Open Access Article
Open Access ArticleDNA-encoded chemical libraries enable the discovery of potent PSMA-ligands with substantially reduced affinity towards the GCPIII anti-target†
Laura
Lucaroni‡
 a,
Sebastian
Oehler
a,
Sebastian
Oehler
 a,
Tony
Georgiev
a,
Tony
Georgiev
 a,
Marco
Müller
a,
Marco
Müller
 a,
Matilde
Bocci
a,
Matilde
Bocci
 a,
Roberto
De Luca
a,
Roberto
De Luca
 a,
Nicholas
Favalli
a,
Nicholas
Favalli
 a,
Dario
Neri
abc,
Samuele
Cazzamalli
*a and
Luca
Prati
a,
Dario
Neri
abc,
Samuele
Cazzamalli
*a and
Luca
Prati
 *c
*c
aPhilochem AG, R&D Department, CH-8112 Otelfingen, Switzerland. E-mail: samuele.cazzamalli@philochem.ch; Tel: +41 43 544 88 19
bSwiss Federal Institute of Technology, Department of Chemistry and Applied Biosciences, CH-8093 Zurich, Switzerland
cPhilogen S.p.A., 53100 Siena, Italy. E-mail: luca.prati@philogen.com; Tel: +39 0577 178 16 59
First published on 2nd April 2024
Abstract
Prostate-specific membrane antigen (PSMA) is a tumor-associated protein that has been successfully targeted with small organic ligands and monoclonal antibodies. Pluvicto™ is a PSMA-targeted radioligand therapeutic (RLT) recently approved by the FDA for the treatment of metastatic castration-resistant prostate cancer (2022 FDA marketing authorization). Although a large Phase III clinical trial (VISION trial) demonstrated clinical benefits in patients treated with Pluvicto™, the therapeutic window of the drug is narrowed by its undesired accumulation in healthy organs. Glutamate carboxypeptidase III (GCPIII), an enzyme sharing 70% identity with PSMA, may be responsible for the off-target accumulation of PSMA-RLTs in salivary glands and kidneys. In this work, we designed and synthesized affinity and selectivity maturation DNA-encoded chemical libraries (ASM-DELs) comprising 18′284′658 compounds that were screened in parallel against PSMA and GCPIII with the aim to identify potent and selective PSMA ligands for tumor-targeting applications. Compound A70-B104 was isolated as the most potent and selective ligand (KD of 900 pM for PSMA, KD of 40 nM for GCPIII). 177Lu-A70-B104-DOTA, a radiolabeled derivative of compound A70-B104, presented selective accumulation in PSMA-positive cancer lesions (i.e., 7.4% ID g−1, 2 hour time point) after systemic administration in tumor-bearing mice. The results of autoradiography experiments showed that 177Lu-A70-B104-DOTA selectively binds to PSMA-positive cancer tissues, while negligible binding on human salivary glands was observed.
Introduction
Radioligand therapeutics (RLTs) are innovative radiopharmaceutical products that combine the use of radionuclide payloads with small organic ligands that specifically bind to proteins expressed on cancer cells or in the tumor microenvironment.1 This approach enables the selective accumulation of biocidal radiation within the tumor mass, thus maximizing the therapeutic effect of radioactivity while minimizing unwanted healthy tissue damage.1 Lutathera™,2 Xofigo™,3 and Pluvicto™4 are examples of approved products for the treatment of specific solid tumor entities. Treatment with RLTs is usually paired with companion diagnostics that enables the selection of patients with high ligand uptake in tumor masses.5Prostate-specific membrane antigen (PSMA)6 is one of the few examples of tumor-associated antigens which have been validated through nuclear medicine studies for the development of targeted anti-cancer RLTs.7,8 While the physiological function of PSMA in prostate cells is not yet fully understood,9–12 its pronounced upregulation within prostate cancer lesions led to the development of high-affinity monoclonal antibodies and small organic ligands,13–20 including (2S,2′S)-2,2′-(carbonyldiimino)dipentanedioic acid (DUPA, IC50,PSMA = 47 nM).21 Further chemical elaborations on DUPA prompted the identification of PSMA-11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 and PSMA-61723 as lead compounds for diagnostic and therapeutic applications (respectively) with excellent biodistribution profiles in tumor-bearing mice24,25 and patients.26,27
22 and PSMA-61723 as lead compounds for diagnostic and therapeutic applications (respectively) with excellent biodistribution profiles in tumor-bearing mice24,25 and patients.26,27
Pluvicto™, also known as 177Lu-PSMA-617, is the most recent RLT product gaining marketing authorization for the treatment of metastatic castration-resistant prostate cancer (mCRPC; FDA approval in 2022).4 Pluvicto™ consists of PSMA-617 conjugated to a DOTA-lutetium-177 complex as the radioactive payload (beta-emitter). The registrational phase III clinical trial VISION showed that treatment with Pluvicto™ in combination with the best standard of care (BSoC) enhances the overall response rate from 2% (BSoC alone) to 30%. Only a marginal benefit was observed in overall survival (15.3 months vs. 11.3 months with BSoC alone).4 Despite the efficacy demonstrated, around 12%28 of individuals discontinued Pluvicto™ due to treatment-related adverse events, including xerostomia (“dry mouth”) and renal and urinary disorders. Nuclear medicine studies indicate that these side effects are due to the accumulation of the drug in healthy structures, including the salivary and lacrimal glands, kidneys, spleen, and liver.29,30 The undesired uptake of Pluvicto™ in healthy organs limits the possibility to increase the administered dose, thereby narrowing its therapeutic window.31
Our group has recently hypothesized that the off-target accumulation of Pluvicto™ may be linked to the presence of glutamate carboxypeptidase III (GCPIII) in healthy tissues.31 GCPIII shares 70% sequence identity with PSMA,31 along with a comparable enzymatic activity and analogous structure of the active site.32 We have previously shown that a fluorescent surrogate of Pluvicto™ (PSMA-617-FITC) strongly binds to recombinant GCPIII with a KD of ∼1 nM. This antigen is abundantly expressed in the salivary glands and kidneys (by immunofluorescence studies),31 and this molecular event may be responsible for the accumulation of biocidal radiation in healthy structures. Despite documented efforts to identify ligands specific to PSMA over GCPIII,33,34 PSMA-targeted RLTs which show high tumor-to-salivary gland and tumor-to-kidney ratios have not yet been reported.
DNA-encoded chemical library (DEL) technology is a powerful tool for the discovery of tumor-targeting ligands.33,35–37 DELs are large collections of small organic compounds, individually encoded by unique DNA strands that act as amplifiable identification barcodes. Binders against immobilized proteins of interest (targets) are typically isolated by affinity capture, followed by PCR amplification and high-throughput DNA sequencing (HTDS). The DEL technology has been proven reliable for de novo discovery and affinity maturation strategies.35,38–40 In the latter, a known binder for the target protein is coupled to an existing DEL with the aim of increasing its affinity to the target protein.38–40 In this article, we report the design, screening, and hit-validation of affinity and selectivity maturation DELs (ASM-DELs), aiming at improving ligand affinity to PSMA of previously identified HIT compounds, together with their selectivity against GCPIII (the biological “anti-target” of Pluvicto™).
Results
The structure and design of novel affinity and selectivity maturation-DELs (ASM-DELs) are shown in Fig. 1. PSMA-selective HIT1 and HIT2 compounds chosen for the construction of the final ASM-DELs were derived from de novo DEL selections previously reported by our group33 [Fig. 1B and S33A, B†]. The design of HIT3 (DUPA) was inspired by the seminal work of Kozikowski et al.21 [Fig. 1B]. This compound displays high potency towards PSMA (KD,PSMA = 1 nM), but lacks selectivity for GCPIII (KD,GCPIII = 1 nM) [Fig. S33C†]. A number of ASM-DELs were used in our study. Some libraries featured a linear arrangement of amino acid building blocks41 (LL1, LL2, Fig. 1B). This design may be particularly attractive for the discovery of protease inhibitors, considering the peptidic nature of protease substrates. By contrast, MB libraries featured a central aromatic core with a free carboxylic acid and up to three amino acids as diversity elements (MB1, MB2, and MB3) [Fig. 1B]. Finally, the PRA DEL design35 displayed a linear arrangement of two amino acids coupled to a propargyl glycine scaffold. All these libraries feature a terminal amino moiety suitable for derivatization with the HITs by amide bond formation. Detailed procedures (split-and-pool) followed during the synthesis of DEL-precursors and the final ASM-DELs are reported in ESI Sections S2 and S3.† Eighteen highly pure ASM-DELs were synthesized (>95% purity, calculated from the HPLC peak area after purification of the final pool), comprising 18′284′658 molecules. Underivatized DELs and ASM-DELs (capped with HIT1, HIT2, or HIT3) were screened in parallel against site-specifically biotinylated human recombinant PSMA and GCPIII (immobilized on streptavidin beads), following state of the art screening protocols.42 All DEL selections were performed in duplicate.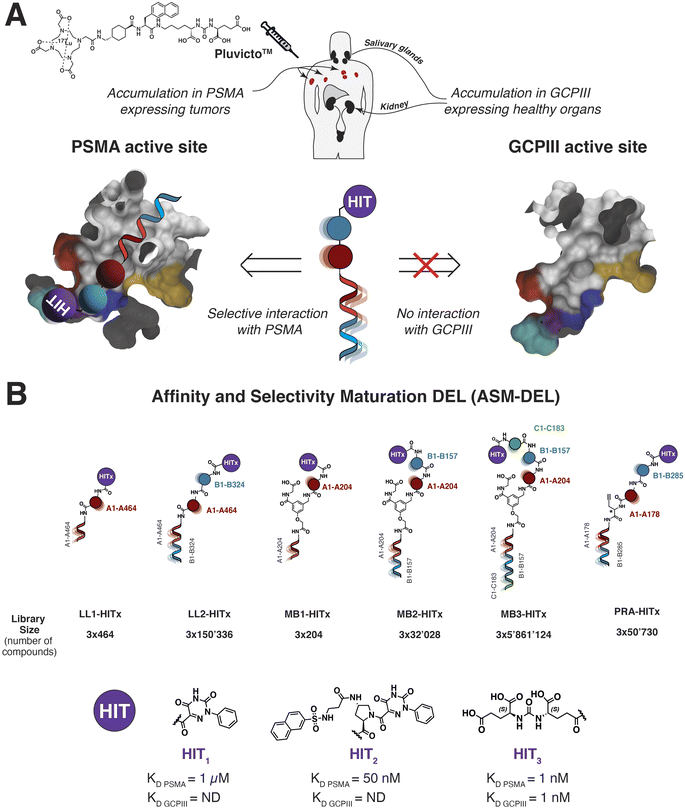 | ||
| Fig. 1 (A) Pluvicto™ accumulates both in PSMA-positive tumors and in GCPIII-positive healthy organs (i.e., salivary glands and kidneys). PSMA (PDB:5O5T) and GCPIII (PDB:3FED) active sites are presented in the figure as follows: S1′ pockets (cyan), zinc-II active sites (purple), arginine patches (blue), S1 accessory sites (red), and arene-binding sites (yellow). (B) Design of the ASM-DELs and the chemical structure of the PSMA-HITs included in the final libraries (affinity constants measured by fluorescence polarization (FP) are reported, ESI 3.1.3, Fig. S33†). HIT1 and HIT2 have been discovered thanks to de novo DEL discovery activities,33 while HIT3 represents the DUPA tumor-targeting headpiece. The asterisk “*” indicates a racemic mixture. | ||
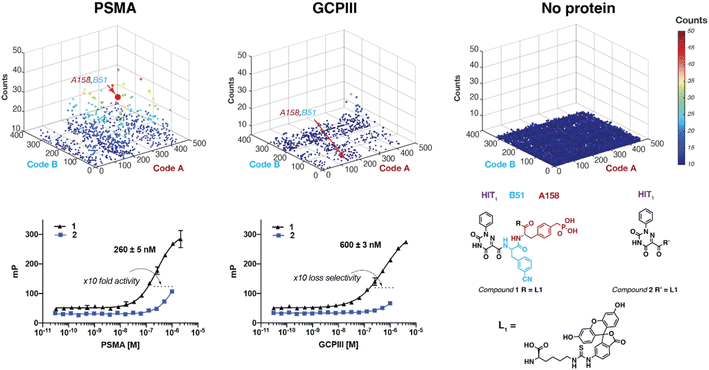
Fig. 2 shows fingerprints of DEL selections, obtained by PCR amplification and high-throughput DNA sequencing of ASM-DEL LL2-HIT1 against PSMA and GCPIII. The combination of A158-B51 building blocks was preferentially enriched in the PSMA selection, with a ∼40-fold enrichment over the background. The combination was resynthesized as a fluorescein lysine derivative (compound 1) and its binding properties to PSMA and GCPIII were studied by fluorescence polarization [Fig. 2]. Compound 1 displayed an increased affinity against PSMA (KD,PSMA = 260 ± 5 nM) as compared to a fluorescein lysine derivative of HIT1 (compound 2, KD,PSMA > 2 μM), but a loss of PSMA-selectivity against the GCPIII anti-target was observed (KD,GCPIII = 600 ± 3 nM). PSMA and GCPIII selections performed with other ASM-DEL LL (LL1 and LL2) did not result in the identification of compounds with preferential PSMA binding properties [see DEL-selection fingerprints reported in ESI Section S5 and Appendix 15.1].†
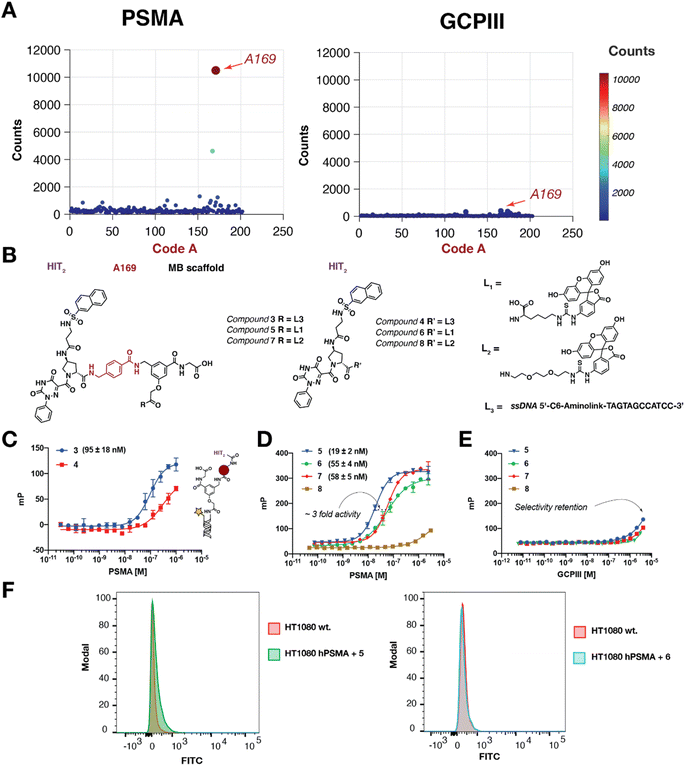
The affinity maturation libraries ASM-DELs MB1, MB2 and MB3 were coupled with HIT1, HIT2, and HIT3 and the resulting compound collections were screened against PSMA and GCPIII. Ligands with preferential PSMA binding were discovered only from ASM-DEL MB1-HIT2 [Fig. 3]. The combination of HIT2 with building block A169 was identified as selective PSMA HIT (enrichment factor of ∼220), while the same building block combination was not enriched in GCPIII selections [Fig. 3A]. Fluorescence polarization studies revealed that combinations of MB-A169-HIT2 (compounds 3, 5, and 7) recognized PSMA with a higher affinity compared to the unmodified HIT2 precursor (compounds 4, 6, and 8). On-DNA fluorescence polarization studies showed that the DNA-MB-A169-HIT2 conjugate (compound 3) bound to PSMA with a dissociation constant KD = 95 nM, which was substantially improved compared to the KD of DNA-HIT2 and compound 4 [Fig. 3C]. Binding studies performed by using fluorescence polarization with compounds devoid of the DNA moiety confirmed that both lysine and pegylated MB-A169-HIT2 derivatives (compounds 5 and 7) were able to bind to PSMA with higher affinities compared to HIT2 (compounds 6 and 8). The best binding constant was measured for Lys-MB-A169-HIT2 (compound 5, KD,PSMA of 19 ± 2 nM) [Fig. 3D]. We observed that the removal of the MB scaffold from Lys-MB-A169-HIT2 slightly reduced the PSMA affinity of the compound (from KD = 19 nM to 41 nM) [ESI 7, Fig. S120†]. Interestingly, the affinity matured MB-A169-HIT2 derivatives (featuring a lysine or a PEG2 linker, respectively) retained the GCPIII selectivity observed for HIT2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33 [Fig. 3E]. Fluorescein derivatives of HIT2 and MB-A169-HIT2 were tested by flow cytometry on PSMA-positive and PSMA-negative tumor cells (HT1080.hPSMA and HT1080.wt). The results, which are shown in Fig. 3F, indicated that both HIT2 and MB-A169-HIT2 were not able to interact with PSMA on tumor cells expressing the human antigen on their cellular membrane, as no significant fluorescence shift between HT1080.hPSMA and HT1080.wt cell samples was observed by flow cytometry. For this reason, the MB-A169-HIT2 combination was not further investigated in in vivo biodistribution studies.
33 [Fig. 3E]. Fluorescein derivatives of HIT2 and MB-A169-HIT2 were tested by flow cytometry on PSMA-positive and PSMA-negative tumor cells (HT1080.hPSMA and HT1080.wt). The results, which are shown in Fig. 3F, indicated that both HIT2 and MB-A169-HIT2 were not able to interact with PSMA on tumor cells expressing the human antigen on their cellular membrane, as no significant fluorescence shift between HT1080.hPSMA and HT1080.wt cell samples was observed by flow cytometry. For this reason, the MB-A169-HIT2 combination was not further investigated in in vivo biodistribution studies.
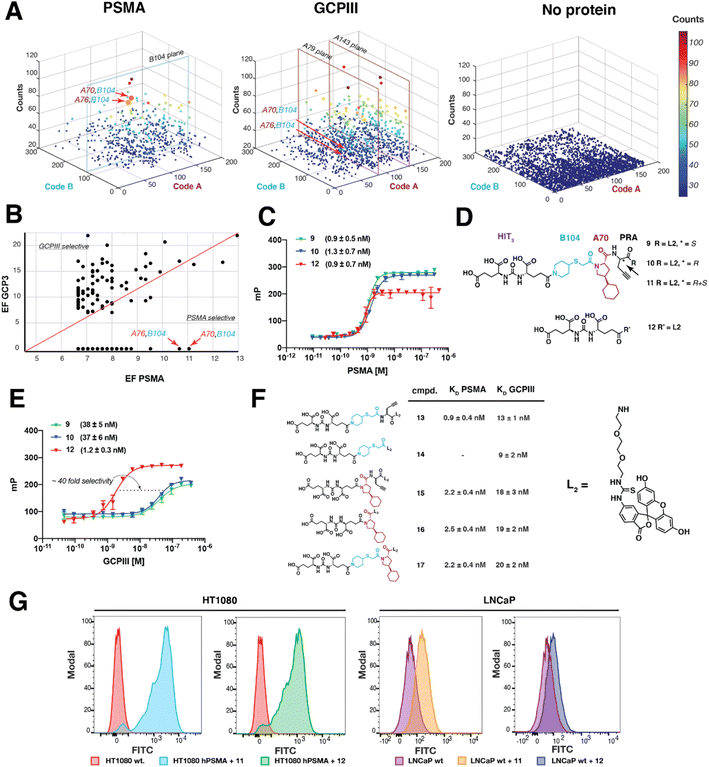
PRA-DEL was also functionalized with HIT1, HIT2, and HIT3 and subjected to affinity-capture selections on PSMA and GCPIII [See ESI Section S5 and Appendix 15.1†]. The selection fingerprint of the PRA-HIT3, displayed in Fig. 4A, revealed A70-B104 and A76-B104 as the most enriched combinations for PSMA, compared to selections performed with GCPIII or in the absence of a target protein. A plot of the enrichment factors towards GCPIII and PSMA (Fig. 4B) revealed that compounds A70-B104 and A76-B104 were preferentially recovered in PSMA selections (bottom right side of the graph in Fig. 4B). Fluorescent derivatives of the diastereoisomers S and R of A70-B104 (compounds 9 and 10) were synthesized and tested by FP against recombinant PSMA and GCPIII. Similar affinity constants against PSMA were observed for the two compounds (compound 9KD,PSMA 0.9 ± 0.5 nM and compound 10KD,PSMA 1.3 ± 0.7 nM) and the original HIT3 (compound 12KD,PSMA 0.9 ± 0.3 nM) [Fig. 4C]. By contrast, the affinity-matured compounds displayed ∼40-fold higher dissociation constants towards GCPIII (compound 9KD,GCPIII 38 ± 6 nM and compound 10KD,GCPIII 37 ± 5 nM) as compared to the original HIT3 (DUPA, KD,GCPIII 1.2 ± 0.3 nM) [Fig. 4E], possibly as a result of a steric clash in the GCPIII binding pocket [Fig. 4F]. Flow cytometry experiments on HT1080.hPSMA and LNCaP tumor cells were performed to assess whether compounds were able to recognize PSMA in a native cellular environment. Both compounds 11 and 12 avidly bound to PSMA, as shown by the cellular staining presented in Fig. 4G. ASM-matured compound 11 showed a more pronounced shift in flow cytometry on both HT1080.hPSMA and LNCaP cells, compared to compound 12 [Fig. 4G].
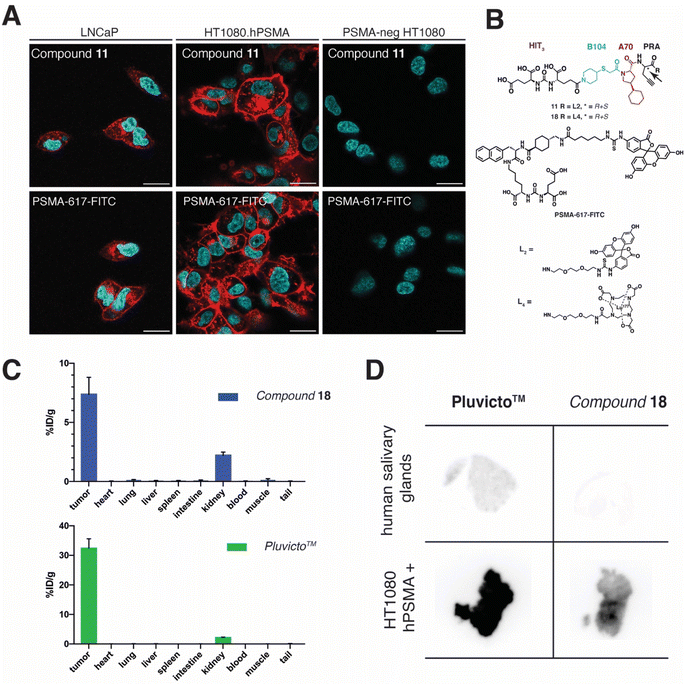
The internalization rate of compound 11 in LNCaP and PSMA-positive HT1080 cancer cells was assessed by confocal microscopy analysis. PSMA-617-FITC was used in the same experiment as a positive control. The results of this experiment indicate that both compounds internalize at a similar rate in both PSMA-positive cancer cell lines, with progressive internalization at 1, 6, and 24 hours after incubation [Fig. 5A and ESI S123†]. No staining was detected in PSMA-negative cancer cells (HT1080.wt).
Finally, we performed quantitative biodistribution studies with radiolabelled compounds to assess whether the favourable PSMA binding properties were associated with a preferential enrichment of the molecules at the tumor site. A head-to-head comparison of A70-B104-HIT3 with Pluvicto™ was performed, as the latter product is routinely used in the clinic [Fig. 5C]. The corresponding DOTA conjugates were labeled with 177Lu (compound 18 and Pluvicto™) and were intravenously injected into BALB/c nude mice (n = 3 per group) bearing HT1080.hPSMA tumors (single dose). Mice were sacrificed two hours after compound administration and the quantitative biodistribution of both molecules was evaluated by counting the radioactivity associated with their lutetium-177 payload. Selective accumulation in PSMA-positive tumor lesions was observed both for compound 18 (i.e., 7.4% ID g−1) and Pluvicto™ (i.e., 32.4% ID g−1). Excellent tumor-to-blood ratios were measured for both molecules (T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B = 145 and 2079, respectively) [Fig. 5C]. In addition, the kidney uptake of Pluvicto™ and compound 18 is comparable (2.16%ID g−1vs. 2.22% ID g−1, respectively) while Pluvicto™ displays a higher tumor-to-kidney ratio than the one reported for 18 as a consequence of its higher tumor uptake.
B = 145 and 2079, respectively) [Fig. 5C]. In addition, the kidney uptake of Pluvicto™ and compound 18 is comparable (2.16%ID g−1vs. 2.22% ID g−1, respectively) while Pluvicto™ displays a higher tumor-to-kidney ratio than the one reported for 18 as a consequence of its higher tumor uptake.
Since mice express GCPIII in immunologically privileged sites such as testes and ovaries,43 we performed a comparative analysis using PSMA-positive tumors and healthy human salivary glands by ex vivo autoradiography. Human salivary glands and HT1080.hPSMA tumor specimens were incubated with 177Lu-PSMA-617 and 18. While Pluvicto™ displayed high binding on tumor samples and moderate staining on salivary glands, compound 18 showed distinctive and selective accumulation only on HT1080.hPSMA samples. No detectable staining on human salivary glands was observed [Fig. 5D].
Discussion
Pluvicto™ is the most recent RLT approved for the treatment of patients affected by metastatic castration-resistant prostate cancer. The ultra-high affinity of the drug to its cognate target antigen (prostate specific membrane antigen, expressed on the surface of prostate cancer cells) enables the efficient and selective targeted delivery of radionuclide payloads to tumors. Despite the clinical benefit demonstrated in the Phase III VISION trial, 12% of the patients discontinued the treatment with the drug due to adverse events (e.g., xerostomia)44 which additionally limited the administration of Pluvicto™ at higher doses. Some of the undesired side effects occur due to the expression of GCPIII in salivary glands and kidneys, which is a highly homologous PSMA protein that causes an off-targeted accumulation of the PSMA-targeted RLT in healthy tissue.31In this article, we have described the affinity and selectivity maturation of nanomolar and sub-nanomolar PSMA ligands, by using DNA-encoded chemical library technology, to improve the affinity to PSMA, while enhancing ligand selectivity for the tumor-associated antigen over GCPIII. Therefore, the occurrence of selective and potent PSMA binders may prevent unwanted off-target accumulation in healthy organs, thus improving the RLT therapeutic window. We synthesized affinity and selectivity DNA-encoded chemical libraries (ASM-DELs) comprising 18′284′658 individually encoded compounds, obtained by derivatization of six different DELs with three elected HITs. Screening against PSMA and GCPIII resulted in the identification of several novel building block combinations with the three HITs. Compound 11 (A70-B104-HIT3) was identified from the PRA-DEL as the most potent and selective PSMA ligand (KD,PSMA = 900 pM; KD,GCPIII = 38 nM). The compound was found to bind to and internalize into PSMA-positive cancer cells by flow cytometry and confocal microscopy experiments. Ligand internalization of PSMA-targeted RLTs may influence their in vivo tumor uptake and tumor residence time, thus having an impact on their anti-cancer efficacy.
Moreover, in vivo biodistribution experiments in tumor-bearing mice, conducted with a lutetium-177 radioactive derivative of compound 11, showed successful and selective accumulation in PSMA-expressing tumors (tumor uptake = ∼7.4% ID g−1, 2 hours after systemic administration). Furthermore, ex vivo autoradiography analysis provided an estimation of the ability of the ligand to selectively target the cancer lesion against human salivary glands, in contrast to Pluvicto™, which exhibited visible staining for both tissues.
The clinical maximum tolerated dose of Pluvicto™ has not yet been formally identified. The dose regimen approved for treating metastatic prostate cancer patients (i.e., 7.4 GBq per dose, for up to 6 administrations) has been established by the early clinical use of the drug under compassionate treatment and similar regulatory frameworks in Germany and Australia. However, adverse events including xerostomia are observed in a good proportion of patients treated at the approved Pluvicto™ dose, thus leading in ∼12% of cases to discontinuation of the treatment.44 Although in the past, medical strategies have been proposed to somewhat alleviate these undesired effects (e.g., cooling of the throat45,46), the generation of PSMA ligands that do not interact with GCPIII in healthy organs could solve the unpleasant burden of salivary gland toxicity.44 Beforehand, medicinal chemistry efforts have highlighted the benefit of introducing “rigid linkers” on DUPA to obtain PSMA-selective ligands.34 Unfortunately, while the described derivatives were presented as highly potent and selective, their ability to target PSMA-positive tumors after in vivo administration was never formally demonstrated. We have utilized our high-throughput DEL platform to obtain potent and selective PSMA-targeting ligands with flexible and linear structures from a large collection of compounds. DEL technology has been previously applied as an effective tool to mature the affinity of small organic ligands for tumor-targeting purposes. Our group has recently published the implementation of DEL strategies to mature the affinity of nanomolar ligands against carbonic anhydrase IX and fibroblast activation protein to obtain ultra-potent binders with affinities in the low picomolar range and with an improved tumor-residence time.35,37 In this new article, we have shown how DELs can be instrumental in enhancing in parallel the affinity to a target protein and the selectivity against an anti-target protein of available HIT compounds. Selectivity is a fundamental property of targeted therapies, with a great impact on minimizing healthy organ uptake and preventing systemic side effects. The PSMA-selective compounds resulting from this DEL selection campaign are “portable” moieties that can be alternatively coupled to DNA-barcodes, fluorophores, or radionuclide chelators without losing their affinity and selectivity for the cognate target.
One of the limitations of this study is the lack of an in vivo preclinical model that allows the assessment of the selectivity of DEL-derived tumor-targeting ligands. Tumor-bearing mice are routinely used to assess the tumor-targeting properties of antibodies and small organic ligands. However, in the case of PSMA, unwanted accumulation in salivary glands cannot be predicted in rodents or minipigs23,24,47 (ESI 13.3, Fig. S127†) as, in those species, GCPIII is not expressed in salivary glands. Autoradiography experiments on human salivary glands and PSMA-positive tumors are presented here as a method of choice to quantify the selectivity of novel PSMA ligands and to benchmark them against Pluvicto™ (Fig. 5D).
Autoradiography studies show no binding of compound 18 on human salivary glands. Our work relies on the assumption that salivary gland uptake of urea-based PSMA ligands is mainly caused by GCPIII expression in the tissue. According to alternative studies, salivary gland accumulation appears to be non-PSMA-related48 or partly non-specific.49 It is evident that the underlying mechanism is still under investigation, and additional work is necessary to advance in the field of PSMA radioligand therapeutics.
Given its high potency, preferential binding to PSMA compared to GCPIII, internalization properties and excellent in vivo tumor-targeting performance, compound A70-B104-HIT3 is a promising HIT compound for the generation of imaging and therapeutic constructs based on radionuclides (e.g., gallium-68 and lutetium-177) and cytotoxic drugs (e.g., monomethyl auristatin E). The anti-cancer efficacy of radioligand therapeutics is directly determined by their tumor uptake and residence time in target neoplastic masses. These two features are strongly influenced by the ligand affinity for the cognate target and by the compounds' physicochemical properties. Medicinal chemistry optimization of A70-B104-HIT3 tumor uptake is needed to increase the tumor-to-kidney ratio and improve the drug's therapeutic window. Nuclear medicine studies that demonstrate the ability of the compound to target tumors in prostate cancer patients with PSMA-positive lesions while sparing healthy organs are still needed to warrant its clinical application.
Conclusion
Salivary gland toxicity remains a critical problem that limits dose escalation of PSMA-targeted RLTs in patients affected by prostate cancer. At the same time, the high similarity between PSMA and GCPIII, expressed in salivary glands, represents a molecular challenge for conventional medicinal chemistry. As demonstrated in this work, the use of DEL technology can be of great utility for the maturation of binders to generate new potent and selective PSMA ligands. Affinity and selectivity matured compound A70-B104-HIT3 is a portable PSMA-selective compound that can be used for the generation of theranostic small molecules resulting in therapeutic agents that are better tolerated and more efficacious in patients with antigen-positive prostate cancer lesions.Data availability
The datasets supporting this article have been uploaded as part of the ESI.†Author contributions
Laura Lucaroni: writing – review & editing, writing – original draft, visualization, validation, methodology, investigation, formal analysis, data curation, conceptualization. Sebastian Oehler: investigation, validation, methodology. Tony Georgiev: investigation. Marco Müller: investigation, validation, methodology. Matilde Bocci: investigation. Roberto de Luca: investigation. Nicholas Favalli: investigation. Dario Neri: writing – review & editing, writing – original draft, visualization, validation, supervision, resources, project administration, methodology, funding acquisition, formal analysis, data curation, conceptualization. Samuele Cazzamalli: writing – review & editing, writing – original draft, visualization, validation, supervision, resources, project administration, methodology, investigation, funding acquisition, formal analysis, data curation, conceptualization. Luca Prati: writing – review & editing, writing – original draft, visualization, validation, supervision, project administration, methodology, investigation, formal analysis, data curation, conceptualization.Conflicts of interest
D. N. is co-founder, CEO, CSO and President of the Scientific Advisory Board of Philogen. L. L., S. O., T. G., M. M., R. D. L., N. F. and S. C. are employed by Philochem AG, and the research and development unit of Philogen. L. P. is employed by Philogen S.p.A.Acknowledgements
The authors thank M. Bigatti and É. M. D. Gorre for the help in DEL synthesis. Furthermore, the authors thank C. Pellegrino for the help in stable cell line development, and C. Comacchio for the support with flow cytometry studies. Moreover, we thank L. Principi for the help with HRMS. The authors received no specific funding for this work.References
- G. Sgouros, L. Bodei, M. R. McDevitt and J. R. Nedrow, Radiopharmaceutical Therapy in Cancer: Clinical Advances and Challenges, Nat. Rev. Drug Discovery, 2020, 19(9), 589–608, DOI:10.1038/s41573-020-0073-9.
- U. Hennrich and K. Kopka, Lutathera®: The First FDA- and EMA-Approved Radiopharmaceutical for Peptide Receptor Radionuclide Therapy, Pharmaceuticals, 2019, 12(3), 114, DOI:10.3390/ph12030114.
- P. G. Kluetz, W. Pierce, V. E. Maher, H. Zhang, S. Tang, P. Song, Q. Liu, M. T. Haber, E. E. Leutzinger, A. Al-Hakim, W. Chen, T. Palmby, E. Alebachew, R. Sridhara, A. Ibrahim, R. Justice and R. Pazdur, Radium Ra 223 Dichloride Injection: U.S. Food and Drug Administration Drug Approval Summary, Clin. Cancer Res., 2014, 20(1), 9–14, DOI:10.1158/1078-0432.CCR-13-2665.
- J. Fallah, S. Agrawal, H. Gittleman, M. H. Fiero, S. Subramaniam, C. John, W. Chen, T. K. Ricks, G. Niu, A. Fotenos, M. Wang, K. Chiang, W. F. Pierce, D. L. Suzman, S. Tang, R. Pazdur, L. Amiri-Kordestani, A. Ibrahim and P. G. Kluetz, FDA Approval Summary: Lutetium Lu 177 Vipivotide Tetraxetan for Patients with Metastatic Castration-Resistant Prostate Cancer, Clin. Cancer Res., 2023, 29(9), 1651–1657, DOI:10.1158/1078-0432.CCR-22-2875.
- L. P. Orellana García, F. Ehmann, P. A. Hines, A. Ritzhaupt and A. Brand, Biomarker and Companion Diagnostics—A Review of Medicinal Products Approved by the European Medicines Agency, Front. Med., 2021, 8, 753187, DOI:10.3389/fmed.2021.753187.
- W. D. W. Heston, Characterization and Glutamyl Preferring Carboxypeptidase Function of Prostate Specific Membrane Antigen: A Novel Folate Hydrolase, Urology, 1997, 49(3), 104–112, DOI:10.1016/S0090-4295(97)00177-5.
- N. Schülke, O. A. Varlamova, G. P. Donovan, D. Ma, J. P. Gardner, D. M. Morrissey, R. R. Arrigale, C. Zhan, A. J. Chodera, K. G. Surowitz, P. J. Maddon, W. D. W. Heston and W. C. Olson, The Homodimer of Prostate-Specific Membrane Antigen Is a Functional Target for Cancer Therapy, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(22), 12590–12595, DOI:10.1073/pnas.1735443100.
- N. M. Donin and R. E. Reiter, Why Targeting PSMA Is a Game Changer in the Management of Prostate Cancer, J. Nucl. Med., 2018, 59(2), 177, DOI:10.2967/jnumed.117.191874.
- C. Lauri, L. Chiurchioni, V. M. Russo, L. Zannini and A. Signore, PSMA Expression in Solid Tumors beyond the Prostate Gland: Ready for Theranostic Applications?, J. Clin. Med., 2022, 11(21), 6590, DOI:10.3390/jcm11216590.
- A. Paschalis, B. Sheehan, R. Riisnaes, D. N. Rodrigues, B. Gurel, C. Bertan, A. Ferreira, M. B. K. Lambros, G. Seed, W. Yuan, D. Dolling, J. C. Welti, A. Neeb, S. Sumanasuriya, P. Rescigno, D. Bianchini, N. Tunariu, S. Carreira, A. Sharp, W. Oyen and J. S. de Bono, Prostate-Specific Membrane Antigen Heterogeneity and DNA Repair Defects in Prostate Cancer, Eur. Urol., 2019, 76(4), 469–478, DOI:10.1016/j.eururo.2019.06.030.
- W. P. Fendler, M. Eiber, M. Beheshti, J. Bomanji, J. Calais, F. Ceci, S. Y. Cho, S. Fanti, F. L. Giesel, K. Goffin, U. Haberkorn, H. Jacene, P. J. Koo, K. Kopka, B. J. Krause, L. Lindenberg, C. Marcus, F. M. Mottaghy, D. E. Oprea-Lager, J. R. Osborne, M. Piert, S. P. Rowe, H. Schöder, S. Wan, H.-J. Wester, T. A. Hope and K. Herrmann, PSMA PET/CT: Joint EANM Procedure Guideline/SNMMI Procedure Standard for Prostate Cancer Imaging 2.0, Eur. J. Nucl. Med. Mol. Imaging, 2023, 50(5), 1466–1486, DOI:10.1007/s00259-022-06089-w.
- A. Ghosh and W. D. W. Heston, Tumor Target Prostate Specific Membrane Antigen (PSMA) and Its Regulation in Prostate Cancer, J. Cell. Biochem., 2004, 91(3), 528–539, DOI:10.1002/jcb.10661.
- T. Liu, C. Liu, Y. Ren, X. Guo, J. Jiang, Q. Xie, L. Xia, F. Wang, H. Zhu and Z. Yang, Development of an Albumin-Based PSMA Probe With Prolonged Half-Life, Front. Mol. Biosci., 2020, 7, 585024, DOI:10.3389/fmolb.2020.585024.
- A. E. Machulkin, Y. A. Ivanenkov, A. V. Aladinskaya, M. S. Veselov, V. A. Aladinskiy, E. K. Beloglazkina, V. E. Koteliansky, A. G. Shakhbazyan, Y. B. Sandulenko and A. G. Majouga, Small-Molecule PSMA Ligands. Current State, SAR and Perspectives, J. Drug Targeting, 2016, 24(8), 679–693, DOI:10.3109/1061186X.2016.1154564.
- S. P. Rowe, K. L. Gage, S. F. Faraj, K. J. Macura, T. C. Cornish, N. Gonzalez-Roibon, G. Guner, E. Munari, A. W. Partin, C. P. Pavlovich, M. Han, H. B. Carter, T. J. Bivalacqua, A. Blackford, D. Holt, R. F. Dannals, G. J. Netto, M. A. Lodge, R. C. Mease, M. G. Pomper and S. Y. Cho, 18F-DCFBC PET/CT for PSMA-Based Detection and Characterization of Primary Prostate Cancer, J. Nucl. Med., 2015, 56(7), 1003, DOI:10.2967/jnumed.115.154336.
- Y. Chen, C. A. Foss, Y. Byun, S. Nimmagadda, M. Pullambhatla, J. J. Fox, M. Castanares, S. E. Lupold, J. W. Babich, R. C. Mease and M. G. Pomper, Radiohalogenated Prostate-Specific Membrane Antigen (PSMA)-Based Ureas as Imaging Agents for Prostate Cancer, J. Med. Chem., 2008, 51(24), 7933–7943, DOI:10.1021/jm801055h.
- S. R. Banerjee, K. P. Maresca, L. Francesconi, J. Valliant, J. W. Babich and J. Zubieta, New Directions in the Coordination Chemistry of 99mTc: A Reflection on Technetium Core Structures and a Strategy for New Chelate Design, Nucl. Med. Biol., 2005, 32(1), 1–20, DOI:10.1016/j.nucmedbio.2004.09.001.
- F. P. Olatunji, M. Pun, J. W. Herman, O. Romero, M. Maniatopoulos, J. D. Latoche, R. A. Parise, J. Guo, J. H. Beumer, C. J. Anderson and C. E. Berkman, Modular Smart Molecules for PSMA-Targeted Chemotherapy, Mol. Cancer Ther., 2022, 21(11), 1701–1709, DOI:10.1158/1535-7163.MCT-22-0160.
- C. P. Leamon, J. A. Reddy, A. Bloomfield, R. Dorton, M. Nelson, M. Vetzel, P. Kleindl, S. Hahn, K. Wang and I. R. Vlahov, Prostate-Specific Membrane Antigen-Specific Antitumor Activity of a Self-Immolative Tubulysin Conjugate, Bioconjugate Chem., 2019, 30(6), 1805–1813, DOI:10.1021/acs.bioconjchem.9b00335.
- M. C. Gong, S. S. Chang, M. Sadelain, N. H. Bander and W. D. W. Heston, Prostate-Specific Membrane Antigen (PSMA)-Specific Monoclonal Antibodies in the Treatment of Prostate and Other Cancers, Cancer Metastasis Rev., 1999, 18(4), 483–490, DOI:10.1023/A:1006308826967.
- A. P. Kozikowski, F. Nan, P. Conti, J. Zhang, E. Ramadan, T. Bzdega, B. Wroblewska, J. H. Neale, S. Pshenichkin and J. T. Wroblewski, Design of Remarkably Simple, Yet Potent Urea-Based Inhibitors of Glutamate Carboxypeptidase II (NAALADase), J. Med. Chem., 2001, 44(3), 298–301, DOI:10.1021/jm000406m.
- M. Eder, M. Schäfer, U. Bauder-Wüst, W.-E. Hull, C. Wängler, W. Mier, U. Haberkorn and M. Eisenhut, 68Ga-Complex Lipophilicity and the Targeting Property of a Urea-Based PSMA Inhibitor for PET Imaging, Bioconjugate Chem., 2012, 23(4), 688–697, DOI:10.1021/bc200279b.
- M. Benešová, M. Schäfer, U. Bauder-Wüst, A. Afshar-Oromieh, C. Kratochwil, W. Mier, U. Haberkorn, K. Kopka and M. Eder, Preclinical Evaluation of a Tailor-Made DOTA-Conjugated PSMA Inhibitor with Optimized Linker Moiety for Imaging and Endoradiotherapy of Prostate Cancer, J. Nucl. Med., 2015, 56(6), 914, DOI:10.2967/jnumed.114.147413.
- E. A. M. Ruigrok, N. van Vliet, S. U. Dalm, E. de Blois, D. C. van Gent, J. Haeck, C. de Ridder, D. Stuurman, M. W. Konijnenberg, W. M. van Weerden, M. de Jong and J. Nonnekens, Extensive Preclinical Evaluation of Lutetium-177-Labeled PSMA-Specific Tracers for Prostate Cancer Radionuclide Therapy, Eur. J. Nucl. Med. Mol. Imaging, 2021, 48(5), 1339–1350, DOI:10.1007/s00259-020-05057-6.
- S. B. Kim, I. H. Song, Y. S. Song, B. C. Lee, A. Gupta, J. S. Lee, H. S. Park and S. E. Kim, Biodistribution and Internal Radiation Dosimetry of a Companion Diagnostic Radiopharmaceutical, [68Ga]PSMA-11, in Subcutaneous Prostate Cancer Xenograft Model Mice, Sci. Rep., 2021, 11(1), 15263, DOI:10.1038/s41598-021-94684-6.
- C. Meyer, V. Prasad, A. Stuparu, P. Kletting, G. Glatting, J. Miksch, C. Solbach, K. Lueckerath, L. Nyiranshuti, S. Zhu, J. Czernin, A. J. Beer, R. Slavik, J. Calais and M. Dahlbom, Comparison of PSMA-TO-1 and PSMA-617 Labeled with Gallium-68, Lutetium-177 and Actinium-225, EJNMMI Res., 2022, 12(1), 65, DOI:10.1186/s13550-022-00935-6.
- G. Ferreira, A. Iravani, M. S. Hofman and R. J. Hicks, Intra-Individual Comparison of 68Ga-PSMA-11 and 18F-DCFPyL Normal-Organ Biodistribution, Canc. Imag., 2019, 19(1), 23, DOI:10.1186/s40644-019-0211-y.
- O. Sartor, J. de Bono, K. N. Chi, K. Fizazi, K. Herrmann, K. Rahbar, S. T. Tagawa, L. T. Nordquist, N. Vaishampayan, G. El-Haddad, C. H. Park, T. M. Beer, A. Armour, W. J. Pérez-Contreras, M. DeSilvio, E. Kpamegan, G. Gericke, R. A. Messmann, M. J. Morris and B. J. Krause, Lutetium-177–PSMA-617 for Metastatic Castration-Resistant Prostate Cancer, N. Engl. J. Med., 2021, 385(12), 1091–1103, DOI:10.1056/NEJMoa2107322.
- A. Afshar-Oromieh, H. Hetzheim, C. Kratochwil, M. Benesova, M. Eder, O. C. Neels, M. Eisenhut, W. Kübler, T. Holland-Letz, F. L. Giesel, W. Mier, K. Kopka and U. Haberkorn, The Theranostic PSMA Ligand PSMA-617 in the Diagnosis of Prostate Cancer by PET/CT: Biodistribution in Humans, Radiation Dosimetry, and First Evaluation of Tumor Lesions, J. Nucl. Med., 2015, 56(11), 1697, DOI:10.2967/jnumed.115.161299.
- F. Bois, C. Noirot, S. Dietemann, I. C. Mainta, T. Zilli, V. Garibotto and M. A. Walter, [(68)Ga]Ga-PSMA-11 in Prostate Cancer: A Comprehensive Review, Am. J. Nucl. Med. Mol. Imaging, 2020, 10(6), 349–374 CAS.
- L. Lucaroni, T. Georgiev, E. Prodi, S. Puglioli, C. Pellegrino, N. Favalli, L. Prati, M. G. Manz, S. Cazzamalli, D. Neri, S. Oehler and G. Bassi, Cross-Reactivity to Glutamate Carboxypeptidase III Causes Undesired Salivary Gland and Kidney Uptake of PSMA-Targeted Small-Molecule Radionuclide Therapeutics, Eur. J. Nucl. Med. Mol. Imaging, 2023, 50(3), 957–961, DOI:10.1007/s00259-022-05982-8.
- B. Vorlova, T. Knedlik, J. Tykvart and J. Konvalinka, GCPII and Its Close Homolog GCPIII: From a Neuropeptidase to a Cancer Marker and Beyond, Front. Biosci.-Landmark, 2019, 24(4), 648–687 CrossRef CAS PubMed.
- S. Oehler, L. Lucaroni, F. Migliorini, A. Elsayed, L. Prati, S. Puglioli, M. Matasci, K. Schira, J. Scheuermann, D. Yudin, M. Jia, N. Ban, D. Bushnell, R. Kornberg, S. Cazzamalli, D. Neri, N. Favalli and G. Bassi, A DNA-Encoded Chemical Library Based on Chiral 4-Amino-Proline Enables Stereospecific Isozyme-Selective Protein Recognition, Nat. Chem., 2023, 15, 1431–1443, DOI:10.1038/s41557-023-01257-3.
- J. Tykvart, J. Schimer, A. Jančařík, J. Bařinková, V. Navrátil, J. Starková, K. Šrámková, J. Konvalinka, P. Majer and P. Šácha, Design of Highly Potent Urea-Based, Exosite-Binding Inhibitors Selective for Glutamate Carboxypeptidase II, J. Med. Chem., 2015, 58(10), 4357–4363, DOI:10.1021/acs.jmedchem.5b00278.
- S. Puglioli, E. Schmidt, C. Pellegrino, L. Prati, S. Oehler, R. De Luca, A. Galbiati, C. Comacchio, L. Nadal, J. Scheuermann, G. M. Manz, D. Neri, S. Cazzamalli, G. Bassi and N. Favalli, Selective Tumor Targeting Enabled by Picomolar Fibroblast Activation Protein Inhibitors Isolated from a DNA-Encoded Affinity Maturation Library, Chem, 2022, 9(2), 411–429, DOI:10.1016/j.chempr.2022.10.006.
- G. Bassi, N. Favalli, C. Pellegrino, Y. Onda, J. Scheuermann, S. Cazzamalli, M. G. Manz and D. Neri, Specific Inhibitor of Placental Alkaline Phosphatase Isolated from a DNA-Encoded Chemical Library Targets Tumor of the Female Reproductive Tract, J. Med. Chem., 2021, 64(21), 15799–15809, DOI:10.1021/acs.jmedchem.1c01103.
- N. Favalli, G. Bassi, C. Pellegrino, J. Millul, R. De Luca, S. Cazzamalli, S. Yang, A. Trenner, N. L. Mozaffari, R. Myburgh, M. Moroglu, S. J. Conway, A. A. Sartori, M. G. Manz, R. A. Lerner, P. K. Vogt, J. Scheuermann and D. Neri, Stereo- and Regiodefined DNA-Encoded Chemical Libraries Enable Efficient Tumour-Targeting Applications, Nat. Chem., 2021, 13(6), 540–548, DOI:10.1038/s41557-021-00660-y.
- M. Wichert, N. Krall, W. Decurtins, R. M. Franzini, F. Pretto, P. Schneider, D. Neri and J. Scheuermann, Dual-Display of Small Molecules Enables the Discovery of Ligand Pairs and Facilitates Affinity Maturation, Nat. Chem., 2015, 7(3), 241–249, DOI:10.1038/nchem.2158.
- G. Bassi, N. Favalli, M. Vuk, M. Catalano, A. Martinelli, A. Trenner, A. Porro, S. Yang, C. L. Tham, M. Moroglu, W. W. Yue, S. J. Conway, P. K. Vogt, A. A. Sartori, J. Scheuermann and D. Neri, A Single-Stranded DNA-Encoded Chemical Library Based on a Stereoisomeric Scaffold Enables Ligand Discovery by Modular Assembly of Building Blocks, Adv. Sci., 2020, 7(22), 2001970, DOI:10.1002/advs.202001970.
- S. Melkko, J. Scheuermann, C. E. Dumelin and D. Neri, Encoded Self-Assembling Chemical Libraries, Nat. Biotechnol., 2004, 22(5), 568–574, DOI:10.1038/nbt961.
- M. Leimbacher, Y. Zhang, L. Mannocci, M. Stravs, T. Geppert, J. Scheuermann, G. Schneider and D. Neri, Discovery of Small-Molecule Interleukin-2 Inhibitors from a DNA-Encoded Chemical Library, Chem. - Eur. J., 2012, 18(25), 7729–7737, DOI:10.1002/chem.201200952.
- W. Decurtins, M. Wichert, R. M. Franzini, F. Buller, M. A. Stravs, Y. Zhang, D. Neri and J. Scheuermann, Automated Screening for Small Organic Ligands Using DNA-Encoded Chemical Libraries, Nat. Protoc., 2016, 11(4), 764–780, DOI:10.1038/nprot.2016.039.
- T. Knedlík, B. Vorlová, V. Navrátil, J. Tykvart, F. Sedlák, Š. Vaculín, M. Franěk, P. Šácha and J. Konvalinka, Mouse Glutamate Carboxypeptidase II (GCPII) Has a Similar Enzyme Activity and Inhibition Profile but a Different Tissue Distribution to Human GCPII, FEBS Open Bio, 2017, 7(9), 1362–1378 CrossRef PubMed.
- O. Sartor, J. de Bono, K. N. Chi, K. Fizazi, K. Herrmann, K. Rahbar, S. T. Tagawa, L. T. Nordquist, N. Vaishampayan, G. El-Haddad, C. H. Park, T. M. Beer, A. Armour, W. J. Pérez-Contreras, M. DeSilvio, E. Kpamegan, G. Gericke, R. A. Messmann, M. J. Morris and B. J. Krause, Lutetium-177–PSMA-617 for Metastatic Castration-Resistant Prostate Cancer, N. Engl. J. Med., 2021, 385(12), 1091–1103, DOI:10.1056/NEJMoa2107322.
- C. A. Taylor, A. Shankar, M. N. Gaze, C. Peet, J. E. Gains, S. Wan, S. Voo, D. Priftakis and J. B. Bomanji, Renal Protection during 177lutetium DOTATATE Molecular Radiotherapy in Children: A Proposal for Safe Amino Acid Infusional Volume during Peptide Receptor Radionuclide Therapy, Nucl. Med. Commun., 2022, 43(2), 242–246, DOI:10.1097/MNM.0000000000001497.
- B. Yilmaz, S. Nisli, N. Ergul, R. U. Gursu, O. Acikgoz and T. F. Çermik, Effect of External Cooling on 177Lu-PSMA Uptake by the Parotid Glands, J. Nucl. Med., 2019, 60(10), 1388–1393, DOI:10.2967/jnumed.119.226449.
- J. Roy, B. M. Warner, F. Basuli, X. Zhang, K. Wong, T. Pranzatelli, A. T. Ton, J. A. Chiorini, P. L. Choyke, F. I. Lin and E. M. Jagoda, Comparison of Prostate-Specific Membrane Antigen Expression Levels in Human Salivary Glands to Non-Human Primates and Rodents, Cancer Biother.Radiopharm., 2020, 35(4), 284–291, DOI:10.1089/cbr.2019.3079.
- N. J. Rupp, C. A. Umbricht, D. A. Pizzuto, D. Lenggenhager, A. Töpfer, J. Müller, U. J. Muehlematter, D. A. Ferraro, M. Messerli, G. B. Morand, G. F. Huber, D. Eberli, R. Schibli, C. Müller and I. A. Burger, First Clinicopathologic Evidence of a Non–PSMA-Related Uptake Mechanism for 68Ga-PSMA-11 in Salivary Glands, J. Nucl. Med., 2019, 60(9), 1270, DOI:10.2967/jnumed.118.222307.
- N. Heynickx, C. Segers, A. Coolkens, S. Baatout and K. Vermeulen, Characterization of Non-Specific Uptake and Retention Mechanisms of [177Lu]Lu-PSMA-617 in the Salivary Glands, Pharmaceuticals, 2023, 16(5), 692, DOI:10.3390/ph16050692.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06668a |
| ‡ First author. |
| This journal is © The Royal Society of Chemistry 2024 |