.
- C. Papagno and L. Trojano, Neurol. Sci., 2018, 39, 215–223 CrossRef PubMed.
- Y. Hou, X. Dan, M. Babbar, Y. Wei, S. G. Hasselbalch, D. L. Croteau and V. A. Bohr, Nat. Rev. Neurol., 2019, 15, 565–581 CrossRef PubMed.
- W. Yang, J. L. Hamilton, C. Kopil, J. C. Beck, C. M. Tanner, R. L. Albin, E. Ray Dorsey, N. Dahodwala, I. Cintina, P. Hogan and T. Thompson, NPJ Parkinsons Dis., 2020, 6, 15 CrossRef PubMed.
- J. Jankovic and L. G. Aguilar, Neuropsychiatr. Dis. Treat., 2008, 4, 743 CrossRef CAS PubMed.
- P. S. Group, N. Engl. J. Med., 2004, 351, 2498–2508 CrossRef PubMed.
- P. A. LeWitt, N. Engl. J. Med., 2008, 359, 2468–2476 CrossRef CAS PubMed.
- K. C. Teo and S.-L. Ho, Transl. Neurodegener., 2013, 2, 1–10 CrossRef PubMed.
- L. Dezsi and L. Vecsei, CNS Neurol. Disord.: Drug Targets, 2017, 16, 425–439 CrossRef CAS PubMed.
- A. Schapira, J. Cooper, D. Dexter, J. Clark, P. Jenner and C. Marsden, J. Neurochem., 1990, 54, 823–827 CrossRef CAS.
- C. Henchcliffe and M. F. Beal, Nat. Clin. Pract. Neurol., 2008, 4, 600–609 CrossRef CAS PubMed.
- D. F. Tardiff and S. Lindquist, Drug Discovery Today: Technol., 2013, 10, e121–e128 CrossRef.
- X. Chen, J. Guo, J. Bao, W. Xu, Y. Huang and Y. Wang, Curr. Psychopharmacol., 2012, 1, 103–110 CAS.
- M. H. Abdel-Wahab, J. Biochem. Mol. Toxicol., 2005, 19, 32–41 CrossRef CAS PubMed.
- H. Q. Liu, X. Z. Zhu and E. Q. Weng, Acta Pharmacol. Sin., 2005, 26, 17–26 CrossRef CAS PubMed.
- M. Mustapha and C. N. Mat Taib, Bosnian J. Basic Med. Sci., 2021, 21, 422–433 CAS.
- J. Kim, H. Kim and S. B. Park, J. Am. Chem. Soc., 2014, 136, 14629–14638 CrossRef CAS.
- J. Kim, J. Koo, J. Jung, W. Cho, W. S. Lee, C. Kim, W. Park and S. B. Park, Nat. Commun., 2016, 7, 13196 CrossRef CAS PubMed.
- S. K. Ko, J. H. Jang, E. Kim and S. B. Park, Chem. Commun., 2006, 28, 2962–2964 RSC.
- S. Lee, Y. Nam, J. Y. Koo, D. Lim, J. Park, J. Ock, K. Suk and S. B. Park, Nat. Chem. Biol., 2014, 10, 1055–1060 CrossRef CAS PubMed.
- Y.-H. Shin, H. Cho, B. Y. Choi, J. Ha, J. Kim, I. Yim, S. W. Suh and S. B. Park, Angew. Chem., Int. Ed., 2021, 60, 1831–1838 CrossRef CAS PubMed.
- J. Yim, J. Lee, S. Yi, J. Y. Koo, S. Oh, H. Park, M. A. Bae, J. Park and S. B. Park, Exp. Mol. Med., 2022, 54, 2200–2209 CrossRef CAS PubMed.
- F. He, X. Ru and T. Wen, Int. J. Mol. Sci., 2020, 21, 4777 CrossRef CAS PubMed.
- H. Kawagishi and T. Finkel, Nat. Med., 2014, 20, 711–713 CrossRef CAS PubMed.
- S. Kasai, S. Shimizu, Y. Tatara, J. Mimura and K. Itoh, Biomolecules, 2020, 10, 320 CrossRef CAS PubMed.
- J. M. Lee, J. H. Lee, M. K. Song and Y. J. Kim, Antioxidant, 2022, 11, 130 CrossRef CAS.
- J. Park, S. Oh and S. B. Park, Angew. Chem., Int. Ed., 2012, 51, 5447–5451 CrossRef CAS PubMed.
- T. Iso, T. Suzuki, L. Baird and M. Yamamoto, Mol. Cell. Biol., 2016, 36, 3100–3112 CrossRef CAS PubMed.
- R. Milo, BioEssays, 2013, 35, 1050–1055 CrossRef CAS PubMed.
- M. McMahon, K. Itoh, M. Yamamoto and J. D. Hayes, J. Biol. Chem., 2003, 278, 21592–21600 CrossRef CAS PubMed.
- J. Ha, H. Park, J. Park and S. B. Park, Cell Chem. Biol., 2021, 28, 394–423 CrossRef CAS PubMed.
- H. Park and S. B. Park, Chem. Sci., 2019, 10, 3449–3458 RSC.
- F. Hong, K. R. Sekhar, M. L. Freeman and D. C. Liebler, J. Biol. Chem., 2005, 280, 31768–31775 CrossRef CAS.
- M. McMahon, D. J. Lamont, K. A. Beattie and J. D. Hayes, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 18838–18843 CrossRef CAS.
- T. Ohnuma, S. Nakayama, E. Anan, T. Nishiyama, K. Ogura and A. Hiratsuka, Toxicol. Appl. Pharmacol., 2010, 244, 27–36 CrossRef CAS.
- Y. Mitsuishi, H. Motohashi and M. Yamamoto, Front. Oncol., 2012, 2, 200 Search PubMed.
- A. T. Dinkova-Kostova, R. V. Kostov and P. Canning, Arch. Biochem. Biophys., 2017, 617, 84–93 CrossRef CAS.
- S. Dayalan Naidu, A. Muramatsu, R. Saito, S. Asami, T. Honda, T. Hosoya, K. Itoh, M. Yamamoto, T. Suzuki and A. T. Dinkova-Kostova, Sci. Rep., 2018, 8, 8037 CrossRef PubMed.
- H. Hara, M. Ohta, K. Ohta, S. Kuno and T. Adachi, Mol. Brain Res., 2003, 119, 125–131 CrossRef CAS PubMed.
- W. Hur, Z. Sun, T. Jiang, D. E. Mason, E. C. Peters, D. D. Zhang, H. Luesch, P. G. Schultz and N. S. Gray, Chem. Biol., 2010, 17, 537–547 CrossRef CAS.
- Y. Katsuragi, Y. Ichimura and M. Komatsu, Curr. Opin. Toxicol., 2016, 1, 54–61 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, supplemental data, and spectral data. See DOI: https://doi.org/10.1039/d3sc06997d |
| ‡ Present address: Material Technology Team, Samsung Electronics Co., Ltd, 1 Samsung-ro Giheung-gu, Yongin-si, 17113, Korea. |
| § Present address: Neuromeka, Inc., 7 Yeonmujang5ga-gil Seongdong-gu, Seoul, 04782, Korea. |
| ¶ Present address: Korea Institute of Toxicology, 141 Gajeong-ro, Yuseong-gu Deajeon, 34114, Korea. |
| || Present address: Department of Neurosurgery, The University of Texas Health Science Center at Houston, 6431 Fannin St, Houston, TX 77030, USA. |
| This journal is © The Royal Society of Chemistry 2024 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) >
>