Open Access Article
Open Access ArticleMn2+-induced structural flexibility enhances the entire catalytic cycle and the cleavage of mismatches in prokaryotic argonaute proteins†
Lirong
Zheng‡
 ae,
Bingxin
Zhou‡
*ac,
Yu
Yang‡
b,
Bing
Zan‡
a,
Bozitao
Zhong
ae,
Bingxin
Zhou‡
*ac,
Yu
Yang‡
b,
Bing
Zan‡
a,
Bozitao
Zhong
 b,
Banghao
Wu
b,
Yan
Feng
b,
Qian
Liu
*b and
Liang
Hong
*abcd
b,
Banghao
Wu
b,
Yan
Feng
b,
Qian
Liu
*b and
Liang
Hong
*abcd
aInstitute of Natural Sciences, Shanghai Jiao Tong University, Shanghai 200240, China. E-mail: hongl3liang@sjtu.edu.cn; bingxin.zhou@sjtu.edu.cn
bState Key Laboratory for Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China. E-mail: liuqian1018@sjtu.edu.cn
cShanghai National Center for Applied Mathematics (SJTU Center), Shanghai Jiao Tong University, Shanghai 200240, China
dZhangjiang Institute for Advanced Study, Shanghai Jiao Tong University, Shanghai 200240, China
eDepartment of Cell and Developmental Biology & Michigan Neuroscience Institute, University of Michigan Medical School, 48105, Ann Arbor, MI, USA
First published on 14th March 2024
Abstract
Prokaryotic Argonaute (pAgo) proteins, a class of DNA/RNA-guided programmable endonucleases, have been extensively utilized in nucleic acid-based biosensors. The specific binding and cleavage of nucleic acids by pAgo proteins, which are crucial processes for their applications, are dependent on the presence of Mn2+ bound in the pockets, as verified through X-ray crystallography. However, a comprehensive understanding of how dissociated Mn2+ in the solvent affects the catalytic cycle, and its underlying regulatory role in this structure–function relationship, remains underdetermined. By combining experimental and computational methods, this study reveals that unbound Mn2+ in solution enhances the flexibility of diverse pAgo proteins. This increase in flexibility through decreasing the number of hydrogen bonds, induced by Mn2+, leads to higher affinity for substrates, thus facilitating cleavage. More importantly, Mn2+-induced structural flexibility increases the mismatch tolerance between guide-target pairs by increasing the conformational states, thereby enhancing the cleavage of mismatches. Further simulations indicate that the enhanced flexibility in linkers triggers conformational changes in the PAZ domain for recognizing various lengths of nucleic acids. Additionally, Mn2+-induced dynamic alterations of the protein cause a conformational shift in the N domain and catalytic sites towards their functional form, resulting in a decreased energy penalty for target release and cleavage. These findings demonstrate that the dynamic conformations of pAgo proteins, resulting from the presence of the unbound Mn2+ in solution, significantly promote the catalytic cycle of endonucleases and the tolerance of cleavage to mismatches. This flexibility enhancement mechanism serves as a general strategy employed by Ago proteins from diverse prokaryotes to accomplish their catalytic functions and provide useful information for Ago-based precise molecular diagnostics.
Introduction
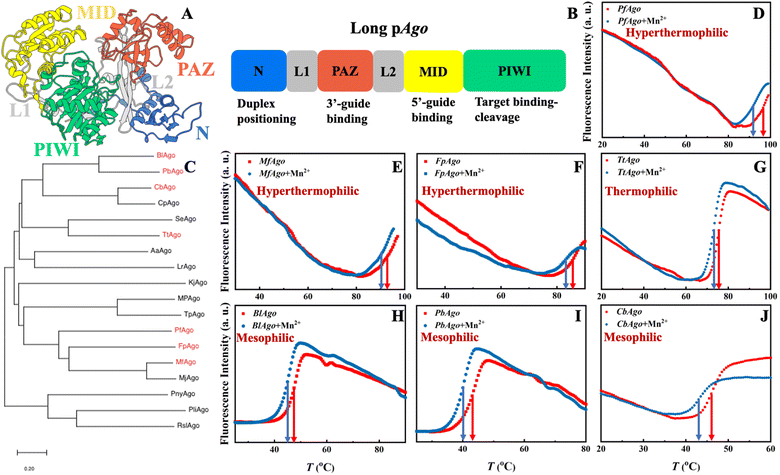
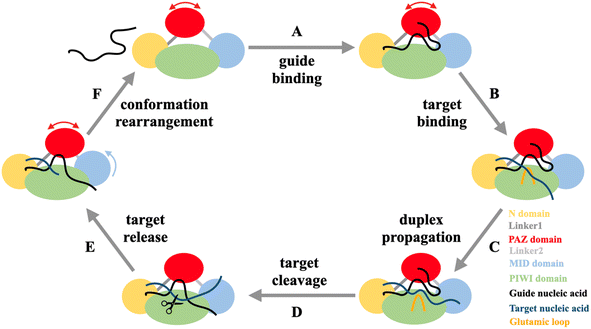
Prokaryotic Argonaute (pAgo) proteins, a class of endonucleases that play a crucial role in DNA interference in prokaryotic organisms,1–3 have gained significant attention in the field of biotechnology and medicine for their ability to target and cleave specific sequences in DNA/RNA.4–11 One of the most important applications is diagnostics, where pAgo proteins can be utilized to design molecular diagnostic assays that detect and quantify specific nucleic acid sequences, such as pathogens or cancer-associated mutations.6,7,12–14 These assays lead to the development of more sensitive and specific diagnostic methods, greatly improving the early detection and accurate treatment of diseases. Another crucial use of pAgo proteins is to label nucleic acids both in vitro and in vivo.15–18 Their high affinity to the substrate, stability in the protein-substrate complex, and specific recognition of the target sequence offer significant advantages over traditional methods that rely on nucleic acid binding, such as DNA-painting and DNA fluorescence in situ hybridization (FISH) technology. Furthermore, the combined action of pAgo proteins and nuclease-deficient RecBC helicase can cleave double-strand DNA,10 potentially leading to the design of gene therapies that specifically target disease-causing genes. These applications of pAgo proteins provide new possibilities for the development of therapeutics and genetic manipulation technologies.pAgo proteins predominantly contain six domains, including the N-terminal domain, Linker1, PIWI-Argonaute-Zwille domain (PAZ), Linker2, middle domain (MID), and P-element Induced Wimpy Testis domain (PIWI).3,19,20 Notably, the structural dynamics of these domains are critical for the biofunction of pAgo proteins.21–26 The pAgo proteins bind to guide nucleic acids, with the 5′-end and 3′-end of the nucleic acid being accommodated in the basic pocket of the MID domain and the PAZ domain, respectively.27 This guide-loaded complex then searches for complementary target strands through base-pairing with the seed region of the guide strand, which is followed by the propagation of the nucleic acid duplex.21,27 Subsequently, the 3′-end of the guide is released from the PAZ domain, and the active site of the Glu-finger is closed for the correct base-pairing.21 Conformational changes of the catalytic tetrad in the PIWI domain then allow for the cleavage of the target strand.21 Finally, the N domain helps to release the cleaved target, and the protein–guide complex is rearranged to its initial conformation for the next round of cleavage.28,29
In particular, the presence of Mn2+ is crucial for the catalytic activity of pAgo proteins, as determined by their crystal structures.21,25,30–34 Most known pAgo proteins utilize one Mn2+ bound to the MID pocket for interactions with the 5′ phosphate in the guide strand, and two Mn2+ ions bound to the catalytic sites (a DEDX motif, where X is K, D, or H) in the PIWI domain for cleavage of the target strand between the 10th and 11th bases, counted from the 5′-end of the guide.21,31 Despite the direct role of these bound Mn2+, a large number of Mn2+ are dissociated and present in the solvent. However, the role of these free ions in the catalytic function of pAgo proteins remains unknown. This situation prompts the question of whether these unbound Mn2+ in the solvent affect the functional flexibility of pAgo proteins and, subsequently, influence the endonuclease activity of pAgo proteins.
In this study, we employed a combination of differential scanning fluorimetry (DSF), nano differential scanning calorimetry (nano DSC), circular dichroism (CD) spectrum, synchrotron small-angle X-ray scattering (SAXS), biochemical assays, and molecular dynamics (MD) simulation to investigate the impact of unbound Mn2+ on the structural dynamics and endonuclease activity of prokaryotic Argonaute (pAgo) proteins from mesophilic (Clostridium butyricum Ago (CbAgo),31Paenibacilllus borealis Ago (PbAgo),35Pseudooceanicola lipolyticus Ago (PliAgo),30 and Brevibacillus laterosporus Ago (BlAgo)35), thermophilic (Thermus thermophilus Ago (TtAgo)) and hyperthermophilic (Pyrococcus furiosus Ago (PfAgo),36Methanocaldococcus fervens Ago (MfAgo)37 and Ferroglobus placidus Ago (FpAgo)38) organisms. Our results indicate that the presence of unbound Mn2+ reduces the hydrogen bonds in pAgo proteins from mesophiles to hyperthermophiles. This reduction in hydrogen bonds grants the protein a large degree of structural flexibility, while the overall packing of the protein remains intact. Furthermore, we demonstrated that Mn2+-dependent dynamics of pAgo proteins show a higher affinity to substrates and thus greatly promote the binding between the guide and target strands, which is key to facilitating the cleavage activity. Remarkably, Mn2+ exerts a significant influence on the precision of target DNA cleavage, which increases mismatch tolerance between guide–target pairs by increasing the conformational states. All-atom MD simulations at the conditions mimicking the Mn2+-present solvent environment revealed that the flexibility of Link 1 and Link 2 is greatly enhanced after incubating with Mn2+, which induces the conformational change of the PAZ domain to recognize various lengths of guide strands. Additionally, the increased flexibility in the glue-finger and the PAZ domain could help the propagation of duplexes. Furthermore, the Mn2+-induced dynamical change shifts the conformation of catalytic sites and the N domain towards its functional form, lowering the energy penalty for target strand cleavage and release, respectively. Overall, our data revealed that the unbound Mn2+-induced flexibility of pAgo proteins is crucial for their catalytic functions, and these findings are generally valid for pAgo proteins obtained from various prokaryotes. Moreover, this study sheds light on the underlying molecular mechanisms of the catalytic cycle of pAgo proteins, providing new avenues for research in the field of prokaryotic immunity systems and precise molecular diagnostics.
Results
Mn2+ enhances the flexibility of pAgo proteins obtained from mesophiles, thermophiles, and hyperthermophiles.The molecular architecture of long pAgo protein is characterized by a distinct two-lobed structure, which is a common feature among members of this family3,22 (Fig. 1A). The first lobe comprises the N domain, Linker 1, and PAZ domain, while the second lobe comprises the Linker 2, MID domain, and PIWI domain (Fig. 1B). The phylogenetic position of pAgo proteins studied herein, as well as their structural characteristics, are presented in Fig. 1C and S1,† respectively. Although the overall packing topology of pAgo proteins obtained from different prokaryotes exhibits a similar tertiary and secondary structure, their functions and optimal physiological temperatures are vastly divergent.3,24
We first examined how the secondary structures of pAgo proteins vary with the concentration of Mn2+ at their respective physiological temperatures. We used CD spectroscopy, a technique that measures the secondary structure of a protein by analyzing its absorption of the circularly polarized light.39 The results indicate that the presence of Mn2+ does not significantly alter the secondary structures of pAgo proteins (Fig. S2A–C†). It should be noted that we use Mn2+ in this study rather than Mg2+ because PfAgo cannot perform its functions when using Mg2+.36
We then characterized the effect of Mn2+ on the thermostability of pAgo proteins by using DSF spectroscopy. The DSF method is a quantitative method that allows for the assessment of thermostability by measuring changes in fluorescence intensity as the temperature is progressively increased, which monitors the tertiary structural changes in proteins.40 The melting temperature (Tm) of the tertiary structure of different pAgo proteins was determined with and without the presence of Mn2+. As shown in Fig. 1D–J, the addition of Mn2+ results in a significant decrease in the Tm of pAgo proteins, reducing the thermostability, whereas other divalent cations (Mg2+, Ca2+, and Zn2+) do not have this effect (Fig. S2D–E†). To further validate this finding, the Tm of pAgo proteins was also estimated by nano DSC, which measures the heat capacity of a protein as a function of temperatures.41 The results obtained through nano DSC were consistent with those obtained through DSF spectroscopy, i.e., a similar trend of increased denaturation in the presence of Mn2+ (Fig. S3†). Interestingly, we found that there is no significant difference observed when the Tm of the secondary structure of pAgo proteins was evaluated under similar conditions (Fig. S4†). Thus, the above results suggest that the presence of Mn2+ could disrupt interactions between amino acids participating in the tertiary structure of the protein but not those involved in the secondary structure (alpha-helix and beta-sheet), thereby reducing the thermostability of the protein.
To gain a deeper understanding of the structural factors that contribute to the observed difference in Tm between pAgo proteins incubated with Mn2+ and those incubated without, we conducted all-atom molecular dynamics (MD) simulations on mesophilic (CbAgo), thermophilic (TtAgo), and hyperthermophilic (FpAgo) pAgo proteins incubated with and without Mn2+. The example of snapshots of MD simulations is shown in Fig. S5† (the example of the potential energy of protein as a function of MD simulation time is shown in Fig. S6†). Our simulations reveal that the pAgo proteins incubated without Mn2+ exhibit a greater number of hydrogen bonds when compared to those incubated with Mn2+, while the number of salt bridges remains constant (Table 1). This disparity in intermolecular interactions, i.e., loss of hydrogen bonds, could reduce the energy barrier for protein unfolding in pAgo proteins incubated with Mn2+, which explains the decreased Tm observed in pAgo proteins (Fig. 1D–J).
By integrating the results of CD, DSF, and MD simulations, it suggests that Mn2+ possesses the capacity to perturb the intermolecular interactions among amino acid residues participating in the building of the tertiary structure, while not for the secondary structure (Table S1†).
It has been reported that structural flexibility is crucial for the biofunction of pAgo proteins.22,24 The intriguing question then arises as to whether the decreased intermolecular interactions resulting from the addition of Mn2+ can increase the flexibility of pAgo proteins. To address this, we conducted a structural analysis of FpAgo, TtAgo, BlAgo, and CbAgo using synchrotron SAXS and Porod–Debye analysis.24,42,43 The Porod–Debye analysis, which is a combination of the scattering intensity and the fourth power law of the scattering wavevector, q4·I(q), can be utilized to reveal the flexibility of biomacromolecules.42 An unambiguous Porod plateau in the scattering profile indicates the protein takes a rigid structure, while a lack of a plateau implies that the biomacromolecule forms dynamic conformations.42 As shown in Fig. 2A–D, the Porod–Debye plots of FpAgo, TtAgo, BlAgo, and CbAgo all displayed a clear Porod plateau. However, after incubation with Mn2+, a significant change in the Porod–Debye region was observed, with a loss of the plateau suggesting the protein flexibility is increased in the presence of Mn2+ (Fig. 2A–D). To further support these findings, we compared the flexibility of pAgo proteins incubated with and without Mn2+ using root mean square fluctuation (RMSF) derived from MD simulations. The RMSF of pAgo proteins incubated with Mn2+ is found to be larger than pAgo proteins incubated without Mn2+, which indicates that Mn2+ confers increased flexibility to pAgo proteins (Fig. 2E–H). Our findings demonstrate that Mn2+ has a universal effect on enhancing the structural dynamics of pAgo proteins from different prokaryotes.
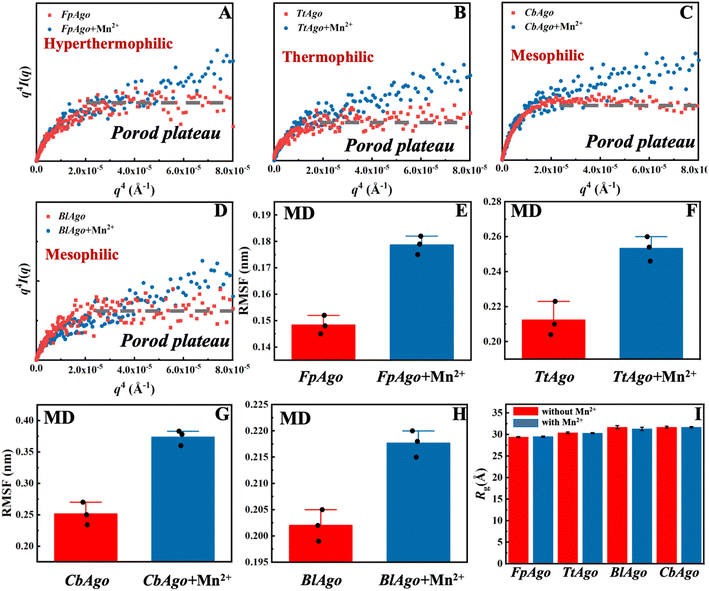 | ||
| Fig. 2 Structures and dynamics of pAgo proteins incubated with and without Mn2+ derived from SAXS and MD simulation. Porod–Debye analysis, q4I(q) vs. q4, of (A) FpAgo, (B) TtAgo, (C) BlAgo, and (D) CbAgo incubated with and without Mn2+. The concentration of Mn2+ is 5 mM. The dashed gray line in each panel indicates the Porod plateau. The details about the SAXS data collection and analysis are summarized in Materials and methods. Comparison of RMSF of (E) FpAgo, (F) TtAgo, (G) CbAgo, and (H) BlAgo incubated with and without Mn2+. RMSF of pAgo proteins is averaged over entire residues. The results from three independent MD simulations were quantified. Error bars represent the standard deviations of three independent simulations. The detailed procedure is presented in the Materials and methods. (I) Rg of FpAgo, TtAgo, BlAgo, and CbAgo incubated with and without Mn2+ derived from SAXS. The Guinier plots of proteins are shown in Fig. S7–S17.† | ||
Furthermore, we compared the structural compactness of pAgo proteins incubated with and without Mn2+. By utilizing Guinier analysis,44,45 one can derive the radius of gyration (Rg) of the protein, and we found that Rg of pAgo proteins incubated with Mn2+ is comparable to that of pAgo proteins incubated without Mn2+ (Fig. 2I, S7–S17, and Table S3†), which is in agreement with the Rg derived from MD simulations (Fig. S18 and S19†). Furthermore, the Porod volume of pAgo proteins incubated with and without Mn2+ was found to be similar, indicating that Mn2+ does not have a significant impact on the overall compactness of pAgo proteins. These findings were also supported by the ab initio low-resolution model reconstructed from SAXS data. As shown in Fig. S20,† the ab initio structure of pAgo proteins incubated with Mn2+ is similar to that of pAgo proteins incubated without Mn2+.
The findings from DSF, CD, nano DSC, SAXS, and MD simulation studies provide a convincing explanation of Mn2+’s impact on the structural flexibility of pAgo proteins, which demonstrates that the presence of Mn2+ greatly enhances the protein flexibility while preserving the protein's secondary structure. The observation that the overall structure of pAgo proteins remains similar to the crystalline form when incubated with Mn2+ further supports this conclusion, indicating that the flexibility changes are not due to significant alterations in the protein's overall structure.
Enzymatic functional role of the Mn2+-induced flexibility in pAgo proteins
The physical characterization of Mn2+ has revealed that it can enhance the flexibility of pAgo proteins. However, the implications of this increase in flexibility on the catalytic function of pAgo proteins remain unclear. As shown in Fig. 3A, the catalytic cycle of pAgo proteins involves several functional steps, including binding guide (gDNA binding), target recognition and annealing (tDNA binding), target cleavage, and release (tDNA cleaving and releasing).22,46 Firstly, we investigated the impact of increased protein structural flexibility on gDNA binding. We employed the fluorescence polarization assay on PliAgo (DSF measurements of PliAgo incubated with and without Mn2+ are shown in Fig. S21A†), a unique protein that does not require Mn2+ for substrate binding.30 The result, as shown in Fig. 3B, indicates that as the concentration of Mn2+ increases, the dissociate constant Kd decreases, thus demonstrating an increased binding of gDNA to the protein and the formation of the more protein-gDNA binary complex. To further verify these results, we conducted gDNA binding experiments on CbAgo using a mutation on the binding site amino acids, resulting in a binding pocket in the MID domain that does not bind Mn2+ (the amino acids sequence is shown in Table S5†).30 We found that the CbAgo mutant (CbAgoM) incubated with Mn2+ has a greater binding affinity to gDNA than it incubated without Mn2+ (Fig. 3C). We also performed the electrophoretic mobility shift assay (EMSA) on CbAgoM binding to gDNA. The result, illustrated in Fig. S22,† indicates that with an increase in the concentration of Mn2+, the amount of free gDNA decreases, while the protein–gDNA binary complex increases. This observation also demonstrates an enhanced binding of gDNA when the protein is incubated with Mn2+. The above results provide additional support for the notion that the increase in protein flexibility caused by Mn2+ can enhance the binding ability of proteins to gDNA (DSF measurements of CbAgoM incubated with and without Mn2+ are shown in Fig. S21B†). Additionally, an increased quantity of protein–gDNA binary complexes can effectively function as templates, thereby facilitating the binding of a greater amount of tDNA.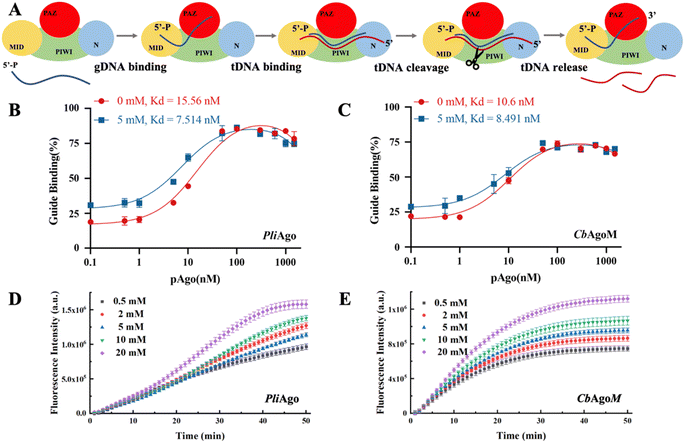 | ||
Fig. 3 The role of the Mn2+-dependent structural dynamics of pAgo proteins for binding guide DNA and cleavage of target DNA. (A) Schematic diagram of DNA-catalytic cycle of pAgo proteins. Fluorescence polarization assay of the binding of (B) PliAgo and (C) CbAgoM to gDNA at different concentrations of Mn2+. Cleavage assays of (D) PliAgo and (E) CbAgoM incubated with different concentrations of Mn2+. The cleavage activity of pAgo proteins is traced by fluorescence intensity as a function of time. In all experiments, protein, guide, and target were mixed at a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio and incubated at 37 °C. The reaction buffer without Mn2+ contains 10 mM EDTA. Three samples were used for each experimental condition. The results from three independent experiments were quantified. Error bars represent the standard deviations of three independent experiments. The detailed procedure is presented in the Materials and methods. The nucleotide sequences of the gDNA and tDNA are presented in Table S4.† 1 molar ratio and incubated at 37 °C. The reaction buffer without Mn2+ contains 10 mM EDTA. Three samples were used for each experimental condition. The results from three independent experiments were quantified. Error bars represent the standard deviations of three independent experiments. The detailed procedure is presented in the Materials and methods. The nucleotide sequences of the gDNA and tDNA are presented in Table S4.† | ||
The process of cleavage and release of tDNA is of paramount importance for the subsequent rounds of tDNA cleavage mediated by pAgo proteins.22 To investigate the impact of Mn2+ on the cleavage efficiency of pAgo proteins on tDNA, we employed a highly sensitive fluorescence-based assay. To this end, we labeled the 5′- and 3′-end of tDNA with the fluorescent group 6′ FAM and quenching group BHQ1, respectively. This allowed us to monitor the cleavage and release process of pAgo proteins on tDNA in real-time, with the absence of a fluorescent signal indicating that tDNA was either not cleaved or had been cleaved but not yet released, and the presence of a fluorescent signal indicating that tDNA had been cleaved and released successfully. As depicted in Fig. 3D and E, the cleavage rate and yield of cleavage products of PliAgo and CbAgoM on tDNA exhibited a clear dependence on the concentration of Mn2+, with tested higher concentrations resulting in increased cleavage products. Our findings provide important insights into the role of Mn2+ in modulating the catalytic activity of pAgo proteins, indicating that the enhancement of protein flexibility brought about by Mn2+ might facilitate conformational changes during the cleavage process and the release of products.
It has been reported that small interfering DNA of varying lengths present inside cells can function as gDNA for proteins, which subsequently cleave invading DNA.47 In light of this, we explored the flexibility of pAgo proteins and their potential roles in facilitating binding to tDNA and gDNA of different lengths. Our analysis, illustrated in Fig. S23,† revealed that when the concentration of Mn2+ increases from 0.1 mM to 5 mM, Kd for the protein with tDNA and gDNA of various lengths decreases, indicating enhanced binding of gDNA and tDNA to the pAgo protein. These findings suggest that the ability of pAgo proteins to adapt their conformation, facilitated by an increase in structural flexibility induced by Mn2+, enables them to bind to tDNA and gDNA of varying lengths.
We then investigated the effect of Mn2+-induced flexibility in proteins on the cleavage and release of tDNA of varying lengths. Our experiments utilized a range of Mn2+ concentrations to measure the rate of tDNA cleavage and release. As shown in Fig. S24,† the results revealed a statistically significant positive correlation between increasing concentrations of Mn2+ and the fluorescence intensity per unit of time. This observation demonstrates an augmentation in the rate of tDNA cleavage and release in response to Mn2+-induced changes in pAgo protein flexibility, thus suggesting that the enhancement of pAgo protein flexibility serves as a facilitative mechanism for the cleavage and release of tDNA.
Mn2+-dependent precision cleavage of target DNA
It's widely recognized that the efficiency of pAgo's cleavage is influenced by mismatches that occur between the guide and the target, particularly within the seed region. The presence of these mismatches can cause “bubbles” to form between the guide and the target, consequently affecting their mutual recognition and the subsequent cleavage by pAgo (Fig. 4A). In this section, we also mutated the amino acids in the binding pocket of pAgo proteins to exclude the impact of Mn2+ on binding gDNA and tDNA.30 In the case of PfAgo, the introduction of a single nucleotide mismatch at varying positions within the guide can cause weakening or even elimination of target cleavage (Fig. 4B). Interestingly, when the concentration of Mn2+ is increased, PfAgo can cleave the target even with a mismatched guide (Fig. 4B). This suggests that Mn2+ enhances the flexibility of PfAgo, thereby increasing its tolerance for mismatches. We then selected three guides that either perfectly matched the target sequence (guide-m0) or contained mismatches at the fourth (guide-m4) and seventh (guide-m7) positions. We performed cleavage assays at gradually increasing Mn2+ concentrations (Fig. 4C and D). Under conditions of 0.1 mM Mn2+, the three guides directed varied catalytic efficiency, ranging from 20% to 70%. Conversely, when the Mn2+ concentration was increased to 1 mM, the cleavage efficiency of all three guides reached a comparable level. Similarly, CbAgo demonstrated enhanced activity with increasing Mn2+ concentrations, as shown in Fig. S25.†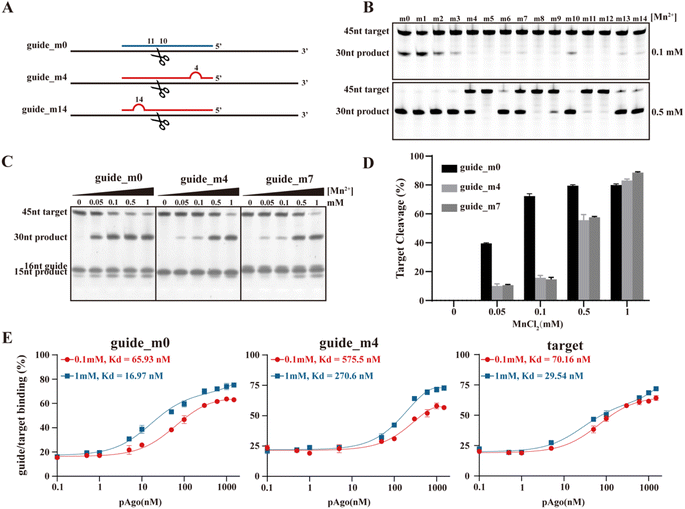 | ||
| Fig. 4 The role of the Mn2+-dependent structural dynamics of pAgo proteins for the designed mismatched guide DNA directed cleavage on target DNA. (A) Schematic representation of the designed mismatched gDNA directed cleavage on tDNA. The introduced mismatched nucleotides are indicated by the red semicircle. The nucleic acids sequence of gDNA and tDNA are shown in Table S6.†PfAgo is used here. (B) and (C) Gel detection results for the designed mismatched gDNA-directed cleavage on tDNA at different concentrations of Mn2+. (D) Quantifying the effects of incubating with Mn2+ on tDNA cleavage obtained from panel (C). (E) Fluorescence polarization assay of the designed mismatched guide and target at different concentrations of Mn2+. The results from three independent experiments were quantified. Error bars represent the standard deviations of three independent experiments. It should be noted that we mutated the amino acids in the binding pocket of pAgo proteins to exclude the impact of Mn2+ on binding gDNA and tDNA. | ||
To further investigate the role of Mn2+, we conducted fluorescence polarization experiments to measure the apparent dissociation constants (Kd) of PfAgo with either the guide or the target at different Mn2+ concentrations. We observed that an increase in the concentration of Mn2+ enhances the affinity of PfAgo for both the guide and the target. For the guides, the Kd values increased up to 4-fold and 2-fold for guide-m0 and guide-m4, respectively. For the target, the Kd values increased 3-fold (Fig. 4E). These findings suggest an increased binding affinity of the protein for mismatched gDNA and tDNA, which is attributed to increased protein flexibility under elevated Mn2+ concentrations. This provides further support for our hypothesis that Mn2+ may play a crucial role in enhancing the tolerance of DNA cleavage to mismatches.
Thus, the data obtained indicate that the enhancement of flexibility in pAgo proteins, induced by Mn2+, broadens the tolerance for mismatches between the gDNA and the tDNA. This heightened tolerance, however, incurs a trade-off, leading to a consequent decrease in the precision of tDNA cleavage orchestrated by the protein. This pivotal finding underscores the profound implications of the interplay between protein flexibility and nucleic acid detection accuracy, highlighting the necessity for maintaining an optimal balance between these two critical factors.
Mn2+-dependent dynamical states in pAgo protein derived from MD simulations
To gain a more atomic-level understanding of the impact of Mn2+ on the conformational dynamics related to the functionality of pAgo proteins, we conducted MD simulations of pAgo protein both incubated with and without Mn2+ mimicking the experimental conditions. As can be seen in Fig. S27,† the SAXS profile calculated from MD simulation agrees quantitatively with the experimental profile, demonstrating that the CbAgo obtained experimentally after incubation with Mn2+ can be described by the MD-derived structures.Firstly, we analyzed the overall conformational changes in proteins in the presence of Mn2+. Fig. 5A compares the conformation of CbAgo incubated without Mn2+ (upper panel) to that of CbAgo with Mn2+ (lower panel) over the trajectory time. The result shows that the addition of Mn2+ leads to an increase in the conformational dynamics of CbAgo. This finding is further validated by the observation that the root mean square fluctuation (RMSF) of proteins increases in the presence of Mn2+ (Fig. 5B). Although the flexibility of the protein is enhanced by Mn2+, the secondary structure and the radius of gyration remain consistent regardless of the presence of Mn2+, aligning with the SAXS and CD results presented in Fig. 1 and 2. Additionally, when analyzing the conformational phase space of CbAgo sampled in the MD simulation, we observed that the presence of Mn2+ allows the protein to explore a significantly larger conformational space (Fig. 5C and D). We also analyzed the locations of Mn2+ on protein at the molecular level. As shown in Fig. S24,† Mn2+ on the surfaces of different protein domains (N, Linker 1, PAZ, Linker 2, MID, and PIWI) engages in electrostatic interactions with amino acids. These interactions disrupt hydrogen bonds in the protein surface (Table 1), which leads to an enhancement in the structural flexibility of the protein. This finding aligns with the increased RMSF of the protein after incubating with Mn2+ (Fig. 5B). Specifically, Mn2+ plays a crucial role in interacting with Linker 1 and Linker 2 (Fig. S24†), contributing to the motion of the PAZ domain, which is known to be essential for the functionality of pAgo proteins.22
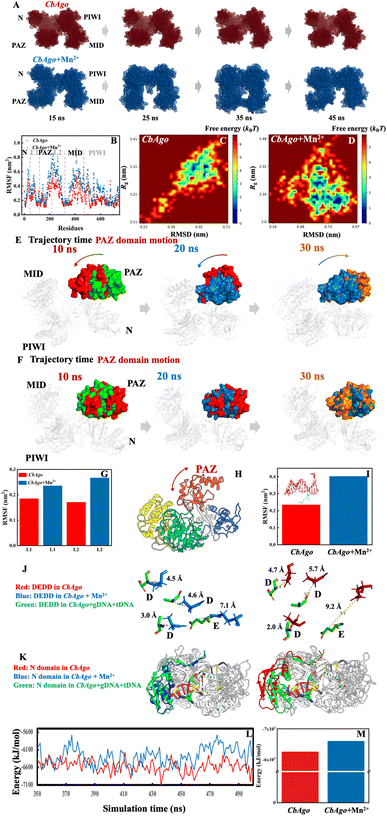 | ||
| Fig. 5 The effect of Mn2+ on the structural dynamics of pAgo protein derived from MD simulations. (A) The MD-derived structure of CbAgo incubated with (blue) and without (red) the presence of Mn2+ at different simulation times. (B) Comparison of RMSF of the overall structure. Free energy landscape of CbAgo incubated (C) without and (D) with Mn2+. RMSD is the root mean squared deviation of the structure in the first MD snapshot after equilibration. Conformational change of PAZ domain in CbAgo incubated (E) with and (F) without Mn2+ obtained from MD simulation. Comparison of RMSF of (G) linkers and (I) catalytic loop of CbAgo incubated with and without Mn2+. (H) Scheme of the motion of PAZ domain in pAgo protein. (J) Structural alignment of catalytic sites of CbAgo incubated without (left panel) and with (right panel) Mn2+ with respect to that of the experimental functional conformation (PDB ID: 6qzk). Catalytic sites, DEDD, in CbAgo incubated without Mn2+, with Mn2+, and in the experimental functional form are highlighted in red, blue, and green, respectively. (K) Structural alignment of N domain of CbAgo incubated without (left panel) and with (right panel) Mn2+ with respect to that of the experimental functional conformation (PDB ID: 6QZK). N domain in CbAgo incubated without Mn2+, with Mn2+, and in the experimental functional form is highlighted in red, blue, and green, respectively. Here, the protein structure is aligned by PyMOL. The alignment on Calpha is achieved by using PyMOL software. (L) The binding energy of CbAgo to gDNA/tDNA duplex over the entire simulation time derived from MD simulations. (M) The averaged binding energy of CbAgo to gDNA/tDNA duplex. The initial structure of MD simulation is the protein–gDNA–tDNA tertiary complex (PDB ID: 6QZK). | ||
Secondly, we analyzed the conformational changes related to the specific functionality of proteins.
Specifically, the glutamic loop, as shown in Fig. 5I, can adopt distinct conformations in the presence of the target strand. It inserts into the catalytic sites and binds metal ions during the formation of the extended guide–target duplex. Therefore, the enhanced conformational dynamics in the PAZ domain (Fig. 5G) and catalytic loop (Fig. 5I) promote the closure of the binding cleft and activate the cleavage of the target (Fig. 6C).21,23
Furthermore, we also observed that the catalytic tetrads adjust their alignments to functional ones, thereby reducing the energy barrier for conformational change (Fig. 5J), which accelerates the cleavage of target nucleic acid (Fig. 6D).
Furthermore, we analyzed the binding energy49 of protein to DNA in both the presence and absence of Mn2+. The results show that the binding energy of the protein to DNA is reduced upon the addition of Mn2+ throughout the simulation time (Fig. 5L and M). This finding also validates that the presence of Mn2+ can facilitate the release of cleaved target nucleic acids and promote the re-adjustment of conformations.
Discussion
Divalent cations play a fundamental role in biomacromolecule dynamics, primarily situated in ribosomes,50,51 enzymes,30,52,53 and structural proteins.54 The proper function and stability of such proteins necessitate the presence of ions. Divalent ions can impart consequential effects on the conformation and functionality of proteins, which is integral to numerous biological processes like enzymatic catalysis,31 regulation of protein–protein interactions,54 and signal transduction.55 Divalent ions also play a pivotal role in the regulation of membrane transport phenomena like ion transit across the plasma membrane and intracellular movement of ions and molecules.56 In ion channels and pumps, divalent ions serve as indispensable regulators, controlling the opening and closing of these channels, thereby regulating the flux of ions into and out of cells.56 Additionally, it has been demonstrated that ions impact the activity of endonucleases. For instance, the Mg2+-dependent conformational rearrangements of the CRISPR-Cas12a R-loop complex are crucial for complete double-stranded DNA cleavage.52In the case of pAgo proteins, we unraveled the molecular mechanisms of unbound Mn2+ regulating the binding, cleavage, and release of products through the use of spectroscopy, biochemical assays, and MD simulation. Initially, the presence of Mn2+ can disrupt certain hydrogen bonds within non-secondary structures, thereby increasing the flexibility of proteins and the associated entropy, without compromising the structural integrity. Next owing to the increased flexibility of the L1 linker and L2 linker, the conformational number of the PAZ domain is significantly increased. This enhanced structural flexibility facilitates the binding of the pAgo protein to gDNA, and helps the PAZ domain recognize gDNA of varying lengths. Furthermore, the enhanced structural variability of the glutamic loop and PAZ domain aids in the propagation of the DNA duplex, preparing it for tDNA cleavage. In addition, once the holo-form arrangement is achieved by the catalytic sites and the N domain, the catalytic center in the PIWI domain can be exposed to the tDNA. This exposure allows for the cleavage of the tDNA, facilitating the release of the cleaved product. Our simulation data indicates that the energy barrier for releasing the target in this step is also lowered by the presence of Mn2+. The analysis of the conformational dynamics of this catalytic cycle reveals that the presence of Mn2+ reduces the energy barrier for conformational changes between different functional conformations. Furthermore, this result unveiled the unexpected dynamical role of unbound Mn2+ in increasing the conformation states of pAgo proteins, which is crucial for their catalytic functions.
Previous structural research, based on crystallography, has demonstrated that pAgo proteins must undergo significant conformational changes to implement their functions, including the rearrangement of catalytic tetrads, propagation of duplexes through shifting loops in the PIWI domain, and motion of the PAZ domain, and release of cleaved targets by adjusting the PAZ and N domains.21 Therefore, structural flexibility is crucial for the conformational arrangement needed to achieve the functionality of pAgo proteins at different stages. In our study, we found that pAgo proteins from mesophiles, thermophiles, and hyperthermophiles exhibit a significant increase in flexibility when unbounded Mn2+ is present.
We found that Mn2+ can induce the structural flexibility of pAgo proteins from various prokaryotes, thereby promoting the rapid binding of the protein to gDNA and the formation of a stable protein-guide binary complex. This complex serves as a template to recognize various target nucleic acids in a short period. Furthermore, we observed that an increase in Mn2+ concentration can enhance the tolerance of cleavage to mismatches between guide DNA and target DNA. In prokaryotes, it has been demonstrated that pAgo proteins widely participate in hosts to defend against the invasion of nucleic acids2,47 According to our results, we can speculate that the increase in protein structural flexibility induced by Mn2+ can assist microorganisms in efficiently resisting external viral invasions. Firstly, the Mn2+-enhanced flexibility of protein structure can aid pAgo proteins in binding to plasmids, quickly chopping the plasmids to generate guide nucleic acids for recognition of invading nucleic acids. Next, the increased structural flexibility of pAgo proteins facilitates rapid conformational modifications. This allows them to quickly bind with different types (sequences and lengths) of nucleic acids in hosts and identify various invading nucleic acids, such as phages. Finally, the increased flexibility in pAgo proteins can aid in the protein's cleavage of mismatched nucleic acids, suggesting that the pAgo proteins in the cell may utilize a sequence of guide nucleic acids to bind to target nucleic acids with different sequences or lengths more efficiently. This, in turn, enhances the recognition and cleavage of invading DNA, preventing the replication of invader nucleic acids and ultimately safeguarding the host.
pAgo proteins have been widely used in nucleic acid detection.6,7,12 pAgo protein can bind to different gDNAs, which allows it to specifically recognize various complementary tDNAs. This capability enables the simultaneous detection of various diseases in argonaute-mediated nucleic acid diagnostics.7 In this study, we found that Mn2+-induced structural flexibility in the protein significantly enhances the binding affinity of pAgo protein to gDNAs of different lengths and sequences, without exhibiting a preference for a specific gDNA. This expanded improvement in the gDNA binding affinity profile of pAgo proteins suggests that elevating the concentration of Mn2+ in the detection system can trigger rapid binding of pAgo proteins to diverse gDNAs. This results in the formation of stable protein–gDNA binary complexes, facilitating the swift identification of various tDNAs and thereby accomplishing the goal of fast nucleic acid detection. Moreover, the current regulation of protein binding affinity is primarily accomplished through the mutation of amino acids in the protein—a method that is both time-consuming and costly.57,58 Nevertheless, our results indicate that it is possible to adjust the binding affinity of pAgo to gDNA by regulating ion concentrations, whether increasing or decreasing them, to meet specific requirements in nucleic acid detection. This approach could offer a simpler and more cost-effective alternative for argonaute-based nucleic acid detection.
The precision of nucleic acid detection methodologies is critically reliant on the accurate cleavage by pAgos. The high fidelity of such assays is underpinned by the exactness of the initial DNA cleavage process. Any deviations or inaccuracies within this fundamental step can substantially diminish the efficiency of subsequent amplification and signal readout stages.6 Our findings shed light on the role of Mn2+-induced flexibility in augmenting the proficiency of the pAgo protein in handling mismatches between gDNA and tDNA. We observed a correlation between increased concentrations of Mn2+ and an enhanced capacity of the pAgo protein to accommodate gDNA and tDNA mismatches, facilitating tDNA cleavage. The elevated presence of Mn2+ could enhance the protein's conformational flexibility and various functional states, which is verified by the MD simulations. This increased structural flexibility enables the protein to adapt its conformational state to cater to a variety of mismatch scenarios, thus improving the efficiency of tDNA cleavage. These findings carry significant implications for the effectiveness of various recently developed nucleic acid detection technologies.5,6
Conclusion
In this study, we investigated how the unbound Mn2+ affects the dynamics, structure, and endonuclease activity of pAgo proteins obtained from mesophilic (CbAgo, BlAgo, PliAgo, and PbAgo), thermophilic (TtAgo), and hyperthermophilic (PfAgo, FpAgo, and MfAgo) microorganisms using a combination of experimental and computational methods. Our results indicated that the presence of these unbound Mn2+ reduces the hydrogen bonds in pAgo proteins to release a great degree of structural flexibility while maintaining the overall structural integrity. Biochemical assays revealed that this Mn2+-induced structural flexibility in pAgo proteins significantly increases the substrate affinity and facilitates the binding of guide and target strands. Complementary computer simulations confirmed these experimental findings and revealed that the increased flexibility of Linker 1 and Linker 2 promotes the conformational mobility of the PAZ domain, allowing for the recognition of guide and target strands of varying lengths. Additionally, changes in the conformations of the PAZ domain and glutamic finger promote the propagation of the duplex. By comparing the structures of pAgo proteins incubated with Mn2+ to those of the protein–guide–target ternary complex, we observed that the conformation of catalytic sites and N domain shifts towards a functional state, which reduces the energy cost of conformational changes required for the cleavage of target strand and the release of the products.Our findings reveal that unbound Mn2+ plays an important role in the endonuclease functions of pAgo proteins, and this mechanism is a general strategy employed by pAgo proteins to enhance their catalytic functions. Unlike previous structural studies, which highlight the importance of the bound Mn2+ in directly participating in guide binding and target cleavage, our work discovers a role of unbound Mn2+ in an indirect way. The unbound free Mn2+ plays a crucial part in promoting the structural flexibility of pAgo proteins. This benefit extends not only to the binding of the protein to guide and target DNA, but also to the cleavage and release processes.
More importantly, by harnessing the influence of protein flexibility induced by unbound Mn2+ on the degree of mismatch between guide and target nucleic acids, we can finely tune the precision of target recognition by pAgo proteins through the modulation of Mn2+ concentration. This crucial characteristic expands the scope of molecular diagnostic applications, further highlighting the significance of structural flexibility in optimizing the performance of pAgo proteins.
Materials and methods
Protein expression and purification
A codon-optimized version of the PfAgo, TtAgo, FpAgo, MfAgo, BlAgo, PbAgo, CbAgo, CbAgoM, and PliAgo gene was synthesized by Sangon Biotech (Shanghai, China), and was cloned into the pET28a plasmid to construct pEX-Ago with an N terminal His-tag. The expression plasmid was transformed into Escherichia coli BL21 (DE3) cells. A 10 ml seed culture was grown at 37 °C in LB medium with 50 μg ml−1 kanamycin and was subsequently transferred to 1 L of LB in a shaker flask containing 50 μg ml−1 kanamycin. The cultures were incubated at 37 °C until the OD600 reached 0.6–0.8, and protein expression was then induced by the addition of isopropyl-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM, followed by incubation for 20–24 h at 18 °C. Cells were harvested by centrifugation for 30 min at 6000 rpm, and the cell pellets were collected for later purification. The cell pellets were resuspended in lysis buffer (15 mM Tris–HCl, 500 mM NaCl, pH 7.4) and then disrupted using a High-Pressure Homogenizer at 700–800 bar for 5 min (Gefran, Italy). The lysates were centrifuged for 30 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C, after which the supernatants were subjected to Ni-NTA affinity purification with elution buffer (15 mM Tris–HCl, 500 mM NaCl, 250 mM imidazole, pH 7.4). Further gel filtration purification using a Superdex 200 (GE Tech, USA) was carried out with an elution buffer. The fractions resulting from gel filtration were analyzed by SDS-PAGE, and fractions containing the protein were flash frozen at −80 °C in buffer (15 mM Tris–HCl pH 7.4, 200 mM NaCl, 10% glycerin).
000 rpm at 4 °C, after which the supernatants were subjected to Ni-NTA affinity purification with elution buffer (15 mM Tris–HCl, 500 mM NaCl, 250 mM imidazole, pH 7.4). Further gel filtration purification using a Superdex 200 (GE Tech, USA) was carried out with an elution buffer. The fractions resulting from gel filtration were analyzed by SDS-PAGE, and fractions containing the protein were flash frozen at −80 °C in buffer (15 mM Tris–HCl pH 7.4, 200 mM NaCl, 10% glycerin).
Differential scanning fluorimetry
Each pAgo protein sample containing 1 μM of protein in a buffer containing 15 mM Tris–HCl (pH = 7.4) and 200 mM NaCl was prepared in triplicate and added to PCR tubes. To examine the effect of Mn2+ on pAgo proteins, pAgo proteins were incubated in Tris–HCl buffer (pH = 7.4) with 5 mM Mn2+ or 5 mM EDTA for 20 min. SYPRO Orange dye available as 5000× stock (Sigma-Aldrich) was added just before the measurement of the pAgo proteins in an appropriate amount to achieve a final concentration of the dye of 5×. The thermal denaturation of the pAgo proteins was monitored by exciting the SYPRO Orange dye at 470 nm and monitoring its fluorescence emission at 570 nm using Q-PCR (Analytikjena, Germany). The baseline correction is used by the Opticon Monitor software available on the PCR instrument.Nano differential scanning calorimetry
Nano differential scanning calorimetry (nanoDSC) measurements were performed by using Nano DSC instruments (TA, USA). The concentration of pAgo proteins was 0.1 mg ml−1 in a buffer containing 15 mM Tris–HCl (pH 7.4) and 200 mM NaCl. To examine the effect of Mn2+ on pAgo proteins, pAgo proteins were incubated in 15 mM Tris–HCl buffer (pH = 7.4) with 5 mM Mn2+ or 5 mM EDTA for 20 min. All the experiments were carried out at temperatures ranging from 10 to 110 °C with a heating rate of 1 °C min−1 and under a pressure of 3 atm. The melting curves of pAgo proteins were subtracted from the buffer scans.Circular dichroism spectroscopy
The CD measurements on the evolution of the secondary structure of pAgo proteins were performed in a Jasco J-1500 spectropolarimeter with a 1 mm pathlength cell. The signals in the far-UV CD region (222 nm) were monitored as a function of temperature to determine the thermal unfolding of pAgo proteins. The concentrations of pAgo proteins were 0.1 mg ml−1 in 1× PBS buffer (pH = 7.4) for the various measurements. The pAgo proteins were heated in the range of 15 °C to 100 °C with a 1 °C min−1 heating rate and equilibrated for 3 min at each temperature, and the CD data were collected at 0.5 °C intervals. For CD spectra, we used PBS buffer as the solvent instead of Tris–HCl buffer because the Tris–HCl buffer has a strong CD background, which will significantly affect the analysis of the CD signal of proteins.24Small-angle X-ray scattering
Small-angle X-ray scattering (SAXS) measurement was employed to monitor the structural evolution of pAgo proteins incubated with and without Mn2+. In situ synchrotron SAXS measurements were conducted at BL19U2 beamline in Shanghai Synchrotron Radiation Facility (SSRF). The X-ray wavelength was 0.103 nm. Protein samples were dissolved in a buffer containing 15 mM Tris–HCl (pH 7.4), 200 mM NaCl, and 5 mM MnCl2 or EDTA. The concentration of samples is 0.5 mg ml−1. Protein solutions were loaded into the silica cell and then gently refreshed with a syringe pump to prevent X-ray damage. The measurements were carried out at 25 °C. To calculate the absolute intensity of protein, the empty cell and buffer were also measured at corresponding temperatures. Two-dimensional (2D) diffraction patterns were collected by the Pilatus 2 M detector with a resolution of 1043 × 981 pixels of 172 μm × 172 μm. Twenty sequential 2D images were collected with 0.5 s exposure time per frame. The 2D scattering patterns were then integrated into one-dimensional (1D) intensity curves by using Fit2D software from the European Synchrotron Radiation Facility (ESRF). Frames with no radiation damage were used for further processing. The SAXS data were compared to the atomic model by using the CRYSOL software.59ab initio reconstruction of protein structure by a chain-like ensemble of dummy residues is generated by GASBOR.60 P1 (no symmetry) and monodisperse data mode were used in generating the GASBOR model. At least 10 iterations of GASBOR programs were independently performed to validate the models. A presentative of the most typical model obtained was used in the figures. The one-dimensional data is processed by using ScÅtter and ATSAS software.61 The details of SAXS data collection and analysis are summarized in Table S3.†Molecular dynamics simulations
In MD simulations, a standard simulation box typically contains approximately ∼55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules, encompassing water molecules, proteins, and Mn2+ in our study. Thus, there are two ways to establish the simulation system for atomic-level analysis of protein dynamics. One approach (System 1, with a ratio of 1 protein: 6 Mn2+: 55
000 molecules, encompassing water molecules, proteins, and Mn2+ in our study. Thus, there are two ways to establish the simulation system for atomic-level analysis of protein dynamics. One approach (System 1, with a ratio of 1 protein: 6 Mn2+: 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 water in a simulation box) involves the ratio between Mn2+ and water to mimic biophysical experiments, while the other approach (System 2, with a ratio of 1 protein: 5000 Mn2+: 50
000 water in a simulation box) involves the ratio between Mn2+ and water to mimic biophysical experiments, while the other approach (System 2, with a ratio of 1 protein: 5000 Mn2+: 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 water in a simulation box) considers the ratio between Mn2+ and protein to mimic biophysical experiments. To determine which simulation system better agrees with the experimental system, we compared the SAXS profiles calculated from the MD protein structure with the SAXS profile calculated from experiments by using CRYSOL.62 As shown in Fig. S27,† we observed that the Chi2 values of the SAXS profile obtained from System 1 (Chi2 = 1.506) are much closer to the experimental SAXS profile compared to the one calculated from System 2 (Chi2 = 30.964). This result indicates that the MD structure from System 1 better aligns with the experimental structure. Therefore, we selected System 1 for further atomic-level analysis of pAgo protein dynamics.
000 water in a simulation box) considers the ratio between Mn2+ and protein to mimic biophysical experiments. To determine which simulation system better agrees with the experimental system, we compared the SAXS profiles calculated from the MD protein structure with the SAXS profile calculated from experiments by using CRYSOL.62 As shown in Fig. S27,† we observed that the Chi2 values of the SAXS profile obtained from System 1 (Chi2 = 1.506) are much closer to the experimental SAXS profile compared to the one calculated from System 2 (Chi2 = 30.964). This result indicates that the MD structure from System 1 better aligns with the experimental structure. Therefore, we selected System 1 for further atomic-level analysis of pAgo protein dynamics.
The initial structures of CbAgo and TtAgo for simulations were taken from PDB crystal structures 6QZK and 4N47, respectively. The initial structures of BlAgo and FpAgo for simulations were taken from AlphaFold2.63 The initial structure of CbAgo–gDNA–tDNA for simulations was taken from PDB crystal structures 6QZK. Protein and a large number of water molecules were filled in a cubic box (see Fig. S5†). For proteins and complexes incubated with the Mn2+ system, 6 Mn2+ were added into the simulation box to mimic the experimental concentration, and 32 chlorine counter ions were added to keep the system neutral in charge. For proteins and complexes in an aqueous solution, 16 chlorine counter ions were added to keep the system neutral in charge. The CHARMM36m force field64 was used for proteins, complex, and Mn2+, and the CHARMM-modified TIP3P model was chosen for water. The simulations were carried out at 298 K. After the 4000-step energy-minimization procedure, the systems were heated and equilibrated for 100 ps in the NVT ensemble and 500 ps in the NPT ensemble. The 100 ns production simulations were carried out at 1 atm with the proper periodic boundary condition, and the integration step was set to 2 fs. Fig. S6† shows the 100 ns profile of potential energy as a function of MD trajectory time for CbAgo incubated with and without Mn2+. It was clear that the equilibration procedure was sufficient for minimizing the energy of protein structures. The covalent bonds with hydrogen atoms were constrained by the LINCS algorithm.65 Lennard-Jones interactions were truncated at 12 Å with a force-switching function from 10 to 12 Å. The electrostatic interactions were calculated using the particle mesh Ewald method66 with a cutoff of 12 Å on an approximately 1 Å grid with a fourth-order spline. The temperature and pressure of the system are controlled by the velocity rescaling thermostat67 and the Parrinello–Rahman algorithm,68 respectively. All MD simulations were performed using GROMACS 2020.4 software packages. Representative simulation snapshots of the systems are given in Fig. S5.†
Single-strand DNA cleavage assay
For standard activity assays, cleavage experiments were performed in a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio (protein
1 molar ratio (protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) guide
guide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) target). First, 5 μM pAgo protein was mixed with a synthetic 1 μM gDNA guide in the reaction buffer (15 mM Tris–HCl (pH 7.4), 200 mM NaCl, different concentrations of MnCl2 or 5 mM EDTA). The solution was then pre-incubated at 37 °C for 20 min. After pre-incubation, 1 μM tDNA, which was labeled with the fluorescent group 6-FAM at the 5′-end and the quencher BHQ1 at the 3′-end, was added to the mixture. The cleavage experiments were performed at 37 °C. All experiments were performed in triplicate, and the fluorescence signals were traced by the quantitative real-time PCR QuantStudio 5 (Thermo Fisher Scientific, USA) with λex = 495 nm and λem = 520 nm. The results were analyzed by QuantStudioTM Design & Analysis Software v1.5.1. All the gDNA and tDNA used for cleavage are listed in Table S4.†
target). First, 5 μM pAgo protein was mixed with a synthetic 1 μM gDNA guide in the reaction buffer (15 mM Tris–HCl (pH 7.4), 200 mM NaCl, different concentrations of MnCl2 or 5 mM EDTA). The solution was then pre-incubated at 37 °C for 20 min. After pre-incubation, 1 μM tDNA, which was labeled with the fluorescent group 6-FAM at the 5′-end and the quencher BHQ1 at the 3′-end, was added to the mixture. The cleavage experiments were performed at 37 °C. All experiments were performed in triplicate, and the fluorescence signals were traced by the quantitative real-time PCR QuantStudio 5 (Thermo Fisher Scientific, USA) with λex = 495 nm and λem = 520 nm. The results were analyzed by QuantStudioTM Design & Analysis Software v1.5.1. All the gDNA and tDNA used for cleavage are listed in Table S4.†
In this work, 5 mM Mn2+ is used in the majority of our study. 5 mM Mn2+ is widely used in the mechanistic studies of pAgo proteins and pAgo-mediate nucleic acid diagnostics.7,69–74 Therefore, to ensure the practical relevance of our experimental results, we set the Mn2+ concentration to 5 mM in the majority of our experiments.
Fluorescence polarization assay
To determine the apparent dissociation constant (Kd) for proteins binding to either gDNA or tDNA, a fluorescence polarization assay was conducted using a multifunctional enzyme-linked immunosorbent assay plate reader (Spark, Tecan). A solution was prepared by combining 5 nM of 3′ 6-FAM labeled guide or target DNA with proteins across a concentration range of 0 to 1500 nM in a reaction buffer (15 mM Tris–HCl at pH 7.4, 200 mM NaCl, and 5 mM MnCl2). This mixture was incubated at 37 °C for 1 hour and subsequently transferred to a light-protected 96-well ELISA plate. The degree of polarization was measured using the Spark Tecan plate reader, employing an excitation wavelength of 485 nm and an emission wavelength of 525 nm. All experiments were independently conducted three times. The binding percentages were analyzed using Microsoft Excel and Prism 8 (GraphPad) software. The data was fitted with the Hill equation, incorporating a Hill coefficient of 2 to 2.5.Electrophoretic mobility shift assay
To examine the loading of gDNA onto pAgo proteins, proteins and 3′ end FAM-labeled guide were incubated in 20 μL of reaction buffer containing 15 mM Tris–HCl pH 7.4, 200 mM NaCl for 20 min at 37 °C. The concentration of 3′ end FAM labeled guide and protein was fixed as 1 μM and 5 μM, respectively, whereas the concentration of MnCl2 varied. Then the samples were mixed with 5 μL 5× loading buffer (Tris–HCl (pH 7.4), 25% glycerin, Bromophenol blue) and resolved by 12% native PAGE. Nucleic acids were visualized using Gel Doc™ XR+.Data availability
All the data are presented in ESI.†Author contributions
Lirong Zheng, Bingxin Zhou: conceptualization. Lirong Zheng, Yu Yang, Bing Zan, Bozitao Zhong, Banghao Wu: methodology. Lirong Zheng, Bingxin Zhou, Qian Liu, Yan Feng, Liang Hong: writing-reviewing and editing.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by NSF China (62302291, 11974239 and 22063007) and the Innovation Program of Shanghai Municipal Education Commission, National Key Research & Development Program of China (2018YFA0900403), Shanghai Pilot Program for Basic Research-Shanghai Jiao Tong University (21TQ1400204). We would like to thank the Instrumental Analysis Center of Shanghai Jiao Tong University for assistance with thermal performance tests via nano DSC and CD. We would like to thank Dr Na Li from BL19U2 beamline of Shanghai Synchrotron Radiation Facility (SSRF) for the help with synchrotron X-ray measurements on No. h20pr0008. Lirong Zheng would like to thank the Michigan Advancement Postdoctoral Program.References
- B. Koopal, et al., Short prokaryotic Argonaute systems trigger cell death upon detection of invading DNA, Cell, 2022, 185, 1471–1486.e1419 CrossRef CAS PubMed.
- D. C. Swarts, et al., DNA-guided DNA interference by a prokaryotic Argonaute, Nature, 2014, 507, 258–261 CrossRef CAS PubMed.
- D. C. Swarts, et al., The evolutionary journey of Argonaute proteins, Nat. Struct. Mol. Biol., 2014, 21, 743–753 CrossRef CAS PubMed.
- J. W. Hegge, D. C. Swarts and J. van der Oost, Prokaryotic Argonaute proteins: novel genome-editing tools?, Nat. Rev. Microbiol., 2018, 16, 5–11 CrossRef CAS PubMed.
- J. Song, et al., Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics, Nucleic Acids Res., 2019, 48, e19 CrossRef PubMed.
- Q. Liu, et al., Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations, Nucleic Acids Res., 2021, 49, e75 CrossRef CAS PubMed.
- R. He, et al., Pyrococcus furiosus Argonaute-mediated nucleic acid detection, Chem. Commun., 2019, 55, 13219–13222 RSC.
- Y. Qin, Y. Li and Y. Hu, Emerging Argonaute-based nucleic acid biosensors, Trends Biotechnol., 2022, 40, 910–914 CrossRef CAS PubMed.
- X. Li, et al., Mesophilic Argonaute-based isothermal detection of SARS-CoV-2, Front. Microbiol., 2022, 13 Search PubMed.
- R. Vaiskunaite, J. Vainauskas, J. J. L. Morris, V. Potapov and J. Bitinaite, Programmable cleavage of linear double-stranded DNA by combined action of Argonaute CbAgo from Clostridium butyricum and nuclease deficient RecBC helicase from E. coli, Nucleic Acids Res., 2022, 50, 4616–4629 CrossRef CAS PubMed.
- G. Xun, S. T. Lane, V. A. Petrov, B. E. Pepa and H. Zhao, A rapid, accurate, scalable, and portable testing system for COVID-19 diagnosis, Nat. Commun., 2021, 12, 2905 CrossRef CAS PubMed.
- J. Song, et al., Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics, Nucleic Acids Res., 2020, 48, e19 CrossRef PubMed.
- F. Wang, et al., PfAgo-based detection of SARS-CoV-2, Biosens. Bioelectron., 2021, 177, 112932 CrossRef CAS PubMed.
- G. Xun, et al., Argonaute with stepwise endonuclease activity promotes specific and multiplex nucleic acid detection, Bioresour. Bioprocess., 2021, 8, 46 CrossRef.
- M. Filius, et al., High-Speed Super-Resolution Imaging Using Protein-Assisted DNA-PAINT, Nano Lett., 2020, 20, 2264–2270 CrossRef CAS PubMed.
- L. Chang, et al., AgoFISH: cost-effective in situ labelling of genomic loci based on DNA-guided dTtAgo protein, Nanoscale Horiz., 2019, 4, 918–923 RSC.
- A. Toudji-Zouaz, V. Bertrand and A. Barrière, Imaging of native transcription and transcriptional dynamics in vivo using a tagged Argonaute protein, Nucleic Acids Res., 2021, 49, e86 CrossRef CAS PubMed.
- S. Shin, et al., Quantification of purified endogenous miRNAs with high sensitivity and specificity, Nat. Commun., 2020, 11, 6033 CrossRef CAS PubMed.
- K. S. Makarova, Y. I. Wolf, J. van der Oost and E. V. Koonin, Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements, Biol. Direct, 2009, 4, 29 CrossRef PubMed.
- S. Willkomm, K. S. Makarova and D. Grohmann, DNA silencing by prokaryotic Argonaute proteins adds a new layer of defense against invading nucleic acids, FEMS Microbiol. Rev., 2018, 42, 376–387 CrossRef CAS PubMed.
- G. Sheng, et al., Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 652–657 CrossRef CAS PubMed.
- L. Lisitskaya, A. A. Aravin and A. Kulbachinskiy, DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins, Nat. Commun., 2018, 9, 5165 CrossRef PubMed.
- Y. Wang, et al., Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes, Nature, 2009, 461, 754–761 CrossRef CAS PubMed.
- L. Zheng, et al., Loosely-packed dynamical structures with partially-melted surface being the key for thermophilic argonaute proteins achieving high DNA-cleavage activity, Nucleic Acids Res., 2022, 50, 7529–7544 CrossRef CAS PubMed.
- S. Willkomm, et al., Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein, Nat. Microbiol., 2017, 2, 17035 CrossRef CAS PubMed.
- S. Willkomm, et al., Single-molecule FRET uncovers hidden conformations and dynamics of human Argonaute 2, Nat. Commun., 2022, 13, 3825 CrossRef CAS PubMed.
- Y. Wang, G. Sheng, S. Juranek, T. Tuschl and D. J. Patel, Structure of the guide-strand-containing argonaute silencing complex, Nature, 2008, 456, 209–213 CrossRef CAS PubMed.
- P. B. Kwak and Y. Tomari, The N domain of Argonaute drives duplex unwinding during RISC assembly, Nat. Struct. Mol. Biol., 2012, 19, 145–151 CrossRef CAS PubMed.
- T. Miyoshi, K. Ito, R. Murakami and T. Uchiumi, Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute, Nat. Commun., 2016, 7, 11846 CrossRef CAS PubMed.
- L. Lisitskaya, et al., Programmable RNA targeting by bacterial Argonaute nucleases with unconventional guide binding and cleavage specificity, Nat. Commun., 2022, 13, 4624 CrossRef CAS PubMed.
- J. W. Hegge, et al., DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute, Nucleic Acids Res., 2019, 47, 5809–5821 CrossRef CAS PubMed.
- J. S. Parker, E. A. Parizotto, M. Wang, S. M. Roe and D. Barford, Enhancement of the Seed-Target Recognition Step in RNA Silencing by a PIWI/MID Domain Protein, Mol. Cell, 2009, 33, 204–214 CrossRef CAS PubMed.
- A. Zander, et al., Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii, Nat. Microbiol., 2017, 2, 17034 CrossRef CAS PubMed.
- E. Kaya, et al., A bacterial Argonaute with noncanonical guide RNA specificity, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 4057–4062 CrossRef CAS PubMed.
- H. Dong, et al., Characterization of Argonaute nucleases from mesophilic bacteria Paenibacillus borealis and Brevibacillus laterosporus, Bioresour. Bioprocess., 2021, 8, 133 CrossRef.
- D. C. Swarts, et al., Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA, Nucleic Acids Res., 2015, 43, 5120–5129 CrossRef CAS PubMed.
- Y. Chong, Q. Liu, F. Huang, D. Song and Y. Feng, Characterization of a recombinant thermotolerant argonaute protein as an endonuclease by broad guide utilization, Bioresour. Bioprocess., 2019, 6, 21 CrossRef.
- X. Guo, et al., A Hyperthermophilic Argonaute From Ferroglobus placidus With Specificity on Guide Binding Pattern, Front. Microbiol., 2021, 12, 654345 CrossRef PubMed.
- R. W. Woody, “[4] Circular dichroism”, in Methods in Enzymology, Academic Press, 1995, vol. 246, pp. 34–71 Search PubMed.
- F. H. Niesen, H. Berglund and M. Vedadi, The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability, Nat. Protoc., 2007, 2, 2212–2221 CrossRef CAS PubMed.
- G. M. D. Ferreira, et al., Lactoferrin denaturation induced by anionic surfactants: The role of the ferric ion in the protein stabilization, Int. J. Biol. Macromol., 2018, 117, 1039–1049 CrossRef CAS PubMed.
- R. P. Rambo and J. A. Tainer, Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law, Biopolymers, 2011, 95, 559–571 CrossRef CAS PubMed.
- R. P. Rambo and J. A. Tainer, Super-Resolution in Solution X-Ray Scattering and Its Applications to Structural Systems Biology, Annu. Rev. Biophys., 2013, 42, 415–441 CrossRef CAS PubMed.
- K. Sharma, I. L. Antunes, V. Rajulapati and A. Goyal, Low-resolution SAXS and comparative modeling based structure analysis of endo-β-1,4-xylanase a family 10 glycoside hydrolase from Pseudopedobacter saltans comb. nov, Int. J. Biol. Macromol., 2018, 112, 1104–1114 CrossRef CAS PubMed.
- C. Putnam, Guinier peak analysis for visual and automated inspection of small-angle X-ray scattering data, J. Appl. Crystallogr., 2016, 49, 1412–1419 CrossRef CAS PubMed.
- W. E. Salomon, S. M. Jolly, M. J. Moore, P. D. Zamore and V. Serebrov, Single-Molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of Its Nucleic Acid Guides, Cell, 2015, 162, 84–95 CrossRef CAS PubMed.
- D. C. Swarts, et al., Autonomous Generation and Loading of DNA Guides by Bacterial Argonaute, Mol. Cell, 2017, 65, 985–998.e986 CrossRef CAS PubMed.
- K. W. Doxzen and J. A. Doudna, DNA recognition by an RNA-guided bacterial Argonaute, PLoS One, 2017, 12, e0177097 CrossRef PubMed.
- C. Wang, D. A. Greene, L. Xiao, R. Qi and R. Luo, Recent Developments and Applications of the MMPBSA Method, Front. Mol. Biosci., 2018, 4, 87 CrossRef PubMed.
- J. Li, X. Zhang, L. Hong and Y. Liu, Entropy Driving the Mg2+-Induced Folding of TPP Riboswitch RNA, J. Phys. Chem. B, 2022, 126, 9457–9464 CrossRef CAS PubMed.
- J. L. Fiore, E. D. Holmstrom and D. J. Nesbitt, Entropic origin of Mg2+-facilitated RNA folding, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2902–2907 CrossRef CAS PubMed.
- H. Son, et al., Mg2+-dependent conformational rearrangements of CRISPR-Cas12a R-loop complex are mandatory for complete double-stranded DNA cleavage, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2113747118 CrossRef CAS PubMed.
- C. Andreini, I. Bertini, G. Cavallaro, G. L. Holliday and J. M. Thornton, Metal ions in biological catalysis: from enzyme databases to general principles, JBIC, J. Biol. Inorg. Chem., 2008, 13, 1205–1218 CrossRef CAS PubMed.
- C. Hureau and P. Faller, Aβ-mediated ROS production by Cu ions: Structural insights, mechanisms and relevance to Alzheimer's disease, Biochimie, 2009, 91, 1212–1217 CrossRef CAS PubMed.
- F. H. Yu and W. A. Catterall, The VGL-Chanome: A Protein Superfamily Specialized for Electrical Signaling and Ionic Homeostasis, Sci. STKE, 2004, re15 Search PubMed.
- Y. Nevo and N. Nelson, The NRAMP family of metal-ion transporters, Biochim. Biophys. Acta, Mol. Cell Res., 2006, 1763, 609–620 CrossRef CAS PubMed.
- B. P. Kleinstiver, et al., Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing, Nat. Biotechnol., 2019, 37, 276–282 CrossRef CAS PubMed.
- M. H. Tran, et al., A more efficient CRISPR-Cas12a variant derived from Lachnospiraceae bacterium MA2020, Mol. Ther.–Nucleic Acids, 2021, 24, 40–53 CrossRef CAS PubMed.
- D. Svergun, C. Barberato and M. H. J. Koch, CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates, J. Appl. Crystallogr., 1995, 28, 768–773 CrossRef CAS.
- D. I. Svergun, M. V. Petoukhov and M. H. J. Koch, Determination of Domain Structure of Proteins from X-Ray Solution Scattering, Biophys. J., 2001, 80, 2946–2953 CrossRef CAS PubMed.
- K. Manalastas-Cantos, et al., ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis, J. Appl. Crystallogr., 2021, 54, 343–355 CrossRef CAS PubMed.
- D. Franke, et al., ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions, J. Appl. Crystallogr., 2017, 50, 1212–1225 CrossRef CAS PubMed.
- J. Jumper, et al., Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- J. Huang, et al., CHARMM36m: an improved force field for folded and intrinsically disordered proteins, Nat. Methods, 2017, 14, 71–73 CrossRef CAS PubMed.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS PubMed.
- U. Essmann, et al., A smooth particle mesh Ewald method, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- M. Parrinello and A. Rahman, Polymorphic transitions in single crystals: A new molecular dynamics method, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- Y. Wang, et al., Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex, Nature, 2008, 456, 921–926 CrossRef CAS PubMed.
- Y. Liu, et al., A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis, Nucleic Acids Res., 2021, 49, 1597–1608 CrossRef CAS PubMed.
- Y. Cao, et al., Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single-and double-stranded DNA at 37° C, Cell discovery, 2019, 5, 38 CrossRef PubMed.
- L. Wang, et al., A bacterial Argonaute with efficient DNA and RNA cleavage activity guided by small DNA and RNA, Cell Rep., 2022, 41, 111533 CrossRef CAS PubMed.
- X. Jiang, Y. Liu, Q. Liu and L. Ma, Characterization of a Programmable Argonaute Nuclease from the Mesophilic Bacterium Rummeliibacillus suwonensis, Biomolecules, 2022, 12, 355 CrossRef CAS PubMed.
- Q. Liu, et al., A programmable pAgo nuclease with RNA target-cleavage specificity from the mesophilic bacterium Verrucomicrobia: A pAgo with RNA target cleavage specificity, Acta Biochim. Biophys. Sin., 2023, 55, 1204 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06221j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |