Open Access Article
Open Access ArticleCombinatorial discovery of antibacterials via a feature-fusion based machine learning workflow†
Cong
Wang‡
ab,
Yuhui
Wu‡
ab,
Yunfan
Xue‡
a,
Lingyun
Zou
a,
Yue
Huang
a,
Peng
Zhang
 *abc and
Jian
Ji
*abc and
Jian
Ji
 *abc
*abc
aMOE Key Laboratory of Macromolecule Synthesis and Functionalization, Department of Polymer Science and Engineering, Zhejiang University, Hangzhou, Zhejiang 310027, PR China. E-mail: zhangp7@zju.edu.cn; jijian@zju.edu.cn
bInternational Research Center for X Polymers, International Campus, Zhejiang University, Haining, Zhejiang 314400, PR China
cState Key Laboratory of Transvascular Implantation Devices, Zhejiang University, Hangzhou, Zhejiang 311202, P. R. China
First published on 26th March 2024
Abstract
The discovery of new antibacterials within the vast chemical space is crucial in combating drug-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA). However, the traditional approach of screening the entire chemical library in an ergodic manner can be laborious and time-consuming. Machine learning-assisted screening of antibacterials alleviates the exploration effort but suffers from the lack of reliable and related datasets. To address these challenges, we devised a combinatorial library comprising over 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 candidates based on the Ugi reaction. A focused library was subsequently generated through uniform sampling of the entire library to narrow down the preliminary screening scale. A novel feature-fusion architecture called the latent space constraint neural network was developed which incorporated both fingerprint and physicochemical molecular descriptors to predict the antibacterial properties. This integration allowed the model to leverage the complementary information provided by these descriptors and improve the accuracy of predictions. Three lead compounds that demonstrated excellent efficacy against MRSA while alleviating drug resistance were identified. This workflow highlights the integration of machine learning with the combinatorial chemical library to expedite high-quality data collection and extensive data mining for antibacterial screening.
000 candidates based on the Ugi reaction. A focused library was subsequently generated through uniform sampling of the entire library to narrow down the preliminary screening scale. A novel feature-fusion architecture called the latent space constraint neural network was developed which incorporated both fingerprint and physicochemical molecular descriptors to predict the antibacterial properties. This integration allowed the model to leverage the complementary information provided by these descriptors and improve the accuracy of predictions. Three lead compounds that demonstrated excellent efficacy against MRSA while alleviating drug resistance were identified. This workflow highlights the integration of machine learning with the combinatorial chemical library to expedite high-quality data collection and extensive data mining for antibacterial screening.
Introduction
Antibacterial discovery continues to pose a dire challenge as drug-resistant bacteria proliferate globally,1,2 while new antibacterial compounds hide within the vast chemical space. Over the past few decades, small molecular libraries have been meticulously designed based on existing antibacterials to identify novel hit compounds.3–6 However, these methods substantially rely on empirical knowledge, resulting in a restricted exploration of the total chemical space with a lack of structural diversity. Virtual screening approaches thrive as promising alternatives due to their capability of generating millions of candidates with diverse motifs in a single trial.7 Nevertheless, the limited accessibility of synthesis routes for library candidates and the scarcity of rapid evaluation tools for virtual candidates impede their widespread application.8Libraries constructed through combinatorial chemistry such as multi-component reactions provide a valuable solution for accessing a broad chemical space with favorable synthesis accessibility.9 The inherent tolerance of numerous building blocks10 and high conversion efficiency make combinatorial chemistry the most acclaimed method for diverse library constructions.11,12 These libraries enable the exploration of abundant possibilities within the chemical space and have found potential in large-scale molecular data storage,13 self-assembly dipeptide hydrogel generation,14 and protein–protein interaction inhibitor development.15 The Ugi reaction (UR) merges equivalent carboxylic acid, amine, aldehyde, and isonitrile components into a peptoid backbone with pendant functional groups.16 The bioactivity of Ugi products has been demonstrated in areas such as antiviral17 and analgesic18 research. However, combinatorial libraries for target screening are typically limited in size ranging from tens to hundreds of compounds and tend to follow the pre-existing molecular scaffolds to raise the hit rate.18,19 The attempt to exponentially expand the potential chemical space necessitates high-throughput pipelines to manage the huge pool of candidates such as DNA-encoded libraries15,20 and micro-dispensers,21 which has arisen concerns regarding cost efficiency. Furthermore, the process of analyzing the collected data and extracting meaningful insights remains a laborious task.
Machine learning emerges as a promising route to handle the massive data,22–24 and it has demonstrated success in screening antibacterials from candidate libraries.25–27 Its application in screening natural product libraries is particularly relevant,28 as most clinically applied antibiotics are derived from natural sources such as vancomycin.29 Recently, machine learning classifiers, including random forest, support vector machine, and logistic regression, have been employed to predict the antibiotic activity of products from biosynthetic gene clusters,30 which encode and govern the production of natural metabolites.31 However, obtaining such products under standard laboratory conditions can be challenging.32 Moreover, the exploration of the broader chemical space beyond natural products using machine learning remains a formidable task that holds the potential for discovering entirely new chemical scaffolds. For instance, Collins' group developed a graph neural network that leverages multiple chemical libraries, leading to the discovery of several highly effective antibiotics against the deadly strains of Acinetobacter baumannii.25,33 Nevertheless, these pipelines heavily rely on commercial and stationary libraries, which can lead to duplicate discoveries. Another approach employed by Das' group utilizes guidance from classifiers trained on the latent space of generative autoencoders to screen antimicrobial peptides against diverse pathogens.34 It is worth noting that all the aforementioned models require a substantial amount of labeled data to train the models. Consequently, most studies gather training data from published literature or open-source databases. However, for target compounds such as antimicrobial peptides, obtaining relevant data directly from the literature can be challenging due to variations in measurement conditions. This often leads to out-of-distribution problems and undesirable generalization errors. Therefore, the integration of machine learning models with quantitative data from a combinatorial library offers a compelling approach to unveil novel antibacterials concealed within the intricate possibilities.
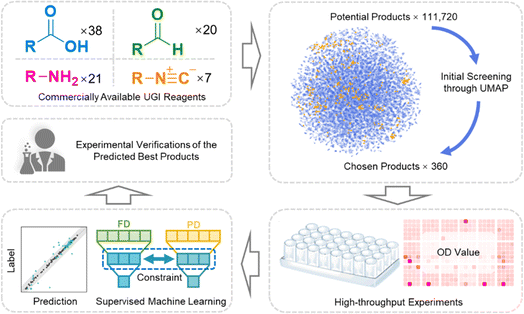
Herein, we proposed a new workflow, which fused the combinatorial library and machine learning to expedite the screening of antibacterials. To reduce the scope of preliminary screening, a uniform manifold approximation and projection algorithm (UMAP) was employed to uniformly sample the chemical space. Subsequently, 360 combinations were synthesized and their antibacterial properties were characterized parallelly. The data were input into the specially designed latent space constraint neural network (LSCNN) model. The antibacterial performance of the whole 111![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 720 potential products in the library was predicted and ranked by the LSCNN model. The top batch of compounds with the best antimicrobial properties was selected for further validation. Remarkably, three leads exhibited excellent antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) with reduced drug resistance development.
720 potential products in the library was predicted and ranked by the LSCNN model. The top batch of compounds with the best antimicrobial properties was selected for further validation. Remarkably, three leads exhibited excellent antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) with reduced drug resistance development.
Results and discussion
Commercially available carboxylic acids, amines, aldehydes and isonitriles were collected respectively (Table S1†) to generate a whole library with 111![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 720 candidates (Fig. 1 and 2A). In order to avoid the high cost accompanied with the laborious synthesis and purification process in pursuit of traversing the entire compound library, a focused library was created to represent the whole chemical space. UMAP was applied to reduce high-dimensional representations of the overall library to two-dimensional representations. Based on the reduced two-dimensional distribution map (Fig. S1†), 360 representative combinations were carefully selected to cover the distribution as uniformly as possible, thereby reducing the redundancy of the training dataset and aligning its distribution consistently with the whole library. The target bacterial strain chosen for evaluation was MRSA, given its high frequency and lethal nature in hospital-acquired infections.35 All individual components were excluded from exhibiting any antibacterial activity (Fig. S2†). Subsequently, 360 combinations were synthesized in parallel and tested against MRSA. The result of the initial test was summarized as a heatmap in Fig. 2B. Optical density (OD) values at 595 nm were tagged as antibacterial activities for each combination. It was observed that combinations with desired antibacterial effects (depicted by dark red color) were rare amidst the majority of combinations showing negligible activity (depicted by pale red color). It could indeed be anticipated as a laborious endeavor to uncover the few hit compounds within the vast library.
720 candidates (Fig. 1 and 2A). In order to avoid the high cost accompanied with the laborious synthesis and purification process in pursuit of traversing the entire compound library, a focused library was created to represent the whole chemical space. UMAP was applied to reduce high-dimensional representations of the overall library to two-dimensional representations. Based on the reduced two-dimensional distribution map (Fig. S1†), 360 representative combinations were carefully selected to cover the distribution as uniformly as possible, thereby reducing the redundancy of the training dataset and aligning its distribution consistently with the whole library. The target bacterial strain chosen for evaluation was MRSA, given its high frequency and lethal nature in hospital-acquired infections.35 All individual components were excluded from exhibiting any antibacterial activity (Fig. S2†). Subsequently, 360 combinations were synthesized in parallel and tested against MRSA. The result of the initial test was summarized as a heatmap in Fig. 2B. Optical density (OD) values at 595 nm were tagged as antibacterial activities for each combination. It was observed that combinations with desired antibacterial effects (depicted by dark red color) were rare amidst the majority of combinations showing negligible activity (depicted by pale red color). It could indeed be anticipated as a laborious endeavor to uncover the few hit compounds within the vast library.
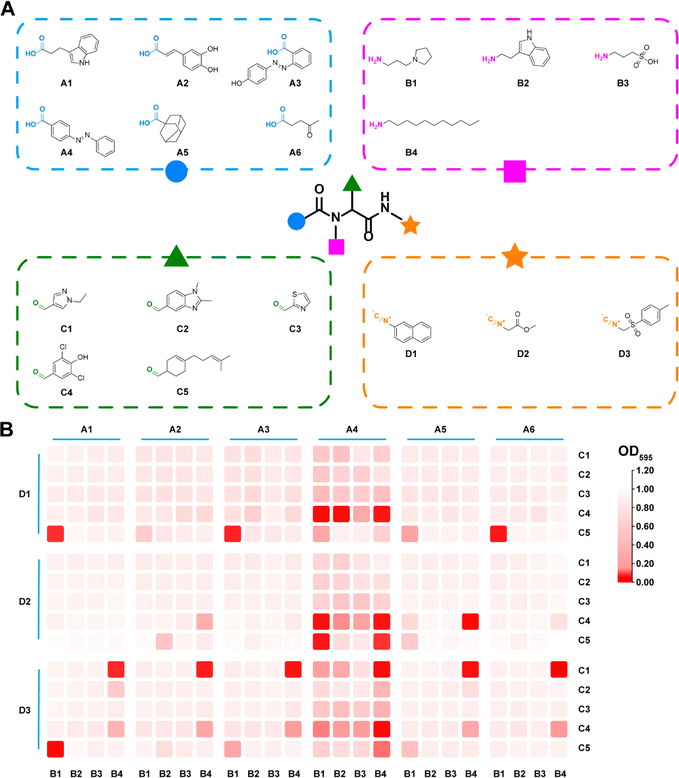 | ||
| Fig. 2 (A) The combinatorial library based on the Ugi reaction. (B) The heatmap for antibacterial activity of the synthesized preliminary library. | ||
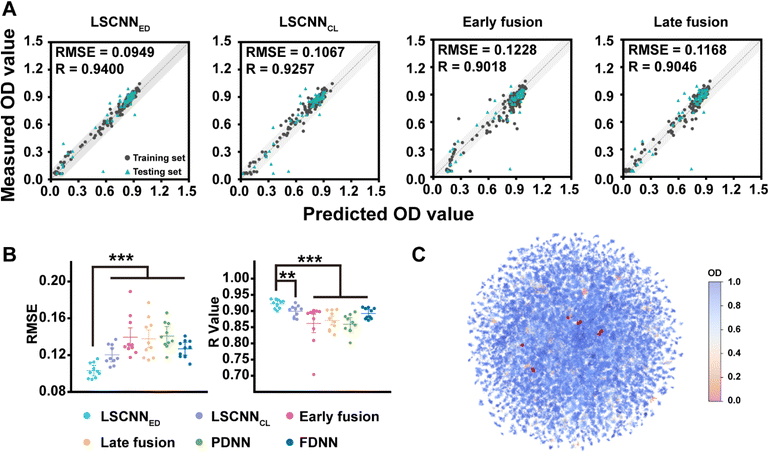
Machine learning was introduced to analyze the preliminary data. The extraction of meaningful molecular features played an essential role in developing an accurate machine learning model for antibacterial property prediction. One widely accepted molecular feature was the fingerprint descriptor (FD),36 which utilized binary encoding to indicate the presence or absence of specific chemical structures. However, it tended to neglect the physicochemical properties of molecules to some extent. Conversely, the physicochemical descriptor (PD) focused primarily on the physicochemical properties of molecules,37 overlooking the structural information. Considering their complementary nature, the fusion of PD and FD was a reasonable approach to enhance the model's accuracy. In addition, training models on small datasets could be challenging as the process was inclined to be unstable and different random seeds usually led to distinct models. Hence, improving the robustness of the model was also crucial when dealing with small datasets. Recently, there had been successful attempts which utilized multi-modal data, such as images, texts and audios38–40 to learn shared embedding representation spaces. These approaches leveraged the rich multi-modal information and achieved impressive zero-shot performance. Motivated by these remarkable outcomes,40 we proposed a novel feature-fusion architecture for antibacterial property prediction called LSCNN (Fig. S3†). LSCNN contained two multilayer perceptrons (MLP) with PD and FD as inputs, respectively. The outputs of both MLPs were OD values. Importantly, LSCNN imposed constraints on the hidden layers of the two MLPs as part of the loss function to learn the shared embedding space and facilitate interactions between different features. During the testing and prediction process, the averaged output of the two MLPs was used as the final output of LSCNN. We explored different feature fusion architectures (LSCNNED and LSCNNCL denoted Euclidean distance loss and contrastive loss, early fusion denoted feature-level fusion, and late fusion denoted the concatenation of PD and FD representations at the hidden layer, see ESI† for details). As demonstrated in Fig. 3A and B, the higher Pearson correlation coefficient (R) and lower root mean square error (RMSE) of LSCNN on the test set outperformed other commonly used feature fusion methods. Ablation experiments demonstrated that imposing constraints in the latent space produced better results than directly averaging the outputs of two separate MLPs (Fig. S4†). Moreover, the variance of LSCNN training results was significantly smaller than that of other feature fusion methods. We speculated that enforcing constraints in the hidden layer could stabilize the training process and reduce the fluctuation caused by the difference in weight initialization on small datasets. Subsequently, the OD predictions for the entire library were visualized as a heatmap (Fig. 3C) against the reduced UMAP distribution, with the top-10 combinations (represented by red points) clearly separated.
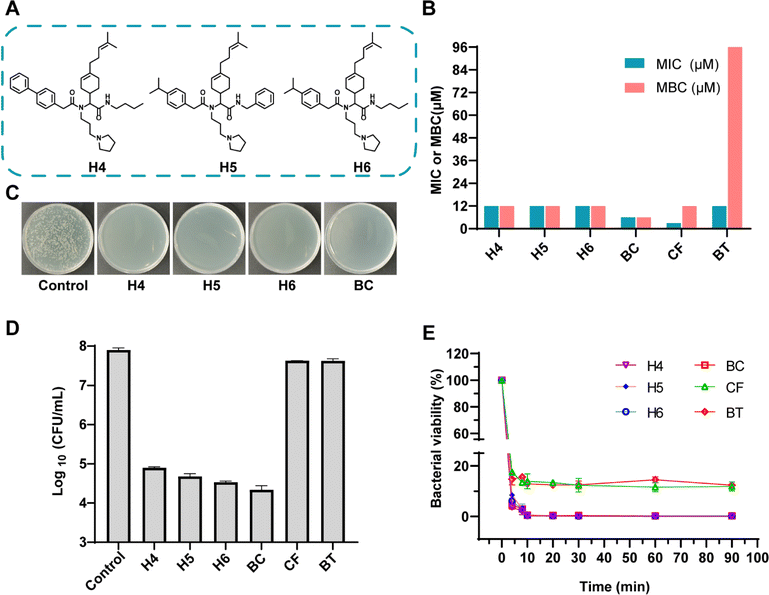
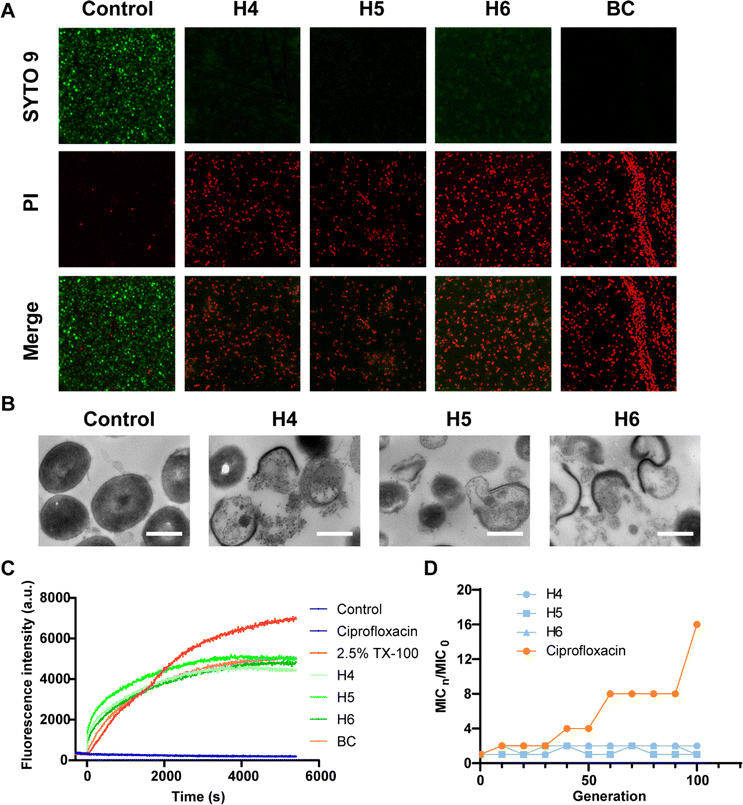
To validate predictions from LSCNN, a set of top-10 combinations was synthesized and subjected to antibacterial tests. The components were confirmed to have no inherent antibacterial activity (Fig. S5†). Remarkably, 6 out of 10 combinations (60% hit rate) demonstrated effective antibacterial properties against MRSA (Fig. S6†). In contrast, only 19 out of the initial 360 combinations (5.3% hit rate) showed potential antibacterial activity when an OD value below 0.1 was set as the cutoff. This significant increase in hit rate clearly indicated the crucial improvement achieved by our LSCNN model. Further purification was performed and the hit Ugi products (H1–6) were subjected to antibacterial assays (Fig. S7–S19 and Table S2†). Notably, H4–6 exhibited excellent antibacterial activity with both minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values measured at 12 μM (Fig. 4). In comparison, benzalkonium chloride (BC), a quaternary ammonium which was commonly applied as hospital biocides against nosocomial pathogens,41 displayed MIC and MBC values at 6 μM. Two antibiotics with a broad antibacterial spectrum, ciprofloxacin (CF) and bacitracin (BT), presented MIC at 3 and 12 μM respectively (Fig. 4B). The bacterial population was reduced by three orders of magnitude through incubation with H4–6 at 2× MIC within a 6 hours incubation period (Fig. 4C and D). However, CF and BT (96 μM) failed to effectively kill MRSA at 108 CFU mL−1 within 6 hours. Rapid killing effect were preferred in clinical, while normal antibiotics took a longer time to exhibit antibacterial performance.42,43 Moreover, bacterial killing kinetic assays revealed that all three hit compounds exhibited a rapid bactericidal capacity, effectively eliminating 99% of MRSA within just 10 minutes, which prevailed over CF and BT (Fig. 4E). A live/dead bacterial kit was employed to stain MRSA cells incubated with the hit compounds. Propidium iodide (PI) could penetrate impaired bacterial membranes and integrate with DNA to emit red fluorescence. All stained samples except control presented prominent red fluorescence, which presented excellent antibacterial activity of H4–6 (Fig. 5A). In addition, the molecular structures of H4–6 showed significant differences from the preliminary dataset (Fig. S20†), which demonstrated the successful generalization of the workflow.
The antibacterial mechanism of the hit compounds was further investigated. Transmission electron microscopy (TEM) images of MRSA cells revealed severe membrane damage, which implied the membrane-associated bactericidal mechanism of our products (Fig. 5B). This evidence was further supported by the dying experiment with DiSC3(5), which was a membrane potential sensitive probe.44 Triton X-100 (TX-100) was set as the positive control. DiSC3(5) fluorescence rapidly quenched in intact membranes due to the high concentration and exhibited enhanced fluorescence upon release in cell membranes with an imbalance in membrane potential. Intense membrane potential depolarization was observed in assays treated with the hit compounds, exhibiting a similar terminus to benzalkonium chloride. In contrast, another widely applied antibiotic ciprofloxacin failed to induce membrane potential depolarization (Fig. 5C), which typically inhibited DNA synthesis and replication to exert antibacterial effect. In light of the growing drug resistance among bacteria in clinical cases, the resistance development of MRSA against H4–6 was further evaluated. Within 100 generations, no drug resistance was observed for H4–6, whereas the MIC of ciprofloxacin increased 16 times (Fig. 5D). These findings underscored the ability of our hit compounds to effectively combat drug-resistant bacterial strains and address the urgent need for new antibacterial agents.
Conclusions
In summary, our study involved the construction of an unbiased combinatorial library with broad chemical space through the Ugi reaction. We employed the UMAP algorithm to visualize the high-dimensional distribution of the candidate pool in a two-dimensional map. The exhaustive synthesis and evaluation of the entire library chemical space were spared by uniform map sampling. The preliminary library was synthesized and screened against MRSA with the OD values tagged for each combination. To accurately predict the antibacterial activity of the entire library, a special LSCNN model was developed which incorporated both FD and PD of the molecules. After training the model with the quantitative data collected from a relatively focused library, the model was capable of ranking the antibacterial activity of the whole library. The validation experiments confirmed the activity of 6 hit combinations against MRSA, demonstrating the efficiency of our approach compared to the blind screening of the entire library. Additionally, three purified compounds exhibited rapid killing kinetics against MRSA and interfered intensely with the membrane potential, leading to significant membrane damage. This bactericidal mechanism might effectively suppress the emergence of antibacterial resistance commonly developed in clinical occasions. Our workflow integrated the massive data from the combinatorial library and the powerful generalization capability of the feature-fusing LSCNN model, which presented a promising paradigm for the discovery of new antibacterials.Data availability
The computational method and additional experimental data are available in the ESI.†Author contributions
C. Wang, Y. Wu, and Y. Xue conceived the idea, conducted the experiments, and wrote the manuscript together. L. Zou, and Y. Huang participated in the antibacterial evaluation experiments. P. Zhang, and J. Ji guided the whole project. All authors proof read the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (52293381 and 22175152), the National Key Research and Development Program of China (2022YFB3807300), the Huadong Medicine Joint Funds of the Zhejiang Provincial Natural Science Foundation of China (LHDMZ22H300011), and the Fundamental Research Funds for the Central Universities (226-2022-00146).Notes and references
- C. J. L. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. Robles Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne, M. G. Chipeta, F. Fell, S. Hackett, G. Haines-Woodhouse, B. H. Kashef Hamadani, E. A. P. Kumaran, B. McManigal, S. Achalapong, R. Agarwal, S. Akech, S. Albertson, J. Amuasi, J. Andrews, A. Aravkin, E. Ashley, F.-X. Babin, F. Bailey, S. Baker, B. Basnyat, A. Bekker, R. Bender, J. A. Berkley, A. Bethou, J. Bielicki, S. Boonkasidecha, J. Bukosia, C. Carvalheiro, C. Castañeda-Orjuela, V. Chansamouth, S. Chaurasia, S. Chiurchiù, F. Chowdhury, R. Clotaire Donatien, A. J. Cook, B. Cooper, T. R. Cressey, E. Criollo-Mora, M. Cunningham, S. Darboe, N. P. J. Day, M. De Luca, K. Dokova, A. Dramowski, S. J. Dunachie, T. Duong Bich, T. Eckmanns, D. Eibach, A. Emami, N. Feasey, N. Fisher-Pearson, K. Forrest, C. Garcia, D. Garrett, P. Gastmeier, A. Z. Giref, R. C. Greer, V. Gupta, S. Haller, A. Haselbeck, S. I. Hay, M. Holm, S. Hopkins, Y. Hsia, K. C. Iregbu, J. Jacobs, D. Jarovsky, F. Javanmardi, A. W. J. Jenney, M. Khorana, S. Khusuwan, N. Kissoon, E. Kobeissi, T. Kostyanev, F. Krapp, R. Krumkamp, A. Kumar, H. H. Kyu, C. Lim, K. Lim, D. Limmathurotsakul, M. J. Loftus, M. Lunn, J. Ma, A. Manoharan, F. Marks, J. May, M. Mayxay, N. Mturi, T. Munera-Huertas, P. Musicha, L. A. Musila, M. M. Mussi-Pinhata, R. N. Naidu, T. Nakamura, R. Nanavati, S. Nangia, P. Newton, C. Ngoun, A. Novotney, D. Nwakanma, C. W. Obiero, T. J. Ochoa, A. Olivas-Martinez, P. Olliaro, E. Ooko, E. Ortiz-Brizuela, P. Ounchanum, G. D. Pak, J. L. Paredes, A. Y. Peleg, C. Perrone, T. Phe, K. Phommasone, N. Plakkal, A. Ponce-de-Leon, M. Raad, T. Ramdin, S. Rattanavong, A. Riddell, T. Roberts, J. V. Robotham, A. Roca, V. D. Rosenthal, K. E. Rudd, N. Russell, H. S. Sader, W. Saengchan, J. Schnall, J. A. G. Scott, S. Seekaew, M. Sharland, M. Shivamallappa, J. Sifuentes-Osornio, A. J. Simpson, N. Steenkeste, A. J. Stewardson, T. Stoeva, N. Tasak, A. Thaiprakong, G. Thwaites, C. Tigoi, C. Turner, P. Turner, H. R. van Doorn, S. Velaphi, A. Vongpradith, M. Vongsouvath, H. Vu, T. Walsh, J. L. Walson, S. Waner, T. Wangrangsimakul, P. Wannapinij, T. Wozniak, T. E. M. W. Young Sharma, K. C. Yu, P. Zheng, B. Sartorius, A. D. Lopez, A. Stergachis, C. Moore, C. Dolecek and M. Naghavi, Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, Lancet, 2022, 399, 629–655 CrossRef CAS PubMed
.
- Z. Shang, S. Y. Chan, Q. Song, P. Li and W. Huang, The Strategies of Pathogen-Oriented Therapy on Circumventing Antimicrobial Resistance, Research, 2020, 2020, 2016201 CAS
.
- X.-Q. Kong, B.-Y. Wei, C.-X. Yu, X.-N. Guan, W.-P. Ma, G. Liu, C.-G. Yang and F.-J. Nan, Design, Synthesis and Biological Evaluation of Bengamide Analogues as ClpP Activators, Chin. J. Chem., 2020, 38, 1111–1115 CrossRef CAS
.
- L. Ferrazzano, A. Viola, E. Lonati, A. Bulbarelli, R. Musumeci, C. Cocuzza, M. Lombardo and A. Tolomelli, New isoxazolidinone and 3,4-dehydro-β-proline derivatives as antibacterial agents and MAO-inhibitors: A complex balance between two activities, Eur. J. Med. Chem., 2016, 124, 906–919 CrossRef CAS PubMed
.
- J. Liu, C. Du, H. T. Beaman and M. B. B. Monroe, Characterization of Phenolic Acid Antimicrobial and Antioxidant Structure–Property Relationships, Pharmaceutics, 2020, 12, 419 CrossRef CAS PubMed
.
- I. B. Seiple, Z. Zhang, P. Jakubec, A. Langlois-Mercier, P. M. Wright, D. T. Hog, K. Yabu, S. R. Allu, T. Fukuzaki, P. N. Carlsen, Y. Kitamura, X. Zhou, M. L. Condakes, F. T. Szczypiński, W. D. Green and A. G. Myers, A platform for the discovery of new macrolide antibiotics, Nature, 2016, 533, 338–345 CrossRef CAS PubMed
.
- K. Kranthiraja and A. Saeki, Experiment-Oriented Machine Learning of Polymer:Non-Fullerene Organic Solar Cells, Adv. Funct. Mater., 2021, 31, 2011168 CrossRef CAS
.
- R. Gómez-Bombarelli, J. Aguilera-Iparraguirre, T. D. Hirzel, D. Duvenaud, D. Maclaurin, M. A. Blood-Forsythe, H. S. Chae, M. Einzinger, D.-G. Ha, T. Wu, G. Markopoulos, S. Jeon, H. Kang, H. Miyazaki, M. Numata, S. Kim, W. Huang, S. I. Hong, M. Baldo, R. P. Adams and A. Aspuru-Guzik, Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach, Nat. Mater., 2016, 15, 1120–1127 CrossRef PubMed
.
- Á. Furka, Forty years of combinatorial technology, Drug Discov. Today, 2022, 27, 103308 CrossRef PubMed
.
- A. Volkov, J. Mi, K. Lalit, P. Chatterjee, D. Jing, S. L. Carnahan, Y. Chen, S. Sun, A. J. Rossini, W. Huang and L. M. Stanley, General Strategy for Incorporation of Functional Group Handles into Covalent Organic Frameworks via the Ugi Reaction, J. Am. Chem. Soc., 2023, 145, 6230–6239 CrossRef CAS PubMed
.
- L. Rotolo, D. Vanover, N. C. Bruno, H. E. Peck, C. Zurla, J. Murray, R. K. Noel, L. O'Farrell, M. Arainga, N. Orr-Burks, J. Y. Joo, L. C. S. Chaves, Y. Jung, J. Beyersdorf, S. Gumber, R. Guerrero-Ferreira, S. Cornejo, M. Thoresen, A. K. Olivier, K. M. Kuo, J. C. Gumbart, A. R. Woolums, F. Villinger, E. R. Lafontaine, R. J. Hogan, M. G. Finn and P. J. Santangelo, Species-agnostic polymeric formulations for inhalable messenger RNA delivery to the lung, Nat. Mater., 2023, 22, 369–379 CrossRef CAS PubMed
.
- D. Chan, J.-C. Chien, E. Axpe, L. Blankemeier, S. W. Baker, S. Swaminathan, V. A. Piunova, D. Y. Zubarev, C. L. Maikawa, A. K. Grosskopf, J. L. Mann, H. T. Soh and E. A. Appel, Combinatorial Polyacrylamide Hydrogels for Preventing Biofouling on Implantable Biosensors, Adv. Mater., 2022, 34, 2109764 CrossRef CAS PubMed
.
- C. E. Arcadia, E. Kennedy, J. Geiser, A. Dombroski, K. Oakley, S.-L. Chen, L. Sprague, M. Ozmen, J. Sello, P. M. Weber, S. Reda, C. Rose, E. Kim, B. M. Rubenstein and J. K. Rosenstein, Multicomponent molecular memory, Nat. Commun., 2020, 11, 691 CrossRef CAS PubMed
.
- F. Li, J. Han, T. Cao, W. Lam, B. Fan, W. Tang, S. Chen, K. L. Fok and L. Li, Design of self-assembly dipeptide hydrogels and machine learning via their chemical features, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 11259–11264 CrossRef CAS PubMed
.
- V. B. K. Kunig, M. Potowski, M. Akbarzadeh, M. Klika Škopić, D. dos Santos Smith, L. Arendt, I. Dormuth, H. Adihou, B. Andlovic, H. Karatas, S. Shaabani, T. Zarganes-Tzitzikas, C. G. Neochoritis, R. Zhang, M. Groves, S. M. Guéret, C. Ottmann, J. Rahnenführer, R. Fried, A. Dömling and A. Brunschweiger, TEAD–YAP Interaction Inhibitors and MDM2 Binders from DNA-Encoded Indole-Focused Ugi Peptidomimetics, Angew. Chem., Int. Ed., 2020, 59, 20338–20342 CrossRef CAS PubMed
.
- B.-X. Quan, H. Shuai, A.-J. Xia, Y. Hou, R. Zeng, X.-L. Liu, G.-F. Lin, J.-X. Qiao, W.-P. Li, F.-L. Wang, K. Wang, R.-J. Zhou, T. T.-T. Yuen, M.-X. Chen, C. Yoon, M. Wu, S.-Y. Zhang, C. Huang, Y.-F. Wang, W. Yang, C. Tian, W.-M. Li, Y.-Q. Wei, K.-Y. Yuen, J. F.-W. Chan, J. Lei, H. Chu and S. Yang, An orally available Mpro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron, Nat. Microbiol., 2022, 7, 716–725 CrossRef CAS PubMed
.
- D. Yamane, S. Onitsuka, S. Re, H. Isogai, R. Hamada, T. Hiramoto, E. Kawanishi, K. Mizuguchi, N. Shindo and A. Ojida, Selective covalent targeting of SARS-CoV-2 main protease by enantiopure chlorofluoroacetamide, Chem. Sci., 2022, 13, 3027–3034 RSC
.
- M. Nami, P. Salehi, M. Dabiri, M. Bararjanian, S. Gharaghani, M. Khoramjouy, A. Al-Harrasi and M. Faizi, Synthesis of novel norsufentanil analogs via a four-component Ugi reaction and in vivo, docking, and QSAR studies of their analgesic activity, Chem. Biol. Drug Des., 2018, 91, 902–914 CrossRef CAS PubMed
.
- K. Liu, W. M. McCue, C.-w. Yang, B. C. Finzel and X. Huang, Combinatorial synthesis of a hyaluronan based polysaccharide library for enhanced CD44 binding, Carbohydr. Polym., 2023, 300, 120255 CrossRef CAS PubMed
.
- R. Kita, T. Osawa and S. Obika, Conjugation of oligonucleotides with activated carbamate reagents prepared by the Ugi reaction for oligonucleotide library synthesis, RSC Chem. Biol., 2022, 3, 728–738 RSC
.
- A. Osipyan, S. Shaabani, R. Warmerdam, S. V. Shishkina, H. Boltz and A. Dömling, Automated, Accelerated Nanoscale Synthesis of Iminopyrrolidines, Angew. Chem., Int. Ed., 2020, 59, 12423–12427 CrossRef CAS PubMed
.
- D. Reker, Y. Rybakova, A. R. Kirtane, R. Cao, J. W. Yang, N. Navamajiti, A. Gardner, R. M. Zhang, T. Esfandiary, J. L'Heureux, T. von Erlach, E. M. Smekalova, D. Leboeuf, K. Hess, A. Lopes, J. Rogner, J. Collins, S. M. Tamang, K. Ishida, P. Chamberlain, D. Yun, A. Lytton-Jean, C. K. Soule, J. H. Cheah, A. M. Hayward, R. Langer and G. Traverso, Computationally guided high-throughput design of self-assembling drug nanoparticles, Nat. Nanotechnol., 2021, 16, 725–733 CrossRef CAS PubMed
.
- H. Hao, Y. Xue, Y. Wu, C. Wang, Y. Chen, X. Wang, P. Zhang and J. Ji, A paradigm for high-throughput screening of cell-selective surfaces coupling orthogonal gradients and machine learning-based cell recognition, Bioact. Mater., 2023, 28, 1–11 CAS
.
- J. Westermayr, J. Gilkes, R. Barrett and R. J. Maurer, High-throughput property-driven generative design of functional organic molecules, Nat. Comput. Sci., 2023, 3, 139–148 CrossRef CAS PubMed
.
- J. M. Stokes, K. Yang, K. Swanson, W. Jin, A. Cubillos-Ruiz, N. M. Donghia, C. R. MacNair, S. French, L. A. Carfrae, Z. Bloom-Ackermann, V. M. Tran, A. Chiappino-Pepe, A. H. Badran, I. W. Andrews, E. J. Chory, G. M. Church, E. D. Brown, T. S. Jaakkola, R. Barzilay and J. J. Collins, A Deep Learning Approach to Antibiotic Discovery, Cell, 2020, 180, 688–702 CrossRef CAS PubMed
.
- J. Huang, Y. Xu, Y. Xue, Y. Huang, X. Li, X. Chen, Y. Xu, D. Zhang, P. Zhang, J. Zhao and J. Ji, Identification of potent antimicrobial peptides via a machine-learning pipeline that mines the entire space of peptide sequences, Nat. Biomed. Eng., 2023, 7, 797–810 CrossRef CAS PubMed
.
- X. M. Deng, K. H. Chen, K. Pang, X. T. Liu, M. S. Gao, J. Ren, G. W. Yang, G. P. Wu, C. J. Zhang, X. F. Ni, P. Zhang, J. Ji, J. Z. Liu, Z. W. Mao, Z. L. Wu, Z. Xu, H. K. Zhang and H. Y. Li, Key progresses of MOE key laboratory of macromolecular synthesis and functionalization in 2022, Chin. Chem. Lett., 2024, 35, 108861 CrossRef CAS
.
- Y. Sugimoto, F. R. Camacho, S. Wang, P. Chankhamjon, A. Odabas, A. Biswas, P. D. Jeffrey and M. S. Donia, A metagenomic strategy for harnessing the chemical repertoire of the human microbiome, Science, 2019, 366, eaax9176 CrossRef CAS PubMed
.
- C. Walsh, Where will new antibiotics come from?, Nat. Rev. Microbiol., 2003, 1, 65–70 CrossRef CAS PubMed
.
- A. S. Walker and J. Clardy, A Machine Learning Bioinformatics Method to Predict Biological Activity from Biosynthetic Gene Clusters, J. Chem. Inf. Model., 2021, 61, 2560–2571 CrossRef CAS PubMed
.
- D. Zhang, J. Zhang, S. Kalimuthu, J. Liu, Z.-M. Song, B.-b. He, P. Cai, Z. Zhong, C. Feng, P. Neelakantan and Y.-X. Li, A systematically biosynthetic investigation of lactic acid bacteria reveals diverse antagonistic bacteriocins that potentially shape the human microbiome, Microbiome, 2023, 11, 91 CrossRef CAS PubMed
.
- G. Liu and J. M. Stokes, A brief guide to machine learning for antibiotic discovery, Curr. Opin. Microbiol., 2022, 69, 102190 CrossRef CAS PubMed
.
- G. Liu, D. B. Catacutan, K. Rathod, K. Swanson, W. Jin, J. C. Mohammed, A. Chiappino-Pepe, S. A. Syed, M. Fragis, K. Rachwalski, J. Magolan, M. G. Surette, B. K. Coombes, T. Jaakkola, R. Barzilay, J. J. Collins and J. M. Stokes, Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii, Nat. Chem. Biol., 2023, 19, 1342 CrossRef CAS PubMed
.
- P. Das, T. Sercu, K. Wadhawan, I. Padhi, S. Gehrmann, F. Cipcigan, V. Chenthamarakshan, H. Strobelt, C. dos Santos, P.-Y. Chen, Y. Y. Yang, J. P. K. Tan, J. Hedrick, J. Crain and A. Mojsilovic, Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations, Nat. Biomed. Eng., 2021, 5, 613–623 CrossRef CAS PubMed
.
- N. A. Turner, B. K. Sharma-Kuinkel, S. A. Maskarinec, E. M. Eichenberger, P. P. Shah, M. Carugati, T. L. Holland and V. G. Fowler, Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research, Nat. Rev. Microbiol., 2019, 17, 203–218 CrossRef CAS PubMed
.
- D. Rogers and M. Hahn, Extended-Connectivity Fingerprints, J. Chem. Inf. Model., 2010, 50, 742–754 CrossRef CAS PubMed
.
- J. Yang, L. Tao, J. He, J. R. McCutcheon and Y. Li, Machine learning enables interpretable discovery of innovative polymers for gas separation membranes, Sci. Adv., 2022, 8, eabn9545 CrossRef CAS PubMed
.
- A. Radford, J. W. Kim, C. Hallacy, A. Ramesh, G. Goh, S. Agarwal, G. Sastry, A. Askell, P. Mishkin, J. Clark, G. Krueger and I. Sutskever, Learning Transferable Visual Models From Natural Language Supervision, arXiv, 2021, preprint, arXiv:2103.00020, DOI:10.48550/arXiv.2103.00020.
- C. Jia, Y. F. Yang, Y. Xia, Y. T. Chen, Z. Parekh, H. Pham, Q. V. Le, Y. H. Sung, Z. Li and T. Duerig, Scaling Up Visual and Vision-Language Representation Learning With Noisy Text Supervision, arXiv, 2021, preprint, arxiv:2102.05918, DOI:10.48550/arXiv.2102.05918.
- J. N. Li, R. R. Selvaraju, A. D. Gotmare, S. Joty, C. M. Xiong and S. C. H. Hoi, Align before Fuse: Vision and Language Representation Learning with Momentum Distillation, arXiv, 2021, preprint, arXiv:2107.07651, DOI:10.48550/arXiv.2107.07651.
- K. Smith and I. S. Hunter, Efficacy of common hospital biocides with biofilms of multi-drug resistant clinical isolates, J. Med. Microbiol., 2008, 57, 966–973 CrossRef CAS PubMed
.
- P. S. Cham, Deepika, R. Bhat, D. Raina, D. Manhas, P. Kotwal, D. P. Mindala, N. Pandey, A. Ghosh, S. Saran, U. Nandi, I. A. Khan and P. P. Singh, Exploring the Antibacterial Potential of Semisynthetic Phytocannabinoid: Tetrahydrocannabidiol (THCBD) as a Potential Antibacterial Agent against Sensitive and Resistant Strains of Staphylococcus aureus, ACS Infect. Dis., 2024, 10, 64–78 CrossRef CAS PubMed
.
- M. Zhou, Y. Qian, J. Xie, W. Zhang, W. Jiang, X. Xiao, S. Chen, C. Dai, Z. Cong, Z. Ji, N. Shao, L. Liu, Y. Wu and R. Liu, Poly(2-Oxazoline)-Based Functional Peptide Mimics: Eradicating MRSA Infections and Persisters while Alleviating Antimicrobial Resistance, Angew. Chem., Int. Ed., 2020, 59, 6412–6419 CrossRef CAS PubMed
.
- H. Zhang, Q. Chen, J. Xie, Z. Cong, C. Cao, W. Zhang, D. Zhang, S. Chen, J. Gu, S. Deng, Z. Qiao, X. Zhang, M. Li, Z. Lu and R. Liu, Switching from membrane disrupting to membrane crossing, an effective strategy in designing antibacterial polypeptide, Sci. Adv., 2023, 9, eabn0771 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06441g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |