Open Access Article
Open Access ArticleModular, multi-robot integration of laboratories: an autonomous workflow for solid-state chemistry†
Amy. M.
Lunt
ab,
Hatem
Fakhruldeen
a,
Gabriella
Pizzuto
 a,
Louis
Longley
a,
Alexander
White
a,
Nicola
Rankin
ab,
Rob
Clowes
a,
Ben
Alston
ab,
Lucia
Gigli
a,
Louis
Longley
a,
Alexander
White
a,
Nicola
Rankin
ab,
Rob
Clowes
a,
Ben
Alston
ab,
Lucia
Gigli
 c,
Graeme M.
Day
c,
Graeme M.
Day
 c,
Andrew I.
Cooper
c,
Andrew I.
Cooper
 *ab and
Samantha Y.
Chong
*ab and
Samantha Y.
Chong
 *ab
*ab
aDepartment of Chemistry and Materials Innovation Factory, University of Liverpool, L7 3NY, UK. E-mail: aicooper@liverpool.ac.uk; schong@liverpool.ac.uk
bLeverhulme Research Centre for Functional Materials Design, University of Liverpool, Liverpool L7 3NY, UK
cComputational Systems Chemistry, School of Chemistry, University of Southampton, SO17 1BJ, UK
First published on 26th December 2023
Abstract
Automation can transform productivity in research activities that use liquid handling, such as organic synthesis, but it has made less impact in materials laboratories, which require sample preparation steps and a range of solid-state characterization techniques. For example, powder X-ray diffraction (PXRD) is a key method in materials and pharmaceutical chemistry, but its end-to-end automation is challenging because it involves solid powder handling and sample processing. Here we present a fully autonomous solid-state workflow for PXRD experiments that can match or even surpass manual data quality, encompassing crystal growth, sample preparation, and automated data capture. The workflow involves 12 steps performed by a team of three multipurpose robots, illustrating the power of flexible, modular automation to integrate complex, multitask laboratories.
Robots can carry out a range of repetitive and iterative laboratory tasks, particularly those involving liquid handling,1 such as for organic synthesis,2–5 but many experiments remain hard to automate. One example is powder X-ray diffraction (PXRD), which is a central tool for characterizing the structure of ordered solids6 , including functional materials7,8 and pharmaceutical polymorphs.9 PXRD can be easier to implement than methods that require growing and harvesting a single diffractable crystal,10 and it provides important information about structure and purity.11–13 PXRD is used for the rapid identification of crystal forms and for detecting the existence of polymorphs;14–16 this is valuable in both materials research7,8,17,18 and in pharmaceutical chemistry.19,20 Indeed, pharmaceutically active molecules must undergo exhaustive and expensive screening experiments to fully understand their crystal form landscapes before they can be approved for clinical trials.21–23
Previously, high-throughput crystallization screens have used robots and other automated platforms16,17,24–26 to accelerate the discovery of materials such as pharmaceuticals,21–26 porous organic cages27 and photovoltaics,28 but these workflows tend to be only partially automated. For closed-loop autonomous workflows, we must automate and connect all stages in the PXRD experiment. This starts with crystal growth and is followed by sample preparation, often by mechanical grinding to reduce the crystal size to allow better orientational averaging. The resulting powders are then transferred into a sample holder, such as a multi-well plate, followed by loading the PXRD instrument and data collection. At present, this sample preparation and transfer is typically done by hand, even for ‘high throughput’, automated workflows,17 and this is laborious. Likewise, there is a plethora of other materials workflows where similar solid-handling operations are required, such as preparing samples for conductivity analysis or for microscopy.
Here, we present a fully autonomous, modular robotic workflow that prepares crystalline materials and then collects their powder diffraction data. This modular approach integrates three separate robotic platforms—a liquid handling platform for the crystallization stage (Chemspeed FLEX LIQUIDOSE, Fig. 1, step 1, ESI Fig. S1 & S2†), a mobile manipulator for sample transport and equipment manipulation (KUKA KMR iiwa; Fig. 1, steps 1, 2, 8 & 9–12), and a dual-arm robot for sample preparation (ABB YuMi; Fig. 1, steps 3–7). The workflow uses a standard powder X-ray diffractometer, which is used by the mobile manipulator in an anthropomorphic way without any substantial modification. These heterogenous robotic and automation platforms work together synchronously to achieve the multiple steps in the workflow (Fig. 1), orchestrated by our system architecture, ARChemist (ESI, Fig. S3†).29 We exemplify this approach with two different organic compounds and show that PXRD data collected by the autonomous robotic workflow are of comparable quality, or in some cases better, to data collected for samples prepared by hand. Hence, these data are suitable for identifying compounds and distinguishing between their polymorphs. We also demonstrate matching crystalline powders against sets of putative polymorphs generated by crystal structure prediction (CSP) methods, which is a key step in integrating predictive computational methods into closed-loop materials discovery.
Description of the modular robotic workflow
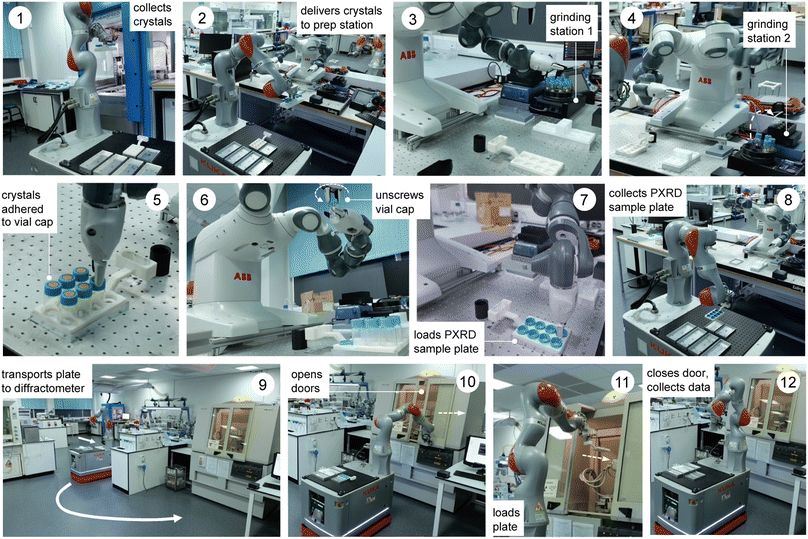
The workflow comprises three robots and 12 steps, as outlined in Fig. 1 and the videos in the ESI (Videos 1 & 2).† First, crystals are grown using a Chemspeed platform (ESI, Fig. S1 and S2†), whereby the material of interest is dispensed in a variety of solvents or solvent mixtures and these solvents are allowed to evaporate, thus growing crystals (step 0, not shown). Quite often this leads to large crystals that adhere strongly to the sides of the sample vials (ESI, Fig. S4†) and these must therefore be reduced in size and removed from the vial prior to diffraction, as described below. In step 1 (Fig. 1), a rack of eight crystal samples is collected from the Chemspeed platform using a mobile KUKA robot;30 the Chemspeed platform was modified with an automated vertical sash door to facilitate this. Each of the eight vials is then capped with a sample lid sealed with an adhesive Kapton polymer film that will ultimately receive the ground crystalline powder (ESI, Fig. S4 and S5†). In step 2, the mobile KUKA manipulator delivers the rack of eight samples to the preparation station, which involves a dual-arm ABB YuMi robot. In step 3, the dual-arm YuMi robot transfers the eight samples to grinding station 1, where they are reduced in size using mechanical attrition by magnetic stirring with preloaded Teflon stir bars.In step 4, the YuMi robot inverts the eight samples and transfers them to grinding station 2, where they are agitated using a shaker plate to reduce the particle size further and to transfer the sample onto the adhesive Kapton polymer film in the cap of each vial. In step 5, the YuMi robot inverts the samples again and transfers them to the X-ray diffraction plate; at this point, the sample is adhered to the Kapton film in the vial cap (ESI, Fig. S4†) and any excess sample and the Teflon stir bar falls back into the vial. In step 6, the YuMi robot unscrews each vial cap, inverts it, and places it back into the PXRD plate (step 7, ESI, Fig. S6†), which is then collected by the KUKA KMR iiwa robot (step 8) and transported to the diffractometer (step 9). In step 10, the KUKA robot opens the sliding doors of the diffractometer, loads the plate into the instrument (step 11), and then closes the doors and X-ray data is collected for the eight samples (step 12). The sample rack can then be retrieved from the PXRD instrument by the KUKA robot and another rack of eight samples processed and analysed, as required. A full loop of sample preparation, transport, and data collection takes around nine hours for a rack of eight samples with the data acquisition settings used here, although this time depends on the scan parameters; here, 1 hour for sample processing plus eight 1 hour PXRD scans. We estimate that the timescale for the equivalent human operations (i.e., sample preparation, plate loading and PXRD sample loading) is approximately the same, at least for one rack of samples. However, the potential for 24/7 continuous operation30 means that the long-term sample throughput could be significantly faster while maintaining higher consistency between experiments by using these robots. For example, processing 3 racks per day, 7 days a week would give a throughput of 168 samples per week, whereas a realistic human researcher throughput might be 1 rack per day, 5 days per week; that is, 40 samples. Also, automation frees researchers from relatively mundane tasks to focus on more intellectual activity, such as data analysis and interpretation.
The overall workflow is detailed by the videos in the ESI (Videos 1 & 2†) and associated process flow diagrams (Fig. S7 & S8†). As shown in Fig. 2, this autonomous PXRD workflow is part of a larger laboratory that contains several other workflows—for example, our autonomous photocatalysis workflow30 is in the same area as the X-ray diffractometer (labelled C in Fig. 2).
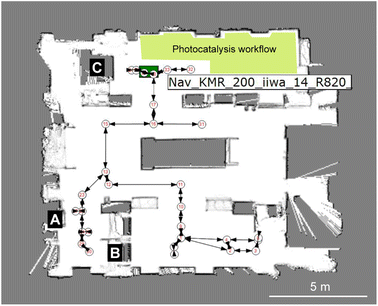 | ||
| Fig. 2 Heterogeneous modular integration of laboratories using multiple robots. Image from the KUKA Sunrise software showing the robot-generated map of the lab with numbered nodes and edges that correspond to taught paths and way points. For the experiments described here, the key modules are the Chemspeed liquid handling platform (A), the YuMi dual-arm sample preparation station (B) and the powder X-ray diffractometer (C). The location of the KUKA KMR iiwa robot is shown by the green rectangle, in this case approaching the X-ray diffractometer, C. The location of our photocatalysis workflow (not discussed here) is shaded in yellow.30 | ||
The layout of the workflow is, to some extent, arbitrary: for example, the location of the X-ray diffractometer is fixed by proximity to its cooling water supply, and there was insufficient space to locate the Chemspeed FLEX LIQUIDOSE robot (A, Fig. 2) or the ABB YuMi preparation station (B) adjacent to the diffractometer. This does not matter under a modular paradigm that uses mobile manipulators to integrate stations because the transport time between the stations is a small fraction of the overall workflow cycle time compared to the slow steps, which in this case are solvent evaporation for crystallization and PXRD data acquisition. The approach is therefore inherently scalable: for example, one can envisage coupling two workflows together, whereby the most crystalline materials are selected for testing as photocatalysts30 and those powders transferred automatically into that workflow, which is housed in the same laboratory (yellow shaded area in Fig. 2). Also, because we use collaborative robots, or ‘cobots’, the laboratory space can be shared with human researchers, and we do this daily.
Automated data collection for benzimidazole
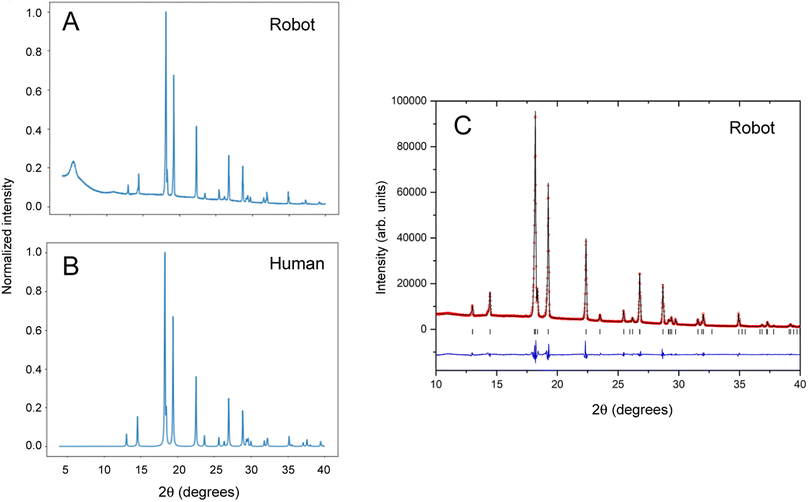
Benzimidazole is an organic heterocyclic compound that typically exists as its alpha polymorph in the form of solid white crystals. Benzimidazole derivatives are used in pharmaceuticals such as antacids, antiparasitic drugs and opioids.31–33 A stock solution of benzimidazole in methanol (0.1 g mL−1) was used for the automated experiment. The input station in the Chemspeed FLEX LIQUIDOSE platform was loaded with a rack of eight sample vials (20 mL vial volume) preloaded with magnetic Teflon stir bars and capped with Kapton film vial lids, as described above. The stock solutions were dispensed into the eight sample vials and left to evaporate inside the Chemspeed platform. Once the solid samples were dry, the vials were capped by the Chemspeed platform. Like many organic compounds, benzimidazole often crystallizes as large, blocky crystals that are hard to recover and unsuitable for PXRD analysis without further preparation, as shown in the ESI (Fig. S4†).Next, the mobile KUKA manipulator collects the samples from the Chemspeed platform and delivers them to the preparation station for processing, as described above, followed by automated PXRD analysis. To compare our autonomous method with the traditional manual approach, 2 mL of the stock solution was also dispensed into a sample vial and left to evaporate, whereafter the solid was recovered and ground by hand using a mortar and pestle. This sample was mounted, also by hand, in a 96-well aluminium plate prior to data collection. Data collected under the same scan conditions for robot-prepared and human-prepared benzimidazole crystals are compared in Fig. 3A and B.
The broad peak at around 2θ = 6° in the robot-collected data results from the adhesive Kapton tape on which the robot-prepared samples are loaded. Apart from this artefact, the two datasets are comparable in terms of peak width, peak positions, and relative peak intensities. The PXRD pattern generated by the autonomous robotic workflow shows good signal to noise and is of sufficient quality to extract structural information (Fig. 3C). The unit cell parameters that were determined (a = 13.6000(5) Å; b = 6.8564(2) Å; c = 6.9905(2) Å) confirmed the formation of the alpha polymorph by comparison to reference structures in the CSD database (ESI, Table S1†). The analysis showed that the sample prepared by the automated process had better homogeneity in terms of crystallite size, resulting in more consistent peak profiles that could be better modelled. This enabled more precise lattice parameters with smaller standard uncertainties to be determined than from the conventional, manually prepared sample (ESI, Table S1†).
Automated data collection for polymorphic ROY
The archetypal example of a polymorphic organic molecule is 5-methyl-2-[(2-nitrophenyl) amino]-3-thiophenecarbonitrile, commonly referred to as ROY.34–38 It is an intermediate in the synthesis of the antipsychotic drug olanzapine and it is named ROY because of its red, orange, and yellow polymorphs. There have been many crystallization studies conducted for ROY, and various polymorphs have been observed, which often occur concomitantly with each other as mixtures. Here, samples of ROY were prepared from solid 5-methyl-2-[(2-nitrophenyl) amino]-3-thiophenecarbonitrile by dissolving it in acetone with the addition of different percentage volumes of water as an antisolvent (ESI, Table S2†). In this case, the sample solutions were prepared manually because of the slow evaporation time for water, which would require the Chemspeed robot to be idle over long periods of time, although this step could have been automated as for benzimidazole, above. After a dry, solid product had formed, the samples were loaded into the input station for the workflow for processing and PXRD analysis, as before (Fig. 4). This experiment produced two different polymorphs of ROY, and the data obtained by the robot was of sufficient quality to identify them, even when appearing as a mixture (e.g., sample 4 in Fig. 4C and D). As for benzimidazole, unit cell parameters extracted from the PXRD data for sample 1 confirmed the effective homogenization of the powder and its phase purity (ESI, Fig. S14, Table S3†); again, the robot-prepared data was superior to the manually produced samples, resulting in improved fit statistics and uncertainties. The data quality across the batch of eight samples can be seen in Fig. 4D.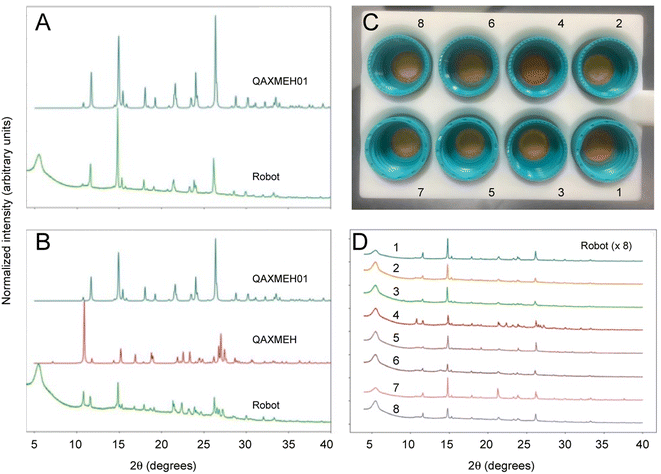 | ||
| Fig. 4 PXRD patterns collected autonomously for ROY polymorphs. (A) Comparison of the diffraction pattern for ROY processed using the robotic workflow (sample 1 in (C) and (D)), as compared with the simulated PXRD pattern for the published monoclinic yellow (Y) polymorph (CSD reference code QAXMEH); (B) PXRD patterns for ROY processed using the autonomous robotic workflow (sample 4 in (C) and (D)), as compared with the simulated patterns for two published forms: the monoclinic Y polymorph (QAXMEH) and the monoclinic orange needle (ON) polymorph (QAXMEH01), suggesting that a phase mixture is formed under these conditions; (C) photograph of ROY processed using the robotic workflow at various concentrations and solvent ratios (see ESI, Table S1,† for crystallization conditions); (D) diffraction patterns for the ROY samples shown in (C). | ||
Polymorph matching against crystal structure prediction datasets
Crystal structure prediction (CSP) is valuable for anticipating polymorphism of active pharmaceutical ingredients39 and in guiding the discovery of molecular crystals with targeted properties.40 We therefore assessed the possibility of comparing the powder X-ray diffraction patterns generated by the automated robotic workflow against structure sets of putative crystal structures generated by CSP. From sample 1 for benzimidazole and ROY, we found that comparison of the experimental data against the low energy CSP structures using the variable-cell experimental powder difference (VC-xPWDF) method41 identifies the predicted crystal structure as the observed polymorph; that is, alpha benzimidazole (Fig. 5A) has the most similar simulated powder X-ray diffraction pattern. Likewise, sample 1 of ROY can be identified as matching the predicted low-energy Y polymorph most closely (Fig. 5B). This demonstrates the possibility of identifying newly discovered crystal structures in an automated manner by comparison against pre-computed libraries of predicted crystal structures, which would create an important feedback mechanism between computational screening of materials and automated crystallization in the laboratory.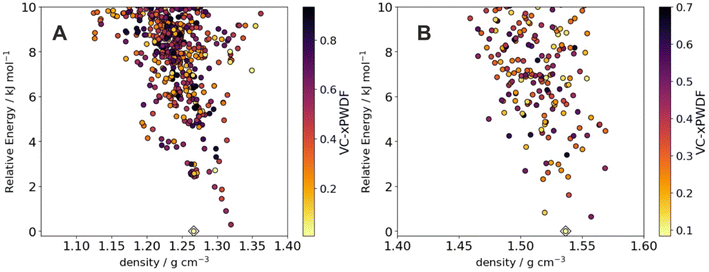 | ||
| Fig. 5 Matching robotic PXRD data with computationally predicted crystal structures. Energy-density distribution of low-energy crystal structure prediction (CSP) structures of (A) benzimidazole and (B) ROY. Each point corresponds to a distinct predicted crystal structure; points are colored by dissimilarity of their simulated powder X-ray diffraction patterns compared to the pattern collected from the robot workflow. For ROY, we show results using pattern 1 from Fig. 4. The CSP structures corresponding to the alpha polymorph of benzimidazole and the Y polymorph of ROY are indicated with diamonds; in both cases, these correspond to the lowest dissimilarity (greatest similarity) to the experimental data. | ||
Outlook
This work illustrates the potential of modular and flexible robots42 to accelerate PXRD experiments, thus integrating a key materials characterization method into a ‘self-driving laboratory’.43 While the videos presented here show a single cycle (one rack of eight samples), it should be possible in the future to operate this workflow 24/7 over extended periods in a closed-loop way, as demonstrated in our earlier implementation of mobile robots for photocatalyst optimization.30 Likewise, we will need to introduce a suitable database and data standards; this should be relatively straightforward for PXRD data, where there are existing standards, but may be more challenging for other solid-state measurements. Some method improvements will be necessary for fully autonomous matching with predicted crystal structures, which are sensitive to sample preparation and polymorphic purity. For example, for the three ROY samples that we identified by eye as pure polymorph Y, PXRD matching was able to identify the correct CSP structure. However, the matching algorithm identifies incorrect structures when presented with PXRD of samples of a polymorphic mixture, such as sample 4 in Fig. 4D. Even then, however, the CSP structure corresponding to polymorph Y was identified among the best matches to the experimental PXRD (9th out of 264 CSP structures), albeit not as the best match. Likewise, some samples of benzimidazole that we tested had different relative peak intensities their PXRD pattern, probably because of a non-uniform distribution of crystallite orientations, which led to less reliable matching to the CSP structure that corresponds to alpha benzimidazole (ESI, Table S4†). These challenges should be addressable via improvements to the sample preparation methods and the PXRD matching algorithms.The introduction of sample processing and the use of three separate robots rather than one makes this solid-state workflow significantly more complex than our earlier photocatalysis study,30 which comprised a single, easily automated measurement (gas chromatography) and no sample processing. Indeed, this workflow ranks among the most complex autonomous systems reported for chemistry to date,2–5,42–45 and its modular nature offers unique scope for expansion and diversification. An automated platform of comparable complexity is AMANDA,44 which is being developed for photovoltaics research; another impressive example is the AutoBASS platform that can assemble up to 64 CR2023 battery cells.45 However, both of those platforms involve large, custom-built integrated robotic systems. By contrast, our workflow uses commercially available, ‘off the shelf’ robots and other common laboratory hardware with little or no modification.
While this manuscript was under review, another study was published describing an autonomous laboratory for the accelerated synthesis of materials, focusing on inorganic oxides and phosphates.46 This study also presented a complex, multistep solid-state workflow that encompassed PXRD analysis, in this case using synthesis recipes that were trained on natural-language models.
Our modular robotic integration approach is adaptable and scalable. By using a mobile manipulator to integrate the various stations, the workflow can be arranged in almost any configuration, and it is easily extended by adding other stations, subject only to available laboratory space (Fig. 2). As such, we see the general concept of integrating stationary and mobile robots using a core software architecture29 as a powerful strategy for automating a range of research activities beyond diffraction experiments. This will also allow us to guide autonomous closed-loop robotic experiments using computational predictions and artificial intelligence.47 To give just one example, it should be possible to use this autonomous PXRD workflow to identify crystallization conditions that produce polymorphs that are predicted to have certain desirable functional properties,40 and then to automatically take those materials forward for property testing.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper or the ESI Materials† and the repository links therein. The crystal structure prediction data can be found at: https://doi.org/10.5258/SOTON/D2924.Author contributions
S. Y. C. and A. I. C. conceived the project. A. M. L. led the experimental work, the design of the dual-arm robotic processing station, and the overall construction of the workflow. L. L. and A. W. also contributed to the design of the processing station. N. R. assisted with design and 3D printing of components in the workflow. R. C. assisted with the PXRD and Chemspeed integrations. B. M. A. was involved in the overall workflow design. H. F. and G. P. contributed to the robotics aspects. H. F. led the design and implementation of the ARChemist software suite. A. M. L. and S. Y. performed the LeBail fits for the PXRD data. G. M. D. and L. G. carried out the crystal structure prediction (CSP) work and structural matches with CSP data. All authors contributed to the writing of the paper.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This project has received funding from the European Research Council under the European Union's Horizon 2020 research and innovation program (grant agreement no. 856405). The authors also received funding from the Engineering and Physical Sciences Research Council (EPSRC) (EP/V026887/1, EP/T031263/1) and the Leverhulme Trust via the Leverhulme Research Centre for Functional Materials Design. AIC thanks the Royal Society for a Research Professorship (RSRP\S2\232003). We thank Prof. K. Thurow (University of Rostock) for advice and for inspiring us to work with mobile robots. We thank Dr Isaiah Bourne for assistance with microscopy measurements.References
- F. Kong, L. Yuan, Y. F. Zheng and W. Chen, Automatic liquid handling for life science: a critical review of the current state of the art, J. Lab. Autom., 2012, 17, 169–185 CrossRef CAS PubMed.
- J. Li, et al., Synthesis of many different types of organic small molecules using one automated process, Science, 2015, 347, 1221–1226 CrossRef CAS PubMed.
- A. Adamo, et al., On-demand continuous-flow production of pharmaceuticals in a compact, reconfigurable system, Science, 2016, 352, 61–67 CrossRef CAS PubMed.
- D. T. Ahneman, J. G. Estrada, S. D. Dreher and A. G. Doyle, Predicting reaction performance in C–N cross-coupling using machine learning, Science, 2018, 360, 186–190 CrossRef CAS PubMed.
- S. Steiner, et al., Organic synthesis in a modular robotic system driven by a chemical programming language, Science, 2019, 363, eaav2211 CrossRef CAS PubMed.
- A. A. Bunaciu, E. Gabriela Udriştioiu and H. Y. Aboul-Enein, X-ray diffraction: Instrumentation and applications, Crit. Rev. Anal. Chem., 2015, 45, 289–299 CrossRef CAS PubMed.
- C. Gropp, T. Q. Ma, N. Hanikel and O. M. Yaghi, Design of higher valency in covalent organic frameworks, Science, 2020, 370, 484 CrossRef PubMed.
- C. Baerlocher, et al., Unraveling the perplexing structure of the Zeolite SSZ-57, Science, 2011, 333, 1134–1137 CrossRef CAS PubMed.
- R. Censi and P. Di Martino, Polymorph impact on the bioavailability and stability of poorly soluble drugs, Molecules, 2015, 20, 18759 CrossRef CAS PubMed.
- K. D. M. Harris and E. Y. Cheung, How to determine structures when single crystals cannot be grown: opportunities for structure determination of molecular materials using powder diffraction data, Chem. Soc. Rev., 2004, 33, 526–538 RSC.
- K. D. M. Harris, M. Tremayne and B. M. Kariuki, Contemporary advances in the use of powder X-ray diffraction for structure determination, Angew. Chem., Int. Ed., 2001, 40, 1626–1651 CrossRef CAS PubMed.
- R. Černý, Crystal structures from powder diffraction: Principles, difficulties and progress, Crystals, 2017, 7, 142 CrossRef.
- C. F. Holder and R. E. Schaak, Tutorial on powder X-ray diffraction for characterizing nanoscale materials, ACS Nano, 2019, 13, 7359–7365 CrossRef CAS PubMed.
- X. Yao, R. F. Henry and G. G. Z. Zhang, Ritonavir Form III: A new polymorph after 24 years, J. Pharm. Sci., 2023, 112, 237–242 CrossRef CAS PubMed.
- J. Li, S. A. Bourne and M. R. Caira, New polymorphs of isonicotinamide and nicotinamide, Chem. Commun., 2011, 47, 1530–1532 RSC.
- V. W. Rosso, et al., High-throughput crystallization screening technique with transmission PXRD analysis, Org. Process Res. Dev., 2023, 27, 1437–1444 CrossRef CAS.
- P. Cui, et al., Mining predicted crystal structure landscapes with high throughput crystallization: old molecules, new insights, Chem. Sci., 2019, 10, 9988–9997 RSC.
- P. Zhang, et al., Polymorphism, phase transformation and energetic properties of 3-nitro-1,2,4-triazole, RSC Adv., 2018, 8, 24627–24632 RSC.
- S. L. Morissette, S. Soukasene, D. Levinson, M. J. Cima and Ö. Almarsson, Elucidation of crystal form diversity of the HIV protease inhibitor ritonavir by high-throughput crystallization, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 2180–2184 CrossRef CAS PubMed.
- E. H. Lee, A practical guide to pharmaceutical polymorph screening & selection, Asian J. Pharm. Sci., 2014, 9, 163–175 CrossRef.
- S. L. Morissette, et al., High-throughput crystallization: Polymorphs, salts, co-crystals and solvates of pharmaceutical solids, Adv. Drug Delivery Rev., 2004, 56, 275–300 CrossRef CAS PubMed.
- A. J. Alvarez, A. Singh and A. S. Myerson, Polymorph screening: Comparing a semi-automated approach with a high throughput method, Cryst. Growth Des., 2009, 9, 4181–4188 CrossRef CAS.
- A. Newman, Specialized solid form screening techniques, Org. Process Res. Dev., 2013, 17, 457–471 CrossRef CAS.
- A. J. Florence, A. Johnston, P. Fernandes, N. Shankland and K. Shankland, An automated platform for parallel crystallization of small organic molecules, J. Appl. Crystallogr., 2006, 39, 922–924 CrossRef CAS.
- C. J. Brown, et al., Enabling precision manufacturing of active pharmaceutical ingredients: workflow for seeded cooling continuous crystallizations, Mol. Syst. Des. Eng., 2018, 3, 518–549 RSC.
- M. L. Peterson, et al., Iterative high-throughput polymorphism studies on acetaminophen and an experimentally derived structure for form III, J. Am. Chem. Soc., 2002, 124, 10958–10959 CrossRef CAS PubMed.
- R. L. Greenaway, et al., High-throughput discovery of organic cages and catenanes using computational screening fused with robotic synthesis, Nat. Commun., 2018, 9, 2849 CrossRef CAS PubMed.
- Z. Li, et al., Robot-accelerated perovskite investigation and discovery, Chem. Mater., 2020, 32, 5650–5663 CrossRef CAS.
- H. Fakhruldeen, G. Pizzuto, J. Glowacki and A. I. Cooper, ARChemist: Autonomous Robotic Chemistry System Architecture, IEEE Int. Conf. Robot. Autom., 2022, 6013–6019 Search PubMed.
- B. Burger, et al., A mobile robotic chemist, Nature, 2020, 583, 237–241 CrossRef CAS PubMed.
- Y. T. Lee, Y. J. Tan and C. E. Oon, Benzimidazole and its derivatives as cancer therapeutics: The potential role from traditional to precision medicine, Acta Pharm. Sin. B, 2023, 13, 478 CrossRef CAS PubMed.
- W. Zieliński and A. Katrusiak, Hydrogen bonds NH⋯N in compressed benzimidazole polymorphs, Cryst. Growth Des., 2013, 13, 696–700 CrossRef.
- S. Krawczyk and M. Gdaniec, Polymorph β of 1H-benzimidazole, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2005, 61, o4116–o4118 CrossRef CAS.
- L. Yu, et al., Thermochemistry and conformational polymorphism of a hexamorphic crystal system, J. Am. Chem. Soc., 2000, 122, 585–591 CrossRef CAS.
- J. A. Foster, et al., Pharmaceutical polymorph control in a drug-mimetic supramolecular gel, Chem. Sci., 2017, 8, 78–84 RSC.
- S. Chen, I. A. Guzei and L. Yu, New polymorphs of ROY and new record for coexisting polymorphs of solved structures, J. Am. Chem. Soc., 2005, 127, 9881–9885 CrossRef CAS PubMed.
- M. Tan, et al., ROY revisited, again: the eighth solved structure, Faraday Discuss., 2018, 11, 477–491 RSC.
- L. Yu, Polymorphism in molecular solids: An extraordinary system of red, orange, and yellow crystals, Acc. Chem. Res., 2010, 43, 1257–1266 CrossRef CAS PubMed.
- C. R. Taylor, et al., Minimizing polymorphic risk through cooperative computational and experimental exploration, J. Am. Chem. Soc., 2020, 142, 16668–16680 CrossRef CAS PubMed.
- A. Pulido, et al., Functional materials discovery using energy–structure–function maps, Nature, 2017, 543, 657–664 CrossRef CAS PubMed.
- R. A. Mayo, K. M. Marczenko and E. R. Johnson, Quantitative matching of crystal structures to experimental powder diffractograms, Chem. Sci., 2023, 14, 4777–4785 RSC.
- B. P. MacLeod, et al., Flexible automation accelerates materials discovery, Nat. Mater., 2022, 21, 722–726 CrossRef CAS PubMed.
- B. P. MacLeod, et al., Self-driving laboratory for accelerated discovery of thin-film materials, Sci. Adv., 2020, 6, eaaz8867 CrossRef CAS PubMed.
- J. Wagner, et al., The evolution of Materials Acceleration Platforms: toward the laboratory of the future with AMANDA, J. Mater. Sci., 2021, 56, 16422–16446 CrossRef CAS.
- B. Zhang, L. Merker, A. Sanin and H. S. Stein, Robotic cell assembly to accelerate battery research, Digital Discovery, 2022, 1, 755–762 RSC.
- N. J. Szymanski, et al., An autonomous laboratory for the accelerated synthesis of novel materials, Nature, 2023, 624, 86–91 CrossRef CAS PubMed.
- Z. Ren, Z. Ren, Z. Zhang, T. Buonassisi and J. Li, Autonomous experiments using active learning and AI, Nat. Rev. Mater., 2023, 8, 563–564 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06206f |
| This journal is © The Royal Society of Chemistry 2024 |